Introduction
Glioma originates from glial cells of the brain and
is one of the most commonly diagnosed intracranial tumors,
accounting for ~40 to 50% of central nervous system tumors
(1,2). Malignant gliomas (stages III or IV,
according to the World Health Organization) account for ~77.5% of
all gliomas (3). Malignant gliomas,
especially glioblastoma (GBM), have a high incidence rate, high
recurrence rate post-operation, high mortality rate and low cure
rate (3). The most detrimental
biological feature of malignant gliomas is the infiltrative growth
of cancer cells, and the tumor tissues that cannot frequently be
completely removed through surgery (4). Although advancements have been made in
the pathogenesis, diagnosis and treatment of glioma recently, the
prognosis of glioma, especially glioblastoma, is still not
satisfying; with a 2 year survival rate of only 9% (5,6). At
present, the clinical diagnosis of glioma depends on the results of
histopathological examination, however the targets are usually
difficult to identify (7).
Therefore, it is of great importance to seek targets for early
diagnosis, to assess therapeutic effects and for prognostic
prediction of glioma. Although some markers have been recognized
for glioma, such as human cartilage glycoprotein 39 (YKL-40),
EphA2, CD133 and matrix metalloproteinase (MMP)-9, identification
of more accurate and specific targets are still required for
patients with glioma (8,9).
The kinesin protein superfamily (KIF) is a type of
molecular motor, which plays an important role in mitosis,
organelle transport, meiosis, signal transduction, protein
transport and several other cellular functions (10). Following the first naming of kinesin
by Hirokawa and Tanaka (11),
dozens of kinesin proteins have been identified up to date. Two
types, namely conventional kinesin and unconventional kinesin, can
be determined according to the different functions of kinesin
proteins. The former has intracellular transport activity, such as
KIF1A and KIF5B, whereas the latter does not exhibit ATP enzyme
activity and cannot play a role in intracellular material transport
(12,13). Previous studies have revealed that
kinesin proteins are associated with neurodegenerative diseases
such as Alzheimer's disease (14)
and Huntington's disease (15),
diabetes (16) and nephropathy
(17). Moreover, aberrant kinesin
expression could induce the alteration of the distribution of
intracellular genetic material, which could progressively lead to
tumorigenesis, through chromosomal over-agglutination, spindle
formation abnormalities, cell division defects or aneuploidy, and
mitotic arrest (18). Accumulating
evidence has revealed that kinesin proteins are widely involved in
the occurrence and development of a variety of tumors, and their
expression levels are also directly associated with the occurrence
and development of many tumors (19,20).
Kinesin-12, also known as KIF15, is a
microtubule-dependent motor protein and participates in mitosis and
neuronal development (21). A
previous study reported that KIF15 was a critical regulator in the
promotion of pancreatic cancer proliferation through MEK/ERK
signaling pathway (22). Milic
et al revealed that KIF15-IN-1, an inhibitor of KIF15, could
suppress cancer cell growth (23).
However, the role of KIF15 in promoting the development of glioma
and its potential as a therapeutic target need to be determined.
Therefore, the present study demonstrated for the first time that
the expression of KIF15 was upregulated in glioma tissues and was
positively associated with the pathological staging, recurrence
risk and poor prognosis. Moreover, the knockdown (KD) of KIF15
could significantly inhibit the development and stemness of glioma,
indicating that KIF15 may be a potential therapeutic target for the
treatment of glioma.
Materials and methods
Materials
The following materials were used: U87 MG cell line
(glioblastoma of unknown origin; Shanghai Fuheng Biological
Technology), U251 (BeNa Technology). The cells were both
authenticated by short tandem repeat profiling. BR-V-108 (Shanghai
Bioscienceres) was used as a plasmid vector from TOP10 E.
coli competent cells (cat. no. CB104-03; Tiangen Biotech Co.,
Ltd.). D-Hanks and trypsin were obtained from Shanghai Chemical
Reagent Company) and the KIF15 antibody was purchased from Fine
Test (cat. no. FNab04551).
Six-week-old male BALB/c nude mice were purchased
from Shanghai Jake BIO Technology Co., Ltd. and divided into two
groups randomly with 6 mice in each group. All mice were housed
under specific pathogen-free housing conditions.
Collection of clinical samples
The use of human tissues was approved by the
Institutional Review Board of Changzhou No. 2 People's Hospital.
The formalin-fixed, paraffin-embedded tissue microarray of
glioblastoma containing tissue samples collected from 164 patients
were purchased from Shanghai Outdo Biotech Company (Table I). The written informed consents
were collected from all patients.
 | Table I.Relationship between KIF15 expression
and tumor characteristics in patients with glioma. |
Table I.
Relationship between KIF15 expression
and tumor characteristics in patients with glioma.
|
|
| KIF15
expression |
|
|---|
|
|
|
|
|
|---|
| Features | No. of
patients | Low | High | P-value |
|---|
| All patients | 164 | 75 | 89 |
|
| Age (years) |
|
|
| <0.0001 |
|
<41 | 73 | 46 | 27 |
|
|
≥41 | 91 | 29 | 62 |
|
| Sex |
|
|
| 0.138 |
|
Male | 106 | 53 | 53 |
|
|
Female | 58 | 22 | 36 |
|
| Recurrence |
|
|
| <0.0001 |
|
Yes | 88 | 24 | 64 |
|
| No | 76 | 51 | 25 |
|
| Stage |
|
|
| <0.0001 |
| I | 22 | 22 | 0 |
|
| II | 71 | 61 | 10 |
|
|
III | 49 | 6 | 43 |
|
| IV | 22 | 0 | 22 |
|
Cell culture
U87 MG and U251 cells were grown in six-well plates
using DMEM (Invitrogen: Thermo Fisher Scientific, Inc.)
supplemented with 10% FBS at 37°C in humidified air containing 5%
CO2. Cell culture media was replaced every 72 h. The
cells were subcultured at 80% confluence using 0.05% trypsin with
0.02% EDTA. The subsequent experiments were then performed on cells
following a 24 h incubation in DMEM medium without FBS. All
experiments were performed under serum-free conditions, in which
the cells remained viable in a non-proliferating state.
Bioinformatics of KIF15 expression in
glioma from a public database
The Cancer Genome Atlas (TCGA) database (https://cancergenome.nih.gov/) TCGA-GBM was utilized
to collect the expression of KIF15 in 5 normal tissues and 169 GBM
tissues.
Target gene RNA interference
lentiviral vector preparation
Using the KIF15 gene as the template, the RNA
interference targeting sequences (Pbr-13015/shKIF15-1,
5′-CAGGATCGTTTGCTCTCAGAA-3′; Pbr-13016/shKIF15-2,
5′-AGGCAGCTAGAATTGGAATCA-3′; Pbr-00141/shKIF15-3,
5′-GCTGAAGTGAAGAGGCTCAAA-3′) were designed and single-stranded DNA
oligo were synthesized (Generay Biotech Co., Ltd.). The synthesized
single-stranded DNA oligo was dissolved in an annealing buffer,
using a water bath at 90°C for 15 min to form double-strand DNA.
The BR-V-108 vector was linearized using AgeI and
EcoRI. A 20 µl reaction system was prepared according to the
Fermentas T4 DNA Ligase instructions, and the double-stranded DNA
oligo was ligated to the linearized vector (100 ng/µl). The
ligation product was transferred into the prepared TOP10E. coli
competent cells (100 µl) with 500 µl antibiotic-free lysogeny broth
(LB) liquid medium and incubated at 37°C for 1 h. A bacterial
solution (150 µl) was spread on LB solid medium containing Amp and
cultured overnight in a 37°C incubator. A 20 µl PCR reaction system
was prepared, and a single colony was selected as a template with a
sterile tip to perform PCR amplification. The correctly sequenced
bacterial solution was transferred to 150 ml LB liquid medium
containing Amp antibiotics, and cultured overnight at 37°C. The
plasmid was subsequently purified according to the EndoFree Maxi
Plasmid Kit instructions (cat. no. DP117; Tiangen Biotech Co.,
Ltd.). The quality-qualified plasmids were transferred for virus
packaging. Notably, the infection protocol included the temperature
(37°C), duration (18 h) and MOI (10) of infection. Subsequent experiments
were performed after further culturing the cells for 72 h.
Reverse transcription-quantitative
(RT-q)PCR
Total RNA were isolated from U87 MG and U251 cells
using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.) and
Direct-zol™ RNA MiniPrep (Zymo Research), according to
manufacturer's instructions. The RNA was quantified using NanoDrop
2000 (Thermo Fisher Scientific, Inc.). First-strand cDNA was
obtained using 1 µg total RNA, random primers and M-MLV reverse
transcriptase (Promega Corporation). qPCR was conducted using AceQ
qPCR SYBR Green Master mix (Vazyme). The 2−∆∆Cq method
was utilized for the quantification of gene expression, with
β-actin as an endogenous control. The following PCR primers were
used: KIF15 forward, 5′-CTCTCACAGTTGAATGTCCTTG-3′ and reverse,
5′-CTCCTTGTCAGCAGAATGAAG-3′; β-actin primers forward,
5′-CCTATTTCCCATGATTCCTTCATA-3′ and reverse,
5′-GTAATACGGTTATCCACGCG-3′. The PCR protocol was: 3 min at 95°C; 1
min at 94°C, 1 min at 60°C, 1 min at 72°C for 35 cycles; and 10 min
at 72°C. PCR products were separated on a 20 g/l agarose gel
stained with ethidium bromide and viewed under ultraviolet
light.
Western blot analysis
In order to investigate the expression level of
KIF15, cells were collected and lysed using RIPA lysis buffer (Cell
Signal Technology, Inc.), including protease inhibitors, according
to the manufacturer's instructions. The concentration of protein
was determined using the BCA Protein Assay kit (cat. no. 23225;
Thermo Fisher Scientific Pierce). Subsequently, the total cellular
proteins were subjected to SDS-PAGE (10% gel) for separation (20 µg
protein per lane). After transferring to polyvinylidene difluoride
(PVDF) membranes, blots were incubated with 5% BSA in Tris-buffered
saline containing 0.5% Tween-20 for 60 min and incubated overnight
at 4°C on a rocker with the following primary antibodies: KIF15
antibody (1:1,000; cat. no. FNab04551; Fine Test), GAPDH antibody
(1:3,000; cat. no. AP0063; Bioworld), Bax antibody (1:1,000; cat.
no. ab32503; Abcam), p21 antibody (1:400; cat. no. BM3990; Boster),
Survivin antibody (1:500; cat. no. ab469; Abcam), MEK1/2 antibody
(1:500; cat. no. ab178876; Abcam), p-MEK1/2 (1:500; cat. no.
ab194754; Abcam), ERK1/2 (1:500; cat. no. 4695; CST) and p-ERK1/2
(1:500; cat. no. 4370; CST). Following three washes with TBST for 5
min, the membranes were incubated with horseradish peroxidase
(HRP)-conjugated goat anti-rabbit IgG polyclonal secondary antibody
(1:3,000; cat. no. A0208; Beyotime Institute of Biotechnology) at
room temperature for 1 h. Amersham ECL + plus™ western blotting
system kit (cat. no. RPN3352; GE Healthcare) was used for color
developing. Each membrane was visualized using the ECL-Plus™
Western blotting system (GE Healthcare Life Sciences), and proteins
were detected with an X-ray imaging analyzer (Kodak). Densitometric
analysis was performed using ImageJ (version 1.8.0; National
Institutes of Health).
MTT assay
An MTT assay was applied to analyze in vitro
cell viability. Following the trypsinization of cells in the
logarithmic growth phase, glioma cells (2,000 cells/well) were
seeded into a 96-well (100 µl/well) (cat. no. 3599; Corning Inc.)
overnight. MTT solution (20 µl/well, 5 mg/ml; cat. no. JT343;
Genview) was added to cells and incubated for 4 h. DMSO (100
µl/well) was added to dissolve the formazan crystals. Absorbance
values were measured at 490 nm using a microplate reader (cat. no.
M2009PR; Tecan Group, Ltd.) after 24, 48, 72, 96, and 120 h of
growth; 570 nm was used as the reference wavelength. The cell
viability ratio was calculated using the following formula: cell
viability (%) = OD (treated)/OD (control) ×100%.
Apoptotic assay
Lentivirus-infected cells were seeded in 6 cm
dishes. The cells were subsequently digested with trypsin and
resuspended in the same medium, then 10 µl Annexin
V-allophycocyanin (APC) was added for staining the cells for 15 min
in a dark at room temperature. Cell apoptosis analysis was
performed using an Annexin V-Allophycocyanin/Propidium Iodide kit
(eBioscience; Thermo Fisher Scientific, Inc.) The apoptotic rate of
cells was measured using a FACScan analyzer (Merck KGaA). The
results were visualized using GuavaSoft software (version 3.1.1;
EMD Millipore).
Detection of cell cycle by
fluorescence-activated cells sorting (FACS)
Cells in the exponential growth phase were
collected. The cells were subsequently washed with cold PBS and
fixed in 950 µl of cold 70% ethanol for 1 h. Finally, cells were
stained by propidium iodide (PI) and kept away from light for 10–15
min at room temperature. The cell cycle was detected by BD
FACSCalibur flow cytometer (BD Biosciences) with the throughput of
cells ~200–350 cells.
Immunohistochemistry analysis
The expression of KIF15 was detected by
immunohistochemistry in glioblastoma tissues. Tumor and
para-carcinoma tissue sections from glioblastoma patients were
deparaffinized. Following citrate antigen repair and blocking by 3%
H2O2 for 10 min at room temperature, the
samples were incubated with the KIF15 antibody (1:50; Fine Test) at
4°C overnight in an incubator. The sections were subsequently
incubated with goat anti-rabbit IgG H&L horseradish peroxidase
(HRP)-conjugated secondary antibody (1:400; cat. no. ab6721; Abcam)
at 37°C for 1 h. Images were captured by a light microscope
(Olympus Corp.). Ki-67 antibody (1:100; cat. no. ab16667; Abcam),
Bax antibody (1:250; cat. no. ab32503; Abcam), p21 antibody (1:50;
cat. no. BM3990; BOSTER) and Survivin antibody (1:100; cat. no.
ab469; Abcam) were used in the immunohistochemistry analysis of
tumor sections removed from mice models.
Caspase 3/7 activity assay
Cell apoptosis was also quantified by the
Caspase-Glo® 3/7 assay kit (Promega Corp.), according to
the manufacturer's protocol.
Ethics approval and tumor-bearing
animal model
All the animal experimental protocols were approved
by the Ethics committee at The Affiliated Changzhou No. 2 People's
Hospital of Nanjing Medical University. BALB/c male nude mice
(10–15 g, 6 weeks old) were cultured in SPF-class housing in the
laboratory with a 12 day/night environment and fed with a standard
diet. U87 MG cells (5×106), transfected with shKIF5 or
shCtrl, were implanted into the right underarm of mice (BALB/c male
nude mice) subcutaneously. Volumes of the xenograft tumors were
measured by calipers after 7, 13, 19, 22 and 28 days
post-infection. Although the humane endpoint in our protocol was a
tumor >15 mm, all mice were sacrificed before reaching the
humane endpoint. After 28 days of culture, the mice were sacrificed
by injecting 1% pentobarbital sodium (100 mg/kg body weight), and
the tumors were collected for the measurement of weight and for
immunohistochemistry (IHC) analysis.
Human apoptosis antibody array
The expression profiles of a series of
apoptosis-associated proteins induced by KIF15 knockdown in U87MG
cells were detected by human apoptosis antibody array (RayBiotech),
according to manufacturer's instructions. Briefly, the array
membrane was placed into a dish and cell lysates were added to each
well for incubation at 4°C with gentle shaking overnight. The
membranes were washed and then incubated with lyophilized
biotinylated antibodies for 1 h on a rocking platform shaker. After
washing away the excess molecules, the membranes were further
incubated with horseradish peroxidase-conjugated streptavidin for
30 min. The expression levels of proteins were analyzed using the
Gel-Pro Analyzer software (version 6.3; Media Cybernetics).
Statistical analysis
All experiments were carried out in at least 3
independent experiments. Data were expressed as the mean ± standard
deviation for continuous variables and analyzed using GraphPad
Prism 6 software (GraphPad Software, Inc.). Student's t-test and
χ2 test were used to analyze the statistical difference.
Statistical analysis between multiple groups was performed using
ANOVA followed by Tukey's HSD post hoc test. The survival analysis
was performed by the Kaplan-Meier method and analyzed by the
log-rank test. P<0.05 was considered to indicate a statistically
significant difference.
Results
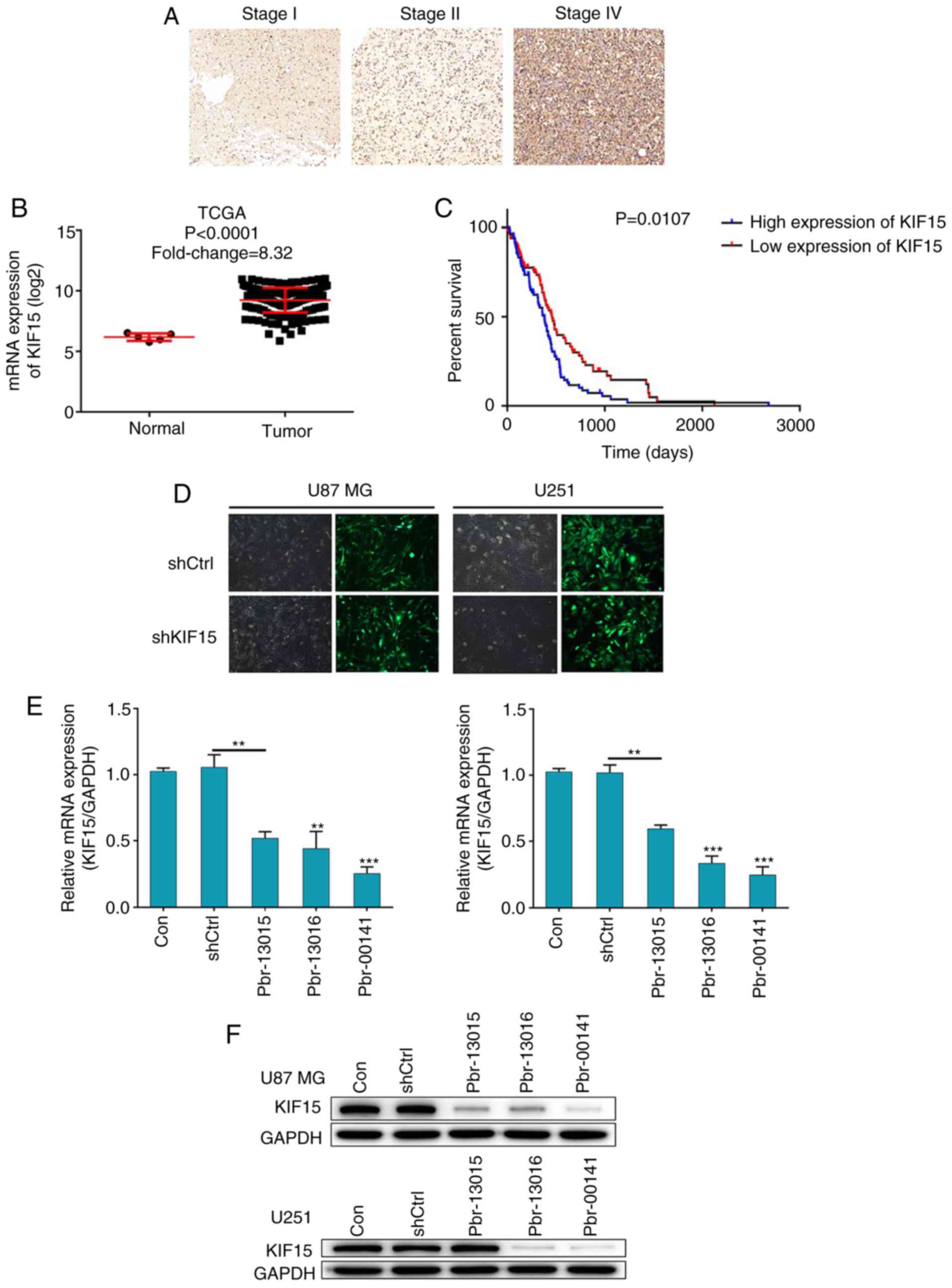
KIF15 is upregulated in glioma tissues
and is associated with poor prognosis
In order to investigate the role of KIF15 in the
prognosis of glioma, clinical specimens of glioma tissues and the
corresponding follow-up information were collected. The protein
expression level of KIF15 was detected by IHC analysis in glioma
tissues at different pathological stages. The results revealed that
the expression of KIF15 was significantly higher at the later stage
(Fig. 1A). RNA-seq data mining of
The Cancer Genome Atlas (TCGA) database confirmed that KIF15
expression was significantly increased in glioma tissues compared
with normal tissues (Fig. 1B).
Furthermore, high expression of KIF15 in glioma tissues was
associated with older age, late pathological stage and risk of
recurrence (Table I). Moreover, the
Kaplan-Meier survival analysis revealed that patients with
relatively high expression of KIF15 suffered from notably shorter
overall survival, as well as poor prognosis (Fig. 1C). The aforementioned results
indicated the involvement of KIF15 in the development and
progression of glioma and its potential role as a prognostic
indicator.
Construction of KIF15 KD cell
models
In order to clarify the role of KIF15 in glioma
further, two glioma cell lines U87 MG and U251 were selected for
the construction of KIF15-knockdown (KD) cell models. Artificially
prepared lentivirus plasmid shKIF15, which expressed 3 different
RNAi sequences (Pbr-03015, Pbr-13016 and Pbr-00141) for silencing
KIF15, were transfected into cells to downregulate the levels of
KIF15 expression. The cells transfected with the corresponding
empty vector were used as the negative control (shCtrl). The
transduction efficiency in both U87 MG and U251 cells was assessed
through the detection of green fluorescent protein (GFP), which was
tagged on the lentivirus vector; >80% efficiency was revealed in
both cell lines for both the shCtrl and shKIF15 groups (Fig. 1D). Subsequently, through the
detection of mRNA and protein levels of KIF15 in each group by qPCR
and western blotting, respectively, Pbr-00141 was identified as the
most efficient sequence for KIF15 knockdown in both cell lines; and
was thus used in all subsequent experiments for the shKIF15 group
(Fig. 1E and F). In summary, given
the upregulated expression level of KIF15 in clinical specimens,
the KIF15-KD cell models were successfully constructed for the
subsequent studies.
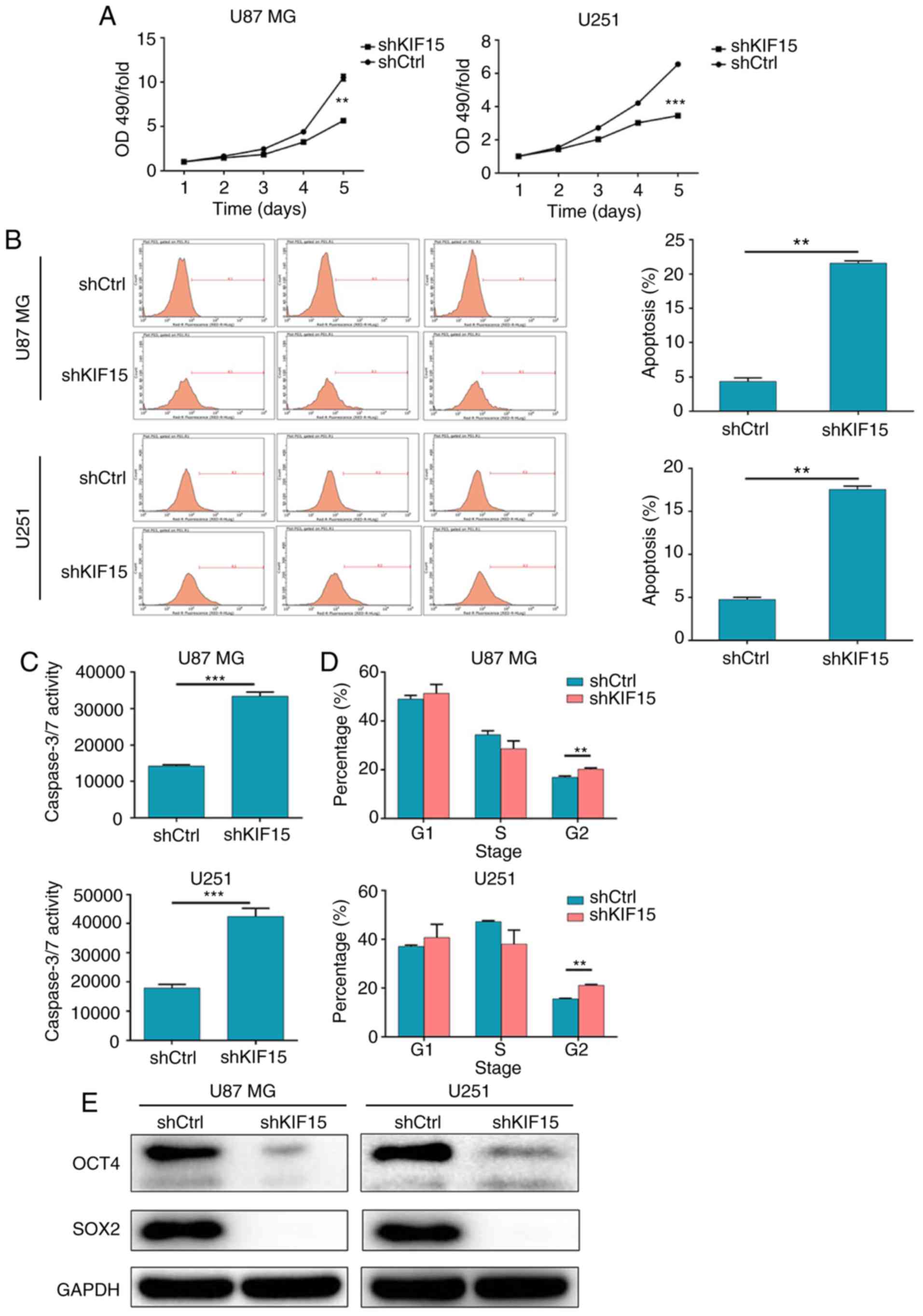
Silencing of KIF15 inhibits cell
proliferation and stemness, induces cell apoptosis and affects the
cell cycle in glioma cells
The effect of KIF15 KD on the functions of glioma
cells was evaluated. The results of the MTT assay revealed that the
downregulation of KIF15 in U87 MG and U251 cells significantly
inhibited cell proliferation (Fig.
2A). Moreover, flow cytometric analysis was performed to
explore the effect of KIF15 on cell apoptosis in U87 MG and U251
cells. As revealed in Fig. 2B, the
silencing of KIF15 notably enhanced cell apoptosis in both cell
lines. The apoptosis percentages for KIF15-KD U87 MG and KIF15-KD
U251 cells were 5- and 3.8-fold higher compared with the shCtrl
groups, respectively. Moreover, KIF15 KD-induced cell apoptosis was
also assessed by the detection of caspase 3/7 activity (Fig. 2C). Furthermore, as revealed in
Fig. 2D, cell cycle analysis
indicated an increased percentage of cells in the G2
phase in the KIF15-KD groups for both cell lines; this demonstrated
the G2-arresting ability of KIF15 KD in glioma cells.
Furthermore, the expression levels of stemness-associated proteins
OCT4 and SOX2 were both inhibited in U87 MG and U251 cells
following the silencing of KIF15 (Fig.
2E). Overall, the silencing of KIF15 inhibited cell
proliferation and stemness, induced cell apoptosis and promoted
G2-arrest in glioma cells.
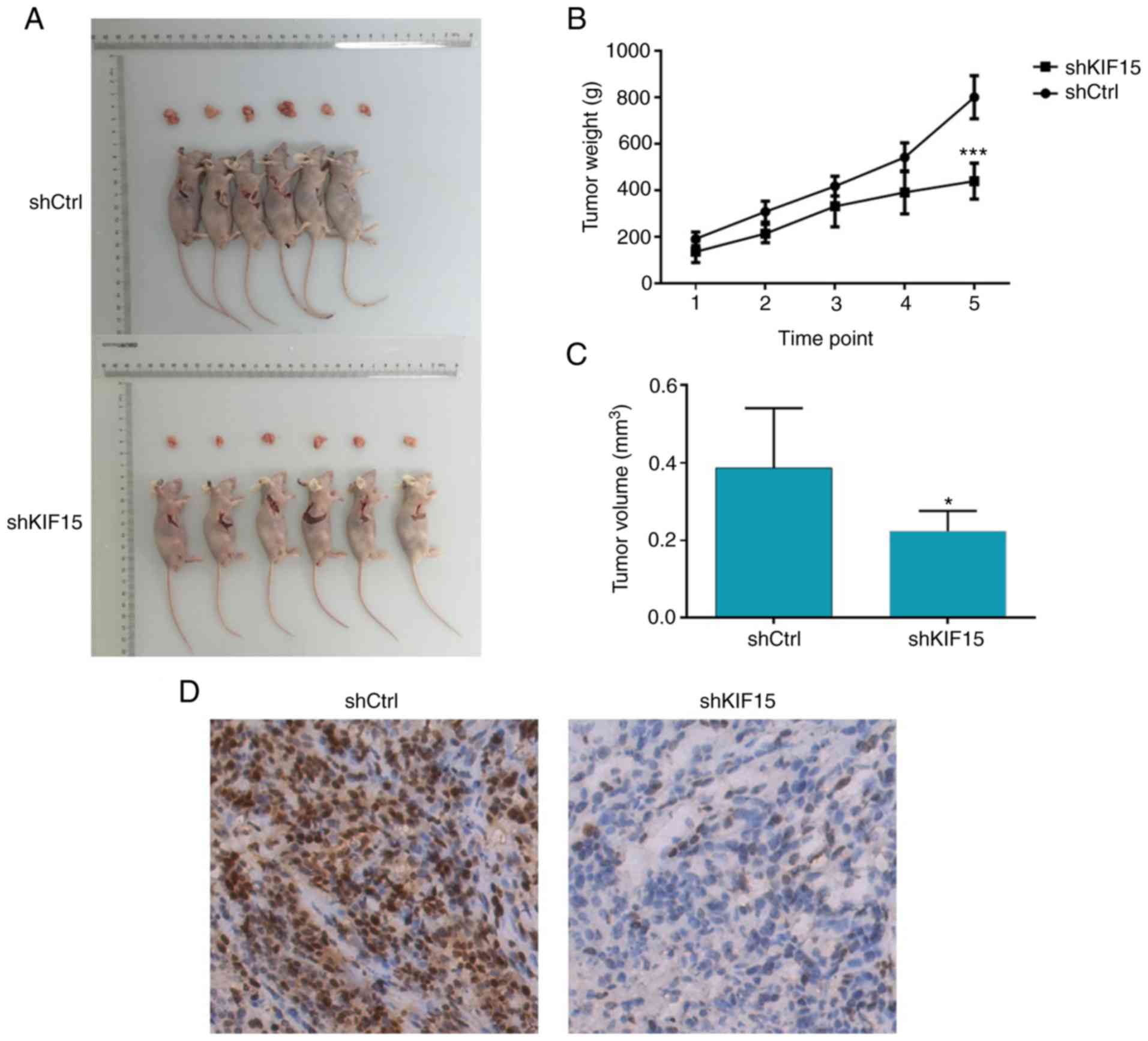
Silencing of KIF15 impairs tumor
growth in vivo
The ability of KIF15 KD to impair tumor growth was
assessed in vivo. Therefore, a mouse xenograft model was
constructed by subcutaneously injecting U87 MG cells with or
without KIF15 KD into nude mice and monitoring tumor growth. The
weight and volume of the tumors removed from mice in the shKIF15
group were significantly lower compared with the shCtrl group
(time-point 1: 7 days; 2: 13 days; 3: 19 days; 4: 22 days; 5: 28
days following inoculation; P<0.001; Fig. 3A-C). Moreover, the tumors formed in
the shKIF15 group displayed lower Ki-67 index compared with tumors
removed from the shCtrl group, as detected by IHC analysis
(Fig. 3D). Overall, these results
indicated that the silencing of KIF15 resulted in impaired
tumorigenicity of glioma cells in vivo, which was consistent
with the data obtained in the proliferation and apoptosis assays
in vitro.
Mechanistic study of KIF15 in
glioma
In order to explore the potential mechanism of the
regulatory ability of KIF15 in glioma, human apoptosis antibody
array was performed to identify the differentially expressed
proteins induced by KIF15 KD in U87 MG cells compared with the
shCtrl group. The upregulated expression of CD40 and CD40L in
shKIF15 group revealed in Fig. 4A,
indicated the potential participation of the CD40/CD40L signaling
pathway in KIF15-induced inhibition in glioma. Moreover, several
proteins associated with apoptosis and cell cycle were selected for
verification in U251 cells. As revealed in Fig. 4B, the protein expression of
pro-apoptotic proteins Bax and p21 were also upregulated in U251
cells with KIF15 KD, whereas the anti-apoptosis protein Survivin
was downregulated; thus demonstrating the ability of KIF15 KD to
induce cell apoptosis, which is consistent with the aforementioned
cellular experiments. In addition, the upregulated expression
levels of Bax and p21, and downregulated expression of Survivin
detected by IHC were also observed in the tumor tissues from mice
in the shKIF15 group (Fig. 4C).
Conversely, the effects of KIF15 on the activation of the MEK/ERK
signaling pathway were investigated, based on the detection of
MEK1/2, p-MEK1/2, ERK1/2, p-ERK1/2 by western blot analysis. The
results indicated that, the expression of p-MEK1/2 and p-ERK1/2 was
downregulated by KIF15 KD, whereas the expression of MEK1/2 and
ERK1/2 remained unchanged, indicating the inhibition of the MEK/ERK
signaling pathway by KIF15 knockdown (Fig. 4D).
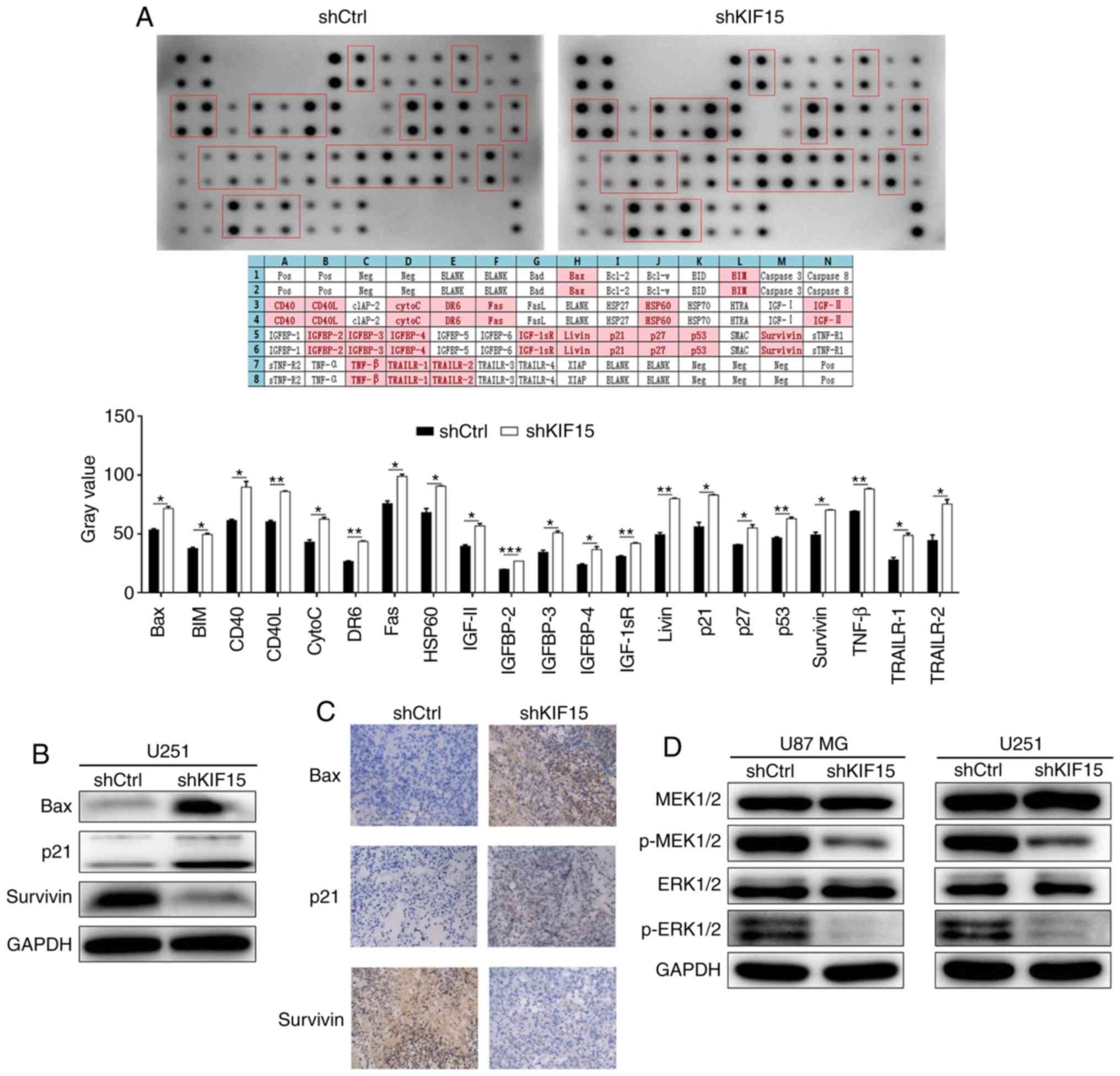 | Figure 4.Mechanism study of KIF15 KD in
glioma. (A) Human apoptosis antibody array analysis was performed
and analyzed in U87 MG cells with or without KIF15 KD. (B) The
protein expression levels of Bax, p21 and Survivin in U251 cells
were detected by western blot analysis. (C) The expression levels
of Bax, p21 and Survivin were detected by IHC in tumor sections of
a mouse xenograft model (magnification, ×200). (D) The expression
levels of MEK1/2, p-MEK1/2, ERK1/2, p-ERK1/2 were detected by
western blot analysis in U87 MG and U251 cells with or without
KIF15 KD. *P<0.05, **P<0.01, ***P<0.001. KIF15,
kinesin-12; KD, knockdown. |
Discussion
Glioma is the most common primary intracranial
malignant tumor in adults, and also the most common intracranial
malignancy in children (3,24). It has the characteristics of rapid
growth, strong infiltration and high recurrence rate (25). At present, among the various
treatment strategies for cancer, the most commonly used treatment
for gliomas is the combination of surgical resection and
radiotherapy/chemotherapy. However, the median survival period of
patients with glioma was revealed to be only 12 to 18 months, and
most patients fail to survive the next 2 years following diagnosis
(26). In recent years, the
development of molecular biology has deepened the understanding of
the molecular characteristics of malignant glioma and promoted the
discovery of some glioma markers. For example, the epidermal growth
factor receptor signaling pathway plays an important role in the
pathogenesis, proliferation, invasion and mediation of drug
resistance in glioma, and its expression increases with the
increasing tumor grade (27).
Moreover, Yan et al (28)
reported that miR-96 could promote the development and progression
of glioma, through the enhancement of the activation of the
Wnt/β-catenin signaling pathway. As a member of the Eph subfamily
screened from the human epithelial keratinocyte cDNA library by
Wykosky et al (29), EphA2
was specifically reported to be highly expressed in nearly 90% of
GBM tissues and cell lines, whereas its expression remains normal
in brain tissues. Furthermore, several proteins, such as YKL-40 and
CD133 and long non-coding (lnc)RNAs such as lncRNA MALAT1, have
been demonstrated to be involved in the promotion or suppression of
glioma (30,31). However, more accurate and specific
biomarkers for glioma still need to be explored for the better
understanding of its mechanism and for the development of novel
treatment strategies.
Kinesin is a protein superfamily that plays an
important role in eukaryotic intracellular trafficking and cell
division (10). Kinesins and the
intracellular transport of substances participated by kinesins play
an important role in maintaining the basic functions of cells
(13). Previous studies have
revealed that kinesins were directly related to several types of
diseases such as neurodegenerative diseases. Notably, accumulating
evidence revealed that some members of the kinesin superfamily were
associated with human malignancies. Wang et al (32) reported that the downregulation of
KIF2A could inhibit the proliferation and migration of breast
cancer cells and has the potential to be used as an independent
prognostic marker of outcome in breast cancer. The expression of
KIF23 was also reported to be associated with the prognosis of
patients undergoing lung cancer surgery and has the potential as a
therapeutic target for the development of lung cancer treatment
(33). Furthermore, some kinesin
members were also revealed to be involved in glioma. For example,
the mitotic kinesin KIF11 could promote the proliferation, invasion
and self-renewal of GBM (34–36).
Chen et al (37) revealed
that KIF1B could be a target for anti-invasive therapies of glioma
due to its ability to promote migration and invasion in glioma,
through the cell surface localization of MT1-MMP.
KIF15, also named kinesin-12, is a
microtubule-dependent plus-end-directed motor protein, which is
best known for its role in mitosis. A study by Liu et al
(38) revealed that KIF15 was
highly expressed in both the cortex and ganglia at embryonic
stages, whereas its expression diminished progressively with the
development of neurons. Furthermore, KIF15 was recognized as a
modulator of axonal development (39). Moreover, the structure and function
of KIF15 was also studied in recent years due to the speculation
that it may be a useful target for cancer therapy. Studies have
revealed that KIF15 plays an important role in the maintenance of
spindle bipolarity and is a breast cancer tumor antigen (40). Recently, KIF15 was reported to be
overexpressed in both breast cancer and lung adenocarcinoma, and is
potentially involved in the regulation of the cell cycle (41,42).
Moreover, Wang et al (22)
demonstrated that KIF15 could promote pancreatic cancer cell
proliferation via the regulation of the MEK/ERK pathway. To the
best of our knowledge, the potential of KIF15 as a target for
anti-growth therapies in glioma has not been reported and is still
unclear.
In the present study, clinical specimens were
collected to detect the expression of KIF15 by IHC, which revealed
relatively high KIF15 expression at the later-stage glioma tissues.
The high expression of KIF15 in glioma tissues compared with normal
tissues was demonstrated by the RNA-seq data of TCGA. Moreover, the
statistical analysis revealed that patients with high KIF15
expression were associated with older age, late pathological stage,
high recurrence risk and shorter survival period. The results
obtained from the clinical samples indicated the potential
association between KIF15 expression and the development and
prognosis of glioma.
To verify and further explore the role of KIF15 in
the development and progression of glioma, U87 MG and U251 cell
lines were selected to construct KIF15-KD cell models. According to
the different capabilities of the 3 shRNAs designed for silencing
KIF15 to knockdown KIF15, inhibit cell proliferation and promote
cell apoptosis (data not shown), the most efficient one was
screened for all in vitro and in vivo studies. The
subsequent studies revealed that the silencing of KIF15 inhibited
cell proliferation, induced cell apoptosis and promoted the arrest
of glioma cells at the G2 phase. Notably, the stemness
of glioma cells, which was demonstrated to be the fundamental
source of drug resistance and recurrence of gliomas, was also
significantly inhibited by KIF15 KD, revealed by the downregulation
of OCT4 and SOX2 expression levels. In addition, the generally
similar expression of the internal reference (GAPDH) also
demonstrated that the downregulation of OCT4 and SOX2 did not
result from enhanced cell death. Moreover, the mouse xenograft
model revealed that KIF15 KD inhibited tumor growth and the
expression of Ki-67 in vivo. Moreover, a human apoptosis
antibody array was performed, which revealed the potential role of
the CD40/CD40L signaling pathways. Furthermore, the changes in the
expression of various apoptosis-associated proteins, including Bax,
p21 and Survivin, in the shKIF15 group were verified in both the
cellular and animal experiments, and were consistent with the
results of cell apoptosis. Moreover, consistent with the reported
ability of KIF15 to regulate the MEK/ERK signaling pathway
(22), the present study
demonstrated that the knockdown of KIF15 markedly downregulated the
expression of p-MEK1/2 and p-ERK1/2, as well as deactivated the
MEK/ERK signaling pathway. The aforementioned results indicated the
involvement of KIF15 in the development and progression of
glioma.
In conclusion, the present study indicated that
KIF15 may act as tumor promoter in glioma and could be a future
therapeutic target and prognostic indicator. Further studies
focusing on the regulatory mechanism of KIF15 in glioma could
benefit the better understanding of glioma carcinogenesis.
Acknowledgements
Not applicable.
Funding
The present study was financially supported by the
National Natural Science Foundation of China (No. 81302181), the
Jiangsu Provincial Medical Youth Talent Programme and Key Research
and Development Program of Jiangsu Province (BE2019652).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
FL made substantial contributions to the concept and
design of the present study. QW, WH and CQ conducted the
experiments. CQ and BH conducted data analysis. QW, WH and FL
produced the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The use of human tissues was approved by the
Institutional Review Board of Changzhou No. 2 People's Hospital.
The written informed consents were collected from all patients. All
the animal experimental protocols were approved by the Ethics
committee at The Affiliated Changzhou No. 2 People's Hospital of
Nanjing Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors have no conflicts of interest to
disclose.
References
|
1
|
Weller M, Pfister SM, Wick W, Hegi ME,
Reifenberger G and Stupp R: Molecular neuro-oncology in clinical
practice: A new horizon. Lancet Oncol. 14:e370–e379. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Maher EA, Furnari FB, Bachoo RM, Rowitch
DH, Louis DN, Cavenee WK and DePinho RA: Malignant glioma: Genetics
and biology of a grave matter. Genes Dev. 15:1311–1333. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wen PY and Kesari S: Malignant gliomas in
adults. New Engl J Med. 359:492–507. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nutt CL, Mani DR, Betensky RA, Tamayo P,
Cairncross JG, Ladd C, Pohl U, Hartmann C, Mclaughlin ME, Batchelor
TT, et al: Gene expression-based classification of malignant
gliomas correlates better with survival than histological
classification. Cancer Res. 63:1602–1607. 2003.PubMed/NCBI
|
|
5
|
Nayak L and Reardon DA: High-Grade
gliomas. Continuum (Minneap Minn). 23:1548–1563. 2017.PubMed/NCBI
|
|
6
|
Stupp R, Mason WP, van den Bent MJ, Weller
M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn
U, et al: Radiotherapy plus concomitant and adjuvant temozolomide
for glioblastoma. New Engl J Med. 352:987–996. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Khasraw M and Lassman AB: Advances in the
treatment of malignant gliomas. Curr Oncol Rep. 12:26–33. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cohen AL and Colman H: Glioma biology and
molecular markers. Cancer Treat Res. 163:15–30. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kros JM, Mustafa DM, Dekker LJ, Sillevis
Smitt PA, Luider TM and Zheng PP: Circulating glioma biomarkers.
Neuro Oncol. 17:343–360. 2015.PubMed/NCBI
|
|
10
|
Hirokawa N and Tanaka Y: Kinesin
superfamily proteins (KIFs): Various functions and their relevance
for important phenomena in life and diseases. Exp Cell Res.
334:16–25. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hirokawa N, Bloom GS and Vallee RB:
Cytoskeletal architecture and immunocytochemical localization of
microtubule-associated proteins in regions of axons associated with
rapid axonal transport: The beta,
beta′-iminodipropionitrile-intoxicated axon as a model system. J
Cell Biol. 101:227–239. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hirokawa N, Niwa S and Tanaka Y: Molecular
motors in neurons: Transport mechanisms and roles in brain
function, development, and disease. Neuron. 68:610–638. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Okada Y, Yamazaki H, Sekine-Aizawa Y and
Hirokawa N: The neuron-specific kinesin superfamily protein KIF1A
is a unique monomeric motor for anterograde axonal transport of
synaptic vesicle precursors. Cell. 81:769–780. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ari C, Borysov SI, Wu J, Padmanabhan J and
Potter H: Alzheimer amyloid beta inhibition of Eg5/kinesin 5
reduces neurotrophin and/or transmitter receptor function.
Neurobiol Aging. 35:1839–1849. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mcguire JR, Rong J, Li SH and Li XJ:
Interaction of huntingtin- associated protein-1 with kinesin light
chain: Implications in intracellular trafficking in neurons. J Biol
Chem. 281:3552–3559. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang W, Tanaka Y, Bundo M and Hirokawa N:
Antioxidant signaling involving the microtubule motor KIF12 is an
intracellular target of nutrition excess in beta cells. Dev Cell.
31:202–214. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mrug M, Li R, Cui X, Schoeb TR, Churchill
GA and Guay-Woodford LM: Kinesin family member 12 is a candidate
polycystic kidney disease modifier in the cpk mouse. J Am Soc
Nephrol. 16:905–916. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mazumdar M, Lee JH, Sengupta K, Ried T,
Rane S and Misteli T: Tumor formation via loss of a molecular motor
protein. Curr Biol. 16:1559–1564. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chandrasekaran G, Tátrai P and Gergely F:
Hitting the brakes: Targeting microtubule motors in cancer. Br J
Cancer. 113:693–698. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang W, Shi Y, Li J, Cui W and Yang B:
Up-Regulation of KIF14 is a predictor of poor survival and a novel
prognostic biomarker of chemoresistance to paclitaxel treatment in
cervical cancer. Bioscience Rep. 36:e3152016. View Article : Google Scholar
|
|
21
|
Sturgill EG, Norris SR, Guo Y and Ohi R:
Kinesin-5 inhibitor resistance is driven by kinesin-12. J Cell
Biol. 213:213–227. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang J, Guo X, Xie C and Jiang J: KIF15
promotes pancreatic cancer proliferation via the MEK-ERK signalling
pathway. Brit J Cancer. 117:245–255. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Milic B, Chakraborty A, Han K, Bassik MC
and Block SM: KIF15 nanomechanics and kinesin inhibitors, with
implications for cancer chemotherapeutics. Proc Natl Acad Sci USA.
115:E4613–E4622. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Pollack IF, Finkelstein SD, Woods J,
Burnham J, Holmes EJ, Hamilton RL, Yates AJ, Boyett JM, Finlay JL,
Sposto R, et al: Expression of p53 and prognosis in children with
malignant gliomas. New Engl J Med. 346:420–427. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Omuro A and Deangelis LM: Glioblastoma and
other malignant gliomas: A clinical review. JAMA. 310:1842–1850.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tafani M, De Vito M, Frati A, Pellegrini
L, De Santis E, Sette G, Eramo A, Sale P, Mari E, Santoro A, et al:
Pro-inflammatory gene expression in solid glioblastoma
microenvironment and in hypoxic stem cells from human glioblastoma.
J Neuroinflamm. 8:322011. View Article : Google Scholar
|
|
27
|
Sampson JH, Choi BD, Sanchez-Perez L,
Suryadevara CM, Snyder DJ, Flores CT, Schmittling RJ, Nair SK, Reap
EA, Norberg PK, et al: EGFRvIII mCAR-modified T-cell therapy cures
mice with established intracerebral glioma and generates host
immunity against tumor-antigen loss. Clin Cancer Res. 20:972–984.
2016. View Article : Google Scholar
|
|
28
|
Yan Z, Wang J, Wang C, Jiao Y, Qi W and
Che S: MiR-96/HBP1/Wnt/β-catenin regulatory circuitry promotes
glioma growth. FEBS Lett. 588:3038–3046. 2016. View Article : Google Scholar
|
|
29
|
Wykosky J, Gibo DM, Stanton C and Debinski
W: Interleukin-13 receptor alpha 2, EphA2, and Fos-related antigen
1 as molecular denominators of high-grade astrocytomas and specific
targets for combinatorial therapy. Clin Cancer Res. 14:199–208.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Han Y, Wu Z, Wu T, Huang Y, Cheng Z, Li X,
Sun T, Xie X, Zhou Y and Du Z: Tumor-suppressive function of long
noncoding RNA MALAT1 in glioma cells by downregulation of MMP2 and
inactivation of ERK/MAPK signaling. Cell Death Dis. 7:e21232016.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cao S, Wang Y, Li J, Lv M, Niu H and Tian
Y: Tumor-suppressive function of long noncoding RNA MALAT1 in
glioma cells by suppressing miR-155 expression and activating FBXW7
function. Am J Cancer Res. 6:2561–2574. 2016.PubMed/NCBI
|
|
32
|
Wang J, Ma S, Ma R, Qu X, Liu W, Lv C,
Zhao S and Gong Y: KIF2A silencing inhibits the proliferation and
migration of breast cancer cells and correlates with unfavorable
prognosis in breast cancer. BMC Cancer. 14:4612014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Iltzsche F, Simon K, Stopp S, Pattschull
G, Francke S, Wolter P, Hauser S, Murphy DJ, Garcia P, Rosenwald A
and Gaubatz S: An important role for Myb-MuvB and its target gene
KIF23 in a mouse model of lung adenocarcinoma. Oncogene.
36:110–121. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Venere M, Horbinski C, Crish JF, Jin X,
Vasanji A, Major J, Burrows AC, Chang C, Prokop J, Wu Q, et al: The
mitotic kinesin KIF11 is a driver of invasion, proliferation, and
self-renewal in glioblastoma. Sci Transl Med. 7:143r–304r. 2015.
View Article : Google Scholar
|
|
35
|
Matsuda M, Yamamoto T, Matsumura A and
Kaneda Y: Highly efficient eradication of intracranial glioblastoma
using Eg5 siRNA combined with HVJ envelope. Gene Ther.
16:1465–1476. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Exertier P, Javerzat S, Wang B, Franco M,
Herbert J, Platonova N, Winandy M, Pujol N, Nivelles O and Ormenese
S: Impaired angiogenesis and tumor development by inhibition of the
mitotic kinesin Eg5. Oncotarget. 4:2302–2316. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen S, Han M, Chen W, He Y, Huang B, Zhao
P, Huang Q, Gao L, Qu X and Li X: KIF1B promotes glioma migration
and invasion via cell surface localization of MT1-MMP. Oncol Rep.
35:971–977. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liu M, Nadar VC, Kozielski F, Kozlowska M,
Yu W and Baas PW: Kinesin-12, a mitotic microtubule-associated
motor protein, impacts axonal growth, navigation and branching. J
Neurosci. 30:14896–14906. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Xu M, Liu D, Dong Z, Wang X, Wang X, Liu
Y, Baas PW and Liu M: Kinesin-12 influences axonal growth during
zebrafish neural development. Cytoskeleton (Hoboken). 71:555–563.
2015. View Article : Google Scholar
|
|
40
|
Scanlan MJ, Gout I, Gordon CM, Williamson
B, Stockert E, Gure AO, Jäger D, Chen YT, Mackay A, O'Hare MJ and
Old LJ: Humoral immunity to human breast cancer: Antigen definition
and quantitative analysis of mRNA expression. Cancer Immun.
1:42001.PubMed/NCBI
|
|
41
|
Zou JX, Duan Z, Wang J, Sokolov A, Xu J,
Chen CZ, Li JJ and Chen HW: Kinesin family deregulation coordinated
by bromodomain protein ANCCA and histone methyltransferase MLL for
breast cancer cell growth, survival, and tamoxifen resistance. Mol
Cancer Res. 12:539–549. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Bidkhori G, Narimani Z, Hosseini Ashtiani
S, Moeini A, Nowzari-Dalini A and Masoudi-Nejad A: Reconstruction
of an integrated genome-scale co-expression network reveals key
modules involved in lung adenocarcinoma. PLoS One. 8:e675522013.
View Article : Google Scholar : PubMed/NCBI
|