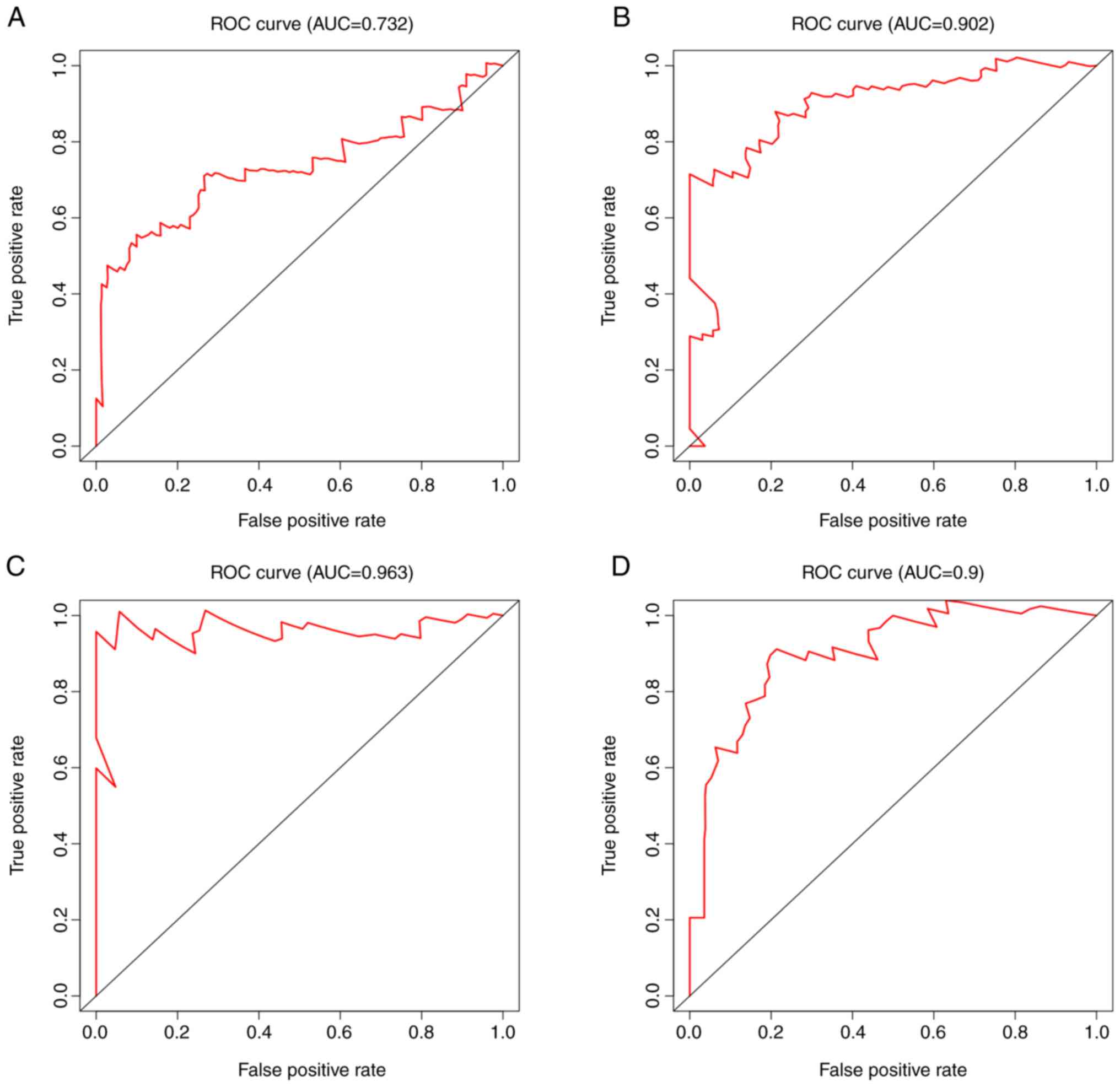
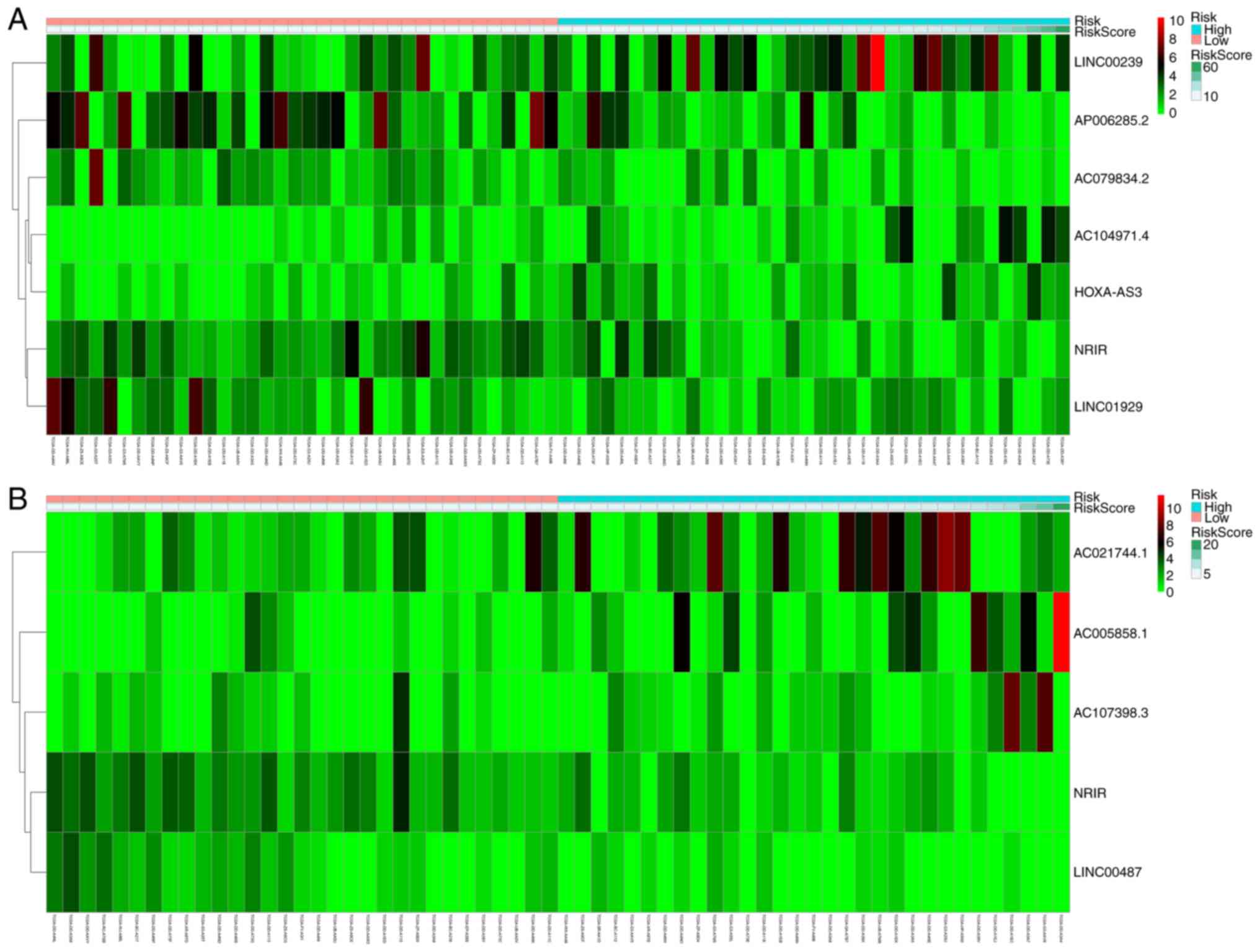
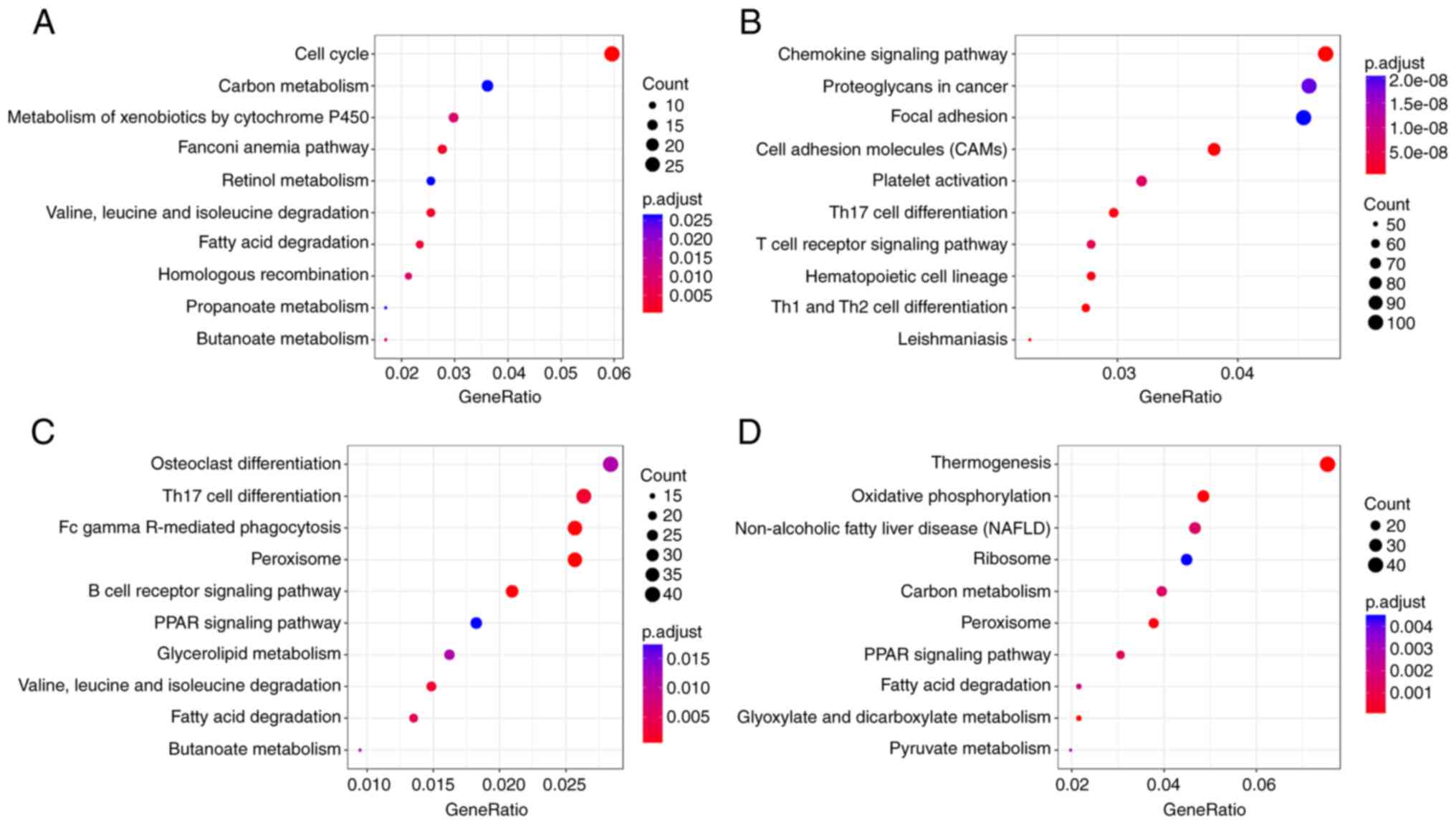
|
1
|
Forner A, Reig M and Bruix J:
Hepatocellular carcinoma. Lancet. 391:1301–1314. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fong ZV and Tanabe KK: The clinical
management of hepatocellular carcinoma in the United States,
Europe, and Asia: A comprehensive and evidence-based comparison and
review. Cancer. 120:2824–2838. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bruix J, Gores GJ and Mazzaferro V:
Hepatocellular carcinoma: Clinical frontiers and perspectives. Gut.
63:844–855. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wu L, Pan C, Wei X, Shi Y, Zheng J, Lin X
and Shi L: lncRNA KRAL reverses 5-fluorouracil resistance in
hepatocellular carcinoma cells by acting as a ceRNA against
miR-141. Cell Commun Signal. 16:472018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Huang Y, Xiang B, Liu Y, Wang Y and Kan H:
LncRNA CDKN2B-AS1 promotes tumor growth and metastasis of human
hepatocellular carcinoma by targeting let-7c-5p/NAP1L1 axis. Cancer
Lett. 437:56–66. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Schmitt AM and Chang HY: Long noncoding
RNAs in cancer pathways. Cancer Cell. 29:452–463. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Song YX, Sun JX, Zhao JH, Yang YC, Shi JX,
Wu ZH, Chen XW, Gao P, Miao ZF and Wang ZN: Non-coding RNAs
participate in the regulatory network of CLDN4 via ceRNA mediated
miRNA evasion. Nat Commun. 8:2892017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu Y, Feng J, Sun M, Yang G, Yuan H, Wang
Y, Bu Y, Zhao M, Zhang S and Zhang X: Long non-coding RNA HULC
activates HBV by modulating HBx/STAT3/miR-539/APOBEC3B signaling in
HBV-related hepatocellular carcinoma. Cancer Lett. 454:158–170.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jiang R, Tang J, Chen Y, Deng L, Ji J, Xie
Y, Wang K, Jia W, Chu WM and Sun B: The long noncoding RNA lnc-EGFR
stimulates T-regulatory cells differentiation thus promoting
hepatocellular carcinoma immune evasion. Nat Commun. 8:151292017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Friedman SL: Evolving challenges in
hepatic fibrosis. Nat Rev Gastroenterol Hepatol. 7:425–436. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Takaya H, Kawaratani H, Tsuji Y, Nakanishi
K, Saikawa S, Sato S, Sawada Y, Kaji K, Okura Y, Shimozato N, et
al: von Willebrand factor is a useful biomarker for liver fibrosis
and prediction of hepatocellular carcinoma development in patients
with hepatitis B and C. United European Gastroenterol J.
6:1401–1409. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
O'Rourke JM, Sagar VM, Shah T and Shetty
S: Carcinogenesis on the background of liver fibrosis: Implications
for the management of hepatocellular cancer. World J Gastroenterol.
24:4436–4447. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ko CJ, Lin PY, Lin KH, Lin CC and Chen YL:
Presence of fibrosis is predictive of postoperative survival in
patients with small hepatocellular carcinoma.
Hepatogastroenterology. 61:2295–2300. 2014.PubMed/NCBI
|
|
16
|
Kadri HS, Blank S, Wang Q, Kim KW, Fiel
MI, Luan W and Hiotis SP: Outcomes following liver resection and
clinical pathologic characteristics of hepatocellular carcinoma
occurring in patients with chronic hepatitis B and minimally
fibrotic liver. Eur J Surg Oncol. 39:1371–1376. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hung HH, Su CW, Lai CR, Chau GY, Chan CC,
Huang YH, Huo TI, Lee PC, Kao WY, Lee SD and Wu JC: Fibrosis and
AST to platelet ratio index predict post-operative prognosis for
solitary small hepatitis B-related hepatocellular carcinoma.
Hepatol Int. 4:691–699. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Abdalla AF, Zalata KR, Ismail AF, Shiha G,
Attiya M and Abo-Alyazeed A: Regression of fibrosis in paediatric
autoimmune hepatitis: Morphometric assessment of fibrosis versus
semiquantiatative methods. Fibrogenesis Tissue Repair. 2:22009.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A Bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. Omics. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Artinyan A, Mailey B, Sanchez-Luege N,
Khalili J, Sun CL, Bhatia S, Wagman LD, Nissen N, Colquhoun SD and
Kim J: Race, ethnicity, and socioeconomic status influence the
survival of patients with hepatocellular carcinoma in the United
States. Cancer. 116:1367–1377. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shebl FM, Capo-Ramos DE, Graubard BI,
McGlynn KA and Altekruse SF: Socioeconomic status and
hepatocellular carcinoma in the United States. Cancer Epidemiol
Biomarkers Prev. 21:1330–1335. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu H, Cen D, Yu Y, Wang Y, Liang X, Lin H
and Cai X: Does fibrosis have an impact on survival of patients
with hepatocellular carcinoma: Evidence from the SEER database? BMC
Cancer. 18:11252018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yan J, Zhou C, Guo K, Li Q and Wang Z: A
novel seven-lncRNA signature for prognosis prediction in
hepatocellular carcinoma. J Cell Biochem. 120:213–223. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhao QJ, Zhang J, Xu L and Liu FF:
Identification of a five-long non-coding RNA signature to improve
the prognosis prediction for patients with hepatocellular
carcinoma. World J Gastroenterol. 24:3426–3439. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wu Y, Wang PS, Wang BG, Xu L, Fang WX, Che
XF, Qu XJ, Liu YP and Li Z: Genomewide identification of a novel
six-LncRNA signature to improve prognosis prediction in resectable
hepatocellular carcinoma. Cancer Med. 7:6219–6233. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sui J, Miao Y, Han J, Nan H, Shen B, Zhang
X, Zhang Y, Wu Y, Wu W, Liu T, et al: Systematic analyses of a
novel lncRNA-associated signature as the prognostic biomarker for
hepatocellular carcinoma. Cancer Med. May 15–2018.(Epub ahead of
print). doi: 10.1002/cam4.1541. View Article : Google Scholar
|
|
28
|
Shi YM, Li YY, Lin JY, Zheng L, Zhu YM and
Huang J: The discovery of a novel eight-mRNA-lncRNA signature
predicting survival of hepatocellular carcinoma patients. J Cell
Biochem. Nov 28–2018.(Epub ahead of print). doi:
10.1002/jcb.28028.
|
|
29
|
Ma Y, Luo T, Dong D, Wu X and Wang Y:
Characterization of long non-coding RNAs to reveal potential
prognostic biomarkers in hepatocellular carcinoma. Gene.
663:148–156. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liao X, Yang C, Huang R, Han C, Yu T,
Huang K, Liu X, Yu L, Zhu G, Su H, et al: Identification of
potential prognostic long non-coding RNA biomarkers for predicting
survival in patients with hepatocellular carcinoma. Cell Physiol
Biochem. 48:1854–1869. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gu J, Zhang X, Miao R, Ma X, Xiang X, Fu
Y, Liu C, Niu W and Qu K: A three-long non-coding
RNA-expression-based risk score system can better predict both
overall and recurrence-free survival in patients with small
hepatocellular carcinoma. Aging (Albany NY). 10:1627–1639. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
El-Serag HB, Siegel AB, Davila JA, Shaib
YH, Cayton-Woody M, McBride R and McGlynn KA: Treatment and
outcomes of treating of hepatocellular carcinoma among medicare
recipients in the United States: A population-based study. J
Hepatol. 44:158–166. 2006. View Article : Google Scholar : PubMed/NCBI
|