Introduction
Ovarian cancer (OC), a cancer of the genital system,
is a common malignancy worldwide (1),
and its incidence has been increasing in recent years (2). Radical surgery along with
radio-chemotherapy is employed for treating OC (3). For most OC patients, the outcome of
radical surgery followed by either chemotherapy or radiotherapy is
not favorable (4,5). Thus, there is an urgent need to find
more effective therapeutic targets for treating OC.
Circular RNAs (circRNAs) are a type of non-coding
RNAs (ncRNAs), which do not have a potential to code for proteins
(6,7).
Some of them are critically dysregulated in cells and are
associated with disease progression in humans (8–12).
Recently, disordered circRNAs were identified in many tumors,
including OC (13–16). For instance, circP4HB promoted
aggressiveness and metastasis of non-small-cell lung carcinoma
(NSCLC) by absorbing miR-133a-5p (17). Regarding OC, circRNA profiling
revealed circRNA1656 as a novel biomarker in high-grade serous OC
(18). To date, various circRNAs have
been identified, but only a few circRNAs have been explored in the
context of OC. An in-depth study of the regulation of circRNAs
would provide a new theory for the development and progression of
OC.
There are abundant unexplored circRNAs related to
OC, and circ-FAM53B is a newly discovered circRNA addressed in the
present study. circ-FAM53B, also known as hsa_circ_0000267, is
located on chr 10: 126370175-126370948, having a spliced sequence
length of 773 bp. Previously, it was identified as an upregulated
circRNA in hepatocellular carcinoma that contributes to oncogenesis
by sponging miR-646 (19). In our
study, the expression of circ-FAM53B in OC was evaluated, and
upregulation of circ-FAM53B was identified in OC specimens and
cells. In addition, a survival study demonstrated that the overall
survival of patients with high circ-FAM53B expression was
significantly lower than that of patients with low circ-FAM53B
expression. Moreover, the regulatory effects of
circ-FAM53B/miRNA-646/vesicle-associated membrane protein 2 (VAMP2)
and circ-FAM53B/miRNA-647/mouse double minute 2 (MDM2) on inducing
the progression of OC were investigated. These findings may provide
new clues for OC treatment.
Materials and methods
Experimental tissues
Fifty-four pairs of OC tissues/non-tumor tissues
were harvested from patients attending the Department of
Gynecology, Heilongjiang Provincial Hospital, from November 2012 to
June 2014. The mean age of the 54 OC patients included in the study
was 51.2. None of the patients received intervention before
surgery. An informed consent form was signed by each patient.
Approval from the Ethics Committee of Heilongjiang Provincial
Hospital was obtained to conduct the present study.
Cell lines and culture
OC cells, HO8910, SKOV3, OVCAR3, and A2780 and one
normal cell line (IOSE80) were obtained from Type Culture of
Chinese Academy of Sciences (Shanghai, China) or preserved in our
laboratory. The cells were maintained in a medium (HyClone; GE
Healthcare Life Sciences) containing 10% fetal bovine serum (FBS;
Gibco; Thermo Fisher Scientific. Inc.) in a 37°C, 5% CO2
incubator, and the medium was replaced every 2–3 days.
RNA extraction and quantification
Total RNA was extracted using TRIzol (Invitrogen;
Thermo Fisher Scientific, Inc.) and quantified. The extracted RNA
was reverse-transcribed to cDNA. Next, cDNA was amplified using the
SYBR Premix Ex Taq II kit (Takara Biotechnology Co., Ltd.).
Reaction conditions were as follows: 95°C/5 min, 95°C/15 sec,
60°C/30 sec, for 40 cycles. The primers for circ-FAM53B and GAPDH
were as follows: circ-FAM53B forward, 5′-ACGACAAGAAGGTCGGTGTT-3′
and reverse, 5′-ATTCCCAGATGCTGGTGCTC-3′; GAPDH forward,
5′-GGGAGCCAAAAGGGTCAT-3′ and reverse, 5′-GAGTCCTTCCACGATACCAA-3′.
The 2−ΔΔCq method was applied to quantify the relative
gene expression (20).
Cell transfection
Cells were transfected with
siRNA-circ-FAM53B/VAMP2/MDM2, vector-circ- FAM53B/VAMP2/MDM2, and
miRNA-646/647 mimics and inhibitors using Lipofectamine 3000
(Invitrogen; Thermo Fisher Scientific, Inc.). The siRNAs
specifically targeting the back-spliced junction site of
circ-FAM53B were obtained from Shanghai GenePharma Co., Ltd. The
targeted sequences were as follows: si-circ-FAM53B-1,
5′-CCAAGATGGCACAGAAAATGA-3′; si-circ-FAM53B-2,
5′-GGCACAGAAAATGACAGATG-3′; miR-646 mimics,
5′-AAGCAGCUGCCUCUGAGGC-3′ (sense), and 5′-CUCAGAGGCAGCUGCUUUU-3′
(antisense); miR-646 inhibitor, 5′-GCCUCAGAGGCAGCUGCUU-3′; miR-647
mimics, 5′-GUGGCUGCACUCACUUCCUUC-3′ (sense), and
5′-AGGAAGUGAGUGCAGCCACUU-3′ (antisense); and miR-647 inhibitor:
GAAGGAAGUGAGUGCAGCCAC.
Cell Counting Kit-8 (CCK-8)
Cells were placed in a 96-well plate at a
concentration of 2×103 cells/well. At the selected
time-points (0, 24, 48, 72 and 96 h), 10 µl of reagent (Dojindo
Molecular Technologies, Inc.) was added to each well. After 2 h,
the OD value at 450 nm was read by a microplate reader (Tecan
Group, Ltd.), and cell viability curves were plotted.
Cell apoptosis assay
Cells at logarithmic growth phase were digested and
centrifuged at 188 × g for 5 min. The cells were resuspended in 400
µl of binding buffer, then stained with 5 µl of Annexin V-FITC and
5 µl of PI (Beyotime Institute of Biotechnology). A flow cytometer
(CyFlow® Cube 6; Sysmex Partec GmbH) was utilized to
assess cell apoptosis.
Migration and invasion assays
For the wound healing assay, 5×105 cells
were added to each well and incubated overnight. Cells were
detached from the wells using the tip of a 20 µl pipette and
incubated in serum-free medium for 36 h (A2780 cells) and 30 h
(HO8910 cells). A light microscope (Leica Microsystems GmbH) was
used to photograph the wound closure area.
For the Transwell assay, the cells were resuspended
in serum-free medium and seeded in the upper unit of
Matrigel-coated (BD Biosciences) Transwell unit (8 µm; EMD
Millipore), whereas 600 µl of complete medium were added into the
lower chamber. After 48 h of incubation, the invasive cells were
fixed in 4% paraformaldehyde (Beijing Solarbio Science &
Technology Co., Ltd.) for 20 min, crystal violet (Beyotime
Institute of Biotechnology) staining was performed for 20 min, and
the cells were counted using a microscope (Leica Microsystems
GmbH). A Transwell migration assay was carried out using the same
protocol except for the Matrigel pre-coating.
Western blot assay
Total protein was isolated by RIPA buffer (Beyotime
Institute of Biotechnology, Beijing, China). The extracted proteins
were quantified with a bicinchoninic acid (BCA) Protein Assay
Reagent Kit (Beyotime Institute of Biotechnology). In brief, BCA
reagent A and reagent B were mixed in a ratio of 50:1 (v/v). Then,
200 µl of BCA working solution was added to each well and incubated
at 37°C for ~30 min. The OD value of each well at 562 nm was
detected using a microplate reader (Tecan Group, Ltd.), and the
results were calculated employing a standard curve, plotted using
an appropriate standard. Proteins (40 µg) were subjected to 10%
SDS-PAGE and transferred to polyvinylidene difluoride (PVDF)
membranes (Merck KGaA). The membranes were blocked with 5% skim
milk diluted in Tris-buffered saline containing 0.05% Tween-20 for
2 h at room temperature. The membranes were then incubated with
primary antibodies against VAMP2 (dilution 1:10,000; cat. no.
ab181869), MDM2 (dilution 1:1,000; cat. no. ab38618) or GAPDH
(dilution 1:10,000; cat. no. ab181602) (Abcam, Cambridge, MA, USA)
at 4°C overnight. Next, the blots were incubated in secondary
antibody for 2 h at room temperature (dilution 1:5,000; cat. no.
SE134; Solarbio, Beijing, China). The protein bands were obtained
using a BeyoECL Plus Kit (Beyotime Institute of Biotechnology) and
detected using a chemiluminescence system (Tanon Science and
Technology Co., Ltd.). Protein expression levels were normalized to
GAPDH. ImageJ 1.48 (National Institutes of Health, Bethesda, MD,
USA) software was used to determine the gray value of the protein
band.
Subcellular fractionation
To assess the cellular localization of circ-FAM53B,
cytosolic and nuclear fractions were harvested employing a
Nuclear/Cytoplasmic Isolation Kit (Life Technologies; Thermo Fisher
Scientific, Inc.). In brief, the cells were lysed and centrifuged
(16,000 × g) at 4°C, and the supernatant was utilized as a
cytosolic fraction. Then, the pellets were washed and incubated
with buffer at 4°C for 10 min and used as a nuclear fraction.
Target prediction and dual luciferase
reporter gene assay
The possibility of interaction between circ-FAM53B
and miR-646/miR-647 was predicted using the Circular RNA
Interactome online database (https://circinteractome.nia.nih.gov). The downstream
targets of miR-646 and miR-647 were analyzed by TargetScan Human
7.2 database (http://www.targetscan.org/vert_72/). To verify these
predicted interactions, a dual luciferase reporter gene assay was
carried out. circ- FAM53B/VAMP2/MDM2 Wt and circ-FAM53B/VAMP2/MDM2
Mut were constructed based on the predicted binding sequences to
miRNA-646 and miRNA-647. Cells were co-transfected with
circ-FAM53B/VAMP2/MDM2 Wt or circ-FAM53B/VAMP2/MDM2 Mut and
miRNA-646-NC or miRNA-646 mimic for 48 h. Cells were also
co-transfected with circ-FAM53B/VAMP2/MDM2 Wt or
circ-FAM53B/VAMP2/MDM2 Mut and miRNA-647-NC or miRNA-647 mimic for
48 h. After cell lysis, relative luciferase activity was determined
(Promega Corp.).
Data analysis
The data are displayed as the mean ± standard
deviation (SD) based on three independent experiments. Statistical
analyses were carried out using GraphPad Prism 5.01 software
(GraphPad, Inc.). The differences between groups were analyzed by
Student's t-test or one-way ANOVA (followed by Tukey's
multiple comparison test). Correlation between the circ-FAM53B
level and pathological indexes of OC patients was analyzed through
the Fisher's exact test. The Kaplan-Meier methods were applied to
analyze the survival probability of OC patients. Valuation was
performed using the log-rank test. Cox regression analysis was
utilized to explore the independent prognostic factors for the
patients with OC. A P-value <0.05 was considered to indicate a
statistically significant difference.
Results
Overexpression of circ-FAM53B is
identified in OC tissue samples and cell lines and is associated
with adverse prognosis
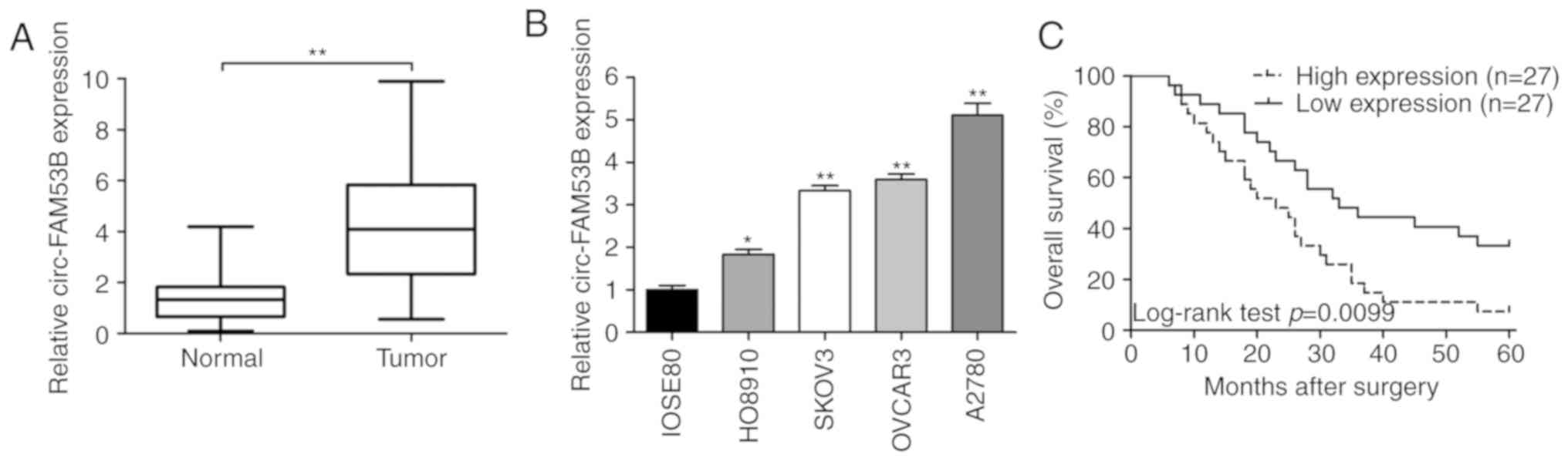
The expression of circ-FAM53B in 54 cases of OC
tissues and paired noncancerous samples was assessed by
quantitative real-time polymerase chain reaction (qRT-PCR). As
displayed in Fig. 1A, circ-FAM53B was
upregulated in OC tissues compared to that in their counterparts.
Similarly, upregulation of circ-FAM53B was revealed in OC cell
lines, including HO8910, SKOV3, OVCAR3, and A2780, compared to that
in IOSE80 cells (Fig. 1B). As
depicted in Table I, the circ-FAM53B
level was related to tumor size (P=0.028), FIGO stages (P=0.013),
and lymph node invasion (P=0.006) of OC (Table I). Moreover, Kaplan-Meier curves
revealed a poor survival rate in OC patients with upregulated
circ-FAM53B (Fig. 1C). Univariate
analysis indicated FIGO stage (P=0.049) and circ-FAM53B expression
(P=0.013) as two factors related to adverse prognosis of patients.
Further multivariate analysis demonstrated that circ-FAM53B could
be regarded as an independent prognostic indicator (P=0.049;
Table II).
 | Table I.circ-FAM53B expression and
clinicopathologic characteristics of OC patients. |
Table I.
circ-FAM53B expression and
clinicopathologic characteristics of OC patients.
|
|
| Circ-FAM53B
expression |
|
|---|
|
|
|
|
|
|---|
| Clinicopathologic
variables | No. of
patients | High (%) | Low (%) | P-value |
|---|
| Age (years) |
|
|
| 0.158 |
|
<50 | 34 | 14 (25.93) | 20 (37.04) |
|
|
≥50 | 20 | 13 (24.07) | 7 (12.96) |
|
| Tumor size
(cm) |
|
|
| 0.028 |
|
<3 | 29 | 10 (18.52) | 19 (35.19) |
|
| ≥3 | 25 | 17 (31.48) | 8 (14.81) |
|
| Histology |
|
|
| 0.773 |
|
Serous | 36 | 19 (35.19) | 17 (31.48) |
|
|
Mucinous | 18 | 8 (14.81) | 10 (18.52) |
|
| FIGO stage |
|
|
| 0.013 |
|
I–II | 24 | 7 (12.96) | 17 (31.48) |
|
|
III–IV | 30 | 20 (37.04) | 10 (18.52) |
|
| Lymph node
metastasis |
|
|
| 0.006 |
|
Negative | 29 | 9 (16.67) | 20 (37.04) |
|
|
Positive | 25 | 18 (33.33) | 7 (12.96) |
|
| Differentiation
grade |
|
|
| 0.577 |
|
Well/Moderately | 33 | 15 (27.78) | 18 (33.33) |
|
|
Poorly | 21 | 12 (22.22) | 9 (16.67) |
|
 | Table II.Univariate and multivariate analysis
of prognostic factors for overall survival in OC patients. |
Table II.
Univariate and multivariate analysis
of prognostic factors for overall survival in OC patients.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variables | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Overall
survival |
|
|
|
|
|
|
| Age (≥50 vs.
<50) | 0.978 | 0.527–1.816 | 0.944 |
|
|
|
| Tumor size (≥3 cm
vs. <3 cm) | 1.319 | 0.724–2.403 | 0.366 |
|
|
|
| Histology (serous
vs. mucinous) | 1.307 | 0.681–2.507 | 0.420 |
|
|
|
| FIGO stage (III–IV
vs. I–II) | 1.857 | 1.003–3.438 | 0.049 |
|
| 0.218 |
| Lymph node
metastasis (positive vs. negative) | 1.700 | 0.930–3.106 | 0.085 |
|
|
|
| Differentiation
grade (poorly vs. well/moderately) | 1.179 | 0.633–2.196 | 0.604 |
|
|
|
| circ-FAM53B
expression (high vs. low) | 2.187 | 1.181–4.050 | 0.013 | 1.915 | 1.002–3.666 | 0.049 |
circ-FAM53B plays an oncogenic role in
OC cells
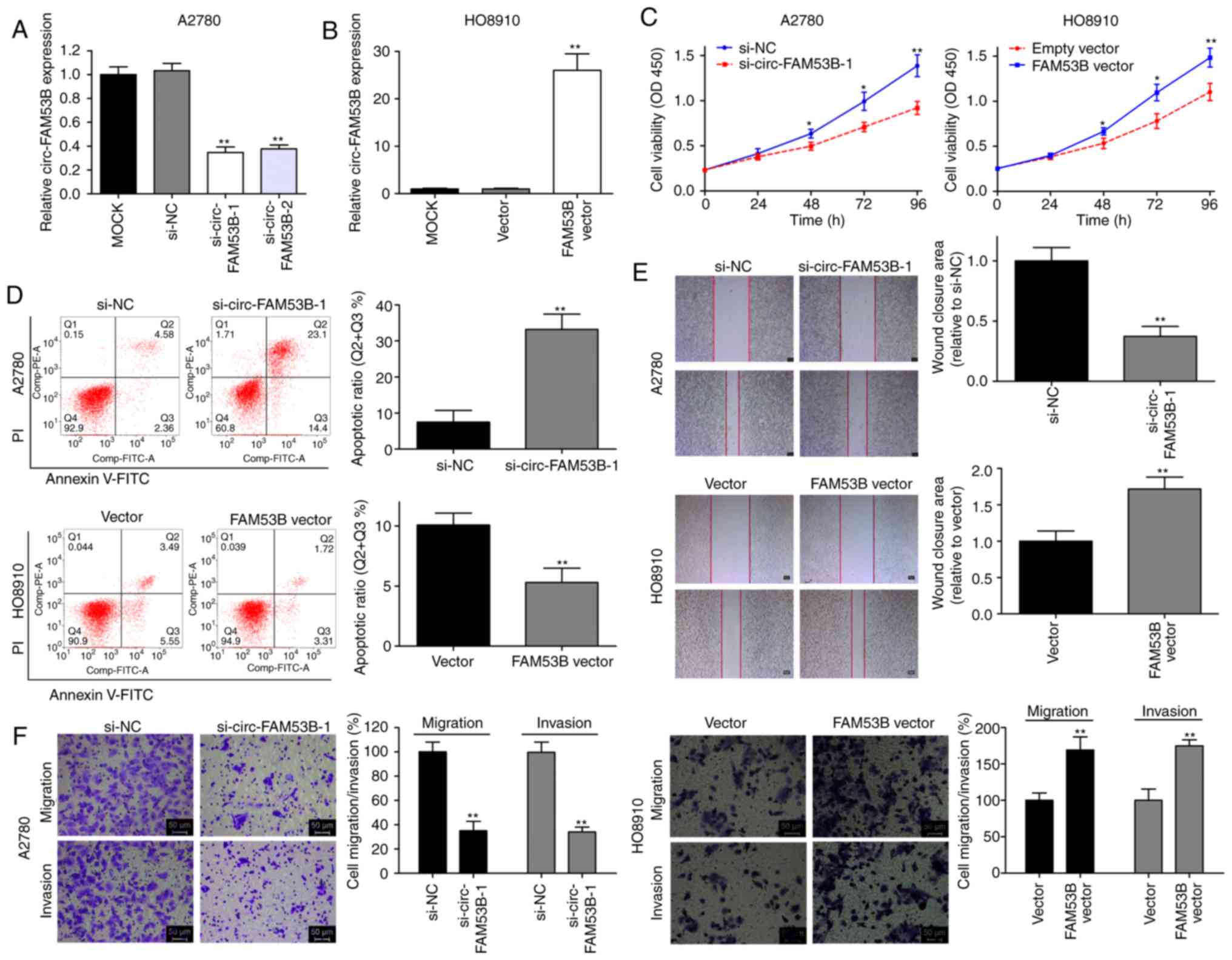
To verify the oncogenic role of circ-FAM53B in OC
progression, functional assays were conducted. We harvested
optimally transfected cells after 48 h and detected the relative
expression of circ-FAM53B. The high circ-FAM53B expression in the
A2780 cell line was decreased by transfection with si-circ-FAM53B-1
or si-circ-FAM53B-2 (Fig. 2A).
si-circ-FAM53B-1 had a more effective knockdown efficiency than
si-circ-FAM53B-2; thus, it was selected for further assays. Plasmid
transfection technology was applied using a low-expression
circ-FAM53B cell line, HO8910. As revealed in Fig. 2B, the overexpression efficiency was
satisfactory. Cell viability in the si-circ-FAM53B-1 group was
significantly decreased in the A2780 cell line (Fig. 2C), whereas overexpression of
circ-FAM53B enhanced the viability of HO8910 cells (Fig. 2C). Flow cytometric analysis also
demonstrated that the low expression of circ-FAM53B markedly
increased the apoptosis rate of A2780 cells (Fig. 2D). In addition, ectopically expressed
circ-FAM53B significantly decreased HO8910 cell apoptosis (Fig. 2D). Downregulation of circ-FAM53B
expression decreased the wound closure area in A2780 cells
(Fig. 2E), whereas upregulation of
circ-FAM53B increased the HO8910 cell migratory potential (Fig. 2E). Moreover, downregulation of
circ-FAM53B expression could decrease the migratory and invasive
rates in the A2780 cell line (Fig.
2F). An increased level of circ-FAM53B enhanced metastatic
properties in HO8910 cells (Fig. 2F).
These data indicated that high levels of circ-FAM53B contribute to
cell progression in OC cell lines.
circ-FAM53B sponges miR-646 and
miR-647 to increase VAMP2 and MDM2 expression, respectively
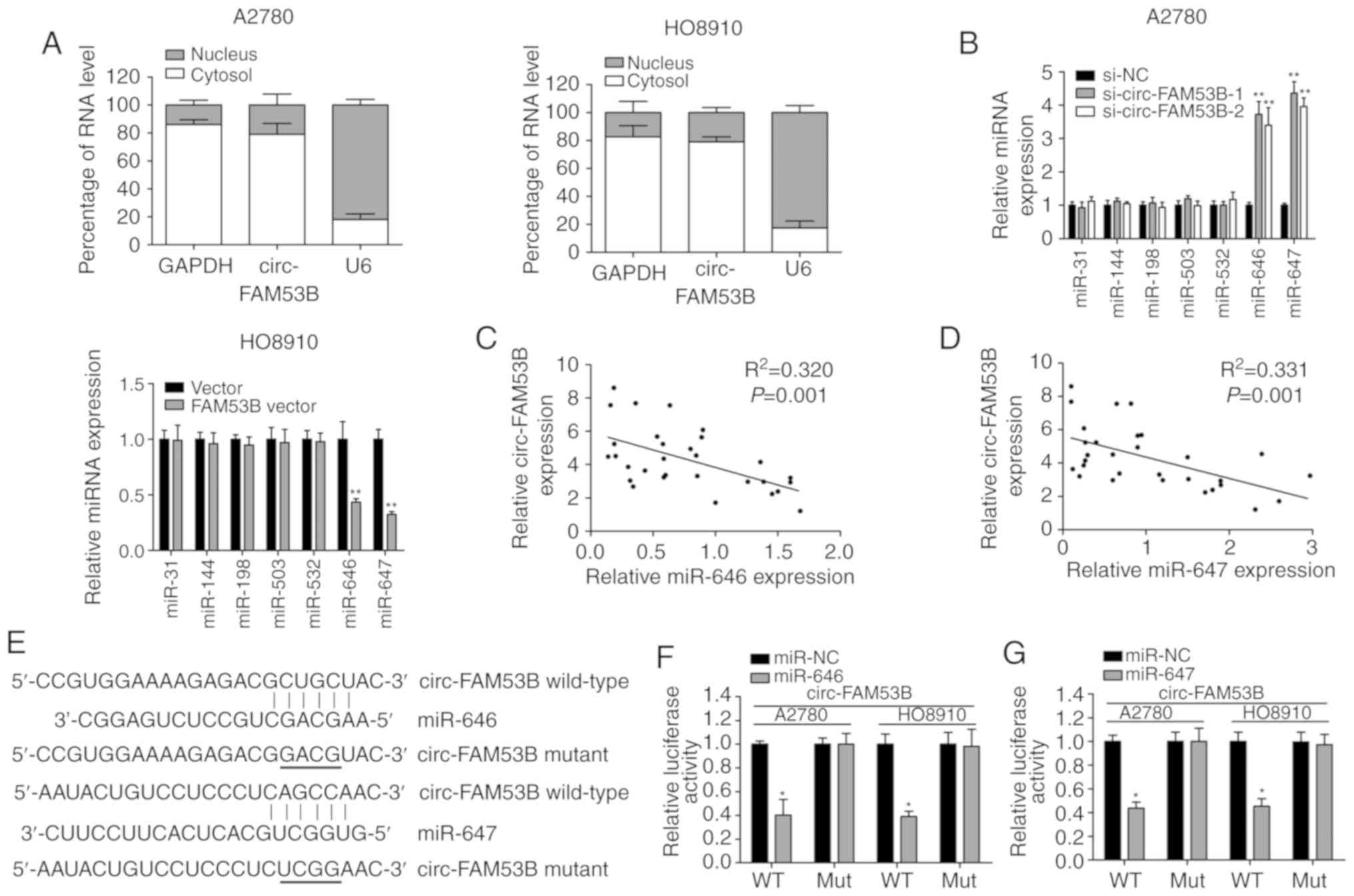
To explore the mechanisms of action of circ-FAM53B
in OC cells, the localization of circ-FAM53B in OC cells was first
analyzed by a subcellular fractionation assay. The results
indicated that circ-FAM53B was enriched in the cytoplasm (Fig. 3A), indicating that circ-FAM53B is
capable of modulating gene expression at the post-transcriptional
level. Thus, the miRNAs that may interact with circ-FAM53B were
further predicted, using the Circular RNA Interactome online
database. From the seven predicted miRNAs (miR-31, miR-144,
miR-198, miR-503, miR-532, miR-646, and miR-647), it was revealed
that the expression of miR-646 and miR-647 were modulated by
circ-FAM53B levels in A2780 and HO8910 cell lines (Fig. 3B). A negative correlation between
circ-FAM53B and miR-646/miR-647 was also identified in the OC
tissue samples (Fig. 3C and D). In
addition, miR-646 and miR-647 were predicted to bind to the
sequence of circ-FAM53B. Overexpression of miR-646 decreased the
luciferase intensity of circ-FAM53B Wt, indicating its binding
condition (Fig. 3E and F). Similarly,
miR-647 could bind to the seed sequence of circ-FAM53B, as verified
by the luciferase reporter gene assay (Fig. 3E and G). Therefore, it was concluded
that circ-FAM53B sponges miR-646 and miR-647 in OC cells.
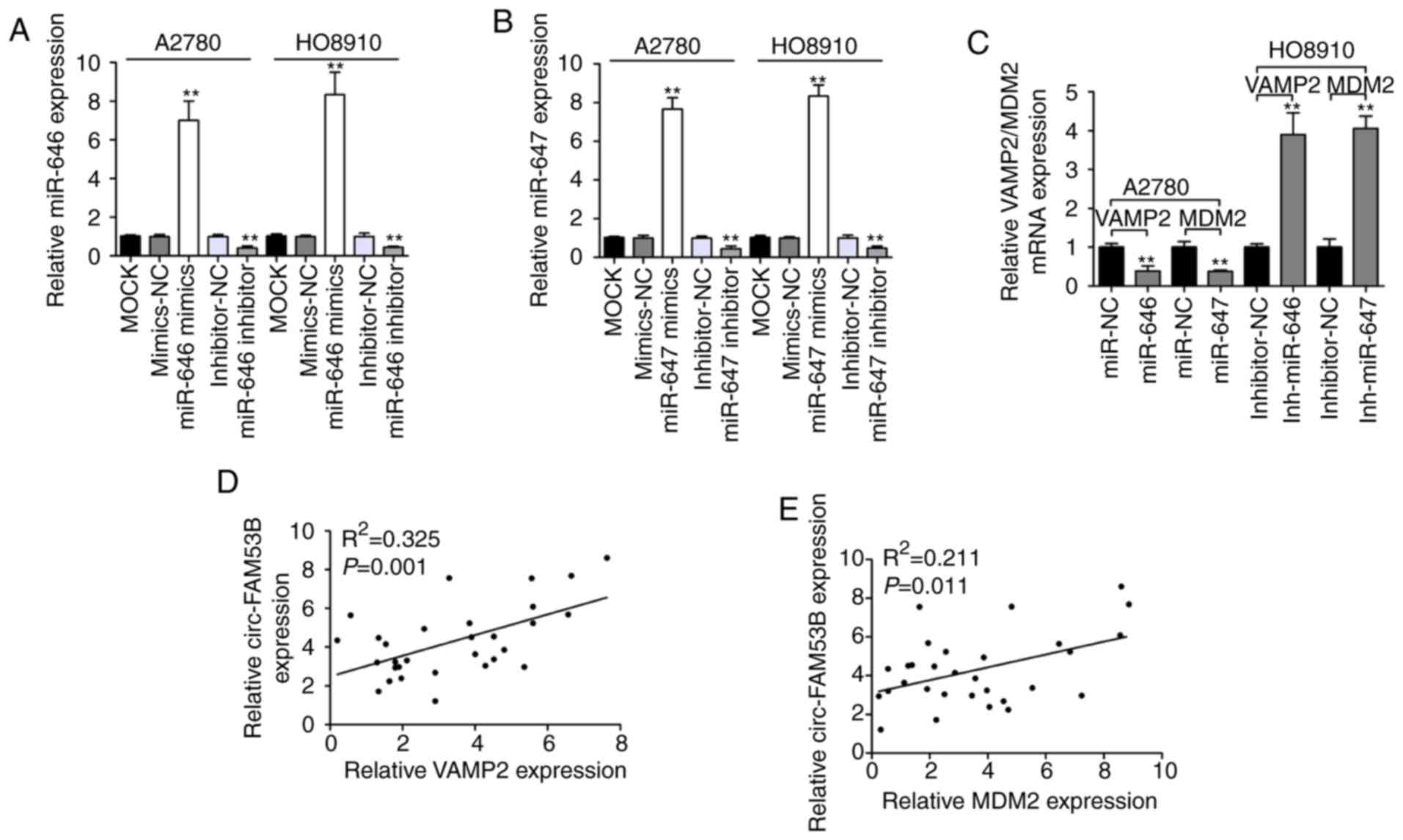
Subsequently, the downstream targets of miR-646 and miR-647 with an
oncogenic role were predicted and analyzed by TargetScan Human 7.2
database. miR-646 and miR-647 were predicted to have a potential to
interact with VAMP2 3′-UTR and MDM2 3′-UTR, respectively. As
revealed in Fig. 4A and B, miR-646
and miR-647 mimics could significantly enhance the expression
levels of miR-646 and miR-647, respectively. In addition, qRT-PCR
data revealed that miR-646 and miR-647 inhibitor significantly
downregulated miR-646 and miR-647 expression, respectively.
Transfection with miR-646 or miR-647 mimics could significantly
downregulate the expression of VAMP2 and MDM2, respectively
(Fig. 4C). In addition, transfection
with miR-646 or miR-647 inhibitor enhanced VAMP2 and MDM2
expression, respectively (Fig. 4C).
Moreover, it was revealed that circ-FAM53B expression was
positively correlated with the levels of VAMP2 and MDM2 in OC
tissues (Fig. 4D and E). Furthermore,
the dual-luciferase reporter gene assay confirmed an interaction
between miR-646 and VAMP2 3′-UTR (Fig. 4F
and G). A similar result was also revealed with miR-647 and
MDM2 3′-UTR (Fig. 4F and H). These
data indicated that VAMP2 and MDM2 are the downstream targets of
miR-646 and miR-647, respectively. The expression of VAMP2 and MDM2
was decreased after the silencing of circ-FAM53B, while
co-transfection with inh-miR-646/VAMP2 vector or inh-miR-647/MDM2
vector partially restored VAMP2 and MDM2 expression, respectively.
Additionally, miR-646 mimics or si-VAMP2 could partially inhibit
the high VAMP2 expression revealed by the circ-FAM53B vector.
Co-transfection with either miR-647 mimics or si-MDM2 partially
decreased the high MDM2 expression induced by the circ-FAM53B
vector (Fig. 4I).
The oncogenic functions of circ-FAM53B
are partially attributed to its modulation of VAMP2 and MDM2
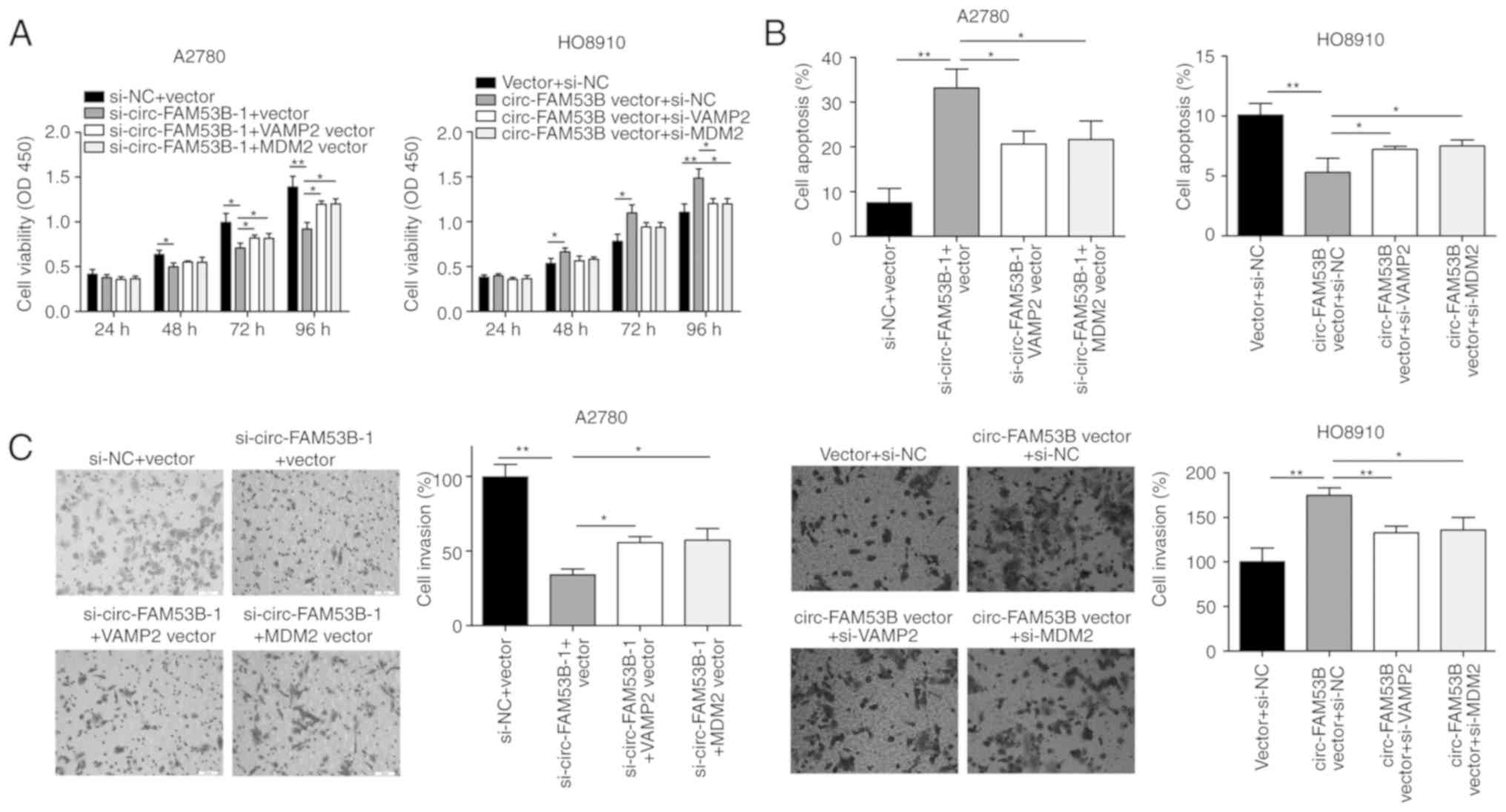
In the present experiment, circ-FAM53B silencing
attenuated the viability of A2780 cells, which was partly reversed
after co-transfection with VAMP2 or MDM2 vector (Fig. 5A). In HO8910 cells transfected with
circ-FAM53B vector, cell viability was significantly increased,
which was reversed by transfection with si-VAMP2 or si-MDM2
(Fig. 5A). Similar results were also
observed in the flow cytometric experiment (Fig. 5B). The inhibited invasive capacity of
A2780 cells with circ-FAM53B knockdown was partially reversed by
VAMP2 or MDM2 vector (Fig. 5C).
Similarly, the increased invasion of HO8910 cells with
overexpressed circ-FAM53B was reversed by si-VAMP2 and si-MDM2
(Fig. 5C).
Discussion
OC is developing into a serious disease, thereby
posing a great burden on human health. RNA sequencing provides a
new strategy for the treatment of human cancers, including OC
(18,21,22).
circ-FAM53B is located on chr 10: 126370175-126370948, and its
spliced length is 773 bp. It was identified as an upregulated
circRNA in hepatocellular carcinoma, contributing to oncogenesis by
sponging miR-646 (19). However, its
expression pattern, clinical significance, functions, and molecular
mechanisms in other cancers remain unclear. In the present study,
circ-FAM53B was revealed to be elevated in OC and was closely
related to clinical severity of OC patients. Consequently, it can
be used as an independent prognostic predictor for OC patients.
Overexpression of circ-FAM53B accelerated OC cells to proliferate,
migrate, and invade the ovaries, whereas silencing of circ-FAM53B
caused the opposite effect. Furthermore, circ-FAM53B was mainly
localized in the cytoplasm, which indicated its mechanism of gene
regulation at the post-transcriptional level.
miRNAs are also identified as a class of noncoding
RNAs (23,24). A growing number of studies have
revealed that miRNAs play a significant role in regulating gene
expression patterns during physiological and cell biological
processes (25). Some identified
miRNAs can be recognized as effectors of oncogenesis. For example,
miR-21 is an anti-apoptotic factor in gliomas (26). A majority of animal microRNAs, which
have been characterized thus far, affect protein synthesis of their
target mRNAs (25). A hypothesis
related to competing endogenous RNA (ceRNA) has been recently
proposed, and various studies have reported this mechanism in the
field of RNA research. For example, hsa_circ_0076248 can promote
the progression of glioma by affecting the miR-181a/SIRT1 pathway
(27). Therefore, circRNAs and miRNAs
and the correlation between them should be considered as a
significant measure to study OC. Several studies have demonstrated
that circRNAs interact with target miRNAs and suppress the
downstream targets, thus regulating the behaviors of tumor cells
(23). For example, circ-ERBB2 can
promote gastric cancer development and progression through the
regulation of miR-503/CACUL1 and miR-637/MMP-19 signaling (24). Previously, Pan et al revealed
an interaction between miR-646 and circ-FAM53B in hepatocellular
carcinoma (19). In the present
study, miR-646 and miR-647 were identified as two miRNAs which can
be sponged by circ-FAM53B in OC cells. miR-646 and miR-647 have
been reported to act as tumor suppressors in certain human cancers.
It is reported that miR-646 could be sponged by oncogenic lncRNA
HOTAIR to mediate estrogen-induced metastasis of endometrial cancer
cells (28). Another study revealed
that miR-646 exerts a tumor-suppressive role in gastric cancer by
modulating FOXK1 (29). In addition,
miR-647 was revealed to suppress progression in gastric cancer and
NSCLC (30,31). Additionally, SRF-MYH9 and TRAF2 were
identified as downstream targets of miR-647 in gastric cancer and
NSCLC, respectively (30,31).
In the present study, it was further revealed that
VAMP2 and MDM2 are the downstream targets of miR-646 and miR-647,
respectively. VAMP2 was first identified in synaptic vesicles from
rat brain, and has been revealed to be implicated in synaptic
vesicle docking and fusion with plasma membrane proteins (32). In addition, it has been revealed to be
significantly upregulated in a wide range of malignancies,
including bladder cancer (33),
osteosarcoma (34), and other
cancers. MDM2 is an oncoprotein that facilitates rapid degradation
of the tumor suppressor, p53 (35).
In contrast to other tumor suppressors, such as Rb, p16, or PTEN,
p53 is frequently overexpressed in tumors, although its function is
ablated. Basically, mutant p53 is frequently upregulated in ovarian
cancer cells; however, wild-type p53 is diminished in these cells.
The function of wild-type p53 is frequently decreased due to
different alterations, which result in an enhanced activity of its
negative regulators, such as MDM2. Suppression of the interaction
between MDM2 and p53 could stabilize the p53 protein, thus leading
to suppression of cancer progression (36). Various studies have indicated its
oncogenic function in tumorigenesis, including esophageal squamous
cell carcinoma (37), breast cancer
(38), and lung adenocarcinoma
(39). However, the role and
mechanism of VAMP2 and MDM2 in OC have not been studied to date. In
the present study, circ-FAM53B was identified as a ceRNA to
competitively bind to miR-646 and miR-647 to upregulate VAMP2 and
MDM2 expression, thus mediating the behaviors of OC cells. OC is a
multi-factorial, multi-step, complex disease with many dysregulated
circRNAs/miRNAs. This study merely reveals the tip of the iceberg
in the development of OC and its progression.
In summary, circ-FAM53B was markedly overexpressed
in tissues and cells of OC, and it was associated with clinical
severity and adverse prognosis in OC patients. Additionally,
circ-FAM53B contributed to OC cell progression by sponging miR-646
and miR-647, thus increasing the expression of VAMP2 and MDM2,
respectively.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets supporting the project are available
from the corresponding author upon reasonable request.
Authors' contributions
JL designed the study and provided the final
approval of the version to be published. DS and LZ performed the
experiments. DS and JL analyzed the data. All authors have approved
the final version of the publication and agree to be accountable
for all aspects of the research in ensuring that the accuracy or
integrity of any part of the work are appropriately investigated
and resolved.
Ethics approval and consent to
participate
Approval from the Ethics Committee of Heilongjiang
Provincial Hospital was obtained to conduct the present study and
an informed consent form was signed by each patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bast RC Jr, Hennessy B and Mills GB: The
biology of ovarian cancer: New opportunities for translation. Nat
Rev Cancer. 9:415–428. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Timmermans M, Sonke GS, Slangen BFM,
Baalbergen A, Bekkers RLM, Fons G, Gerestein CG, Kruse AJ, Roes EM,
Zusterzeel PLM, et al: Outcome of surgery in advanced ovarian
cancer varies between geographical regions; opportunities for
improvement in the netherlands. Eur J Surg Oncol. 45:1425–1431.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Morice P, Gouy S and Leary A: Mucinous
ovarian carcinoma. N Engl J Med. 380:1256–1266. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Stewart C, Ralyea C and Lockwood S:
Ovarian cancer: An integrated review. Semin Oncol Nurs. 35:151–156.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zang J, Lu D and Xu A: The interaction of
circRNAs and RNA binding proteins: An important part of circRNA
maintenance and function. J Neurosci Res. Dec 21–2018.(Epub ahead
of print). View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hao L, Rong W, Bai L, Cui H, Zhang S, Li
Y, Chen D and Meng X: Upregulated circular RNA circ_0007534
indicates an unfavorable prognosis in pancreatic ductal
adenocarcinoma and regulates cell proliferation, apoptosis, and
invasion by sponging miR-625 and miR-892b. J Cell Biochem.
120:3780–3789. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xu Y, Yao Y, Zhong X, Leng K, Qin W, Qu L,
Cui Y and Jiang X: Downregulated circular RNA hsa_circ_0001649
regulates proliferation, migration and invasion in
cholangiocarcinoma cells. Biochem Biophys Res Commun. 496:455–461.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Aufiero S, Reckman YJ, Pinto YM and
Creemers EE: Circular RNAs open a new chapter in cardiovascular
biology. Nat Rev Cardiol. 16:503–514. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li KS, Pan F, Mao XD, Liu C and Chen YJ:
Biological functions of circular RNAs and their roles in occurrence
of reproduction and gynecological diseases. Am J Transl Res.
11:1–15. 2019.PubMed/NCBI
|
|
13
|
Bach DH, Lee SK and Sood AK: Circular RNAs
in cancer. Mol Ther Nucleic Acids. 16:118–129. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang W, Liu Y, Gao R, Xiu Z and Sun T:
Knockdown of cZNF292 suppressed hypoxic human hepatoma SMMC7721
cell proliferation, vasculogenic mimicry, and radioresistance. Cell
Signal. 60:122–135. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Matboli M, Shafei AE, Ali MA, Ashry AM,
Kamal KM, Agag MA, Reda I, Tash EF and Ali M: circRNAs
(hsa_circ_00156, hsa_circ_000224, and hsa_circ_000520) are novel
potential biomarkers in hepatocellular carcinoma. J Cell Biochem.
Nov 13–2018.(Epub ahead of print).
|
|
16
|
Guo J, Duan H, Li Y, Yang L and Yuan L: A
novel circular RNA circ-ZNF652 promotes hepatocellular carcinoma
metastasis through inducing snail-mediated epithelial-mesenchymal
transition by sponging miR-203/miR-502-5p. Biochem Biophys Res
Commun. 513:812–819. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang T and Wang X, Du Q, Wu N, Liu X, Chen
Y and Wang X: The circRNA circP4HB promotes NSCLC aggressiveness
and metastasis by sponging miR-133a-5p. Biochem Biophys Res Commun.
513:904–911. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gao Y, Zhang C, Liu Y and Wang M: Circular
RNA profiling reveals circRNA1656 as a novel biomarker in high
grade serous ovarian cancer. Biosci Trends. 13:204–211. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pan H, Tang L, Jiang H, Li X, Wang R, Gao
J and Li Q: Enhanced expression of circ_0000267 in hepatocellular
carcinoma indicates poor prognosis and facilitates cell progression
by sponging miR-646. J Cell Biochem. Feb 5–2019.(Epub ahead of
print). View Article : Google Scholar :
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yang C, Wei Y, Yu L and Xiao Y:
Identification of altered circular RNA expression in serum exosomes
from patients with papillary thyroid carcinoma by high-throughput
sequencing. Med Sci Monit. 25:2785–2791. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Teng F, Xu J, Zhang M, Liu S, Gu Y, Zhang
M, Wang X, Ni J, Qian B, Shen R and Jia X: Comprehensive circular
RNA expression profiles and the tumor-suppressive function of
circHIPK3 in ovarian cancer. Int J Biochem Cell Biol. 112:8–17.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mao W, Huang X, Wang L, Zhang Z, Liu M, Li
Y, Luo M, Yao X, Fan J and Geng J: Circular RNA hsa_circ_0068871
regulates FGFR3 expression and activates STAT3 by targeting
miR-181a-5p to promote bladder cancer progression. J Exp Clin
Cancer Res. 38:1692019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li X, He M, Guo J and Cao T: Upregulation
of circular RNA circ-ERBB2 predicts unfavorable prognosis and
facilitates the progression of gastric cancer via miR-503/CACUL1
and miR-637/MMP-19 signaling. Biochem Biophys Res Commun.
511:926–930. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chan JA, Krichevsky AM and Kosik KS:
MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells.
Cancer Res. 65:6029–6033. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lei B, Huang Y, Zhou Z, Zhao Y, Thapa AJ,
Li W, Cai W and Deng Y: Circular RNA hsa_circ_0076248 promotes
oncogenesis of glioma by sponging miR-181a to modulate SIRT1
expression. J Cell Biochem. 120:6698–6708. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhou YX, Wang C, Mao LW, Wang YL, Xia LQ,
Zhao W, Shen J and Chen J: Long noncoding RNA HOTAIR mediates the
estrogen-induced metastasis of endometrial cancer cells via the
miR-646/NPM1 axis. Am J Physiol Cell Physiol. 314:C690–C701. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang P, Tang WM, Zhang H, Li YQ, Peng Y,
Wang J, Liu GN, Huang XT, Zhao JJ, Li G, et al: MiR-646 inhibited
cell proliferation and EMT-induced metastasis by targeting FOXK1 in
gastric cancer. Br J Cancer. 117:525–534. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang YS, Chen T, Cai YJ, Dong J, Bai F,
Gao X, Tian L, Duan N and Liu D: MicroRNA-647 promotes the
therapeutic effectiveness of argon-helium cryoablation and inhibits
cell proliferation through targeting TRAF2 via the NF-κB signaling
pathway in non-small cell lung cancer. Onco Targets Ther.
11:6777–6784. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ye G, Huang K, Yu J, Zhao L, Zhu X, Yang
Q, Li W, Jiang Y, Zhuang B, Liu H, et al: MicroRNA-647 targets
SRF-MYH9 axis to suppress invasion and metastasis of gastric
cancer. Theranostics. 7:3338–3353. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ralston E, Beushausen S and Ploug T:
Expression of the synaptic vesicle proteins VAMPs/synaptobrevins 1
and 2 in non-neural tissues. J Biol Chem. 269:15403–15406.
1994.PubMed/NCBI
|
|
33
|
Raja SA, Abbas S, Shah STA, Tariq A, Bibi
N, Yousuf A, Khawaja A, Nawaz M, Mehmood A, Khan MJ and Hussain A:
Increased expression levels of Syntaxin 1A and Synaptobrevin
2/vesicle-associated membrane protein-2 are associated with the
progression of bladder cancer. Genet Mol Biol. 42:40–47. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li L, Wang X and Liu D: MicroRNA-185
inhibits proliferation, migration and invasion in human
osteosarcoma MG63 cells by targeting vesicle-associated membrane
protein 2. Gene. 696:80–87. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wade M, Li YC and Wahl GM: MDM2, MDMX and
p53 in oncogenesis and cancer therapy. Nat Rev Cancer. 13:83–96.
2013. View
Article : Google Scholar : PubMed/NCBI
|
|
36
|
Vassilev LT, Vu BT, Graves B, Carvajal D,
Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, et
al: In vivo activation of the p53 pathway by small-molecule
antagonists of MDM2. Science. 303:844–848. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sawada R, Maehara R, Oshikiri T, Nakamura
T, Itoh T, Kodama Y, Kakeji Y and Zen Y: MDM2 copy number increase:
A poor prognostic, molecular event in esophageal squamous cell
carcinoma. Hum Pathol. 89:1–9. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang W, Wu J, Fei X, Chen W, Li Y, Shen K
and Zhu L: CHD1L promotes cell cycle progression and cell motility
by up-regulating MDM2 in breast cancer. Am J Transl Res.
11:1581–1592. 2019.PubMed/NCBI
|
|
39
|
Tang Y, Xuan Y, Qiao G, Ou Z, He Z, Zhu Q,
Liao M and Yin G: MDM2 promotes epithelial-mesenchymal transition
through activation of Smad2/3 signaling pathway in lung
adenocarcinoma. Onco Targets Ther. 12:2247–2258. 2019. View Article : Google Scholar : PubMed/NCBI
|