Introduction
Head and neck squamous cell carcinoma (HNSCC),
arising in the oral cavity as well as in the pharyngeal and
laryngeal regions of the neck, is the sixth most commonly diagnosed
cancer worldwide. In general, complete cure of HNSCC is less than
50% (1). Despite advanced treatment
strategies, the outcome of head and neck cancer has not
significantly improved to date (2).
For this reason, there is still a demand for new substances
suppressing tumor growth and invasion.
Chelidonium majus, the greater celandine, has
been historically used to fight cancer and other diseases. It was
described in detail in Jonathan Hartwell's compendium (3) ‘Plants used against Cancer’. The greater
celandine gained overwhelming interest when Ukrain, a drug derived
from C. majus, was selected for use in cancer treatment
during the 1990s. The drug is, according to the manufacturer, a
semi-synthetic derivative of the purified alkaloid chelidonine
modified with thiophosphoric acid (Thiotepa). Moreover, it has been
proposed to kill tumor cells selectively without damaging primary
cells and to exhibit an immune modulatory effect (4–6). There are
many studies reporting the treatment of different cancers using
Ukrain. During the 1990s, a ‘hype’ about Ukrain arose and numerous
case reports appeared in the no longer published journal ‘Drugs
Under Experimental and Clinical Research’. Studies reported the
benefits of Ukrain such as a prolonged life span or even complete
remission, without having adverse side effects (7–9). Ukrain
was shown to be cytotoxic for a variety of tumor cell lines in
vitro (10–13). Furthermore, several groups proved the
induction of apoptosis by Ukrain in diverse tumor systems in
vitro (10,14).
As it has been shown that chelidonine is not very
effective in HNSCC cell lines (15)
and there is still a lack of studies of other C. majus
alkaloids as possible therapeutic agents in HNSCC, the aqueous
extract NSC-631570 was chosen to be tested in vitro for its
action in HNSCC.
Combined liquid chromatography and mass spectroscopy
of NSC-631570 was applied. The effects of NSC-631570 and its major
alkaloid chelerythrine on drug-sensitive and drug-resistant HNSCC
cell lines and on primary mucosal keratinocytes and fibroblasts
were investigated in the present study. Apoptosis as well as the
influence of the extract on gene expression and on the motility of
HNSCC cells in a 3-dimensional, spheroid-based invasion assay were
analyzed. The results are discussed critically with respect to
previously published research.
Materials and methods
Reagents
NSC-631570 was kindly provided by Dr Wassil Novicky
(Vienna, Austria), who is the inventor of Ukrain. Reference
substances chelidonine, allocryptopine, chelerythrine and
sanguinarine were purchased from Sigma-Aldrich; Merck KGaA.
Liquid chromatography-mass
spectroscopy of NSC-631570
LC-MS analysis was performed using a Shimadzu
LC-MS-2020 mass spectrometer (Shimadzu Deutschland GmbH) containing
a DGU-20A3R degassing unit, a LC20AB liquid chromatograph and
SPD-20A UV/Vis detector. A Synergi 4 U fusion-RP column (150×4.6
mm; Phenomonex) was used as a stationary phase, and a gradient of
MeOH/water was applied as a mobile phase: Solvent A: water with
0.1% formic acid, solvent B: MeOH with 0.1% formic acid. Solvent A
ranged from 0 to 100% in 8 min, and then remained at 100% for 5
min, before reducing from 100 to 5% in 1 min, and then being held
5% for 4 min, The method was performed with a flow rate of 1.0
ml/min, and UV detection was measured at 245 nm. Allocryptopine,
chelerythrine, sanguinarine and chelidonine were used as reference
substances.
Cell lines and cell culture
Squamous cell carcinoma cell lines, originating from
laryngeal or hypopharyngeal tumors were used for the study,
comprising the major proportion of cases treated at the ENT,
Wuerzburg University Hospital. The cell line FaDu (LG standards)
originating from a hypopharyngeal carcinoma was grown in RPMI-1640
medium (Seromed), supplemented with 10% foetal bovine serum (FBS).
HLaC78 and HLaC79 cell lines derived from larynx carcinoma
(16) were maintained in RPMI-1640
medium. HLaC79-Tax was obtained by isolation and the selective
cultivation of a paclitaxel-resistant HLaC79 clone. It was grown as
the original cell line supplemented with 10 nM (HLaC79-Tax)
paclitaxel. As primary cells, human mucosal fibroblasts and
keratinocytes were used. Fibroblast and keratinocyte cultures used,
were thawed from frozen samples, generated from a tonsil surgery
specimen in 2007. Fibroblast cultures were established by explant
cultures in DMEM/10% FBS. The isolation of keratinocytes was
performed as previously described (17). Keratinocytes were maintained in
Keratinocyte Medium 2 (PromoCell). Patient informed written consent
was obtained prior to the study. The present study was approved by
the Institutional Ethics Committee on Human Research of The Julius
Maximilian University of Wuerzburg (study approval no. 16/06).
Cell viability and proliferation
assay
Cells were seeded at 5,000 cells/well in 96-well
plates. The cells were then treated with increasing concentrations
of NSC-631570, chelerythrine or allocryptopine for 48 h. Cell
proliferation was measured 48 h later by administration of MTT at a
concentration of 1 mg/ml. After a 4-h incubation, MTT-staining
solution was replaced by isopropanol and the cells were incubated
at 37°C for 45 min. The color change of yellow MTT to a blue
formazan dye was measured with an ELISA reader at a wavelength of
570 nm. Relative toxicity was calculated as the surviving cell % by
setting solvent-treated control cells to 100%.
Apoptosis
FACS analysis was performed using the BD Pharmingen
Annexin V-APC kit (BD Biosciences) according to the kit manual. In
brief, HLaC78 and FaDu cells were treated with EC50
concentrations of NSC-631570 or chelerythrine, respectively for 24
h, harvested and washed twice with cold PBS. The shortened
incubation time was chosen in order to enrich early apoptotic
stages within the cell populations (18). Cells were then resuspended in 1X
binding buffer (0.1 M HEPES, pH 7.4, 1.4 M NaCl, 25 mM
CaCl2) at a concentration of 1×106 cells/ml.
To 100 µl of this cell suspension, 5 µl Annexin V-APC and 5 µl
7-AAD (included in the kit) were added, the cells were vortexed and
incubated for 15 min in the dark. An amount of 400 µl of 1X binding
buffer was added. Within 1 h, FACS analysis was performed at an
excitation wavelength of 650 nm.
In vitro invasion assay
Tumor spheroids were established by dispending 5,000
cells/well of HLaC78 cells on ultra-low-attachment (ULA) 96-well
round-bottomed plates (Corning, Inc.). For the migration assay,
spheroids of HLaC78 were transferred manually to different
extracellular matrix substrates. For this reason, the surface of
flat-bottomed 96-well plates was coated with 0.1% gelatine, 5 µg/ml
fibronectin, 50 µg/ml laminin, 50 µg/ml collagen I (all purchased
at Sigma Aldrich; Merck KGaA) or 125 µg/ml Matrigel®
(Becton Dickinson) for 2 h at room temperature. Wells were washed
twice with PBS and blocked with 1% bovine serum albumin in PBS for
1 h; 72-h old spheroids were then manually transferred to the
coated wells. Spheroids were incubated with or without NSC-631570
or chelerythrine. Migration was analyzed by photographing spheroids
after 1 and 24 h with a Leica DMI 4000 inverted fluorescence
microscope (Leica Microsystems) at 5-fold magnification. Incubation
time was chosen due to the fact, that the cell line HLaC78 needs
approximately 24 h to invade completely without inhibiting
substances (personal observation). Quantification of migrated areas
was performed with ImageJ software (version 1.52a; National
Institutes of Health, NIH, Bethesda, MD, USA).
RNA extraction and RNA quality
control
RNA of HNSCC cell cultures was isolated with the
RNeasy kit (Qiagen) according to the manufacturer's instructions.
Purity and concentration were determined photometrically. In
expression arrays, RNA quality was determined with the RNA 6000
Nano kit and a Bioanalyzer 2100 instrument (Agilent). RNA integrity
numbers (RINs) of our samples ranged between 9.4 and 9.7.
Microarray analysis
For microarray hybridization, 100 ng total RNA was
amplified and labelled using the IVT Express kit and hybridized to
GeneChip PrimeView Human Gene Expression arrays (both from
Affymetrix; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. Raw microarray data were background
corrected, normalized and summarized to probe set expression values
using the Robust Microarray Average (RMA) algorithm (19,20). Data
pre-processing and calculation of fold-changes between treated and
untreated expression values was performed with the Affymetrix
Transcriptome analysis Console 4.0.1 (Affymetrix; Thermo Fisher
Scientific, Inc.). Microarray data were deposited in
MIAME-compliant form at Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) with the
identifier GSE115874.
TaqMan real-time PCR
In order to confirm the strong increase in
CYP1A1 expression caused by NSC-631570 in FADU cells and
other HNSCC cell lines, FaDu and HLaC78 cell lines were treated for
48 h with either chelerythrine, allocryptopine or NSC-631570 at
their EC50 concentrations. RNA was isolated (see above
section) and real-time TaqMan PCR (Applied Biosystems; Thermo
Fisher Scientific, Inc.) was performed in triplicates on a
real-time PCR cycler (Applied Biosystems; Thermo Fisher Scientific,
Inc.) using the TaqMan gene expression assay for CYP1A1.
Relative quantification was calculated according to the
2−ΔΔCq method (21).
Expression values were normalized to the expression of GAPDH as an
endogenous control, stably expressed in HNSCC cell lines.
Tube formation assay
Using tube formation assays, the ability of
endothelial cells to form three-dimensional capillary-like
structures was analyzed. Ibidi angiogenesis slides (15-well, Ibidi
GmbH) were coated with growth factor reduced basement membrane
extract (BME; Trevigen). BME gels were overlaid with
1×104 human umbilical vein endothelial cells (HUVECs) in
growth medium, with and without the addition of NSC-631570 or
chelerythrine, respectively. Cells were incubated for 6 h and
images were captured. Analysis of the images was performed by
Wimasis. For quantification, four parameters were analyzed: tube
length, number of branching points, covered area and number of
loops.
Statistical analysis
All statistical analyses and graphs were performed
with Graph Pad Prism 6 (GraphPad Software, La Jolla, CA, USA). Data
are presented as the mean of three independent experiments or 8
measured spheroids ± standard deviation. As statistical tests
unpaired t-test (angiogenesis, migration measurements) or ANOVA
Dunnett's multiple comparison test (RT-qPCR) were used. Differences
were considered to be significant as indicated in the figures and
legends: ****P<0.0001, ***P<0.001, **P<0.01,
*P<0.05.
Results
LC-MS analysis
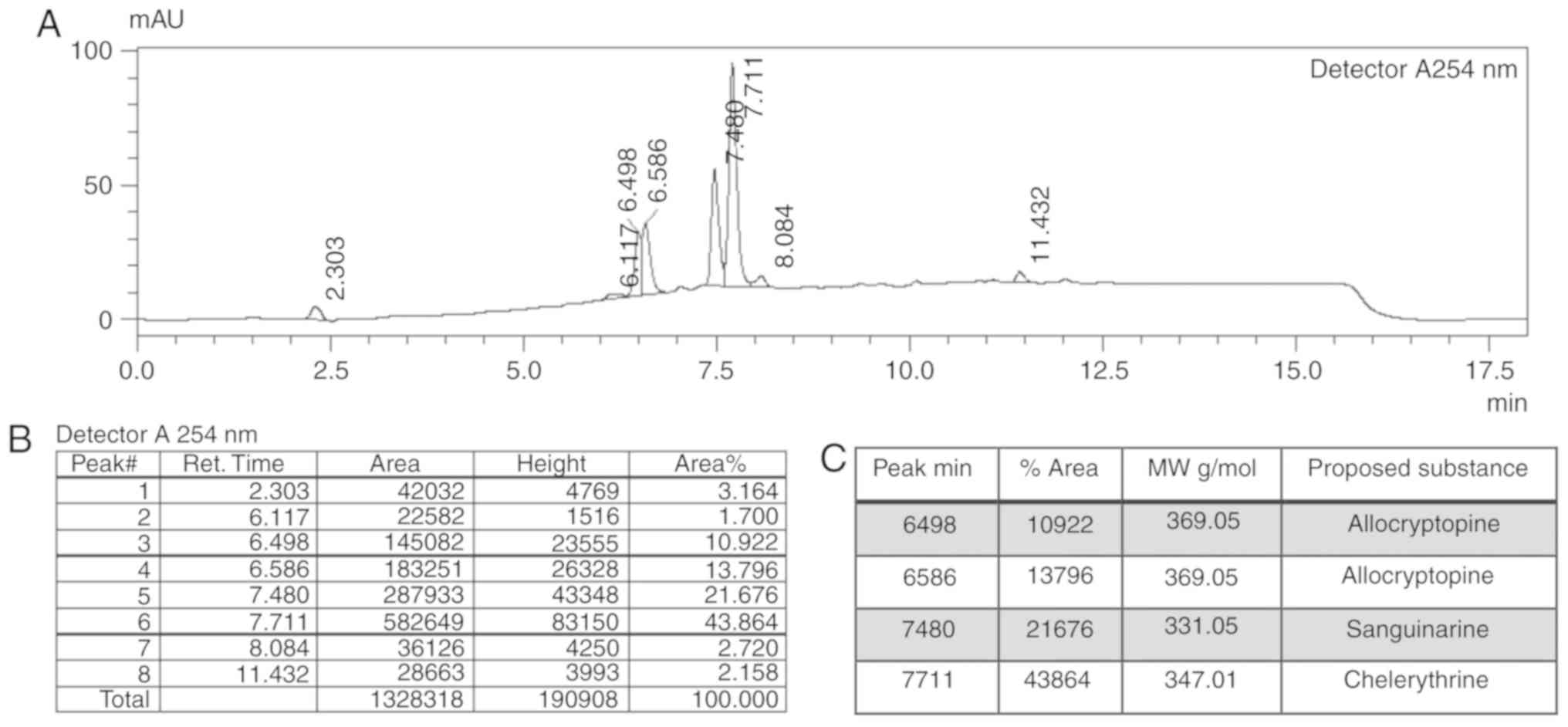
LC-MS analysis of the NSC-631570 extract displayed
four major peaks at 6.5, 6.6, 7.5 and 7.7 min in the UV
chromatogram at 254 nm (Fig. 1A) with
the percentage area of 10.9, 13.8, 21.7 and 43.9%, respectively
(Fig. 1B). Corresponding molecular
masses are shown in Fig. 1C.
LC-MS of the reference substances revealed molecular
masses of 369.05 for allocryptopine, 347.01 for chelerythrine,
331.05 for sanguinarine and 352.95 for chelidonine. According to
the present analysis, the most abundant constituents of NSC-631570
are chelerythrine (43.86%) and allocryptopine (24.72%). Their
effective concentrations in an extract solution of 10 µg/ml were
calculated to be 2.47 µg/ml for allocryptopine and 4.39 µg/ml for
chelerythrine.
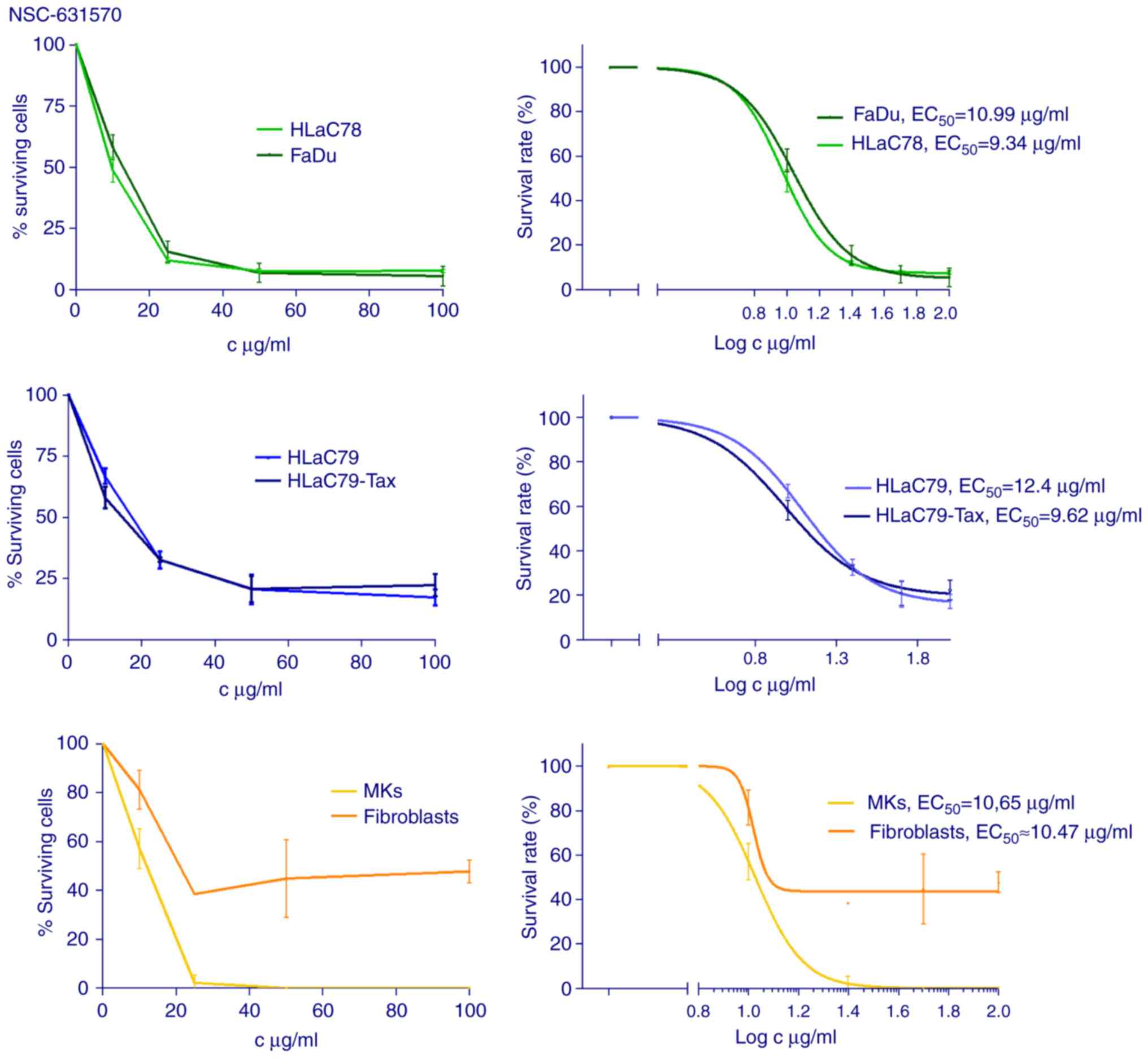
Cytotoxicity
The cell lines FaDu, HLaC78, HLaC79, HLaC79-Tax,
primary mucosal keratinocytes (MKs) and fibroblasts were incubated
with increasing concentrations of NSC-631570 (10, 25, 50, 100
µg/ml) for 48 h. The MTT assay was used to measure cell viability
and cytotoxicity (Fig. 2). For the
calculation of inhibition rates, at least three independent
experiments were carried out.
NSC-631570 treatment showed clear dose-response
curves with calculated EC50 concentrations varying
around an average concentration of 10 µg/ml (FaDu 10.99, HLaC78
9.34, HLaC79 12.4, HLaC79-Tax 9.62, MKs 10.65, fibroblasts ~10.47
µg/ml).
P-glycoprotein-overexpressing HLaC79-Tax cells were
similarly affected by NSC-631570 as their original
paclitaxel-sensitive cell line HLaC79. MKs were also sensitive to
NSC-631570, while fibroblasts proved to be much more resistant,
even at high concentrations (Fig.
2).
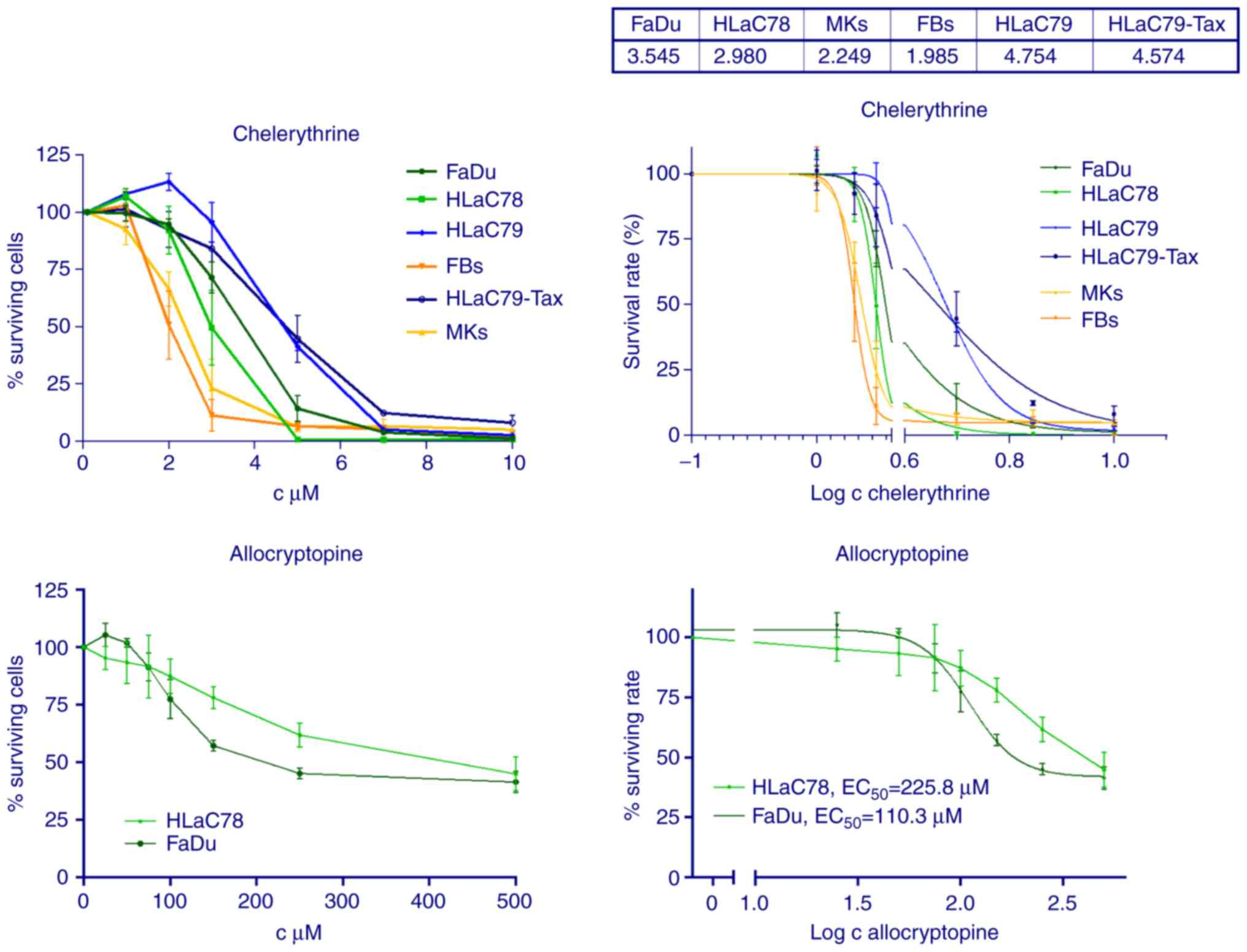
Chelerythrine exerted strong cytotoxicity on HNSCC
cell lines FaDu and HLaC78 with an EC50 dose of
approximately 3 µM (Fig. 3).
Allocryptopine, however, revealed only weak cytotoxic effects on
HNSCC cell lines, even at non-physiological doses up to 500 µM
(Fig. 3).
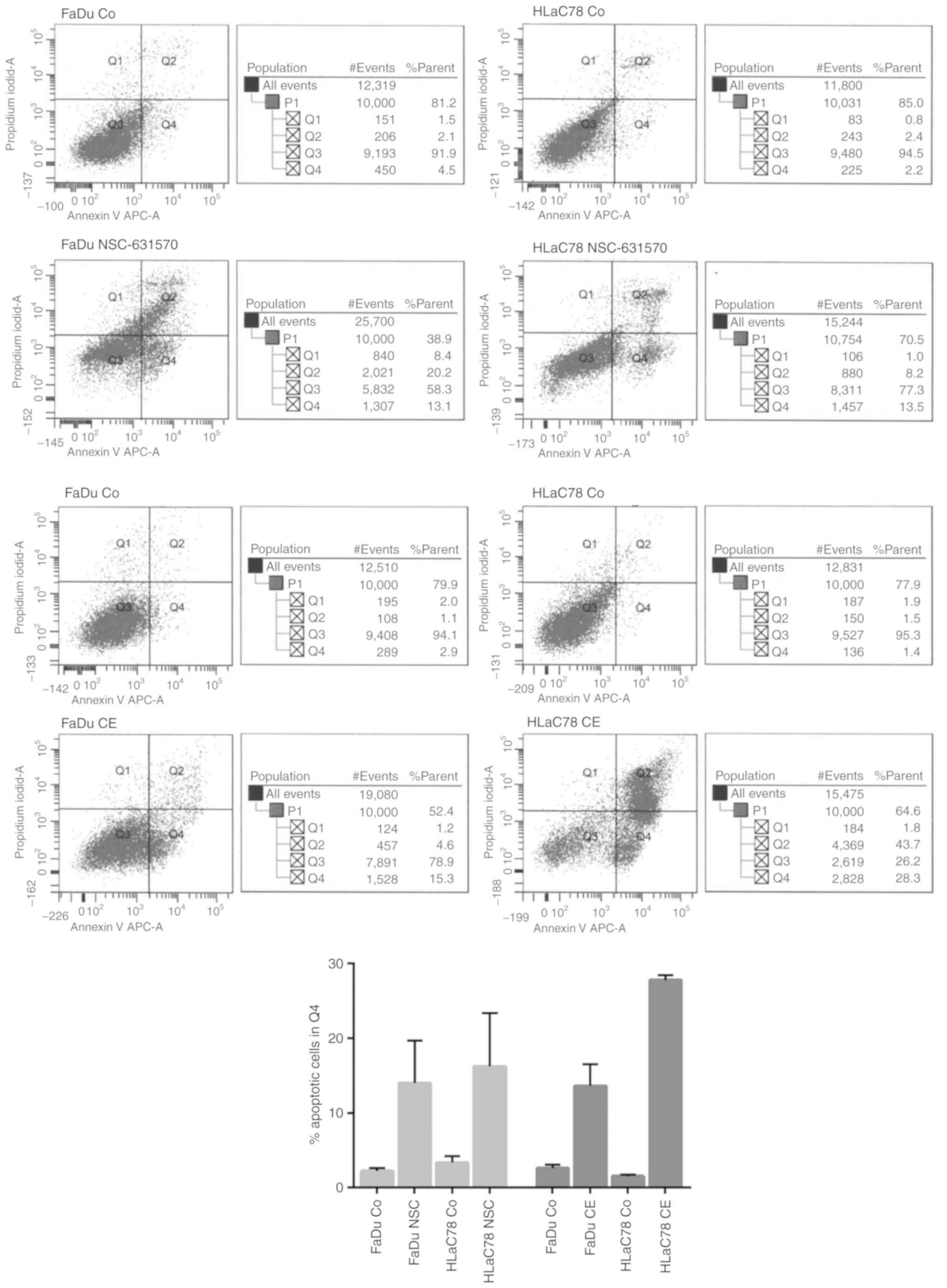
Apoptosis
Apoptosis of NSC-631570 (NSC) or chelerythrine
treated (CE) and untreated (Co) FaDu and HLaC78 cells was
determined after 24 h incubation by FACS analysis. Cell lines were
incubated with the calculated EC50 doses of each drug.
Flow cytometry analysis using the APC Annexin V kit showed an
apoptosis-promoting effect for NSC-631570 in both cell lines. With
the EC50 dose of 10 µg/ml, NSC-631570 revealed a
discrete apoptotic cell fraction of 13.1% in FaDu and 13.5% in
HLaC78 cells, respectively (Fig.
4).
Chelerythrin had a stronger effect with regard to
triggering apoptosis, especially in HLaC78 cells. The corresponding
EC50 concentrations achieved 15.3% (FaDu) and 28.3%
(HLaC78) of cells at the pre-apoptotic stages (Fig. 4).
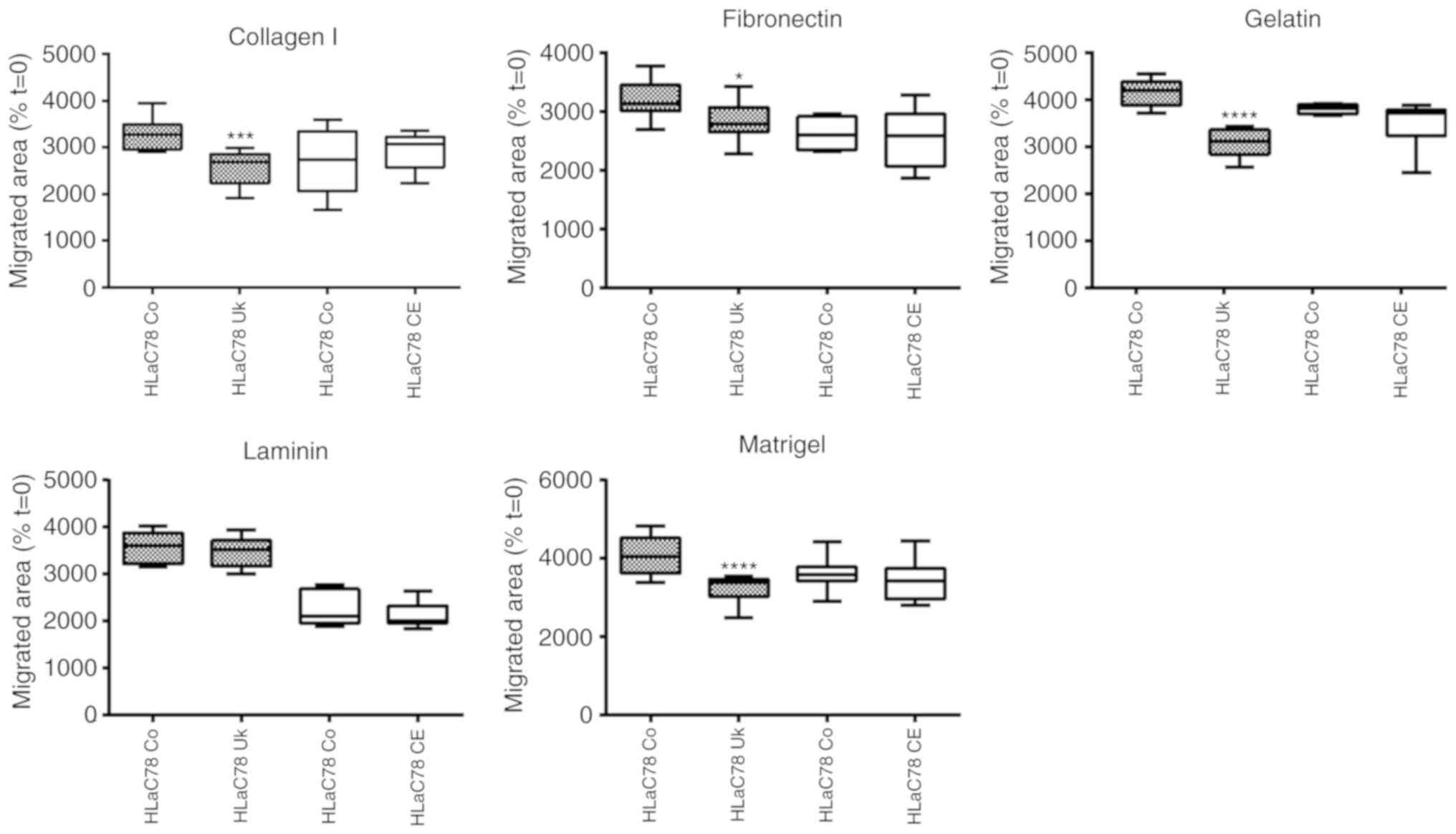
Cell motility on extracellular matrix
(ECM) proteins
Spheroid-based experiments were used to investigate
the influence of NSC-631570 on invasion on different ECM
substrates.
Spheroids of FaDu and HLaC78 cells were grown in
ultra-low-attachment-plate wells. After 72 h, they were pipetted
into wells, coated with gelatine, fibronectin, laminin, collagen I
and Matrigel and incubated with (Uk) or without (Co) NSC-631570 or
chelerythrine (CE) for 18 h. Cell migration was quantified by
photographing native and outgrown spheroids at t=0 and t=18.
Outgrown areas were measured with ImageJ software (area
calculation). For each condition, eight spheroids were measured.
Calculation of cell motility was achieved by setting the area at
t=0 at 100%. Results are displayed in Fig. 5.
The comparison of migrated areas revealed a strong
suppression of motility in FaDu cells induced by NSC-631570 on all
tested ECM surfaces. Invasion of the highly invasive HLaC78 cell
line was also significantly suppressed with the exception of
laminin (Fig. 5). Chelerythrine
showed no anti-invasive influence in the spheroid-based migration
assay (Fig. 5).
Gene expression
To generate a gene expression profile of the
NSC-631570-treated cells, microarray analysis was performed using
FaDu cells which were treated for 48 h with the EC50 of
NSC-631570 (10 µg/ml). Of a total number of 49,372 genes tested,
223 (0.45%) were upregulated and 696 (1.41%) were downregulated by
NSC-631570. The top 50 upregulated and downregulated genes are
summarized in Table IA and B.
Complete array results have been deposited at Gene Expression
Omnibus (http://www.ncbi.nlm.nih.gov/geo) with the identifier
GSE115874. Pathway analysis revealed major expression changes
affecting apoptosis, cell cycle, integrin-mediated cell adhesion,
RNA processing and translation factors, as summarized in Table II.
 | Table I.The top 50 upregulated and
downregulated genes in FaDu cells triggered by treatment with 10
µg/ml NSC-631570 for 48 h. |
Table I.
The top 50 upregulated and
downregulated genes in FaDu cells triggered by treatment with 10
µg/ml NSC-631570 for 48 h.
| A, Upregulated
genes |
|---|
|
|---|
| FC | Gene symbol | Description |
|---|
| 23.44 | CYP1A1 | Cytochrome P450,
family 1, subfamily A, polypeptide 1 |
| 8.00 | CYP1B1 | Cytochrome P450,
family 1, subfamily B, polypeptide 1 |
| 5.61 | XAF1 | XIAP associated
factor 1 |
| 5.35 | MX2 | MX dynamin-like
GTPase 2 |
| 5.01 | ISG20 | Interferon
stimulated exonuclease gene 20 kDa |
| 4.61 | OTUB2 | OTU deubiquitinase,
ubiquitin aldehyde binding 2 |
| 4.37 | OAS2 |
2′-5′-Oligoadenylate synthetase 2 |
| 4.26 | NFATC4 | Nuclear factor of
activated T-cells, calcineurin-dependent 4 |
| 4.12 | OAS1 |
2′-5′-Oligoadenylate synthetase 1 |
| 4.07 | SPOCK1 |
Sparc/osteonectin |
| 3.99 | IRF9 | Interferon
regulatory factor 9 |
| 3.94 | ALDH3A1 | Aldehyde
dehydrogenase 3 family, member A1 |
| 3.80 |
SERPINB2 | Serpin peptidase
inhibitor, clade B (ovalbumin), member 2 |
| 3.78 | IFI44L | Interferon-induced
protein 44-like |
| 3.51 | RHCG | Rh family, C
glycoprotein |
| 3.44 | NAV3 | neuron navigator
3 |
| 3.43 | COL4A6 | Collagen. type IV,
α 6 |
| 3.39 | FA2H | Fatty acid
2-hydroxylase |
| 3.31 | MDK | Midkine (neurite
growth-promoting factor 2) |
| 3.29 | DHX58 | DEXH
(Asp-Glu-X-His) box polypeptide 58 |
| 3.28 | PCDHB2 | Protocadherin β
2 |
| 3.26 | MYEOV | Myeloma
overexpressed |
| 3.25 | CORO2A | Coronin, actin
binding protein, 2A |
| 3.23 | GCHFR | GTP cyclohydrolase
I feedback regulator |
| 3.23 |
ZNF436-AS1 | ZNF436 antisense
RNA 1 |
| 3.21 | KRT6A | Keratin 6A, type
II |
| 3.20 | S100P | S100 calcium
binding protein P |
| 3.18 | VAT1 | Vesicle amine
transport 1 |
| 3.16 | RARRES3 | Retinoic acid
receptor responder (tazarotene induced) 3 |
| 3.16 | GALNT12 | Polypeptide
N-acetylgalactosaminyltransferase 12 |
| 3.16 | LAMP3 |
Lysosomal-associated membrane protein
3 |
| 3.09 | IRF7 | Interferon
regulatory factor 7 |
| 3.06 | PRR15 | Proline rich
15 |
| 3.01 | SLFN5 | Schlafen family
member 5 |
| 2.97 |
C1orf116 | Chromosome 1 open
reading frame 116 |
| 2.96 | PTPRM | Protein tyrosine
phosphatase, receptor type, M |
| 2.96 | ABCG1 | ATP binding
cassette subfamily G member 1 |
| 2.95 | SQRDL | Sulfide quinone
reductase-like (yeast) |
| 2.93 | ADAM8 | ADAM
metallopeptidase domain 8 |
| 2.92 | SH3KBP1 | SH3-domain kinase
binding protein 1 |
| 2.92 | RSAD2 | Radical S-adenosyl
methionine domain containing 2 |
| 2.91 | KHDC1L | KH homology domain
containing 1-like |
| 2.91 | WFDC2 | WAP four-disulfide
core domain 2 |
| 2.90 | PLA2G4A | Phospholipase A2,
group IVA (cytosolic, calcium-dependent) |
| 2.87 |
SYNE3 | Long intergenic
non-protein coding RNA 341 |
| 2.85 | STC1 | Stanniocalcin
1 |
| 2.85 | GALC |
Galactosylceramidase |
| 2.83 | NOV | Nephroblastoma
overexpressed |
| 2.82 | CPEB4 | Cytoplasmic
polyadenylation element binding protein 4 |
|
| B, Downregulated
genes |
|
| FC | Gene
symbol |
Description |
|
| −17.74 | ZNF664 | Zinc finger protein
664 |
| −11.35 | COTL1 | Coactosin-like
F-actin binding protein 1 |
| −9.95 | DDB1 | Damage-specific DNA
binding protein 1 |
| −9.31 | PLK2 | Polo-like kinase
2 |
| −8.11 | DHFR | Dihydrofolate
reductase |
| −7.32 | FKBP9 | FK506 binding
protein 9 |
| −6.83 | CTGF | Connective tissue
growth factor |
| −6.72 | DYNLL2 | Dynein, light
chain, LC8-type 2 |
| −6.43 | EIF4H | Eukaryotic
translation initiation factor 4H |
| −6.24 | CBX5 | Chromobox homolog
5 |
| −6.14 | EMP1 | Epithelial membrane
protein 1 |
| −6.10 | FCF1 | FCF1
rRNA-processing protein |
| −6.07 | TMPO | Thymopoietin |
| −6.01 | CHP1 | Calcineurin-like
EF-hand protein 1 |
| −5.99 |
TOR1AIP2 | Torsin A
interacting protein 2 |
| −5.94 | SAFB | Scaffold attachment
factor B |
| −5.92 | ACTN4 | Actinin, α 4 |
| −5.80 | OS9 | Osteosarcoma
amplified 9, endoplasmic reticulum lectin |
| −5.76 | SART1 | Squamous cell
carcinoma antigen recognized by T-cells 1 |
| −5.69 | HNRNPA1 | Heterogeneous
nuclear ribonucleoprotein A1 |
| −5.68 | IST1 | Increased sodium
tolerance 1 homolog (yeast) |
| −5.52 | SF3B3 | Splicing factor 3b
subunit 3 |
| −5.50 | CYB5R3 | Cytochrome b5
reductase 3 |
| −5.40 | PIM1 | Pim-1
proto-oncogene, serine/threonine kinase |
| −5.36 | YY1AP1 | YY1 associated
protein 1 |
| −5.31 | HSPA5 | Heat shock 70kDa
protein 5 (glucose-regulated protein, 78 kDa) |
| −5.27 | CNN2 | Calponin 2 |
| −5.23 | FER | fer (fps/fes
related) tyrosine kinase |
| −5.20 | MRPS27 | Mitochondrial
ribosomal protein S27 |
| −5.08 | ARFGAP2 | ADP-ribosylation
factor GTPase activating protein 2 |
| −5.05 | LAMP1 |
Lysosomal-associated membrane protein
1 |
| −5.05 | RELA | v-rel avian
reticuloendotheliosis viral oncogene homolog A |
| −5.01 | KRT5 | Keratin 5, type
II |
| −4.90 | QKI | QKI, KH domain
containing, RNA binding |
| −4.84 | STIP1 | Stress-induced
phosphoprotein 1 |
| −4.82 | AP1G1 | Adaptor-related
protein complex 1, γ 1 subunit |
| −4.74 | MCM7 | minichromosome
maintenance complex component 7 |
| −4.68 | AKT3 | v-akt murine
thymoma viral oncogene homolog 3 |
| −4.63 | CTTN | Cortactin |
| −4.6 | FAM98B | Family with
sequence similarity 98, member B |
| −4.57 | ZDHHC7 | Zinc finger,
DHHC-type containing 7 |
| −4.45 | EEF2 | Eukaryotic
translation elongation factor 2 |
| −4.45 | HMGA1 | High mobility group
AT-hook 1 |
| −4.41 | COPB2 | Coatomer protein
complex subunit β 2 (β prime) |
| −4.4 | MYH9 | Myosin, heavy chain
9, non-muscle |
| −4.39 | MCM4 | Minichromosome
maintenance complex component 4 |
| −4.39 | POLDIP3 | Polymerase
(DNA-directed), delta interacting protein 3 |
| −4.37 |
RAB1A/1B | RAB1A, RAB1B,
members RAS oncogene family |
| −4.32 | CNOT1 | CCR4-NOT
transcription complex subunit 1 |
 | Table II.Major pathways concerned in FaDu
cells, triggered by treatment with 10 µg/ml NSC-631570 for 48
h. |
Table II.
Major pathways concerned in FaDu
cells, triggered by treatment with 10 µg/ml NSC-631570 for 48
h.
| Pathway/FC | Gene symbol | Description |
|---|
| Apoptosis |
|
|
|
3.09 | IRF7 | Interferon
regulatory factor 7 |
|
2.53 | IRF5 | Interferon
regulatory factor 5 |
|
−2.10 | IKBKB | Inhibitor of kappa
light polypeptide gene enhancer in B-cells, kinase β |
|
−5.05 | RELA | v-rel avian
reticuloendotheliosis viral oncogene homolog A |
|
−2.18 | BAK1 |
BCL2-antagonist/killer 1 |
|
−3.69 | CASP2 | Caspase 2 |
|
−2.78 | CASP8 | Caspase 8,
apoptosis-related cysteine peptidase |
|
−2.34 | BCL2L1 | BCL2-like 1 |
|
−2.56 | TP53 | Tumor protein
p53 |
|
−3.49 | MCL1 | Myeloid cell
leukemia 1 |
|
−2.47 | BCL2L2 | BCL2-like 2 |
| Cell cycle |
|
|
|
−2.17 | CDH1 | Cadherin 1, type
1 |
|
−2.18 | CDC14B | Cell division cycle
14B |
|
2.35 | TBC1D8 | TBC1 domain family,
member 8 (with GRAM domain) |
|
−2.53 | HDAC1 | Histone deacetylase
1 |
|
−2.09 | CDC20 | Cell division cycle
20 |
|
−2.67 | PRKDC | Protein kinase,
DNA-activated, catalytic polypeptide |
|
−2.09 | HDAC8 | Histone deacetylase
8 |
|
−3.47 | PLK1 | Polo-like kinase
1 |
|
−3.36 | SKP2 | S-phase
kinase-associated protein 2, E3 ubiquitin protein ligase |
|
−4.39 | MCM4 | Minichromosome
maintenance complex component 4 |
|
−2.62 | MCM6 | Minichromosome
maintenance complex component 6 |
|
−4.74 | MCM7 | Minichromosome
maintenance complex component 7 |
|
−2.89 | E2F4 | E2F transcription
factor 4, p107/p130-binding |
|
2.63 | CCNE1 | Cyclin E1 |
|
−3.49 | CCND2 | Cyclin D2 |
| Integrin-mediated
cell adhesion |
|
|
|
−2.28 | PDPK1 | 3-phosphoinositide
dependent protein kinase 1 |
|
−3.09 | CRK | v-crk avian sarcoma
virus CT10 oncogene homolog |
|
−2.02 | SHC1 | SHC (Src homology 2
domain containing) transforming protein 1 |
|
2.17 | SEPP1 | selenoprotein P,
plasma, 1 |
|
2.29 | ITGA2 | Integrin, α 2
(CD49B, α 2 subunit of VLA-2 receptor) |
|
−2.77 | CAPN1 | Calpain 1, (mu/I)
large subunit |
|
−3.52 | PXN | Paxillin |
|
−2.73 | PDPK1 | 3-Phosphoinositide
dependent protein kinase 1 |
|
−4.68 | AKT3 | v-akt murine
thymoma viral oncogene homolog 3 |
| mRNA
processing |
|
|
|
−2.05 | RPS28 | Ribosomal protein
S28 |
|
−2.12 | DHX16 | DEAH
(Asp-Glu-Ala-His) box polypeptide 16 |
|
−2.83 | CPSF4 | Cleavage and
polyadenylation specific factor 4 |
|
−3.77 | RNPS1 | RNA binding protein
S1, serine-rich domain |
|
−3.13 | SRRM1 | Serine/arginine
repetitive matrix 1 |
|
−2.02 | SFSWAP | Splicing factor,
suppressor of white-apricot family |
|
−3.14 | SUGP1 | SURP and G-patch
domain containing 1 |
|
−2.40 | RNPS1 | RNA binding protein
S1, serine-rich domain |
|
−2.07 | NCBP2 | Nuclear cap binding
protein subunit 2 |
|
−3.85 | NXF1 | Nuclear RNA export
factor 1 |
|
−2.72 | HNRNPL | Heterogeneous
nuclear ribonucleoprotein L |
|
−2.88 | HNRNPU | Heterogeneous
nuclear ribonucleoprotein U |
|
−2.45 | TAF13 | TAF13 RNA
polymerase II |
|
−2.82 | POLR1A | Polymerase (RNA) I
polypeptide A |
|
−2.79 | GTF2E1 | General
transcription factor IIE subunit 1 |
|
−2.71 | POLR2E | Polymerase (RNA) II
(DNA directed) polypeptide E, 25 kDa |
|
−2.76 | ERCC2 | Excision repair
cross-complementation group 2 |
| Translation
factors |
|
|
|
−4.45 | EEF2 | Eukaryotic
translation elongation factor 2 |
|
−3.58 | EIF2S3 | Eukaryotic
translation initiation factor 2, subunit 3 γ, 52kDa |
|
−2.65 | EIF3F | Eukaryotic
translation initiation factor 3, subunit F |
|
−6.43 | EIF4H | Eukaryotic
translation initiation factor 4H |
|
−4.44 | EEF2 | Eukaryotic
translation elongation factor 2 |
|
−2.75 | EIF4B | Eukaryotic
translation initiation factor 4B |
|
−2.57 |
EIF5A/5AL1 | Eukaryotic
translation initiation factor 5A; 5A-like 1 |
|
−2.24 | EIF2AK1 | Eukaryotic
translation initiation factor 2-α kinase 1 |
|
−2.10 | EIF4A2 | Eukaryotic
translation initiation factor 4A2 |
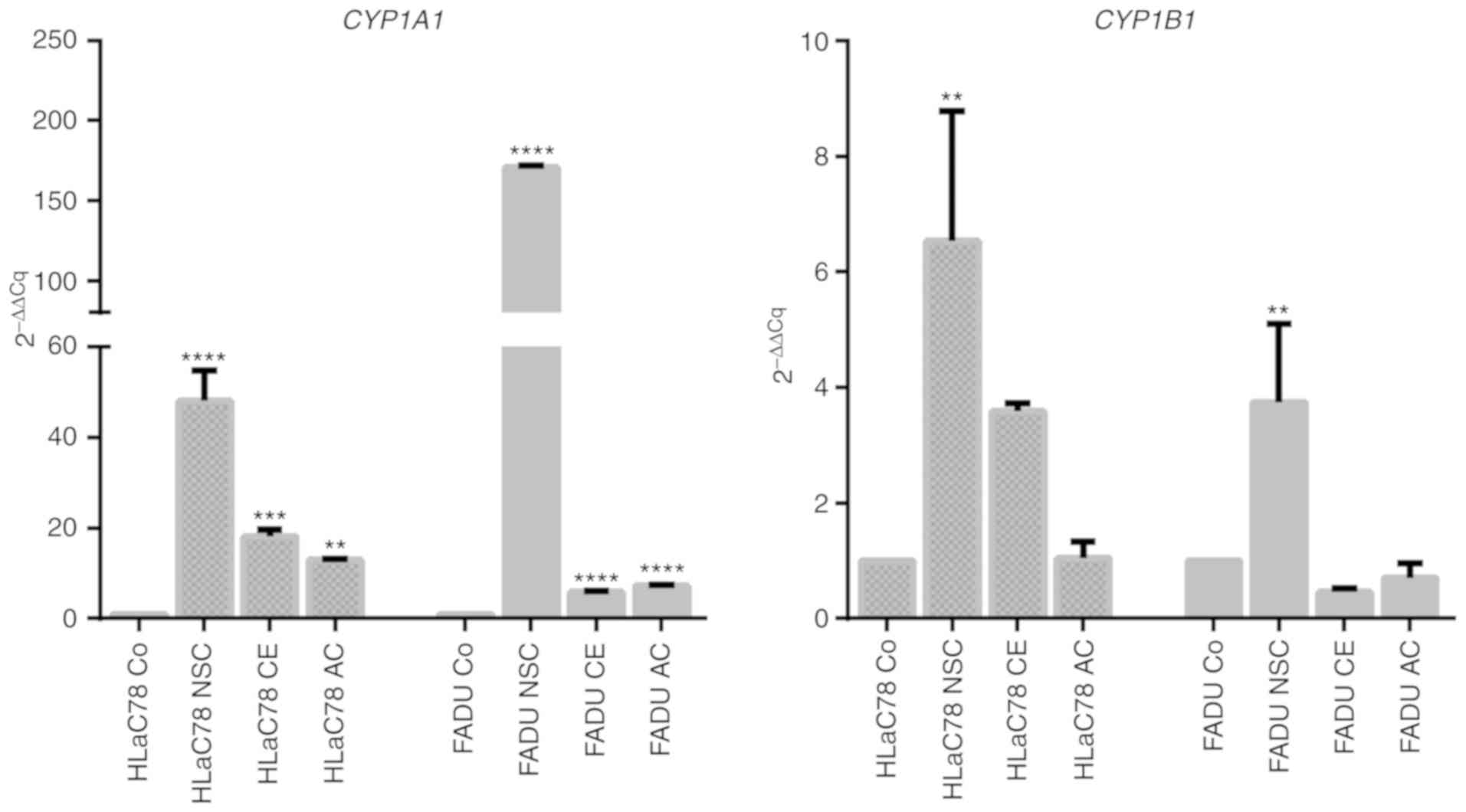
Microarray data validation by TaqMan
RT-PCR
Real-time PCR was performed for NSC-631570-treated
cell lines FaDu and HLaC78 to verify array results and to analyze
whether differential gene expression is a cell-type independent
process. For RT-qPCR, TaqMan probes for CYP1A1 and
Cyp1B1 were chosen, as these genes were the most upregulated
genes and with respect to a previously published study,
downregulation of these genes was observed following treatment with
a Chelidonium majus extract (22). FaDu and HLaC78 cells were treated with
(NSC) or without (Co) NSC-631570 or EC50 concentrations
of chelerythrine (CE) or allocryptopine (AC) respectively, for 48
h.
CYP1A1 and CYP1B1 expression was
upregulated in both cell lines in response to treatment with
NSC-631570, verifying array data and indicating a specific effect
of the Chelidonium majus extract, which was not restricted
to one cell line (Fig. 6).
Chelerythrine and allocryptopine at least contributed to
CYP1A1 upregulation in both cell lines as compared to the
control (Co). For CYP1B1, upregulation was confirmed in both
cell lines following treatment with NSC-631570. Treatment with
chelerythrine or allocryptopine showed no significant difference in
expression changes compared to the untreated control.
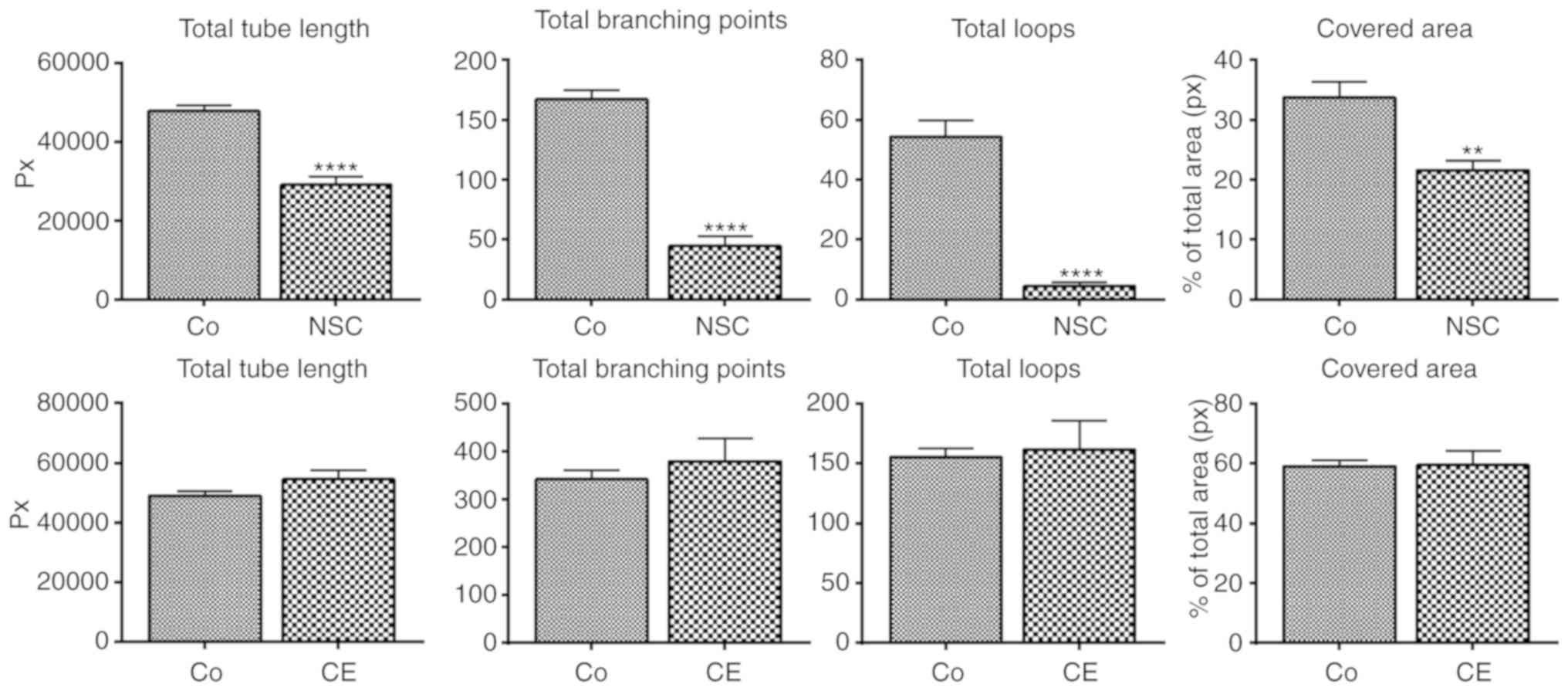
Angiogenesis
NSC-631570 was tested for anti-angiogenic properties
with the tube formation assay. Tube formation of endothelial cells,
naturally occurring on reconstituted basement membrane, was
obviously inhibited by the Chelidonium majus extract
NSC-631570 (Fig. 7, upper panels).
The quantification of inhibition showed significant decreases in
all parameters tested (Fig. 7). There
was no significant inhibition of any tube formation parameter by
chelerythrine (CE) (Fig. 7, lower
panels).
Discussion
In the present study, the Chelidonium majus
extract NSC-631570 was examined for its effect on HNSCC cell lines
in vitro. Particularly, the influence of the drug with
respect to cell growth, apoptosis, migration and angiogenesis on
tumor cells as well as its cytotoxicity against primary cells was
investigated.
LC-MS analysis revealed chelerythrine and
allocryptopine to be the most abundant alkaloids of the
Chelidonium majus extract. Composition of the extract was
found to be essentially comparable to the LC/MS analysis data of
Ukrain published by Jesionek et al in 2016 (23).
NSC-631570 was demonstrated to exhibit strong
cytotoxic activity on HNSCC tumor cells, originating from laryngeal
or hypopharyngeal subsites, even on those overexpressing
p-glycoprotein. Results may differ for head and neck cell lines,
derived from other locations. Primary cells were also affected by
NSC-631570; however, sensitivity differed between generally
sensitive epithelial keratinocytes and mucosal fibroblasts.
Fibroblasts were much more resistant against NSC-631570 compared to
the HNSCC cell lines tested in this study. Therefore, this study
cannot support the advertised selective toxicity of Ukrain on
cancer cells without restriction. In 2013, a study was published,
which analyzed the uptake and intracellular effects of celandine
alkaloids in normal and malignant cells (24). Obviously, the toxicity of alkaloid
mixtures depends on the differential uptake into the different cell
types as well as various targets within the cells (e.g.
post-translational modified tubulin isotypes), also differing
between cell types. Therefore, the exact effect of an alkaloid
mixture in differential cell systems seems to be hardly
predictable. Chelerythrine showed a comparable toxicity profile on
tumor cell lines, but also a harsher effect on primary cells, also
on fibroblasts. This is in agreement with the study of Malíková
et al (25), who also observed
increased sensitivity of primary cells against chelerythrine.
Chelerythrine is a known protein kinase C (PKC) inhibitor and in
general PKC inhibitors such as staurosporin and others have failed
to be effective in cancer therapy, although anticancer activity was
exhibited in several in vitro systems (reviewed in ref.
26). Recently, however, new
mechanisms of action beyond PKC inhibition have been published for
chelerythrine, concerning the influence on gene expression in
cancer cells (27,28). These results, together with the
observation, that chelerythrine is able to delay the growth of
xenografted tumors in vivo without considerable toxic side
effects (29), shed new light on the
potential use of chelerythrine as an anticancer therapeutic
agent.
HNSCC cell death induced by allocryptopine required
concentrations up to 1 mM, excluding this alkaloid from further
studies. Certain changes in gene expression, however (e.g.
upregulation of CYP1A1), triggered by NSC-631570, may be
caused or at least supported by the action of allocryptopine. The
upregulation of cytochrome P4501A in response to allocryptopine and
protopine has been shown in human hepatocytes and hepatic cancer
cells (29).
Cell migration, one of the hallmarks of metastasis,
is crucial for tumor formation and progression towards metastatic
phenotypes. Especially in HNSCC, this process can be
life-threatening, as surgical removal of locally invaded tumor
cells in surrounding tissue is difficult without severe functional
impairment. To evaluate the influence of NSC-631570 on the invasion
on gelatine, laminin, fibronectin, collagen type I and Matrigel, a
spheroid-based invasion assay was performed. FaDu and HLaC78
spheroids were treated with or without the EC50 of
NSC-631570 for 24 h. Invasion of FaDu cells was significantly
inhibited on all tested ECM substrates. For the highly invasive
cell line HLaC78, significance was not reached for the invasion
inhibition on laminin.
Chelerythrine failed to inhibit invasion in our
invasion model, although it has been published to have
anti-metastatic properties in hepatocellular carcinoma cell lines
(30).
Tube formation assays using HUVECs, cultivated with
or without NSC-631570, revealed a significant inhibition of
angiogenesis, which was previously presented at a congress in
Brazil in 1998 (31) by Wassil
Nowicky, the manufacturer of NSC-631570. The role of chelerythrine
in angiogenesis inhibition has not been analyzed to date, although
it has been shown that it was able to downregulate the expression
of vascular endothelial growth factor A (VEGF-A) in MCF-7 breast
cancer cells. In the present study, chelerythrine clearly failed to
inhibit angiogenesis. However, it has to be taken into
consideration, that the tube formation assay solely comprises
inhibition of endothelial cell tube formation. It does not comprise
tumor cell-derived angiogenic signals.
The high impact of NSC-631570 was found to be
accompanied by gene expression changes. Microarray analysis showed
differential expression of 1,195 genes caused by NSC-631570,
whereby 268 genes were upregulated and 927 were downregulated. The
most striking changes in gene expression revealed by microarray
analysis occurred in the genes CYP1A1 and CYP1B1,
which were strongly upregulated. CYP1A1 and CYP1B1
are involved in the xenobiotic metabolism of poly-aromatic
carcinogens and steroid hormones (among others), as well as
anticancer drugs. Therefore, they have been associated with drug
resistance in cancers (32).
Microarray data captured for CYP1A1 and CYP1B1 were
verified by RT-PCR for FaDu and HLaC78 cells, treated either with
NSC-631570, chelerythrine or allocryptopine alone.
The upregulation of CYP1A1 and CYP1B1
occurred in both cell lines, indicating that this effect is not
specific for the FaDu cell line and may be a general effect in
HNSCC cells. Chelerythrine and allocryptopine seem to contribute to
the upregulation of CYP1A1, while CYP1B1 expression
was not influenced significantly by both active ingredients. These
results are not in agreement with previous studies of El-Readi
et al (22). They observed a
downregulation of CYP1A1 and CYP1B1 following
treatment with a C. majus extract, concluding that the
tested colon cancer cells overcame drug resistance upon treatment.
The role of the mainly extrahepatic occurring CYP1 family, which
belongs to the cytochrome P450 system, and especially that of
CYP1A1, is controversially discussed. CYP1A1 has been shown
to be involved in carcinogenesis by activating pro-carcinogens,
such as N-nitrosamines. On the other hand it has been shown to be
cancer-preventive by metabolizing and thus activating dietary
compounds, such as flavonoids (reviewed in ref 33). Possibly, a
certain homeostasis of the diverse functions of that enzyme
determines its role in cancer. The observed upregulation of
CYP1A1 in response to treatment with NSC-631570 or the
isolated celandine alkaloids chelerythrine and allocryptopine in
HNSCC cell lines therefore cannot be assigned to the cytotoxicity
of the drug or to a progression in drug resistance. Interestingly,
similar results were published by Orland et al (34). The authors likewise reported an
upregulation of CYP1 family genes in HepG2 liver cancer cell lines,
caused by a Chelidonium majus extract.
In summary, NSC-631570 showed a high anticancer
impact in HNSCC cell lines. The extract exhibited cytotoxic,
anti-invasive and pro-apoptotic activities on HNSCC cell lines.
Furthermore, it acted in an anti-angiogenic manner on endothelial
cells. Compared to its major ingredient chelerythrine, it showed
clear advantages concerning the aqueous formulation, the toxicity
on primary cells and anti-migratory properties.
Acknowledgements
We would like to thank Dr Wassil Nowicky for
providing NSC-631570.
Funding
This research received no specific grant from any
funding agency in the public, commercial, or not-for-profit
sectors.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
RH performed all cell culture experiments, apoptosis
and RT-qPCR. JS performed LC-MS analysis. JR assisted in molecular
biological experiments. CP was responsible for cell culture
maintenance and tube formation assays. UH provided material and
equipment for LC-MS analysis. MS collected the data and evaluated
the results and assisted in writing the manuscript. All authors
read and approved the manuscript and agree to be accountable for
all aspects of the research in ensuring that the accuracy or
integrity of any part of the work are appropriately investigated
and resolved.
Ethics approval and consent to
participate
Mucosal keratinocytes and fibroblasts were obtained
from tonsil surgery. Informed written consent was obtained prior to
enrolment in the study. The study was approved by the Institutional
Ethics Committee on Human Research of the Julius Maximilian
University Würzburg (study approval no. 16/06).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests. There is no other active or previous relation or
collaboration with Dr W. Nowicky, who generously provided
NSC-631570.
Glossary
Abbreviations
Abbreviations:
|
ECM
|
extracellular matrix
|
|
HNSCC
|
head and neck squamous cell
carcinoma
|
|
MDR-1
|
gene locus multi-drug resistance-1,
coding for p-glycoprotein
|
|
HUVECs
|
human umbilical vein endothelial
cells
|
|
PKC
|
protein kinase C
|
|
VEGF
|
vascular endothelial growth factor
|
|
CYP
|
cytochrome P450
|
References
|
1
|
Kamangar F, Dores GM and Anderson WF:
Patterns of cancer incidence, mortality, and prevalence across five
continents: Defining priorities to reduce cancer disparities in
different geographic regions of the world. J Clin Oncol.
24:2137–2150. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Goerner M, Seiwert TY and Sudhoff H:
Molecular targeted therapies in head and neck cancer-an update of
recent developments. Head Neck Oncol. 2:82010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hartwell JL: Plants Used Against Cancer: A
Survey. Bioactive Plants Vol II. Quarterman Publications; Lawrence,
MA: 1982
|
|
4
|
Danilos J, Zbroja-Sontag W, Baran E,
Kurylcio L, Kondratowicz L and Jusiak L: Preliminary studies on the
effect of Ukrain
(Tris(2-([5bS-(5ba,6b,12ba)]-5b,6,7,12b,13,14-hexahydro-13-methyl[1,3]
benzodioxolo[5,6-v]-1-3-dioxolo[4,5-i]phenanthridinium-6-ol]-ethaneaminyl)phosphinesulfide.6HCl)
on the immunological response in patients with malignant tumours.
Drugs Exp Clin Res. 18 (Suppl):S55–S62. 1992.
|
|
5
|
Hohenwarter O, Strutzenberger K, Katinger
H, Liepins A and Nowicky JW: Selective inhibition of in vitro cell
growth by the anti-tumour drug Ukrain. Drugs Exp Clin Res. 18
(Suppl):S1–S4. 1992.
|
|
6
|
Sotomayor EM, Rao K, Lopez DM and Liepins
A: Enhancement of macrophage tumouricidal activity by the alkaloid
derivative Ukrain. In vitro and in vivo studies. Drugs Exp Clin
Res. 18 (Suppl):S5–S11. 1992.
|
|
7
|
Kadan P, Korsh OB and Hiesmayr W: Ukrain
in the treatment of urethral recurrent carcinoma (case report).
Drugs Exp Clin Res. 22:271–273. 1996.PubMed/NCBI
|
|
8
|
Lohninger A, Korsh OB and Melnyk A:
Combined therapy with Ukrain and chemotherapy in ovarian cancer
(case report). Drugs Exp Clin Res. 22:259–262. 1996.PubMed/NCBI
|
|
9
|
Schramm E, Nowicky JW and Godysh Y:
Biophysiological effects of Ukrain therapy in a patient with breast
cancer (case report). Drugs Exp Clin Res. 22:247–254.
1996.PubMed/NCBI
|
|
10
|
Habermehl D, Kammerer B, Handrick R, Eldh
T, Gruber C, Cordes N, Daniel PT, Plasswilm L, Bamberg M, Belka C
and Jendrossek V: Proapoptotic activity of Ukrain is based on
Chelidonium majus L. alkaloids and mediated via a
mitochondrial death pathway. BMC Cancer. 6:142006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lanvers-Kaminsky C, Nolting DM, Köster J,
Schröder A, Sandkötter J and Boos J: In-vitro toxicity of Ukrain
against human Ewing tumor cell lines. Anticancer Drugs.
17:1025–1030. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Panzer A, Hamel E, Joubert AM, Bianchi PC
and Seegers JC: Ukrain(TM), a semisynthetic Chelidonium
majus alkaloid derivative, acts by inhibition of tubulin
polymerization in normal and malignant cell lines. Cancer Lett.
160:149–157. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Panzer A, Joubert AM, Bianchi PC and
Seegers JC: The antimitotic effects of Ukrain, a Chelidonium
majus alkaloid derivative, are reversible in vitro. Cancer
Lett. 150:85–92. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gagliano N, Moscheni C, Torri C, Donetti
E, Magnani I, Costa F, Nowicky W and Gioia M: Ukrain modulates
glial fibrillary acidic protein, but not connexin 43 expression,
and induces apoptosis in human cultured glioblastoma cells.
Anticancer Drugs. 18:669–676. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Herrmann R, Roller J, Polednik C and
Schmidt M: Effect of chelidonine on growth, invasion, angiogenesis
and gene expression in head and neck cancer cell lines. Oncol Lett.
16:3108–3116. 2018.PubMed/NCBI
|
|
16
|
Zenner HP, Lehner W and Herrmann IF:
Establishment of carcinoma cell lines from larynx and submandibular
gland. Arch Otorhinolaryngol. 225:269–277. 1979. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Schmidt M, Grünsfelder P and Hoppe F:
Induction of matrix metalloproteinases in keratinocytes by
cholesteatoma debris and granulation tissue extracts. Eur Arch
Otorhinolaryngol. 257:425–429. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Vermes I, Haanen C, Steffens-Nakken H and
Reutelingsperger C: A novel assay for apoptosis. Flow cytometric
detection of phosphatidylserine expression on early apoptotic cells
using fluorescein labelled Annexin V. J Immunol Methods. 184:39–51.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Irizarry RA, Hobbs B, Collin F,
Beazer-Barclay YD, Antonellis KJ, Scherf U and Speed TP:
Exploration, normalization, and summaries of high density
oligonucleotide array probe level data. Biostatistics. 4:249–264.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bolstad BM, Irizarry RA, Astrand M and
Speed TP: A comparison of normalization methods for high density
oligonucleotide array data based on variance and bias.
Bioinformatics. 19:185–193. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
El-Readi MZ, Eid S, Ashour ML, Tahrani A
and Wink M: Modulation of multidrug resistance in cancer cells by
chelidonine and Chelidonium majus alkaloids. Phytomedicine.
20:282–294. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jesionek W, Fornal E, Majer-Dziedzic B,
Móricz AM, Nowicky W and Choma IM: Investigation of the composition
and antibacterial activity of Ukrain™ drug using liquid
chromatography techniques. J Chromatogr A. 1429:340–347. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kulp M and Bragina O: Capillary
electrophoretic study of the synergistic biological effects of
alkaloids from Chelidonium majus L. in normal and cancer
cells. Anal Bioanal Chem. 405:3391–3397. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Malíková J, Zdarilová A, Hlobilková A and
Ulrichová J: The effect of chelerythrine on cell growth, apoptosis,
and cell cycle in human normal and cancer cells in comparison with
sanguinarine. Cell Biol Toxicol. 22:439–453. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mochly-Rosen D, Das K and Grimes KV:
Protein kinase C, an elusive therapeutic target? Nat Rev Drug
Discov. 11:937–957. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jana J, Mondal S, Bhattacharjee P,
Sengupta P, Roychowdhury T, Saha P, Kundu P and Chatterjee S:
Chelerythrine downregulates expression of VEGFA, BCL2 and KRAS by
arresting G-Quadruplex structures at their promoter regions. Sci
Rep. 7:407062017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Banerjee A, Sanyal S, Dutta S, Chakraborty
P, Das PP, Jana K, Vasudevan M, Das C and Dasgupta D: The plant
alkaloid chelerythrine binds to chromatin, alters H3K9Ac and
modulates global gene expression. J Biomol Struct Dyn.
35:1491–1499. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chmura SJ, Dolan ME, Cha A, Mauceri HJ,
Kufe DW and Weichselbaum RR: In vitro and in vivo activity of
protein kinase C inhibitor chelerythrine chloride induces tumor
cell toxicity and growth delay in vivo. Clin Cancer Res. 6:737–742.
2000.PubMed/NCBI
|
|
30
|
Zhu Y, Pan Y, Zhang G, Wu Y, Zhong W, Chu
C, Qian Y and Zhu G: Chelerythrine inhibits human hepatocellular
carcinoma metastasis in vitro. Biol Pharm Bull. 41:36–46. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Koshelnick J, Moskvina E, Binder BR and
Nowicky JW: Ukrain (NSC-631570) inhibits angiogenic differentiation
of human endothelial cells in vitro. 17th International Cancer
Congress Brazil: pp. 91–95. 1998
|
|
32
|
Rodriguez M and Potter DA: CYP1A1
regulates breast cancer proliferation and survival. Mol Cancer Res.
11:780–792. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Androutsopoulos VP, Tsatsakis AM and
Spandidos DA: Cytochrome P450 CYP1A1: Wider roles in cancer
progression and prevention. BMC Cancer. 9:1872009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Orland A, Knapp K, König GM,
Ulrich-Merzenich G and Knöß W: Combining metabolomic analysis and
microarray gene expression analysis in the characterization of the
medicinal plant Chelidonium majus L. Phytomedicine.
21:1587–1596. 2014. View Article : Google Scholar : PubMed/NCBI
|