Introduction
Cervical cancer is the most common female malignancy
worldwide (1,2). The incidence and mortality rates
associated with cervical cancer are particularly higher in
developing countries (3,4). It has been estimated that 9.89/100,000
cervical cancer cases and 3.05/100,000 cancer-related deaths
occurred in China in 2015 (5). Almost
all cervical cancers are caused by persistent infection with
high-risk human papillomavirus (HPV), most commonly, HPV16 and
HPV18 (6–8).
During carcinogenesis, immune checkpoint pathways
are often exploited to evade immune surveillance (9,10),
particularly the development of cervical cancer. Hepatitis A virus
cellular receptor 2 (HAVCR2) is also known as T-cell immunoglobulin
and mucin-domain containing-3 (Tim-3) (11–13). The
most important role of Tim-3 is to negatively regulate Th1
immunity. Once Tim-3 binds to its ligand, galectin-9, Tim-3
inhibits Th1 and Th17 responses by hampering their expansion. The
Tim-3/galectin-9 pathway contributes to the suppressive tumor
microenvironment (TME) in the human body through Treg promotion
upon T cell receptor (TCR) activation (14,15). The
DNA methylation of the Tim-3 promoter cooperates with
lineage-specific transcription factors in the control of Th cell
development (16,17). Cao et al found that the
expression of Tim-3 in tumor cells may be an independent prognostic
factor for patients with cervical cancer; moreover, Tim-3
expression may promote metastatic potential in cervical cancers
(18). Liang et al found that
decreased galectin-9 expression was inversely associated with the
malignant potential or differentiation of cervical intraepithelial
neoplasia (CIN) and cervical squamous cell carcinoma (SCC) as a
differentiation biomarker (19).
Enhancer of zeste homolog 2 (EZH2) mRNA expression has been
shown to be increased in cancer tissues compared to normal tissues,
and the overexpression of EZH2 has been shown to be associated with
the FIGO stage, histological type and lymph node metastasis in
cervical cancer (20). The inhibition
of DNA (cytosine-5)-methyltransferase 3A (DNMT3A) promotes cervical
cancer cell apoptosis, which further demonstrates that DNMT3A is
involved in cervical carcinogenesis (21). However, whether the abnormal
methylation of the promoter regions of Tim-3/galectin-9 plays a
role in the carcinogenesis of cervical cancer remains unclear.
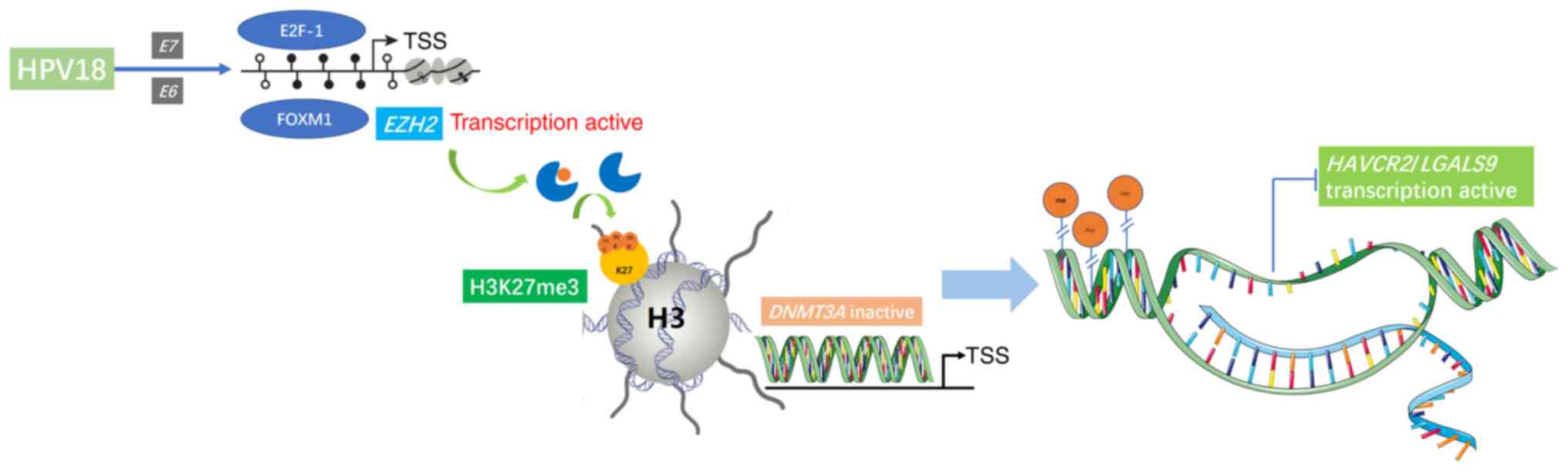
In the present study, it was demonstrated that the
promoter regions of HAVCR2/LGALS9 were partially methylated,
and that the protein expression of Tim-3/galectin-9 was increased
when their methylation level decreased in accordance with the
decreased DNMT3A expression. Our previous study found that HPV18
was the second most carcinogenic type of HPV in the Shaanxi
Province of China (22). Moreover,
HPV18 E6 and E7 participate in the upregulation of EZH2 and
H3K27me3 expression, which inhibits DNMT3A expression to
downregulate the HAVCR2/LGALS9 methylation levels. We thus
hypothesized that the HPV18 oncoproteins E6 and E7 may play an
important role in regulating the expression of Tim-3/galectin-9 in
the immune microenvironment of cervical cancer through DNA
methylation mediated by EZH2/H3K27me3/DNMT3A.
Materials and methods
Patients and samples
A total of 24 cervical cancer tissue specimens, 24
matched peri-carcinomatous tissue specimens and 16 normal cervical
tissue specimens were obtained from the First Affiliated Hospital
of Xi'an Jiaotong University between January, 2014 and December,
2017. All patients were diagnosed by two senior pathologists, and
no patient had received chemotherapy or radiotherapy prior to
surgery; details of the patient characteristics are presented in
Table I. Each patient enrolled in
this study had volunteered and provided written informed consent.
This study was approved by the Ethics Committee of the First
Affiliated Hospital of Xi'an Jiaotong University (G-272) in
Shaanxi, China.
 | Table I.Patient characteristics (n=24). |
Table I.
Patient characteristics (n=24).
| Item | No. |
|---|
| Age (years) |
|
|
≤44 | 10 |
|
>44 | 14 |
| Clinical
stages |
|
| Ia | 0 |
| Ib | 8 |
|
IIa | 11 |
|
IIb | 5 |
| Pathological
pattern |
|
|
Squamous cell carcinoma | 21 |
|
Adenocarcinoma | 3 |
| Pathological
grading |
|
| I | 1 |
| II | 19 |
|
III | 4 |
| Lymph nodes
metastasis |
|
|
Yes | 3 |
| No | 21 |
| HPV infection |
|
|
Positive | 16 |
|
Negative | 8 |
The cervical cancer samples were collected as
previous described (23). After the
tissues were dissected, each sample was washed with sterilized PBS
and stored at −80°C. All procedures were performed on ice.
Data mining
Data obtained from the Gene Expression Profiling
Interactive Analysis (GEPIA; http://gepia.cancer-pku.cn/) database was utilized to
analyze the mRNA expression level of HAVCR2/LGALS9 in
cervical cancer and normal cervical tissues. The correlation
between HAVCR2 and LGALS9 expression was determined
by Pearson's correlation analysis. In addition, the association
between a high and low HAVCR2/LGALS9 mRNA expression with
the overall survival (OS) of patients with cervical cancer was
analyzed by Kaplan-Meier analysis, and the hazard ratio (HR) and
log-rank P-value were also computed.
HPV-DNA testing
HPV-DNA of 24 cervical cancer tissue samples was
examined by polymerase chain reaction (PCR) and flow-through
hybridization. In total, 21 HPV genotypes were qualitatively
examined using the HPV genotyping test kit according to alkaline
phosphatase system (HybriBio), including high-risk HPV types,
namely HPV16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59 and 68,
and common HPV types in China, namely HPV53, 66 and CP8304.
Cell lines, culture conditions,
overexpression and construction of cell lines with gene
silencing
The two human cervical cancer cell lines, HeLa and
C33A, were obtained from the Cell Bank, Shanghai Institutes for
Biological Sciences, Chinese Academy of Sciences, Shanghai. All
these cells were cultured in high-glucose Dulbecco's modified
Eagle's medium (DMEM; HyClone) supplemented with 10% fetal bovine
serum (FBS; Biological Industries) at 37°C in an atmosphere of 5%
CO2. For the preparation of Plenti-CMV-puro-Dest vector
containing the HPV18 E6/E7/E67 fragment, the HPV18 E6/E7/E67
fragment was cloned from the genomic DNA of the HPV18(+) cell line,
HeLa. The DNA fragment was treated with Kpn1 and
Pst1, and the target gene was then ligated into the entry
vector pENTR-MCS. The plasmid Plenti-CMV-puro-Dest and pENTR-MCS
recombination reactions were performed using LR Clonase II
(Invitrogen; Thermo Fisher Scientific). Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific) was used to transfect the
plasmid into the HPV(−) C33A cell line. The transduced cells were
then selected with puromycin. Stably transduced cells were
maintained in culture in the presence of puromycin. The cell lines
were named C33A-18E6, C33A-18E7, and C33A-18E6/E7. The expression
of the genes was determined by RT-qPCR.
The HeLa tumor cells were transfected with scramble,
HPV18-E6, HPV18-E7, EZH2 and DNMT3A-specific siRNA (GenePharma),
the following siRNA oligos for EZH2, HPV18-E6, HPV18-E7 and DNMT3A
are listed in Table II. The siRNAs
were transfected using X-tremeGENE siRNA Transfection Reagent
(Roche), and the transfected cells were analyzed for HPV18-E6,
HPV18-E7, EZH2 and DNMT3A expression levels by RT-qPCR and western
blot analysis. All cell lines were named 18E6-siRNA, 18E7-siRNA,
18E6/E7-siRNA, EZH2-siRNA and DNMT3A-siRNA.
 | Table II.Primer sequences. |
Table II.
Primer sequences.
| Name | Application | Sequence |
|---|
|
DNMT3A-ChIP-F1 | ChIP-qPCR |
ATCATCAGTAGGGCGGGGTGGCCAC |
|
DNMT3A-ChIP-R1 | ChIP-qPCR |
CTCCAATGCTTCCAGGTCCCTCCGT |
|
DNMT3A-ChIP-F2 | ChIP-qPCR |
TTGGAGAACCTCCCGAAGGAAAACC |
|
DNMT3A-ChIP-R2 | ChIP-qPCR |
GCCACCCTTTTAGCGTCACAGAACC |
|
DNMT3A-ChIP-F3 | ChIP-qPCR |
CGTTGGGGGGGCGGGTGCTGGGCTG |
|
DNMT3A-ChIP-R3 | ChIP-qPCR |
TGACTGGCACAGGACATGGCGTGCT |
|
DNMT3A-ChIP-F4 | ChIP-qPCR |
CATGGGGAAGGAGAACAGCCCCCAC |
|
DNMT3A-ChIP-R4 | ChIP-qPCR |
GCACTGGAAGACTGAAAGATTTCAT |
|
EZH2-ChIP-F1 | ChIP-qPCR |
GCTCACGCCTGTAATCCCAGCACTT |
|
EZH2-ChIP-R1 | ChIP-qPCR |
GGAGTTTCGCTCTGGTTGTCCAGGC |
|
EZH2-ChIP-F2 | ChIP-qPCR |
GGCTGAGGCATGAGAATCGCTTGAA |
|
EZH2-ChIP-R2 | ChIP-qPCR |
TGAGACGGAGTTTCGCTCTGGTTGT |
|
EZH2-ChIP-F3 | ChIP-qPCR |
GCCTGCACACCGCCTTCCTGAGAGG |
|
EZH2-ChIP-R3 | ChIP-qPCR |
GGGGTTCGCTGTAAGGGACGCCACT |
|
EZH2-ChIP-F4 | ChIP-qPCR |
CCACACGGCCAGTGGCGTCCCTTAC |
|
EZH2-ChIP-R4 | ChIP-qPCR |
CACGCAGAGTGCGCTCAGGGCTCGT |
|
HAVCR2-ChIP-F1 | ChIP-qPCR |
GTGGAAAAAATCTGTCACTTAGGGG |
|
HAVCR2-ChIP-R1 | ChIP-qPCR |
ATTTTTAGTAGAGACGGGGTTTCTC |
|
HAVCR2-ChIP-F2 | ChIP-qPCR |
CCTGTAATCCCAGCTACTCAGGAGG |
|
HAVCR2-ChIP-R2 | ChIP-qPCR |
CTTGTTCAATGTGTGTACTTCCCAT |
|
HAVCR2-ChIP-F3 | ChIP-qPCR |
CCCAATGCATTTAATGGCATAAATG |
|
HAVCR2-ChIP-R3 | ChIP-qPCR |
CAGCCACACTCCCATAACTGAGGTA |
|
HAVCR2-ChIP-F4 | ChIP-qPCR |
GGAACTCAACACTTTCTGATCATTC |
|
HAVCR2-ChIP-R4 | ChIP-qPCR |
GACTTTGACCTTCAAACTTCCAACT |
|
LGALS9-ChIP-F1 | ChIP-qPCR |
GGTAGAGTAAAATGTACAGATCCTG |
|
LGALS9-ChIP-R1 | ChIP-qPCR |
GCGAGACCTTGTCTCTACTAAAAAT |
|
LGALS9-ChIP-F2 | ChIP-qPCR |
TCAGCCTCCCAATGTGCTGAATTAC |
|
LGALS9-ChIP-R2 | ChIP-qPCR |
CCAGATCCAAACTTGACTTGAAGTG |
|
LGALS9-ChIP-F3 | ChIP-qPCR |
TCCTGTGGCCTAGCTCCTTTTTATT |
|
LGALS9-ChIP-R3 | ChIP-qPCR |
AGAAAAACTGCTTGGTGAGTTGTAA |
|
LGALS9-ChIP-F4 | ChIP-qPCR |
CACATATGTTTTCCTTTCTCTTGGG |
|
LGALS9-ChIP-R4 | ChIP-qPCR |
ACACCTGTGGTCTCAGCTACATGGG |
|
HPV18-E6-F | RT-qPCR |
CAACACGGCGACCCTACAAG |
|
HPV18-E6-R | RT-qPCR |
GCTGGATTCAACGGTTTCTGG |
|
HPV18-E7-F | RT-qPCR |
ACATTTACCAGCCCGACG |
|
HPV18-E7-R | RT-qPCR |
CAAAGGACAGGGTGTTCAGA |
| GAPDH-F | RT-qPCR |
GCACCGTCAAGGCTGAGAAC |
| GAPDH-R | RT-qPCR |
TGGTGAAGACGCCAGTGGA |
|
HAVCR2-ML | MS-PCR |
TATAAAATGAGAAATTGGTCGGGCG |
|
HAVCR2-MR | MS-PCR |
TTACAAACATATACCACCACCCCGA |
|
HAVCR2-UL | MS-PCR |
GAAATTGGTTGGGTGTGGTGGTTAT |
|
HAVCR2-UR | MS-PCR |
TATACCACCACCCCAAATAATTTTA |
|
LGALS9-ML | MS-PCR |
TTTTCGAGATAGGTTTGCGATTTTG |
|
LGALS9-MR | MS-PCR |
AATACCGACACCCTTCAATCACCAC |
|
LGALS9-UL | MS-PCR |
GAGTTTTTGAGATAGGTTTGTGATT |
|
LGALS9-UR | MS-PCR |
ATACCAACACCCTTCAATCACCACA |
|
EZH2-sense | Gene silencing |
CGGCUUCCCAAUAACAGUATT |
|
EZH2-anti-sense | Gene silencing |
UACUGUUAUUGGGAAGCCGTT |
|
DNMT3A-sense | Gene silencing |
GCCAAGGUCAUUGCAGGAATT |
|
DNMT3A-anti-sense | Gene silencing |
UUCCUGCAAUGACCUUGGCTT |
|
HPV18-E6-sense | Gene silencing |
CGCAGAGAAACACAAGUAUTT |
|
HPV18-E6-anti-sense | Gene silencing |
AUACUUGUGUUUCUCUGCGTT |
|
HPV18-E7-sense | Gene silencing |
GUCACACAAUGUUGUGUAUTT |
|
HPV18-E7-anti-sense | Gene silencing |
AUACACAACAUUGUGUGACTT |
| Negative
control-sense | Gene silencing |
UUCUCCGAACGUGUCACGUTT |
| Negative
control-sense | Gene silencing |
ACGUGACACGUUCGGAGAATT |
5-Aza-2′-deoxycytidine (5-Aza-CdR)
treatment
A total of 1.0×105 HeLa and C33A cells
per well were cultured separately in 6-well plates in DMEM with 10%
FBS. After 24 h, the medium was replaced with fresh medium
containing 0, 2.5 or 5 µM 5-Aza-CdR (Sigma-Aldrich; Merck KGaA).
The medium containing 5-Aza-CdR was replaced every 24 h during a
72-h period, as previously described (23).
DNA extraction, bisulfite modification
and methylation- specific PCR (MS-PCR)
Genomic DNA was isolated from the cells and tissues
using a Takara Mini BEST Universal Genomic DNA Extraction kit
(Takara) according to the manufacturer's instructions. DNA
modification was performed as previously described (23). A total of 500 ng of the extracted DNA
was bisulfite-modified with the EZ DNA Methylation-Gold™ kit (Zymo
Research). Modified DNA templates were used for MS-PCR with Zymo
TaqTM PreMix (E2003; Zymo Research) following the instructions of
the manufacturer. The online software MethPrimer (http://www.urogene.org/methprimer/) profiled CpG
islands in the region that is located from −2,000 to −200 bp
upstream of ATG in the HAVCR2/LGALS9 promoters. One
pair of primers was designed to amplify the HAVCR2/LGALS9
promoter regions. The primer pairs used for MS-PCR are listed in
Table II. The thermocycling
conditions were as follows: 95°C for 10 min and 40 cycles of 95°C
for 30 sec. The annealing temperature for the methylated primer
pairs of HAVCR2 and LGALS9 was 60°C, respectively,
while that for the unmethylated primer pairs was 55°C and 56.3°C,
respectively for 30 sec and 72°C for 30 sec, followed by an
incubation step at 72°C for 7 min. The MS-PCR products were
separated on a 2% agarose gel, stained with Gelview and visualized
under ultraviolet illumination (Bio-Rad). Methylation level was
calculated by the ratio of methylated and unmethylated levels. The
calculation method was as follows: Methylation(M)=M/(M + U);
unmethylation=U/(M + U). The grey value of each band represented
its relative expression and was measured using ImageJ software.
Each reaction was performed in triplicate.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from cells using TRIzol
Reagent (Life Technologies; Thermo Fisher Scientific) according to
the manufacturer's instructions and as previously described
(24). The RNA was then subjected to
reverse transcription with a cDNA PrimeScript™ RT Master Mix
(Takara). The primers used for qPCR are listed in Table II. qPCR was carried out using
SYBR-Green II Premix (Takara) by a two-step amplification procedure
according to the manufacturer's protocol. The reaction conditions
were as follows: 95°C for 30 sec, 40 cycles of 95°C for 5 sec and
60°C for 30 sec,72°C for 5 sec. We used the cycle threshold (CT) as
the representative point. GAPDH was used as an internal
reference gene. The relative expression of HPV18 E6 and
E7 in each group (fold change compared with the control) was
calculated using the formula: RQ=2−∆∆Cq (25). Each reaction was performed in
triplicate.
Western blot analysis
The cells were harvested in RIPA lysis buffer
containing 1 mM protease inhibitor cocktail and 1 mM PMSF. The
protein determination was assessed using a BCA detection kit
(Beyotime Biotechnology, China). Proteins were resolved by 12%
SDS-PAGE and electroblotted onto a polyvinylidene fluoride membrane
(Millipore), 40 µg of protein loaded per lane, which was blocked
for 1 h at room temperature in 5% skim milk, 1X TBS, 0.1% Tween-20.
After blocking, the membrane was incubated with primary antibodies
overnight at 4°C followed by incubation with HRP-conjugated
secondary antibodies for 1 h at room temperature. The antibodies
used were as follows: Rabbit polyclonal antibody against human
Tim-3 (1:500 dilution, ab185703; Abcam), rabbit polyclonal antibody
against human galectin-9 (1:100 dilution, ab123712; Abcam), mouse
monoclonal antibody against human DNMT3A (1:200 dilution,
sc-365769; Santa Cruz Biotechnology), rabbit monoclonal antibody
against human EZH2 (1:1,000 dilution, 5246; Cell Signaling
Technology), rabbit monoclonal antibody against human H3K27me3
(1:1,000 dilution, 9733; Cell Signaling Technology), mouse
monoclonal antibody against HPV18 E6 (1:250 dilution, NB100-2729;
Novus Biologicals) and mouse monoclonal antibody against HPV18 E7
(1:250 dilution, NB110-17215; Novus Biologicals), β-actin mouse
monoclonal antibody (1:500 dilution, 60008-1-lg; Proteintech). The
secondary antibodies were as follows: HRP-conjugated rabbit
anti-mouse IgG (1:5,000 dilution, D110273-0100; BBI Life Sciences),
HRP-conjugated goat anti-rabbit IgG (1:5,000 dilution, D110058; BBI
Life Sciences). All antibodies were diluted by 1X TBST. The
chemiluminescence signal was detected following incubation with
enhanced chemiluminescence reagent (Millipore, Mass). The grey
value of each band was measured with ImageJ software (1.47v).
Chromatin immunoprecipitation (ChIP)
assay and ChIP-qPCR
ChIP assays were carried out using the Simple ChIP
Enzymatic Chromatin IP kit (9003; Cell Signaling Technology).
Anti-H3K27me3 antibody (1:50 dilution, 9733; Cell Signaling
Technology), anti-EZH2 antibody (1:100 dilution, 5246; Cell
Signaling Technology), anti-DNMT3A antibody (1:50 dilution, ab2850;
Abcam), anti-E2F-1 antibody (1:100 dilution, 3742; Cell Signaling
Technology), anti-histone H3 antibody (1:50 dilution, 4620; Cell
Signaling Technology), anti-IgG antibody (1:1,000 dilution, 2729;
Cell Signaling Technology) and anti-FOXM1 antibody (1:100 dilution,
20459; Cell Signaling Technology) were used according to the
manufacturer's instructions. The antibodies were incubated
overnight at 4°C. The DNMT3A, EZH2 and HAVCR2/LGALS9
promoters DNA were detected by RT-qPCR using the promoter
DNA-specific primers listed in Table
II. RT-qPCR was performed as described above. IgG was used as a
negative control and histone H3 was used as a positive control.
Co-immunoprecipitation (co-IP)
assays
Immunoprecipitation was carried out to assess the
interaction between EZH2, DNMT3A and HPV18 E6 and E7. After
harvesting the total protein from the HeLa cells, the supernatants
were incubated overnight at 4°C with rabbit anti-EZH2 (1:300
dilution, 5246; Cell Signaling Technology) and rabbit anti-DNMT3A
(1:50 dilution, ab13888; Abcam) and protein G-Dynabeads (Thermo
Fisher Scientific) were then conjugated to EZH2 and DNMT3A. The
samples were then electrophoresed through gradient 12%
SDS-polyacrylamide gels and transferred to membranes that were
probed with mouse anti-HPV18 E6 and E7 antibodies, respectively
(1:250 dilution; Novus Biologicals, NB100-2729/NB110-17215) and
antibodies were incubated overnight at 4°C. This was followed by
incubation with horseradish peroxidase-conjugated secondary
antibodies for 1 h at room temperature. The secondary antibodies:
HRP-conjugated rabbit anti-mouse IgG (1:5,000 dilution,
D110273-0100; BBI Life Sciences), HRP-conjugated goat anti-rabbit
IgG (1:5,000 dilution, D110058; BBI Life Sciences). All antibodies
were diluted using 1X TBST. Western blot analysis was performed as
described above.
Statistical analysis
Statistical analyses were performed using GraphPad
Prism 7 software (GraphPad Software). A paired t-test and one-way
ANOVA were carried out on samples within groups, Dunnett's test was
used as the post hoc test after one-way ANOVA. A P-value <0.05
was considered to indicate a statistically significant difference.
The data are presented as the means ± standard error of the mean
(SEM). All experiments were independently repeated at least thrice,
with consistent results.
Results
Protein expression of Tim-3/galectin-9
is increased in cervical cancer tissues, and the gene methylation
level is decreased
The protein expression of Tim-3/galectin-9 in the 24
cervical cancer tissues, 24 matched pericarcinomatous tissues and
16 normal cervical tissues was detected by western blot analysis.
Compared to that in normal cervical tissues, the average relative
expression of Tim-3/galectin-9 was higher in the cervical cancer
and pericarcinomatous tissues, although no significant difference
was observed between the tumor tissues and pericarcinomatous
tissues (Fig. 1A-C). The relative
protein expression of Tim-3/galectin-9 in the HPV-positive cervical
cancer tissues was higher than that in the HPV-negative cervical
cancer tissues (Fig. 1D). Moreover,
Tim-3 is encoded by HAVCR2, and galectin-9 is encoded by
LGALS9. Using the GEPIA database (http://gepia.cancer-pku.cn/), we compared the mRNA
expression of HAVCR2/LGALS9 between the cervical cancer and
normal cervical samples. The results indicated that the average
mRNA expression levels of HAVCR2 and LGALS9 were
higher in the cancer than in the normal cervical samples (Fig. 1E). We then investigated the effects of
HAVCR2/LGALS9 on patient prognosis using the GEPIA database.
In particular, an increased LGALS9 expression was associated
with a poor overall survival (OS). Of note, no significant
association was observed between the expression of HAVCR2
and patient prognosis by the GEPIA database (Fig. 1F). Furthermore, HAVCR2
positively correlated with LGALS9 (r=0.26, P<0.05)
(Fig. 1G). In addition, the results
of immunohistochemical analysis revealed that Tim-3 and galectin-9
were expressed in the tumor cells of the cervical cancer tissues
(Fig. 1H). Furthermore, Tim-3 and
galectin-9 were expressed in the HeLa and C33A cells (Fig. 1I).
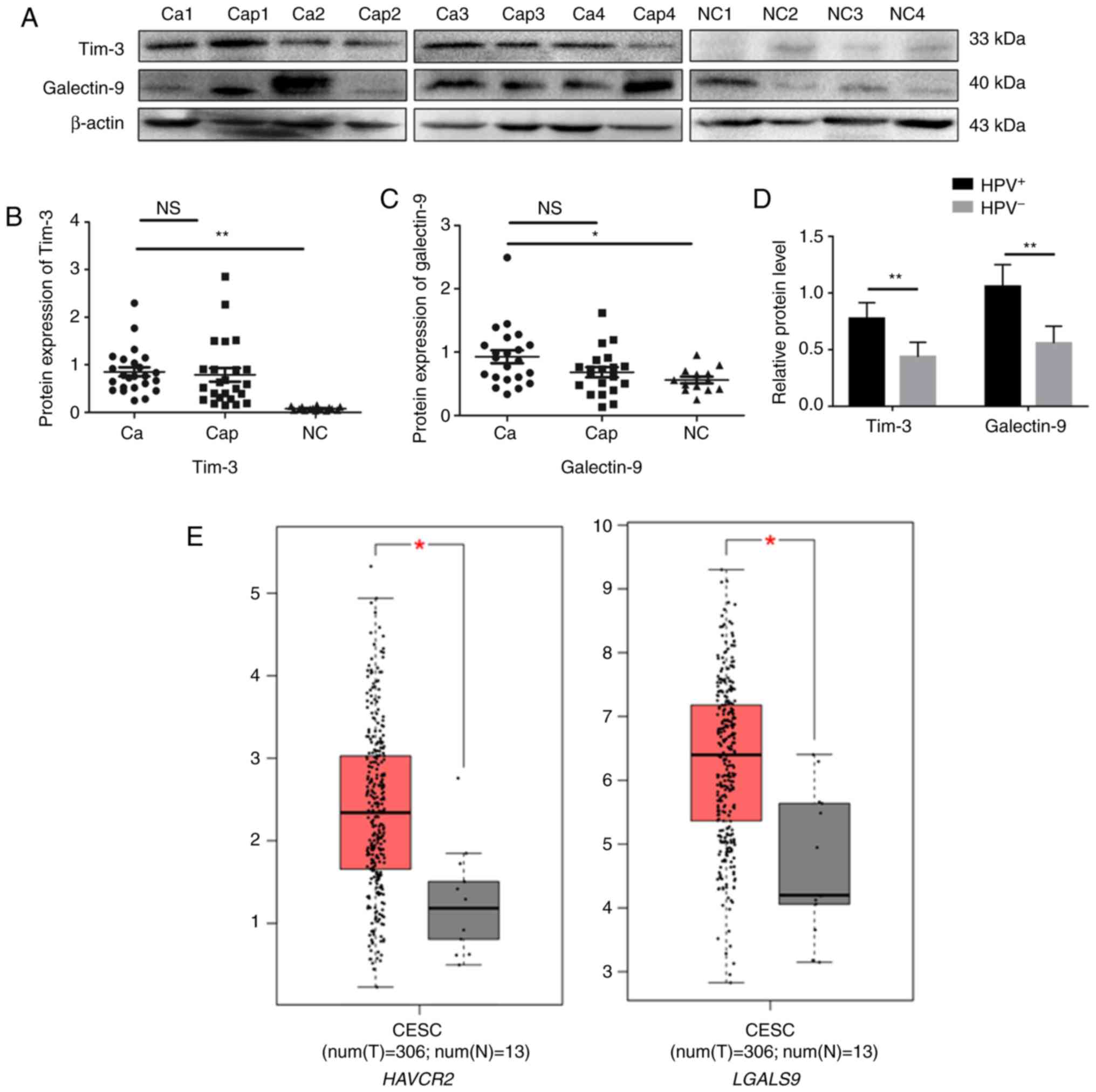 | Figure 1.Tim-3/galectin-9 protein and gene
methylation levels in human cervical cancer tissues. (A) The
protein levels of Tim-3 and galectin-9 in cervical cancer (Ca),
para-carcinoma (Cap) (n=24) and normal cervical tissues (NC) (n=16)
detected by western blot analysis. Blot images of 4 representative
samples are shown from each group. (B) Densitometric analysis of
Tim-3 and (C) galectin-9 protein levels in cervical cancer (n=24),
para-carcinoma (n=24) and normal cervical tissues (n=16). (D) Gray
level analysis of Tim-3/galectin-9 protein expression in
HPV-positive (n=16) and -negative (n=8) cervical cancer tissues.
(E) The mRNA expression level of HAVCR2/LGALS9 in cervical
cancer, analyzed by GEPIA. (M, methylated; U, unmethylated);
methylated and unmethylated levels were quantified as M/M+U% and
U/M+U%, respectively. *P<0.05; **P<0.01; ns, not significant.
(F) The prognostic value of mRNA level of HAVCR2/LGALS9 in
cervical cancer patients, analyzed by GEPIA. (G) The correlation
between HAVCR2/LGALS9 in cervical cancer, analyzed by GEPIA.
(H) Positive immunohistochemistry staining of Tim-3 and galectin-9
in representative cervical cancer samples. Magnification, ×400. (I)
Tim-3 and galectin-9 expression in HeLa and C33A cells detected by
western blot analysis. (J) Methylation level of
HAVCR2/LGALS9 promoter regions in cervical cancer (n=9) and
normal cervical tissues (n=9) detected by methylation-specific PCR
(MS-PCR). (K) Gray level analysis of HAVCR2/LGALS9
methylation level in cervical cancer and normal cervical tissues.
(M, methylated; U, unmethylated); methylated and unmethylated
levels were quantified as M/M+U% and U/M+U%, respectively.
*P<0.05; **P<0.01; ns, not significant. |
Further experiments revealed that the
HAVCR2/LGALS9 promoters in 9 cervical cancer tissues and 9
normal cervical tissues displayed a hypermethylated status in
normal cervical tissues compared with cervical cancer tissue
samples, possibly leading to the inhibition of gene expression
(Fig. 1J and K). Taken together,
these results suggest that Tim-3/galectin-9 expression is increased
in cervical cancer tissues.
Methylation status in the promoter
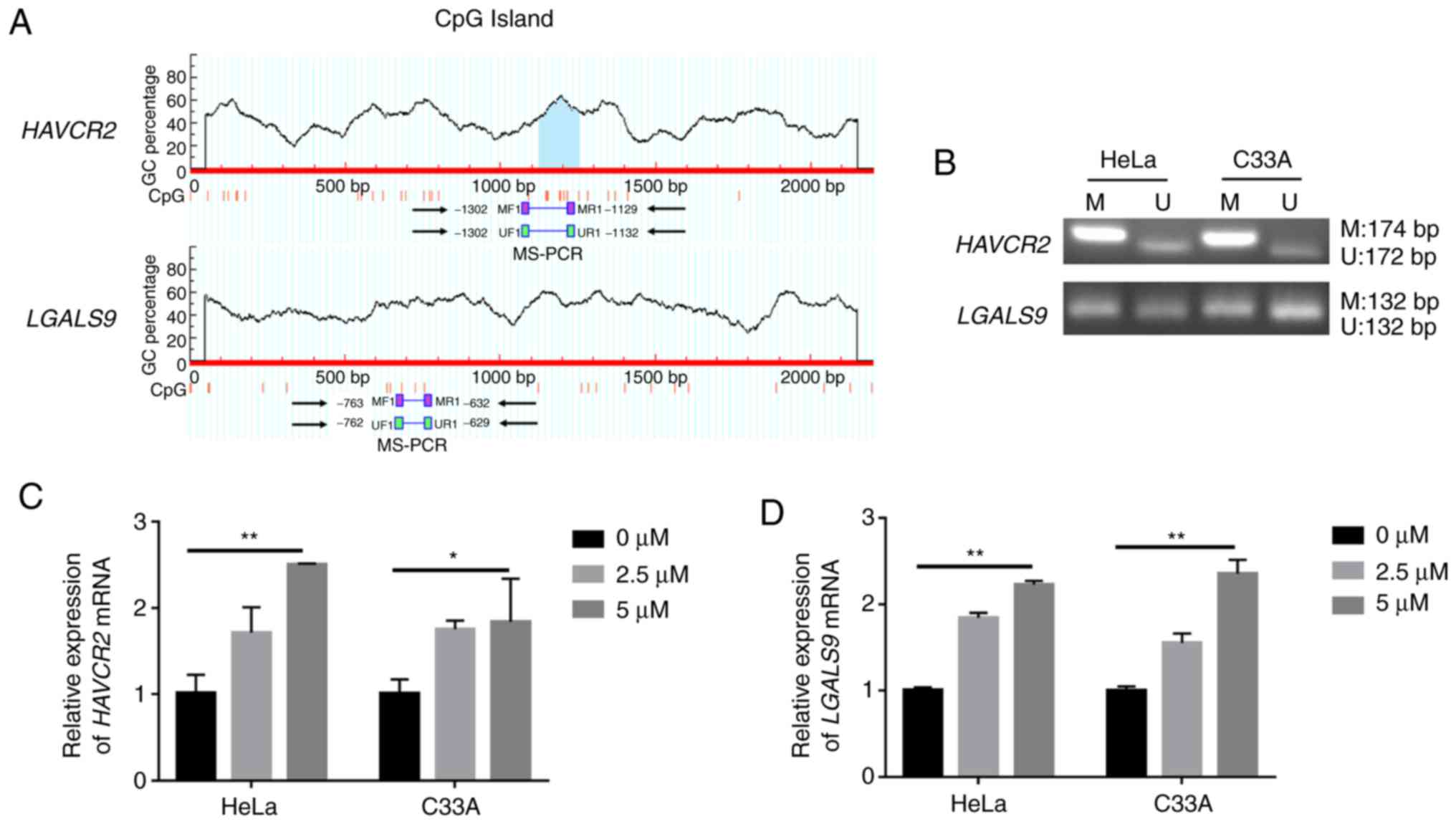
regions of HAVCR2/LGALS9 in cervical cancer cell lines
The online software, MethPrimer, profiled CpG
islands in the region that is located from −2,000 to −200 bp
upstream of ATG and the transcription starts site (TSS) in the
HAVCR2/LGALS9 promoters (Fig.
2A). One pair of primers was designed to amplify the
HAVCR2/LGALS9 promoter regions. MS-PCR analysis revealed
that these regions upstream of ATG were partially methylated in the
HeLa and C33A cells (Fig. 2B).
The mRNA expression level of HAVCR2/LGALS9 in
the HeLa and C33A cell lines following treatment with the
DNA-demethylating reagent, 5-Aza-CdR, was determined to identify
whether the methylation status in the promoter regions regulates
the expression of HAVCR2/LGALS9 genes at the transcriptional
level. The results suggested that the mRNA expression of
HAVCR2/LGALS9 in the HeLa and C33A cells increased in a
dose-dependent manner following cellular DNA demethylation
(Fig. 2C and D). These findings
illustrate that HAVCR2/LGALS9 expression was reversed by
5-Aza-CdR, which promoted the expression of the
HAVCR2/LGALS9 genes at the transcriptional level.
Methylation levels in the promoter
regions of HAVCR2/LGALS9 are regulated by DNMT3A in cervical cancer
cell lines
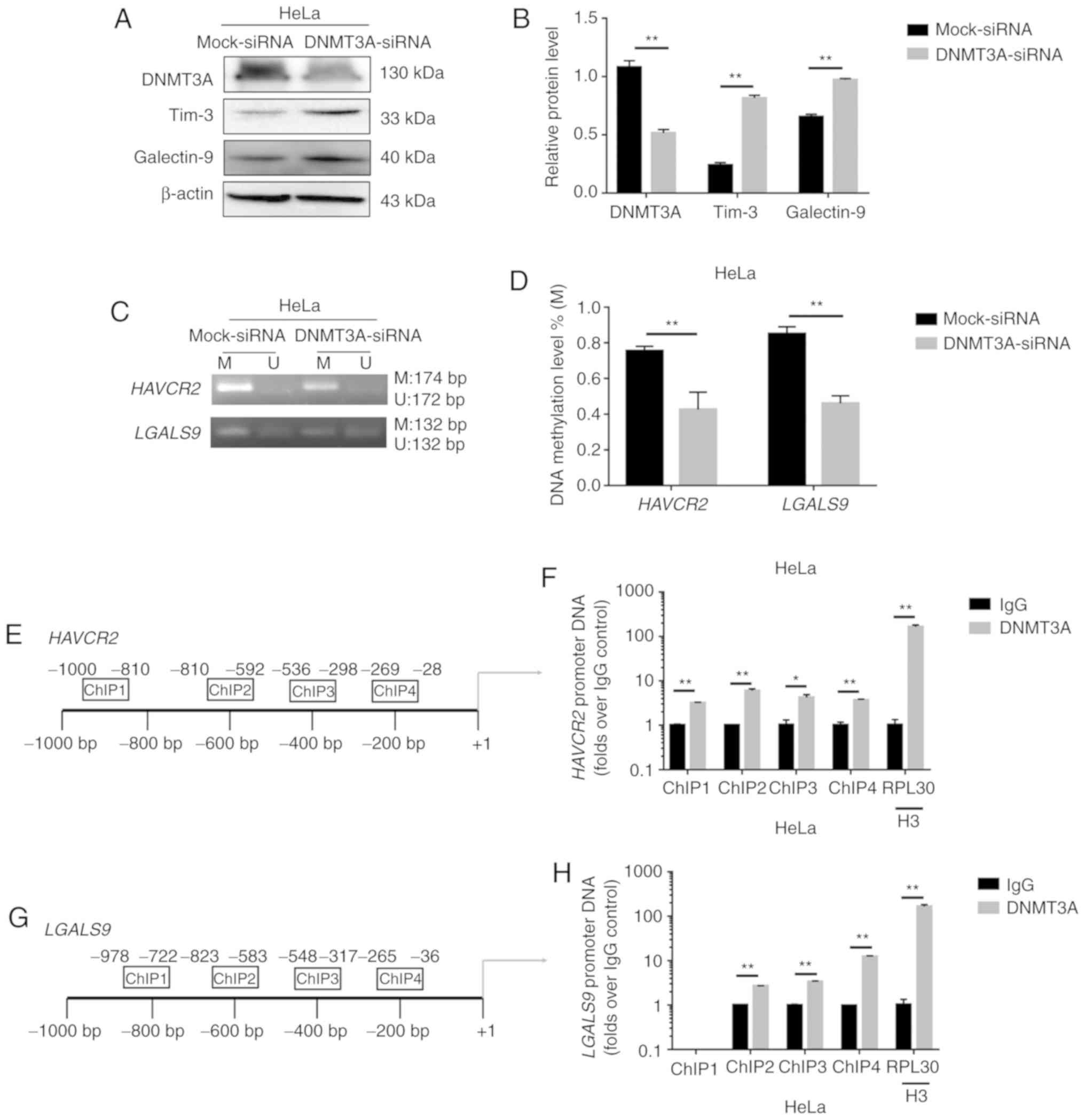
Following the knockdown of DNMT3A by siRNA, the
expression level of Tim-3/galectin-9 was increased (Fig. 3A and B), and in turn, the degree of
methylation of the genes encoding these proteins was decreased
(Fig. 3C and D). These results
suggest that DNMT3A plays an important role in regulating
Tim-3/galectin-9 expression. To confirm this causal association, we
performed ChIP analysis to evaluate the mechanisms underlying the
DNA methylation-mediated regulation of Tim-3/galectin-9. ChIP
analysis revealed the enhanced binding of DNMT3A (Fig. 3F and H) to the HAVCR2/LGALS9
promoters (Fig. 3E and G) in the HeLa
cells. Taken together, the results indicated that DNMT3A
downregulated expression of the negative costimulatory molecules
Tim-3/galectin-9 by upregulating the HAVCR2/LGALS9
methylation levels in cervical cancer.
EZH2 and H3K27me3 suppress DNMT3A
expression in HeLa cells
The protein expression of EZH2, H3K27me3 and DNMT3A
was detected by western blot analysis in 24 cervical cancer
tissues, 24 matched pericarcinomatous tissues and 16 normal
cervical tissues. The average relative expression levels of EZH2
and H3K27me3 were higher in the cervical cancer tissues than the
controls, and DNMT3A expression was lower in the cancer tissues
(Fig. 4A and B).
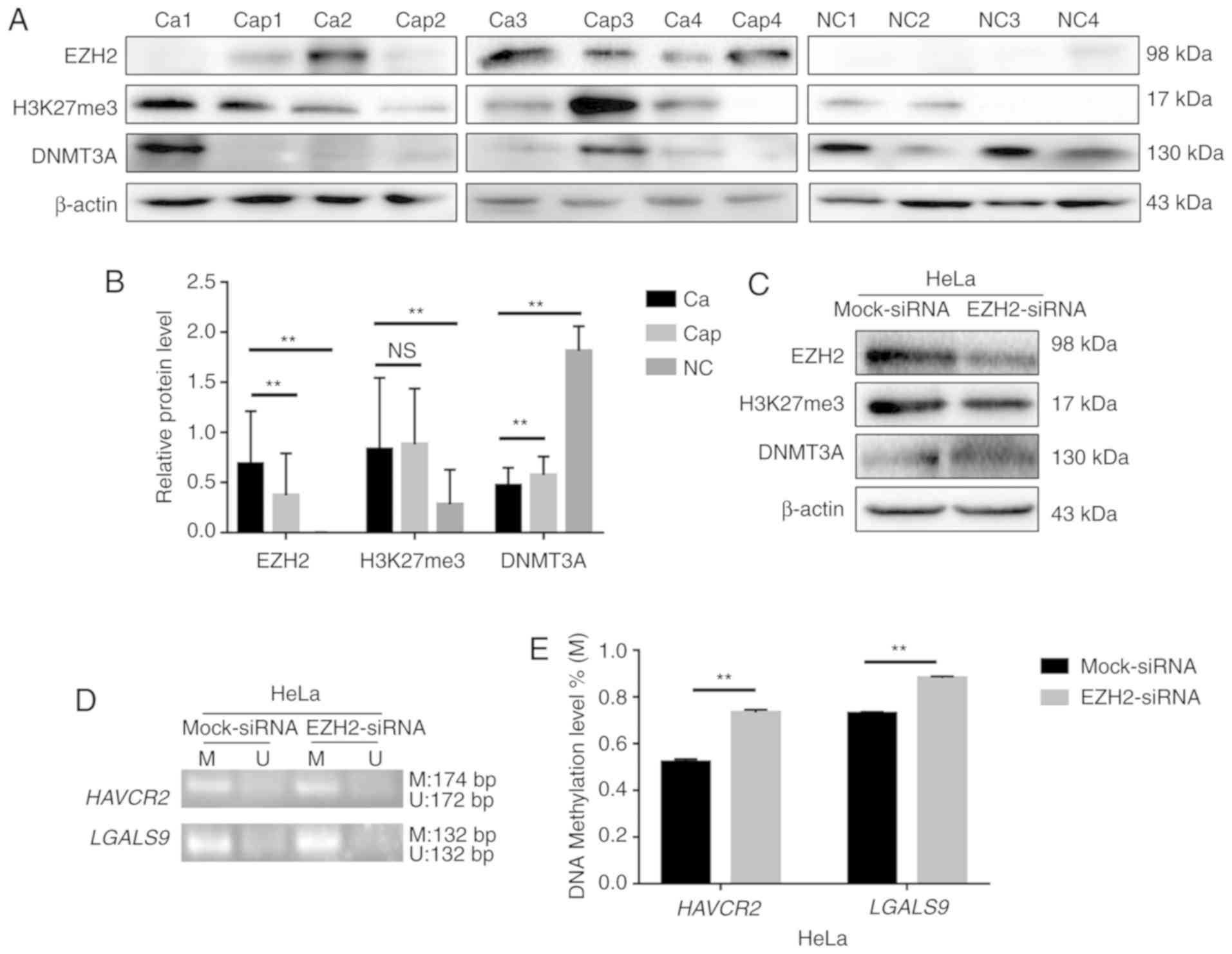 | Figure 4.EZH2 and H3K27me3 regulate
Tim-3/galectin-9 expression through DNMT3A. (A) The protein levels
of EZH2, H3K27me3 and DNMT3A in cervical cancer (Ca), paracarcinoma
(Cap) (n=24) and normal cervical tissues (NC) (n=16) detected by
western blot analysis. Blot images of 4 representative samples are
shown from each group. (B) Densitometric analysis of EZH2, H3K27me3
and DNMT3A protein levels in cervical cancer (n=24), paracarcinoma
(n=24) and normal cervical tissues (n=16). (C and F) Western blot
analysis of HeLa cells 48 h following transfection with siRNA
against EZH2. β-actin protein was used as a loading control between
lanes. (D) MS-PCR analysis was used to examine the methylation
level of HAVCR2/LGALS9. (E) Gray level analysis of
HAVCR2/LGALS9 methylation levels in HeLa cells in which EZH2
was knocked down (M, methylated; U, unmethylated); the methylated
and unmethylated levels were quantified as M/M+U% and U/M+U%,
respectively. (G) Schematic representation of the 4 regions of the
DNMT3A promoter amplified in the chromatin
immunoprecipitation (ChIP)-quantitative PCR (qPCR) experiment. (H
and I) Chromatin was cross-linked, fragmented and
immunoprecipitated with either IgG (mock) or anti-EZH2 and
anti-H3K27me3 ChIP-grade antibody and the purified DNA was used to
amplify with respective primer pairs for indicated four regions in
the DNMT3A promoters in qPCR. The enrichment of EZH2 and
H3K27me3 on DNMT3A promoter relative to IgG in HeLa cells,
and H3 against RPL30 was used as a positive control.
**P<0.01; ns, not significant. |
H3K27me3 expression was downregulated and DNMT3A
expression was upregulated when EZH2 was knocked down by siRNA in
the HeLa cells (Fig. 4C). Moreover,
the methylation levels of the HAVCR2/LGALS9 genes were
upregulated (Fig. 4D and E), and the
expression of Tim-3/galectin-9 was downregulated (Fig. 4F). H3K27me3 can thus regulate the
methylation level of HAVCR2/LGALS9 by altering the
expression of DNMT3A.
EZH2 targets H3K27me3 as part of polycomb repressive
complex 2 (PRC2), and its increased expression has been linked to a
number of malignancies, with the highest EZH2 protein levels being
associated with advanced disease and a poor prognosis (27). In this study, in a screen for
epigenetic mechanisms that regulate DNMT3A expression in HeLa
cells, ChIP analysis revealed that the HeLa cells exhibited the
highest EZH2 and H3K27me3 densities in a region upstream of the
transcription initiation region (Fig.
4G). EZH2 and H3K27me3 negatively regulated the expression of
DNMT3A by acting on the −1,000 to +1 region in the promoter of
DNMT3A (Fig. 4H and I). These
results suggest that EZH2 and H3K27me3 can alter the expression of
the negative costimulatory molecules Tim-3/galectin-9 by
synergistically regulating the expression of DNMT3A.
HPV18 oncoproteins E6/E7 can alter the
methylation and expression level of Tim-3/galectin-9 in cervical
cancer cells
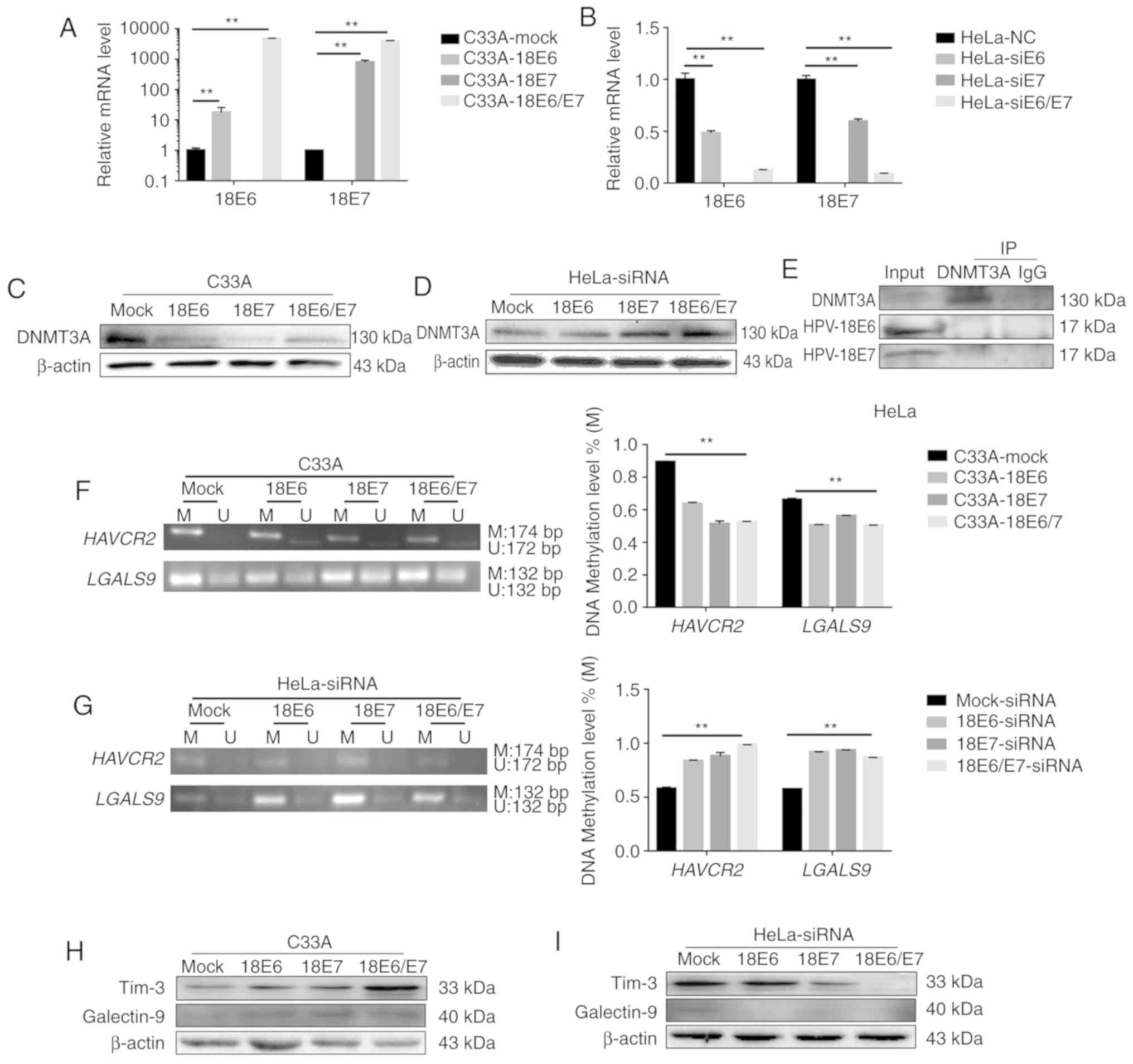
The mRNA expression of HPV18 E6 and E7 was increased
following the overexpression of HPV18 E6 and E7 (Fig. 5A). The protein expression of DNMT3A
was decreased when HPV18 E6 and/or E7 were overexpressed in the
C33A cells (Fig. 5C). At the same
time, the knockdown of E6 and/or E7 in the HeLa cells significantly
decreased HPV18 E6 and E7 expression (Fig. 5B). In addition, the knockdown of E6
and/or E7 in the HeLa cells increased DNMT3A protein expression
(Fig. 5D). These results revealed
that HPV18 E6 and/or E7 can lead to a reduction in DNMT3A
expression. Co-IP assay also revealed that neither HPV18 E6 nor E7
combined with DNMT3A directly (Fig.
5E).
Furthermore, the data revealed that HPV18
oncoproteins E6 and/or E7 downregulated the methylation level of
HAVCR2/LGALS9 significantly (Fig.
5F), and these altered methylation levels contributed to an
increased Tim-3/galectin-9 expression among the cell lines with the
overexpression of the HPV18 oncoproteins, E6 and/or E7, compared to
the C33A cells (Fig. 5H). The
knockdown of E6 and/or E7 yielded the opposite results in the HeLa
cells (Fig. 5G and I). These results
suggest that the HPV18 E6/E7 oncoproteins reduce the methylation
level of the HAVCR2/LGALS9 promoters and in turn promote
translation of the protein products.
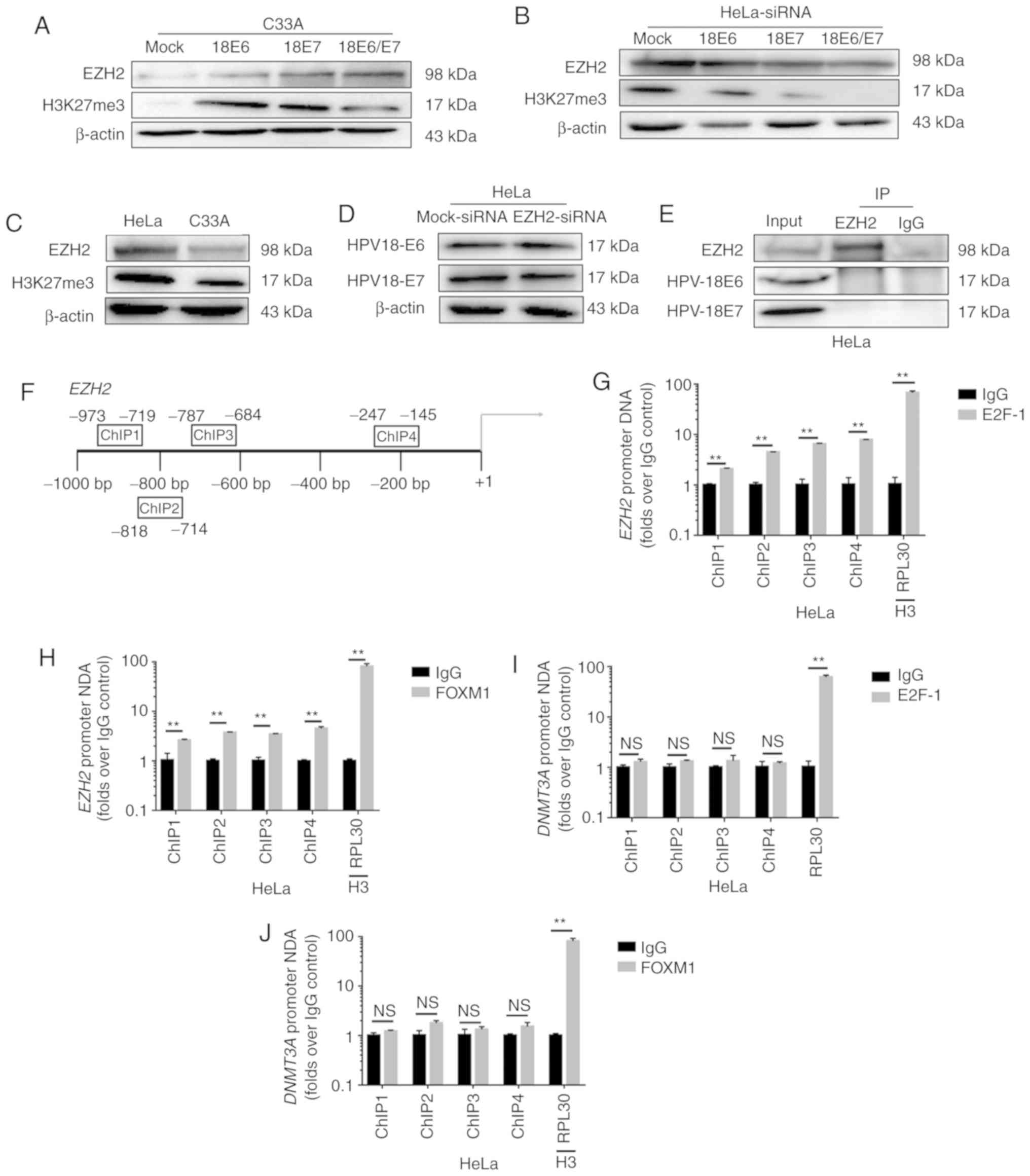
HPV18 oncoproteins E6/E7 participate
in the regulation of EZH2 and H3K27me3
EZH2 participates in H3K27me3-mediated gene
silencing (27,28). In this study, the EZH2 and H3K27me3
protein levels were strongly decreased when HPV18 oncoproteins were
knocked down in the HeLa cells, and the overexpression of HPV18 E6
or/and E7 led to an increase in EZH2 and H3K27me3 expression
(Fig. 6A and B). The expression of
EZH2 and H3K27me3 in the HeLa cells was higher than that in the
C33A cells (Fig. 6C). These results
suggest that EZH2 and H3K27me3 are novel activation targets of
HPV18 oncogenes.
However, the expression of HPV oncoproteins E6 or E7
was not altered when EZH2 was knocked down in the HeLa cells
(Fig. 6D). Co-IP assays revealed that
neither HPV18 E6 nor HPV18 E7 bound EZH2 directly (Fig. 6E). E6 oncoprotein induces FOXM1
expression via the MZF1/NKX2-1 axis in HPV-associated tumorigenesis
(29). The viral oncoprotein HPV E7
targets pRb, and the pRb/E2F-1 pathway directly regulates Kv10.1
transcription during G2/M (30–32). E2F-1
is enriched at proximal promoter of EZH2 in MCF-7 cells
(33). FOXM1 is enriched on the
promoter of EZH2 in HCT116 cells (34). In this study, in a screening for the
epigenetic mechanisms that regulated EZH2 expression in HeLa cells,
ChIP analysis revealed that the highest FOXM1 and E2F-1 density was
found in a region upstream of the transcription initiation region
in HeLa cells (Fig. 6F). It has also
been shown that FOXM1 and E2F-1 regulates the expression of EZH2 by
acting on the −1,000 to +1 region of the EZH2 promoter
(Fig. 6G and H). Moreover, ChIP
analysis did not reveal the enhanced binding of FOXM1 and E2F-1
(Fig. 6I and J) to the DNMT3A
promoters in the HeLa cells. Taken together, these findings suggest
that HPV18 E6 and E7 promote EZH2 and H3K27me3 expression through
the interaction of FOXM1 and E2F-1 with the promoter region of
EZH2, and inhibit DNMT3A expression.
Discussion
Epigenetic alterations, such as DNA methylation are
essential for the carcinogenesis of cervical cancer, which results
in the activation or exclusion of certain genes (35,36).
Cicchini et al (37) found
that HPV infection distinctly altered the methylation patterns in
HPV-associated cancer, and that HPV E7-dependent promoter
hypermethylation led to the downregulation of the chemokine CXCL14
and the suppression of antitumor immune responses. Epigenetically
regulated cytokines mediate the expression of tumor-associated
genes and manipulate their biological role in cancer (38–40).
The Tim-3/galectin-9 pathway, which plays a pivotal
role in immune regulation, is differentially regulated in a variety
of tumors and is a potential therapeutic target. Tim-3 and
galectin-9 have been found to be significantly overexpressed in
gastric cancer (41). Additionally,
high Tim-3 and low galectin-9 expression levels have been shown to
be associated with a poor prognosis of patients with esophageal
squamous cell carcinoma (42). A
higher expression level of Tim-3 has been shown to be positively
associated with a shorter progression-free survival of patients
with clear cell renal cell carcinoma (43). Patients with glioma with a high
expression of galectin-9 have been shown to exhibit a worse overall
survival (44).
In this study, the expression level of
Tim-3/galectin-9 was higher in human cervical cancer samples than
in normal cervical tissue. The high expression of LGALS9,
but not HAVCR2 was negatively associated with overall
survival. Thus, it can be considered that the increased expression
of secreted galectin-9 from cancer cells binds with Tim-3, causing
immunosuppression in the tumor microenvironment; this leads to a
poor overall survival in cervical cancer patients, suggesting that
Tim-3 and galectin-9 promote the occurrence and development of
cervical cancer. The levels of Tim-3 and galectin-9 were similar in
cancer and para-cancerous tissues; thus, the role of Tim-3 and
galectin-9 in paracancerous tissues requires further research,
which is a limitation of the present study.
We found that there was partial methylation in the
promoter regions of the HAVCR2/LGALS9 genes, which encode
negative costimulatory molecules Tim-3/galectin-9 in HeLa and C33A
cells and in cervical cancer tissues; these methylation patterns in
cancer cells could be reversed by 5-Aza-CdR treatment, leading to
an upregulated Tim-3/galectin-9 expression. Tim-3/galectin-9
expression was promoted and the methylation levels of the CpG
island in the HAVCR2/LGALS9 promoters were reduced when
DNMT3A was knocked down. ChIP analysis revealed that DNMT3A
directly bound to the HAVCR2/LGALS9 promoter regions in HeLa
cells.
EZH2 is an essential catalytic subunit of PRC2,
which silences gene expression by generating a methylated
epigenetic mark at H3K27me3 (27,45,46).
DNMT3A cooperates with EZH2, and H3K27me3 contributes to the
transcriptional regulation of gene expression (47–49). A
number of epigenetic alterations occur in cellular genomes during
HPV-associated carcinogenesis, and these alterations include
histone modifications (50–53). Some researchers have also demonstrated
that EZH2, H3K27me3 and DNMT1 cooperatively orchestrate epigenetic
modification of the wwc1 gene promoter in breast cancer
(54). In this study, we found that
EZH2 and H3K27me3 was overexpressed and DNMT3A was expressed at low
levels in cervical cancer tissues. Therefore, we hypothesized that
EZH2 and H3K27me3 promote DNMT3A expression in cervical cancer. The
downregulation of EZH2 inhibited H3K27me3 expression in HeLa cells
and was accompanied by increased DNMT3A protein levels,
demonstrating that DNMT3A is a downstream target gene of EZH2 and
H3K27me3. Both EZH2 and H3K27me3 were enriched at CpG loci within
the DNMT3A gene promoter in HeLa cells, as demonstrated by
ChIP assays.
The knockdown of EZH2 and H3K27me3 expression was
also associated with an increased HAVCR2/LGALS9 methylation,
which in turn caused the downregulation of Tim-3/galectin-9
expression. The EZH2-catalyzed trimethylation of H3K27 may be a
prerequisite for promoter DNA methylation by recruiting DNMT3A.
EZH2 and H3K27me3 synergistically regulated DNMT3A promoter
expression and cooperatively orchestrated the epigenetic
modification of the DNMT3A gene promoter.
HPV16 E6 and E7 can promote the secretion of soluble
Tim-3 in oropharyngeal squamous cell carcinoma, resulting in a poor
patient prognosis (55). Notably, in
this study, Tim-3/galectin-9 was expressed at higher levels in
HPV-positive cancer tissues than in HPV-negative tissues. Altering
the expression of HPV18 oncoproteins E6 and/or E7 also altered the
Tim-3/galectin-9 protein expression level and the methylation
status of their promoters. It has been suggested that HPV18 E6 and
E7 regulate the expression of Tim-3/galectin-9 in cervical cancer
by affecting the methylation level of the genes encoding these
proteins. HPV18 E6 and E7 induced the downregulation of DNMT3A
protein. The expression of Tim-3/galectin-9 was increased, and the
methylation level of the CpG islands in the HAVCR2/LGALS9
promoters was reduced when DNMT3A was knocked down. These results
indicate that the HPV18 oncoproteins regulate Tim-3/galectin-9
expression through DNMT3A. However, the co-IP assay found that
neither HPV18 E6 nor HPV18 E7 directly interacted with DNMT3A.
The results of this study illustrate that HPV18 E6
and/or E7 can elevate EZH2 expression in cervical cancer cells. In
addition, the expression of EZH2 and H3K27me3 proteins in cervical
cancer tissues was higher than that in normal tissues. However, the
co-IP assay found that neither HPV18 E6 nor HPV18 E7 directly
interacted with EZH2, and EZH2 knock down did not affect the
expression of HPV18 E6 and E7. E6 oncoprotein can induce the
expression of FOXM1 through the MZF1/NKX2-1 axis (29). HPV mediates EZH2 expression through
the transcriptional activation of the EZH2 promoter via
E7-mediated release of E2F factors from inhibitory pocket proteins
(56). The most well-studied cellular
target of the viral oncogene E7 is pRb, and E2F-1 is the main
target that is regulated by pRb. HPV E7 oncoprotein also leads to
increased gene expression through the direct binding of the E2F-1
transcription factor to the gene promoter (30,57–59). EZH2
alterations in HPV16 E6/E7 HFKs lead to a loss of H3K27me3 and to
the transcriptional depression of H3K27me3-targeted HOX genes
(51). In this study, we used a ChIP
assay to illustrate that the transcription factors FOXM1 and E2F-1
can directly bind to the promoter of EZH2, but do not
directly bind to the promoter of DNMT3A. Therefore, we
assert the preliminary conclusion that EZH2 and H3K27me3 expression
is regulated by HPV18 E6 and E7 through the transcription factors
FOXM1 and E2F-1.
Our data reveal a key role for EZH2-H3K27me3-DNMT3A
in regulating the costimulatory molecules Tim-3/galectin-9 in
cervical cancer that involves the HPV18 oncoproteins E6/E7
(Fig. 7). EZH2 and H3K27me3 may
represent therapeutic targets, and an epigenetic agent or inhibitor
aimed at these proteins could decrease Tim-3/galectin-9 expression.
The inhibition of the expression of EZH2 and H3K27me3 may be an
effective approach with which to augment the efficacy of negative
costimulatory molecules against cervical cancer. DNMT1 and DNMT3B
are also associated with the methylation of genes. In this study,
we focused on the role of DNMT3A in the regulation of Tim-3 and
galectin-9, which is another limitation of this study; therefore,
in the future, we aim to study the role of DNMT1 and DNMT3B in the
regulation of Tim-3 and galectin-9 expression.
Acknowledgements
The authors would like to thank all the teachers at
the Center for Translational Medicine of the First Affiliated
Hospital of Xi'an Jiaotong University for providing technical
assistance.
Funding
This research was supported by the National Natural
Science Foundation of China (no. 81472428), a Fundamental Research
Funds for the Central Universities and Nutrition Asia Research
Grant by BASF and National Natural Science Foundation of China
(grant nos. 81672350, 81872225).
Availability of data and materials
The dataset analyzed during the current study are
publicly available from the online database: GEPIA database
(http://gepia.cancer-pku.cn/).
Authors' contributions
LZ, ST and YJ conducted the experiments. LZ, LS, MZ,
LW, JZ and XY participated in the data analysis. LZ, LS, TY and XY
designed the experiments. MZ, LW, MP, YJ, TY and JZ collected the
samples from the cervical cancer patients. LZ, ST, TY, JZ and XY
wrote and edited the manuscript. All authors have read and approved
the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
the First Affiliated Hospital of Xi'an Jiaotong University (G-272)
in Shaanxi, China. Written informed consent was obtained from all
patients to participate in this study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Senkomago V, Duran D, Loharikar A, Hyde
TB, Markowitz LE, Unger ER and Saraiya M: CDC activities for
improving implementation of human papillomavirus vaccination,
cervical cancer screening, and surveillance worldwide. Emerg Infect
Dis. 23:2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fader AN: Surgery in cervical cancer. N
Engl J Med. 379:1955–1957. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jiang X, Tang H and Chen T: Epidemiology
of gynecologic cancers in China. J Gynecol Oncol. 29:e72018.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tsu V and Jeronimo J: Saving the World's
Women from Cervical Cancer. The New England J Med. 374:2509–2511.
2016. View Article : Google Scholar
|
|
5
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Schlecht NF, Kulaga S, Robitaille J,
Ferreira S, Santos M, Miyamura RA, Duarte-Franco E, Rohan TE,
Ferenczy A, Villa LL and Franco EL: Persistent human papillomavirus
infection as a predictor of cervical intraepithelial neoplasia.
JAMA. 286:3106–3114. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Saraiya M, Unger ER, Thompson TD, Lynch
CF, Hernandez BY, Lyu CW, Steinau M, Watson M, Wilkinson EJ,
Hopenhayn C, et al: US assessment of HPV types in cancers:
Implications for current and 9-valent HPV vaccines. J Natl Cancer
Inst. 107:djv0862015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kahn JA: HPV vaccination for the
prevention of cervical intraepithelial neoplasia. N Engl J Med.
361:271–278. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Baumeister SH, Freeman GJ, Dranoff G and
Sharpe AH: Coinhibitory pathways in immunotherapy for cancer. Annu
Rev Immunol. 34:539–573. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Topalian SL, Taube JM, Anders RA and
Pardoll DM: Mechanism-driven biomarkers to guide immune checkpoint
blockade in cancer therapy. Nat Rev Cancer. 16:275–287. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu F, Liu Y and Chen Z: Tim-3 expression
and its role in hepatocellular carcinoma. J Hematol Oncol.
11:1262018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
de Mingo Pulido A, Gardner A, Hiebler S,
Soliman H, Rugo HS, Krummel MF, Coussens LM and Ruffell B: TIM-3
regulates CD103(+) dendritic cell function and response to
chemotherapy in breast cancer. Cancer Cell. 33:60–74.e6. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
He Y, Cao J, Zhao C, Li X, Zhou C and
Hirsch FR: TIM-3, a promising target for cancer immunotherapy. Onco
Targets Ther. 11:7005–7009. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li X, Hu W, Zheng X, Zhang C, Du P, Zheng
Z, Yang Y, Wu J, Ji M, Jiang J and Wu C: Emerging immune
checkpoints for cancer therapy. Acta Oncol. 54:1706–1713. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Moriyama K, Kukita A, Li YJ, Uehara N,
Zhang JQ, Takahashi I and Kukita T: Regulation of
osteoclastogenesis through Tim-3: Possible involvement of the
Tim-3/galectin-9 system in the modulation of inflammatory bone
destruction. Lab Invest. 94:1200–1211. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chou FC, Kuo CC, Chen HY, Chen HH and
Sytwu HK: DNA demethylation of the TIM-3 promoter is critical for
its stable expression on T cells. Genes Immun. 17:179–186. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sasidharan Nair V, El Salhat H, Taha RZ,
John A, Ali BR and Elkord E: DNA methylation and repressive H3K9
and H3K27 trimethylation in the promoter regions of PD-1, CTLA-4,
TIM-3, LAG-3, TIGIT, and PD-L1 genes in human primary breast
cancer. Clin Epigenetics. 10:782018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cao Y, Zhou X, Huang X, Li Q, Gao L, Jiang
L, Huang M and Zhou J: Tim-3 expression in cervical cancer promotes
tumor metastasis. PLoS One. 8:e538342013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liang M, Ueno M, Oomizu S, Arikawa T,
Shinonaga R, Zhang S, Yamauchi A and Hirashima M: Galectin-9
expression links to malignant potential of cervical squamous cell
carcinoma. J Cancer Res Clin Oncol. 134:899–907. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Azizmohammadi S, Azizmohammadi S, Safari
A, Kaghazian M, Sadrkhanlo M, Behnod V and Seifoleslami M:
High-level expression of RIPK4 and EZH2 contributes to lymph node
metastasis and predicts favorable prognosis in patients with
cervical cancer. Oncol Res. 25:495–501. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sun J, Ji J, Huo G, Song Q and Zhang X:
miR-182 induces cervical cancer cell apoptosis through inhibiting
the expression of DNMT3a. Int J Clin Exp Pathol. 8:4755–4763.
2015.PubMed/NCBI
|
|
22
|
Cao D, Zhang S, Zhang Q, Wei X, Zhao M, Ma
Q, Li Y, Wang L, Pei M, Yang T, et al: Prevalence of high-risk
human papillomavirus infection among women in Shaanxi province of
China: A hospital-based investigation. J Med Virol. 89:1281–1286.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wei X, Zhang S, Cao D, Zhao M, Zhang Q,
Zhao J, Yang T, Pei M, Wang L, Li Y and Yang X: Aberrant
hypermethylation of SALL3 with HPV involvement contributes to the
carcinogenesis of cervical cancer. PLoS One. 10:e01457002015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhao M, Li Y, Wei X, Zhang Q, Jia H, Quan
S, Cao D, Wang L, Yang T, Zhao J, et al: Negative immune factors
might predominate local tumor immune status and promote
carcinogenesis in cervical carcinoma. Virol J. 14:52017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Simon JA and Lange CA: Roles of the EZH2
histone methyltransferase in cancer epigenetics. Mutat Res.
647:21–29. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fu Y, Chen J, Pang B, Li C, Zhao J and
Shen K: EZH2-induced H3K27me3 is associated with epigenetic
repression of the ARHI tumor-suppressor gene in ovarian cancer.
Cell Biochem Biophys. 71:105–112. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ler LD, Ghosh S, Chai X, Thike AA, Heng
HL, Siew EY, Dey S, Koh LK, Lim JQ, Lim WK, et al: Loss of tumor
suppressor KDM6A amplifies PRC2-regulated transcriptional
repression in bladder cancer and can be targeted through inhibition
of EZH2. Sci Transl Med. 9:eaai83122017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen PM, Cheng YW, Wang YC, Wu TC, Chen CY
and Lee H: Up-regulation of FOXM1 by E6 oncoprotein through the
MZF1/NKX2-1 axis is required for human papillomavirus- associated
tumorigenesis. Neoplasia. 16:961–971. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Urrego D, Movsisyan N, Ufartes R and Pardo
LA: Periodic expression of Kv10.1 driven by pRb/E2F1 contributes to
G2/M progression of cancer and non-transformed cells. Cell Cycle.
15:799–811. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Finzer P, Krueger A, Stohr M, Brenner D,
Soto U, Kuntzen C, Krammer PH and Rösl F: HDAC inhibitors trigger
apoptosis in HPV-positive cells by inducing the E2F-p73 pathway.
Oncogene. 23:4807–4817. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
McCance DJ: Human papillomaviruses and
cell signaling. Science's STKE. 2005:pe292005.
|
|
33
|
Ghosh K, Chatterjee B, Maheswari U, Athifa
M and Kanade SR: 4-Nonylphenol-enhanced EZH2 and RNF2 expression,
H3K27me3 and H2AK119ub1 marks resulting in silencing of p21(CDKN1A)
in vitro. Epigenomics. 11:899–916. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang Y, Wu M, Lei Z, Huang M, Li Z, Wang
L, Wang L, Cao Q, Han D, Chang Y, et al: Dysregulation of
miR-6868-5p/FOXM1 circuit contributes to colorectal cancer
angiogenesis. J Exp Clin Cancer Res. 37:2922018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mersakova S, Nachajova M, Szepe P,
Kasajova PS and Halasova E: DNA methylation and detection of
cervical cancer and precancerous lesions using molecular methods.
Tumor Biol. 37:23–27. 2016. View Article : Google Scholar
|
|
36
|
Steenbergen RD, Snijders PJ, Heideman DA
and Meijer CJ: Clinical implications of (epi)genetic changes in
HPV-induced cervical precancerous lesions. Nat Rev Cancer.
14:395–405. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cicchini L, Blumhagen RZ, Westrich JA,
Myers ME, Warren CJ, Siska C, Raben D, Kechris KJ and Pyeon D:
High-risk human papillomavirus E7 alters Host DNA methylome and
represses HLA-E expression in human keratinocytes. Sci Rep.
7:36332017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yasmin R, Siraj S, Hassan A, Khan AR,
Abbasi R and Ahmad N: Epigenetic regulation of inflammatory
cytokines and associated genes in human malignancies. Mediators
Inflamm. 2015:2017032015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Micevic G, Thakral D, McGeary M and
Bosenberg MW: PD-L1 methylation regulates PD-L1 expression and is
associated with melanoma survival. Pigment Cell Melanoma Res.
32:435–440. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang T, Wu J, Ungvijanpunya N,
Jackson-Weaver O, Gou Y, Feng J, Ho TV, Shen Y, Liu J, Richard S,
et al: Smad6 Methylation Represses NFκB activation and periodontal
inflammation. J Dent Res. 97:810–819. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang Y, Zhao E, Zhang Z, Zhao G and Cao H:
Association between Tim3 and Gal9 expression and gastric cancer
prognosis. Oncol Rep. 40:2115–2126. 2018.PubMed/NCBI
|
|
42
|
Hou N, Ma J, Li W, Zhao L, Gao Q and Mai
L: T-cell immunoglobulin and mucin domain-containing protein-3 and
galectin-9 protein expression: Potential prognostic significance in
esophageal squamous cell carcinoma for Chinese patients. Oncol
Lett. 14:8007–8013. 2017.PubMed/NCBI
|
|
43
|
Komohara Y, Morita T, Annan DA, Horlad H,
Ohnishi K, Yamada S, Nakayama T, Kitada S, Suzu S, Kinoshita I, et
al: The Coordinated Actions of TIM-3 on cancer and myeloid cells in
the regulation of tumorigenicity and clinical prognosis in clear
cell renal cell carcinomas. Cancer Immunol Res. 3:999–1007. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Liang T, Wang X, Wang F, Feng E and You G:
Galectin-9: A predictive biomarker negatively regulating immune
response in glioma patients. World Neurosurg. Aug 27–2019.(Epub
ahead of print). doi: 10.1016/j.wneu.2019.08.117. View Article : Google Scholar
|
|
45
|
Zhou T, Sun Y, Li M, Ding Y, Yin R, Li Z,
Xie Q, Bao S and Cai W: Enhancer of zeste homolog 2-catalysed H3K27
trimethylation plays a key role in acute-on-chronic liver failure
via TNF-mediated pathway. Cell Death Dis. 9:5902018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Sharma V, Malgulwar PB, Purkait S, Patil
V, Pathak P, Agrawal R, Kulshreshtha R, Mallick S, Julka PK, Suri
A, et al: Genome-wide ChIP-seq analysis of EZH2-mediated H3K27me3
target gene profile highlights differences between low- and
high-grade astrocytic tumors. Carcinogenesis. 38:152–161.
2017.PubMed/NCBI
|
|
47
|
Cui H, Hu Y, Guo D, Zhang A, Gu Y, Zhang
S, Zhao C, Gong P, Shen X, Li Y, et al: DNA methyltransferase 3A
isoform b contributes to repressing E-cadherin through cooperation
of DNA methylation and H3K27/H3K9 methylation in EMT-related
metastasis of gastric cancer. Oncogene. 37:4358–4371. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Mochizuki D, Misawa Y, Kawasaki H, Imai A,
Endo S, Mima M, Yamada S, Nakagawa T, Kanazawa T and Misawa K:
Aberrant epigenetic regulation in head and neck cancer due to
distinct EZH2 overexpression and DNA hypermethylation. Int J Mol
Sci. 19:E37072018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Manzo M, Wirz J, Ambrosi C, Villasenor R,
Roschitzki B and Baubec T: Isoform-specific localization of DNMT3A
regulates DNA methylation fidelity at bivalent CpG islands. EMBO J.
36:3421–3434. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Hyland PL, Mcdade SS, Mccloskey R, Dickson
GJ, Arthur K, McCance DJ and Patel D: Evidence for alteration of
EZH2, BMI1, and KDM6A and epigenetic reprogramming in human
papillomavirus type 16 E6/E7-expressing keratinocytes. J Virol.
85:10999–11006. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Lindsay CD, Kostiuk MA, Harris J,
O'Connell DA, Seikaly H and Biron VL: Efficacy of EZH2 inhibitory
drugs in human papillomavirus-positive and human
papillomavirus-negative oropharyngeal squamous cell carcinomas.
Clin Epigenetics. 9:952017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Biron VL, Mohamed A, Hendzel MJ, Alan
Underhill D and Seikaly H: Epigenetic differences between human
papillomavirus-positive and -negative oropharyngeal squamous cell
carcinomas. J Otolaryngol Head Neck Surg. 41 (Suppl 1):S65–S70.
2012.PubMed/NCBI
|
|
53
|
Leonard S, Pereira M, Fox R, Gordon N, Yap
J, Kehoe S, Luesley D, Woodman C and Ganesan R: Over-expression of
DNMT3A predicts the risk of recurrent vulvar squamous cell
carcinomas. Gynecol Oncol. 143:414–420. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Liu X, Li C, Zhang R, Xiao W, Niu X, Ye X,
Li Z, Guo Y, Tan J and Li Y: The EZH2- H3K27me3-DNMT1 complex
orchestrates epigenetic silencing of the wwc1 gene, a Hippo/YAP
pathway upstream effector, in breast cancer epithelial cells. Cell
Signal. 51:243–256. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Hladikova K, Partlova S, Koucky V, Boucek
J, Fonteneau JF, Zabrodsky M, Tachezy R, Grega M, Špíšek R and
Fialová A: Dysfunction of HPV16-specific CD8+ T cells
derived from oropharyngeal tumors is related to the expression of
Tim-3 but not PD-1. Oral Oncol. 82:75–82. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Holland D, Hoppe-Seyler K, Schuller B,
Lohrey C, Maroldt J, Dürst M and Hoppe-Seyler F: Activation of the
enhancer of zeste homologue 2 gene by the human papillomavirus E7
oncoprotein. Cancer Res. 68:9964–9972. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Yeo-Teh NSL, Ito Y and Jha S: High-risk
human papillomaviral oncogenes E6 and E7 target key cellular
pathways to achieve oncogenesis. Int J Mol Sci. 19:E17062018.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Vuillier C, Lohard S, Fetiveau A, Allegre
J, Kayaci C, King LE, Braun F, Barillé-Nion S, Gautier F, Dubrez L,
et al: E2F1 interacts with BCL-xL and regulates its subcellular
localization dynamics to trigger cell death. EMBO Rep. 19:234–243.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Youn HS, Kim TY, Park UH, Moon ST, An SJ,
Lee YK, Hwang JT, Kim EJ and Um SJ: Asxl1 deficiency in embryonic
fibroblasts leads to cellular senescence via impairment of the
AKT-E2F pathway and Ezh2 inactivation. Sci Rep. 7:51982017.
View Article : Google Scholar : PubMed/NCBI
|