Introduction
Colorectal cancer (CRC) is one of the most common
cancers in world, ranking third overall in terms of incidence rates
and second in terms of mortality rates, with >1.8 million new
cases and 861,663 death cases reported worldwide in 2018 (1). Both the incidence and mortality rates
of CRC have increased in China in the past decade; in 2018, the
latest epidemiological statistics of Globocan reported that the
incidence and mortality rates of CRC were 23.7 and 10.9,
respectively, per 100,000 (1).
Unfortunately, in the majority of patients, CRC is diagnosed at an
advanced stage, following the metastasis to adjacent or distant
organs (2); however, the mechanisms
regulating metastasis in CRC remain largely unknown. Therefore,
there is an urgent requirement to identify the molecular mechanisms
of CRC metastasis to provide novel therapeutic targets for the
treatment of the disease.
Endoplasmic reticulum (ER) stress is reportedly
involved in CRC metastasis (3). The
ER has established unique signaling pathways to combat stress,
which are collectively known as the unfolded protein response (UPR)
(4); glucose regulated protein 78
(GRP78) initiates the UPR and it has been demonstrated to promote
the resistance of CRC cells to oxaliplatin (5). Depending on the status of GRP78, the
ER transmembrane sensors, inositol-requiring enzyme 1 (IRE1),
protein kinase RNA activated-like ER kinase (PERK) and activating
transcription factor 6 (ATF6) are also involved in initiating
signaling pathways involved in the UPR (4). IRE1 catalyzes a unique splicing event
that removes 26 nucleotides from X-box-binding protein 1 (XBP1)
mRNA, and the activation of the IRE1/XBP1 pathway has been observed
to induce CRC cell invasion (3);
however, the mechanism underlying the IRE1/XBP1 pathway induction
of CRC cell invasion is not fully elucidated. The phosphorylation
of PERK activates the downstream signaling molecule, α-subunit of
eukaryotic initiation factor-2 (eIF2α), which effectively inhibits
protein synthesis (4), and has been
associated with the hypoxia-induced metastasis of cervical cancer
(6). Finally, the proteolytic
processing of ATF6 activates the ATF6 pathway, and ATF6 activation
was reported to be involved in pancreatic cancer stem cell
migration (7). However, the roles
of the PERK/eIF2α and ATF6 pathway in CRC migration are unknown. In
the present study, thapsigargin (TG) was used as an ER stress
inducer to irreversibly inhibit the sarco/ER Ca2+ ATPase
and promote rapid ER Ca2+ depletion (8).
Long non-coding RNAs (lncRNAs) are non-coding
transcripts of >200 nucleotides in length and certain lncRNAs
serve important roles in CRC metastasis (9,10).
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1),
also known as nuclear enriched abundant transcript 2 or LINC00047,
is a lncRNA (11). MALAT1 is found
to be overexpressed in colorectal cancer patients (12) and multiple studies have reported an
association between MALAT1 expression and CRC metastasis (9,13). The
first study demonstrating the UPR-induced regulation of lncRNA
expression was in a study of the flavivirus infection, whereby
MALAT1 expression was increased through the PERK pathway of the UPR
(14). However, the mechanisms
underlying increased MALAT1 expression levels in CRC are not clear,
in addition to whether the UPR pathway is involved in upregulating
MALAT1 expression in CRC. It is hypothesized that the ER stress
pathway regulates MALAT1 expression in CRC; thus, the present study
aimed to identify the association between the ER stress pathway,
MALAT1 expression and cell migration in CRC, in addition to
elucidating the roles of ER stress in CRC development.
Materials and methods
Patient studies
The present study was approved by the Ethics
Committee of The First Hospital of Hebei Medical University (no.
2016004). Written informed consent prior to the study was obtained
from all patients (n=38; 18 males, 20 females; average age=61.5
years). Patients were informed that they could withdraw from study
participation at any time. Thirty-eight CRC tissue samples were
collected from the First Hospital of Hebei Medical University
between October 2016 and March 2017. After surgical removal,
tissues were immediately frozen in liquid nitrogen then immediately
stored in a freezer at −80°C. Patients had not received local or
systemic treatment prior to the operation. The pathological
diagnosis of all cancer tissue samples was adenocarcinoma and the
clinicopathological features of patients are presented in Table I.
 | Table I.Clinicopathological characteristics
of patients with CRC. |
Table I.
Clinicopathological characteristics
of patients with CRC.
| Number of total
patients | 38 |
|---|
| Age, years | 61.5±15.2 |
| Sex, number (%) of
patients |
|
|
Male | 18 |
|
Female | 20 |
| Dukes, number (%)
of patients |
|
|
A,B | 22 |
|
C,D | 16 |
| Depth of invasion,
number (%) of patients |
|
|
T1,T2 | 18 |
|
T3,T4 | 20 |
| Location, number
(%) of patients |
|
|
Colon | 17 |
|
Rectum | 21 |
| Lymph node
metastasis, number (%) of patients |
|
|
Absent | 22 |
|
Present | 16 |
| Distant metastasis,
number (%) of patients |
|
|
Absent | 32 |
|
Present | 6 |
| Differentiation,
number (%) of patients |
|
|
Poor | 6 |
|
Moderate | 30 |
|
Well | 2 |
Reagents and plasmids
TG, a non-competitive inhibitor of the sarco/ER
Ca2+ ATPase that promotes rapid ER Ca2+
depletion and the elevation of cytoplasmic Ca2+
concentrations to induce ER stress (8), was purchased from BioVision, Inc. The
inhibitors, 4 µ8C (cat. no. S7272; IRE1/XBP1 pathway inhibitor, 1
µM), GSK2606414 (cat. no. S7307; PERK/eIF2α/ATF4 pathway inhibitor,
1 µM) and AEBSF (cat. no. S7378; ATF6 pathway inhibitor, 0.3 µM),
were purchased from Selleck Chemicals; anti-GRP78 (product no.
3177), anti-eIF2α (product no. 5324) and anti-phospho-eIF2α
(product no. 3398) primary antibodies were obtained from Cell
Signaling Technology, Inc.; anti-XBP1 (product code ab198999) and
anti-ATF6 (product code ab122897) primary antibodies were purchased
from Abcam; the anti-ATF4 primary antibody (WL02330) was purchased
from Wanleibio Co., Ltd.; the anti-β-actin primary antibody (cat.
no. 60008-1-Ig) was purchased from Wuhan Sanying Biotechnology
(ProteinTech Group, Inc.); and secondary antibodies (anti-mouse IgG
cat no. A23910; anti-rabbit IgG catalogue no: A23720) were
purchased from Abbkine Scientific Co., Ltd. All primers and small
interfering RNAs (siRNAs) were synthesized and purchased from
Sangon Biotech Co., Ltd.
Cell culture and reagents
The human CRC cell line HCT116 was obtained from
Professor Xiaofeng Sun at the Division of Oncology, Department of
Clinical and Experimental Medicine, Linköping University, Sweden.
SW620, SW1116 and HT29 cells were obtained from Professor Jun Yu at
the Department of Medicine and Therapeutics, The Chinese University
of Hong Kong, Hong Kong. HCT116 and HT29 cells were cultured in
McCoy's 5A medium (Gibco; Thermo Fisher Scientific, Inc.), and
SW620 and SW1116 cells were cultured in DMEM (Gibco; Thermo Fisher
Scientific, Inc.), supplemented with 10% FBS (Gibco; Thermo Fisher
Scientific, Inc.) and 1% penicillin-streptomycin (Invitrogen;
Thermo Fisher Scientific, Inc.). All cells were maintained in a
humidified atmosphere at 37°C and 5% CO2.
Cell Counting Kit-8 assay
Five thousands cells were seeded into 96-well plates
and treated with TG (0–10 µM) for 24 h at 37°C. Following
incubation, 10 µl CCK-8 reagent (Dojindo Molecular Technologies,
Inc.) was added to each well, according to the manufacturer's
protocol. Following incubation for 2 h, the absorbance was
determined using a Promega GloMax Luminescence detector, with each
experiment performed in triplicate.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from frozen tissues (~50 µg)
using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol, and
subsequently resuspended in 50 µl nuclease-free water. A total of 1
µg RNA was reverse-transcribed into cDNA using the PrimeScript RT
kit (Takara Bio Inc., RT temperature protocol: 37°C for 15 min,
85°C for 5 sec). qPCR was subsequently performed using the Power
SYBR® Green Master mix (Applied Biosystems; Thermo
Fisher Scientific, Inc.), according to the manufacturer's protocol
(step 1: 95°C for 10 min; step 2 for 40 cycles: 95°C for 15 sec,
60°C for 1 min). The following primer pairs were used for the qPCR:
MALAT1 forward, 5′-GTTACTCTTTTTTCCCCCCACCCCC-3′ and reverse,
5′-TTCTCCCCCACCCTCTCTCTTCCCT-3′; GRP78 forward,
5′-GCCTGTATTTCTAGACCTGCC-3′ and reverse,
5′-TTCATCTTGCCAGCCAGTTG-3′; XBP1 forward, 5′-AATGAAGTGAGGCCAGTGG-3′
and reverse, 5′-TCAATACCGCCAGAATCCATG-3′; ATF4 forward,
5′-CCTTCACCTTCTTACAACCT-3′ and reverse, 5′-GTAGTCTGGCTTCCTATCTC-3′;
and GAPDH forward, 5′-ACCCACTCCTCCACCTTTG-3′ and reverse,
5′-CTCTTGTGCTCTTGCTGGG-3′ (15).
The expression levels were normalized to the internal reference
gene GAPDH and quantified using the 2−ΔΔCq method
(16).
Migration assay
For migration assays, Transwell plates with 8-µm
pores (BD Biosciences) were used. Briefly, a total of
2×105 cells were plated in the upper chambers of
Transwell plates in serum-free medium (McCoy's 5A or DMEM medium,
supplemented with 1% penicillin-streptomycin). Culture medium
(McCoy's 5A or DMEM medium, supplemented with 10% FBS and 1%
penicillin-streptomycin) was plated in the lower chambers.
Following incubation at 37°C for 24 h, the non-invasive cells
remaining in the upper chamber of the Transwell plate were removed
with a cotton swab. Migratory cells were stained with a Diff-Quick
stain kit according to the manufacturer's protocol and counted
using a light microscope (magnification, ×100).
Western blotting
Following 24 h of 0.01 µM TG treatment, total
protein was extracted from 106 cells using an SDS sample
buffer [50 mM Tris-HCl, 2% SDS, 1% glycerol, 6% β-mercaptoethanol,
1% protease inhibitor cocktail (cat. no. HY-K0010; MCE)] and
processed for western blotting analysis as previously described
(17). Briefly, 5–10 µg proteins
(measured by BCA protein assay kit and detected by Promega Glomax
Luminometer) were separated via 10% SDS-PAGE and separated proteins
were transferred onto a PVDF membrane (EMD Millipore). The membrane
was blocked (5% skim milk at 4°C overnight) and probed (4°C
overnight) with the following primary antibodies: Anti-GRP78
(1:1,000), anti-eIF2α (1:1,000), anti-phospho-eIF2α (1:1,000),
anti-XBP1 (1:350), anti-ATF6 (1:500), anti-ATF4 (1:500) and
anti-β-actin (1:3,500). Following the primary antibody incubation,
the membrane was washed with 1X TBST and incubated at room
temperature with DyLight fluorescent dyes-conjugated secondary
antibodies (1:2,500) for 1 h. Protein bands were visualized using
the Odyssey CLx Imaging System (LI-COR Biosciences). ImageJ
software (National Institutes of Health) was used for quantitative
analysis.
Cell transfection
To confirm the effects of MALAT1 knockdown on
migration, MALAT1 gene expression was knocked down using siRNA.
Cells (5×105) were transfected with 50 nM siRNA-MALAT1
(si-MALAT1 forward, 5′-GGAAGUAAUUCAAGAUCAATT-3′ and reverse,
5′-UUGAUCUUGAAUUACUUCCTT-3′; si-MALAT1-2 forward,
5′-GGGCUUCUCUUAACAUUUAUU-3′ and reverse,
5′-UAAAUGUUAAGAGAAGCCCUU-3′; or si-control forward,
5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse,
5′-ACGUGACACGUUCGGAGAATT-3′) using Effectene transfection reagent
(Qiagen GmbH), according to the manufacturer's protocol. Following
24 h of transfection at 37°C in an incubator, cells were treated
with 0.01 µM TG for 24 h. si-MALAT1 was selected for use in future
experiments.
Statistical and bioinformatics
analysis
Statistical analysis was performed using GraphPad
Prism 7 (GraphPad Software, Inc.) and SPSS Statistics version 21
(IBM Corp.). All data are expressed as the mean ± SD. Statistical
differences between groups were determined using two-way ANOVA and
corrected for multiple comparisons using Sidak statistical
hypothesis test (Figs. 1, 3, 4D and
E, 5B-F and 6A), one-way ANOVA and corrected for
multiple comparisons using Dunnett's statistical hypothesis test
(Fig. 4A and B), Student's t-tests
(Fig. 2), and non-parametric
Spearman's rank correlation coefficient (Fig. 7). P<0.05 was considered to
indicate a statistically significant difference. The binding site
sequences were identified using bioinformatics analysis platform
(the JASPAR 2018 database, http://jaspar.genereg.net/).
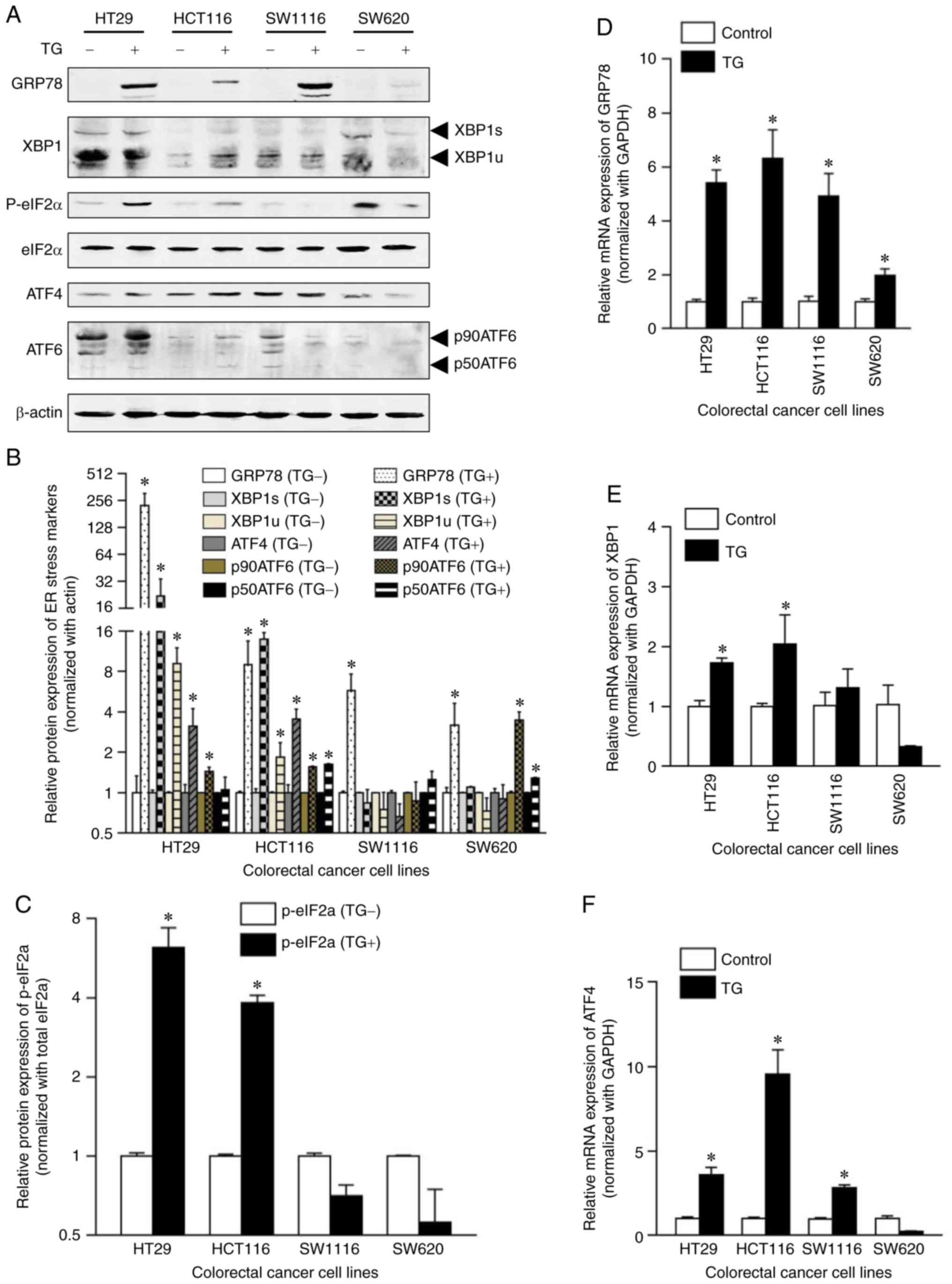 | Figure 5.Effect of TG treatment on the
expression levels of unfolded protein response-associated molecules
in human CRC cell lines. (A) Western blot analysis of the
expression levels of GRP78, XBP1s, XBP1u, ATF4, p90ATF6, p50ATF6,
eIF2α, phosphorylated eIF2α and β-actin in CRC cell lines with or
without 24 h treatment with 0.01 µM TG. (B) Protein expression
levels of GRP78, XBP1s, XBP1u, ATF4, p90ATF6 and p50ATF6 were
semi-quantified and the data was normalized to the loading control
β-actin. Data are presented as the mean ± SD. (C) Expression levels
of phosphorylated eIF2α were semi-quantified and the data was
normalized to total eIF2α expression levels. Data are presented as
the mean ± SD. (D-F) Following 24-h treatment with 0.01 µM TG, (D)
GRP78, (E) XBP1 and (F) ATF4 mRNA expression levels were detected
by reverse transcription quantitative-PCR. The ratios of
GRP78/GAPDH, XBP1/GAPDH and ATF4/GAPDH were presented as induction
(n-fold) relative to the control. Data are presented as the mean ±
SD. *P<0.05. GRP78, glucose regulated protein 78; XBP1,
X-box-binding protein 1; ATF, activating transcription factor;
eIF2α, α-subunit of eukaryotic initiation factor-2; TG,
thapsigargin; CRC, colorectal cancer. |
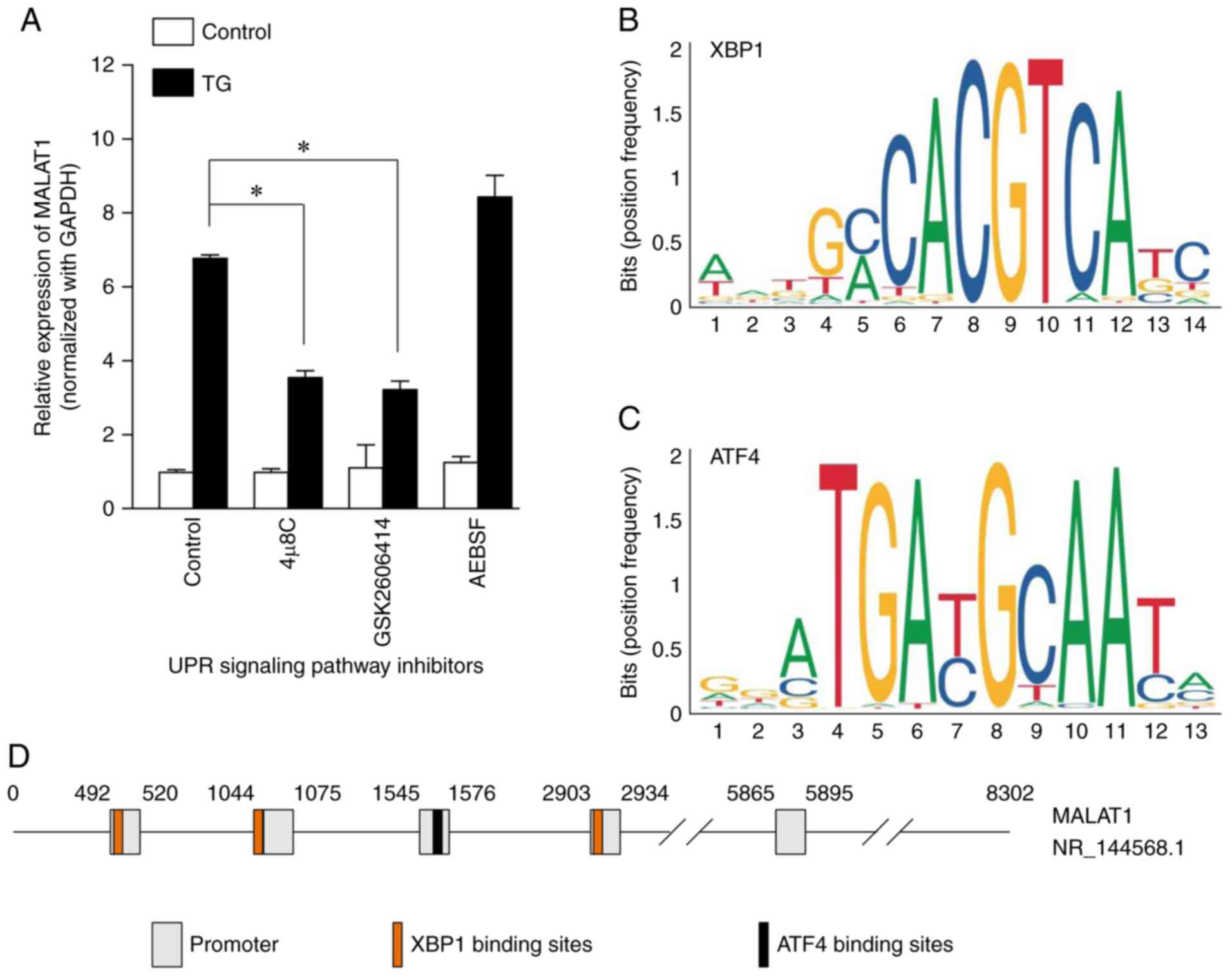 | Figure 6.Effect of UPR signaling pathway
inhibitors on MALAT1 expression levels and prediction of MALAT1
transcription regulation sites. (A) Effect of UPR signaling pathway
inhibitors on MALAT1 expression levels. UPR signaling pathway
inhibitors, 4 µ8C (IRE1/XBP1 pathway inhibitor; 1 µM; 24 h),
GSK2606414 (PERK/eIF2α/ATF4 pathway inhibitor; 1 µM; 24 h) and
AEBSF (ATF6 pathway inhibitor; 0.3 µM; 24 h), were used to inhibit
the activation of their respective signaling pathways in HCT116
cells. Expression levels of long non-coding RNA MALAT1 were
subsequently detected using reverse transcription quantitative-PCR.
The ratio of MALAT1/GAPDH was presented as induction (n-fold)
relative to the control. Data are expressed as the mean ± SD.
*P<0.05. (B and C) The binding sites sequences of (B) XBP1 and
(C) ATF4 transcription factors were presented as the position
frequency matrices in humans. (D) MALAT1 promoter regions (gray
frame; predicted by FPROM data sites) and binding sites of XBP1
(orange frame) and ATF4 (black frame) are presented. UPR, unfolded
protein response; MALAT1, metastasis-associated lung adenocarcinoma
transcript 1; IRE1, inositol-requiring enzyme 1; XBP1,
X-box-binding protein 1; PERK, protein kinase R (PKR)-like ER
kinase; eIF2α, α-subunit of eukaryotic initiation factor-2; ATF,
activating transcription factor. |
Results
Effect of TG on cell viability in the
human CRC cell lines
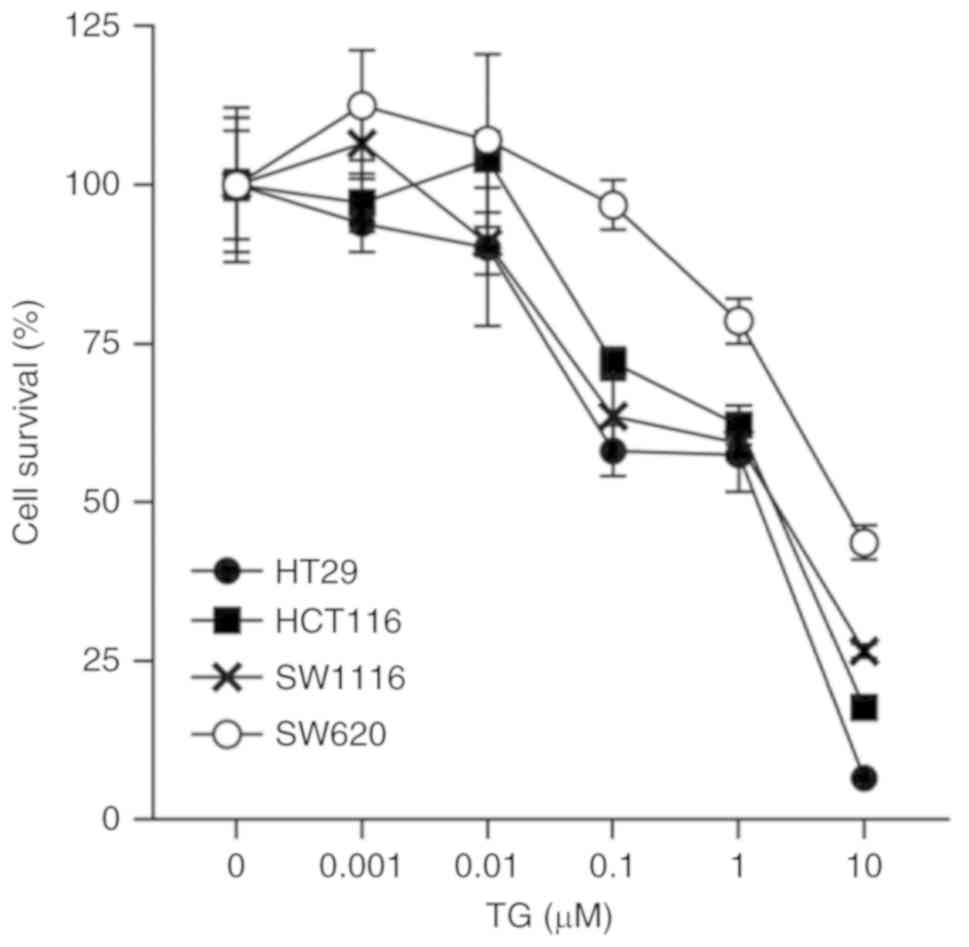
The effect of TG (0–10 µM) treatment for 24 h on the
cell viability of CRC lines was determined (Fig. 1). Cell death was successfully
induced by 0.1 µM TG in HT29, HCT116 and SW1116 cell lines
(58.12±4.03, 72.03±2.37 and 63.59±5.7, respectively, P<0.0001),
but not in the SW620 cells (96.84±3.92; P=0.98). Cell death
occurred at a higher rate when treated with 10 µM TG in the HT29,
HCT116, SW1116 and SW620 cells (6.59±0.31, 17.72±1.31, 26.55±1.1
and 43.66±2.73, respectively; P<0.0001). Cell death was not
induced by 0.01 µM TG in all four CRC cell lines. Thus, 0.01 µM TG
was selected to use in subsequent studies (Fig. 1).
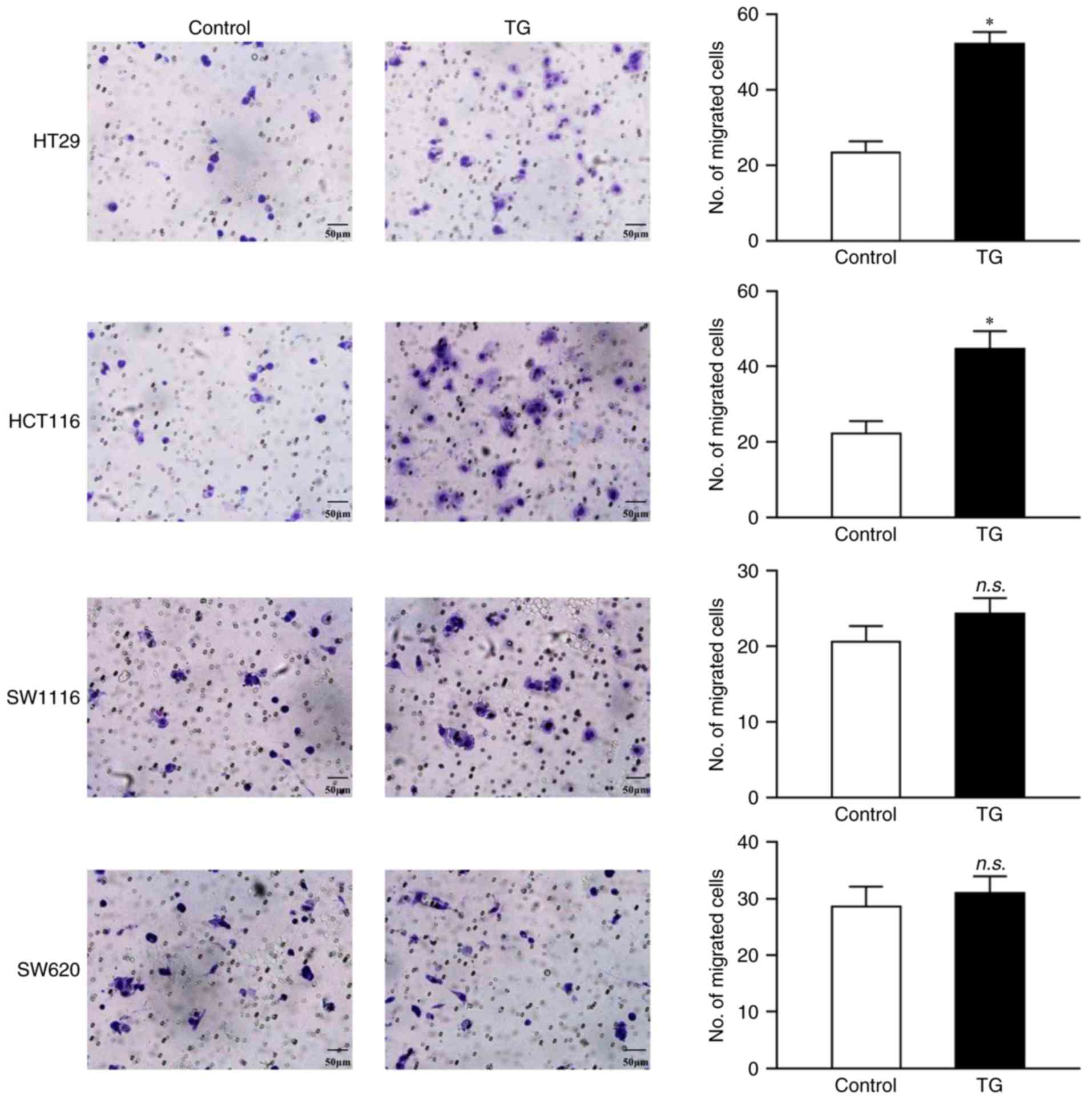
Effects of low dose TG on human CRC
migration
Next, the effect of TG on CRC cell migration was
investigated. Treatment with 0.01 µM TG for 24 h increased cell
migration in HT29 (52.33±3.06 vs. 23.33±3.05; P=0.0003) and HCT116
(44.67±4.73 vs. 22.33±3.21; P=0.0025) cells, but not in SW1116
(24.33±2.08 vs. 20.67±2.10; P=0.097) or SW620 (31.00±3.00 vs.
28.67±3.51; P=0.43; Fig. 2)
cells.
TG-induced cell migration is
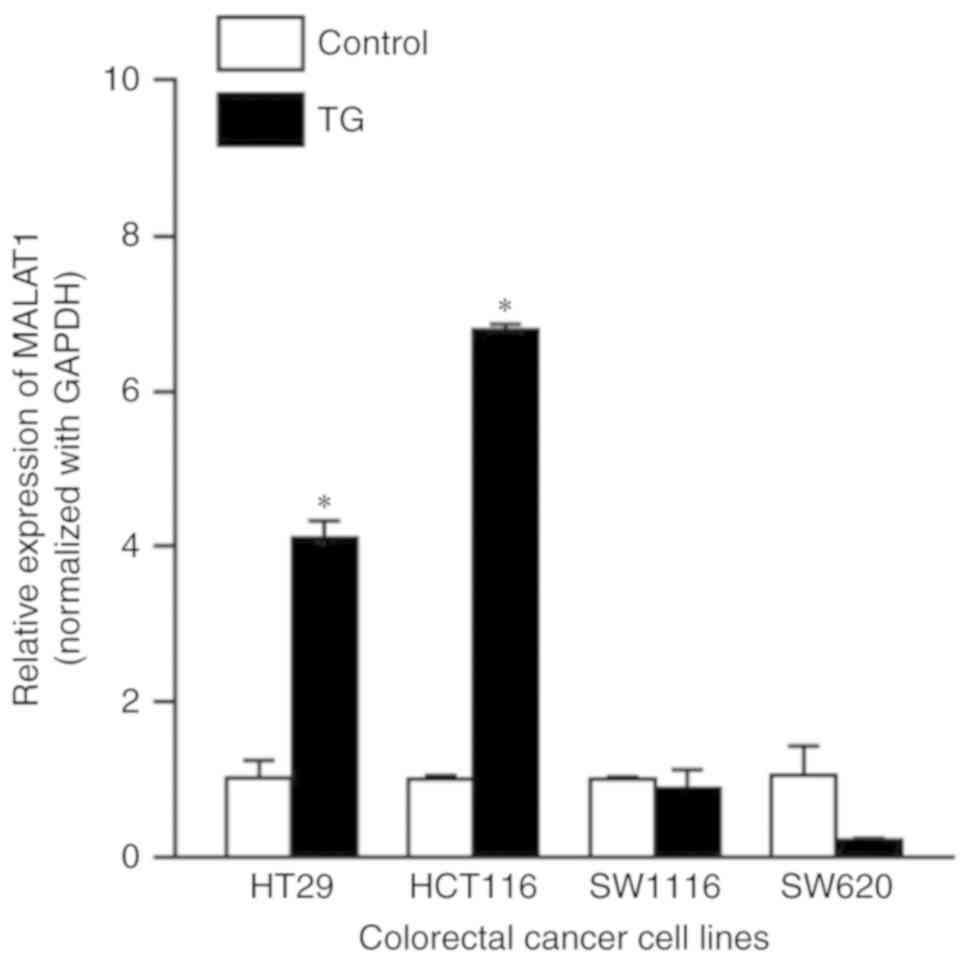
associated with increased expression levels of MALAT1
Following TG treatment, MALAT1 expression levels
were significantly increased in HT29 (4.11±0.22 vs. 1±0.23;
P<0.0001) and HCT116 (6.79±0.07 vs. 1±0.05; P<0.0001) cells,
but not in SW1116 (0.89±0.24 vs. 1±0.03; P=0.99) or SW620
(0.23±0.02 vs. 1±0.38; P=0.002; Fig.
3) cells.
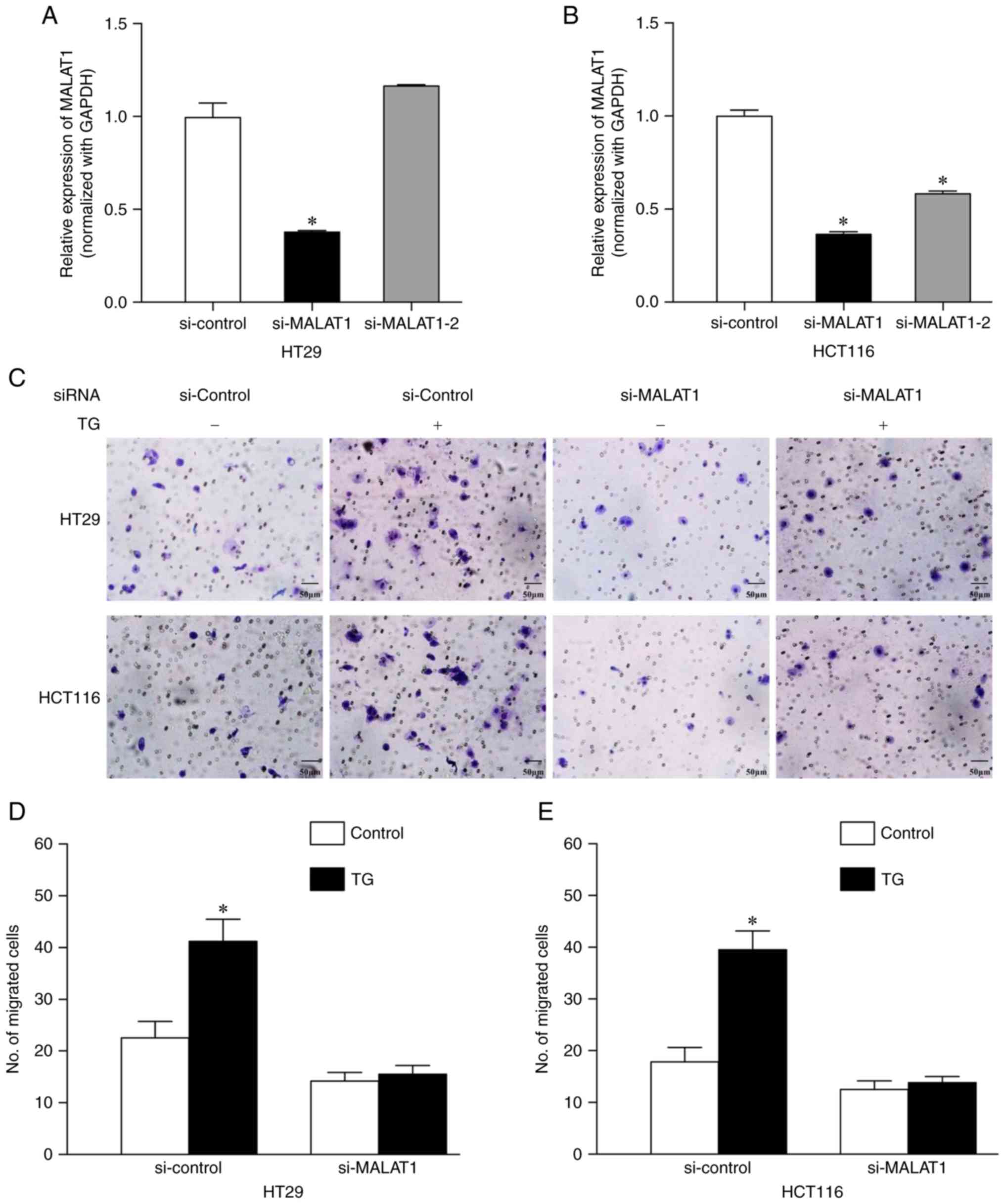
Knockdown of MALAT1 reverses
TG-induced cell migration
MALAT1 gene expression levels were knocked down
using siRNA and rescue experiments of migration were subsequently
performed to identify the role of MALAT1 in CRC. The expression
levels of MALAT1 were significantly decreased by si-MALAT1 in HT29
[0.38±0.004 vs. 1±0.07 (si-control group); P=0.0001] and HCT116
[0.37±0.010 vs. 1±0.03 (si-control group); P<0.0001] cell lines
(Fig. 4A and B). si-MALAT1-2
reduced MALAT1 expression levels in HCT116 cells [0.58±0.012 vs.
1±0.031 (si-control group); P<0.0001], but not in HT29 cells
[1.17±0.003 vs. 1±0.072 (si-control group); P>0.05;
Fig. 4A and B]. Thus, si-MALAT1 was
selected as the siRNA to use for subsequent experiments.
The knockdown of MALAT1 reversed TG-induced cell
migration in HT29 [22.7±3.1 (si-control) vs. 41.3±4.2 (si-control
and TG+); P=0.0002; 14.3±1.5 (si-MALAT1) vs. 15.7±1.5
(si-MALAT1 and TG+); P=0.93] and HCT116 [18.0±2.6
(si-control) vs. 39.7±3.5 (si-control and TG+);
P<0.0001; 12.7±1.5 (si-MALAT1) vs. 14.0±1.0 (si-MALAT1 and
TG+); P=0.89] cells (Fig.
4C-E).
Effects of TG on the expression levels
of UPR-associated molecules in human CRC cell lines
To analyze the mechanism of TG-induced MALAT1
expression, the expression levels or activation of UPR-associated
molecules (GRP78, XBP1s and XBP1u of the IRE1 signaling pathway,
eIF2α, phospho-eIF2α, ATF4 of the PERK signaling pathway, p90ATF6
and p50ATF6 of the ATF6 signaling pathway) were analyzed using
western blotting and RT-qPCR. GRP78 (226.38±80.20 vs. 1±0.34;
P=0.006), XBP1s (21.95±11.98 vs. 1±0.048; P=0.0198), XBP1u
(9.19±2.84 vs. 1±0.02; P=0.0052), ATF4 (3.12±1.12 vs. 1±0.15;
P=0.0155) and p90ATF6 (1.44±0.10 vs. 1±0.004; P=0.0017) protein
expression levels were significantly increased in TG-treated HT29
cells compared with the control group, and TG treatment
successfully induced the phosphorylation of eIF2α (6.20±1.18 vs.
1±0.030; P<0.0001; Fig. 5A-C).
In TG-treated HCT116 cells, the expression levels of GRP78
(9.00±4.55 vs. 1±0.023; P=0.0195), XBP1s (13.89±1.70 vs. 1±0.07;
P=0.00031), XBP1u (1.84±0.53 vs. 1±0.015; P=0.024), ATF4 (3.55±0.66
vs. 1±0.15; P=0.0017), p90ATF6 (1.56±0.024 vs. 1±0.004;
P<0.0001), p50ATF6 (1.63±0.018 vs. 1±0.003; P<0.0001) and
phosphorylated eIF2α (3.83±0.25 vs. 1±0.017; P<0.0001) were
increased compared with the control group (Fig. 5A-C). In TG-treated SW1116 cells, the
expression level of GRP78 (5.75±1.93 vs. 1±0.028; P=0.008) was
significantly increased compared with the control group (Fig. 5A-C). In TG-treated SW620 cells, the
expression levels of GRP78 (3.19±1.46 vs. 1±0.094; P=0.029),
p90ATF6 (3.47±0.53 vs. 1±0.026; P=0.0013) and p50ATF6 (1.29±0.017
vs. 1±0.007; P<0.0001) were significantly increased compared
with the control group (Fig.
5A-C).
Following TG treatment, GRP78 mRNA expression levels
were significantly increased in HT29, HCT116, SW1116 and SW620
cells (5.41±0.48, 6.32±1.06, 4.92±0.84 and 1.97±0.25, respectively;
P<0.0001) compared with the control group (Fig. 5D). XBP1 mRNA expression levels were
significantly increased in HT29 (1.73±0.08 vs. 1±0.096; P=0.081)
and HCT116 (2.04±0.49 vs. 1±0.05; P=0.0034) cells compared with the
control group (Fig. 5E) and ATF4
mRNA expression levels were significantly increased in HT29
(3.59±0.45 vs. 1±0.1; P=0.0008), HCT116 (9.53±1.46 vs. 1±0.084;
P<0.0001) and SW1116 (2.82±0.17 vs. 1±0.053; P=0.026) cells
compared with the control group (Fig.
5F). These findings indicated that TG may induce the activation
of the IRE1 and PERK UPR signaling pathways in HT29 and HCT116
cells; however, TG cannot induce the activation of these two
signaling pathways in SW1116 and SW620 cells. Thus, it is
hypothesized that TG-induced MALAT1 overexpression may be
associated with the IRE1 and PERK signaling pathways.
Effects of UPR signaling pathway
inhibitors on MALAT1 expression
To further confirm the hypothesis, the UPR signaling
pathway inhibitors 4 µ8C (IRE1 pathway inhibitor), GSK2606414 (PERK
pathway inhibitor) and AEBSF (ATF6 pathway inhibitor) were used to
inhibit the activation of their respective signaling pathways.
HCT116 cell were selected to carry out the rescue experiment
because the IRE1, PERK and ATF6 signaling pathways were activated
by TG (Fig. 5). TG-induced MALAT1
overexpression was significantly inhibited by 4µ8C [6.79±0.072
(control and TG+) vs. 3.57±0.16 (4 µ8C and
TG+); P<0.0001] and GSK2606414 [6.79±0.072 (control
and TG+) vs. 3.24±0.21 (GSK2606414 and TG+);
P<0.0001], but not by AEBSF [6.79±0.072 (control and
TG+) vs. 8.45±1.56 (AEBSF and TG+);
P<0.05; Fig. 6A]. These data
indicated that TG-induced MALAT1 expression was associated with the
IRE1 and PERK signaling pathways. To further analyze the molecular
mechanisms of MALAT1 upregulation, the binding sites of XBP1 and
ATF4, two transcription factors, were determined. The binding sites
sequences of XBP1 and ATF4 were identified using the JASPAR 2018
database (http://jaspar.genereg.net/) and
presented as position frequency matrices in humans (Fig. 6B and C). The promoter of MALAT1 was
predicted by FPROM data sites (18), with 5 promoter regions (Fig. 6D) predicted. Near the promoter
regions of MALAT1, three binding sites for XBP1 and one ATF4
binding site were successfully predicted (Fig. 6D).
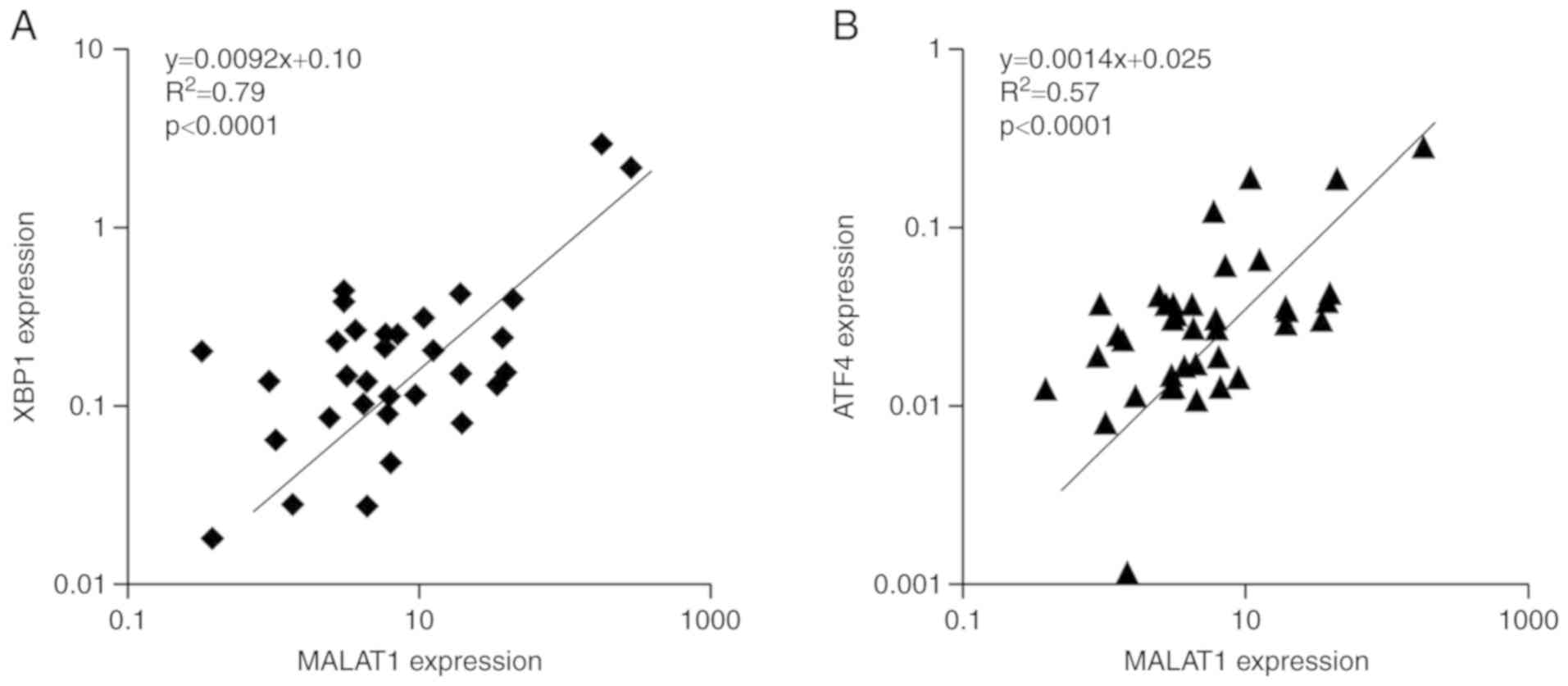
Correlation analysis between XBP1,
ATF4 and MALAT1 mRNA expression levels in patients with CRC
XBP1, ATF4 and MALAT1 mRNA expression levels were
detected by RT-qPCR and the correlation between the genes was
analyzed using Spearman's rank correlation coefficient. The
expression of lncRNA MALAT1 was positively correlated with XBP1
(R2=0.79; P<0.0001; Fig.
7A) and ATF4 (R2=0.57; P<0.0001; Fig. 7B) in CRC tissue samples. Thus, the
relationship between MALAT1 expression and ER stress was further
verified.
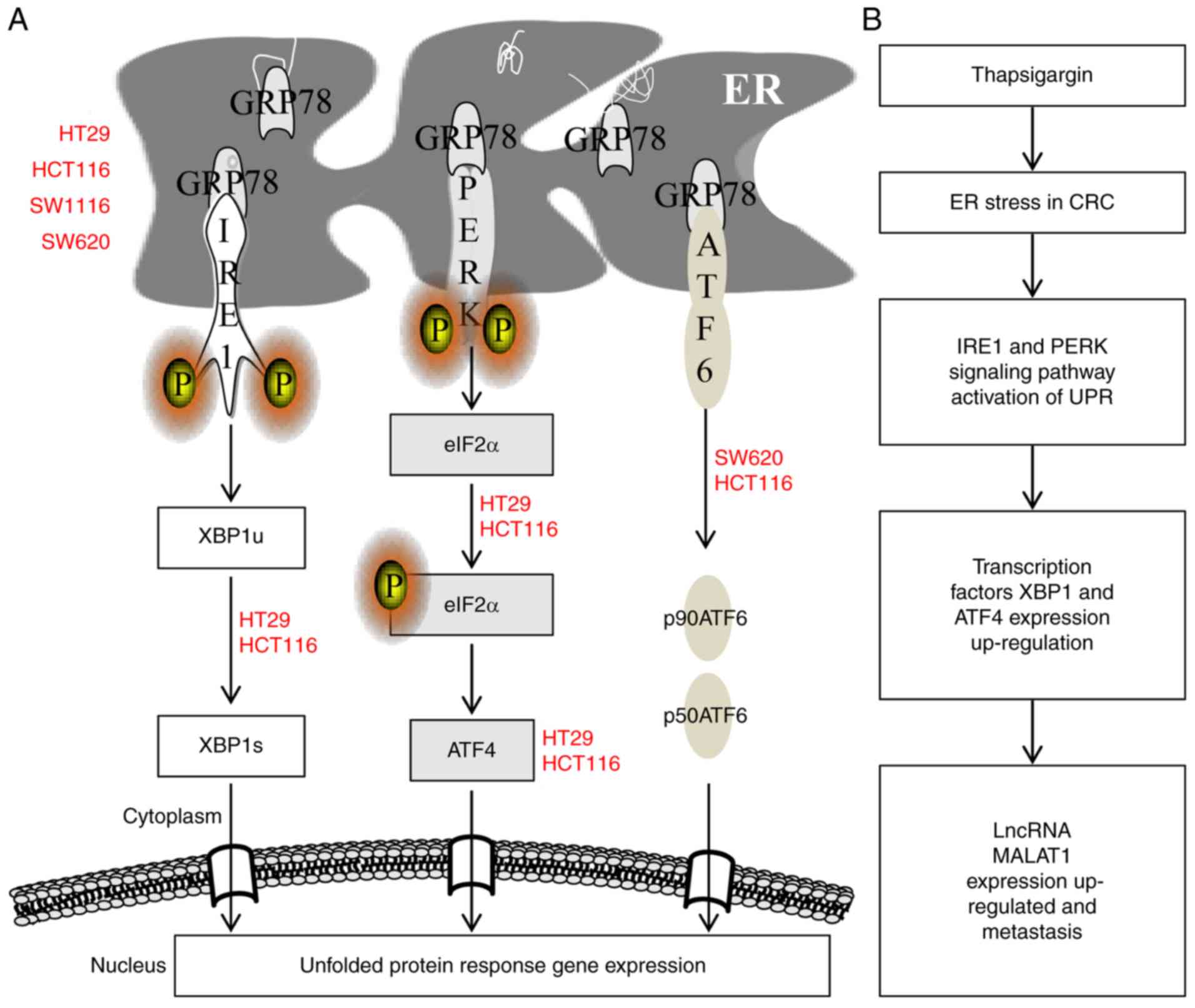
The overall activation of the ER stress pathway in
the four CRC cell lines was summarized in a schematic diagram
(Fig. 8A) and the hypothesized
mechanism of TG-induced increases in lncRNA MALAT1 expression
levels is presented in Fig. 8B.
Discussion
In the present study, it was demonstrated that: i)
Low dose TG induced migration in HT29 and HCT116 cells, but not
SW1116 and SW620 cells, the effect of which was linked to the
enhanced expression levels of MALAT1; ii) the knockdown of MALAT1
using siRNA reversed this TG-induced promotion of cell migration;
iii) TG-induced MALAT1 expression was associated with the
activation of the IRE1/XBP1 and PERK/eIF2α/ATF4 signaling pathways;
and iv) the XBP1 and ATF4 binding sites were found within MALAT1
gene promoter regions. To the best of our knowledge, this study was
the first to report that lncRNA MALAT1 expression levels were
regulated by the IRE1 signaling pathway of the UPR in CRC.
The ER stress activation and the unfolded protein
response (UPR) triggers, response to cancer cellular stress
conditions including glucose deprivation, hypoxia, proteins folding
and secretion of proteins, the denouement is either the restoration
of homeostasis or cell death (19).
Some studies ascertained that ER stress was correction with CRC
progression. ER stress-related ATF6 upregulated cancerous inhibitor
of protein phosphatase 2A contributing to poor prognosis of colon
cancer (20). ATF6 induced
intestinal dysbiosis to promote colorectal tumorigenesis through
innate immune response (21).
The oncogenic roles of XBP1 in CRC and other types
of cancer have been reported in numerous studies; the activation of
the IRE1/XBP1 pathway induced cell proliferation and invasion in
CRC (3), whereby XBP1 was
demonstrated to promote CRC invasion through VEGFR2 (22). In prostate cancer, the IRE1/XBP1
signaling pathway promoted carcinogenesis by activating c-Myc
signaling (23); in oral squamous
cell carcinoma, XBP1 promoted cancer invasion and it was associated
with poor prognosis (24); in
breast cancer, the expression levels of XBP1s in the nucleus were
correlated with shorter survival (25); whereas in ovarian cancer, the
IRE1/XBP1 signaling pathway controlled T-cell functions, thus,
mediating ER stress or targeting the IRE1/XBP1 pathway may restore
the antitumor ability of T-cells (26). In addition, XBP1 positively
regulated the cytolytic activity of human natural killer cells
against leukemia cells (27); in
hepatocellular carcinoma, the IRE1/XBP1 pathway controlled the
expression of interleukin-6 (IL-6) and promoted hepatocarcinoma
progression (28); and in
oropharyngeal carcinoma without papillomavirus, the IRE1/XBP1
pathway induced resistance to radiotherapy by mediating IL-6
production (29). Thus, the
transcription factor XBP1 may be a potential target to mediate
tumor immunology and block cancer progression.
ATF4 has been observed to serve important roles in
ER stress-induced apoptosis (30)
and radiotherapy (31) or
chemotherapy sensitivity (32). In
CRC cells, the activation of the PERK/ATF4 signaling pathway
promoted resistance to 5-fluorouracil (33) and increased expression levels of
ATF4 were associated with glucose deprivation-induced
chemoresistance (34). In prostate
cancer, ATF4 protein expression levels were increased in the cancer
tissue compared with benign prostate tissue (35) and similarly, in breast cancer, ATF4
expression levels were increased in HER2+ breast cancer,
which promoted cell migration through the activation of zinc finger
E-box binding homeobox 1 (ZEB1) and the downregulation of
E-cadherin (36). ATF4 was
associated with cell cycle progression in estrogen receptor
negative breast cancer, which was due to its regulation over the
GSK3β/β-catenin/cyclin D1 pathway (37). Thus, suggesting that the
transcription factor ATF4 may be associated with cancer progression
through regulation of the cell cycle and cell migration.
Overall, the results of the present study indicated
that low dose TG may promote CRC cell migration by upregulating the
expression levels of lncRNA MALAT1; and the TG-induced increased
expression levels of MALAT1 were associated with the activation of
the IRE1/XBP1 and PERK/eIF2α/ATF4 signaling pathways. The XBP1 and
ATF4 binding sites were predicted to be located in the MALAT1 gene
promoter regions; however, the direct interaction between MALAT1
and XBP1 or ATF4 was not verified in this study and will require
further investigation in the future. In conclusion, ER stress may
provide reasoning for the upregulated MALAT1 expression and
metastasis observed in CRC, and ER stress-associated genes,
especially XBP1 and ATF4, may represent potential targets for
controlling metastasis in CRC.
Acknowledgements
The authors thank Professor Xiaofeng Sun and
Professor Jun Yu for providing cells and technical support.
Funding
The study was supported by The National Natural
Science Foundation of China (grant no. 81572758), The Natural
Science Foundation of Hebei (grant no. H2017206286), The Foundation
for Distinguished Young Talents in Higher Education of Hebei (grant
no. BJ2018042), The International Science and Technology
Cooperation Program of China (grant no. 2014DFA31150), the
Latitudinal Projects Foundation from Hebei province (grant nos.
CY201614, zh2018002 and 162777271) and The Spark Program of the
First Hospital of Hebei Medical University (grant no.
XH201701).
Availability of data and materials
The binding site sequences analyzed in the present
study are publicly available from the JASPAR 2018 database
(http://jaspar.genereg.net/).
Authors' contributions
XJ and ZZ conceived and designed the experiments.
DL, WY, GW, JL, YL and XS performed the experiments. YW, CZ and JL
collected and analyzed the data. XJ and ZZ interpreted the findings
and wrote the manuscript. All authors read and approved the
manuscript and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
The present study was performed in accordance with
standard guidelines and was approved by the Ethics Committee of The
First Hospital of Hebei Medical University (no. 2016004).
Patient consent for publication
All patients written informed consent prior to the
study, and all identifying information (including names, initials,
date of birth or hospital numbers) was removed.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Derry MM, Raina K, Agarwal R and Agarwal
C: Characterization of azoxymethane-induced colon tumor metastasis
to lung in a mouse model relevant to human sporadic colorectal
cancer and evaluation of grape seed extract efficacy. Exp Toxicol
Pathol. 66:235–242. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jin C, Jin Z, Chen NZ, Lu M, Liu CB, Hu WL
and Zheng CG: Activation of IRE1alpha-XBP1 pathway induces cell
proliferation and invasion in colorectal carcinoma. Biochem Biophys
Res Commun. 470:75–81. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cox JS and Walter P: A novel mechanism for
regulating activity of a transcription factor that controls the
unfolded protein response. Cell. 87:391–404. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Xi J, Chen Y, Huang S, Cui F and Wang X:
Suppression of GRP78 sensitizes human colorectal cancer cells to
oxaliplatin by downregulation of CD24. Oncol Lett. 15:9861–9867.
2018.PubMed/NCBI
|
|
6
|
Mujcic H, Nagelkerke A, Rouschop KM, Chung
S, Chaudary N, Span PN, Clarke B, Milosevic M, Sykes J, Hill RP, et
al: Hypoxic activation of the PERK/eIF2a arm of the unfolded
protein response promotes metastasis through induction of LAMP3.
Clin Cancer Res. 19:6126–6137. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Limia CM, Sauzay C, Urra H, Hetz C, Chevet
E and Avril T: Emerging roles of the endoplasmic reticulum
associated unfolded protein response in cancer cell migration and
invasion. Cancers (Basel). 11(pii): E6312019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Vieyra A, Mintz E, Lowe J and Guillain F:
Ca2+ binding to sarcoplasmic reticulum ATPase
phosphorylated by Pi reveals four thapsigargin-sensitive
Ca2+ sites in the presence of ADP. Biochim Biophys Acta.
1667:103–113. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ji Q, Cai G, Liu X, Zhang Y, Wang Y, Zhou
L, Sui H and Li Q: MALAT1 regulates the transcriptional and
translational levels of proto-oncogene RUNX2 in colorectal cancer
metastasis. Cell Death Dis. 10:3782019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Barbagallo C, Brex D, Caponnetto A,
Cirnigliaro M, Scalia M, Magnano A, Caltabiano R, Barbagallo D,
Biondi A, Cappellani A, et al: LncRNA UCA1, Upregulated in CRC
biopsies and downregulated in serum exosomes, controls mRNA
expression by RNA-RNA interactions. Mol Ther Nucleic Acids.
12:229–241. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang C, Zhang Q, Hu Y, Zhu J and Yang J:
Emerging role of long non-coding RNA MALAT1 in predicting clinical
outcomes of patients with digestive system malignancies: A
meta-analysis. Oncol Lett. 17:2159–2170. 2019.PubMed/NCBI
|
|
12
|
Li P, Zhang X, Wang H, Wang L, Liu T, Du
L, Yang Y and Wang C: MALAT1 is associated with poor response to
oxaliplatin-based chemotherapy in colorectal cancer patients and
promotes chemoresistance through EZH2. Mol Cancer Ther. 16:739–751.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sun Z, Ou C, Liu J, Chen C, Zhou Q, Yang
S, Li G, Wang G, Song J, Li Z, et al: YAP1-induced MALAT1 promotes
epithelial-mesenchymal transition and angiogenesis by sponging
miR-126-5p in colorectal cancer. Oncogene. 38:2627–2644. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bhattacharyya S and Vrati S: The Malat1
long non-coding RNA is upregulated by signalling through the PERK
axis of unfolded protein response during flavivirus infection. Sci
Rep. 5:177942015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jiang X, Kanda T, Nakamoto S, Miyamura T,
Wu S and Yokosuka O: Involvement of androgen receptor and
glucose-regulated protein 78 kDa in human hepatocarcinogenesis. Exp
Cell Res. 323:326–336. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jiang X, Kanda T, Nakamoto S, Haga Y,
Sasaki R, Nakamura M, Wu S, Mikata R and Yokosuka O: Knockdown of
glucose-regulated protein 78 enhances poly(ADP-ribose) polymerase
cleavage in human pancreatic cancer cells exposed to endoplasmic
reticulum stress. Oncol Rep. 32:2343–2348. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Solovyev VV, Shahmuradov IA and Salamov
AA: Identification of promoter regions and regulatory sites.
Methods Mol Biol. 674:57–83. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang Y, Wang JH, Zhang XL, Wang XL and
Yang L: Endoplasmic reticulum chaperone glucose-regulated protein
78 in gastric cancer: An emerging biomarker. Oncol Lett.
15:6087–6093. 2018.PubMed/NCBI
|
|
20
|
Liu CY, Hsu CC, Huang TT, Lee CH, Chen JL,
Yang SH, Jiang JK, Chen WS, Lee KD and Teng HW: ER stress-related
ATF6 upregulates CIP2A and contributes to poor prognosis of colon
cancer. Mol Oncol. 12:1706–1717. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Coleman OI, Lobner EM, Bierwirth S, Sorbie
A, Waldschmitt N, Rath E, Berger E, Lagkouvardos I, Clavel T, McCoy
KD, et al: Activated ATF6 induces intestinal dysbiosis and innate
immune response to promote colorectal tumorigenesis.
Gastroenterology. 155:1539–1552.e12. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mhaidat NM, Alzoubi KH and Abushbak A:
X-box binding protein 1 (XBP-1) enhances colorectal cancer cell
invasion. J Chemother. 27:167–173. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sheng X, Nenseth HZ, Qu S, Kuzu OF,
Frahnow T, Simon L, Greene S, Zeng Q, Fazli L, Rennie PS, et al:
IRE1a-XBP1s pathway promotes prostate cancer by activating c-MYC
signaling. Nat Commun. 10:3232019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sun Y, Jiang F, Pan Y, Chen X, Chen J,
Wang Y, Zheng X and Zhang J: XBP1 promotes tumor invasion and is
associated with poor prognosis in oral squamous cell carcinoma.
Oncol Rep. 40:988–998. 2018.PubMed/NCBI
|
|
25
|
Wang M, Ruan S, Ming J and Dong F: Nuclear
expression of XBP1s is correlated with breast cancer survival: A
retrospective analysis based on tissue microarray. Onco Targets
Ther. 10:5927–5934. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Song M, Sandoval TA, Chae CS, Chopra S,
Tan C, Rutkowski MR, Raundhal M, Chaurio RA, Payne KK, Konrad C, et
al: IRE1a-XBP1 controls T cell function in ovarian cancer by
regulating mitochondrial activity. Nature. 562:423–428. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang Y, Zhang Y, Yi P, Dong W, Nalin AP,
Zhang J, Zhu Z, Chen L, Benson DM, Mundy-Bosse BL, et al: The
IL-15-AKT-XBP1s signaling pathway contributes to effector functions
and survival in human NK cells. Nat Immunol. 20:10–17. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fang P, Xiang L, Huang S, Jin L, Zhou G,
Zhuge L, Li J, Fan H, Zhou L, Pan C and Zheng Y: IRE1a-XBP1
signaling pathway regulates IL-6 expression and promotes
progression of hepatocellular carcinoma. Oncol Lett. 16:4729–4736.
2018.PubMed/NCBI
|
|
29
|
Lyu X, Zhang M, Li G, Cai Y, Li G and Qiao
Q: Interleukin-6 production mediated by the IRE1-XBP1 pathway
confers radioresistance in human papillomavirus-negative
oropharyngeal carcinoma. Cancer Sci. 110:2471–2484. 2019.PubMed/NCBI
|
|
30
|
Chakraborty S, Ghosh S, Banerjee B, Santra
A, Bhat J, Adhikary A, Chatterjee S, Misra AK and Sen PC:
Mephebrindole, a synthetic indole analog coordinates the crosstalk
between p38MAPK and eIF2a/ATF4/CHOP signalling pathways for
induction of apoptosis in human breast carcinoma cells. Apoptosis.
21:1106–1124. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zong Y, Feng S, Cheng J, Yu C and Lu G:
Up-regulated ATF4 expression increases cell sensitivity to
apoptosis in response to radiation. Cell Physiol Biochem.
41:784–794. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Notte A, Rebucci M, Fransolet M, Roegiers
E, Genin M, Tellier C, Watillon K, Fattaccioli A, Arnould T and
Michiels C: Taxol-induced unfolded protein response activation in
breast cancer cells exposed to hypoxia: ATF4 activation regulates
autophagy and inhibits apoptosis. Int J Biochem Cell Biol. 62:1–14.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Shi Z, Yu X, Yuan M, Lv W, Feng T, Bai R
and Zhong H: Activation of the PERK-ATF4 pathway promotes
chemo-resistance in colon cancer cells. Sci Rep. 9:32102019.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hu YL, Yin Y, Liu HY, Feng YY, Bian ZH,
Zhou LY, Zhang JW, Fei BJ, Wang YG and Huang ZH: Glucose
deprivation induces chemoresistance in colorectal cancer cells by
increasing ATF4 expression. World J Gastroenterol. 22:6235–6245.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Pällmann N, Livgård M, Tesikova M, Zeynep
Nenseth H, Akkus E, Sikkeland J, Jin Y, Koc D, Kuzu OF, Pradhan M,
et al: Regulation of the unfolded protein response through ATF4 and
FAM129A in prostate cancer. Oncogene. 38:6301–6318. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zeng P, Sun S, Li R, Xiao ZX and Chen H:
HER2 upregulates ATF4 to promote cell migration via activation of
ZEB1 and downregulation of E-cadherin. Int J Mol Sci. 20:22232019.
View Article : Google Scholar :
|
|
37
|
Gao S, Ge A, Xu S, You Z, Ning S, Zhao Y
and Pang D: PSAT1 is regulated by ATF4 and enhances cell
proliferation via the GSK3β/β-catenin/cyclin D1 signaling pathway
in ER-negative breast cancer. J Exp Clin Cancer Res. 36:1792017.
View Article : Google Scholar : PubMed/NCBI
|