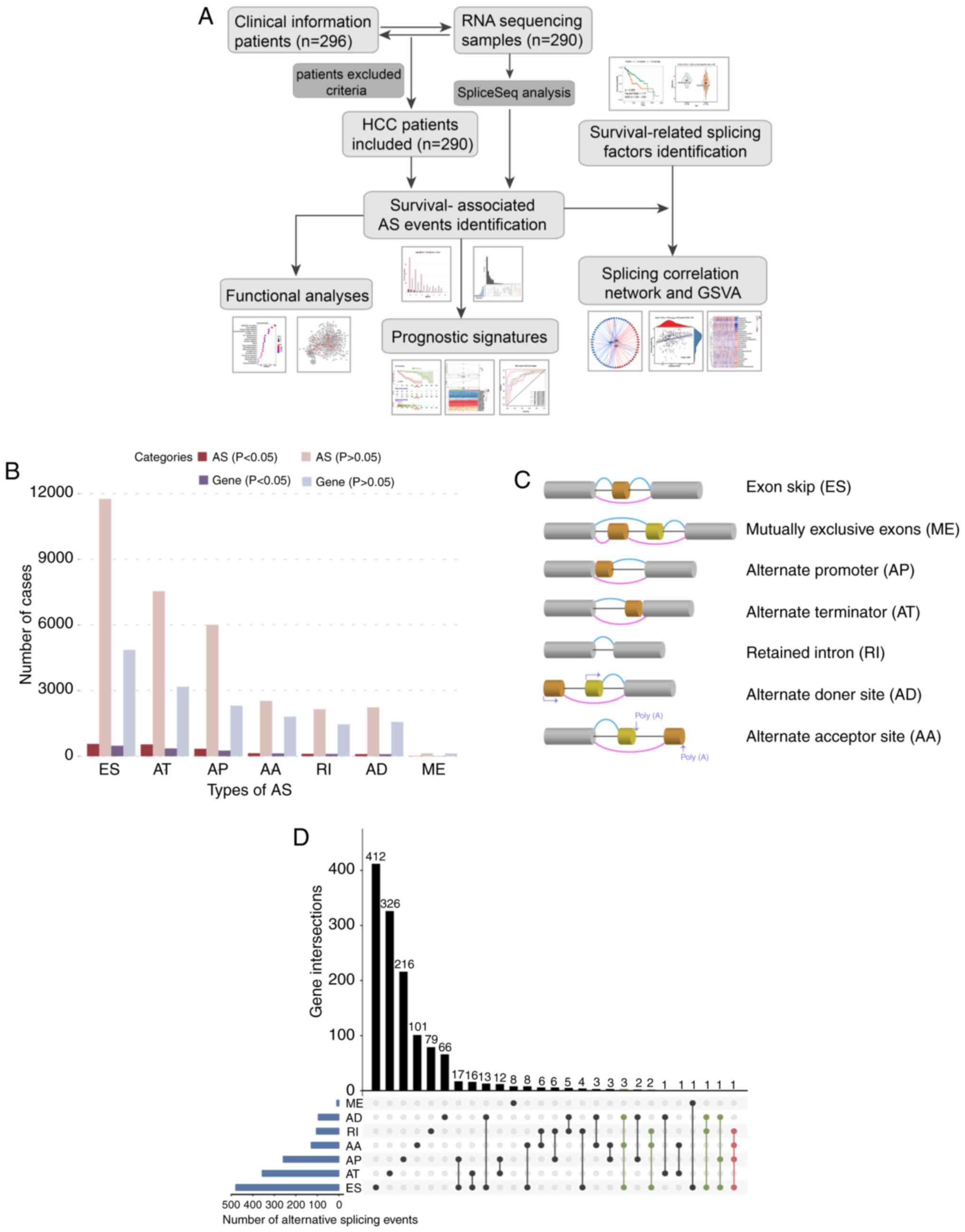
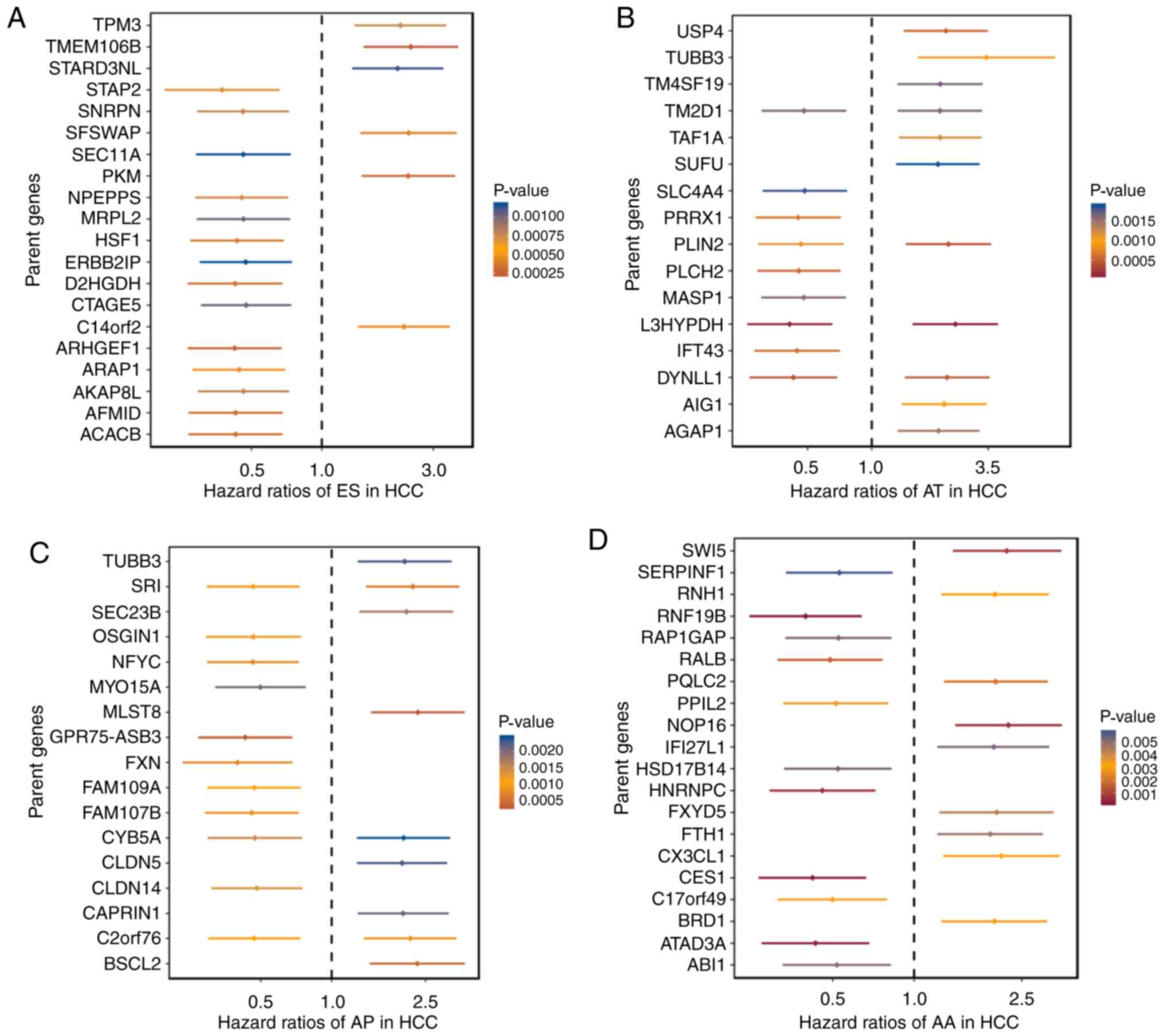
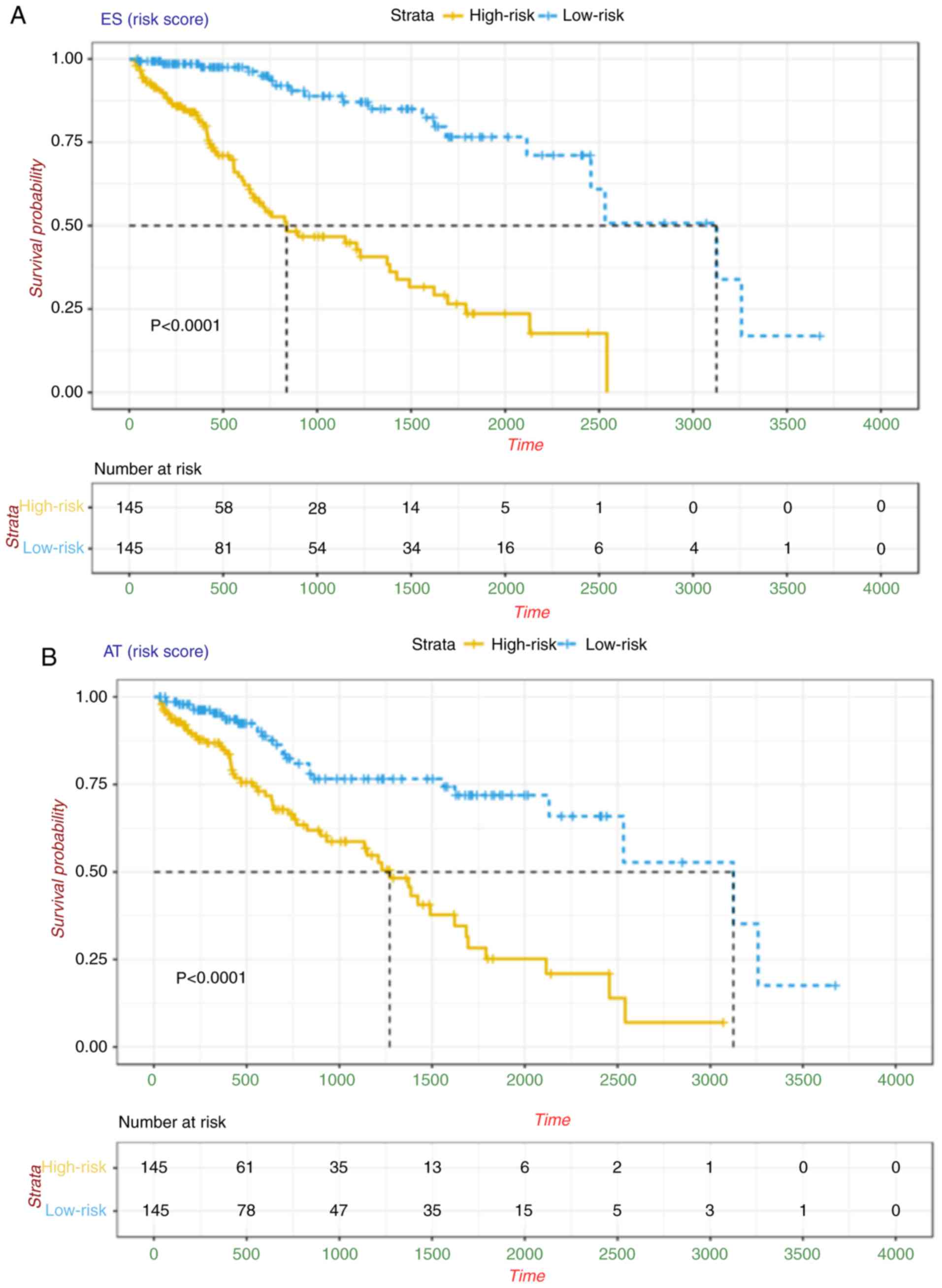
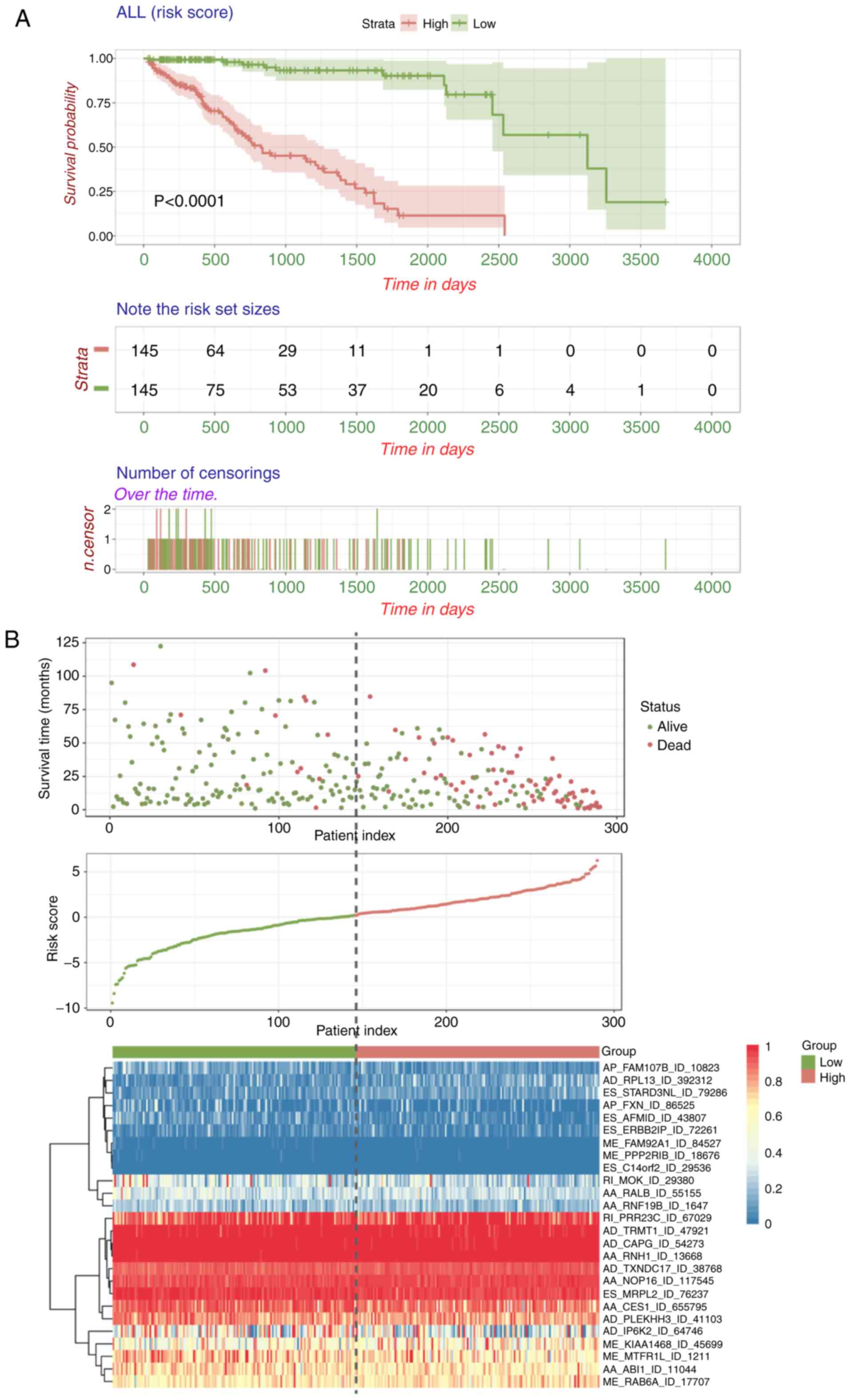
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Vitale A, Peck-Radosavljevic M, Giannini
EG, Vibert E, Sieghart W, Van Poucke S and Pawlik TM: Personalized
treatment of patients with very early hepatocellular carcinoma. J
Hepatol. 66:412–423. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ulahannan SV, Duffy AG, McNeel TS, Kish
JK, Dickie LA, Rahma OE, McGlynn KA, Greten TF and Altekruse SF:
Earlier presentation and application of curative treatments in
hepatocellular carcinoma. Hepatology. 60:1637–1644. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tabrizian P, Jibara G, Shrager B, Schwartz
M and Roayaie S: Recurrence of hepatocellular cancer after
resection: Patterns, treatments, and prognosis. Ann Surg.
261:947–955. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang JH, Wang CC, Hung CH, Chen CL and Lu
SN: Survival comparison between surgical resection and
radiofrequency ablation for patients in BCLC very early/early stage
hepatocellular carcinoma. J Hepatol. 56:412–418. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ueno M, Hayami S, Shigekawa Y, Kawai M,
Hirono S, Okada K, Tamai H, Shingaki N, Mori Y, Ichinose M and
Yamaue H: Prognostic impact of surgery and radiofrequency ablation
on single nodular HCC 5 cm: Cohort study based on serum HCC
markers. J Hepatol. 63:1352–1359. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hiraoka A, Kumada T, Kudo M, Hirooka M,
Tsuji K, Itobayashi E, Kariyama K, Ishikawa T, Tajiri K, Ochi H, et
al: Albumin-bilirubin (ALBI) grade as part of the evidence-based
clinical practice guideline for HCC of the japan society of
hepatology: A comparison with the liver damage and child-pugh
classifications. Liver Cancer. 6:204–215. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li S and Mao M: Next generation sequencing
reveals genetic landscape of hepatocellular carcinomas. Cancer
Lett. 340:247–253. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Huang W, Skanderup AJ and Lee CG: Advances
in genomic hepatocellular carcinoma research. Gigascience.
7:112018. View Article : Google Scholar
|
|
10
|
Zhang YC, Zhou Q and Wu YL: The emerging
roles of NGS-based liquid biopsy in non-small cell lung cancer. J
Hematol Oncol. 10:1672017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cancer Genome Atlas Research Network.
Electronic address, . simplewheeler@bcm.edu; Cancer
Genome Atlas Research Network: Comprehensive and integrative
genomic characterization of hepatocellular carcinoma. Cell.
169:1327–1341.e23. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Villanueva A, Portela A, Sayols S,
Battiston C, Hoshida Y, Mendez-Gonzalez J, Imbeaud S, Letouzé E,
Hernandez-Gea V, Cornella H, et al: DNA methylation-based prognosis
and epidrivers in hepatocellular carcinoma. Hepatology.
61:1945–1956. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Qiu J, Peng B, Tang Y, Qian Y, Guo P, Li
M, Luo J, Chen B, Tang H, Lu C, et al: CpG Methylation signature
predicts recurrence in early-stage hepatocellular carcinoma:
Results from a multicenter study. J Clin Oncol. 35:734–742. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Long J, Wang A, Bai Y, Lin J, Yang X, Wang
D, Yang X, Jiang Y and Zhao H: Development and validation of a
TP53-associated immune prognostic model for hepatocellular
carcinoma. EBioMedicine. 42:363–374. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Long J, Bai Y, Yang X, Lin J, Yang X, Wang
D, He L, Zheng Y and Zhao H: Construction and comprehensive
analysis of a ceRNA network to reveal potential prognostic
biomarkers for hepatocellular carcinoma. Cancer Cell Int.
19:902019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Baralle FE and Giudice J: Alternative
splicing as a regulator of development and tissue identity. Nat Rev
Mol Cell Biol. 18:437–451. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gamazon ER and Stranger BE: Genomics of
alternative splicing: Evolution, development and pathophysiology.
Hum Genet. 133:679–687. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang ET, Sandberg R, Luo S, Khrebtukova I,
Zhang L, Mayr C, Kingsmore SF, Schroth GP and Burge CB: Alternative
isoform regulation in human tissue transcriptomes. Nature.
456:470–476. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Montes M, Sanford BL, Comiskey DF and
Chandler DS: RNA Splicing and disease: Animal models to therapies.
Trends Genet. 35:68–87. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Climente-Gonzalez H, Porta-Pardo E, Godzik
A and Eyras E: The functional impact of alternative splicing in
cancer. Cell Rep. 20:2215–2226. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tao Y, Ma C, Fan Q, Wang Y, Han T and Sun
C: MicroRNA-1296 facilitates proliferation, migration and invasion
of colorectal cancer cells by targeting SFPQ. J Cancer.
9:2317–2326. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
David CJ and Manley JL: Alternative
pre-mRNA splicing regulation in cancer: Pathways and programs
unhinged. Genes Dev. 24:2343–2364. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Song X, Zeng Z, Wei H and Wang Z:
Alternative splicing in cancers: From aberrant regulation to new
therapeutics. Semin Cell Dev Biol. 75:13–22. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lu Y, Xu W, Ji J, Feng D, Sourbier C, Yang
Y, Qu J, Zeng Z, Wang C, Chang X, et al: Alternative splicing of
the cell fate determinant numb in hepatocellular carcinoma.
Hepatology. 62:1122–1131. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Berasain C, Goni S, Castillo J, Latasa MU,
Prieto J and Avila MA: Impairment of pre-mRNA splicing in liver
disease: Mechanisms and consequences. World J Gastroenterol.
16:3091–3102. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ratnadiwakara M, Mohenska M and Anko ML:
Splicing factors as regulators of miRNA biogenesis-links to human
disease. Semin Cell Dev Biol. 79:113–122. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen M and Manley JL: Mechanisms of
alternative splicing regulation: Insights from molecular and
genomics approaches. Nat Rev Mol Cell Biol. 10:741–754. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wahl MC, Will CL and Luhrmann R: The
spliceosome: Design principles of a dynamic RNP machine. Cell.
136:701–718. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Prochazka L, Tesarik R and Turanek J:
Regulation of alternative splicing of CD44 in cancer. Cell Signal.
26:2234–2239. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yoshida K and Ogawa S: Splicing factor
mutations and cancer. Wiley Interdiscip Rev RNA. 5:445–459. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
El Marabti E and Younis I: The cancer
spliceome: Reprograming of alternative splicing in cancer. Front
Mol Biosci. 5:802018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Anczukow O and Krainer AR: Splicing-factor
alterations in cancers. RNA. 22:1285–1301. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tomczak K, Czerwinska P and Wiznerowicz M:
The cancer genome atlas (TCGA): An immeasurable source of
knowledge. Contemp Oncol (Pozn). 19:A68–A77. 2015.PubMed/NCBI
|
|
34
|
Ryan MC, Cleland J, Kim R, Wong WC and
Weinstein JN: SpliceSeq: A resource for analysis and visualization
of RNA-Seq data on alternative splicing and its functional impacts.
Bioinformatics. 28:2385–2387. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liao SG, Lin Y, Kang DD, Chandra D, Bon J,
Kaminski N, Sciurba FC and Tseng GC: Missing value imputation in
high-dimensional phenomic data: Imputable or not, and how? BMC
Bioinformatics. 15:3462014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lex A, Gehlenborg N, Strobelt H, Vuillemot
R and Pfister H: UpSet: Visualization of intersecting sets. IEEE
Trans Vis Comput Graph. 20:1983–1992. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ye J, Zhou G, Zhang Z, Sun L, He X and
Zhou J: Poly (C)-binding protein 2 (PCBP2) promotes the progression
of esophageal squamous cell carcinoma (ESCC) through regulating
cellular proliferation and apoptosis. Pathol Res Pract.
212:717–725. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kamarudin AN, Cox T and Kolamunnage-Dona
R: Time-dependent ROC curve analysis in medical research: Current
methods and applications. BMC Med Res Methodol. 17:532017.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yu G, Wang LG, Han Y and He QY:
ClusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xiong Y, Deng Y, Wang K, Zhou H, Zheng X,
Si L and Fu Z: Profiles of alternative splicing in colorectal
cancer and their clinical significance: A study based on
large-scale sequencing data. EBioMedicine. 36:183–195. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Piva F, Giulietti M, Burini AB and
Principato G: SpliceAid 2: A database of human splicing factors
expression data and RNA target motifs. Hum Mutat. 33:81–85. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15:5502014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hänzelmann S, Castelo R and Guinney J:
GSVA: Gene set variation analysis for microarray and RNA-Seq data.
BMC Bioinformatics. 14:72013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: Limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Lin DC, Mayakonda A, Dinh HQ, Huang P, Lin
L, Liu X, Ding LW, Wang J, Berman BP, Song EW, et al: Genomic and
epigenomic heterogeneity of hepatocellular carcinoma. Cancer Res.
77:2255–2265. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wang BD and Lee NH: Aberrant RNA splicing
in cancer and drug resistance. Cancers (Basel). 10:112018.
View Article : Google Scholar :
|
|
47
|
Shi YM, Li YY, Lin JY, Zheng L, Zhu YM and
Huang J: The discovery of a novel eight-mRNA-lncRNA signature
predicting survival of hepatocellular carcinoma patients. J Cell
Biochem. Nov 28–2018.(Epub ahead of print). doi:
10.1002/jcb.28028.
|
|
48
|
Liu G, Wang H, Fu JD, Liu JY, Yan AG and
Guan YY: A five-miRNA expression signature predicts survival in
hepatocellular carcinoma. APMIS. 125:614–622. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Gu JX, Zhang X, Miao RC, Xiang XH, Fu YN,
Zhang JY, Zhang JY, Liu C and Qu K: Six-long non-coding RNA
signature predicts recurrence-free survival in hepatocellular
carcinoma. World J Gastroenterol. 25:220–232. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Dayton TL, Gocheva V, Miller KM, Israelsen
WJ, Bhutkar A, Clish CB, Davidson SM, Luengo A, Bronson RT, Jacks T
and Vander Heiden MG: Germline loss of PKM2 promotes metabolic
distress and hepatocellular carcinoma. Genes Dev. 30:1020–1033.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Lee JH, Park SR, Chay KO, Seo YW, Kook H,
Ahn KY, Kim YJ and Kim KK: KAI1 COOH-terminal interacting
tetraspanin (KITENIN), a member of the tetraspanin family,
interacts with KAI1, a tumor metastasis suppressor, and enhances
metastasis of cancer. Cancer Res. 64:4235–4243. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chen H, Gao F, He M, Ding XF, Wong AM, Sze
SC, Yu AC, Sun T, Chan AW, Wang X and Wong N: Long-read RNA
sequencing identifies Alternative splice variants in hepatocellular
carcinoma and tumor-specific isoforms. Hepatology. Jan
13–2019.(Epub ahead of print). doi: 10.1002/hep.30500.
|
|
53
|
Li S, Hu Z, Zhao Y, Huang S and He X:
Transcriptome-wide analysis reveals the landscape of aberrant
alternative splicing events in liver cancer. Hepatology.
69:359–375. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Li Y, Sun N, Lu Z, Sun S, Huang J, Chen Z
and He J: Prognostic alternative mRNA splicing signature in
non-small cell lung cancer. Cancer Lett. 393:40–51. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zhu J, Chen Z and Yong L: Systematic
profiling of alternative splicing signature reveals prognostic
predictor for ovarian cancer. Gynecol Oncol. 148:368–374. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Lin P, He RQ, Ma FC, Liang L, He Y, Yang
H, Dang YW and Chen G: Systematic analysis of survival-associated
alternative splicing signatures in gastrointestinal
pan-adenocarcinomas. EBioMedicine. 34:46–60. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Mao S, Li Y, Lu Z, Che Y, Sun S, Huang J,
Lei Y, Wang X, Liu C, Zheng S, et al: Survival-associated
alternative splicing signatures in esophageal carcinoma.
Carcinogenesis. 40:121–130. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Jiang Z, Zhou Q, Ge C, Yang J, Li H, Chen
T, Xie H, Cui Y, Shao M, Li J and Tian H: Rpn10 promotes tumor
progression by regulating hypoxia-inducible factor 1 alpha through
the PTEN/Akt signaling pathway in hepatocellular carcinoma. Cancer
Lett. 447:1–11. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Wang D, Zhang X, Lu Y, Wang X and Zhu L:
Hypoxia inducible factor 1α in hepatocellular carcinoma with
cirrhosis: Association with prognosis. Pathol Res Pract.
214:1987–1992. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Jones RG and Thompson CB: Tumor
suppressors and cell metabolism: A recipe for cancer growth. Genes
Dev. 23:537–548. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Zheng L, Yang W, Wu F, Wang C, Yu L, Tang
L, Qiu B, Li Y, Guo L, Wu M, et al: Prognostic significance of AMPK
activation and therapeutic effects of metformin in hepatocellular
carcinoma. Clin Cancer Res. 19:5372–5380. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Zhou X, Li X, Cheng Y, Wu W, Xie Z, Xi Q,
Han J, Wu G, Fang J and Feng Y: BCLAF1 and its splicing regulator
SRSF10 regulate the tumorigenic potential of colon cancer cells.
Nat Commun. 5:45812014. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Liu F, Dai M, Xu Q, Zhu X, Zhou Y, Jiang
S, Wang Y, Ai Z, Ma L, Zhang Y, et al: SRSF10-mediated IL1RAP
alternative splicing regulates cervical cancer oncogenesis via
mIL1RAP- NF-κB-CD47 axis. Oncogene. 37:2394–2409. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Chen C, Lei J, Zheng Q, Tan S, Ding K and
Yu C: Poly(rC) binding protein 2 (PCBP2) promotes the viability of
human gastric cancer cells by regulating CDK2. FEBS Open Bio.
8:764–773. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Luo K and Zhuang K: High expression of
PCBP2 is associated with progression and poor prognosis in patients
with glioblastoma. Biomed Pharmacother. 94:659–665. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Li F, Bullough KZ, Vashisht AA,
Wohlschlegel JA and Philpott CC: Poly(rC)-binding protein 2
regulates hippo signaling to control growth in breast epithelial
cells. Mol Cell Biol. 36:2121–2131. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Hu CE, Liu YC, Zhang HD and Huang GJ: The
RNA-binding protein PCBP2 facilitates gastric carcinoma growth by
targeting miR-34a. Biochem Biophys Res Commun. 448:437–442. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Wan C, Gong C, Zhang H, Hua L, Li X, Chen
X, Chen Y, Ding X, He S, Cao W, et al: β2-adrenergic receptor
signaling promotes pancreatic ductal adenocarcinoma (PDAC)
progression through facilitating PCBP2-dependent c-myc expression.
Cancer Lett. 373:67–76. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Ghanem LR, Kromer A, Silverman IM, Ji X,
Gazzara M, Nguyen N, Aguilar G, Martinelli M, Barash Y and
Liebhaber SA: Poly(C)-binding protein Pcbp2 enables differentiation
of definitive erythropoiesis by directing functional splicing of
the runx1 transcript. Mol Cell Biol. 38:162018. View Article : Google Scholar
|
|
70
|
Liu H, Chen Z, Jin W, Barve A, Wan YY and
Cheng K: Silencing of α-complex protein-2 reverses alcohol- and
cytokine-induced fibrogenesis in hepatic stellate cells. Liver Res.
1:70–79. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Qin Y, Xue B, Liu C, Wang X, Tian R, Xie
Q, Guo M, Li G, Yang D and Zhu H: NLRX1 mediates MAVS degradation
to attenuate the hepatitis C virus-induced innate immune response
through PCBP2. J Virol. 91:232017. View Article : Google Scholar
|
|
72
|
Mora Gallardo C, Sánchez de Diego A,
Gutiérrez Hernández J, Talavera-Gutiérrez A, Fischer T, Martínez-A
C and van Wely KHM: Dido3-dependent SFPQ recruitment maintains
efficiency in mammalian alternative splicing. Nucleic Acids Res.
47:5381–5394. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Ray D, Kazan H, Cook KB, Weirauch MT,
Najafabadi HS, Li X, Gueroussov S, Albu M, Zheng H, Yang A, et al:
A compendium of RNA-binding motifs for decoding gene regulation.
Nature. 499:172–177. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Ru Y, Chen XJ, Guo WZ, Gao SG, Qi YJ, Chen
P, Feng XS and Zhang SJ: NEAT1_2-SFPQ axis mediates cisplatin
resistance in liver cancer cells in vitro. Onco Targets Ther.
11:5695–5702. 2018. View Article : Google Scholar : PubMed/NCBI
|