Introduction
Glioblastoma multiforme (GBM) is the most common
primary brain cancer and is associated with dismal prognosis, with
reported 2- and 5-year survival rates of 26–33 and 4–5%,
respectively (1). Pathognomonic
features of GBM include the presence of microvascular proliferation
and necrosis (2), both indicative
of hypoxia. As hypoxic cancer cells are genetically unstable and
metastasize frequently, defensive mechanisms responsible for cancer
cell survival under hypoxia could shed the light on factors
responsible for tumor aggressiveness and recurrence (3,4).
Previous studies have shown that GBM biopsy- and xenograft-derived
cell lines exhibit regional variation in oxygen consumption
(5–7). Using cell lines from hypoxia-sensitive
and hypoxia-tolerant GBM cells, substantial differences in their
responses to hypoxia were observed; the hypoxia-tolerant cell lines
(M006× and M059K) significantly reduced their oxygen consumption
rates and maintained their clonogenic potential after 4 days of
hypoxia (0.6% oxygen), whereas the hypoxia-sensitive cells (M010b)
failed to reduce their oxygen consumption rates or to maintain
their clonogenic potential (5).
Additional studies (3,8–11) also
suggest that understanding the molecular mechanism underlying the
adaptation of GBM cells or cross-talk with the hypoxic
microenvironment can lead to defining novel therapeutic
modalities.
The heme family of proteins includes myoglobin (MB),
neuroglobin (Ngb), hemoglobin (Hb) and cytoglobin (Cygb). The
monomeric oxygen-binding hemoprotein MB was the first protein with
a determined three-dimensional molecular structure to be reported
(12). Despite the fact that MB is
one of the most studied proteins (13), its physiological functions in normal
and cancer cells remain to be fully explored. Although MB was
previously considered to be expressed exclusively in cardiac and
skeletal muscle of vertebrates (14), its expression has been reported in
human smooth muscles (15),
non-muscle cells (liver, brain and gills) of hypoxia-tolerant
common carp and goldfish (16,17),
normal human (breast, colon, head and neck) tissues (18,19),
and different human cancers and cancer cells (18–26).
More notably, following extensive systemic research studies on the
human MB gene structure, transcripts and promoters, Bicker et
al (19) reported 16 novel
alternatively spliced variants predominantly expressed in cancer
tissue or cell lines, in addition to the three previously annotated
ones. Out of 19 transcript variants, nine encode the standard MB
protein found in muscle. In contrast to the standard MB transcript
variant 2 (muscle transcript, NM_005368.2) (27), the alternative protein-coding
cancer-associated MB splice variants 9,10, 11 and 13 (NM_203377.1)
are hypoxia-inducible, transcribed from an alternate predicted
promoter different from the one used by variant 2 (20,21,27).
However, whether these MB splice variants are expressed or
regulated by hypoxia in GBM cells has not been investigated.
Our previous studies reported that Ngb, Cygb, and Hb
(α, β, γ, δ, ζ and ε) are expressed in human GBM cell lines, human
GBM tissue microarrays and different tissue tumors (3,8–11);
thus, it was hypothesized that MB may be similarly expressed. In
the present study, it was investigated as to whether MB coding
splice variants (2, 9, 10, 11 and 13) and protein are expressed in
GBM cell lines (hypoxia-sensitive and hypoxia-tolerant) and GBM
tumor xenografts, respectively, and whether their expression
correlates with the hypoxia markers lactate dehydrogenase A (LDHA),
glucose transporter 1 (GLUT1), vascular endothelial growth factor
(VEGF) and carbonic anhydrase IX (CAIX) under hypoxia.
Materials and methods
Cell lines and in vitro culture
conditions
The cell lines used in the study were previously
established in-house from diagnostic biopsies obtained from
patients with diagnoses of GBM; details of their establishment and
characterization have been published previously (7,28).
M059J and M010b cell lines are hypoxia-sensitive (29,30);
M059K and M006×Lo cell lines are hypoxia-tolerant (5,31). All
cells were maintained as monolayer cultures in DMEM/F12 media
(Lonza Group, Ltd.) supplemented with 10% fetal bovine serum (cat.
no. 10270106; Gibco; Thermo Fisher Scientific, Inc.) in a
humidified atmosphere of 5% CO2 at 37°C. All tissue
culture plasticwares were obtained from Greiner Bio-One.
RNA extraction and reverse
transcription (RT)
An RNeasy Plus Mini Kit with gDNA Eliminator Spin
Columns (cat. no. 74136; Qiagen, Inc.) was used to isolate total
RNA from GBM cell lines. RT was conducted with 2 µg total RNA per
20 µl reaction volume using a high-capacity cDNA Reverse
Transcription Kit (cat. no. 4368814; Applied Biosystems; Thermo
Fisher Scientific, Inc.) with RNase Inhibitor (cat. no. N8080119;
Applied Biosystems; Thermo Fisher Scientific, Inc.).
RT-PCR
RT-PCR was conducted with RNA from the M006×Lo cell
line using a Veriti™ Thermal Cycler (Applied Biosystems; Thermo
Fisher Scientific, Inc.) using Platinum Pfx DNA polymerase (cat.
no. 11708-021; Invitrogen; Thermo Fisher Scientific, Inc.) for 40
cycles. Oligonucleotide primer pairs (Integrated DNA Technologies,
Inc.) for three previously annotated MB variants [variant 2
(NM_005368.2), variant 5 (NM_203378.1) and variant 13
(NM_203377.1); formerly variants 1, 2 and 3, respectively]
(26,27,32),
as well as for the novel variants 2 (muscle transcript,
NM_005368.2), 9, 10, 11 and 13 (NM_203377.1) (19,21)
were used (Table SI). The template
DNA was initially denatured for 5 min at 94°C, and each cycle of
PCR amplification was conducted as follows: Denaturation for 15 sec
at 94°C, annealing [different annealing temperature was used for
each variant (Table SI)] for 30
sec and extension for 1 min at 68°C. A final extension was carried
out for 5 min at 68°C, and then the reaction was maintained at 4°C.
Amplified RT-PCR products along with a DNA ladder (1 Kb Plus DNA
ladder; cat. no. 10787026; Invitrogen; Thermo Fisher Scientific,
Inc.) were separated on 2% agarose (cat. no. BP1356-500; Fisher
Scientific; Thermo Fisher Scientific, Inc.) gels and visualized by
ethidium bromide (cat. no. E/P800/10; Fisher Scientific; Thermo
Fisher Scientific, Inc.).
DNA sequencing
Bands of RT-PCR amplicons were excised from the
agarose gel and purified using a MinElute® Gel
Extraction Kit (cat. no. 28604; Qiagen, Inc.). The purified PCR
products were sequenced at Colors Medical Laboratories using a 3500
Genetic Analyzer (Applied Biosystems; Thermo Fisher Scientific,
Inc.). Sequencing data were analyzed using the National Center for
Biotechnology Information (NCBI) Basic Local Alignment Search Tool
(BLAST; version 4) engine (33).
Generation of hypoxia in vitro
To study the effect of hypoxia on the protein-coding
MB mRNA variants compared with hypoxia markers, hypoxia-sensitive
and hypoxia-tolerant cell lines were incubated under moderate
hypoxic conditions that simulate the brain tumor microenvironment.
Generation of hypoxia in vitro was conducted as previously
described with modifications (9,34),
with a hypoxia chamber (Biospherix, Ltd.) connected to a ProOx C21
controller unit (Biospherix, Ltd.) to control the levels of
O2 and CO2 inside the hypoxia chamber. Cells
(~4×105) in the exponential phase of growth were seeded
onto 100-mm tissue culture dishes and then incubated at 37°C under
standard laboratory culture conditions (5% CO2 in air).
Then, 2 days later, cells of each cell line were incubated under
hypoxia (0.6% oxygen and 5% CO2) for 12, 24 or 48 h with
no media change during the course of the experiment. The aerobic
control of each cell line was incubated under normoxic conditions
in the standard CO2 incubator. Four replicate
experiments were carried out for each cell line.
RT-quantitative PCR (qPCR)
RT-qPCR analysis was performed with a Quant Studio
12K Flex Real-Time PCR System (Applied Biosystems; Thermo Fisher
Scientific, Inc.). TaqMan™ Fast Universal PCR Master Mix (cat. no.
4366072; Applied Biosystems; Thermo Fisher Scientific, Inc.) and
validated TaqMan Gene Expression Assays (Applied Biosystems; Thermo
Fisher Scientific, Inc.) were used for human LDHA (assay no.
Hs01378790_g1), GLUT1 (assay no. Hs00892681_m1), VEGF (assay no.
Hs00900055_m1), CAIX (assay no. Hs00154208_m1) and β-actin (cat.
no. 4333762F). Primers (Integrated DNA Technologies, Inc.) for
protein-coding MB mRNA variants 2, 9, 10, 11 and 13 (19–21)
and β-actin (Table SII) were mixed
with Power-Up™ SYBR™ Green Master Mix (cat. no. A25778; Applied
Biosystems; Thermo Fisher Scientific, Inc.) and qPCR was conducted
as previously described (19–21).
The relative fold change in mRNA of target genes for each cell line
was calculated from four experimental replicates (four different
passages) in duplicate assays. In each experimental replica, the
mean quantification cycle (Cq) of the endogenous housekeeping gene
(β-actin) was subtracted from the mean Cq of the target gene for
aerobic controls, and cells treated with hypoxia for 12, 24 and 48
h. Then, the resulting value (∆Cq) of the aerobic control was
subtracted from itself and from the resulting values (∆Cq) of the
12, 24 and 48-h hypoxia-treated cells. The fold change in target
gene expression normalized to endogenous housekeeping gene and
relative to aerobic control was quantified using the
2−ΔΔCq method (35).
Mining the mutation status of
isocitrate dehydrogenase (IDH) in GBM cell lines
The open access ‘DepMap’ portal of the Broad
Institute (https://depmap.org/portal/) (36) was used to check for any identified
somatic mutations in IDH1/2 in the cell lines used in the present
study. The search results can be retrieved online for the M059J
cell line (DepMap ID no. ACH-001118) and for the M059K cell line
(DepMap ID no. ACH-000152).
Hypoxia labeling of tumor
xenografts
Sections used in this study of ectopically-induced
glioma tissue from mice injected with M006×Lo cells were archived
samples generated during a previous study published in 1998
(6). All previous animal
experimentation was reviewed and approved by the Animal Care
Committee at the Cross Cancer Institute in accordance with
guidelines established by the Canadian Council for Animal Care.
Briefly, the tumor tissues were initially obtained from tumor
xenografts initiated in NOD/SCID mice at 6–10 weeks of age, when
106−107 tumor cells were injected
intradermally into both flanks of mice. When the tumor volumes
reached 100–300 mm3, mice received a single
intraperitoneal injection of the hypoxia marker, pimonidazole HCl
(100 mg/kg; Natural Pharmacia International). Mice were euthanized
90 min later, and tumor xenografts were excised, fixed in buffered
formalin (24–48 h at room temperature), embedded in paraffin and
then sectioned at 5-µm intervals. The M006×Lo cell line used for
these experiments has been extensively characterized with respect
to its hypoxia-tolerant phenotype (5–7,29) and
produces xenografted tumors that are consistent in growth rate.
Tumor tissue immunostaining
Pimonidazole adducts were detected as previously
described (37) with modifications.
Xenograft slides were incubated in a dry oven at 62°C for 2 h,
deparaffinized, rehydrated and washed twice with ddH2O
(2 min each), and then antigen retrieval was carried out using a
Bio SB TintoRetrieval Pressure Cooker (Bio SB, Inc.) at high
pressure for 15 min. Xenograft slides were washed, endogenous
peroxidase activity was blocked with Dako REAL peroxidase blocking
buffer (cat. no. S2023; Dako; Agilent Technologies, Inc.) for 5 min
at room temperature and then washed twice. Tissue sections were
blocked for 30 min at room temperature with diluted 5% normal goat
serum (cat. no. ES 1028; Biomeda Corporation). Tissue sections were
incubated for 1 h at room temperature with diluted (Dako Antibody
Diluent; cat. no. S0809; Dako; Agilent Technologies, Inc.) rabbit
primary antibodies against: Pimonidazole (1:200; cat. no.
PAB2627AP; Hypoxyprobe, Inc.), MB (1:100; cat. no. ab77232; Abcam),
CAIX (1:150; cat. no. 5648; Cell Signaling Technology, Inc.), LDHA
(1:100; cat. no. 3582; Cell Signaling Technology, Inc.) or VEGF
(1:100; cat. no. sc-507; Santa Cruz Biotechnology, Inc.). Negative
controls of tissue sections were incubated without primary
antibody. Tissue sections were washed twice with PBS (cat. no.
18912-014; Gibco; Thermo Fisher Scientific, Inc.) for 5 min each at
room temperature, and then incubated with horseradish
peroxidase-conjugated goat anti-rabbit immunoglobulin (Dako
EnVision+; cat. no. K4002; Dako; Agilent Technologies,
Inc.), and then Liquid DAB+ 2-Component system (cat. no. K3467;
Dako; Agilent Technologies, Inc.) was added. The reaction was
stopped by placing xenograft slides in ddH2O. Slides
were counterstained with Mayer's hematoxylin (cat. no. S3309; Dako;
Agilent Technologies, Inc.) for 30 sec at room temperature and then
rinsed in ddH2O. Slides were placed in lithium carbonate
(cat. no. 62470-100G-F; Sigma-Aldrich; Merck KGaA) for 1 min,
washed and dehydrated, and then coverslips were mounted with Dako
Ultramount Aqueous Permanent Mounting Medium (cat. no. S1964; Dako;
Agilent Technologies, Inc.).
For slides stained with hematoxylin and eosin,
slides were stained with eosin (cat. no. RRBD92-W; Atom Scientific
Ltd.) for 2 min at room temperature after hematoxylin staining
prior to the dehydration step.
Evaluation of slides was performed by two
pathologists, with ≥5 fields analyzed per sample. Photomicrographs
were obtained at ×10 and ×40 magnification (Axio Scope.A1; Carl
Zeiss AG)
Statistical analysis
Data are expressed as the mean ± SE of four
replicate experiments. Statistical analyses were performed using
SigmaPlot 13 software (Systat Software Inc.). Differences between
groups were assessed using one-way ANOVA followed by Dunnett's test
for multiple comparisons with the control group. Additionally,
Spearman rank correlation analysis was used to assess the
relationship between MB expression and different hypoxia markers.
The level of significance was set at P<0.05.
Results
Expression of the protein-coding MB
mRNA splice variants 2, 9, 10, 11 and 13 in the M006×Lo cell
line
Amplified RT-PCR products separated on agarose gels
showed clear bands that matched their predicted amplicon size
(Fig. 1). Sequences analysis of the
DNA extracted from agarose bands using the NBCI BLAST engine
revealed perfect alignments to the corresponding mRNA sequences for
the MB variants 2, 9, 10, 11 and 13 (Table SIII).
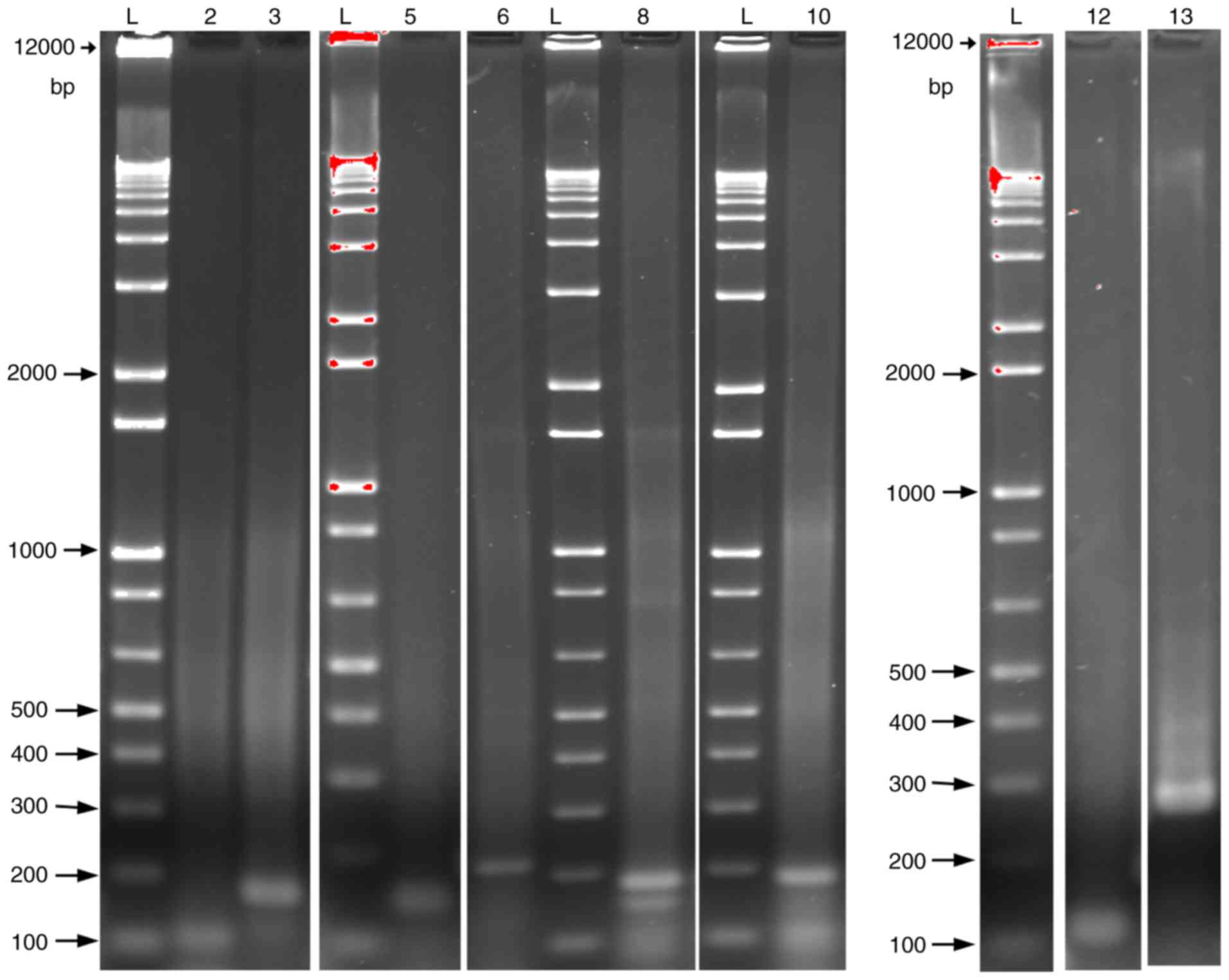 | Figure 1.Protein-coding MB mRNA variant (2, 9,
10, 11 and 13) expression in M006×Lo cells. Amplified reverse
transcription-PCR products formed using specific sequence primer
pairs were separated on 2% agarose gels and visualized using
ethidium bromide. The amplicons of the predicted size were
confirmed by DNA sequencing. Lanes: 2, MB (111 bp); 3, MB (179 bp);
5, MB variant 2 (151 bp); 6, MB variant 2 (219 bp); 8, MB variant 9
(189 bp); 10, MB variant 10 (188 bp); 12, MB variant 11 (121 bp);
13, MB variant 13 (275 bp). For MB and variant 2, two primer pairs
were used to confirm their presence. L, DNA ladder; MB,
myoglobin. |
Quantification of the protein-coding
MB mRNA splice variants 2, 9, 10, 11 and 13 in GBM cell lines under
normoxic and hypoxic conditions
Hypoxia-sensitive (M059J, M010b) and
hypoxia-tolerant (M059K, M006×Lo) human GBM cell lines (5,29–31)
were used for these experiments. Transcripts of all tested MB
variants (Fig. 2A-E) were detected
in all of the investigated cells, with the exception of MB
muscle-type variant 2 transcript, which was not detected in M010b
cells under either normoxic or hypoxic conditions. However,
interestingly, this variant was downregulated by ~46% in the M059J
cell line as early as 12 h after exposure to hypoxia (Fig. 2A). A non-significant trend towards a
reduction (~60%) of variant 2 mRNA was observed in the
hypoxia-tolerant M006×Lo cell at 24 h (P=0.053) and 48 h (P=0.053)
of hypoxia, with its expression in M059K being not significantly
altered in response to hypoxia (Fig.
2A). Significant upregulation (3-fold) of MB variant 9 was
observed in M059K at 48 h of hypoxia, with a trend toward increased
expression being also observed at 24 h (2.3-fold; P=0.085; Fig. 2B). No significant changes in variant
9 gene expression were observed in the rest of the tested GBM cell
lines. MB variants 10, 11 and 13 showed trends of increased
expression under hypoxia (Fig.
2C-E). It is worth mentioning that the expression of these
variants was previously shown to be induced by hypoxia in
MDA-MB-468 and DLD-1 cell lines (19). However, in contrast to the range of
durations of hypoxia employed here, the previous study assessed the
hypoxic response of these variants only after 72 h of hypoxia.
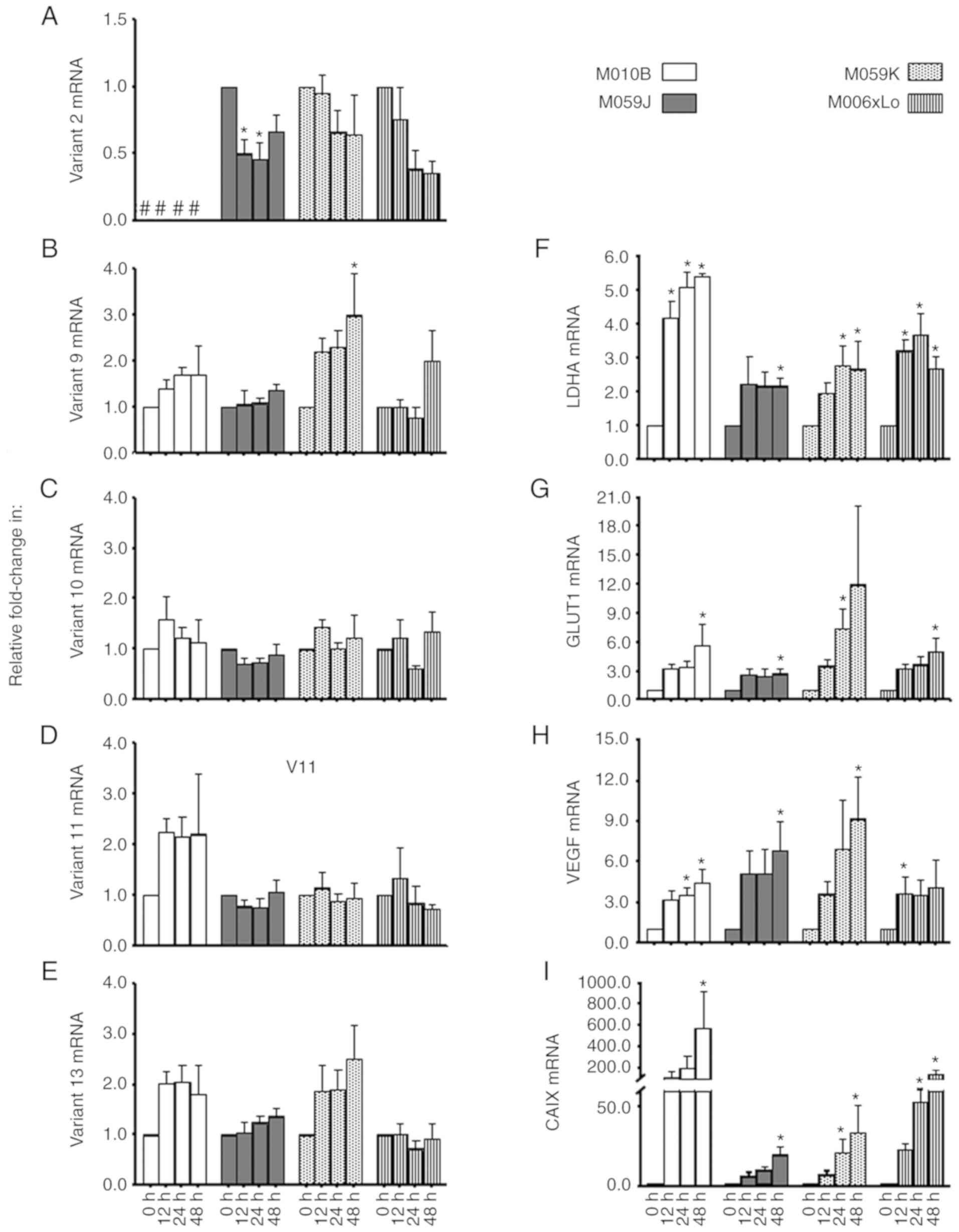 | Figure 2.Expression of protein-coding MB mRNA
splice variants in human GBM cell lines. Expression levels of (A)
MB variant 2, (B) variant 9, (C) variant 10, (D) variant 11 and (E)
variant 13, as well as the hypoxia markers (F) LDHA, (G) GLUT1, (H)
VEGF and (I) CAIX were assessed in the human GBM cell lines, M010b,
M059J, M059K and M06×Lo, following exposure to hypoxia (0.6%
O2) for 0 (aerobic control), 12, 24 and 48 h. Expression
was analyzed via reverse transcription-quantitative PCR, and data
were expressed as fold increases relative to the aerobic control
(n=4). *P<0.05 vs. 0 h. #, MB variant 2 was not detectable in
M010b cells; MB, myoglobin; GBM, glioblastoma multiforme; LDHA,
lactate dehydrogenase A; GLUT1, glucose transporter 1; VEGF,
vascular endothelial growth factor; CAIX, carbonic anhydrase
IX. |
Quantification of hypoxia markers in
GBM cell lines
Hypoxia markers (LDHA, GLUT1, VEGF, and CAIX) were
expressed in all cell lines (Fig.
2F-I) under normoxic conditions. Under hypoxic conditions, LDHA
mRNA levels were significantly (P<0.05) upregulated at 12, 24
and 48 h (M010b and M006×Lo), at 48 h (M059J), or at 24 and 48 h
(M059K) in the different cell lines (Fig. 2F). GLUT1 mRNA levels exhibited
2.5–11.9-fold increases in all cell lines, and were significantly
increased (P<0.05) at 24 h in M059K cells, and at 48 h in M010b,
M059J and M006×Lo cells (Fig. 2G).
VEGF mRNA showed significant increases (P<0.05) at 12 h
(M006×Lo), 24 h (M010b) or 48 h (M010b, M059J and M059K) in
different cells lines; however, the 3.2–6.9-fold upregulation at
other time points was not statistically significant (Fig. 2H). Hypoxic upregulation of CAIX mRNA
levels ranged from 19- to 579-fold; significant increases were
observed at 24 h in M059K and M006×Lo cells, and at 48 h in all
four cell lines (Fig. 2I). As there
were no specific differences in the manner that hypoxia markers
were altered in hypoxia-sensitive (M010b and M059J) compared with
hypoxia-tolerant (M006×Lo and M059K) GBM cells, it can be concluded
that early hypoxia defensive mechanisms may be similar across these
cell lines.
Correlation between protein-coding MB
mRNA variants and hypoxia markers in GBM cell lines
To further characterize the effects of hypoxia on
the expression of MB variants, Spearman rank-order correlation
analysis was performed; the correlation coefficients and
corresponding P-values were calculated (Table I). MB variant 2 expression exhibited
significant moderate-to-strong negative correlation with all of the
tested hypoxia markers in M059J and M006×Lo cells, with a trend of
weak negative correlation with GLUT1 and CAIX also observed in
M059K. Positive correlations between MB variant 9 and nearly all of
the tested hypoxia markers were observed in the tested cells, with
strong (r ≥0.785) and significant (P<10−6)
correlations in M059K cell line suggesting that this variant may
contribute to this cell line's survival under hypoxia. MB variant
10 and variant 11 gene expression did not correlate significantly
with any of the tested hypoxia markers in M010b, M059K or M006×Lo
cells. In the M059J cell line, moderate significant negative
correlations between MB variant 10 or MB variant 11, and GLUT1 and
VEGF were observed, with a trend towards a weakly negative
correlation between variant 10 and CAIX. This finding, although
unexpected, is of potential interest, as it indicates that hypoxia
may negatively impact MB cancer-related variants, 10 and 11, in
that particular cell line, which may potentially contribute to this
cell line's vulnerability to hypoxic conditions. As for MB variant
13, moderate borderline significant correlation with LDHA was
observed in M010b, with no association detected with any of the
tested hypoxia markers in M006×Lo cell lines. In M059J and M059K
cell lines, moderate-strong positive correlations between MB
variant 13 and all tested hypoxia markers were observed.
Collectively, these findings indicate that variant 13 is regulated
by hypoxia in M010b, M059J and M059K cell lines, and may provide a
protective or survival advantage in these cell lines.
 | Table I.Spearman rank correlation analysis of
correlations between protein-coding MB mRNA variants and hypoxia
markers in GBM cell lines. |
Table I.
Spearman rank correlation analysis of
correlations between protein-coding MB mRNA variants and hypoxia
markers in GBM cell lines.
|
|
| MB variant |
|---|
|
|
|
|
|---|
| Cell line | Marker | V2 | V9 | V10 | V11 | V13 |
|---|
| M010B | LDHA | NA | 0.522a | 0.107 | 0.388 | 0.501a |
|
| GLUT1 | NA | 0.310 | 0.200 | 0.233 | 0.233 |
|
| VEGF | NA | 0.0776 | 0.0836 | −0.0687 | 0.257 |
|
| CAIX | NA | 0.164 | 0.0836 | −0.0328 | 0.418 |
| M059J | LDHA | −0.603a | 0.218 | −0.358 | −0.251 | 0.693a |
|
| GLUT1 | −0.788c | 0.191 | −0.642b | −0.555a | 0.567a |
|
| VEGF | −0.809c | 0.179 | −0.713b | −0.582a | 0.540a |
|
| CAIX | −0.579a | 0.460 | −0.424 | −0.397 | 0.690b |
| M059K | LDHA | −0.304 | 0.848c | 0.254 | 0.275 | 0.734c |
|
| GLUT1 | −0.424 | 0.875c | 0.266 | 0.119 | 0.618a |
|
| VEGF | −0.343 | 0.785c | 0.221 | 0.0507 | 0.696b |
|
| CAIX | −0.430 | 0.845c | 0.0896 | 0.0328 | 0.696b |
| M006×Lo | LDHA | −0.615a | −0.122 | −0.266 | −0.296 | −0.158 |
|
| GLUT1 | −0.698b | 0.131 | −0.0776 | −0.179 | 0.0209 |
|
| VEGF | −0.567a | −0.0985 | −0.212 | −0.230 | 0.0119 |
|
| CAIX | −0.738b | 0.119 | −0.119 | −0.397 | −0.227 |
Mutation status of IDH in GBM cell
lines
Mining the mutation status of IDH revealed no
somatic mutations in IDH1/2 in M059J and M059K cell lines (data not
shown) (36).
Expression of MB protein and hypoxia
markers protein in GBM xenografts
To investigate whether the expression of MB is
upregulated under physiologically relevant hypoxic conditions that
mimic the tumor microenvironment in patients with GBM, tumor
xenografts were developed in NOD/SCID mice using M006×Lo cells.
Serial tumor sections were immunostained with antibodies specific
for MB, and hypoxia markers LDHA, VEGF, GLUT1, CAIX and
pimonidazole. In tumor sections, distinct regions of necrosis were
used as landmarks to compare serial tumor sections. Tumor cells
adjacent to necrotic areas were hypoxic, as shown by positive
pimonidazole staining (Fig. 3).
Similar patterns of staining with various degrees of
immunoreactivity were shown in matched tumor regions adjacent to
necrosis that were stained for MB, CAIX, LDHA and VEGF. Other area
of xenograft sections that lacked pimonidazole staining exhibited
weak staining for MB and other hypoxia markers. These parts may
represent well-oxygenated areas. These results indicating the
presence of an association between the expression of MB and hypoxia
marker proteins in xenografts generated using M006×Lo cells
contrast with in vitro results reporting a trend towards a
reduction of variant 2 mRNA, and a lack of correlation between
cancer-related MB variants and hypoxia markers in M006×Lo cells. It
should be noted that the xenograft tissues used in the present
study were archived samples obtained during a previous study
(6). However, in a study
investigating antigen preservation in formalin-fixed,
paraffin-embedded blocks dating from several previous decades, it
was concluded that antigenicity is maintained in FFPE material for
>60 years (38), indicating that
the results in the present study accurately reflect the expression
of globins and pimonidazole-protein adducts in these cells.
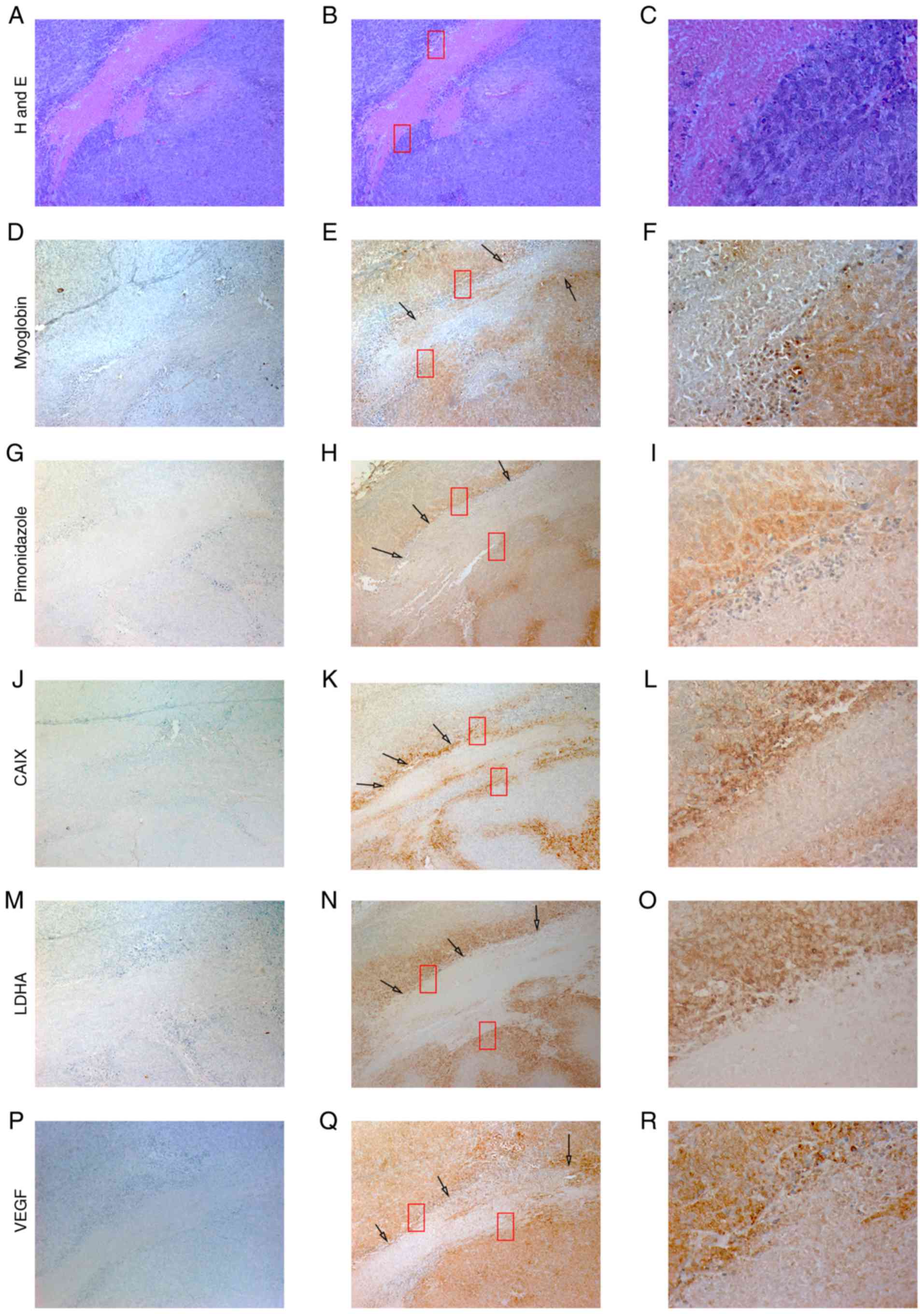 | Figure 3.Expression of MB, CAIX, LDHA and VEGF
in a M006×Lo tumor xenograft. Ectopically-induced glioblastoma
tumor xenografts in mice were generated via injection of M006×Lo
cells. Serial sections of tumor xenografts were stained with (A-C)
H&E, and antibodies specific for (D-F) MB, (G-I) pimonidazole,
(J-L) CAIX, (M-O) LDHA and (P-R) VEGF. Regions of necrosis were
used as tissue landmarks (red boxes). Hypoxic cells identified by
pimonidazole staining also exhibited strong positive staining for
MB, CAIX, LDHA and VEGF (black arrows). MB staining was weakly
positive throughout other parts of tumor sections; however, these
cells showed either weak or no staining for pimonidazole, CAIX,
LDHA and VEGF. Image panels in the left column represent negative
controls of tissue sections where, instead of primary antibody,
slides were incubated with normal goat serum. Photomicrographs were
obtained at ×10 (left and middle panels) and ×40 (right panels)
magnification. |
Discussion
The aim of the present study was to investigate
whether MB is expressed in GBM cells and, if so, whether its
expression is modulated by hypoxia levels that simulate the hypoxic
tumor microenvironment. Both standard and alternative
cancer-associated MB variants have been reported be expressed in
cancer tissue and cell lines, as well as in healthy breast,
skeletal and heart muscle (19,21).
Our previous studies demonstrated that GBM ectopically expresses
three of the globin family members; Ngb, Cygb and Hb, and that
these oxygen-binding molecules are upregulated under hypoxic
conditions (3,9–11).
These observations potentially explain the ability of GBM to grow
and progress under hypoxia, as well as suggesting a reason for the
poor prognosis and frequent recurrence associated with these
cancers. In the present study, it was reported that different
variants of another member of the globin family, MB, are
ectopically expressed in four GBM cell lines (M010b, M059J, M059K
and M006×Lo). The cellular responses of these cell lines to hypoxia
have been characterized previously. In contrast to
hypoxia-sensitive and radiosensitive (M010b and M059J) cells
(29,30), M059K and M006×Lo cells are
hypoxia-tolerant and radioresistant (5,31,39).
It was found that the MB transcripts 9, 10, 11 and
13, and the standard MB transcript (variant 2) were expressed in
the four cell lines, except for MB variant 2, which was
undetectable in M010b. This observation is consistent with those of
Kristiansen et al (27), who
reported that MB was not detected in all cells of a breast cancer
cell panel. Under hypoxia, hypoxia inducible factors (HIFs)
orchestrate various cellular responses by binding to the hypoxia
responsive elements (HREs) in their target genes (40). This induces the transcription of
genes required for metabolic adaptations to O2
deficiency and glucose uptake (such as LDHA and GLUT1), tumor
growth and metastasis (VEGF) and intracellular regulation of pH to
maintain a relatively acidic pH extracellularly (CAIX), conditions
that are compatible with cell viability and proliferation, as well
as invasion and metastasis (40–44).
It was shown earlier that GBM cells modify oxygen
consumption rates in response to variations in oxygen supply, which
may contribute to GBM survival in hypoxic microenvironments
(5). The ability to modify oxygen
consumption differs among GBM cells; the hypoxia-tolerant M006× and
M059K cell lines have the ability to lower oxygen utilization
rates, whereas the hypoxia-sensitive M010b cell line does not.
Consequently, and in contrast to the hypoxia-tolerant cell line
M006, the M010b cell line significantly loses its colony-forming
ability when maintained under hypoxia (5). In the present study, it was shown that
hypoxia-tolerant and hypoxia-sensitive GBM cell lines are indeed
hypoxia-responsive; hypoxia markers, including LDHA, GLUT1, VEGF
and CAIX, were upregulated in the four tested cell lines under
hypoxia. Thus, differences in their susceptibility to low oxygen
tension cannot be explained solely on the basis of differential
recruitment of any of the hypoxia responsive genes discussed
above.
In the present study, MB variant 2 exhibited
modestly negative significant correlations with hypoxia markers. In
spite of the absence of canonical binding sites for the
hypoxia-responsive master transcription factors HIF-1α/-2α in its
promoter (45,46), MB variant 2 was significantly
increased in MDA-MB-468 breast cancer cells, but not DLD-1 colon
cancer cells, following 72 h of 1% O2 (19). This variation in variant 2 hypoxia
responses (downregulation, upregulation or no change) underlines
the complexity of cellar responses to hypoxia in different cell
lines of different tumor origins that may reflect differential
regulation of MB variant 2. On the other hand, the expression of
the MB cancer-associated variants 9, 10, 11 and 13 collectively
exhibited trends towards increased expression that were consistent
with previous reports linking this upregulation with an HRE
candidate within the upstream region of the MB gene, along with
other, thus far uncharacterized, enhancer sites (19,32).
Cell line-dependent changes in cancer-related MB
variant gene expression are also apparent. For example, while a
modest increase in MB variant 13 coupled with significant positive
correlations with hypoxia markers was observed in hypoxia-sensitive
(M010b and M059J) and hypoxia-tolerant cells (M059K), this variant
was essentially unaltered and did not correlate with any of the
tested markers of hypoxia in the M006×Lo cell line. Also, while
hypoxia appeared to positively regulate variants 9 and 13 in the
M059K cell line, as indicated by the significant positive
correlations with tested hypoxia markers, it had no effect on these
variants in the other hypoxia-tolerant cell line, M006×Lo. These
results are similar to previously reported observations, where MB
variant 13 was significantly upregulated in MDA-MB-468 breast
cancer cells but not colon-derived DLD-1 subjected to 72 h at 0.6%
O2 (19,32). Similarly, the present data indicated
that low oxygen tension downregulated variants 10 and 11 in the
M059J cell line, as indicated by the significant negative
correlation with markers of hypoxia; however, it had no effect on
these variants in the M059K cell line.
Of the four-tested GBM cell lines, the M059K cell
line showed the strongest association between variants 9 or 13, and
each of the tested hypoxia markers. These associations potentially
suggested that variant 9 and 13 confer some survival advantage in
M059K cells under hypoxia, and thus may be responsible, at least in
part, for the hypoxia-tolerant phenotype of this cell line. In
contrast to M059K, the other hypoxia-tolerant cell line, M006×Lo,
appeared to not rely on any of the MB cancer-related variants to
survive the hypoxic tumor environment. Earlier reports revealed
that other hypoxia-defying globins were activated in that
particular cell line, such as Ngb and Cygb, in addition to
angiogenic factors, such as matrix metalloproteinase (MMP)-2 and
MMP-9 (3,8,11).
This suggests that activation of these globins and/or other
non-globin-dependent survival pathways may be responsible for this
cell line's hypoxia-tolerant phenotype. It is worth noting that Ngb
and Hb were also expressed and upregulated under low-oxygen
conditions in the M059K cell line (9,11);
therefore, they may also contribute to the hypoxia-tolerant status
demonstrated by this cell line.
MB variant 13 activation, as indicated by the
significant positive correlations with markers of hypoxia, in the
hypoxia-sensitive GBM cell lines appear to be minor or ineffective,
particularly in M010b, where a moderate correlation was only
observed with LDHA. Even in the M059J cell line, where a moderate
positive association was observed between variant 13 gene
expression and each of the tested markers of hypoxia, one would
expect that any role for variant 13 may be offset by the
hypoxia-induced downregulation of the other cancer-related
variants, 10 and 11. It is worth mentioning that both of these cell
lines express Ngb and Cygb (in addition to Hb in the M059K cell
line), and their expression is upregulated by hypoxia (3,9,11).
Thus far, it is not clear whether different members
of the globin family interact and, if so, whether possible
interactions may influence GBM cell hypoxia susceptibility. Future
studies aiming to explore the cross-talk among different globins in
hypoxia-sensitive and hypoxia-tolerant GBM cell lines should
provide useful knowledge into the mechanisms of cancer cell
survival under hypoxia. Further, exploring the contribution of
different MB variants to GBM cell hypoxia sensitivity should
provide insights into MB-dependent survival mechanisms.
Tumor xenografts established using the M006×Lo cell
line were used to investigate whether in vivo hypoxic
conditions could enhance MB expression. The M006×Lo cell line was
selected as it displays a robust hypoxia-tolerant phenotype.
Tumor-bearing mice received a single injection of pimonidazole HCl
90 min prior to sacrificing. Pimonidazole is considered to be a
gold standard for the assessment of tumor hypoxia (47), and in hypoxic cells (oxygen tensions
<10 mmHg), pimonidazole protein adducts are only detected
following systemic administration (48). Co-staining of the cells adjacent to
necrotic areas (using tissue landmarks) with pimonidazole and MB
conformed to the classic model of hypoxia (49), and implied that in vivo MB
expression may be regulated by oxygen-dependent mechanisms. Similar
staining patterns for CAIX, LDHA and VEGF in serial sections of
xenografts indicated that hypoxia is potentially involved in
regulating MB expression in cancer tissues. Either MB variant 2, or
MB cancer-related variants 9, 10, 11 and 13 encode the standard MB
protein as found in muscle, where cancer-related variants are more
strongly expressed (374–708-fold) compared with variant 2 (19). This suggests that expressed MB with
a similar staining pattern to pimonidazole and other hypoxia
markers is actually a translated protein of MB variants 9, 10, 11
or 13. The discrepancy between xenografts of M006×Lo cells and
in vitro culture of M006×Lo cells in terms of the expression
and association of MB protein and hypoxia markers could be
explained by several factors. First, all MB transcript variants
encode the same MB protein detected by MB antibody in the
xenografts of M006×Lo cells, which may indicate that MB expression
in this cell line is determined by the contribution of multiple MB
variants, rather than just one of them. Second, it was shown
previously that the oxygen consumption rates of these glioma cells
measured in vitro are poor predictors of changes likely to
occur in vivo (5). Finally,
the differences in nature between cell culture (in vitro)
and cells in xenografts (in vivo) in terms of time (>2
weeks is required to generate xenografts with volume of 200
mm3, compared with 48 h hypoxia in vitro, the
maximum duration of hypoxia exposure of M006×Lo cells in cell
culture), and the varied severity of hypoxia in the tumor
microenvironment in xenografts, as reflected by the distribution of
pimonidazole staining, a widely accepted marker of tumor hypoxia in
tumor specimens (50). These
factors likely contributed to the differences in outcomes observed
between cells and xenograft tissue. It should be noted that the
lack of RNA interference and overexpression experiments in the
studied glioma cell lines is a limitation of the present study;
however, this issue is currently being addressed.
The MB pattern of expression in xenografts is
similar to our previous observations with Ngb, suggesting that MB
and Ngb (and potentially other globins) are co-expressed in the
same cells in hypoxic microregions of M006×Lo-derived GBM tumor
xenografts (9). It should be noted
that neither heterotopic nor orthotopic xenograft models reflect
the tumor microenvironment and cell fate of GBM in human patients.
However, in the past 60 years, animal models have been used
extensively to study brain tumors (51), due in part to the lack of perfect
and reliable preclinical models (52). In the current study, mouse M006×Lo
xenograft tissue was solely used to confirm the associations
between MB and hypoxia markers, which were observed in vitro
using human GBM cell lines. Future work examining the co-expression
of MB variants and hypoxia markers in other xenograft models (for
example, using M059K or M010b cells) would be required to more
fully understand the variation in expression from one tumor to
another.
In previous studies, MB expression in a subset of
invasive breast cancer cases correlated with HIF-2α and CAIX, but
not with HIF-1α or GLUT1 (27), and
breast ductal carcinoma in situ immunostaining of GLUT1
correlated with the degree of MB expression (32). In contrast, ectopic MB expression
was significantly associated with overall survival in head and neck
squamous cell carcinoma, and did not correlate with the hypoxia
marker CAIX (18). Additionally,
overexpression of MB using lentivirus prevented hypoxia in tumor
cells, promoted differentiation and inhibited metastasis (53). These observations, and the very low
level (several hundred-fold lower than in muscle tissue) of
ectopically-expressed MB in healthy and tumorous tissues, and
cancer cell lines (19), question
its biological functions in this context. Apart from the effect of
hypoxia on MB expression, chemically-induced oxidative stress
(S-nitroso-N-acetylpenicillamine or H2O2) was
also found to increase MB expression in MCF7 breast cancer cells
(26).
According to the revised classification of tumors of
the CNS by the World Health Organization, GBMs are divided into IDH
mutant, wild-type or not otherwise specified (54). It is reported that ~90% of GBMs are
of a primary origin, whereas the rest represent secondary GBM that
progresses from low-grade diffuse astrocytoma or anaplastic
astrocytoma (55). Mutations in IDH
were found in <10% of primary glioblastomas (IDH wild-type
tumors constitute ~90% of cases), as well as ~80% of grade II and
III oligodendrogliomas, astrocytomas and secondary glioblastomas
(56). Patients with glioblastoma
carrying mutations in IDH exhibit an improved prognosis reflected
by longer median overall survival associated with these tumors
(54). To put the present findings
into a clinically relevant perspective, DepMap was used to check
for IDH1/2 mutations in the cell lines used in the present study
(36). IDH1/2 mutations were not
among the mutations reported in the M059J and M059K cell lines,
suggesting that they carry the wild-type gene. No data on the other
two cell lines, M010b and M006XLo, were found; however, in future
studies, they will be checked for IDH1/2 mutations via DNA
sequencing to analyze their characteristics from a clinical
perspective.
Finally, it is worth noting that the ability of a
cell to cope with decreased oxygen tension is not dependent on a
single set of genes. Different genes involved in cell growth and
death, and lipid metabolism, including insulin-like growth factor-1
receptor, fatty acid synthase, sterol regulatory element-binding
protein-1c and programmed cell death protein-1/programmed
death-ligand 1, have been shown to contribute significantly to
cancer cell hypoxia adaptation (57–59).
Therefore, differences among the tested GBM cell lines in terms of
their reliance on MB for hypoxia adaptation are not unexpected, and
highlight the overwhelming complexity of these heterogeneous
cancers.
In summary, it was shown for the first time, to our
knowledge, that in GBM cells, MB cancer-associated variants are
expressed under normoxic and hypoxic conditions, and exhibit
modest-to-strong correlations with hypoxia markers at different
time points following exposure to physiologically relevant
O2 tension. In addition, in xenografts, MB protein
exhibited staining patterns that were similar to hypoxia markers.
Therefore, ectopic expression of MB in GBM cells may be a part of
cellular defense mechanisms used by GBM tumors to survive and adapt
to the hypoxic tumor microenvironment, and consequently resist
chemo- and radiotherapy treatments.
Supplementary Material
Supporting Data
Acknowledgements
We thank Ms Maha Ahmed (Center for Aging and
Associated Diseases, Helmy Institute for Medical Sciences, Zewail
City of Science and Technology) for her administrative support.
Funding
This research was funded by Zewail City for Science
and Technology (internal fund), and Science and Technology
Development Fund of the Ministry of Scientific Research (grant. no.
12695).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
RET and ME conceived of the study. RET, IE, MM, OH,
JAT and ME performed experiments. MEE and ME analyzed the data. MEE
and RE wrote the original draft. RET, MEE, IE, JAT, and ME reviewed
and edited the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study used animal tissue obtained during
a previous study that was approved by the Animal Care Committee at
the Cross Cancer Institute.
Patient consent for participation
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Batash R, Asna N, Schaffer P, Francis N
and Schaffer M: Glioblastoma Multiforme, Diagnosis and Treatment;
Recent Literature Review. Curr Med Chem. 24:3002–3009. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pisapia DJ, Magge R and Ramakrishna R:
Improved pathologic diagnosis-forecasting the future in
glioblastoma. Front Neurol. 8:7072018. View Article : Google Scholar
|
|
3
|
Emara M, Turner AR and Allalunis-Turner J:
Hypoxic regulation of cytoglobin and neuroglobin expression in
human normal and tumor tissues. Cancer Cell Int. 10:332010.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Harada H: How can we overcome tumor
hypoxia in radiation therapy? J Radiat Res. 52:545–556. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Allalunis-Turner MJ, Franko AJ and
Parliament MB: Modulation of oxygen consumption rate and vascular
endothelial growth factor mRNA expression in human malignant glioma
cells by hypoxia. Br J Cancer. 80:104–109. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Franko AJ, Parliament MB, Allalunis-Turner
MJ and Wolokoff BG: Variable presence of hypoxia in M006 human
glioma spheroids and in spheroids and xenografts of clonally
derived sublines. Br J Cancer. 78:1261–1268. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Parliament MB, Franko AJ, Allalunis-Turner
MJ, Mielke BW, Santos CL, Wolokoff BG and Mercer JR: Anomalous
patterns of nitroimidazole binding adjacent to necrosis in human
glioma xenografts: Possible role of decreased oxygen consumption.
Br J Cancer. 75:311–318. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Emara M and Allalunis-Turner J: Effect of
hypoxia on angiogenesis related factors in glioblastoma cells.
Oncol Rep. 31:1947–1953. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Emara M, Salloum N and Allalunis-Turner J:
Expression and hypoxic up-regulation of neuroglobin in human
glioblastoma cells. Mol Oncol. 3:45–53. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Emara M, Turner AR and Allalunis-Turner J:
Adult, embryonic and fetal hemoglobin are expressed in human
glioblastoma cells. Int J Oncol. 44:514–520. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Emara M, Turner AR and Allalunis-Turner J:
Hypoxia differentially upregulates the expression of embryonic,
fetal and adult hemoglobin in human glioblastoma cells. Int J
Oncol. 44:950–958. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kendrew JC, Dickerson RE, Strandberg BE,
Hart RG, DAVIES DR, Phillips DC and Shore VC: Structure of
myoglobin: A three-dimensional Fourier synthesis at 2 A.
resolution. Nature. 185:422–427. 1960. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Totzeck M, Hendgen-Cotta UB, Rammos C,
Petrescu AM, Meyer C, Balzer J, Kelm M and Rassaf T: Assessment of
the functional diversity of human myoglobin. Nitric Oxide.
26:211–216. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xie LK and Yang SH: Brain globins in
physiology and pathology. Med Gas Res. 6:154–163. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Qiu Y, Sutton L and Riggs AF:
Identification of myoglobin in human smooth muscle. J Biol Chem.
273:23426–23432. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fraser J, de Mello LV, Ward D, Rees HH,
Williams DR, Fang Y, Brass A, Gracey AY and Cossins AR:
Hypoxia-inducible myoglobin expression in nonmuscle tissues. Proc
Natl Acad Sci USA. 103:2977–2981. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Roesner A, Mitz SA, Hankeln T and
Burmester T: Globins and hypoxia adaptation in the goldfish,
Carassius auratus. FEBS J. 275:3633–3643. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Meller S, VAN Ellen A, Gevensleben H,
Bicker A, Hankeln T, Gorr TA, Sailer V, Dröge F, Schröck F, Bootz
F, et al: Ectopic myoglobin expression is associated with a
favourable outcome in head and neck squamous cell carcinoma
patients. Anticancer Res. 36:6235–6241. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bicker A, Dietrich D, Gleixner E,
Kristiansen G, Gorr TA and Hankeln T: Extensive transcriptional
complexity during hypoxia-regulated expression of the myoglobin
gene in cancer. Hum Mol Genet. 23:479–490. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Meller S, Bicker A, Montani M, Ikenberg K,
Rostamzadeh B, Sailer V, Wild P, Dietrich D, Uhl B, Sulser T, et
al: Myoglobin expression in prostate cancer is correlated to
androgen receptor expression and markers of tumor hypoxia. Virchows
Arch. 465:419–427. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bicker A, Brahmer AM, Meller S,
Kristiansen G, Gorr TA and Hankeln T: The distinct gene regulatory
network of myoglobin in prostate and breast cancer. PLoS One.
10:e01426622015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Behnes CL, Bedke J, Schneider S, Küffer S,
Strauss A, Bremmer F, Ströbel P and Radzun HJ: Myoglobin expression
in renal cell carcinoma is regulated by hypoxia. Exp Mol Pathol.
95:307–312. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Oleksiewicz U, Daskoulidou N, Liloglou T,
Tasopoulou K, Bryan J, Gosney JR, Field JK and Xinarianos G:
Neuroglobin and myoglobin in non-small cell lung cancer:
Expression, regulation and prognosis. Lung Cancer. 74:411–418.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ruck P, Horny HP, Greschniok A, Wehrmann M
and Kaiserling E: Nonspecific immunostaining of blast cells of
acute leukemia by antibodies against nonhemopoietic antigens.
Hematol Pathol. 9:49–56. 1995.PubMed/NCBI
|
|
25
|
Zhang PJ, Goldblum JR, Pawel BR, Fisher C,
Pasha TL and Barr FG: Immunophenotype of desmoplastic small round
cell tumors as detected in cases with EWS-WT1 gene fusion product.
Mod Patho. 16:229–235. 2003. View Article : Google Scholar
|
|
26
|
Flonta SE, Arena S, Pisacane A, Michieli P
and Bardelli A: Expression and functional regulation of myoglobin
in epithelial cancers. Am J Pathol. 175:201–206. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kristiansen G, Rose M, Geisler C,
Fritzsche FR, Gerhardt J, Lüke C, Ladhoff AM, Knüchel R, Dietel M,
Moch H, et al: Endogenous myoglobin in human breast cancer is a
hallmark of luminal cancer phenotype. Br J Cancer. 102:1736–1745.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Allalunis-Turner MJ, Barron GM, Day RS
III, Fulton DS and Urtasun RC: Radiosensitivity testing of human
primary brain tumor specimens. Int J Radiat Oncol Biol Phys.
23:339–343. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Turcotte ML, Parliament M, Franko A and
Allalunis-Turner J: Variation in mitochondrial function in
hypoxia-sensitive and hypoxia-tolerant human glioma cells. Br J
Cancer. 86:619–624. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Catania A, Urban S, Yan E, Hao C, Barron G
and Allalunis-Turner J: Expression and localization of
cyclin-dependent kinase 5 in apoptotic human glioma cells. Neuro
Oncol. 3:89–98. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
DeHaan C, Habibi-Nazhad B, Yan E, Salloum
N, Parliament M and Allalunis-Turner J: Mutation in mitochondrial
complex I ND6 subunit is associated with defective response to
hypoxia in human glioma cells. Mol Cancer. 3:192004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kristiansen G, Hu J, Wichmann D, Stiehl
DP, Rose M, Gerhardt J, Bohnert A, ten Haaf A, Moch H, Raleigh J,
et al: Endogenous myoglobin in breast cancer is hypoxia-inducible
by alternative transcription and functions to impair mitochondrial
activity: A role in tumor suppression? J Biol Chem.
286:43417–43428. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Altschul SF, Madden TL, Schaffer AA, Zhang
J, Zhang Z, Miller W and Lipman DJ: Gapped BLAST and PSI-BLAST: A
new generation of protein database search programs. Nucleic Acids
Res. 25:3389–3402. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Koch CJ, Howell RL and Biaglow JE:
Ascorbate anion potentiates cytotoxicity of nitro-aromatic
compounds under hypoxic and anoxic conditions. Br J Cancer.
39:321–329. 1979. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ghandi M, Huang FW, Jane-Valbuena J,
Kryukov GV, Lo CC, McDonald ER III, Barretina J, Gelfand ET,
Bielski CM, Li H, et al: Next-generation characterization of the
Cancer Cell Line Encyclopedia. Nature. 569:503–508. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kleiter MM, Thrall DE, Malarkey DE, Ji X,
Lee DY, Chou SC and Raleigh JA: A comparison of oral and
intravenous pimonidazole in canine tumors using intravenous
CCI-103F as a control hypoxia marker. Int J Radiat Oncol Biol Phys.
64:592–602. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Grillo F, Bruzzone M, Pigozzi S, Prosapio
S, Migliora P, Fiocca R and Mastracci L: Immunohistochemistry on
old archival paraffin blocks: Is there an expiry date? J Clin
Pathol. 70:988–993. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Murray D, Mirzayans R, Scott AL and
Allalunis-Turner MJ: Influence of oxygen on the radiosensitivity of
human glioma cell lines. Am J Clin Oncol. 26:e169–e177. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Dengler VL, Galbraith M and Espinosa JM:
Transcriptional regulation by hypoxia inducible factors. Crit Rev
Biochem Mol Biol. 49:1–15. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kaluz S, Kaluzova M, Liao SY, Lerman M and
Stanbridge EJ: Transcriptional control of the tumor- and
hypoxia-marker carbonic anhydrase 9: A one transcription factor
(HIF-1) show? Biochim Biophys Acta. 1795:162–172. 2009.PubMed/NCBI
|
|
42
|
Cui XG, Han ZT, He SH, Wu XD, Chen TR,
Shao CH, Chen DL, Su N, Chen YM, Wang T, et al: HIF1/2α mediates
hypoxia-induced LDHA expression in human pancreatic cancer cells.
Oncotarget. 8:24840–24852. 2017.PubMed/NCBI
|
|
43
|
Turner KJ, Crew JP, Wykoff CC, Watson PH,
Poulsom R, Pastorek J, Ratcliffe PJ, Cranston D and Harris AL: The
hypoxia-inducible genes VEGF and CA9 are differentially regulated
in superficial vs. invasive bladder cancer. Br J Cancer.
86:1276–1282. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Chiche J, Brahimi-Horn MC and Pouyssegur
J: Tumour hypoxia induces a metabolic shift causing acidosis: A
common feature in cancer. J Cell Mol Med. 14:771–794. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kanatous SB, Mammen PP, Rosenberg PB,
Martin CM, White MD, Dimaio JM, Huang G, Muallem S and Garry DJ:
Hypoxia reprograms calcium signaling and regulates myoglobin
expression. Am J Physiol Cell Physiol. 296:C393–C402. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wystub S, Ebner B, Fuchs C, Weich B,
Burmester T and Hankeln T: Interspecies comparison of neuroglobin,
cytoglobin and myoglobin: Sequence evolution and candidate
regulatory elements. Cytogenet Genome Res. 105:65–78. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Tamaki N and Hirata K: Tumor hypoxia: A
new PET imaging biomarker in clinical oncology. Int J Clin Oncol.
21:619–625. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Hirakawa Y, Tanaka T and Nangaku M: Renal
Hypoxia in CKD; Pathophysiology and detecting methods. Front
Physiol. 8:992017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Brown JM and Wilson WR: Exploiting tumour
hypoxia in cancer treatment. Nat Rev Cancer. 4:437–447. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ljungkvist AS, Bussink J, Kaanders JH and
van der Kogel AJ: Dynamics of tumor hypoxia measured with
bioreductive hypoxic cell markers. Radiat Res. 167:127–145. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Huszthy PC, Daphu I, Niclou SP, Stieber D,
Nigro JM, Sakariassen PØ, Miletic H, Thorsen F and Bjerkvig R: In
vivo models of primary brain tumors: Pitfalls and perspectives.
Neuro Oncol. 14:979–993. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Patrizii M, Bartucci M, Pine SR and
Sabaawy HE: Utility of glioblastoma patient-derived orthotopic
xenografts in drug discovery and personalized therapy. Front Oncol.
8:232018. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Galluzzo M, Pennacchietti S, Rosano S,
Comoglio PM and Michieli P: Prevention of hypoxia by myoglobin
expression in human tumor cells promotes differentiation and
inhibits metastasis. J Clin Invest. 119:865–875. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Louis DN, Perry A, Reifenberger G, von
Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD,
Kleihues P and Ellison DW: The 2016 World Health Organization
Classification of Tumors of the Central Nervous System: A summary.
Acta Neuropathol. 131:803–820. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Huang J, Yu J, Tu L, Huang N, Li H and Luo
Y: Isocitrate dehydrogenase mutations in glioma: From basic
discovery to therapeutics development. Front Oncol. 9:5062019.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Megova M, Drabek J, Koudelakova V,
Trojanec R, Kalita O and Hajduch M: Isocitrate dehydrogenase 1 and
2 mutations in gliomas. J Neurosci Res. 92:1611–1620. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Liu Q, Guan JZ, Sun Y, Le Z, Zhang P, Yu D
and Liu Y: Insulin-like growth factor 1 receptor-mediated cell
survival in hypoxia depends on the promotion of autophagy via
suppression of the PI3K/Akt/mTOR signaling pathway. Mol Med Rep.
15:2136–2142. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Ezzeddini R, Taghikhani M, Somi MH, Samadi
N and Rasaee MJ: Clinical importance of FASN in relation to HIF-1α
and SREBP-1c in gastric adenocarcinoma. Life Sci. 224:169–176.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Koh YW, Lee SJ, Han JH, Haam S, Jung J and
Lee HW: PD-L1 protein expression in non-small-cell lung cancer and
its relationship with the hypoxia-related signaling pathways: A
study based on immunohistochemistry and RNA sequencing data. Lung
Cancer. 129:41–47. 2019. View Article : Google Scholar : PubMed/NCBI
|

















