Introduction
Glioma is the most common malignant tumor type of
the central nervous system and the 5-year overall survival (OS) is
<10%. According to the biological behavior and malignancy of the
tumor, glioma may be divided into four grades from World Health
Organization (WHO) grade I to IV. Low-grade glioma (LGG) includes
WHO grade-I and -II tumors, while the other two grades III and IV
are classified as high-grade glioma (HGG). Of note, glioblastoma
multiforme (GBM), the most malignant glioma type with WHO grade IV,
accounts for ~50% of glioma cases, and has a median survival time
of 14.2 months and a 5-year survival rate of <5% (1). In the past decades, despite improvements
in surgical, radio- and chemotherapies, the treatment of glioma has
remained a tremendous challenge (2,3). However,
with the development of novel emerging immunotherapies, which aim
to reinvigorate antitumor immune responses, outcomes have been
significantly improved in a variety of advanced hematologic and
solid malignancies (4–8). This points out a new direction in terms
of treatment strategies for glioma. Thus, novel therapeutic
approaches targeting the interaction between the tumor
microenvironment and immune response are urgently required in this
field.
Various preclinical studies have demonstrated the
success of immunotherapy-based approaches in animal models and
numerous phase I and II clinical trials suggested immunotherapy to
be safe and, in certain cases, improve progression-free survival
(PFS) and OS (9–13). Preclinical studies using murine models
with orthotopic-transplanted gliomas have provided a marked benefit
of checkpoint inhibitors used individually or in combination with
other immunotherapeutic strategies (42). Numerous glioma-associated antigens,
including interleukin (IL)-13 receptor subunit α2, human epidermal
growth factor receptor 2, EPH receptor A2, gp100 and AIM-2 are
being targeted in glioma (14–16). In
addition, tumor-specific neoantigens, including epidermal growth
factor receptor variant III, are being used to target tumor cells
(16,17). The successful preclinical studies have
prompted a number of clinical studies using dendritic cell vaccines
(11). Furthermore, considerable
progress has been achieved in immunotherapy with antibodies,
adoptive T-cell transfer and chimeric antigen receptor T cells in
their respective fields (18–20). Among the aforementioned therapeutic
strategies, immune checkpoint blockade appears to be an exciting
avenue that warrants further development based on the preclinical
studies.
Immune checkpoint proteins are surface molecules on
certain immune cell populations that activate or inhibit immune
function when engaged to their ligands. Numerous studies have
indicated an interaction between the expression of the
co-inhibitory protein and tumor immune escape (21). Therefore, immunotherapy based on
blocking the interaction between an immune checkpoint protein and
its ligands has the potential to restore functional immune cells
and inhibit tumor progression.
The B7 family, an important class of the immune
checkpoint superfamily, has exhibited great potential for
regulating T-cell function and participating in the immune
response. The growing B7 family is now comprised of 10 members,
including CD80 (B7-1), CD86 (B7-2), programmed cell death 1 ligand
1 (PD-L1 or B7-H1), PD-L2 (B7-DC), inducible T cell co-stimulator
ligand (B7-H2), CD276 (B7-H3), B7-H4, V-set immunoregulatory
receptor (B7-H5), B7-H6 and human endogenous retrovirus-H long
terminal repeat-associating protein 2 (HHLA2 or B7-H7) (22). Among these ligands, PD-L1 and PD-L2
represent two ligands for the PD-1 receptor. Recent studies have
indicated that upregulation of PD-1 and PD-L1 in tumor tissue was
associated with poor prognosis in certain cancer types,
demonstrating that PD-1 and PD-L1 may inhibit the function of
T-cells and promote the immune escape of tumor cells (23–25). The
clinical application of a specific antibody which inhibits the
PD-1/PD-L1 pathway has achieved satisfactory curative effects
(26,27). A recent study reported that
upregulation of PD-1 in glioma predicted a poor prognosis (28), indicating the potential value of this
immune checkpoint protein as a therapeutic target in glioma.
HHLA2 is the most recently discovered member of the
B7 family. Transmembrane and immunoglobulin domain-containing 2
(TMIGD2, also known as IGPR-1 or CD28H) is the only known receptor
identified for HHLA2 (29). While its
exact function remains elusive, it has been reported to have
co-stimulatory as well as co-inhibitory properties (30,31). Zhu
et al (31) indicated that the
interaction between CD28H and B7-H7 on antigen-presenting cells
(APCs) co-stimulated human T-cell proliferation and cytokine
production via a pathway involving AKT phosphorylation. By
contrast, Zhao et al (30)
proposed the opposite function for B7H7: In the presence of the
T-cell antigen receptor (TCR) signaling pathway, B7-H7 inhibits the
proliferation of CD4+ and CD8+ T cells. In
addition, B7-H7 significantly reduces cytokine production by T
cells, including interferon-γ, tumor necrosis factor-α, IL-5,
IL-10, IL-13, IL-17α and IL-22. Thus, the ligation of B7-H7 to T
cells suppresses T-cell responses. As with B7-H3, a T-cell
co-inhibitory role and a co-stimulatory role have been reported for
this ligand (22). One explanation is
that HHLA2 has two ligands with opposite functions-TMIGD has a
co-stimulatory role, while the other remains elusive. HHLA2 on APCs
or tumor cells may interact with unknown ligands and exert a
co-inhibitory function in the microenvironment of certain cancers.
Furthermore, it may promote angiogenesis within the tumor
microenvironment via its interaction with TMIGD2 expressed in the
endothelium.
The expression of HHLA2 has been reported in a large
proportion of tumor specimens, including breast, lung, thyroid,
melanoma, pancreas, ovary, liver, bladder, colon, prostate, kidney
and esophageal, but not in endometrial, gallbladder, laryngeal,
stomach and uterine cancer or in lymphoma (29). To date, no systematic study on the
expression status and biological function of HHLA2 in patients with
glioma has been performed, to the best of our knowledge. The
present study aimed to examine the expression of HHLA2 in normal
brain specimens and tumor specimens obtained from patients with
glioma. Furthermore, the potential mechanistic role of HHLA2 in
glioma and the association between HHLA2 expression and tumor
behavior were investigated, and its clinical utility as a
prognostic predictor was assessed.
Materials and methods
Sample and data collection
RNA sequencing data from human glioma samples were
obtained from The Cancer Genome Atlas (TCGA) database (http://www.tcga.org/) and downloaded from the GlioVis
database (http://gliovis.bioinfo.cnio.es/). The dataset
contained 515 LGG samples, 152 GBM samples and 2 undefined samples
(Table I). The characteristics of the
patients are listed in Table I.
Furthermore, data regarding IDH mutation, 1p/19q co-deletion and
telomerase reverse transcriptase (TERT) mutation for the TCGA
cohort were obtained by whole-exon sequencing or
pyrosequencing.
 | Table I.Information of patients with
glioma. |
Table I.
Information of patients with
glioma.
| TCGA database
variable | No. of cases
(N=669) |
|---|
| Information of TCGA
patients |
|
| Age (years) |
|
|
<48 | 312 |
|
≥47 | 297 |
| Missing
data | 60 |
| Sex |
|
|
Male | 355 |
|
Female | 254 |
| Missing
data | 60 |
| IDH |
|
|
Mutant | 429 |
|
Wild-type | 232 |
| Missing
data | 8 |
| OS (months) |
|
|
<26 | 442 |
|
≥26 | 225 |
| Missing
data | 2 |
| Status |
|
|
Survival | 428 |
|
Dead | 239 |
| Missing
data | 2 |
Immunohistochemistry (IHC)
The IHC labelling images of normal brain tissue and
tumor tissue were obtained from The Human Protein Atlas (http://www.proteinatlas.org/). The Human Protein Atlas
used anti-HHLA2 (cat. no. HPA055478; Sigma-Aldrich; Merck KGaA) as
a primary antibody and the tumor tissues were obtained from TCGA
database.
Model of tumor-infiltrating immune
cells (TIICs)
xCell (http://xcell.ucsf.edu/) was used to obtain the
expression level of 64 types of immune cells in the tumor
microenvironment of patients with glioma. Then 20 types of immune
cells with significant difference in infiltration ratio were
screened out using the R package limma (http://bioconductor.riken.jp/packages/3.0/bioc/html/limma.html).
Functional enrichment analysis
Gene ontology (GO) and pathway enrichment analysis
[Kyoto Encyclopedia of Genes and Genomes (KEGG)] were performed to
analyze the genes associated with HHLA2 by using Metascape
(http://metascape.org). Enriched ontological terms
and pathways with P<0.05 were selected and presented in a
heatmap using the R package ‘ComplexHeatmap’ (http://www.bioconductor.org/packages/stats/bioc/ComplexHeatmap/).
Cox proportional hazards regression
model
The prognostic value of each factor was first
assessed by univariate Cox proportional hazards regression.
Subsequently, statistically significant genes were used to
construct the multivariate Cox regression model. Glioma samples
were divided into high-expression and low-expression groups based
on the median level of HHLA2 expression. Kaplan-Meier survival
curves were generated to assess the prognostic value of the model
using the R package survival (https://CRAN.R-project.org/package=survival). A
receiver operating characteristic (ROC) curve was generated to
assess the accuracy of the model with the R package ‘survivalROC’
(https://CRAN.R-project.org/package=survivalROC).
Statistical analysis
Statistical analysis was mainly performed with R
(https://www.r-project.org/) with several
publicly available packages. P<0.05 was considered to indicate
statistical significance. (*P<0.05, **P<0.01, ***P<0.001
and ****P<0.0001, respectively, as indicated in the figures and
legends).
Results
HHLA2 expression is absent in GBM
To evaluate the expression level of HHLA2 in glioma,
the IHC staining data of glioma and normal brain tissue were
obtained from the Human Protein Atlas dataset and analyzed
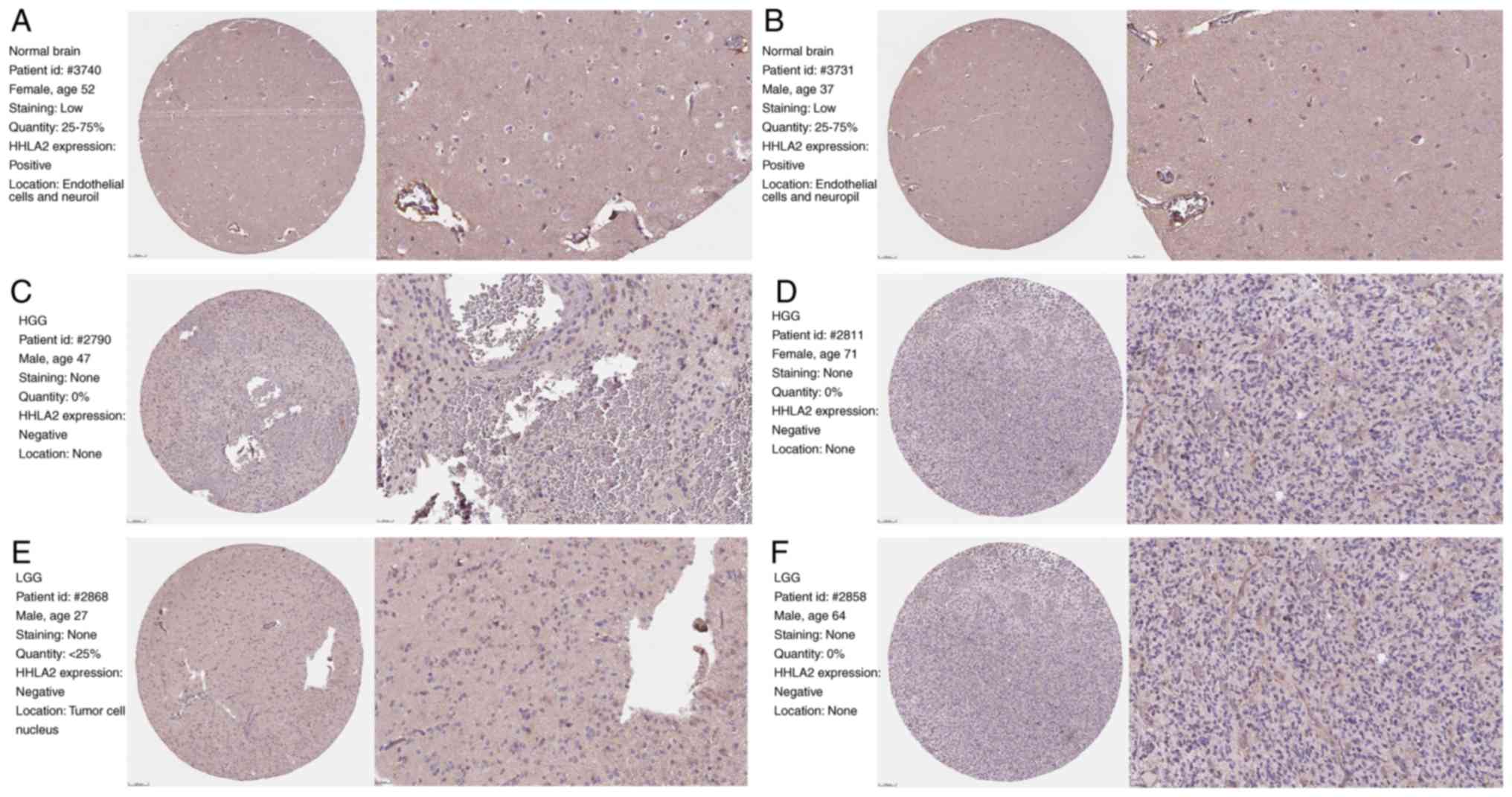
individually. Overall, positive staining for HHLA2 was observed in
endothelial cells and neuropils of normal brain tissue (Fig. 1A and B), while staining was negative
in glial cells and neurons. Furthermore, no tumoral HHLA2
expression was detected in HGG (Fig. 1C
and D). Of note, only a small percentage of LGG samples were
positive for HHLA2 and IHC labeling was observed in tumor cell
nuclei rather than endothelial cells, while other samples with LGG
were negative for HHLA2 (Fig. 1E and
F). This result indicated that with the increasing degree of
tumor malignancy, HHLA2 expression in glioma was gradually reduced
until it was absent.
Downregulated HHLA2 predicts poor
prognosis in glioma
To explore the prognostic role of HHLA2 in glioma,
the association between HHLA2 and several prognostic factors was
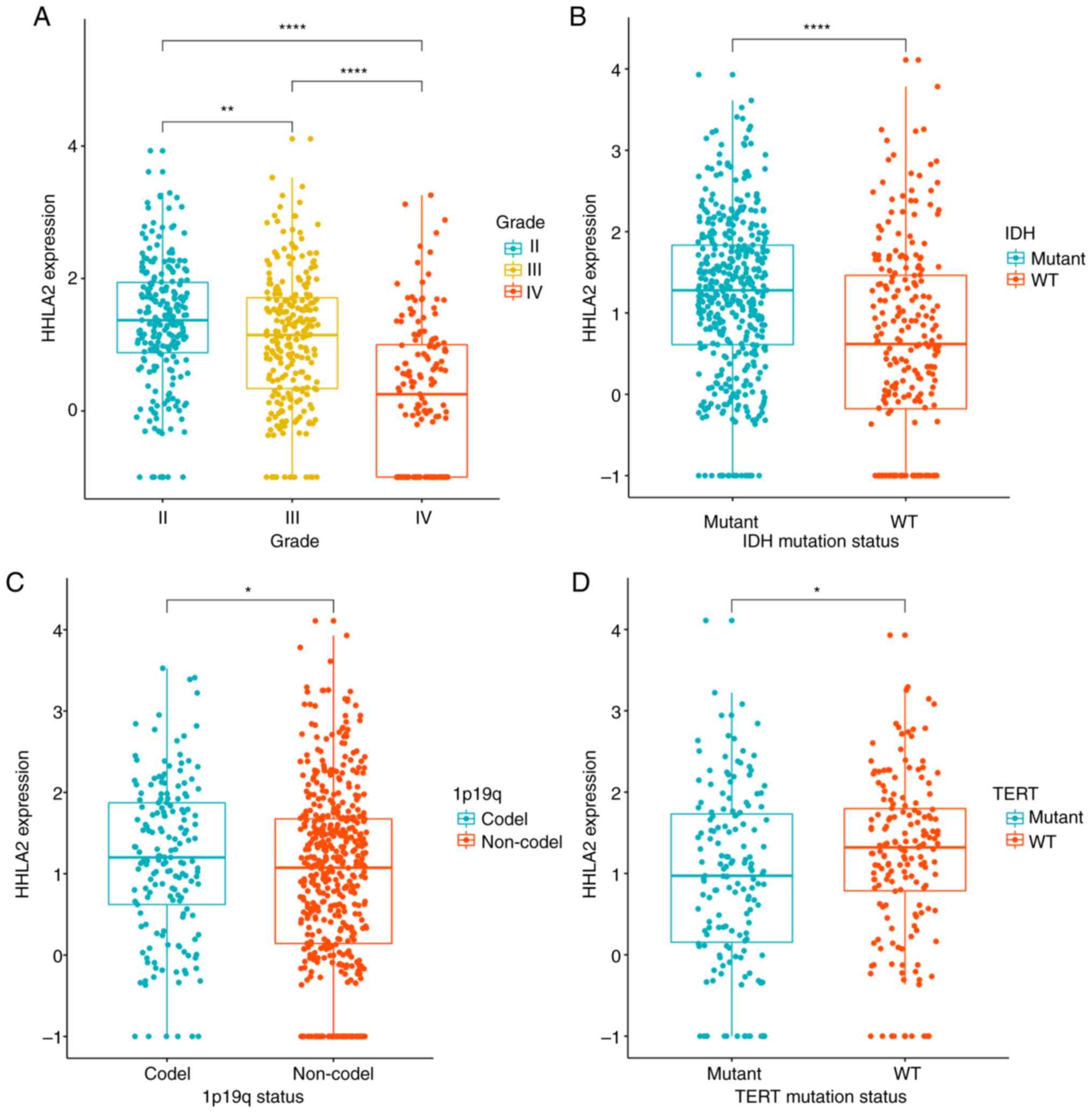
analyzed. The results indicated that the mRNA expression levels of
HHLA2 were significantly decreased with the increase in the grade
of glioma and that the expression level was lowest in GBM
(P<0.0001; Fig. 2A), indicating a
strong correlation between HHLA2 expression and malignancy of
glioma. IDH-mutant and 1p/19q co-deletion types were associated
with a better outcome in glioma. When taking into account the IDH
mutation status, it was indicated that HHLA2 expression was
significantly higher in the IDH mutant group than in the IDH
wild-type group (P<0.0001; Fig.
2B). Furthermore, compared with the 1p/19q no-deletion group,
the 1p/19q co-deletion group had a higher expression of HHLA2
(P<0.05; Fig. 2C). TERT promoter
mutations are usually considered to be associated with poor outcome
(32), and the present results
revealed that in the TERT wild-type group, the expression of HHLA2
was significantly increased (P<0.05; Fig. 2D). These results indicated HHLA2
expression was more prevalent in glioma with lower malignancy.
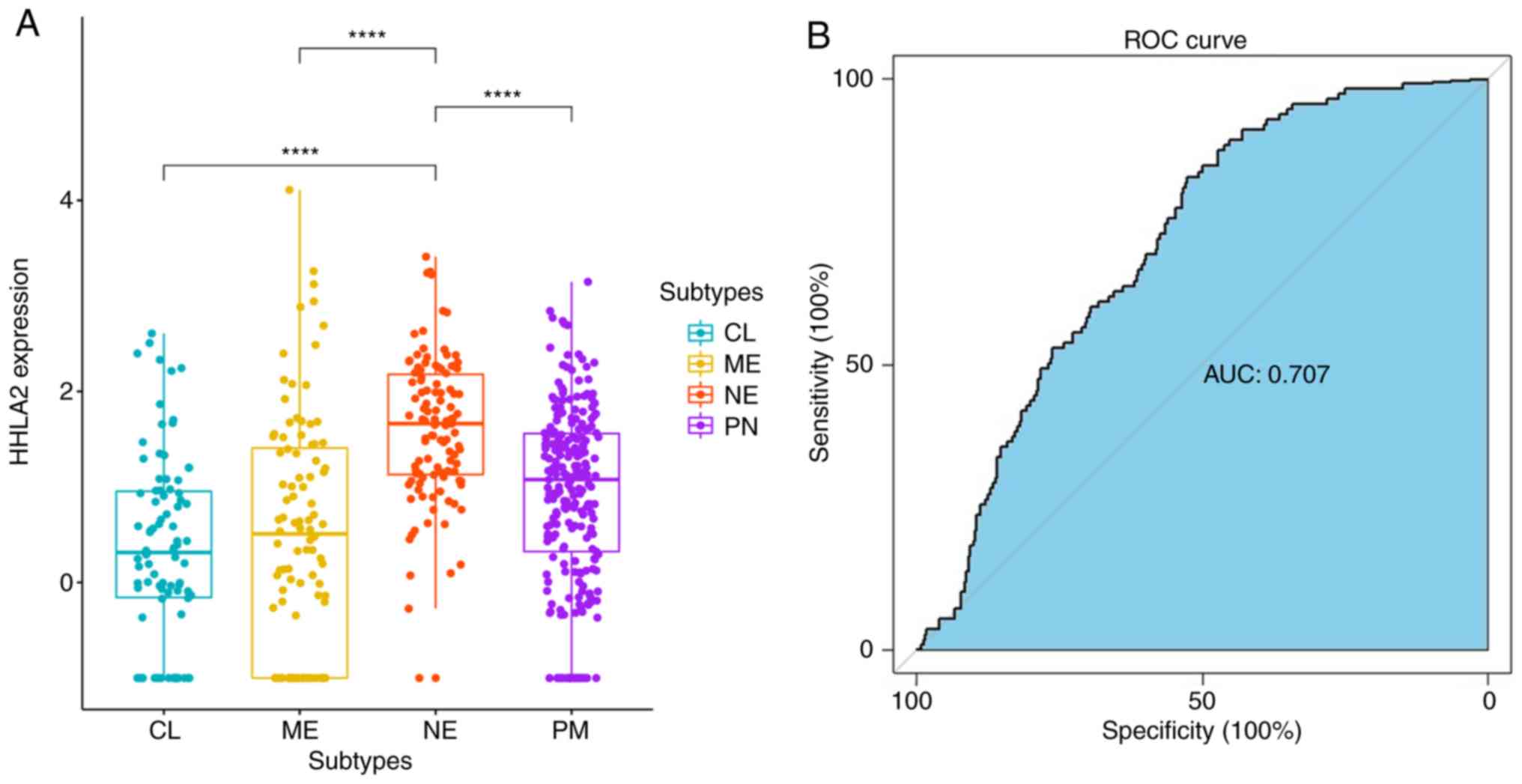
To further elucidate the association between HHLA2
expression and molecular subtypes, patients were divided into four
groups according to subtypes defined by TCGA. Upregulated HHLA2
expression was observed in the neural (NE) subtype rather than in
proneural (PN), classical (CL), and mesenchymal (ME) subtypes
(P<0.0001; Fig. 3A). In addition,
ROC curves were used to evaluate the specificity and sensitivity of
our previous findings, indicating that the expression status of
HHLA2 may serve as a good predictor for the neural subtype of
gliomas [area under curve (AUC)=0.707; Fig. 3B].
Model of tumor-infiltrating immune
cells (TIICs) and tumor-associated macrophages (TAMs) in
glioma
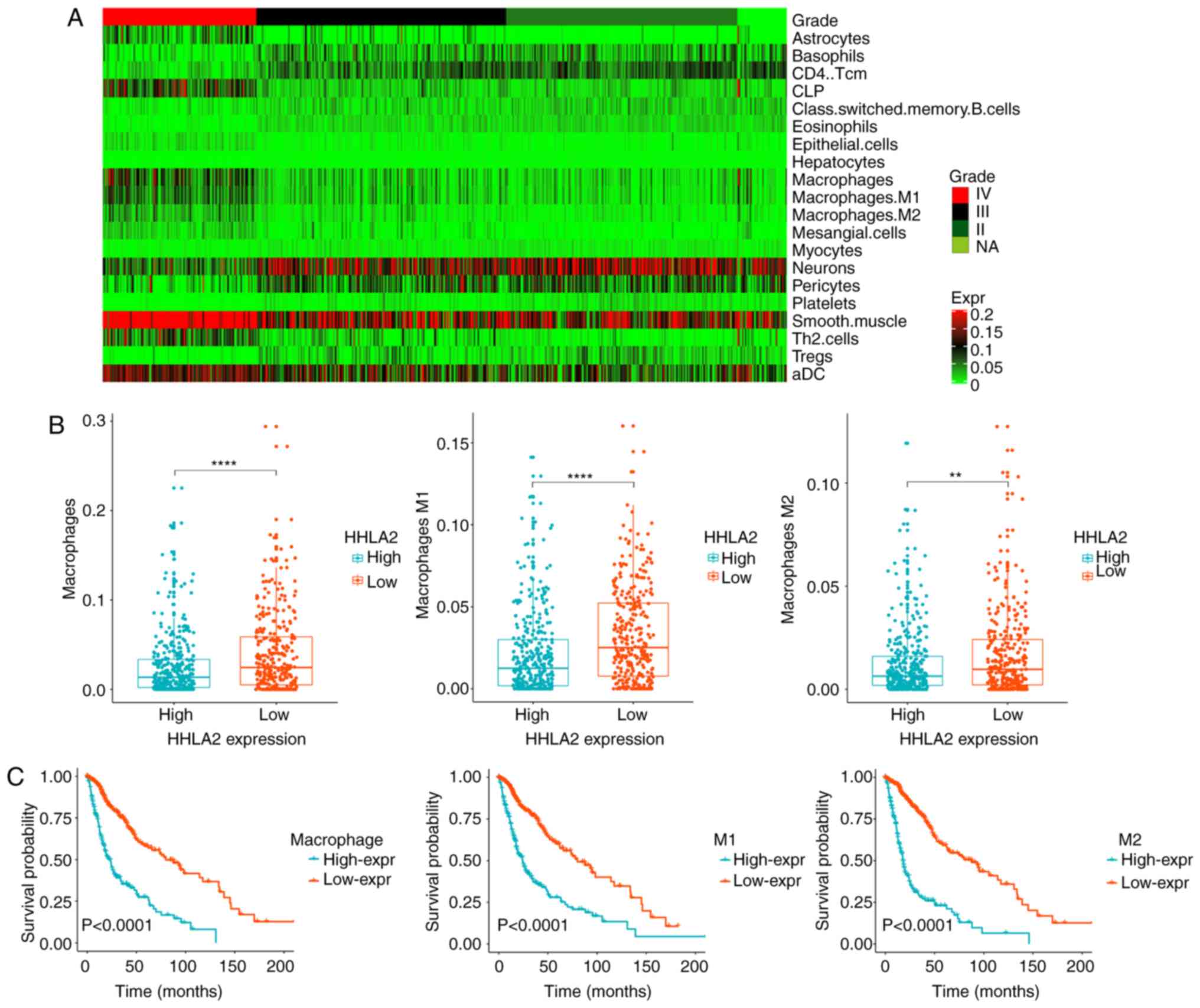
To date, tumor immunotherapy has yielded
significantly improved outcomes in a variety of advanced
hematologic and solid malignancies, including glioma. Hence, to
further understand the function of immune cells in the tumor
microenvironment, the expression model of TIICs in patients with
glioma was explored using xCell (http://xcell.ucsf.edu/) and several immune cells with
significant difference in infiltration ratio were screened out
(Table II). It was revealed that
macrophages were markedly increased in GBM (Fig. 4A). TAMs, developed from monocytes,
have been confirmed to be the most important type of immune cell in
the stroma of tumors, accounting for 50% of the total number of
immune cells, and to have an important role in neoplasia,
metastasis, immune escape and tumor angiogenesis (33,34).
Previous studies have also indicated that HHLA2 is constitutively
expressed on human monocytes and takes part in angiogenesis
(21). In the present study, it was
observed that TAMs were significantly higher in the HHLA2
low-expression group (Fig. 4B) and
predicted a worse prognosis (Fig.
4C). The aforementioned results indicated that HHLA2 may have
an important role in the tumor immune microenvironment, tumor
angiogenesis and the process of monocytes developing into TAMs.
Thus, the association between HHLA2 and TAMs may provide a novel
therapeutic method.
 | Table II.Differentially expressed immune
cells. |
Table II.
Differentially expressed immune
cells.
| Immune cells | Log FD | Adjusted
P-value |
|---|
| Upregulated |
|
|
| Smooth
muscle | −0.162091191 | 3.79E-46 |
|
Macrophages M1 | −0.035218908 | 3.26E-26 |
| DC | −0.047622184 | 4.18E-26 |
|
CLP | −0.064684851 | 4.44E-26 |
| Th2
cells | −0.056711574 | 8.06E-25 |
|
Macrophages | −0.05311427 | 1.73E-22 |
|
Macrophages M2 | −0.022494652 | 2.36E-19 |
|
Mesangial cells | −0.013731035 | 4.96E-19 |
|
Astrocytes | −0.034756525 | 7.22E-17 |
|
Epithelial cells | −0.008775589 | 1.14E-16 |
| Downregulated |
|
|
|
CD4+ Tcm | 0.041603702 | 1.88E-76 |
|
Eosinophils | 0.012507035 | 6.73E-69 |
|
Tregs | 0.018249872 | 9.22E-44 |
|
Neurons | 0.085024979 | 1.87E-40 |
|
Platelets | 0.007315915 | 2.78E-38 |
|
Hepatocytes | 0.001416468 | 1.99E-37 |
|
Basophils | 0.025025816 | 5.25E-29 |
|
Pericytes | 0.040365872 | 8.10E-23 |
|
Class-switched memory
B-cells | 0.010062028 | 2.55E-16 |
|
Myocytes | 0.00361773 | 5.36E-12 |
Correlation of HHLA2 and associated
immune molecules
To further explore the function of HHLA2 in the
immune microenvironment, the correlation between HHLA2 and several
immune-associated molecules was analyzed at the mRNA level. Pearson
correlation analysis indicated that HHLA2 was revealed to be
negatively correlated with co-inhibitory immune checkpoint
molecules, including programmed cell death 1 (PD-1; r=−0.172),
lymphocyte activating 3 (LAG3; r=−0.171), cytotoxic T-lymphocyte
associated protein 4 (CTLA4; r=−0.045) and CD276 (r=−0.434;
Fig. 5A). However, HHLA2 was
positively correlated with CD160 (r=0.261), which is commonly known
as a stimulatory molecule (21,29,30). In
addition, common immune inhibitors, including IL-10 and
transforming growth factor (TGF)-β, were significantly higher in
the HHLA2 low-expression group (Fig. 5C
and D). These results indicated that HHLA2 may co-stimulate the
immune response and have a positive role in tumor immune
microenvironment.
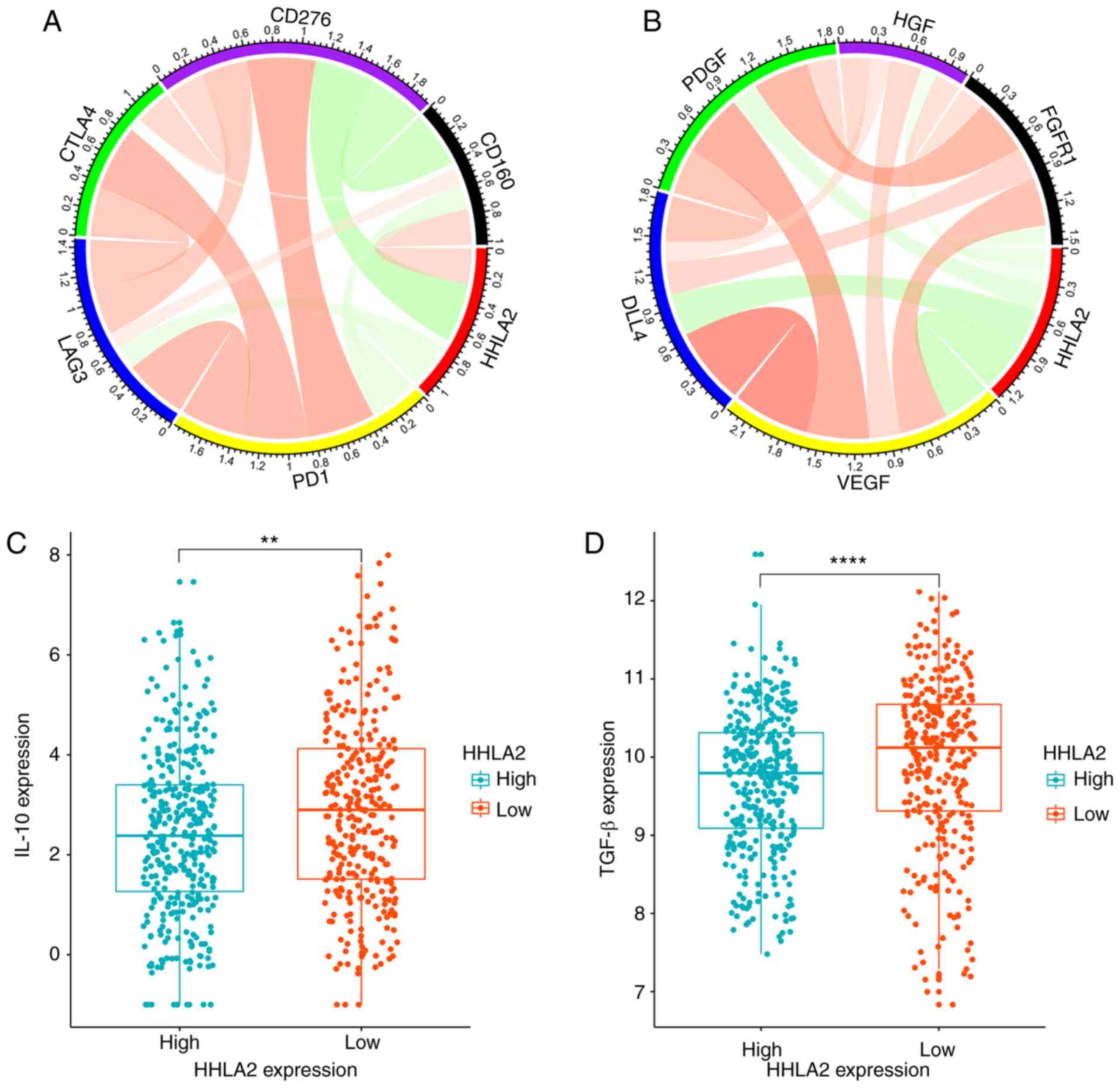 | Figure 5.Correlation of HHLA2 and related
molecules. (A) HHLA2 was positively correlated with CD160 and
negatively correlated with PD-L1, LAG3, CTLA4, and CD276. (B) HHLA2
was negatively correlated with angiogenesis molecules, including
VEGF, DLL4, PDGF, FGFR1 and HGF. Color intensity and the size of
the circle are proportional to the correlation coefficients. (C and
D) IL-10 and TGF-β, representative immune inhibitors, were
significantly higher in the low-HHLA2 expression group (**P<0.01
and ****P<0.0001, respectively). HHLA2, human endogenous
retrovirus-H long terminal repeat-associating protein 2; PD-L1,
programmed cell death 1 ligand 1; LAG3, lymphocyte activating 3;
CTLA4, cytotoxic T-lymphocyte associated protein 4; DLL4, δ-like
canonical Notch ligand 4; FGFR1, fibroblast growth factor receptor
1; HGF, hepatocyte growth factor. |
Mechanism of HHLA2 acting on TAMs
It has been reported that cytokines, including
vascular endothelial growth factor (VEGF) and platelet-derived
growth factor (PDGF), have important roles in the formation of TAMs
and tumor angiogenesis (33,34). To further assess the mechanisms by
which HHLA2 acts on TAMs in malignant glioma, five common molecules
associated with angiogenesis were selected and analyzed
individually (35,36). In the TCGA dataset, it was observed
that HHLA2 was significantly negatively correlated with molecules
including VEGF (r=−0.400), δ-like canonical Notch ligand 4 (DDL4;
r=−0.358), PDGFA (r=−0.228), fibroblast growth factor receptor 1
(FGFR1; r=−0.164) and hepatocyte growth factor (HGF; r=−0.137;
Fig. 5B). These results demonstrated
that overexpression of HHLA2 may have a potential application in
anti-tumor angiogenesis treatment and inhibiting the formation of
TAMs by decreasing VEGF and PDGF.
Enrichment analysis of
HHLA2-associated genes
To further explore the biological function of HHLA2
in glioma, an enrichment analysis with Metascape (http://metascape.org) was also performed. Genes
significantly associated with HHLA2 expression were screened out by
Pearson correlation analysis (Pearson |R|>0.4, P<0.05).
Sequentially, 234 positively correlated genes and 211 negatively
correlated genes were analyzed individually. It was revealed that
positively correlated genes were involved in membrane trafficking,
the glutamate receptor signaling pathway, regulation of neuronal
death, response to toxic substances, regulation of protein
ubiquitination and negative regulation of protein modification
process (Fig. 6A), while negatively
correlated genes were involved in processes that promote abnormal
proliferation, including cell division, DNA replication, DNA
repair, activation of E2F transcription factor 1 (E2F1) target
genes at the G1/S checkpoint, the FOXM1 pathway and the ATR pathway
(Fig. 6B). These results indicated
that HHLA2 may have an important role in preventing normal neurons
from damage, promoting the immune response and inhibiting
neoplastic cell proliferation.
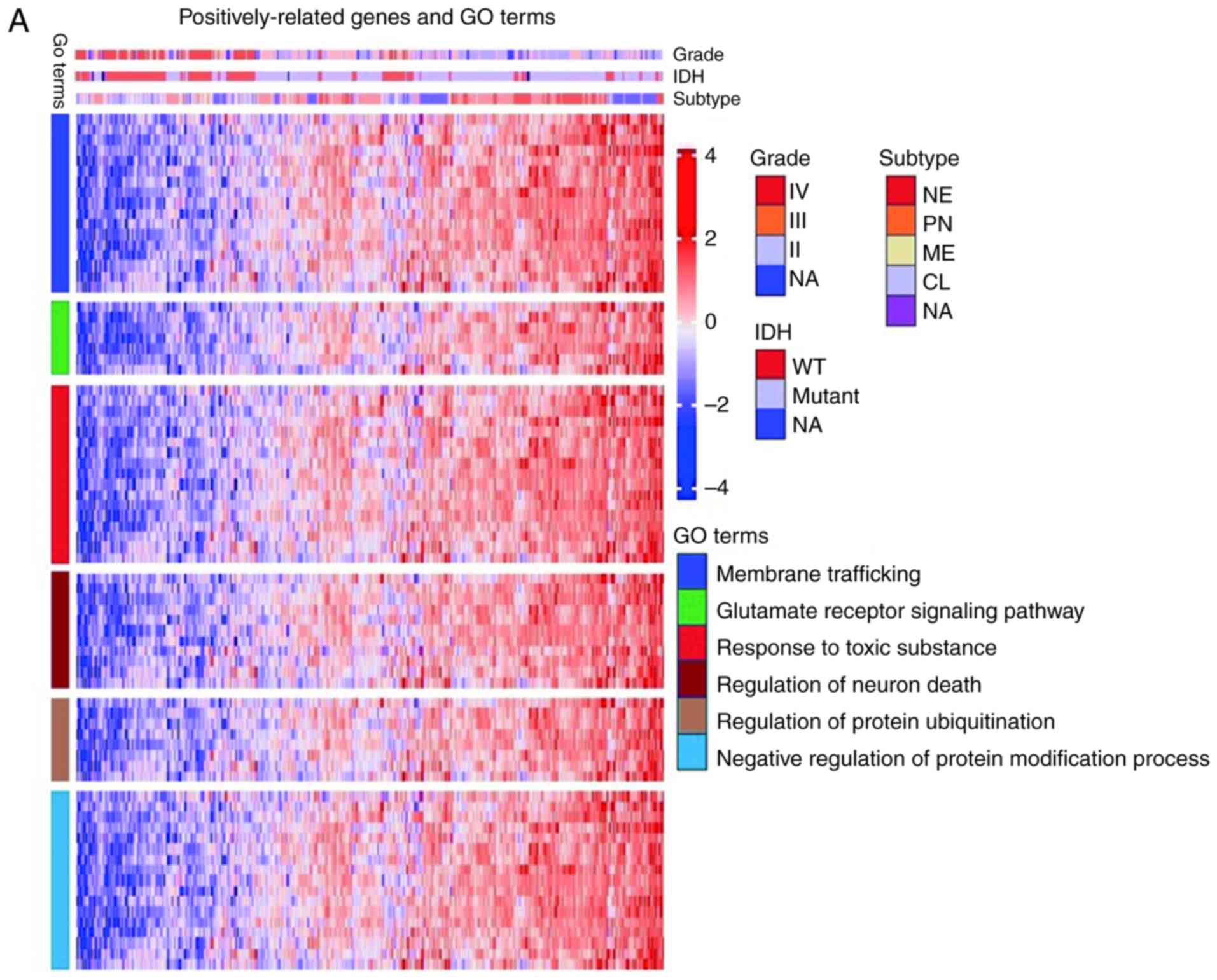 | Figure 6.Enrichment analysis of HHLA2-related
genes. (A) The positively-related genes were involved in biological
process including membrane trafficking, the glutamate receptor
signaling pathway, regulation of neuron death, response to toxic
substance, regulation of protein ubiquitination, and negative
regulation of protein modification process. (B) Negatively-related
genes were involved in biological processes including cell
division, DNA replication, DNA repair, activation of E2F1 target
genes at G1/S, the FOXM1 pathway, and the ATR pathway. HHLA2, human
endogenous retrovirus-H long terminal repeat-associating protein
2. |
Patients with increased HHLA2 have a
favorable survival prognosis
As HHLA2 expression was correlated with favorable
prognostic factors, the prognostic value of HHLA2 expression in
glioma as well as in GBM was then explored. Patients were divided
into a high-expression group and a low-expression group based on
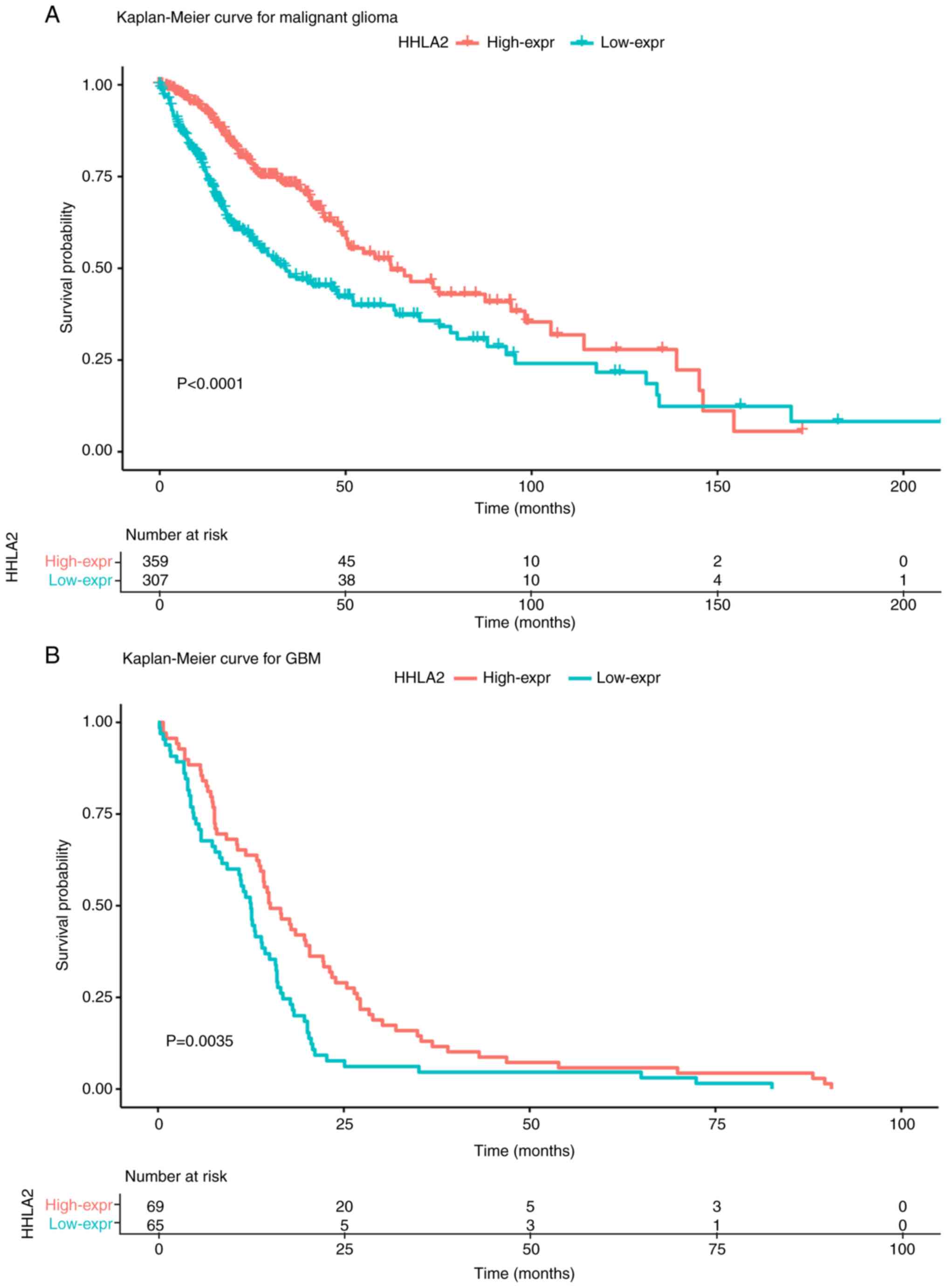
the median HHLA2 level. Kaplan-Meier analysis demonstrated that
higher HHLA2 expression was associated with a better outcome in
patients with glioma of all grades (P<0.0001) as well as in GBM
patients (P=0.021; Fig. 7A and
B).
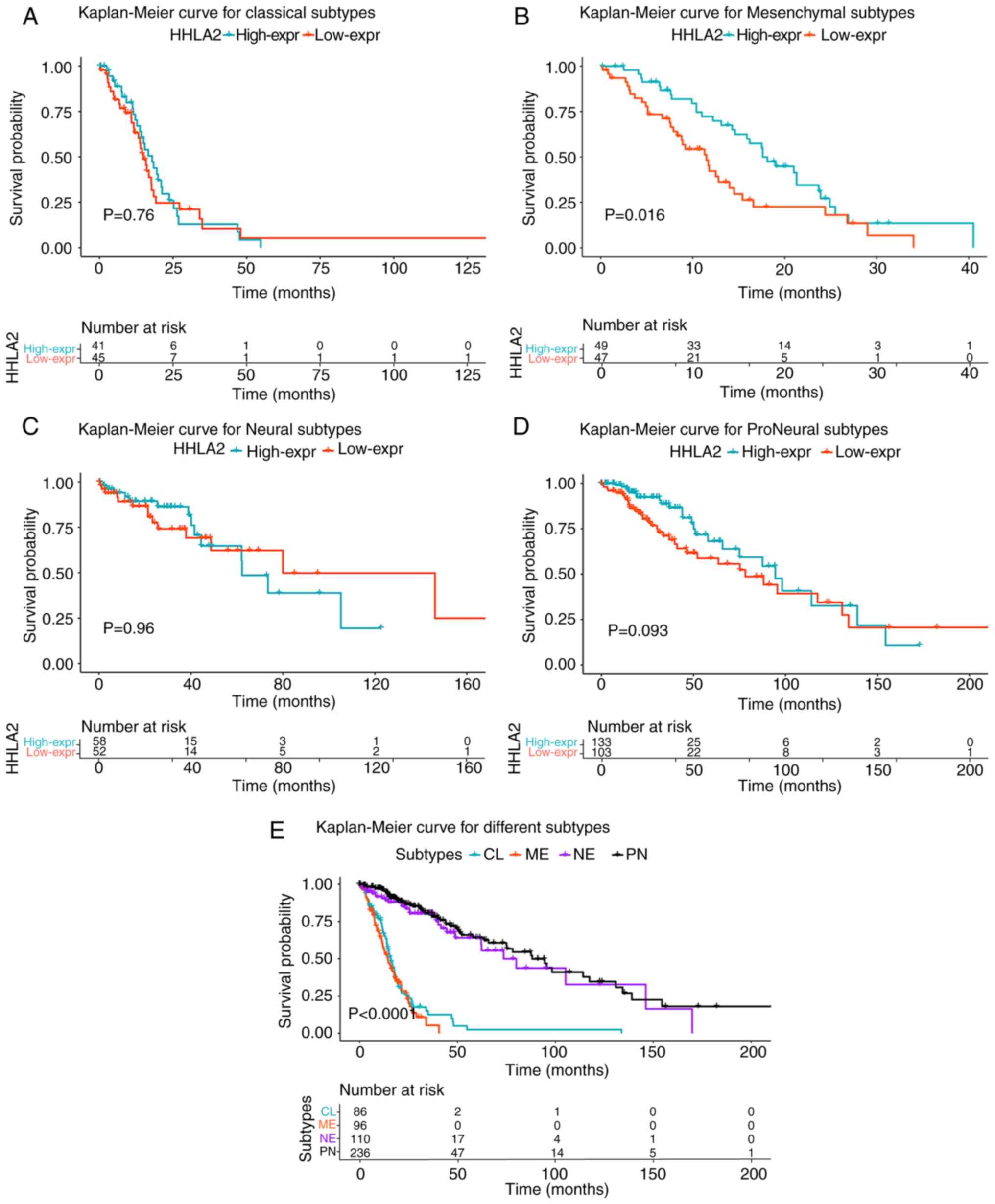
To further comprehend this model, a survival
analysis was performed in four different subtypes of glioma defined
by TCGA (Fig. 8A-D). The results
demonstrated that no statistical significance was detected in the
CL, NE and PN subtypes (Fig. 8A, C and
D). However, higher HHLA2 expression was significantly
associated with a better prognosis in the ME subtype (P=0.013;
Fig. 8B). In addition, compared with
the CL and ME subtypes, patients with the NE and PN subtypes had a
significantly better outcome (P<0.0001; Fig. 8E).
In order to take into account key clinical and
molecular factors, the Cox proportional hazards model was further
applied. Univariate analysis indicated that age, IDH status, grade
and HHLA2 expression were significantly associated with OS
(P<0.0001; Table III).
Furthermore, multivariate analysis indicated that age, IDH status
and grade were independent prognostic factors (P<0.0001;
Table III). However, HHLA2
expression was not an independent prognostic factor according to
the multivariate analysis (P=0.257).
 | Table III.Univariate and multivariate Cox
analysis for OS. |
Table III.
Univariate and multivariate Cox
analysis for OS.
| Characteristic | P-value | HR | 95% CI |
|---|
| Univariate |
|
|
|
|
Age | <0.001 | 4.819 | 3.505–6.625 |
|
Grade | <0.001 | 0.099 | 0.074–0.133 |
| IDH
Wild-type | <0.001 | 8.848 | 6.707–11.670 |
|
HHLA2 | <0.001 | 0.526 | 0.406–0.681 |
| Multivariate |
|
|
|
|
Age | <0.001 | 1.963 | 1.339–2.880 |
|
Grade | <0.001 | 0.193 | 0.133–0.279 |
| IDH
Wild-type | <0.001 | 2.373 | 1.655–3.404 |
|
HHLA2 | 0.257 | 0.854 | 0.650–1.122 |
Discussion
The present study first focused on detecting the
expression level of HHLA2 in normal brain tissue and tumor tissue
obtained from patients with glioma by IHC labeling in the Human
Protein Atlas dataset. By individually analyzing the normal brain
tissue and tumor tissue, it was revealed that HHLA2 was absent in
normal brain cells, including glial cells and neurons, but abundant
in endothelial cells. Furthermore, it was observed that tumoral
HHLA2 expression was absent in HGG, particularly in GBM. Of note,
in contrast to the aforementioned, weak expression of HHLA2 in the
nuclei of tumor cells was observed in part of the patients with
LGG. This result suggested a downward trend in HHLA2 expression
with the increase in the degree of malignancy of the tumor, which
is opposite to previous results according to which HHLA2 expression
was not detected in most organs, but was widely expressed in human
cancers of the breast, lung, thyroid, skin, pancreas, ovary, liver,
bladder, colon, prostate, kidney and esophagus (29). However, Yan et al (37) also reported that HHLA2 was widely
overexpressed in early pancreatic precancerous lesions compared
with pancreatic cancer, although it was not expressed in normal
acinar, islet and ductal cells. Furthermore, overexpression of
HHLA2 was indicated to be significantly associated with a better
outcome. In addition, Zhu et al (31) indicated that HHLA2 engaged with CD28H
to co-stimulate human T-cell proliferation and cytokine production
via a pathway involving AKT phosphorylation. Based on the
aforementioned results, it is reasonable to assume that higher
HHLA2 expression in the early stage of glioma co-stimulated the
immune response, while the expression decreased with the increasing
malignancy of the tumor. It may be speculated that the expression
model of HHLA2 in glioma is similar to that in the pancreas.
However, most patients with glioma only present at the hospital
after evident clinical symptoms have occurred. On this account, it
is difficult to obtain tumor tissue in the early stage of glioma
for IHC labeling and detection of HHLA2 expression. Furthermore,
the present cancer model was validated in the TCGA dataset at the
mRNA level at the same time.
Through analysis of TCGA, the largest cancer
dataset, HHLA2 expression in malignant glioma was assessed at the
transcriptional level. It was revealed that HHLA2 expression in GBM
was significantly lower than that in glioma of other grades.
According to a study by Eckel-Passow et al (32), a single TERT mutation predicted poor
prognosis in glioma, while IDH mutation and 1p/19q co-deletion
predicted a favorable outcome. In the present study, tumors with
IDH mutation, 1p/19q co-deletion and wild-type TERT expressed
higher levels of HHLA2, which was in line with our previous
assumption. Furthermore, the expression levels of HHLA2 were
detected in four different molecular subtypes of glioma defined by
TCGA: PN, NE, CL and ME (38,39). Studies have indicated that PN and NE
subtypes mostly occurred among LGG and were associated with a
favorable prognosis, while the CL and ME subtypes were associated
with a worse outcome (38,40). Of note, in the NE subtype, a
significantly higher expression of HHLA2 compared with that in the
other subtypes was observed, and the HHLA2 expression status was a
good predictor for NE-subtype glioma. These results were in line
with a previous study and indicated that higher expression of HHLA2
may predict a favorable outcome (37).
The present study further explored the expression
model of TIICs in glioma, and it was indicated that macrophages
were markedly increased in GBM vs. LGG. TAMs, developed from
monocytes, have been confirmed to be the most important type of
immune cell in the stroma of tumors, accounting for 50% of all
immune cells, and to have an important role in neoplasia,
metastasis, immune escape and tumoral angiogenesis (33,34).
Immature monocytes migrate to tumor tissues and develop into TAMs
through several cytokines, including VEGF, PDGF, colony-stimulating
factor-1 (CSF-1) and C-C motif chemokine ligand 2. Notably, a
previous study confirmed that HHLA2 was constitutively expressed on
human monocytes (22). Thus, the
association between TAMs and the expression levels of HHLA2 was
explored in the present study. Lower TAMs were observed in the
HHLA2 high-expression group and predicted a better outcome. To
further explore the mechanism of HHLA2 acting on TAMs, several
cytokines linked to angiogenesis and processes that develop
monocytes into TAMs were selected for assessment, revealing that
HHLA2 was negatively correlated with VEGF and PDGF. These results
demonstrated that HHLA2 may inhibit TAM development and tumor
angiogenesis via anti-VEGF and anti-PDGF processes. Kumar et
al (41) reported that anti-CSF1
receptor, specifically targeting TAMs, plus anti-PD1 treatment
significantly improved therapies, compared with anti-PD1 treatment
alone. HHLA2, which may not only inhibit TAM development but also
co-stimulate immune function, has promising potential in immune
therapy for patients with glioma.
The function of HHLA2 in the immune microenvironment
of glioma was then assessed. Taking its role as an immune
stimulator into account, its correlation with several common immune
inhibitors was explored. According to Kamran et al (42), TGF-β and IL-10 are central to
maintaining the immunosuppressive microenvironment of glioma. Thus,
TGF-β and IL-10 were selected as representative immune inhibitors
and included in the analysis. It was observed that IL-10 was
significantly increased in the low HHLA2 expression group, which
was also the case for TGF-β. The function of HHLA2 was then
explored in-depth via GO and KEGG enrichment analysis. The results
demonstrated that HHLA2 was positively associated with response to
toxic substances, regulation of neuronal death, the glutamate
receptor signaling pathway and regulation of protein processes.
This indicated that HHLA2 is able to prevent normal neurons from
damage by negatively regulating the glutamate receptor signaling
pathway. Furthermore, genes which were negatively correlated with
HHLA2 were enriched in cell cycle, DNA replication, DNA repair,
activation of E2F1 target genes at G1/S, the FOXM1 pathway and the
ATR pathway. Activation of E2F1 target genes at G1/S, the FOXM1
pathway and the ATR pathway have been previously confirmed to
promote neoplastic proliferation and tumor progression (43–45). These
results indicated that HHLA2 inhibits tumor progression in the
early stage of glioma by participating in early immune response and
inhibiting neoplastic proliferation and angiogenesis. Hence,
exploring the mechanism of HHLA2 acting on tumor cells may
facilitate the discovery of a cure for this disease.
Notably, the present study was the first to report
on the prognostic significance of HHLA2 in glioma. Kaplan-Meier
survival analysis revealed that higher HHLA2 expression was
consistently and significantly associated with better survival in
glioma and GBM. Furthermore, a subsequent Kaplan-Meier survival
curve analysis for each subtype yielded a similar result for the MΕ
subtype of glioma. Lin et al (40) reported that the CL subtype had the
worst outcome and accounted for a large part of GBM, while the NE
and PN subtypes had a high proportion of LGG with a better outcome.
These results may explain for the absence of a significant
correlation between higher HHLA2 expression and better prognosis in
the CL, NE and PN subtypes of glioma. Furthermore, the univariate
analysis indicated that HHLA2 expression was a significant
prognostic factor, while multivariate analysis demonstrated that
the expression of HHLA2 was not an independent prognostic factor.
Regarding the limitations of the present study, part of the
patients included were lost to follow-up. Thus, the potential of
HHLA2 to be an independent prognostic predictor for glioma should
be further investigated in a larger and more comprehensive
dataset.
However, in other types of human cancer, HHLA2
expression is not always a favourable predictor for patient
survival. The prognostic significance of HHLA2 expression in
osteosarcoma and colorectal carcinoma was identified to be
correlated with metastasis and poor survival (46,47).
Similarly, in triple-negative breast cancer, overexpression of
HHLA2 was associated with lymph node positivity and advanced stage
of the disease at the time of diagnosis, and also with an increased
risk of recurrence (29). HHLA2 has
also been reported to predict a favorable outcome in pancreatic
ductal adenocarcinoma and gastric cancer (37,48). One
explanation is that HHLA2 has two opposite ligands (22), including TMIGD that has a
co-stimulatory role, while the other ligand remains elusive. TMIGD
was indicated to be the major receptor in certain cancer types due
to the tumoral immune microenvironment. It is not uncommon for
members of the B7 family to have a dual function depending on the
immune environment, tumor microenvironment or interaction with
different receptors (22). HHLA2
belongs to group III of the B7 family, which also includes B7-H3
(also known as CD276) and B7×. As with HHLA2, the prognostic
significance of B7-H3 and B7× also remains to be further delineated
(49,50). Wang et al (51) reported that B7-H3 was positively
associated with the Toll-like receptor signaling pathway and
predicted poor survival for glioma patients. Zhou et al
(52) also identified that
overexpression of B7-H3 was associated with the malignancy grade of
brainstem gliomas. However, results demonstrating B7-H3 as a
co-stimulatory molecule associated with prolonged survival in
pancreatic cancer (53) and gastric
cancer (54) have also been reported.
Therefore, the association between HHLA2 and other immune
checkpoint molecules was then analyzed, revealing a negative
correlation with checkpoint inhibitors, including PD-1, LAG3 and
B7-H3. These results indicated that anti-PD-1 plus anti-B7-H3
treatment may be a strategy for glioma treatment, as it may lead to
upregulation of HHLA2. Notably, the present study was the first to
indicate that HHLA2 may act as a co-stimulatory molecule in glioma,
in contrast to other B7 family members, which are commonly
characterized as immune checkpoint inhibitors.
To date, observable progress has been made in the
area of immunotherapy. However, blocking the PD-1/PD-L1 pathway was
only effective in a small number of cases and most patients with
glioma still suffered from disease. Thus, novel therapeutic
strategies targeting other immune checkpoint molecules are in
urgent demand. HHLA2, acting as an immune stimulator and inhibiting
the formation of TAMs, may be a potential therapeutic target.
To the best of our knowledge, the present study was
the first to explore the biological function and clinical roles of
HHLA2 in glioma. The results indicated that HHLA2 acts as an immune
stimulator and inhibits the formation of TAMs. Furthermore, HHLA2
expression was significantly correlated with a favorable outcome.
The present study indicated its potential as a prognostic predictor
and novel therapeutic target.
Acknowledgements
We gratefully acknowledge the TCGA project
organizers as well as all study participants for making data and
results available. In addition, we would like to thank GlioVis
database.
Funding
The present study was supported by the National
Natural Science Foundation of China (nos. 81372683 and 81572489)
(to QC), and (no. 81502175) (to BL).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors contributions
YQ, GD and QC designed this study. YQ, GD, PX, HZ,
FY, RG, HJ and BL performed the data collection and collation. All
the authors were involved in the analysis and interpretation of
data. YQ wrote the paper, with the help of the co-authors. GD, BL
and QC reviewed and revised the manuscript. All authors read and
approved the manuscript and agree to be accountable for all aspects
of the research in ensuring that the accuracy or integrity of any
part of the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
All authors declare that they have no competing
interests.
References
|
1
|
Stupp R, Mason WP, van den Bent MJ, Weller
M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn
U, et al: Radiotherapy plus concomitant and adjuvant temozolomide
for glioblastoma. N Engl J Med. 352:987–996. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jiang T, Mao Y, Ma W, Mao Q, You Y, Yang
X, Jiang C, Kang C, Li X, Chen L, et al: CGCG clinical practice
guidelines for the management of adult diffuse gliomas. Cancer
Lett. 375:263–273. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yang P, Wang Y, Peng X, You G, Zhang W,
Yan W, Bao Z, Wang Y, Qiu X and Jiang T: Management and survival
rates in patients with glioma in China (2004–2010): A retrospective
study from a singleinstitution. J Neurooncol. 113:259–266. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Robert C, Long GV, Brady B, Dutriaux C,
Maio M, Mortier L, Hassel JC, Rutkowski P, McNeil C,
Kalinka-Warzocha E, et al: Nivolumab in previously untreated
melanoma without BRAF mutation. N Engl J Med. 372:320–330. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Robert C, Schachter J, Long GV, Arance A,
Grob JJ, Mortier L, Daud A, Carlino MS, McNeil C, Lotem M, et al:
Pembrolizumab versus ipilimumab in advanced melanoma. N Engl J Med.
372:2521–2532. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Brahmer J, Reckamp KL, Baas P, Crinò L,
Eberhardt WEE, Poddubskaya E, Antonia S, Pluzanski A, Vokes EE,
Holgado E, et al: Nivolumab versus docetaxel in advanced
squamous-cell non-small-cell lung cancer. N Engl J Med.
373:123–135. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Motzer RJ, Escudier B, McDermott DF,
George S, Hammers HJ, Srinivas S, Tykodi SS, Sosman JA, Procopio G,
Plimack ER, et al: Nivolumab versus everolimus in advanced
renal-cell carcinoma. N Engl J Med. 373:1803–1813. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Larkin J, Chiarion-Sileni V, Gonzalez R,
Grob JJ, Cowey CL, Lao CD, Schadendorf D, Dummer R, Smylie M,
Rutkowski P, et al: Combined nivolumab and ipilimumab or
monotherapy in untreated melanoma. N Engl J Med. 373:23–34. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bloch O, Crane CA, Fuks Y, Kaur R, Aghi
MK, Berger MS, Butowski NA, Chang SM, Clarke JL, McDermott MW, et
al: Heat-shock protein peptide complex-96 vaccination for recurrent
glioblastoma: A phase II, single-arm trial. Neuro Oncol.
16:274–279. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mitchell DA, Batich KA, Gunn MD, Huang MN,
Sanchez-Perez L, Nair SK, Congdon KL, Reap EA, Archer GE,
Desjardins A, et al: Tetanus toxoid and CCL3 improve dendritic cell
vaccines in mice and glioblastoma patients. Nature. 519:366–369.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Phuphanich S, Wheeler CJ, Rudnick JD,
Mazer M, Wang H, Nuño MA, Richardson JE, Fan X, Ji J, Chu RM, et
al: Phase I trial of a multi-epitope-pulsed dendritic cell vaccine
for patients with newly diagnosed glioblastoma. Cancer Immunol
Immunother. 62:125–135. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Schuster J, Lai RK, Recht LD, Reardon DA,
Paleologos NA, Groves MD, Mrugala MM, Jensen R, Baehring JM, Sloan
A, et al: A phase II, multicenter trial of rindopepimut (CDX-110)
in newly diagnosed glioblastoma: the ACT III study. Neuro Oncol.
17:854–861. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Vik-Mo EO, Nyakas M, Mikkelsen BV, Moe MC,
Due-Tønnesen P, Suso EM, Sæbøe-Larssen S, Sandberg C, Brinchmann
JE, Helseth E, et al: Therapeutic vaccination against autologous
cancer stem cells with mRNA-transfected dendritic cells in patients
with glioblastoma. Cancer Immunol Immunother. 62:1499–1509. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Reardon DA, Wucherpfennig KW, Freeman G,
Wu CJ, Chiocca EA, Wen PY, Curry WT Jr, Mitchell DA, Fecci PE,
Sampson JH and Dranoff G: An update on vaccine therapy and other
immunotherapeutic approaches for glioblastoma. Expert Rev Vaccines.
12:597–615. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Weller M, Roth P, Preusser M, Wick W,
Reardon DA, Platten M and Sampson JH: Vaccine-based
immunotherapeutic approaches to gliomas and beyond. Nat Rev Neurol.
13:363–374. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Srinivasan VM, Ferguson SD, Lee S,
Weathers SP, Kerrigan BCP and Heimberger AB: Tumor vaccines for
malignant gliomas. Neurotherapeutics. 14:345–357. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Heimberger AB, Suki D, Yang D, Shi W and
Aldape K: The natural history of EGFR and EGFRvIII in glioblastoma
patients. J Transl Med. 3:382005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chandramohan V, Mitchell DA, Johnson LA,
Sampson JH and Bigner DD: Antibody, T-cell and dendritic cell
immunotherapy for malignant brain tumors. Future Oncol. 9:977–990.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Schuessler A, Smith C, Beagley L, Boyle
GM, Rehan S, Matthews K, Jones L, Crough T, Dasari V, Klein K, et
al: Autologous T-cell therapy for cytomegalovirus as a
consolidative treatment for recurrent glioblastoma. Cancer Res.
74:3466–3476. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bonifant CL, Jackson HJ, Brentjens RJ and
Curran KJ: Toxicity and management in CAR T-cell therapy. Mol Ther
Oncolytics. 3:160112016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Janakiram M, Chinai JM, Zhao A, Sparano JA
and Zang X: HHLA2 and TMIGD2: New immunotherapeutic targets of the
B7 and CD28 families. Oncoimmunology. 4:e10265342015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ni L and Dong C: New B7 family checkpoint
in human cancers. Mol Cancer Ther. 16:1203–1211. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zou W, Wolchok JD and Chen L: PD-L1
(B7-H1) and PD-1 pathway blockade for cancer therapy: Mechanisms,
response biomarkers, and combinations. Sci Transl Med.
8:328rv42016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ohaegbulam KC, Assal A, Lazar-Molnar E,
Yao Y and Zang X: Human cancer immunotherapy with antibodies to the
PD-1 and PD-L1 pathway. Trends Mol Med. 21:24–33. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Alsaab HO, Sau S, Alzhrani R, Tatiparti K,
Bhise K, Kashaw SK and Iyer AK: PD-1 and PD-L1 checkpoint signaling
inhibition for cancer immunotherapy: Mechanism, combinations, and
clinical outcome. Front Pharmacol. 8:5612017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sharma P and Allison JP: The future of
immune checkpoint therapy. Science. 348:56–61. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ,
Topalian SL, Hwu P, Drake CG, Camacho LH, Kauh J, Odunsi K, et al:
Safety and activity of antiPD-L1 antibody in patients with advanced
cancer. N Engl J Med. 366:2455–2465. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu S, Wang Z, Wang Y1 Fan X, Zhang C, Ma
W, Qiu X and Jiang T: PD-1 related transcriptome profile and
clinical outcome in diffuse gliomas. Oncoimmunology.
7:e13827922017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Janakiram M, Chinai JM, Fineberg S, Fiser
A, Montagna C, Medavarapu R, Castano E, Jeon H, Ohaegbulam KC, Zhao
R, et al: Expression, clinical significance, and receptor
identification of the newest B7 family member HHLA2 protein. Clin
Cancer Res. 21:2359–2366. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhao R, Chinai JM, Buhl S, Scandiuzzi L,
Ray A, Jeon H, Ohaegbulam KC, Ghosh K, Zhao A, Scharff MD and Zang
X: HHLA2 is a member of the B7 family and inhibits human CD4 and
CD8 T-cell function. Proc Natl Acad Sci USA. 110:9879–9884. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhu Y, Yao S, Iliopoulou BP, Han X,
Augustine MM, Xu H, Phennicie RT, Flies SJ, Broadwater M, Ruff W,
et al: Chen. B7-H5 costimulates human T cells via CD28H. Nat
Commun. 4:20432013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Eckel-Passow JE, Lachance DH, Molinaro AM,
Walsh KM, Decker PA, Sicotte H, Pekmezci M, Rice T, Kosel ML,
Smirnov IV, et al: Glioma groups based on 1p/19q, IDH, and TERT
promoter mutations in tumors. N Engl J Med. 372:2499–2508. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu Y and Cao X: The origin and function
of tumor-associated macrophages. Cell Mol Immunol. 12:1–4. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
de Groot AE and Pienta KJ: Epigenetic
control of macrophage polarization: Implications for targeting
tumor-associated macrophages. Oncotarget. 9:20908–20927. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Arokiaraj MC: A novel targeted
angiogenesis technique using VEGF conjugated magnetic nanoparticles
and in-vitro endothelial barrier crossing. BMC Cardiovasc Disord.
17:2092017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Pitulescu ME, Schmidt I, Giaimo BD,
Antoine T, Berkenfeld F, Ferrante F, Park H, Ehling M, Biljes D,
Rocha SF, et al: Dll4 and Notch signalling couples sprouting
angiogenesis and artery formation. Nat Cell Biol. 19:915–927. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yan H, Qiu W, Koehne de Gonzalez AK, Wei
JS, Tu M, Xi CH, Yang YR, Peng YP, Tsai WY, Remotti HE, et al:
HHLA2 is a novel immune checkpoint protein in pancreatic ductal
adenocarcinoma and predicts post-surgical survival. Cancer Lett.
442:333–340. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Bhat KPL, Balasubramaniyan V, Vaillant B,
Ezhilarasan R, Hummelink K, Hollingsworth F, Wani K, Heathcock L,
James JD, Goodman LD, et al: Mesenchymal differentiation mediated
by NF-κB promotes radiation resistance in glioblastoma. Cancer
Cell. 24:331–346. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Verhaak RG, Hoadley KA, Purdom E, Wang V,
Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al:
Integrated genomic analysis identifies clinically relevant subtypes
of glioblastoma characterized by abnormalities in PDGFRA, IDH1,
EGFR, and NF1. Cancer Cell. 17:98–110. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lin N, Yan W, Gao K, Wang Y, Zhang J and
You Y: Prevalence and clinicopathologic characteristics of the
molecular subtypes in malignant glioma: A multi-institutional
analysis of 941 cases. PLoS One. 9:e948712014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kumar V, Donthireddy L, Marvel D,
Condamine T, Wang F, Lavilla-Alonso S, Hashimoto A, Vonteddu P,
Behera R, Goins MA, et al: Cancer-associated fibroblasts neutralize
the anti-tumor effect of CSF1 receptor blockade by inducing
PMN-MDSC infiltration of tumors. Cancer Cell. 32:654–668.e5. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kamran N, Alghamri MS, Nunez FJ, Shah D,
Asad AS, Candolfi M, Altshuler D, Lowenstein PR and Castro MG:
Current state and future prospects of immunotherapy for glioma.
Immunotherapy. 10:317–339. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Liang YX, Lu JM, Mo RJ, He HC, Xie J,
Jiang FN, Lin ZY, Chen YR, Wu YD, Luo HW, et al: E2F1 promotes
tumor cell invasion and migration through regulating CD147 in
prostate cancer. Int J Oncol. 48:1650–1658. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Gartel AL: FOXM1 in cancer: Interactions
and vulnerabilities. Cancer Res. 77:3135–3139. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Rundle S, Bradbury A, Drew Y and Curtin
NJ: Targeting the ATR-CHK1 axis in cancer therapy. Cancers (Basel).
9(pii): E412017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Koirala P, Roth ME, Gill J, Chinai JM,
Ewart MR, Piperdi S, Geller DS, Hoang BH, Fatakhova YV, Ghorpade M,
et al: HHLA2, a member of the B7 family, is expressed in human
osteosarcoma and is associated with metastases and worse survival.
Sci Rep. 6:311542016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhu Z and Dong W: Overexpression of HHLA2,
a member of the B7 family, is associated with worse survival in
human colorectal carcinoma. Onco Targets Ther. 11:1563–1570. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Shimonosono M, Arigami T, Yanagita S,
Matsushita D, Uchikado Y, Kijima Y, Kurahara H, Kita Y, Mori S,
Sasaki K, et al: The association of human endogenous retrovirus-H
long terminal repeat-associating protein 2 (HHLA2) expression with
gastric cancer prognosis. Oncotarget. 9:22069–22078. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Janakiram M, Shah UA, Liu W, Zhao A,
Schoenberg MP and Zang X: The third group of the B7-CD28 immune
checkpoint family: HHLA2, TMIGD2, B7×, and B7-H3. Immunol Rev.
276:26–39. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Xiao Y and Freeman GJ: A New B7:CD28
family checkpoint target for cancer immunotherapy: HHLA2. Clin
Cancer Res. 21:2201–2203. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wang Z, Wang Z, Zhang C, Liu X, Li G, Liu
S, Sun L, Liang J, Hu H, Liu Y, et al: Genetic and clinical
characterization of B7-H3 (CD276) expression and epigenetic
regulation in diffuse brain glioma. Cancer Sci. 109:2697–2705.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhou Z, Luther N, Ibrahim GM, Hawkins C,
Vibhakar R, Handler MH and Souweidane MM: B7-H3, a potential
therapeutic target, is expressed in diffuse intrinsic pontine
glioma. J Neurooncol. 111:257–264. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Loos M, Hedderich DM, Ottenhausen M, Giese
NA, Laschinger M, Esposito I, Kleeff J and Friess H: Expression of
the costimulatory molecule B7-H3 is associated with prolonged
survival in human pancreatic cancer. BMC Cancer. 9:4632009.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Wu CP, Jiang JT, Tan M, Zhu YB, Ji M, Xu
KF, Zhao JM, Zhang GB and Zhang XG: Relationship between
co-stimulatory molecule B7-H3 expression and gastric carcinoma
histology and prognosis. World J Gastroenterol. 12:457–459. 2006.
View Article : Google Scholar : PubMed/NCBI
|