Introduction
Esophageal cancer is one of the most malignant tumor
types worldwide, ranking third in incidence and fourth in
cancer-related mortality in China (1). According to the latest statistics,
~258,000 new cases of esophageal cancer and ~193,000 associated
deaths were registered in 2014 in China (2). The major pathological type is squamous
cell carcinoma, which progresses rapidly with a poor prognosis, and
the 5-year survival rate is only 10–25% (3,4). To date,
the mechanisms responsible for the occurrence and development of
esophageal squamous cell carcinoma (ESCC) remain to be fully
elucidated, and its treatment lacks specific molecular targets and
effective therapeutic drugs.
Testis expressed 10 (Tex10), a member of the 5
friends of methylated chtop and Rix complexes, has essential roles
in transcriptional regulation and ribosome biogenesis, as well as
the cell cycle (5–7). Most importantly, as a new stemness
factor, Tex10 plays an essential role in the establishment and
maintenance of pluripotency (8,9). A
previous study by our group demonstrated for the first time (to the
best of our knowledge) that Tex10 plays an important role in the
tumorigenesis of hepatocellular carcinoma (HCC) by promoting cancer
stem cell (CSC) properties and chemoresistance (10).
Several studies have demonstrated that epithelial-
mesenchymal transition (EMT) is linked to the acquirement of cancer
stem cell-like phenotypes, which may be a necessary step in the
process of tumor metastasis (11,12).
Cancer cells that have gained the ability to move and invade have
undergone EMT; however, only a small number of these cells may be
transplanted into distant organs and form metastases through
mesenchymal-epithelial transition, and the population of these
cells is considered to be CSCs (13,14). The
acquisition of stem cell-like properties is accompanied by the
activation of EMT during tumor metastasis and the activation of EMT
contributes to the generation and maintenance of CSCs (15). In the present study, it was
hypothesized that Tex10 is involved in ESCC metastasis through the
regulation of EMT and stemness.
Despite the important role of Tex10 in cancer
development, the functional role of Tex10 in ESCC has not yet been
previously investigated, at least to the best of our knowledge. The
present study thus aimed to investigate the expression pattern and
function of Tex10 in ESCC development and to determine the effect
of Tex10 on the migration, invasion, stemness and EMT of ESCC
cells. Furthermore, the present study attempted to unravel the
exact regulatory mechanisms of stemness and EMT, focusing on the
Wnt/β-catenin pathway. The results demonstrated that Tex10 may be a
potential drug target for ESCC therapy and the prevention of
metastasis.
Materials and methods
Patients and tumor samples
The expression levels of Tex10 were measured using
immunohistochemistry (IHC) in human ESCC tissues and their matched
adjacent non-cancerous tissues that were collected from Nanchong
Central Hospital (Nanchong, China) from June, 2017 to June, 2018. A
total of 7 patients (3 females, 4 males) were enrolled in this
study, with a median age of 57 years (range, 48–66 years). The
basic clinical characteristics of the patients are presented in
Table I. Consent was obtained from
all of the patients prior to enrolment. The present study was
approved by the Ethics Committee of the North Sichuan Medical
College [NSMC (Nanchong, China)] and the Ethical Committee of
Nanchong Central Hospital (Nanchong, China), and performed in
accordance with the Declaration of Helsinki.
 | Table I.Baseline characteristics of the
patients included in the study. |
Table I.
Baseline characteristics of the
patients included in the study.
|
|
|
|
|
| TNM categories |
|---|
|
|
|
|
|
|
|
|---|
| Case | Sex | Age (years) | Stage | Tumor grade | T | N | M |
|---|
| 1 | F | 48 | IB | 1 | 1 | 0 | 0 |
| 2 | M | 50 | IIIA | 1 | 3 | 1 | 0 |
| 3 | M | 61 | IIB | 3 | 3 | 0 | 0 |
| 4 | M | 60 | IB | 2 | 1 | 0 | 0 |
| 5 | F | 52 | IIIA | 3 | 3 | 1 | 0 |
| 6 | F | 66 | IIA | 2 | 3 | 0 | 0 |
| 7 | M | 62 | IIIA | 3 | 2 | 2 | 0 |
ESCC cell lines and cell culture
The human ESCC cell line, EC109, used in the present
study was purchased from the Cell Resource Center of the Shanghai
Institute of Life Sciences, Chinese Academy of Sciences and
authenticated by short-tandem repeat testing. EC109 was
mycoplasma-free and cultured in RPMI-1640 medium (Gibco; Thermo
Fisher Scientific, Inc.) containing 10% fetal bovine serum (Gibco;
Thermo Fisher Scientific, Inc.) and 1% penicillin-streptomycin
(Gibco; Thermo Fisher Scientific, Inc.) at 37°C in an incubator
with 5% CO2.
Construction of lentivirus and stable
transfection into cell lines
The lentiviral vector encoding the human Tex10 gene
was constructed by Genechem Co., Ltd. The target sequences of
Tex10-short hairpin (sh)RNA are described in a previous study
(10). The target sequences were as
follows: shTex10, 5′-AGCTACTGCCCTCCGAATTTA-3′; and shRNA control,
5′-TTCTCCGAACGTGTCACGT-3′. Lentiviral vector encoding shTex10 and
non-targeting shRNA were designated as shTex10 and scramble,
respectively. The EC109 cells were transfected with recombinant
lentivirus-transducing units in the presence of polybrene (6 µg/ml)
according to the manufacturer's instructions and stable Tex10
knockdown clones (shTex10-C1, shTex10-C2) were selected with 2.5
µg/ml puromycin (A1113803; Gibco; Thermo Fisher Scientific, Inc.).
The selected pools of knockdown cells were then collected and
cultured within 10 passages for use in the subsequent
experiments.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from the EC109 cells using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and
reverse transcribed using the ThermoScript RT-PCR system (cat. no.
11731015; Invitrogen; Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocol. qPCR with SYBR-Green (cat. no.
1708886; Bio-Rad Laboratories, Inc.) was performed on an iQ5
Multicolor Real-Time PCR Detection system (Bio-Rad Laboratories,
Inc.). According to a previously described method (16), the PCR conditions were as follows:
95°C for 5 min followed by 40 cycles of 95°C for 30 sec and 57°C
for 30 sec. GAPDH was used as an internal control. All reactions
were run in triplicate and quantified with 2-ΔΔCq (17). The sequences of the PCR primers are
listed in Table II.
 | Table II.Sequences of primers used for
RT-qPCR. |
Table II.
Sequences of primers used for
RT-qPCR.
| Gene | Primer sequence
(5′-3′) |
|---|
| Tex10 | F:
TTGCAACTTGCTCATCTTGG |
|
| R:
AGAGTCTGCAGGGAGAACCA |
| GAPDH | F:
TGCACCACCAACTGCTTAGC |
|
| R:
GGCATGGACTGTGGTCATGAG |
| Sox2 | F:
CGCAGACCTACATGAACG |
|
| R:
CCCTGGAGTGGGAGGAA |
| Bmi1 | F:
CAACTGGTTCGACCTTTGCAGATA |
|
| R:
GATGTGCCAATTGCTTCTAATGGA |
| Nanog | F:
AATACCTCAGCCTCCAGCAGATG |
|
| R:
TGCGTCACACCATTGCTATTCTTC |
| Oct4 | F:
CTTGCTGCAGAAGTGGGTGGAGGAA |
|
| R:
CTGCAGTGTGGGTTTCGGGCA |
| c-Myc | F:
GGCCGCTGCCAAACTGGTCT |
|
| R:
TGGGCGAGCTGCTGTCGTTG |
| ABCG2 | F:
ACAACCATTGCATCTTGTCTGTC |
|
| R:
GCTGCAAAGCCGTAAATCCATATC |
| E-cadherin | F:
GGTGAGGGGTTAAGCACAACA |
|
| R:
ACGACGTTAGCCTCGTTCTC |
| Vimentin | F:
ACAACCTGGCCGAGGACATC |
|
| R:
GACGTGCCAGAGACGCATTG |
| Slug | F:
CGGACCCACACATTACCTTGT |
|
| R:
AAAAAGGCTTCTCCCCCGTG |
| Snail | F:
ATGCACATCCGAAGCCACAC |
|
| R:
TGCAGTGGGGACAGGAGAAG |
| Twist1 | F:
GACCTAGATGTCATTGTTTCCAGAG |
|
| R:
CCCACGCCCTGTTTCTTTGAA |
| β-catenin | F:
CGCTGGATTTTCAAAACAGT |
|
| R:
CTGAGGAGCAGCTTCAGTCC |
| CD133 | F:
TGGATGCAGAACTTGACAACGT |
|
| R:
ATACCTGCTACGACAGTCGTGGT |
| CD24 | F:
TGAAGAACATGTGAGAGGTTTGAC |
|
| R:
GAAAACTGAATCTCCATTCCACAA |
| CD44 | F:
GCAGCCTCAGCTCATACCAG |
|
| R:
TGACTGGAGTCCATATCCATCCTT |
IHC
IHC was performed as described in a previous study
by our group (16). ESCC sections of
3 µm thickness were deparaffinized with xylene and antigen
retrieval was then performed with 0.01 M sodium citrate buffer (pH
6.0) in a pressure cooker for 3 min. Endogenous peroxidase was
blocked with 3% H2O2 for 10 min, and the
membrane was then permeated with 0.1% Triton X-100 for 10 min,
followed by the blocking of non-specific binding sites with 3%
bovine serum albumin for 1 h at room temperature. The sections were
then incubated with anti-Tex10 polyclonal antibody (cat. no.
17372-1-AP; 1:100 dilution; Proteintech Group, Inc.) at 4°C
overnight, followed by incubation with a horseradish peroxidase
(HRP)-conjugated secondary antibody [cat. no. PV-6001; Zhongshang
Goldenbridge (ZSGB)-Bio] for 1 h at room temperature. The slides
were developed with diaminobenzidine substrate (cat. no. ZLI-9017;
ZSGB-Bio) and counterstained with hematoxylin for 2 min at room
temperature (cat. no. ZLI-9608; ZSGB-Bio).
Western blot analysis
EC109 cells stably expressing shTex10 or non-target
shRNA were collected and lysed with radioimmunoprecipitation assay
buffer (Keygen Biotech) supplemented with protease inhibitor
cocktail (Roche Applied Science). Proteins from the lysed cells
were fractionated by 10% SDS-PAGE and subsequently transferred onto
nitrocellulose membranes (Hybond C; GE Healthcare Life Sciences).
Non-specific binding sites were blocked with 5% non-fat milk at
room temperature for 1 h and the membranes were incubated with the
following antibodies overnight at 4°C: Anti-Tex10 (cat. no.
17372-1-AP; dilution, 1:250; Proteintech Group, Inc.), Bcl-2 (cat.
no. ET1702-53; dilution, 1:500), Slug (cat. no. EM1706-65;
dilution, 1:1,000), β-catenin (cat. no. ET1601-5; dilution,
1:1,000), transcription factor 4 (TCF4; cat. no. R1401-11;
dilution, 1:500), cyclin D1 (cat. no. ET1601-31; dilution,
1:1,000), CD44 (cat. no. ET1609-74; dilution, 1:1,000), CD24 (cat.
no. 0804-4; dilution, 1:1,000), vimentin (cat. no. M1412-1;
dilution, 1:1,000) (all from HuaBio Inc.), bHLH transcription
factor (c-Myc; cat. no. ab32072; dilution, 1:2,000) and GAPDH (cat.
no. ab9485; dilution, 1:2,000; both from Abcam). The membranes were
then washed and incubated with HRP-conjugated goat anti-rabbit
(cat. no. A4914; dilution, 1:10,000) or goat anti-mouse (cat. no.
A4416; dilution, 1:10,000) antibodies (both from Sigma-Aldrich;
Merck KGaA). The bands were visualized using enhanced
chemiluminescence reagents (cat. no. WP20005; Thermo Fisher
Scientific, Inc.).
Cell proliferation assay
The proliferation of the ESCC cells was measured
using the Cell Counting kit-8 (CCK-8) assay (Keygen Biotech). The
cells (2×103/well) were seeded into 96-well plates and
observed every day, and at specific time-points, 10 µl CCK-8 and
100 µl fresh medium were added to each well. Following incubation
for 1.5 h at 37°C, the absorbance was measured at 450 nm using a
microplate reader (ST-360; KHB). Each condition was assayed in
triplicate.
Flow cytometric evaluation of
apoptosis
The shTex10- and scramble EC109-transfected cells
were treated with 100 µg/ml 5-fluorouracil (F5130; Sigma-Aldrich;
Merck KGaA) for 48 h, and the cells were then collected and washed
with ice-cold PBS. The analysis was performed with an Annexin
V-FITC reagent kit (cat. no. A211-01; Vazyme Biotech Co., Ltd.).
The cells were re-suspended to a final concentration of
1×106 cells/ml in Annexin V-binding buffer and incubated
with AnnexinV-FITC in the presence of propidium iodide for 15 min
at 4°C. Samples were analyzed using a BD FACSCalibur flow cytometer
(BD Biosciences) and subsequent analyses were performed with FlowJo
software (FlowJo 10.5, LLC).
Cell cycle analysis
The EC109 cells cultured in 6-well plates were
digested with trypsin (Gibco; Thermo Fisher Scientific, Inc.),
washed with PBS and then fixed with 75% ethanol at −20°C overnight.
Subsequently, the cells were washed with PBS and incubated with 50
mg/ml propidium iodide containing 80 mg/ml RNase (cat. no. KGA512;
Keygen Biotech) in PBS for 30 min. Fluorescence-assisted cell
sorting data were collected on a BD FACSCalibur flow cytometer (BD
Biosciences) and analyzed with FlowJo software (FlowJo10.5,
LLC).
In vitro migration and invasion
assays
The migration or invasion assays were performed
using polycarbonate Transwell filter chambers (8 µm pore size; cat.
no. 3422; Corning Inc.) and the inserts were coated with or without
Matrigel® (BD Biosciences). The scramble- or Tex10
shRNA-transfected EC109 cells (5×104) in 100 µl
serum-free medium were added to the top chamber, whereas the bottom
chamber was filled with 500 µl medium containing 10% serum.
Following incubation for 48 h at 37°C, all of the non-migrating or
non-invading cells were removed, and cells on the lower membrane of
the inserts were stained with 0.1% crystal violet (KGA229, Keygen
Biotech) at room temperature for 5 min. The number of cells that
had migrated or invaded was counted under a microscope (ECLIPSE
TS100, Nikon) and images were captured. Each experimental condition
was performed with triplicate filters and the experiments were
repeated 3 times.
Sphere formation assay
The spheroid formation assay was based on a
previously described method (10).
The EC109 cells were seeded onto ultralow attachment 6-well plates
(Corning Inc.) and cultured in Dulbecco's modified Eagle's
medium/F12 (cat. no. 10565018; Gibco; Thermo Fisher Scientific,
Inc.) medium containing N2, B27 (Gibco; Thermo Scientific, Inc.),
20 ng/ml epidermal growth factor and 20 ng/ml basic fibroblast
growth factor (EMD Millipore). Following 7 days of incubation at
37°C, the spheres were counted under a microscope (DMi 8;
Leica).
Drug sensitivity assay
The ESCC cells (1×104/well) were seeded
into 96-well plates and a serial dilution of 5-fluorouracil (F5130;
Sigma-Aldrich; Merck KGaA) ranging from 0 to 100 µg/ml was added to
the scrambled or shTex10- transfected EC109 cells after 24 h.
Following incubation for 72 h at 37°C, as previously described
(18,19), cell proliferation was measured with a
CCK-8 assay (KGA317s; Keygen Biotech) as described above.
Clinical dataset analysis
To assess the expression levels of Tex10 mRNA, a
large cohort of patients with ESCC from the public datasets,
GSE23400 and GSE20347, was analyzed in Oncomine (www.oncomine.org) according to the instructions
provided.
Actin cytoskeleton staining
Actin cytoskeleton staining analyses of filamentous
actin (F-actin) was performed as previously described (20). In brief, the EC109 cells stably
expressing shTex10 or scrambled shRNA (1×106/well) were
placed in 6-well plates. At 24 h after seeding, the cells were
washed with PBS and fixed with 4% paraformaldehyde for 24 h at room
temperature. The cells were subsequently incubated for 30 min with
phalloidin-FITC (cat. no. CA1610; Solarbio) and DAPI (cat. no.
32670; Sigma-Aldrich; Merck KGaA). Finally, the cells were washed
with PBS prior to capturing of images under an inverted
fluorescence microscope (DMi 8; Leica).
Statistical analysis
All experiments were performed at least 3
independent experiments independently, and all values were
expressed as the means ± standard deviation. The SPSS 20.0 software
(IBM Corp.) was used for statistical analysis. A two-tailed
unpaired Student's t-test with Welch correction or one-way ANOVA
followed by Dunnett's adjustment (for dose-response effects) or the
Bonferroni test (for multiple comparisons) was used to determine
the statistical significance of the differences in the measured
variables. We have specified the name of the statistical test in
the figure legends. A value of P<0.05 was considered to indicate
a statistically significant difference.
Results
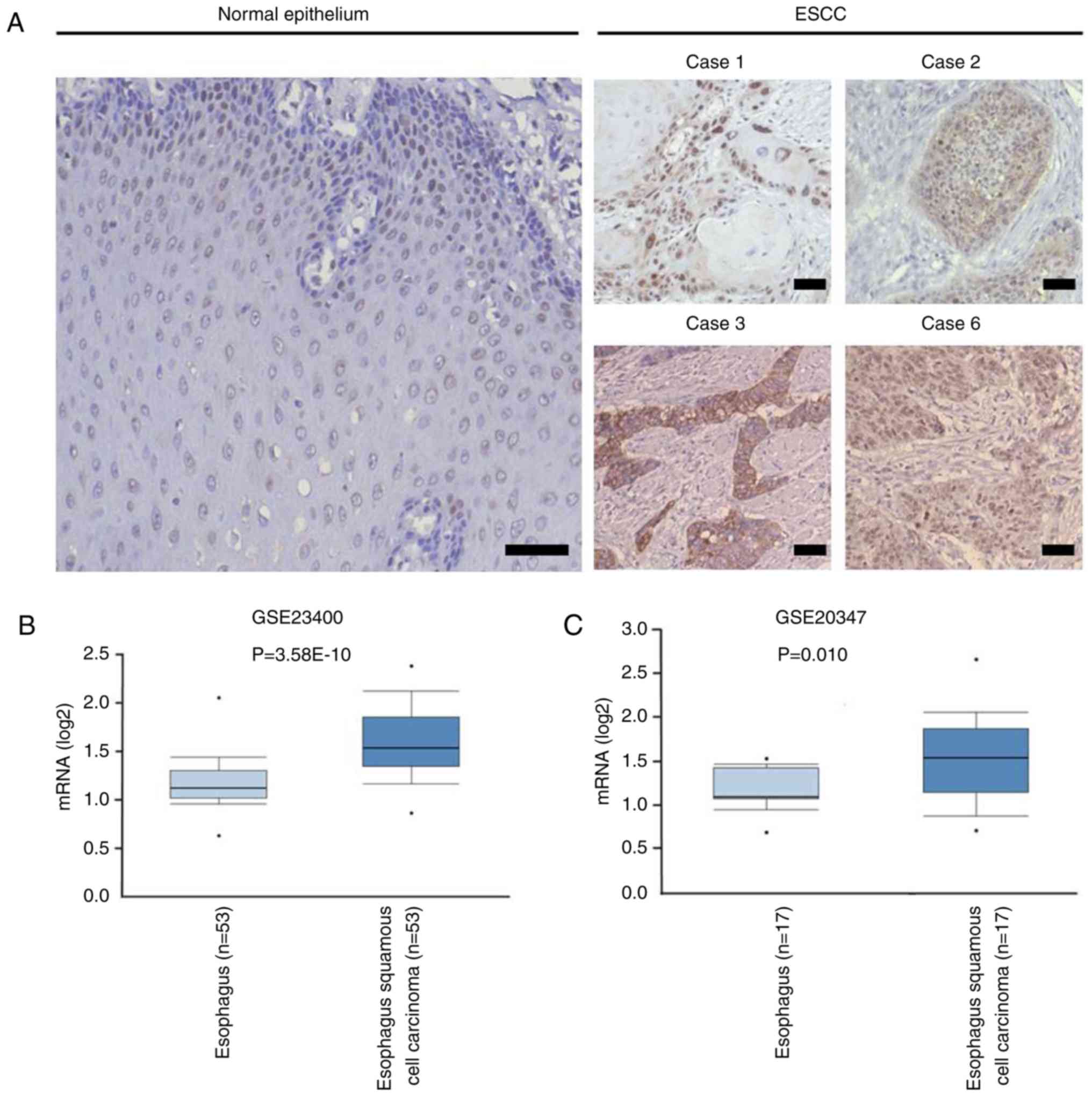
Tex10 expression is elevated in ESCC
tissues
To determine the expression of Tex10 in ESCC and
adjacent normal tissues, IHC was performed, and the results
indicated that the intensity of Tex10 immunostaining in the ESCC
tissues was markedly higher compared with that in the adjacent
non-cancerous tissues (Fig. 1A). The
expression pattern of Tex10 in ESCC was further analyzed in
Oncomine, a publicly accessible cancer informatics database. The
results demonstrated that the mRNA expression of Tex10 in the ESCC
tissues was significantly higher compared with that in the matched
non-cancerous tissues (Fig. 1B and
C). Collectively, these results indicated that Tex10 is
associated with ESCC.
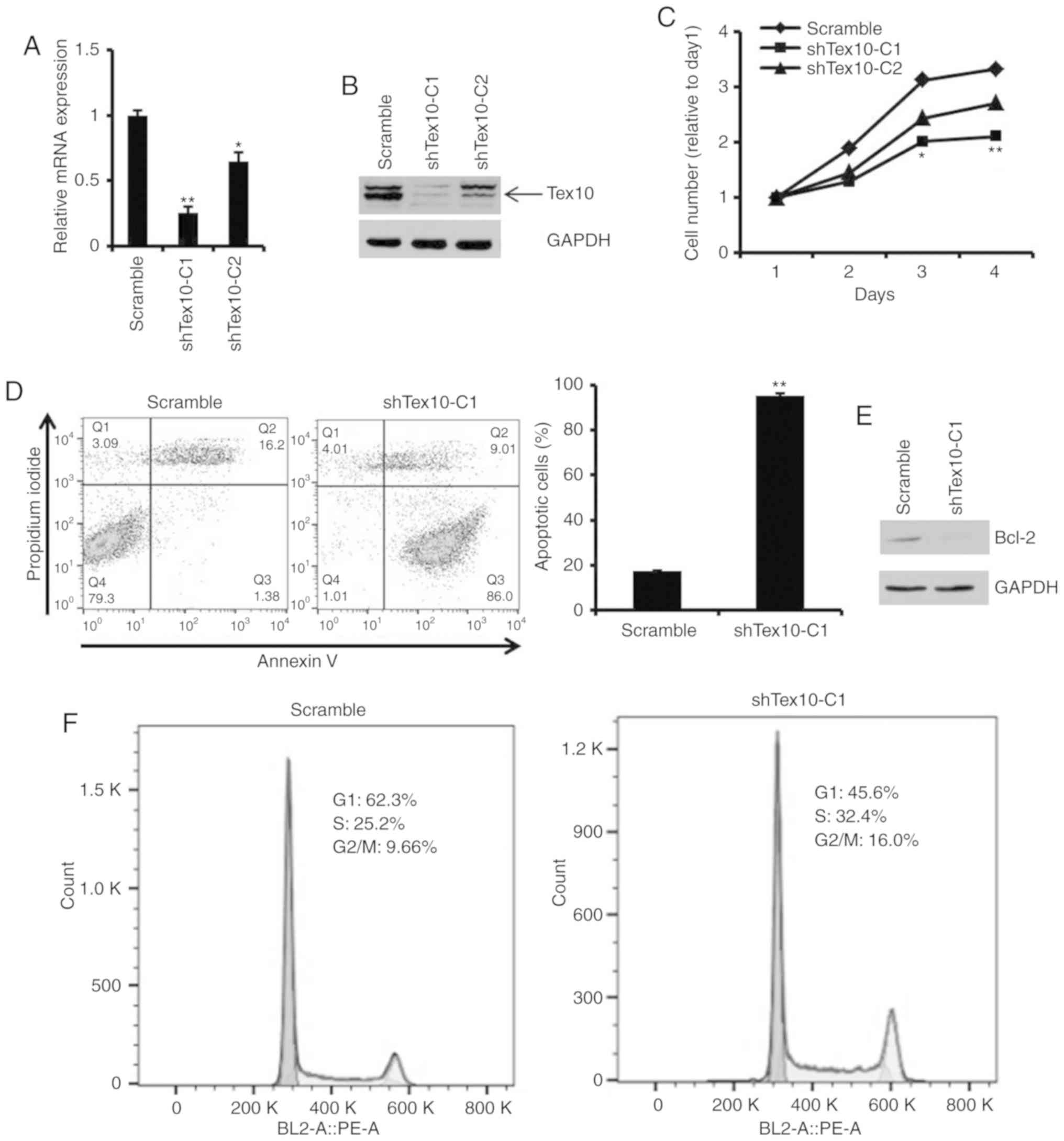
Silencing of Tex10 reduces cell
viability by suppressing proliferation, inducing cell apoptosis and
blocking cell cycle progression
To investigate the biological function of Tex10 in
the ESCC cells, two cell lines with stable Tex10 knockdown
(shTex10-C1, shTex10-C2) were generated using a lentiviral system.
Two shRNAs (scrambled, shTex10) were specifically designed and
constructed, and used to transfect the EC109 cells, followed by
screening with 2.5 µg/ml puromycin for 2 weeks. RT-qPCR and western
blot analysis were performed to measure the knockdown efficiency of
Tex10. The mRNA expression level of Tex10 was markedly decreased in
the shTex10-transfected cells compared with the scramble
shRNA-transfected cells (P<0.05 and P<0.01; Fig. 2A). Consistent with this finding, the
decreased protein expression of Tex10 was observed in the
shTex10-transfected cells (Fig. 2B).
Thus, these results demonstrated that Tex10 was specifically and
effectively inhibited.
Subsequently, it was determined whether Tex10 is
required for the growth and apoptosis of EC109 cells. The results
indicated that the silencing of Tex10 significantly inhibited the
proliferation of the EC109 cells (P<0.05 and P<0.01; Fig. 2C). When Annexin V staining was used to
assess the effects of Tex10 on the apoptosis of the EC109 cells, it
was revealed that the percentages of Annexin V-positive cells were
markedly higher in the shTex10 groups compared with those in the
scramble groups, with ~85% of the shTex10 cells undergoing early
apoptosis (P<0.01; Fig. 2D).
Furthermore, the downregulation of anti-apoptotic factor, Bcl-2,
was observed following the knockdown of Tex10 (Fig. 2E). The effects of Tex10 knockdown on
the cell cycle distribution were then investigated. As shown in
Fig. 2F, cell cycle phase analysis
indicated a decrease in the G0/G1 phase cell population and an
increase in the number of cells in the G2/M and S phase in the
shTex10 group, indicating that the ESCC cells exhibited cell cycle
disorders upon the downregulation of Tex10. Taken together, these
results fully demonstrate that the knockdown of Tex10 suppresses
ESCC cell proliferation, induces cell apoptosis and blocks cell
cycle progression.
Tex10 enhances CSC properties,
including self-renewal and resistance ability
Self-renewal is a prominent feature of CSCs, and to
further determine the effects of Tex10 on the self-renewal
properties of ESCC cells, the sphere-formation assay was employed.
The results indicated that the knockdown of Tex10 led to a
considerable decrease in the number and size of spheroids
(P<0.01; Fig. 3A). Furthermore,
the mRNA levels of stem the cell-specific markers, SRY-Box 2
(Sox2), Nanog and octamer-binding transcription factor 4 (Oct4),
and the CSC-associated markers, c-Myc, polycomb ring finger (Bmi1),
ATP binding cassette subfamily G member 2 (ABCG2), CD133, CD44 and
CD24, decreased significantly in the EC109 cells following the
knockdown of Tex10 (P<0.05 and P<0.01; Fig. 3B). Furthermore, the protein expression
levels of CD44 and CD24 were downregulated following the knockdown
of Tex10 (Fig. 3C). The present study
therefore established that Tex10 silencing severely impairs the
self-renewal capacity of CSCs. The existence of CSCs is the cause
of chemotherapeutic failure (21),
and as one of the major characteristics of CSCs, resistance to
5-fluorouracil is an insuperable barrier for patients with ESCC
(18).
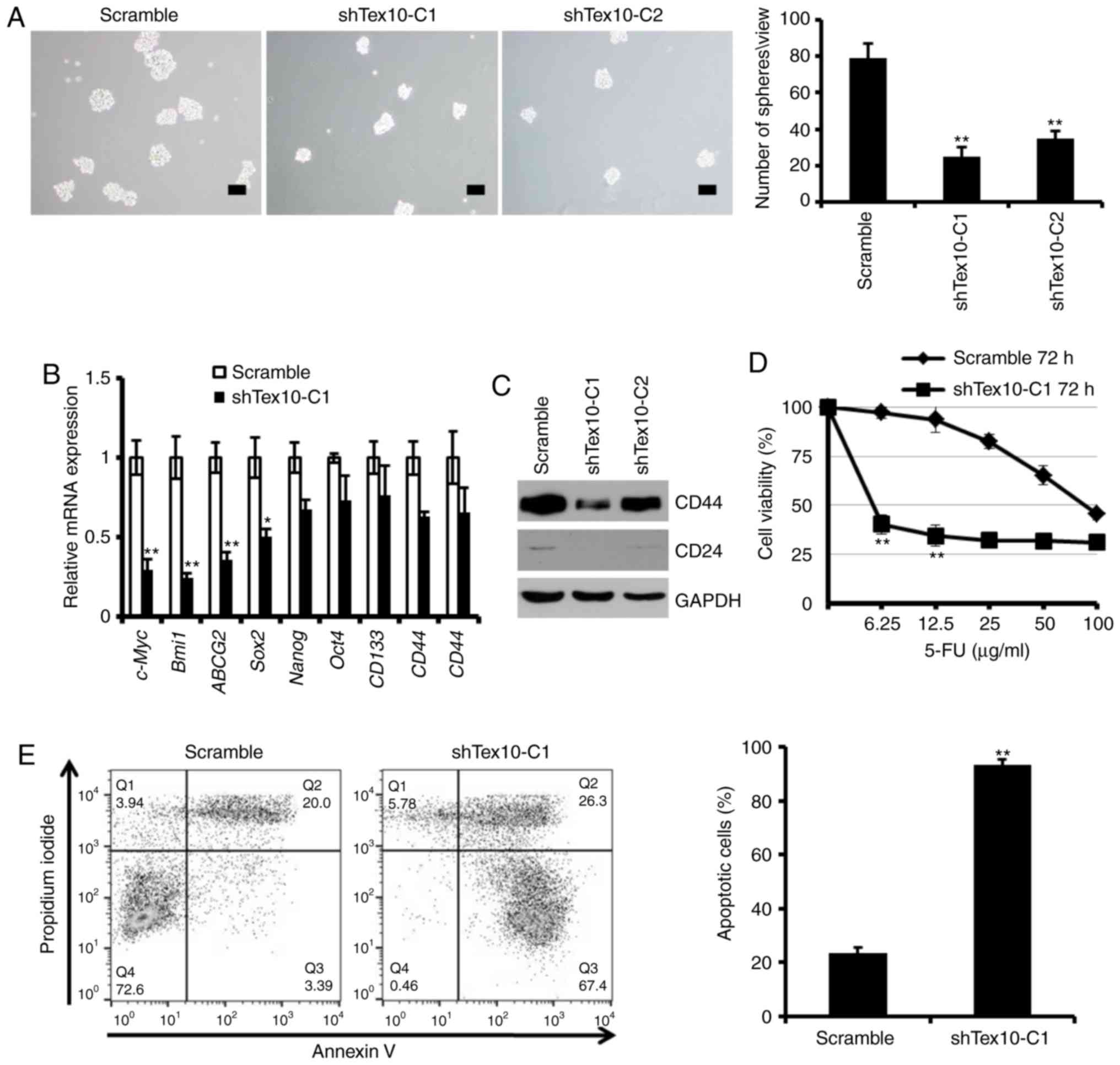 | Figure 3.Tex10 is necessary for the
maintenance of CSC properties. (A) Representative images of spheres
formed by different groups of EC109 cells. Histograms indicate the
mean number of spheres. One-way ANOVA followed by the Bonferroni
test was used to determine the statistical significance of the
differences. **P<0.01, significant difference vs. scramble
group. Scar bar, 100 µm. (B) RT-qPCR analysis of the expression of
pluripotency-associated markers, including Nanog, Oct4, Sox2, Bmi1,
ABCG2, c-Myc, CD133, CD44 and CD24 in different groups of EC109
cells. Error bars represent the means ± SD of 3 independent
experiments. The two-tailed unpaired Student's t-test was used to
determine statistical significance. *P<0.05 and **P<0.01,
significant difference vs. scramble group. (C) Tex10 knockdown
decreased CD44 and CD24 protein expression levels in EC109 cells.
(D) shTex10-C1 and scramble EC109 cells were treated with the
indicated concentrations of 5-fluorouracil (0–100 µg/ml) for 72 h.
Cell viability was analyzed by CCK-8 assay. The OD value of
untreated cells was taken as 100% viable cells. Data are presented
as the means ± SD; n=3; The two-tailed unpaired Student's t-test
was used to determine statistical significance. **P<0.01,
significant difference vs. scramble group. (E) Representative
apoptosis analyses of EC109 cells treated with 5-fluorouracil for
48 h. The values indicate the means ± SD of 3 independent
experiments. The two-tailed unpaired Student's t-test was used to
determine statistical significance. **P<0.01, significant
difference vs. scramble group. Tex10, testis expressed 10; Oct4,
octamer-binding transcription factor 4; Sox2, SRY-Box 2; Bmi1,
polycomb ring finger; ABCG2, ATP binding cassette subfamily G
member 2; c-Myc, bHLH transcription factor. |
Thus, to explore the potential role of Tex10 in
promoting chemoresistance, the effect of 5-fluorouracil on the
survival of EC109 cells in which Tex10 was knocked down was further
investigated. Following treatment with various concentrations of
5-fluorouracil for 72 h, the shTex10 group was observed to be more
sensitive to increasing concentrations of 5-fluorouracil compared
to the scramble group (P<0.01; Fig.
3D), and a significantly lower IC50 value of
5-fluorouracil of 5.3±0.94 µg/ml in the shTex10-transfected EC109
cells compared with 87.5±12.35 µg/ml in the scramble
shRNA-transfected cells was obtained. Furthermore, flow cytometric
analysis of the scramble shRNA- and shTex10-transfected EC109 cells
treated with 100 µg/ml 5-fluorouracil for 48 h indicated that a
larger percentage of shTex10-transfected EC109 cells underwent
apoptosis when exposed to 5-fluorouracil (P<0.01; Fig. 3E). In summary, these results indicate
that Tex10 promotes the self-renewal capacity and drug resistance
of ESCC cells, the two major functional characteristics of
CSCs.
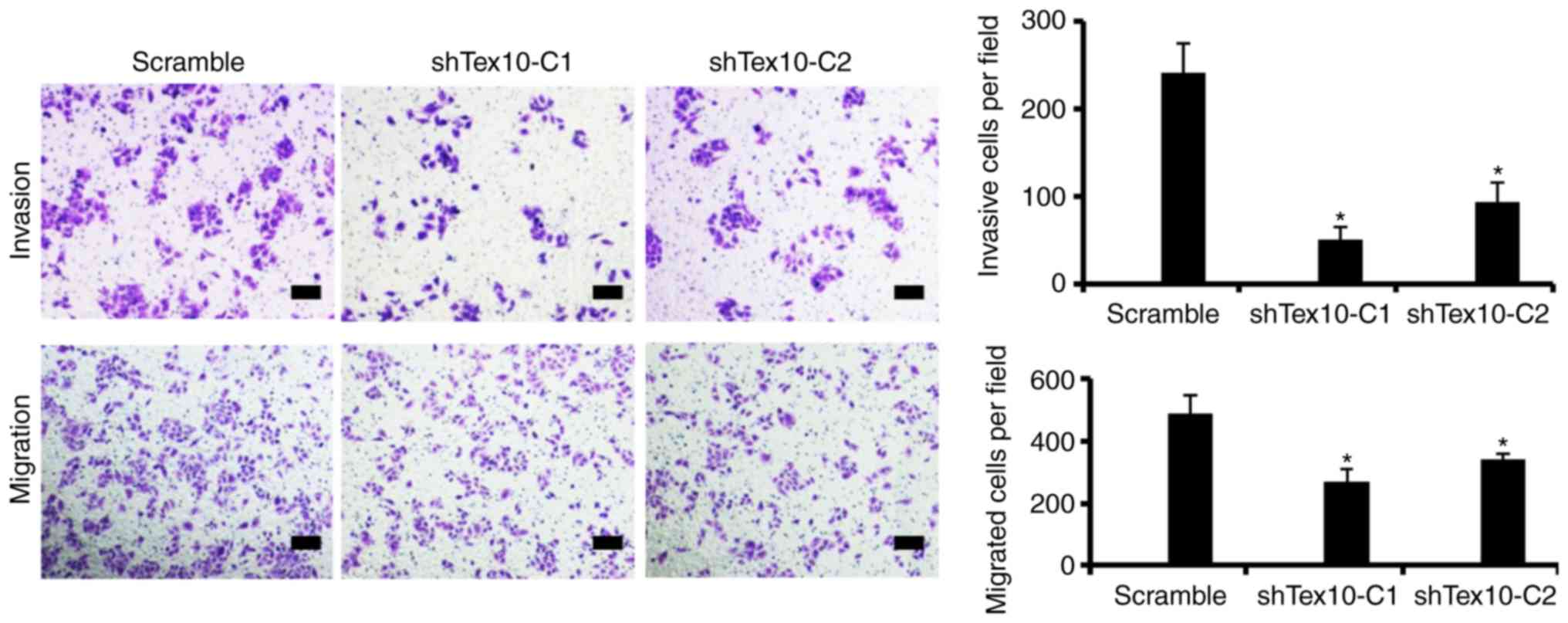
Silencing of Tex10 suppresses the
migration and invasion of ESCC cells in vitro
To further examine the role of Tex10 in invasion and
metastasis in ESCC, the migratory and invasive capacity of the ESCC
cells in which Tex10 was silenced was assessed. Transwell assays
indicated that the silencing of Tex10 markedly suppressed the
migration of the EC109 cells. Furthermore, the invasion of the
EC109 cells significantly decreased following Tex10 knockdown.
These results indicate that Tex10 plays a role in the migration and
invasion of EC109 cells (P<0.05; Fig.
4).
Tex10 promotes EMT by enhancing
Wnt/β-catenin signaling during ESCC tumorigenesis
It was observed that the morphology of the EC109
cells with stable Tex10 knockdown changed from a spindle-like
mesenchymal morphology to an epithelial cobblestone-like appearance
(Fig. 5A). The morphological changes
were further supported by phalloidin staining, indicating that the
EC109 cells in which Tex10 was silenced exhibited a
cobblestone-like shape and shrunken F-actin fibers compared to the
scrambled shRNA-transfected cells (Fig.
5A). Considering the morphological changes of the ESCC cells,
it was hypothesized that an abnormally high level of Tex10 may
trigger EMT. E-cadherin, vimentin, Snail, Slug, Twist1 and
β-catenin have been frequently identified as specific markers for
the EMT process (22,23). Therefore, it was further assessed
whether Tex10 had an impact on the expression of EMT markers by
quantifying the expression levels of these markers by RT-qPCR and
western blot analysis. As presented in Fig. 5B, the mRNA levels of E-cadherin in the
cells in which Tex10 was silenced were significantly increased,
whereas the expression levels of Slug and β-catenin in the
Tex10-silenced cells were significantly decreased; western blot
analysis revealed that the protein levels of Slug and β-catenin
were markedly downregulated, while the expression of vimentin was
not affected (Fig. 5C). These results
thus suggest that the downregulation of Tex10 attenuates EMT in
ESCC.
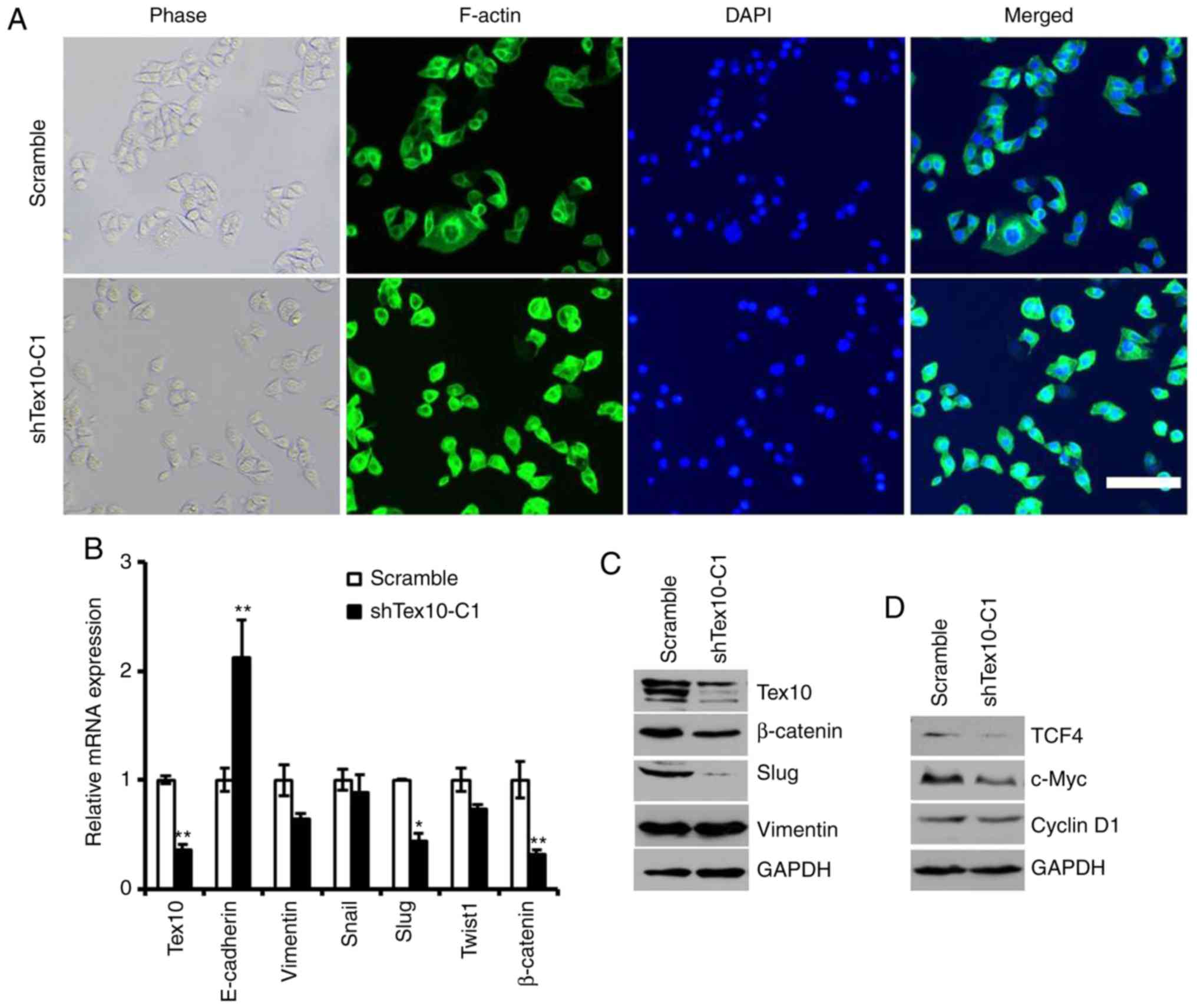 | Figure 5.Tex10-regulated EMT markers. (A)
Representative images of cytoskeleton showeing that Tex10 affected
cellular morphology. Scar bar, 100 µm. (B) The mRNA expression
levels of 5 EMT markers (E-cadherin, Vimentin, Snail, Slug, Twist1
and β-catenin) and Tex10 in EC109 cells were assayed by RT-qPCR.
GAPDH was used as an internal control. The two-tailed unpaired
Student's t-test was used to determine statistical significance.
*P<0.05 and **P<0.01, significant difference vs. scramble
group. (C) The protein levels of EMT marker (Slug, β-catenin,
Vimentin) and Tex10 in different groups of cells were assayed by
western blot analysis. GAPDH was used as an internal control. (D)
The protein levels of targets of the Wnt/β-catenin pathway (TCF4,
c-Myc and Cyclin D1) in different groups of EC109 cells were
assayed by western blot analysis. GAPDH was used as an internal
control. Tex10, testis expressed 10. |
Since Wnt/β-catenin signaling plays an essential
role in promoting tumor invasion and metastasis, and is one of the
major mechanisms regulating EMT, it was investigated whether Tex10
promotes ESCC metastasis via the Wnt/β-catenin pathway. For this,
the expression of downstream target genes of the Wnt/β-catenin
pathway, including cyclin D1, c-Myc and TCF4, was assessed. As
shown in Fig. 5D, the silencing of
Tex10 markedly reduced the protein levels of cyclin D1, c-Myc and
TCF4. Taken together, these results indicate that Tex10 may promote
the EMT in ESCC via activation of the Wnt/β-catenin pathway.
Discussion
Tex10 plays important roles in the regulation of
transcription, the cell cycle and ribosome biogenesis. Most
importantly, Tex10 functions as a key pluripotency factor that
plays an essential role in esophageal squamous cell pluripotency
and early embryonic development (8,9). Although
a previous study by our group indicated that Tex10 plays a distinct
tumorigenic role in HCC by maintaining stem cell-like traits
(10), the potential molecular
mechanisms through which Tex10 exerts its effects on ESCC remain to
be elucidated. To the best of our knowledge, the present study is
the first to report the functional role of Tex10 in the regulation
of the EMT process and stemness by activating Wnt/β-catenin
signaling.
CSCs are heterogeneous populations of tumor cells,
which possess unlimited self-renewal ability and multi-lineage
differentiation potential, and are responsible for tumor
initiation, recurrence and chemotherapy resistance (24,25).
Therefore, CSCs are considered to be an ideal target for cancer
therapy (26). A previous study by
our group reported that Tex10 was involved in maintaining cancer
stem cell properties in HCC (10). In
the present study, it was demonstrated that Tex10 was associated
with CSC-like traits, including chemoresistance and the EMT
phenotype. The knockdown of Tex10 suppressed the sphere-forming
ability of EC109 cells and significantly downregulated the
expression of CSC markers, including c-Myc, Bmi1, ABCG2, CD44 and
CD24. Previous studies have suggested that Tex10 plays a central
role in the establishment and maintenance of ESC pluripotency
(8,9).
The results of the present study demonstrated that the silencing of
Tex10 impaired cell stemness through the reduction of the
expression of Sox2, Nanog and Oct4. In addition, it was indicated
that the knockdown of Tex10 in the EC109 cells enhanced the
sensitivity of the cells to 5-fluorouracil. Taken together, these
results suggest that Tex10 may be a potential target for overcoming
chemoresistance in ESCC.
The activation of EMT contributes to the generation
of CSC-like cells (27,28), which is one of the major molecular
mechanisms responsible for the enhanced migration and invasion
during carcinogenesis (29,30). The present study confirmed that the
silencing of Tex10 substantially reduced ESCC cell migration and
invasion, and Tex10 expression was associated with morphological
changes and the expression of EMT markers; thesilencing of Tex10
upregulated E-cadherin and downregulates vimentin, Slug, β-catenin
expression in ESCC cells, suggesting that Tex10 promotes EMT in
vitro.
During cancer progression, several signaling
pathways are involved in the regulation of EMT, including tumor
growth factor-β, Notch, STAT3, Hedgehog and Wnt/β-catenin (31–35),
leading to the promotion of tumor invasion and metastasis.
Wnt/β-catenin signaling plays an important role in promoting the
self-renewal and tumorigenicity of CSCs in multiple cancer types,
and contributes to EMT during cancer progression (36–38).
However, to the best of our knowledge, there is available study to
date on the association between Tex10 and Wnt/β-catenin signaling
in ESCC. The present study established that the silencing of Tex10
decreased the expression of β-catenin and the target genes, TCF4,
cyclin D1 and c-Myc, suggesting that pharmacological targeting of
Tex10 to block Wnt/β-catenin signaling and inhibit EMT is an
attractive strategy with which to prevent tumor metastasis in ESCC.
The elucidation of the exact roles of Tex10 in the pathogenesis of
ESCC and the molecular mechanisms through which Tex10 activates
Wnt/β-catenin signaling may contribute to a better understanding of
cancer progression. Although the present study revealed the
essential role of Tex10 in positively regulating β-catenin
expression in ESCC, the exact molecular mechanisms underlying this
process remain to be identified; thus, further studies are required
in the future.
Collectively, the present study demonstrated that
Tex10 may serve as a functional marker of CSCs in ESCC, and may
potentially represent a valuable prognostic biomarker and
therapeutic target for ESCC.
Acknowledgements
Not applicable.
Funding
This study was supported by the Project of the
Education Department of Sichuan Province (grant no. 18ZB0214 to
JB), the Innovation Project of Medical Scientific Research Youth in
Sichuan Province (grant no. Q18054 to XX) and the Bureau of Science
and Technology and Intellectual Property Nanchong City, China
(NSMC20170466 to XX, 18SXHZ0211 to XL, 18SXHZ0210 to RX).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
XX and GF conceived the and supervised the study; XX
and RX designed the experiments; RX, XL and CY performed
experiments; YZ, LD and JB developed new software and performed
simulation studies; XL, DX, ZC, CY and KL analyzed the data; XX, RX
and GF wrote the manuscript. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
The study involving the use of human tissue was
approved by the Ethics Committee of the North Sichuan Medical
College (NSMC) and Ethical Committee of the Nanchong Central
Hospital (Sichuan, China). Written informed consent was obtained
from all subjects. All applicable international, national, and/or
institutional guidelines for the care and use of animals were
followed. All procedures performed in the studies were in
accordance with the 1964 Declaration of Helsinki and its later
amendments.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Sun K, Zheng R, Zeng H, Zhang S,
Xia C, Yang Z, Li H, Zou X and He J: Cancer incidence and mortality
in China, 2014. Chin J Cancer Res. 30:1–12. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pennathur A, Gibson MK, Jobe BA and
Luketich JD: Oesophageal carcinoma. Lancet. 381:400–412. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fanis P, Gillemans N, Aghajanirefah A,
Pourfarzad F, Demmers J, Esteghamat F, Vadlamudi RK, Grosveld F,
Philipsen S and van Dijk TB: Five friends of methylated chromatin
target of protein-arginine-methyltransferase[prmt]-1 (chtop), a
complex linking arginine methylation to desumoylation. Mol Cell
Proteomics. 11:1263–1273. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Castle CD, Cassimere EK and Denicourt C:
LAS1L interacts with the mammalian Rix1 complex to regulate
ribosome biogenesis. Mol Biol Cell. 23:716–728. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Finkbeiner E, Haindl M and Muller S: The
SUMO system controls nucleolar partitioning of a novel mammalian
ribosome biogenesis complex. EMBO J. 30:1067–1078. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ding J, Huang X, Shao N, Zhou H, Lee DF,
Faiola F, Fidalgo M, Guallar D, Saunders A, Shliaha PV, et al:
Tex10 coordinates epigenetic control of super-enhancer activity in
pluripotency and reprogramming. Cell Stem Cell. 16:653–668. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhao W, Huang Y, Zhang J, Liu M, Ji H,
Wang C, Cao N, Li C, Xia Y, Jiang Q and Qin J: Polycomb group RING
finger proteins 3/5 activate transcription via an interaction with
the pluripotency factor Tex10 in embryonic stem cells. J Biol Chem.
292:21527–21537. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xiang X, Deng L, Xiong R, Xiao D, Chen Z,
Yang F, Liu K and Feng G: Tex10 is upregulated and promotes cancer
stem cell properties and chemoresistance in hepatocellular
carcinoma. Cell Cycle. 17:1310–1318. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang D, Plukker JTM and Coppes RP: Cancer
stem cells with increased metastatic potential as a therapeutic
target for esophageal cancer. Semin Cancer Biol. 44:60–66. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ouzounova M, Lee E, Piranlioglu R, El
Andaloussi A, Kolhe R, Demirci MF, Marasco D, Asm I, Chadli A,
Hassan KA, et al: Monocytic and granulocytic myeloid derived
suppressor cells differentially regulate spatiotemporal tumour
plasticity during metastatic cascade. Nat Commun. 8:149792017.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pastushenko I and Blanpain C: EMT
transition states during tumor progression and metastasis. Trends
Cell Biol. 29:212–226. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dongre A and Weinberg RA: New insights
into the mechanisms of epithelial-mesenchymal transition and
implications for cancer. Nat Rev Mol Cell Biol. 20:69–84. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ishay-Ronen D, Diepenbruck M, Kalathur
RKR, Sugiyama N, Tiede S, Ivanek R, Bantug G, Morini MF, Wang J,
Hess C and Christofori G: Gain fat-lose metastasis: Converting
invasive breast cancer cells into adipocytes inhibits cancer
metastasis. Cancer Cell. 35:17–32.e16. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xiang X, Deng L, Zhang J, Zhang X, Lei T,
Luan G, Yang C, Xiao ZX and Li Q and Li Q: A distinct expression
pattern of cyclin K in mammalian testes suggests a functional role
in spermatogenesis. PLoS One. 9:e1015392014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Maehara O, Suda G, Natsuizaka M, Ohnishi
S, Komatsu Y, Sato F, Nakai M, Sho T, Morikawa K, Ogawa K, et al:
Fibroblast growth factor-2-mediated FGFR/Erk signaling supports
maintenance of cancer stem-like cells in esophageal squamous cell
carcinoma. Carcinogenesis. 38:1073–1083. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lu YX, Chen DL, Wang DS, Chen LZ, Mo HY,
Sheng H, Bai L, Wu QN, Yu HE, Xie D, et al: Melatonin enhances
sensitivity to fluorouracil in oesophageal squamous cell carcinoma
through inhibition of Erk and Akt pathway. Cell Death Dis.
7:e24322016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Iser IC, Ceschini SM, Onzi GR, Bertoni AP,
Lenz G and Wink MR: Conditioned medium from adipose-derived stem
cells (ADSCs) promotes epithelial-to-mesenchymal-like transition
(EMT-Like) in glioma cells in vitro. Mol Neurobiol. 53:7184–7199.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang HF, Wu C, Alshareef A, Gupta N, Zhao
Q, Xu XE, Jiao JW, Li EM, Xu LY and Lai R: The PI3K/AKT/c-MYC axis
promotes the acquisition of cancer stem-like features in esophageal
squamous cell carcinoma. Stem Cells. 34:2040–2051. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hu X, Zhai Y, Kong P, Cui H, Yan T, Yang
J, Qian Y, Ma Y, Wang F, Li H, et al: FAT1 prevents epithelial
mesenchymal transition (EMT) via MAPK/ERK signaling pathway in
esophageal squamous cell cancer. Cancer Lett. 397:83–93. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen HA, Kuo TC, Tseng CF, Ma JT, Yang ST,
Yen CJ, Yang CY, Sung SY and Su JL: Angiopoietin-like protein 1
antagonizes MET receptor activity to repress sorafenib resistance
and cancer stemness in hepatocellular carcinoma. Hepatology.
64:1637–1651. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Marquardt S, Solanki M, Spitschak A, Vera
J and Pützer BM: Emerging functional markers for cancer stem
cell-based therapies: Understanding signaling networks for
targeting metastasis. Semin Cancer Biol. 53:90–109. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Loureiro R, Mesquita KA, Magalhães-Novais
S, Oliveira PJ and Vega-Naredo I: Mitochondrial biology in cancer
stem cells. Semin Cancer Biol. 47:18–28. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nagle PW, Plukker JTM, Muijs CT, van Luijk
P and Coppes RP: Patient-derived tumor organoids for prediction of
cancer treatment response. Semin Cancer Biol. 53:258–264. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Singh M, Yelle N, Venugopal C and Singh
SK: EMT: Mechanisms and therapeutic implications. Pharmacol Ther.
182:80–94. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen L, Li YC, Wu L, Yu GT, Zhang WF,
Huang CF and Sun ZJ: TRAF6 regulates tumour metastasis through EMT
and CSC phenotypes in head and neck squamous cell carcinoma. J Cell
Mol Med. 22:1337–1349. 2018.PubMed/NCBI
|
|
29
|
Peng JM, Bera R, Chiou CY, Yu MC, Chen TC,
Chen CW, Wang TR, Chiang WL, Chai SP, Wei Y, et al: Actin
cytoskeleton remodeling drives epithelial-mesenchymal transition
for hepatoma invasion and metastasis in mice. Hepatology.
67:2226–2243. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xie K, Ye Y, Zeng Y, Gu J, Yang H and Wu
X: Polymorphisms in genes related to epithelial-mesenchymal
transition and risk of non-small cell lung cancer. Carcinogenesis.
38:1029–1035. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Natsuizaka M, Whelan KA, Kagawa S, Tanaka
K, Giroux V, Chandramouleeswaran PM, Long A, Sahu V, Darling DS,
Que J, et al: Interplay between Notch1 and Notch3 promotes EMT and
tumor initiation in squamous cell carcinoma. Nat Commun.
8:17582017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhao D, Besser AH, Wander SA, Sun J, Zhou
W, Wang B, Ince T, Durante MA, Guo W, Mills G, et al: Cytoplasmic
p27 promotes epithelial-mesenchymal transition and tumor metastasis
via STAT3-mediated Twist1 upregulation. Oncogene. 34:5447–5459.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Huang J, Xiao D, Li G, Ma J, Chen P, Yuan
W, Hou F, Ge J, Zhong M, Tang Y, et al: EphA2 promotes
epithelial-mesenchymal transition through the Wnt/β-catenin pathway
in gastric cancer cells. Oncogene. 33:2737–2747. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kim J, Hyun J, Wang S, Lee C and Jung Y:
MicroRNA-378 is involved in hedgehog-driven
epithelial-to-mesenchymal transition in hepatocytes of regenerating
liver. Cell Death Dis. 9:7212018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Katsuno Y, Qin J, Oses-Prieto J, Wang H,
Jackson-Weaver O, Zhang T, Lamouille S, Wu J, Burlingame A, Xu J
and Derynck R: Arginine methylation of Smad7 by PRMT1 in
TGF-β-induced epithelial-mesenchymal transition and epithelial stem
cell generation. J Biol Chem. 293:13059–13072. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lee SH, Koo BS, Kim JM, Huang S, Rho YS,
Bae WJ, Kang HJ, Kim YS, Moon JH and Lim YC: Wnt/β-catenin
signalling maintains self-renewal and tumourigenicity of head and
neck squamous cell carcinoma stem-like cells by activating Oct4. J
Pathol. 234:99–107. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Xiang D, Cheng Z, Liu H, Wang X, Han T,
Sun W, Li X, Yang W, Chen C, Xia M, et al: Shp2 promotes liver
cancer stem cell expansion by augmenting β-catenin signaling and
predicts chemotherapeutic response of patients. Hepatology.
65:1566–1580. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chai S, Ng KY, Tong M, Lau EY, Lee TK,
Chan KW, Yuan YF, Cheung TT, Cheung ST, Wang XQ, et al: Octamer
4/microRNA-1246 signaling axis drives Wnt/β-catenin activation in
liver cancer stem cells. Hepatology. 64:2062–2076. 2016. View Article : Google Scholar : PubMed/NCBI
|