Introduction
Liver cancer poses an enormous global threat to
human health and life, accounting for approximately 4.7% of all new
cancer cases and 8.2% of all cancer-related deaths in 2018 in the
world (1). Asia and Africa are
high-risk liver cancer areas, with >50% of new liver cancer
cases and deaths occurring in China (1,2). Liver
cancer is often diagnosed at the advanced stage and its prognosis
is also poor (3). Liver cancer can
be cured through surgical resection, liver transplantation and
radiofrequency ablation when the disease is diagnosed at the early
stage, whereas current therapeutic strategies can only hinder
disease progression and extend the length of life for patients with
intermediate and advanced liver cancer (3,4).
Transcatheter arterial embolization (TAE) and transcatheter
arterial chemoembolization (TACE) are often used for the treatment
of liver cancer, including palliative treatments for patients with
unresectable intermediate and advanced liver cancer, and
preoperative treatments for resectable liver cancer (5,6).
Hypoxia, a common phenomenon in the majority of malignant tumors,
has a positive or negative influence on cancer progression and
therapeutic responses depending on duration, environment and
severity (7,8). Severe hypoxia leads to cell death,
while an oxygen level >0.5% inhibits cell death (9).
Long non-coding RNAs (lncRNAs), a group of long
(>200 nucleotides) transcripts without protein-coding potential,
and microRNAs (miRNAs or miRs), a class of short (~20 nucleotides)
transcripts lacking protein-coding potential, have been identified
as vital regulators of gene expression and crucial players in the
tumorigenesis and progression of cancers such as liver cancer
(10,11). Emerging evidence shows that ncRNAs,
including lncRNAs and miRNAs, play vital roles in hypoxia-regulated
cancer processes, including tumor invasion and metastasis (12,13).
The lncRNA myocardial infarction associated transcript (MIAT) has
been revealed to be implicated in the pathogenesis of multiple
diseases, including myocardial infarction, diabetic retinopathy and
schizophrenia (14,15). Previous studies revealed that MIAT
functioned as an oncogenic factor in various cancer types such as
liver cancer (16–18). Additionally, MIAT was involved in
the regulation of hypoxia responses. For instance, MIAT knockdown
enhanced the detrimental effect of hypoxia on cell viability, and
promoted cell apoptosis induced by hypoxia in rat retinal ganglion
cells and retinal Müller cells (19). TACE can induce severe hypoxia, which
leads to the alteration of the tumor microenvironment and
biological responses (20).
Therefore, the roles and molecular basis of MIAT in TACE treatment
for liver cancer were further investigated in hypoxia-treated liver
cancer cells and rat liver tumors in the present study.
Bioinformatics prediction analysis revealed that
MIAT had a probability to bind to miR-203a, and that
hypoxia-inducible factor 1-α (HIF-1α) was a potential target of
miR-203a, which has been reported to be a tumor suppressor in
multiple cancers, including gastric (21), nasopharyngeal (22) and liver cancer (23). Moreover, miR-203a overexpression led
to a notable downregulation of HIF-1α in glioblastoma cells
(24). HIF-1α has been well
documented as a hypoxia-responsive factor in cancers (25,26).
Additionally, previous studies revealed that HIF-1α knockdown
curbed tumor angiogenesis and metastasis, and enhanced the
TAE-mediated antitumor effect in rat liver tumors (27–29).
Consequently, the present study further investigated whether MIAT
could exert its crucial function through regulation of the
miR-203a/HIF-1α axis in TACE treatment for liver cancer in
hypoxia-treated liver cancer cells and rat liver tumors.
Materials and methods
Clinical samples and cell culture
Patients with liver cancer (40–50 years old, male)
were recruited from Henan Provincial People's Hospital (Zhengzhou,
China) from January to May 2017 and divided into a TACE-untreated
group (n=22) and a TACE-treated group (n=20). Necrotic parts of
liver tumors were excluded from our study. The expression levels of
MIAT, miR-203a and HIF-1α were assessed by reverse
transcription-quantitative PCR (RT-qPCR) assay in liver tumors
without necrosis.
Patients in the TACE-treated group received ≥1
session of TACE alone without other treatments prior to resection.
Patients with liver cancer in the TACE-untreated group did not
receive any treatment before surgery. The present experiments were
approved by the Research Ethics Committee of Henan Provincial
People's Hospital. Written informed consent was obtained from all
participants prior to the experiments.
The liver cancer cell line HepG2 (Cell Bank of the
Chinese Academy of Sciences) was cultured in Dulbecco's modified
Eagle's medium (cat. no. 11965092; Thermo Fisher Scientific, Inc.)
supplemented with 10% fetal bovine serum (cat. no. 16140071; Thermo
Fisher Scientific, Inc.) under normoxia (20% O2) or
hypoxia (1% O2, 5% CO2) conditions at 37°C.
This cell line was derived from a 15 year-old Caucasian male with
liver cancer. The cell line was authenticated by STR profiling. The
results were as follows: D5S818: 11,12; D13S317: 9,13; D7S820:
10,10; D16S539: 12,12; vWA: 17,17; THO1: 9,9; Amelogenin: XY; TPOX:
8,9; and CSF1PO: 10,11. Normoxia or hypoxia was created using the
Anoxomat Mark II Anaerobic system (Mart Microbiology B.V.).
Reagents
Small interference (si)RNAs targeting MIAT
(si-MIAT#1, si-MIAT#2) and a scramble control (si-NC), as well as
siRNA targeting HIF-1α (si-HIF-1α) and its negative control (NC)
(si-nc) were obtained from Shanghai GenePharma Co., Ltd. miR-203a
mimic (cat. no. miR10000264-1-5) and its NC (miR-NC, cat. no.
miR1N0000001-1-5), and miR-203a inhibitor (anti-miR-203a; cat. no.
miR20000264-1-5) and its NC (anti-miR-NC; cat. no.
miR2N0000001-1-5) were ordered from Guangzhou RiboBio Co., Ltd. The
HIF-1α coding region was cloned into the pcDNA3.1 vector by
Shanghai Genomeditech Co., Ltd. to obtain a pcDNA-HIF-1α
overexpression plasmid. The target sequences of si-MIAT#1 and
si-MIAT#2 were 5′-CCAGGCTCCTTTAAACCAA-3′ and
5′-GCAGTTCTTAGCTCATATA-3′, respectively. The target sequences of
si-HIF-1α were 5′-GGCCGCTCAATTTATGAAT-3′ and
5′-GCTGGAGACACAATCATAT-3′.
Cell transfection
HepG2 cells were transfected with corresponding
biotin-labeled miRNAs, biotin-labeled probes, miRNA mimics, miRNA
inhibitors, siRNAs or plasmids, and cultured for 24 h under
normoxia or hypoxia conditions. Cell transfection was performed
using DharmaFECT 4 reagent (cat. no. T-2004-0X) or DharmaFECT Duo
transfection reagent (cat. no. T-2010-03) (both from Thermo Fisher
Scientific, Inc.) according to the instructions of the
manufacturer.
RT-qPCR assay
Total RNA was isolated from HepG2 cells and liver
tumors using TRIzol reagent (cat. no. 15596018; Thermo Fisher
Scientific, Inc.) following the protocols of the manufacturer. The
miR-203a level was measured using Bulge-Loop miRNA qRT-PCR Starter
kit (cat. no. C10211-1; Guangzhou RiboBio Co., Ltd.) and Bulge-Loop
miRNA primer sets (cat. no. MQPS0000787-1-100; Guangzhou RiboBio
Co., Ltd.) with small nuclear RNA U6 (cat. no. MQPS0000002-1-100;
Guangzhou RiboBio Co., Ltd.) as the internal control. cDNA first
strand synthesis reaction programs included reverse transcription
at 42°C for 60 min and enzyme inactivation at 70°C for 10 min.
Quantitative PCR reaction programs consisted of pre-denaturation at
95°C for 10 min and 40 cycles of denaturation at 95°C for 2 sec,
annealing at 60°C for 20 sec and extension at 70°C for 10 sec.
For detection of MIAT, HIF-1α and GAPDH levels, cDNA
was synthesized from an RNA template using M-MLV reverse
transcriptase (cat. no. 28025021; Thermo Fisher Scientific, Inc.)
and quantified using Fast SYBR™ Green Master mix (cat. no. 4385617;
Thermo Fisher Scientific, Inc.). GAPDH functioned as the
house-keeping gene to normalize the expression of MIAT and HIF-1α.
The primer sequences for MIAT, HIF-1α and GAPDH were as follows:
HIF-1α forward, 5′-CCACCTATGACCTGCTTGGT-3′ and reverse,
5′-TGTCCTGTGGTGACTTGTC-3′; MIAT forward, 5′-CAAAGAGCCCTCTGCACTAG-3′
and reverse, 5′-ACCTTGGTTACCCCTGTGATG-3′; and GAPDH forward,
5′-GTCTCCTCTGACTTCAACAGCG-3′ and reverse,
5′-ACCACCCTGTTGCTGTAGCCAA-3′. Reverse transcription reaction
samples without M-MLV reverse transcriptase and ribonuclease
inhibitor were incubated at 65°C for 5 min and then immediately
chilled on ice. Next, the reverse transcription reaction was
performed at 37°C for 50 min and the reaction was subsequently
inactivated at 70°C for 15 min. Quantitative PCR reaction programs
included pre-denaturation at 95°C for 5 min and 40 cycles of
denaturation at 95°C for 5 sec and annealing/extension at 60°C for
30 sec. Quantitative PCR reactions were run on Applied Biosystems
7500 Fast Real-Time PCR System (Applied Biosystems; Thermo Fisher
Scientific, Inc.). Relative expression patterns of genes were
analyzed using the 2−ΔΔCT method (30).
Western blot assay
Proteins were extracted from HepG2 cells and liver
tumors by centrifugation using RIPA Lysis Buffer (cat. no. P0013E;
Beyotime Institute of Biotechnology) containing a protease
inhibitor cocktail (cat. no. 5892970001; Roche Diagnostics) and
then quantified using Bio-Rad Bradford Protein Assay kit (cat. no.
5000002; Bio-Rad Laboratories, Inc.). Next, 30 µg protein samples
were separated by 10% SDS-PAGE and transferred to polyvinylidene
fluoride membranes (cat. no. IPVH00010; EMD Millipore). Then, the
membranes were blocked with 5% non-fat milk for 1 h at room
temperature and incubated overnight at 4°C with primary antibodies
against HIF-1α (product code ab16066; 1:2,000 dilution), Ki-67
(product code ab16667; 1:50 dilution), vascular endothelial growth
factor (VEGF; product code ab106580; 1:1,000 dilution) and GAPDH
(product code ab181602; 1:5,000 dilution; all from Abcam).
Subsequently, the membranes were incubated for 1 h at room
temperature with a horseradish peroxidase-conjugated secondary
antibody (product codes ab205718 or ab205719; 1:5,000 dilution;
Abcam). Finally, the protein signals were detected using
SignalFire™ ECL Reagent (product no. 6883; Cell Signaling
Technology, Inc.) on a Bio-Rad ChemiDoc XRS imaging system (Bio-Rad
Laboratories, Inc.) and quantified using Quantity One 1-D Analysis
Software (version 4.6.6; Bio-Rad Laboratories, Inc.).
Cell proliferation detection
The cell proliferative capacity was assessed by
CCK-8 assay kit (cat. no. CK04-01; Dojindo Molecular Technologies,
Inc.) referring to the protocols of the manufacturer. Briefly,
cells (50,000 cells/well) transfected with corresponding
oligonucleotides or plasmids, alone or in combination, were plated
into 96-well plates and maintained under the hypoxia condition for
24 h. Next, 10 µl CCK-8 solution was added into each well and
incubated for 3 h. Finally, the absorbance was determined at 450 nm
using a SpectraMax M3 Multi-Mode microplate reader (Molecular
Devices, LLC).
Transwell migration and invasion
assays
Cell migratory and invasive potential was estimated
using 24-well Transwell chambers (cat. no. 3422; Corning Inc.)
containing 8.0 µm pore filter membrane inserts. For cell invasion
assay, the membranes on the chambers were precoated with Matrigel
(cat. no. 356234; BD Biosciences). At 24 h after transfection, the
transfected cells (3×104) were resuspended into
serum-free medium and then added into the upper chambers. Complete
medium supplemented with 20% FBS was added into the lower chambers.
After 24 h of incubation under hypoxia conditions, cells on the
upper chambers were removed using a cotton swab. The cells attached
on the bottom surface of the membranes were fixed with pre-cold
methanol for 20 min at 4°C, stained with crystal violet solution
(0.1%; cat. no. V5265; Sigma-Aldrich; Merck KGaA) for 30 min at
room temperature and counted in 5 random fields under a light
microscope (magnification ×200). The Transwell migration assay was
performed following the same procedures as described in the cell
invasion assay, i.e., in Transwell chambers, but without
Matrigel.
Luciferase reporter assay
Potential interaction between miR-203a and MIAT or
HIF-1α was predicted through miRcode (http://www.mircode.org/) or TargetScan Human7.2
(http://www.targetscan.org/vert_72/),
respectively. Partial sequences of MIAT and HIF-1α 3′ untranslated
region (UTR) containing the corresponding miR-203a binding sites
were cloned into the psiCHECK-2 vector by Hanbio Biotechnology Co.,
Ltd. to generate MIAT-wild-type (Wt) reporter and HIF-1α-Wt
reporter, respectively. MIAT-mutant (Mut) reporter and HIF-1α-Mut
reporter were also produced by mutating matching miR-203a
complementary sites by Hanbio Biotechnology Co., Ltd. HepG2 cells
were co-transfected with MIAT-Wt reporter, MIAT-Mut reporter,
HIF-1α-Wt reporter, or HIF-1α-Mut reporter and miR-NC or miR-203a
mimic. At 48 h after transfection, luciferase activities were
measured using the Dual-Luciferase Reporter Assay system (cat. no.
E1910; Promega Corporation) according to the instructions of the
manufacturer.
RNA pull-down assay
An RNA pull-down assay was carried out as previously
described (31) to explore whether
MIAT could bind to miR-203a through putative binding sites.
Biotin-labeled wt miR-203a (Bio-miR-203a), biotin-labeled mutant
miR-203a containing mutated MIAT binding sites (Bio-miR-203a-Mut)
and their NC (Bio-miR-NC), as well as biotin-labeled wt MIAT probe
(Bio-MIAT), biotin-labeled mutant MIAT probe (Bio-MIAT-Mut) and a
scramble probe (Bio-NC) were obtained from Sangon Biotech Co., Ltd.
HepG2 cells were transfected with matching biotin-labeled miRNAs or
probes. After 48 h of culture, cells were collected and lysed with
lysis buffer (20 mM Tris-HCl (pH 7.5), 100 mM KCl, 5 mM
MgCl2, 0.3% IGEPAL CA-630 (cat. no. I8896;
Sigma-Aldrich) supplemented with protease inhibitor (cat. no.
5892970001; Roche Diagnostics) and RNase inhibitor (cat. no.
EO0381; Thermo Fisher Scientific, Inc.). Next, cell lysates were
co-incubated overnight at 4°C with pre-treated
Dynabeads® M-270 Streptavidin (cat. no. 11205D; Thermo
Fisher Scientific, Inc.). Subsequently, RNA was isolated and
purified from the beads. Finally, MIAT and miR-203a enrichment
levels were determined by RT-qPCR assay.
RNA immunoprecipitation (RIP)
assay
A RIP assay was carried out in HepG2 cells using the
EZ-Magna RIP kit (cat. no. 17-701; EMD Millipore) and
anti-argonaute 2 (Ago2; cat. no. 03-110; Part #CS204386; EMD
Millipore; 1:200 dilution) or IgG (cat. no. 03-110; Part #CS200621;
EMD Millipore; 1:200 dilution) antibodies following the
instructions of the manufacturer. RNA was isolated and purified
from IgG or Ago2 immunoprecipitation complexes. MIAT and miR-203a
levels were assessed by RT-qPCR assay.
Animal experiments
Adult male Wistar rats (200–220 g) were obtained
from the Laboratory Animal Center of Zhengzhou University and
housed in a pathogen-free environment with a temperature of 22±2°C
and a relative humidity of 50±5% under light-dark cycles (12 h
light/dark). In addition, food and water were provided ad
libitum. Animal experiments were performed with the approval of
the Animal Ethics Committee of Henan Provincial People's Hospital
and the procedures for the Care and Use of Laboratory Animals in
cancer research. Sustained short hairpin RNA against MIAT (sh-MIAT)
and a scramble control (sh-NC) were obtained from Hanbio
Biotechnology Co., Ltd. In this study, a total of 60 rats were used
in the animal experiments. Rat liver cancer cells (106
cells/0.1 ml of normal saline) infected with or without sh-NC or
sh-MIAT lentiviruses were injected into the fascia under the neck
skin of rats (10 rats in each group). All rats were euthanized
using intraperitoneal phenobarbital injection (100 mg/kg; Guangdong
Bangmin Pharmaceutical Co., Ltd.) at 14 days after injection
(experimental animals are euthanized when they are dying or
obsolete due to various factors or they have fulfilled the tasks of
experiments). After breathing arrest, tumors with a size of ~1
cm3 were harvested. Then, fresh tumors (~1 mm in
diameter) without tumor capsules and necrotic tissues were
implanted into the livers of rats as previously described (32). On day 14 after tumor transplantation
(~1 cm3 in tumor volume), TAE was performed following
previously described procedures (32,33).
The NC, TAE+sh-NC, and TAE+sh-MIAT groups contained 10 rats prior
to tumor transplantation. However, 2 rats after sham or TAE
treatment in NC, 1 rat after TAE+sh-NC treatment, and 1 rat after
TAE+sh-MIAT treatment, were revealed to be dead. These deaths may
have been caused by anaesthetization, surgery or liver cancer. Five
rats in the NC, TAE+sh-NC, or TAE+sh-MIAT groups were euthanized
and tumors were resected 2 weeks later, and the tumor volumes were
measured using the formula: V = 0.5 × L (longest tumor diameter) ×
S (shortest tumor diameter) × S (shortest tumor diameter). The
expression levels of MIAT, miR-203a, HIF-1α and VEGF were measured
by RT-qPCR and western blot assay in non-necrotic liver tumors.
Animal health and behavior were monitored every 3 days
post-surgery.
Hematoxylin and eosin (H&E)
staining
H&E staining of liver tumor tissues was
performed 2 weeks after TAE. In brief, formaldehyde-fixed tumor
tissues were embedded in paraffin. Thereafter, the sections (5 µm)
were dewaxed with xylene, hydrated with gradient ethanol solutions,
rinsed with water and stained with hematoxylin staining solution
for 10 min and eosin staining solution for 2 min at room
temperature. Images of the tissue sections were obtained using an
optical microscope (magnification ×200).
Statistical analysis
Data were obtained from >3 independent
experiments and analyzed with GraphPad Prism 7 software (GraphPad
Software, Inc.). Analysis of differences between groups was
conducted using Student's t-test (2-groups data) or one-way ANOVA
with Tukey's post hoc test. (>2-groups data). P<0.05 was
considered to indicate a statistically significant difference.
Results
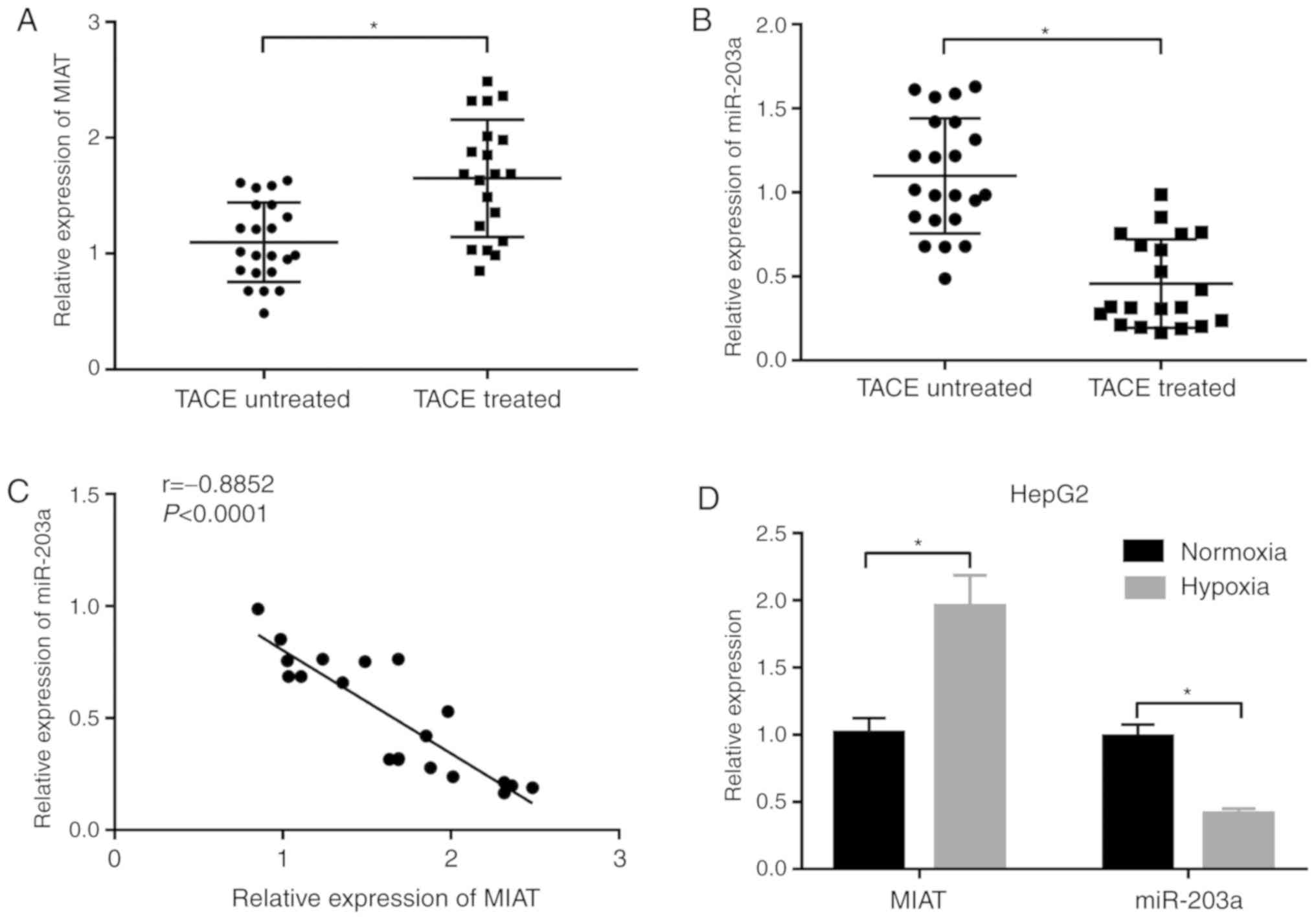
MIAT is highly expressed and miR-203a
is lowly expressed in liver tumors of patients with liver cancer
with TACE treatment and hypoxia-stimulated liver cancer cells
Clinical data revealed that the MIAT level was
notably upregulated and miR-203a expression was markedly
downregulated in the liver tumors of 20 patients with liver cancer
who underwent TACE treatment compared with 22 patients with liver
cancer without TACE treatment (Fig. 1A
and B). Moreover, the miR-203a level was negatively associated
with the MIAT level in the liver tumors of 20 patients with liver
cancer who underwent TACE treatment (Fig. 1C). Consistently, higher MIAT
expression and lower miR-203a expression were observed in HepG2
cells under hypoxia conditions compared with cells under normoxia
conditions (Fig. 1D). These data
indicated that MIAT and miR-203a may play vital roles in TACE
treatment for liver cancer.
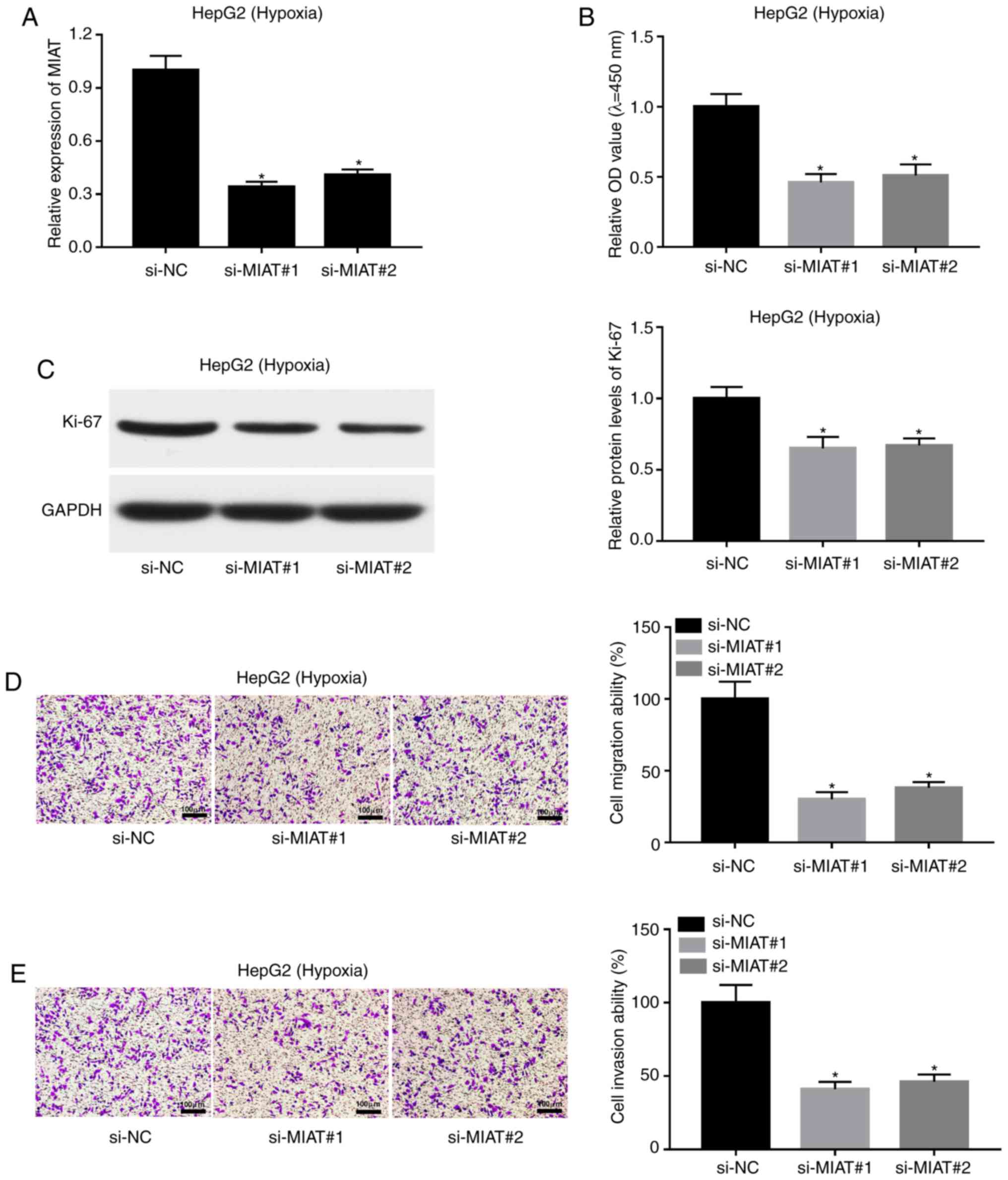
MIAT knockdown suppresses
proliferation, migration and invasion in hypoxia-stimulated HepG2
cells
Next, si-MIAT#1, si-MIAT#2 and a scramble control
(si-NC) were synthesized to evaluate the effect of MIAT loss on the
proliferative, migratory and invasive capacities of HepG2 cells
under hypoxic conditions. Firstly, transfection efficiency analysis
revealed that the transfection of si-MIAT#1 or si-MIAT#2 could
significantly reduce the expression level of MIAT in
hypoxia-treated HepG2 cells compared with cells transfected with
si-NC (Fig. 2A). A CCK-8 assay
revealed that MIAT depletion inhibited proliferation in HepG2 cells
under hypoxic conditions (Fig. 2B).
The present results also demonstrated that the expression of
proliferative markers such as Ki-67 was markedly decreased in
hypoxia-exposed HepG2 cells following MIAT knockdown (Fig. 2C). Moreover, MIAT loss led to a
noticeable reduction in the migratory and invasive abilities of
HepG2 cells exposed to hypoxic conditions (Fig. 2D and E).
MIAT knockdown induces miR-203a
expression by direct interaction
Subsequently, bioinformatics analysis indicated that
MIAT could interact with miR-203a (Fig.
3A). To further demonstrate this prediction, MIAT-Wt reporter
containing wt miR-203a binding sites and MIAT-Mut reporter
containing mutant miR-203a binding sites were constructed.
Following luciferase reporter assay, the results revealed that
miR-203a overexpression led to a significant downregulation of
luciferase activity of the MIAT-Wt reporter, but did not have a
marked effect on the luciferase activity of the MIAT-Mut reporter
(Fig. 3B). Ago2, a core component
of the RNA-induced silencing complex, plays vital roles in
miRNA-mediated gene silencing. Hence, a RIP assay was performed
using an anti-Ago2 antibody in HepG2 cells to explore whether MIAT
could bind miR-203a. The results revealed that MIAT and miR-203a
were significantly enriched in the anti-Ago2 group (Fig. 3C), indicating a possible interaction
between MIAT and miR-203a in HepG2 cells. Moreover, RNA pull-down
assay revealed that MIAT was significantly enriched by Bio-miR-203a
but not by Bio-miR-203a-Mut (Fig.
3D). Similarly, Bio-MIAT could pull down abundant miR-203a in
HepG2 cells, while Bio-MIAT-Mut had little enrichment effect on
miR-203a (Fig. 3E). In summary,
these outcomes revealed that MIAT could directly bind to miR-203a
in HepG2 cells. Next, it was further demonstrated that MIAT loss
mediated by si-MIAT#1 or si-MIAT#2 led to a notable increase in the
expression level of miR-203a in HepG2 cells exposed to hypoxic
conditions (Fig. 3F).
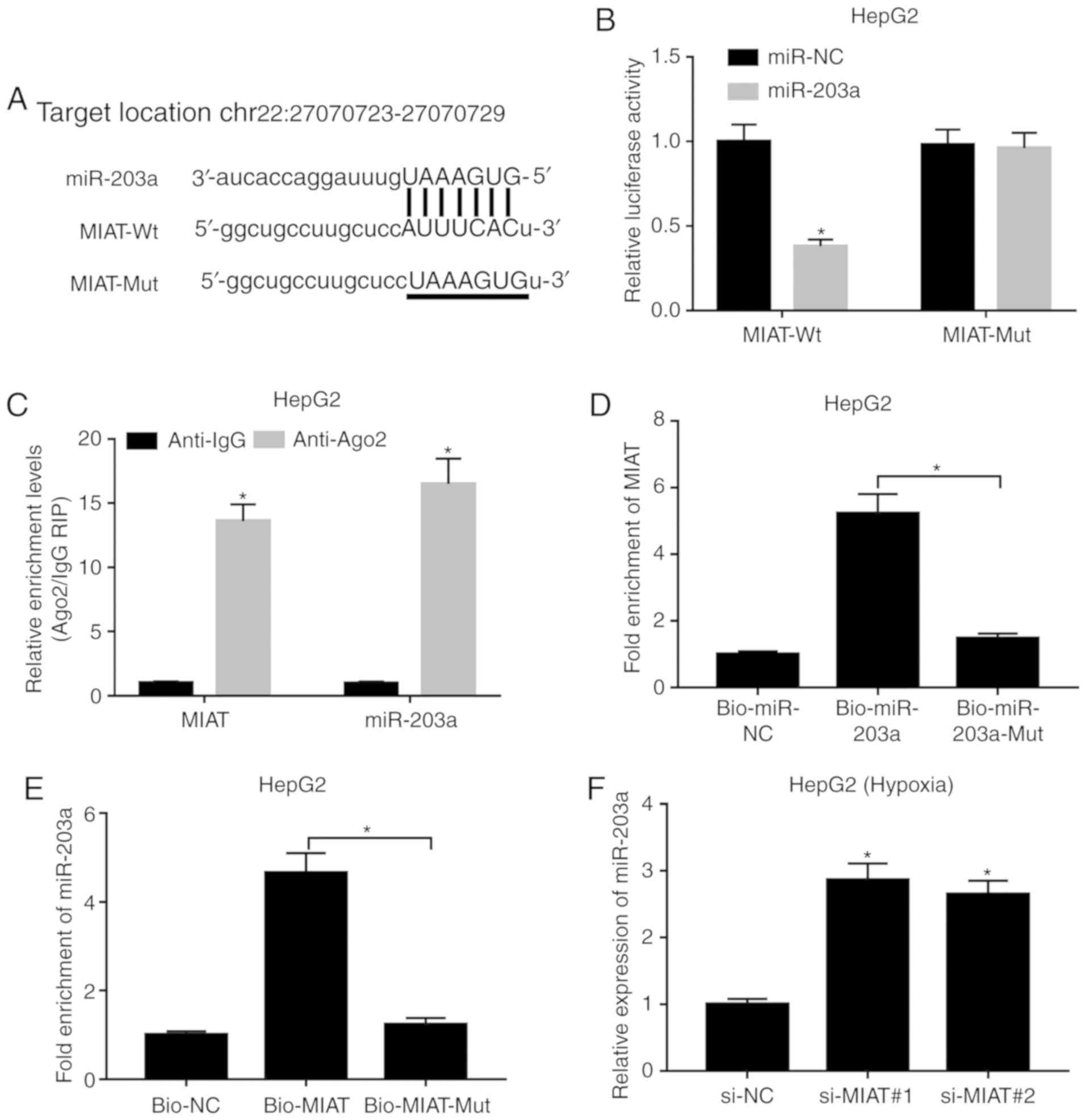 | Figure 3.MIAT knockdown induces miR-203a
expression by direct interaction. (A) Predicted binding sites
between MIAT and miR-203a by miRcode and mutant sites in MIAT-Mut
reporter. (B) HepG2 cells were co-transfected with miR-NC or
miR-203a and MIAT-Wt or MIAT-Mut reporter, followed by detection of
luciferase activities at 48 h after transfection. (C) The
enrichment degree of MIAT and miR-203a in IgG or Ago2
immunoprecipitation complex was assessed by RNA immunoprecipitation
and RT-qPCR assays. (D) HepG2 cells were transfected with
Bio-miR-NC, Bio-miR-203a or Bio-miR-203a-Mut. At 48 h
post-transfection, the MIAT level pulled down by the above
biotin-labeled miRNAs was assessed by RNA pull-down and RT-qPCR
assays. (E) HepG2 cells were transfected with Bio-NC, Bio-MIAT or
Bio-MIAT-Mut. After 48 h of incubation, miR-203a level enriched by
these biotin-labeled probes was detected through RNA pull-down and
RT-qPCR assays. (F) HepG2 cells transfected with si-NC, si-MIAT#1
or si-MIAT#2 were exposed to hypoxic conditions for 24 h. Then, the
expression level of miR-203a was assessed by RT-qPCR assay.
*P<0.05. MIAT, myocardial infarction associated transcript; si,
small interference; miR, microRNA; RT-qPCR, reverse
transcription-quantitative PCR; wt, wild type; mut, mutant; Ago2,
argonaute 2; NC, negative control; Bio, biotin. |
Downregulation of miR-203a weakens the
detrimental effects of MIAT knockdown on proliferation, migration
and invasion of HepG2 cells under hypoxic conditions
Next, an RT-qPCR assay validated that the
transfection of miR-203a mimic led to a marked increase in the
expression level of miR-203a, and the addition of anti-miR-203a
inhibited the increase in the expression level of miR-203a induced
by MIAT knockdown in HepG2 cells under hypoxic conditions (Fig. 4A). Furthermore, miR-203a
overexpression decreased cell proliferation (Fig. 4B), inhibited the expression of
proliferative markers such as Ki-67 (Fig. 4C), and decreased cell migratory and
invasive abilities in HepG2 cells under hypoxic conditions
(Fig. 4D and E). Moreover, it was
further demonstrated that miR-203a loss weakened the detrimental
effects of MIAT knockdown on the proliferation, migration and
invasion of HepG2 cells under hypoxic conditions (Fig. 4B-E).
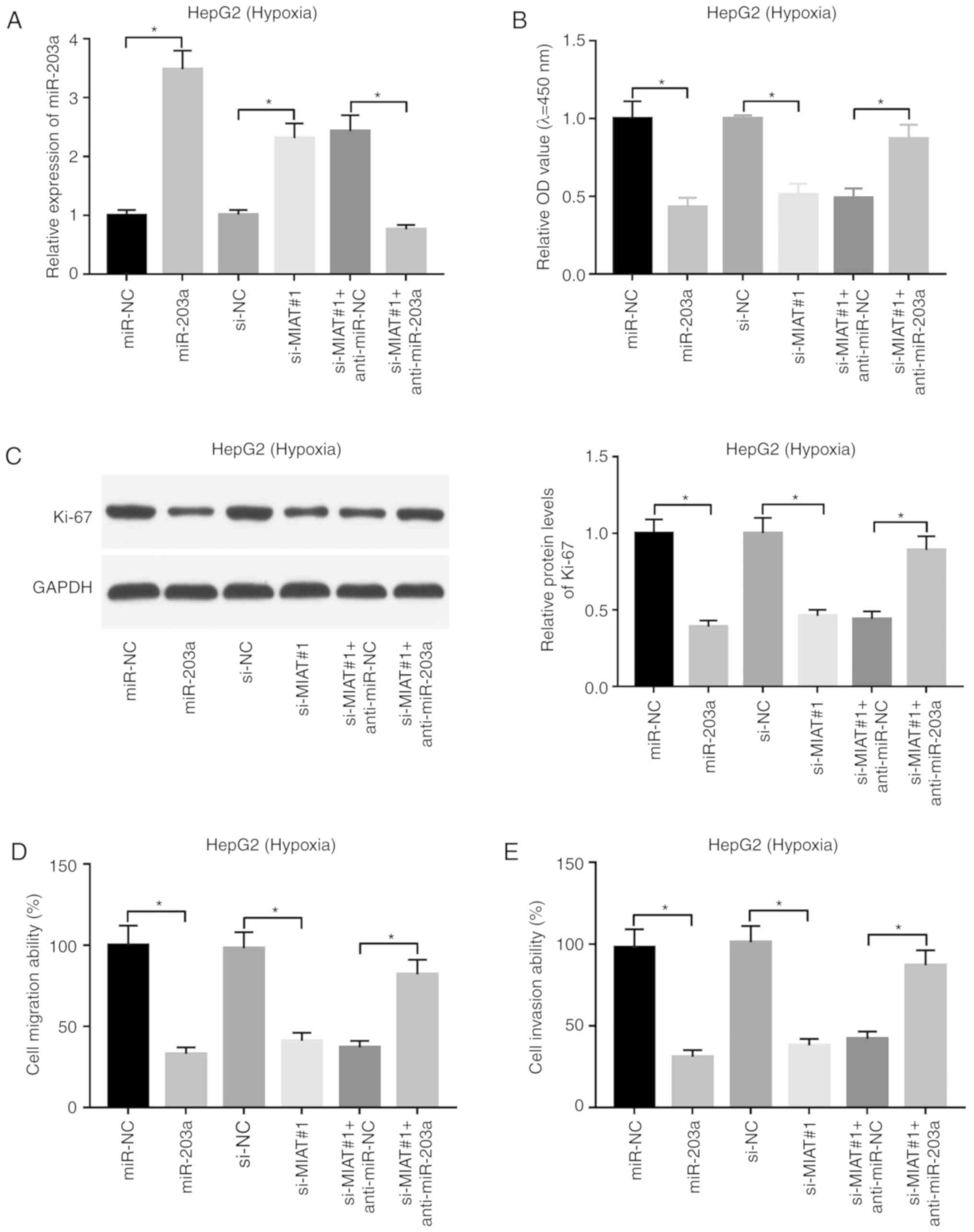 | Figure 4.Downregulation of miR-203a weakens
the detrimental effects of MIAT knockdown on the proliferation,
migration and invasion of HepG2 cells under hypoxic conditions.
(A-E) HepG2 cells transfected with miR-NC, miR-203a, si-NC,
si-MIAT#1, si-MIAT#1 + anti-miR-NC or si-MIAT#1 + anti-miR-203a
were cultured under hypoxic conditions for 24 h. (A) The expression
level of miR-203a was assessed by RT-qPCR assay. (B) Cell
proliferative ability was assessed by CCK-8 assay. (C) The protein
levels of Ki-67 were determined by western blot assay. (D) Cell
migratory and (E) invasive capacities were determined by Transwell
migration and invasion assays. *P<0.05. MIAT, myocardial
infarction associated transcript; si, small interference; miR,
microRNA; NC, negative control; RT-qPCR, reverse
transcription-quantitative PCR. |
HIF-1α is a downstream target of the
MIAT/miR-203a axis
Prediction results revealed the existence of some
complementary bases between HIF-1α 3′UTR and miR-203a (Fig. 5A), indicating that HIF-1α is a
potential target of miR-203a. The results of luciferase reporter
assay revealed that miR-203a overexpression led to a significant
decrease in the luciferase activity of the HIF-1α-Wt reporter,
while miR-203a upregulation did not have a marked effect on the
luciferase activity of the HIF-1α-Mut reporter, which contains
mutant miR-203a binding sites (Fig.
5B), suggesting that miR-203a could directly bind to HIF-1α
3′UTR via putative binding sites. In addition, the present study
demonstrated that the expression level of HIF-1α was significantly
increased in the liver tumors of 20 patients with liver cancer who
underwent TACE treatment compared with the 22 patients with liver
cancer without TACE treatment (Fig.
5C). Consistently, higher HIF-1α expression was observed in
HepG2 cells under hypoxic conditions than in cells under normoxic
conditions (Fig. 5D). Moreover,
RT-qPCR and western blot assays revealed that miR-203a
overexpression or MIAT knockdown led to a significant reduction in
HIF-1α mRNA and protein levels in hypoxia-treated HepG2 cells
(Fig. 5E and F). miR-203 loss
weakened the inhibitory effect of MIAT knockdown on HIF-1α mRNA and
protein expression in hypoxia-stimulated HepG2 cells (Fig. 5E and F).
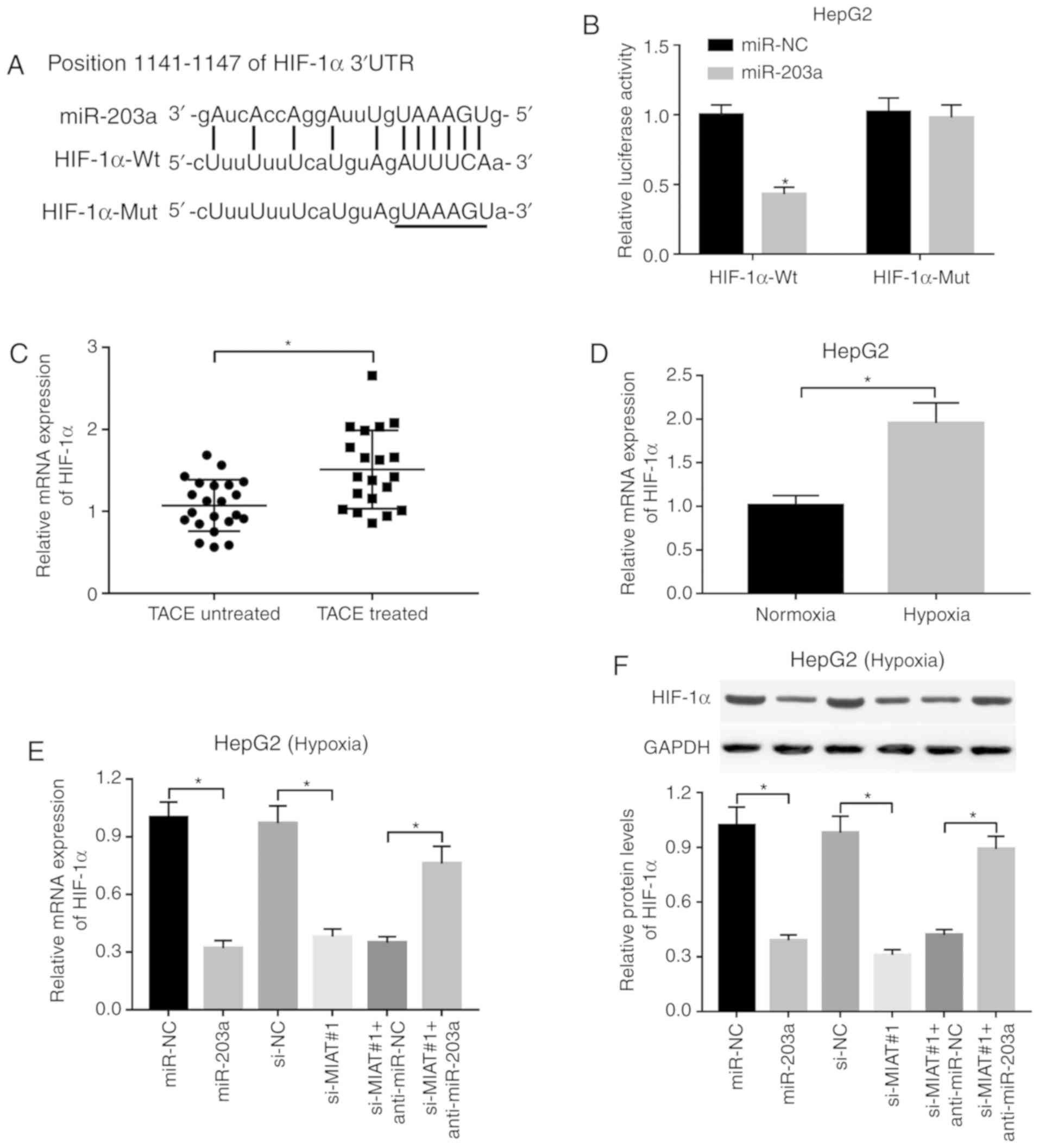 | Figure 5.HIF-1α is a downstream target of the
MIAT/miR-203a axis. (A) Putative complementary sites between
miR-203a and HIF-1α 3′-untranslated region by TargetScan prediction
website and mutant sites in HIF-1α-Mut reporter. (B) The effect of
miR-203a overexpression or no overexpression on the luciferase
activities of HIF-1α-Wt or HIF-1α-Mut reporter was detected by
luciferase reporter assay at 48 h after transfection in HepG2
cells. (C) The mRNA level of HIF-1α was assessed by RT-qPCR assay
in liver tumors of patients with liver cancer with or without TACE
treatment, and (D) HepG2 cells under normoxic or hypoxic
conditions. HepG2 cells transfected with miR-NC, miR-203a, si-NC,
si-MIAT#1, si-MIAT#1 + anti-miR-NC or si-MIAT#1 + anti-miR-203a
were cultured under hypoxic conditions for 24 h. Next, (E) mRNA and
(F) protein levels of HIF-1α were detected by RT-qPCR and western
blot assays, respectively. *P<0.05. MIAT, myocardial infarction
associated transcript; miR, microRNA; HIF-1α, hypoxia-inducible
factor 1-α; Mut, mutant; RT-qPCR, reverse
transcription-quantitative PCR; si, small interference; NC,
negative control. |
Effects of HIF-1α knockdown and
overexpression on the proliferation, migration and invasion of
HepG2 cells under hypoxic conditions
si-HIF-1α and pcDNA-HIF-1α overexpression plasmid
were synthesized to further explore HIF-1α function in liver
cancer. A western blot assay demonstrated that the addition of
si-HIF-1α effectively triggered a reduction in the expression level
of HIF-1α in hypoxia-treated HepG2 cells (Fig. 6A). Conversely, transfection of
pcDNA-HIF-1α led to a significant increase in the protein
expression level of HIF-1α in hypoxia-stimulated HepG2 cells
(Fig. 6A). Subsequent
loss-of-function analysis revealed that depletion of HIF-1α
decreased cell proliferation (Fig.
6B), migration (Fig. 6D) and
invasion (Fig. 6E), and suppressed
Ki-67 expression (Fig. 6C) in
hypoxia-treated HepG2 cells. Conversely, enforced expression of
HIF-1α induced cell proliferation (Fig.
6B), migration (Fig. 6D) and
invasion (Fig. 6E), and increased
Ki-67 expression levels (Fig. 6C)
in hypoxia-stimulated HepG2 cells.
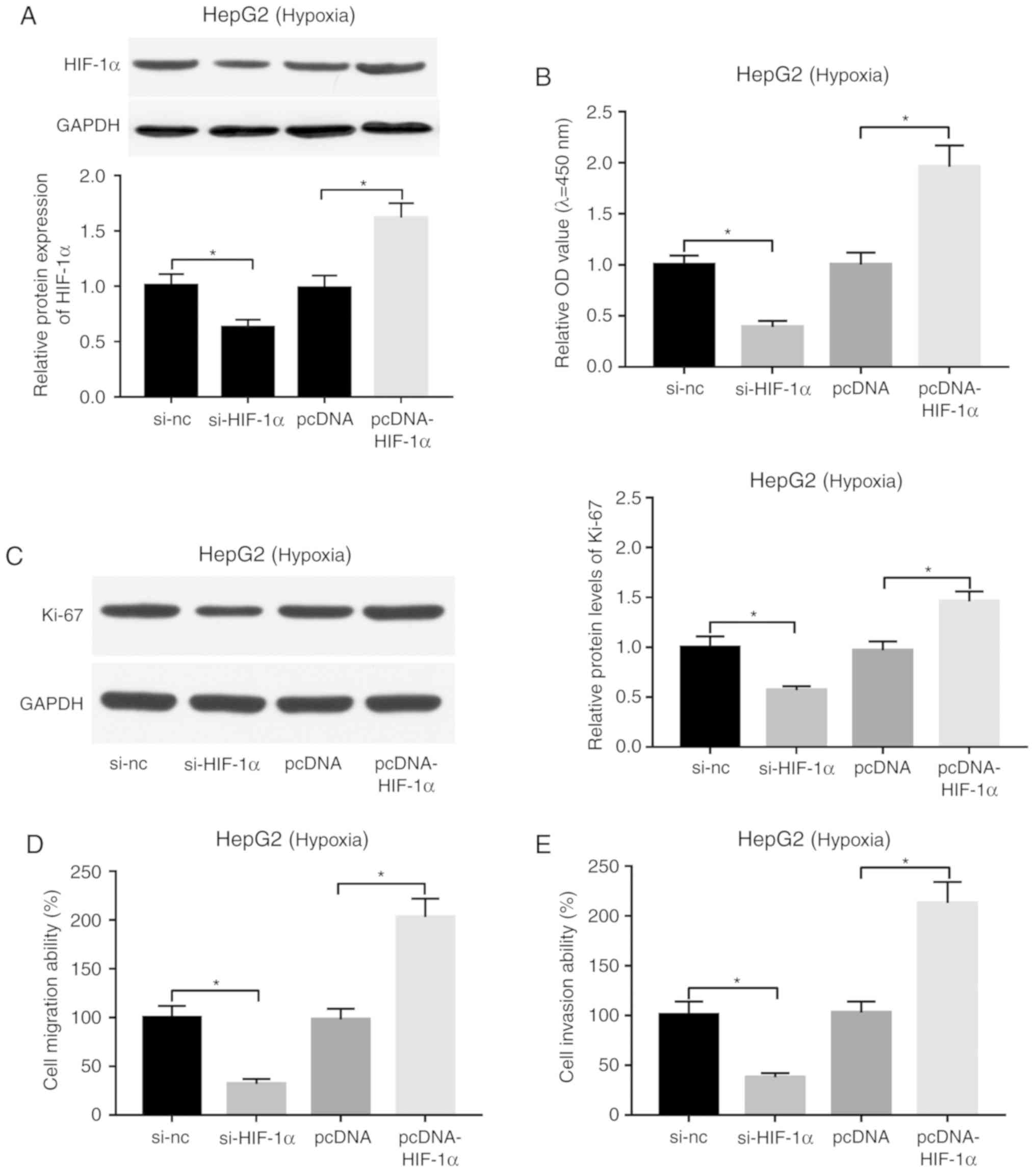 | Figure 6.Effect of HIF-1α knockdown or
overexpression on proliferation, migration and invasion of
hypoxia-treated HepG2 cells. (A-E) HepG2 cells transfected with
si-NC, si-HIF-1α, pcDNA or pcDNA-HIF-1α were cultured under hypoxic
conditions for 24 h, followed by the assessment of (A) HIF-1α
protein level, (B) cell proliferative ability, (C) Ki-67 protein
expression, (D) cell migratory and (E) invasive capacities.
*P<0.05. HIF-1α, hypoxia-inducible factor 1-α; si, small
interference; NC, negative control. |
MIAT knockdown enhances TAE-mediated
antitumor effects by upregulating miR-203a and downregulating
HIF-1α in rat liver tumors
The present study demonstrated that TAE inhibited
the growth of rat liver tumors and MIAT knockdown enhanced the
TAE-mediated antitumor effect in rat liver tumors (Fig. 7A). Lack of MIAT led to markedly
fewer blood vessels (Fig. 7B).
Moreover, TAE treatment led to a significant increase in MIAT and
HIF-1α mRNA levels, and a significant reduction in the expression
level of miR-203a in rat liver tumors (Fig. 7C). Furthermore, the MIAT and HIF-1α
mRNA levels were significantly reduced, while the expression level
of miR-203a was significantly increased, in TAE-treated rat liver
tumors following MIAT knockdown (Fig.
7C). In addition, the protein levels of HIF-1α and VEGF were
significantly increased in TAE-treated rat liver tumors compared
with untreated tumors (Fig. 7C).
Moreover, MIAT knockdown inhibited the increase in HIF-1α and VEGF
protein levels induced by TAE in rat liver tumors (Fig. 7D).
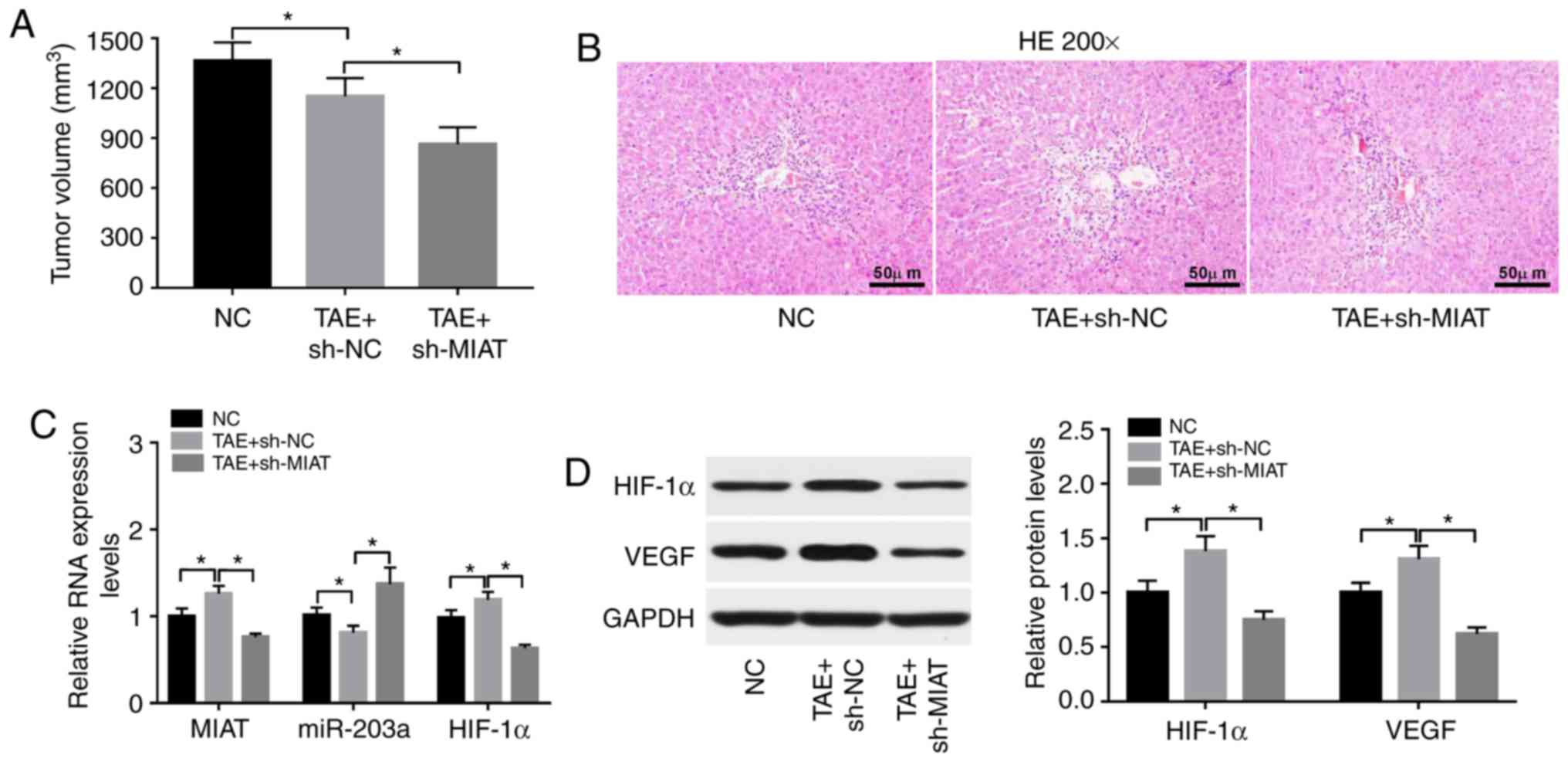 | Figure 7.MIAT knockdown enhances TAE-mediated
antitumor effect by upregulating miR-203a and downregulating HIF-1α
in rat liver tumors. (A) The effect of TAE alone or in combination
with MIAT loss on the volume of rat liver tumors was measured 3
weeks after TAE treatment. The maximum diameter of tumors was 16.1,
14.4 and 12.6 mm in the NC, TAE+sh-NC, and TAE+sh-MIAT groups,
respectively. The maximum volume of tumors was 1502, 1298 or 986
mm3 in the NC, TAE+sh-NC, and TAE+sh-MIAT groups,
respectively. (B) H&E staining of liver tumor tissues. (C)
MIAT, microRNA-203a and HIF-1α mRNA levels were determined by
RT-qPCR assay in rat liver tumors of the sham (NC), TAE and TAE
treatment + MIAT loss groups. (D) Protein levels of HIF-1α and VEGF
were detected by western blot assay in rat liver tumors of the sham
(NC), TAE and TAE treatment + MIAT loss groups. *P<0.05. MIAT,
myocardial infarction associated transcript; TAE, transcatheter
arterial embolization; HIF-1α, hypoxia-inducible factor 1-α;
H&E, hematoxylin and eosin; RT-qPCR, reverse
transcription-quantitative PCR; VEGF, vascular endothelial growth
factor; NC, negative control. |
Discussion
Although surgical resection or liver transplantation
is the principal therapeutic strategy for patients with liver
cancer, only a fraction of patients at the early stage are suitable
for these treatment options (34,35).
Recently, TAE and TACE have emerged as effective locoregional
therapies for each stage of patients with liver cancer, especially
for those with unresectable liver cancer (35,36).
Compared with surgical resection or liver transplantation, TAE and
TACE have advantages such as lesser trauma, slight pain and rapid
recovery. In comparison with chemotherapy, TACE can deliver higher
doses of chemotherapeutic drugs to specific tissues with reduced
systemic drug toxicity (37,38).
The present study demonstrated that MIAT and HIF-1α
were highly expressed, while miR-203a was lowly expressed, in liver
tumors of patients with liver cancer who underwent TACE treatment
and in liver cancer cells upon hypoxia treatment. MIAT expression
was negatively associated with miR-203a expression in liver tumor
samples of patients with liver cancer who underwent TACE treatment.
MIAT could directly bind to miR-203a, and HIF-1α was a direct
target of miR-203a. Moreover, MIAT positively regulated HIF-1α
expression via miR-203a in hypoxia-treated liver cancer cells.
Functional analysis revealed that MIAT knockdown, miR-203a
overexpression or HIF-1α silencing suppressed cell proliferation,
migration and invasion in hypoxia-exposed liver cancer cells.
Conversely, HIF-1α overexpression exerted the opposite function.
Moreover, miR-203a downregulation alleviated the detrimental
effects of MIAT loss on proliferation, migration and invasion in
liver cancer cells under hypoxic conditions. Additionally, MIAT
knockdown enhanced TACE-mediated antitumor effects by increasing
miR-203a, and reducing HIF-1α and VEGF in rat liver tumors.
Prior studies revealed that MIAT knockdown could
hinder tumorigenesis and progression of liver cancer. For example,
MIAT-associated genes were greatly enriched in EMT-linked pathways,
and MIAT knockdown promoted epithelial marker E-cadherin
expression, inhibited mesenchymal marker N-cadherin expression, and
blocked cell migration and invasion in liver cancer (39). MIAT loss weakened the proliferative
and invasive abilities of liver cancer cells and hindered liver
cancer xenograft tumor growth by negatively regulating miR-214
(40). The present data revealed
that MIAT loss impaired cell proliferative, migratory and invasive
potential in hypoxia-stimulated liver cancer cells, and potentiated
TACE-mediated antitumor effects in rat liver tumors.
Following luciferase reporter assay, RIP and RNA
pull-down assays demonstrated that MIAT could directly bind to
miR-203a, and MIAT knockdown led to an increase in miR-203
expression in liver cancer cells. Enforced expression of miR-203a
suppressed cell proliferation, migration and invasion, while
miR-203a downregulation mitigated the inhibitory effects of MIAT
knockdown on cell proliferation, migration and invasion in
hypoxia-treated liver cancer cells.
miR-203a has been revealed to be a tumor-suppressive
miRNA in liver cancer. For instance, miR-203a overexpression
inhibited cell proliferation, hampered cell cycle progression and
promoted cell apoptosis in liver cancer cells (Huh7 and Hep3B)
(41). Ectopic expression of
miR-203a suppressed proliferation, migration and invasion by
targeting BCAT1 in HepG2 cells (42).
Subsequent experiments demonstrated that HIF-1α was
a target of miR-203a. MIAT functioned as a molecular sponge of
miR-203a to sequester miR-203a from its target HIF-1α. HIF-1, which
is composed of HIF-1α and HIF-1β subunits, has been revealed to
induce the expression of hypoxia-responsive genes, which are
implicated in various cancer-related biological events such as
proliferation, angiogenesis and metastasis (38). HIF-1α is easily degraded by the
proteasome under normoxic conditions, whereas HIF-1α is stabilized
and dimerizes with HIF-1β under hypoxic conditions (43). A previous study indicated that the
effect of HIF-1 on the proliferation and apoptosis of liver cancer
cells was controversial (43). A
prior study revealed that HIF-1 improved the invasive and migratory
potential of liver cancer cells by inducing EMT under hypoxic
conditions (44). Consistent with
the outcomes of this study (44),
our results demonstrated that HIF-1α overexpression promoted cell
proliferation, migration and invasion in hypoxia-treated liver
cancer cells. Additionally, the present study revealed that TAE
treatment led to a notable upregulation of MIAT and HIF-1α
expression, and a marked downregulation of miR-203a in rat liver
tumors. In line with our results, Chen et al reported that
the expression level of HIF-1α was notably upregulated in
hypoxia-treated liver cancer cells, and TAE led to a marked
increase in the expression level of HIF-1α in rat liver tumors
(27).
Previous studies revealed that MIAT could facilitate
the development and progression of liver cancer by sponging miR-214
(45) and miR-22-3p (46). Moreover, miR-3662 (47), miR-338-3p (48) and miR-199a (49) inhibited liver cancer tumorigenesis
and progression by directly targeting HIF-1α. These studies
indicated that the aforementioned miRNAs may have potential
competitive effects with miR-203a.
Collectively, the present data revealed that MIAT
knockdown decreased liver cancer cell proliferation, migration and
invasion under hypoxic conditions and, potentiated TACE-mediated
antitumor effects in rat liver tumors by regulating the
miR-203a/HIF-1α axis, thus elucidating the vital roles of MIAT and
miR-203a in TACE treatment for liver cancer, and identifying a
novel and key regulatory pathway (MIAT/miR-203a/HIF-1α) in
hypoxia-associated responses and TACE treatment for liver
cancer.
Acknowledgements
Not applicable.
Funding
No funding was received from the government or
organizations.
Availability of data and materials
The data displayed in the present manuscript are
available from the corresponding author upon reasonable
request.
Authors' contributions
JL conducted the experiments and wrote the
manuscript. GC, JWL and XZ contributed to the experiments. HC
revised the manuscript. All authors participated in the design of
the study and data analysis. All authors read and approved the
manuscript and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
All procedures performed in studies involving human
participants were approved by the Research Ethics Committee of
Henan Provincial People's Hospital, and in accordance with the 1964
Declaration of Helsinki and its later amendments or comparable
ethical standards. Informed consent was obtained from all
individual participants included in the study. All animal
experiments were performed with the approval of the Animal Ethics
Committee of Henan Provincial People's Hospital and the procedures
for the Care and Use of Laboratory Animals in cancer research.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zheng R, Qu C, Zhang S, Zeng H, Sun K, Gu
X and Chen W: Liver cancer incidence and mortality in China:
Temporal trends and projections to 2030. Chin J Cancer Res.
30:571–579. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fu J and Wang H: Precision diagnosis and
treatment of liver cancer in China. Cancer Lett. 412:283–288. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bruix J and Sherman M: Management of
hepatocellular carcinoma: An update. Hepatology. 53:1020–1022.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kudo M, Ueshima K, Kubo S, Sakamoto M,
Tanaka M, Ikai I, Furuse J, Murakami T, Kadoya M, Kokudo N, et al:
Response evaluation criteria in cancer of the liver (RECICL)(2015
revised version). Hepatol Res. 46:3–9. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lanza E, Donadon M, Poretti D, Pedicini V,
Tramarin M, Roncalli M, Rhee H, Park YN and Torzilli G:
Transarterial therapies for hepatocellular carcinoma. Liver Cancer.
6:27–33. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Muz B, de la Puente P, Azab F and Azab AK:
The role of hypoxia in cancer progression, angiogenesis,
metastasis, and resistance to therapy. Hypoxia (Auck). 3:83–92.
2015. View Article : Google Scholar
|
|
8
|
Eales KL, Hollinshead KE and Tennant DA:
Hypoxia and metabolic adaptation of cancer cells. Oncogenesis.
5:e1902016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Greijer AE and Van der Wall E: The role of
hypoxia inducible factor 1 (HIF-1) in hypoxia induced apoptosis. J
Clin Pathol. 57:1009–1014. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liz J and Esteller M: lncRNAs and
microRNAs with a role in cancer development. Biochim Biophys Acta.
1859:169–176. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wong CM, Tsang HC and Ng OL: Non-coding
RNAs in hepatocellular carcinoma: molecular functions and
pathological implications. Nat Rev Gastroenterol Hepatol.
15:137–151. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Choudhry H, Harris AL and McIntyre A: The
tumour hypoxia induced non-coding transcriptome. Mol Aspects Med.
47:35–53. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chang YN, Zhang K, Hu ZM, Qi HX, Shi ZM,
Han XH, Han YW and Hong W: Hypoxia-Regulated lncRNAs in cancer.
Gene. 575:1–8. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liao J, He Q, Li M, Chen Y, Liu Y and Wang
J: LncRNA MIAT: Myocardial infarction associated and more. Gene.
578:158–161. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sun C, Huang L, Li Z, Leng K, Xu Y, Jiang
X and Cui Y: Long non-coding RNA MIAT in development and disease: A
new player in an old game. J Biomed Sci. 25:232018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bountali A, Tonge DP and
Mourtada-Maarabouni M: RNA sequencing reveals a key role for the
long non-coding RNA MIAT in regulating neuroblastoma and
glioblastoma cell fate. Int J Biol Macromol. 130:878–891. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Alipoor FJ, Asadi MH and Torkzadeh-Mahani
M: MIAT lncRNA is overexpressed in breast cancer and its inhibition
triggers senescence and G1 arrest in MCF7 cell line. J Cell
Biochem. 119:6470–6481. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xiang Y, Huang Y, Sun H, Pan Y, Wu M and
Zhang J: Deregulation of miR-520d-3p promotes hepatocellular
carcinoma development via lncRNA MIAT regulation and EPHA2
signaling activation. Biomed Pharmacother. 109:1630–1639. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jiang Q, Shan K, Qun-Wang X, Zhou RM, Yang
H, Liu C, Li YJ, Yao J, Li XM, Shen Y, et al: Long non-coding
RNA-MIAT promotes neurovascular remodeling in the eye and brain.
Oncotarget. 7:49688–49698. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Petrillo M, Patella F, Pesapane F, Suter
MB, Ierardi AM, Angileri SA, Floridi C, de Filippo M and
Carrafiello G: Hypoxia and tumor angiogenesis in the era of
hepatocellular carcinoma transarterial loco-regional treatments.
Future Oncol. 14:2957–2967. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang Z, Zhao Z, Yang Y, Luo M, Zhang M,
Wang X, Liu L, Hou N, Guo Q, Song T, et al: MiR-99b-5p and
miR-203a-3p function as tumor suppressors by targeting IGF-1R in
gastric cancer. Sci Rep. 8:101192018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jiang N, Jiang X, Chen Z, Song X, Wu L,
Zong D, Song D, Yin L, Wang D, Chen C, et al: MiR-203a-3p
suppresses cell proliferation and metastasis through inhibiting
LASP1 in nasopharyngeal carcinoma. J Exp Clin Cancer Res.
36:1382017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fang JF, Zhao HP, Wang ZF and Zheng SS:
Upregulation of RASAL2 promotes proliferation and metastasis, and
is targeted by miR-203 in hepatocellular carcinoma. Mol Med Rep.
15:2720–2726. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chang JH, Hwang YH, Lee DJ, Kim DH, Park
JM, Wu HG and Kim IA: MicroRNA-203 modulates the radiation
sensitivity of human malignant glioma cells. Int J Radiat Oncol
Biol Phys. 94:412–420. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Balamurugan K: HIF-1 at the crossroads of
hypoxia, inflammation, and cancer. Int J Cancer. 138:1058–1066.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Masoud GN and Li W: HIF-1α pathway: Role,
regulation and intervention for cancer therapy. Acta Pharmaceutica
Sinica B. 5:378–389. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen C, Wang J, Liu R and Qian S: RNA
interference of hypoxia-inducible factor-1 alpha improves the
effects of transcatheter arterial embolization in rat liver tumors.
Tumor Biol. 33:1095–1103. 2012. View Article : Google Scholar
|
|
28
|
Sun X, Jiang H, Jiang X, Tan H, Meng Q,
Sun B, Xu R and Krissansen GW: Antisense hypoxia-inducible
factor-1α augments transcatheter arterial embolization in the
treatment of hepatocellular carcinomas in rats. Hum Gene Ther.
20:314–324. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen CS, Zhao Q, Qian S, Li HL, Guo CY,
Zhang W, Yan ZP, Liu R and Wang JH: Ultrasound-guided RNA
interference targeting HIF-1 alpha improves the effects of
transarterial chemoembolization in rat liver tumors. Onco Targets
Ther. 8:3539–3548. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2− ΔΔCT method. Methods. 25:402–408.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Phatak P and Donahue JM: Biotinylated
micro-RNA pull down assay for identifying miRNA targets. Bio
Protoc. 7:2017. View Article : Google Scholar
|
|
32
|
Zhou B, Wang J and Yan Z: Ginsenoside Rg3
attenuates hepatoma VEGF overexpression after hepatic artery
embolization in an orthotopic transplantation hepatocellular
carcinoma rat model. Onco Targets Ther. 7:1945–1954. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kan Z, Sato M, Ivancev K, Uchida B,
Hedgpeth P, Lunderquist A, Rosch J and Yamada R: Distribution and
effect of iodized poppyseed oil in the liver after hepatic artery
embolization: Experimental study in several animal species.
Radiology. 186:861–866. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Daher S, Massarwa M, Benson AA and Khoury
T: Current and future treatment of hepatocellular carcinoma: An
updated comprehensive review. J Clin Transl Hepatol. 6:69–78. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kumar Y, Sharma P, Bhatt N and Hooda K:
Transarterial therapies for hepatocellular carcinoma: A
comprehensive review with current updates and future directions.
Asian Pac J Cancer Prev. 17:473–478. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Transarterial chemoembolization in
hepatocellular carcinoma treatment, . Barcelona clinic liver cancer
staging system. World J Gastroenterol. 21:10327–10335. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Rammohan A, Sathyanesan J, Ramaswami S,
Lakshmanan A, Senthil-Kumar P, Srinivasan UP, Ramasamy R and
Ravichandran P: Embolization of liver tumors: Past, present and
future. World J Radiol. 4:405–412. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Vogl TJ, Naguib NN, Nour-Eldin NE, Rao P,
Emami AH, Zangos S, Nabil M and Abdelkader A: Review on
transarterial chemoembolization in hepatocellular carcinoma:
Palliative, combined, neoadjuvant, bridging, and symptomatic
indications. Eur J Radiol. 72:505–516. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang Z, Wang S and Liu W: EMT-Related
long non-coding RNA in hepatocellular carcinoma: A study with TCGA
database. Biochem Biophys Res Commun. 503:1530–1536. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Huang X, Gao Y, Qin J and Lu S: lncRNA
MIAT promotes proliferation and invasion of HCC cells via sponging
miR-214. Am J Physiol Gastrointest Liver Physiol. 314:G559–G565.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Furuta M, Kozaki KI, Tanaka S, Arii S,
Imoto I and Inazawa J: MiR-124 and miR-203 are epigenetically
silenced tumor-suppressive microRNAs in hepatocellular carcinoma.
Carcinogenesis. 31:766–776. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ji D, Jiang C, Zhang L, Liang N, Jiang T,
Yang B and Liang H: LncRNA CRNDE promotes hepatocellular carcinoma
cell proliferation, invasion, and migration through regulating
miR-203/BCAT1 axis. J Cell Physiol. 234:6548–6560. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Luo D, Wang Z and Wu J, Jiang C and Wu J:
The role of hypoxia inducible factor-1 in hepatocellular carcinoma.
BioMed Res Int. 2014:4092722014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhang L, Huang G, Li X, Zhang Y, Jiang Y,
Shen J, Liu J, Wang Q, Zhu J, Feng X, et al: Hypoxia induces
epithelial-mesenchymal transition via activation of SNAI1 by
hypoxia-inducible factor-1α in hepatocellular carcinoma. BMC
Cancer. 13:1082013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Huang X, Gao Y, Qin J and Lu S: lncRNA
MIAT promotes proliferation and invasion of HCC cells via sponging
miR-214. Am J Physiol Gastrointest Liver Physiol. 314:G559–G565.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhao L, Hu K, Cao J, Wang P, Li J, Zeng K,
He X, Tu PF, Tong T and Han L: lncRNA miat functions as a ceRNA to
upregulate sirt1 by sponging miR-22-3p in HCC cellular senescence.
Aging (Albany NY). 11:7098–7122. 2019.PubMed/NCBI
|
|
47
|
Chen Z, Zuo X, Zhang Y, Han G, Zhang L, Wu
J and Wang X: MiR-3662 suppresses hepatocellular carcinoma growth
through inhibition of HIF-1α-mediated warburg effect. Cell Death
Dis. 9:5492018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Xu H, Zhao L, Fang Q, Sun J, Zhang S, Zhan
C, Liu S and Zhang Y: MiR-338-3p inhibits hepatocarcinoma cells and
sensitizes these cells to sorafenib by targeting hypoxia-induced
factor 1α. PLoS One. 9:e1155652014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Jia XQ, Cheng HQ, Qian X, Bian CX, Shi ZM,
Zhang JP, Jiang BH and Feng ZQ: Lentivirus-Mediated overexpression
of microRNA-199a inhibits cell proliferation of human
hepatocellular carcinoma. Cell Biochem Biophys. 62:237–244. 2012.
View Article : Google Scholar : PubMed/NCBI
|