Introduction
Gastric cancer (GC) is one of the most common
malignant tumors in the digestive system. According to the latest
statistics of GLOBOCAN, there were approximately 1,033,000 new
cases of GC worldwide in 2018, with approximately 783,000
fatalities (1). Based on this
evidence, GC ranks 5th in the incidence of malignant tumors and 2nd
in mortality worldwide (1) Although
various comprehensive treatments including surgical resection,
radiotherapy and chemotherapy have been used at different stages of
the disease, the incidence, mortality and impaired quality of life
thereof are on the increase (2).
Therefore, it is necessary to explore new biomarkers and
therapeutic targets that may aid the development of targeted
therapies for GC.
Metastasis suppressor genes play a key role in tumor
metastasis. A previous study demonstrated that the effects of
metastasis suppressor genes on tumor metastasis were more critical
than those of metastasis promoter genes and that the reduction in
the expression levels of metastasis suppressor genes or their loss
of expression may induce the invasion and metastasis of tumor cells
(3). Kiss-1 was initially
identified as an important tumor metastasis suppressor gene in
human melanoma cells (4).
Kiss-1 is located on the long arm of human chromosome-1. The
protein-encoding gene acts as an endogenous ligand for G-protein
coupled receptor 54 and produces a variety of physiological
effects, including inhibition of tumor cell proliferation,
metastasis, invasion and induction of tumor cell differentiation
and apoptosis (5,6). A decrease in Kiss-1 levels and
the role of this protein in tumor invasion and metastasis have been
evaluated in various tumors, such as those of the bladder (7), colorectum (8) and breast (9). However, the expression levels of
Kiss-1 and its pathogenesis in GC remain to be
elucidated.
The present study examined the methylation status
and expression levels of Kiss-1 in GC tissues and
subsequently assessed the association between Kiss-1
methylation, Kiss-1 expression and clinicopathological
features. The effects of Kiss-1 on the biological function
of specific GC cell lines were also studied. The primary aim of the
study was to investigate the role of Kiss-1 in the
development and progression of GC and whether it could be used for
the prevention or treatment of this disease. The data demonstrated
that a low expression of Kiss-1 played a suppressive role
for the proliferation, migration and invasion of GC cells,
rendering Kiss-1 a potential diagnostic and prognostic
marker.
Materials and methods
Patients and specimens
Samples from GC and non-tumor tissues were collected
at the time of surgical resection at the First Hospital of Hebei
Medical University from June 2014 to June 2016. The samples were
snap-frozen in liquid nitrogen and stored at −80°C.
Paraffin-embedded tissues were prepared at the Department of
Pathology at the same hospital. All diagnoses of GC and gastritis
were confirmed by histopathological examination. The relevant
information regarding patient history and disease characteristics
was extracted from a review of the patients' medical records. The
present study was approved by the Institutional Ethical Review
Committee of the First Hospital of Hebei Medical University and
adhered to the principles of the Declaration of Helsinki. Informed
consent was obtained from each patient prior to the collection of
the tissues.
Cell culture
Human GC cells (AGS and HGC-27) were purchased from
the Culture Collection of the Chinese Academy of Sciences and
cultured in F-12k (AGS) and RPMI-1640 (HGC-27) (Gibco; Thermo
Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum
(FBS), 100 µ/ml penicillin and 50 mg/ml streptomycin. The cells
were incubated at 37°C in a humidified atmosphere of 5%
CO2. The cells were authenticated by STR analysis and no
cross-contamination from other cell lines was found.
Methylation analyses of the promoter
of Kiss-1
The primers used were as follows: Methylated
Kiss-1 forward, 5′-AAAGTTTCGTTTCGGAGGGTTC-3′ and reverse,
5′-CTTTTATAAAACCCGAAATAACG-3′, unmethylated Kiss-1 forward,
5′-AAAGTTTTTTTTGGGGGTTT-3′ and reverse,
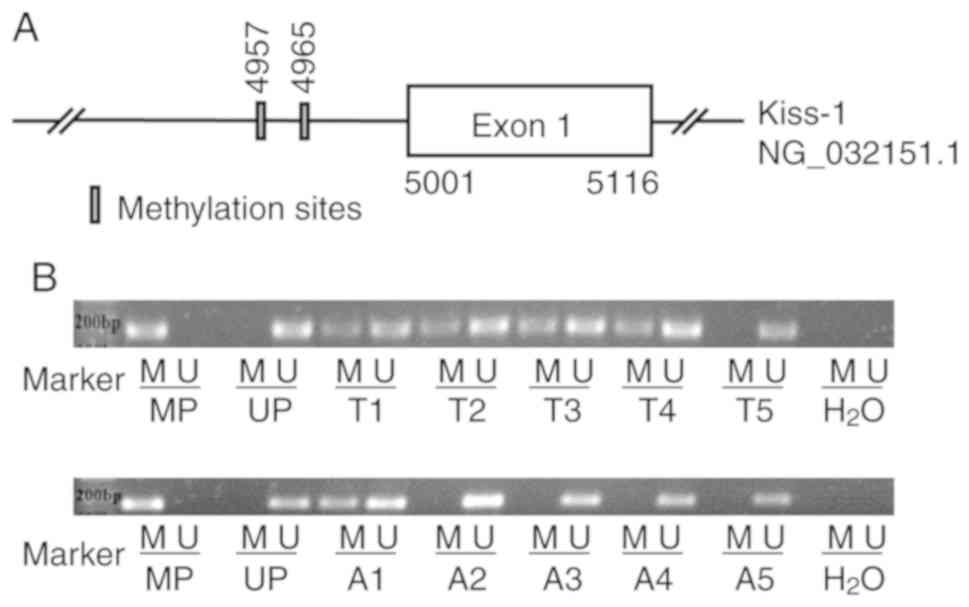
5′-CCTTTTATAAAACCCAAAATAACA-3′ (10). The specific location of methylation
sites in Kiss-1 promoter region is shown in Fig. 1A. Genomic DNA from GC patient tissue
was extracted and modified with sulfite using Universal Genomic DNA
Kit and DNA Methylation Kit (CWBIO, Inc.). Methylation-specific PCR
(MSP) with GoldStar Master Mix (CWBIO, Inc.) was also employed
according to the manufacturer's protocol. The thermocycling
conditions were: Pre-denaturation at 95°C for 10 min, denaturation
at 95°C for 45 sec, annealing for 45 sec (methylation-specific
primer amplification annealing temperature 59°C, non-methylation
specific primer amplification annealing temperature 55°C) and
extension at 72°C for 50 sec. A total of 34 cycles were performed
and the final extension was conducted at 72°C for 7 min. The
reaction products were separated by 2.0% agarose gel
electrophoresis and detected with Ethidium Bromide staining. During
electrophoresis, the methylated positive control (CpG methylation
enzyme modification of DNA extracted from fresh placental tissues
used as a template), the unmethylated positive control (DNA
extracted from fresh placental tissues used as a template) and the
negative control (H2O) were established. The data were
collected using a UV transilluminator (Alpha Innotech Corporation;
ProteinSimple) and subsequently analyzed by AlphaView 3.4 (Alpha
Innotech Corporation; ProteinSimple).
RNA extraction and reverse
transcription PCR (RT-qPCR)
Total RNA was extracted from cell or from GC tissues
using TRIzol® (Invitrogen; Thermo Fisher Scientific,
Inc.) and cDNA was synthesized with the PrimeScript RT reagent kit
(Takara Bio, Inc.) according to the manufacturer's instructions.
Subsequently, Kiss-1 expression levels were quantified by
RT-qPCR using the AceQ qPCR SYBR-Green Master Mix (Vazyme Biotech,
Co. Ltd.) in an ABI-7500 quantitative PCR instrument. Gene
expression was defined based on Cq values and the gene expression
levels were normalized compared with those of the housekeeping gene
GAPDH. The 2−ΔΔCq method (11) was used to calculate the relative
changes in the expression levels. The thermal cycling conditions
used were: 95°C for 5 min, followed by 40 cycles at 95°C for 10
sec, 60°C for 30 sec and 95°C for 15 sec, 60°C for 60 sec and 95°C
for 15 sec. The primers used for Kiss-1 were the following:
Forward: 5′-CTCACTGGTTTCTTGGCAGC-3′; reverse:
5′-CTGGCTTCCTCTCGGTGC-3′. GAPDH was used as an internal
reference control and its detection was performed with the
following primers: Forward: 5′-GAGTCAACGGATTTGGTCGT-3′ and reverse:
5′-CATGGGTGGAATCATATTGGA-3′.
Detection of the expression levels of Kiss-1 protein
by immunohistochemistry. The GC tissue was fixed in 10% neutral
formalin for 24–48 h at room temperature and then cut into paraffin
sections (4 µm). The sections were placed into a 60–65°C box
overnight and deparaffinized in green transparent agent, rehydrated
with an alcohol gradient and washed briefly in distilled water. For
antigen retrieval, the sections were boiled in 0.01 M citrate
buffer (pH 6.0) for 20 min at 100°C and washed in PBS three times
prior to cooling to room temperature. Endogenous peroxidase
activity was blocked with 3% H2O2 (Bohai) for
30 min and the sections were washed in PBS three times.
Subsequently, the sections were incubated with normal sheep serum
for 30 min at room temperature and at 4°C overnight with rat
Anti-Kiss peptin monoclonal antibody (1:100 dilution; cat. no.
ab55384; Abcam). The following day, the sections were rinsed with
fresh PBS and incubated with biotinylated secondary antibody
working solution and horseradish enzyme-labeled streptavidin (cat.
no. SP-9002; ZSGB Biotech) working solution at room temperature for
30 min. Finally, the sections were stained with
3,3′-diaminobenzidine (cat. no. ZLI-9032; ZSGB Biotech) for
visualization. Five fields were randomly selected from the slices
and scored according to the percentage of positive cells in the
field. The scoring system was as follows: <5% corresponded to 0,
6–25% to 1, 26–50% to 2 and ≥51% to 3. The staining intensity score
used was as follows: None corresponded to 0, light yellow to 1,
yellow to 2 and brown to 3. When the two aspects were added
together, a score of ≤3 indicated negative expression, while a
score >3 positive expression.
Vectors and transfection of the target
genes
Kiss-1 m98 and the control m98 vectors were
transfected into AGS GC cells using Lipofectamine 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.). Kiss-1 sh and NC sh were
also transfected into HGC-27 GC cells. The transfection efficacy
was evaluated by RT-qPCR and western blot analyses.
Western blot analysis
Total protein was extracted from cell using RIPA
lysis buffer containing proteinase inhibitor (Solarbio Science
& Technology Co. Ltd.), and quantified using the bicinchoninic
acid protein assay (Solarbio Science & Technology Co. Ltd.) as
recommended by the manufacturers. Approximately 20 µg of total
protein was separated by 10% SDS-PAGE and transferred to a
polyvinylidene difluoride membrane (EMD Millipore). The membrane
was blocked with 5% skimmed milk in PBS-T (10 mmol/l Tris, 145
mmol/NaCl, pH 7.2-7.4) for 2 h at room temperature and subsequently
incubated with mouse monoclonal antibody against Kiss-1
(1:1,000 dilution; cat. no. ab55384; Abcam) or mouse monoclonal
antibody against β-actin (1:3,500 dilution; cat. no. 60008-1-1 g;
ProteinTech Group, Inc.) overnight at 4°C. Anti-mouse IgG (1:2,500
dilution; cat. no. A23910; Abbkine Scientific Co. Ltd.) was used
and the signal was developed with a chemiluminescent substrate. The
images were obtained with an Odyssey CLX infrared fluorescence
scanning imaging system (LICOR) and the intensity of the bands was
analyzed using Image J software (National Institute of Health).
Cell proliferation assay
The transfected cells were independently seeded in
96-well plates and cultured for 24, 48, 72 or 96 h. The Cell
Counting Kit-8 (MedChemExpress) was added to each well and the
cells were incubated at 37°C for 3 h. Absorbance was measured at
450 nm using a microplate reader (Promega Corporation).
Transwell migration and Matrigel
invasion assays
Cell migration and invasion were measured using 8 µm
Transwell chambers (Corning, Inc.). To measure migration,
3.5×104 transfected cells were resuspended in 300 µl
serum-free F-12K or RPMI-1640 medium and added to the upper
chamber, whereas 800 µl F-12K or RPMI-1640 medium containing 10%
FBS was added to the lower chamber. To measure invasion, a chamber
containing Matrigel (Corning, Inc.) was used and the assay was
performed as stated before. Following 24 h of incubation, the
chamber was stained by diff-quick staining and the cells were
counted in five random fields under a light microscope with a
magnification of ×200. The number of cells was expressed as an
average.
Wound healing assay
When the transfected cells reached a growth density
of 85%, the confluent monolayers were scratched with a pipette tip
in order to create a gap to simulate a wound and the non-viable
cells were washed with PBS. The transfected cells were cultured in
RPMI-1640. The images of the plates were obtained under a
microscope (Olympus Corporation) at 0, 24 and 48 h.
Statistical analysis
All results are shown as mean ± SD and were analyzed
using GraphPad Prism 7 (GraphPad Software Inc.). The differences
determined in the in vitro experiments were analyzed using
the unpaired two-tailed t-test and the cellular experiments were
repeated three times. The differences determined in the clinical
tissue experiments were analyzed using the Pearson's Chi-square
test. P<0.05 indicated significant differences.
Results
Methylation-specific PCR (MSP)
analysis of Kiss-1 gene promoter methylation
The MSP data indicated that hypermethylation of the
Kiss-1 gene promoter was observed in 78.43% (40/51) of the
GC tissues, whereas this process was present only in 53.49% (23/43)
of the non-tumor tissues (Fig. 1B).
Chi-square analysis of the patient data collected indicated that
the hypermethylation of the Kiss-1 promoter in GC was
significantly associated with TNM III+IV stage and lymph node
metastasis (P<0.05). However, no significant correlation was
noted between the Kiss-1 promoter hypermethylation and other
clinicopathological variables such as age, sex and histological
grade (P>0.05; Table I).
 | Table I.Association between
clinicopathological features and Kiss-1 methylation in 40
patients with GC. |
Table I.
Association between
clinicopathological features and Kiss-1 methylation in 40
patients with GC.
|
|
| Kiss-1
methylation |
|
|
|
|---|
|
|
|
|
|
|
|
|---|
| Clinicopathological
features | No. of
patients | Positive | Negative | Positive rate
(%) | χ2 | P-value |
|---|
| Adjacent
tissues | 30 | 10 | 20 | 33.33 | 10.658 | 0.001a |
| Gastric
carcinoma | 40 | 29 | 11 | 72.50 |
|
|
| Sex |
|
|
|
|
|
|
|
Male | 32 | 24 | 8 | 75.00 | 0.071 | 0.791 |
|
Female | 8 | 5 | 3 | 62.50 |
|
|
| Age (years) |
|
|
|
|
|
|
|
≤65 | 18 | 15 | 3 | 83.33 | 1.065 | 0.302 |
|
>65 | 22 | 14 | 8 | 63.64 |
|
|
| Histological
grade |
|
|
|
|
|
|
|
Poorly | 14 | 9 | 5 | 64.29 | 0.233 | 0.629 |
| Well,
Moderately | 26 | 20 | 6 | 76.92 |
|
|
| Depth of
infiltration |
|
|
|
|
|
|
| Soaked
in serosa | 19 | 14 | 5 | 73.68 | 0.025 | 0.873 |
| Not
soaked in the film | 21 | 15 | 6 | 71.43 |
|
|
| Lymph node
metastasis |
|
|
|
|
|
|
|
Presence | 24 | 21 | 3 | 87.50 | 5.021 | 0.025a |
|
Absence | 16 | 8 | 8 | 50.00 |
|
|
| TNM stage |
|
|
|
|
|
|
|
I+II | 15 | 7 | 8 | 46.67 | 6.094 | 0.014a |
|
III+IV | 25 | 22 | 3 | 88.00 |
|
|
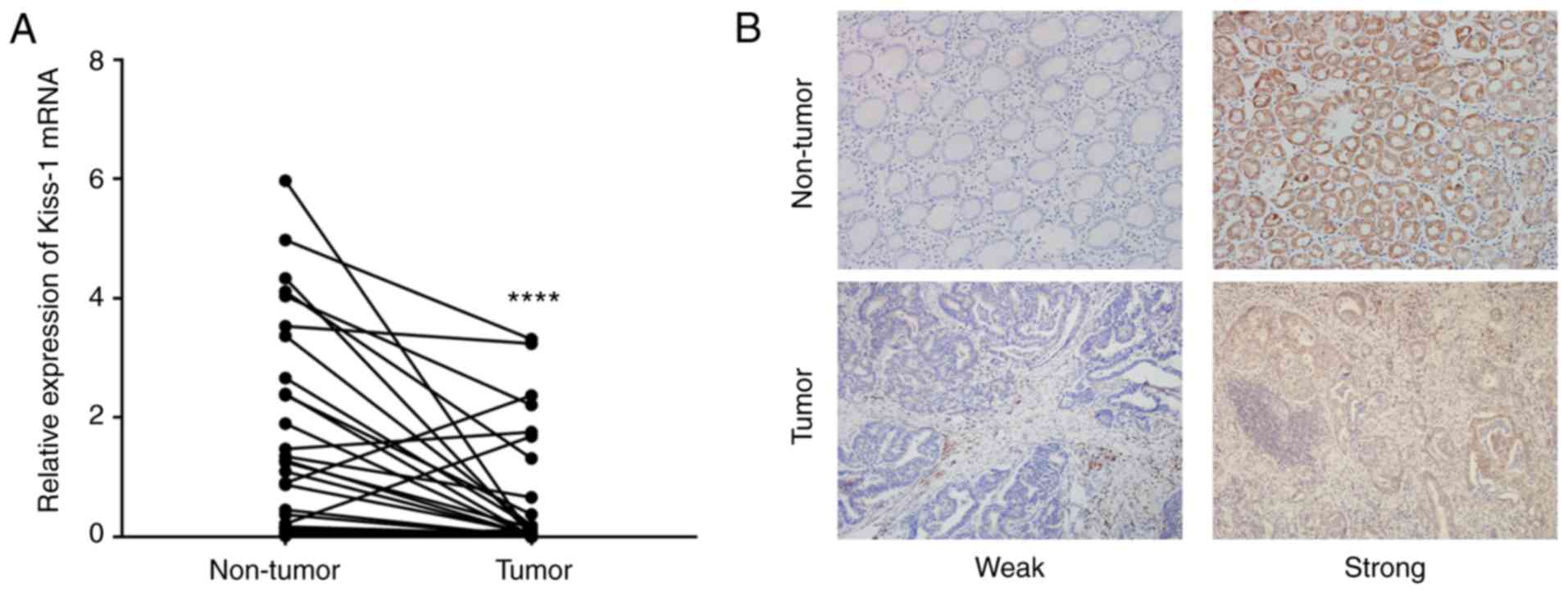
Kiss-1 mRNA and protein expression in
GC
The mRNA expression levels of Kiss-1 were
evaluated in 53 GC tissue samples and 53 non-tumor tissue samples.
The data indicated that the levels of Kiss-1 in the GC
tissues were significantly downregulated compared with those of the
normal tissues (Fig. 2A). The
expression levels of the Kiss-1 protein were assessed in 56
paired GC and adjacent non-tumor tissues by immunohistochemistry.
The data demonstrated that Kiss-1 was expressed in the
cytoplasm of gastric carcinoma cells (Fig. 2B). Kiss-1 staining was
detected in 48.21% (27/56) of GC tissues and its positive
expression was significantly lower in adjacent non-tumor tissues
82.14% (46/56). The correlation between Kiss-1 expression
and clinicopathological features was also analyzed in patients with
GC. Low expression levels of Kiss-1 in GC tissues were
significantly associated with poorly histological grade, lymph node
metastasis and TNM III+IV stage (P<0.05). No significant
association was observed with the remaining variables including
age, sex, tumor size, or depth of invasion (P>0.05; Table II).
 | Table II.Relationship between expression of
Kiss-1 and clinicopathological variables in 56 patients with
GC. |
Table II.
Relationship between expression of
Kiss-1 and clinicopathological variables in 56 patients with
GC.
|
|
| Kiss-1
staining |
|
|
|
|---|
|
|
|
|
|
|
|
|---|
| Clinicopathological
features | No. of
patients | Strong | Weak | Expression rate
(%) | χ2 | P-value |
|---|
| Adjacent
tissues | 56 | 46 | 10 | 82.14 | 14.202 |
<0.001a |
| Gastric
carcinoma | 56 | 27 | 29 | 48.21 |
|
|
| Sex |
|
|
|
|
|
|
|
Male | 38 | 20 | 18 | 52.63 | 0.924 | 0.336 |
|
Female | 18 | 7 | 11 | 38.89 |
|
|
| Age (years) |
|
|
|
|
|
|
|
≤62 | 28 | 13 | 15 | 46.43 | 0.072 | 0.789 |
|
>62 | 28 | 14 | 14 | 50.00 |
|
|
| Tumor size
(cm) |
|
|
|
|
|
|
| ≤4 | 36 | 19 | 17 | 52.78 | 0.841 | 0.359 |
|
>4 | 20 | 8 | 12 | 40.00 |
|
|
| Histological
grade |
|
|
|
|
|
|
|
Poorly | 23 | 7 | 16 | 30.43 | 4.941 | 0.026a |
| Well,
Moderately | 33 | 20 | 13 | 60.61 |
|
|
| Depth of
infiltration |
|
|
|
|
|
|
| Soaked
in serosa | 35 | 13 | 22 | 37.14 | 4.582 | 0.032 |
| Not
soaked in the film | 21 | 14 | 7 | 66.67 |
|
|
| Lymph node
metastasis |
|
|
|
|
|
|
|
Presence | 32 | 8 | 24 | 25.00 | 16.116 |
<0.001a |
|
Absence | 24 | 19 | 5 | 79.17 |
|
|
| TNM stage |
|
|
|
|
|
|
|
I+II | 31 | 23 | 8 | 74.19 | 18.771 |
<0.001a |
|
III+IV | 25 | 4 | 21 | 16.00 |
|
|
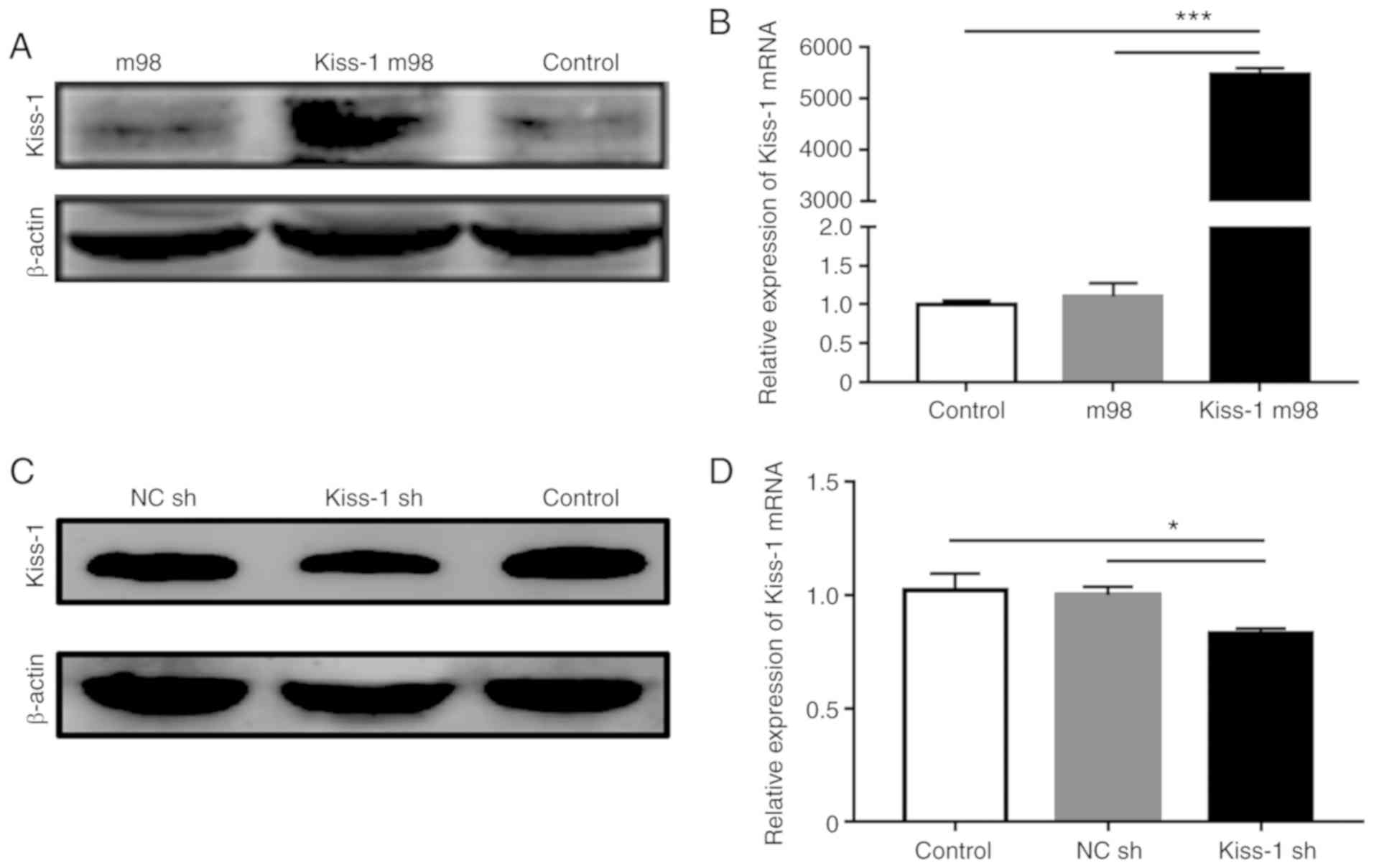
Regulation of the Kiss-1 gene in GC
cells
The AGS and HGC-27 cell lines were selected to
investigate the biological function of Kiss-1 in GC.
Transfection of Kiss-1 m98 vector into AGS cells resulted in
a significant upregulation of the expression of Kiss-1 at
both the mRNA and protein levels (Fig.
3A and B), whereas transfection of Kiss-1 sh
significantly downregulated the transcription and synthesis of
Kiss-1 in HGC-27 cells (Fig. 3C
and D).
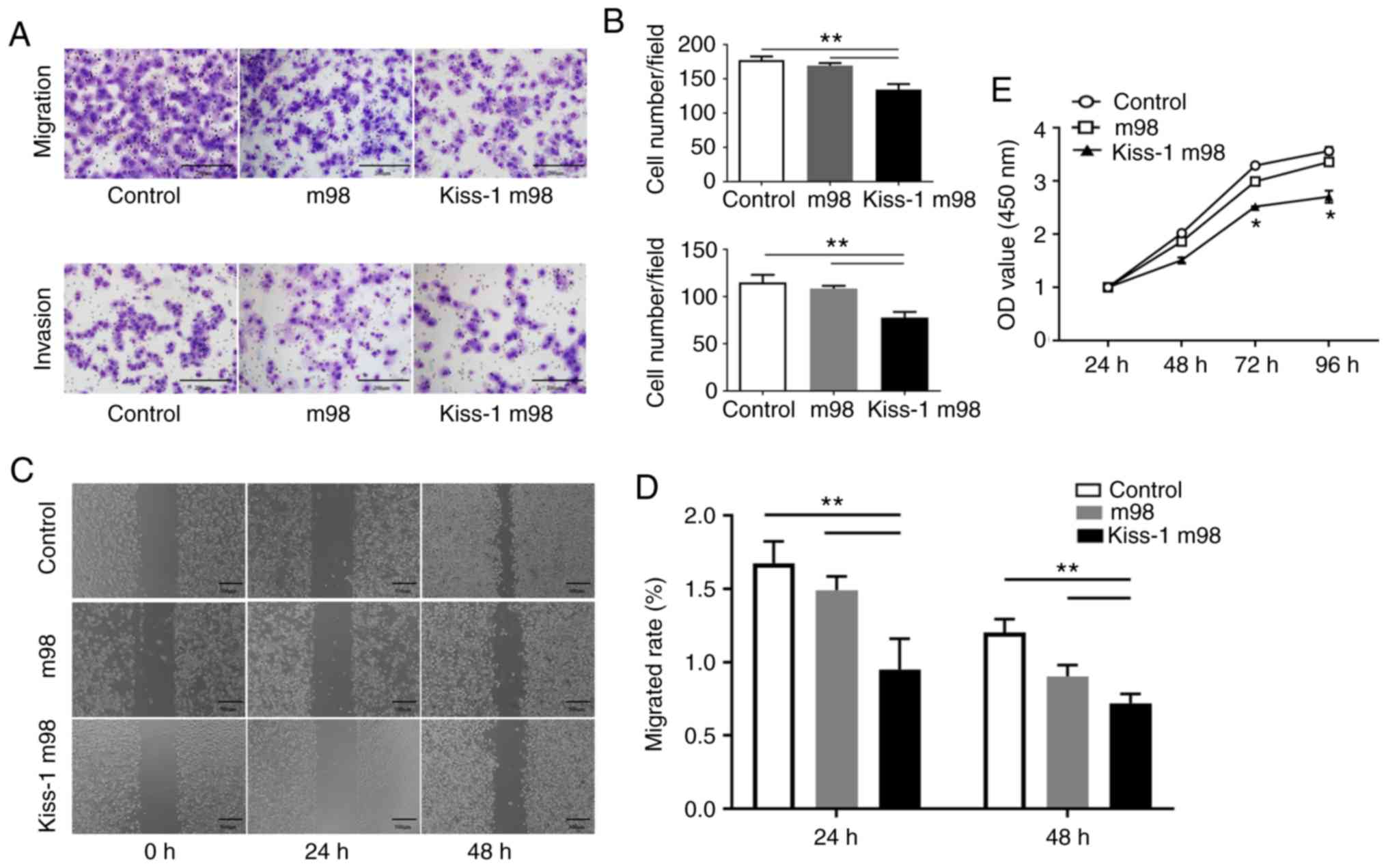
Effects of Kiss-1 overexpression on
migration, invasion and proliferation of AGS cells
The number of cells migrating through the chamber in
Kiss-1 m98 was significantly lower than that noted in the
m98 and control groups (P<0.05). Similarly, the number of cells
invading through the chamber in Kiss-1 m98 was significantly
lower than that of the m98 and control groups (P<0.05) (Fig. 4A and B). The scratch healing rate of
Kiss-1 m98-transfected cells was significantly lower than
that of the m98 and control groups (P<0.05) (Fig. 4C and D). In addition, the cell OD
values were significantly reduced at 48, 72 and 96 h in the
Kiss-1 m98 group (Fig. 4E).
The results indicated that overexpression of Kiss-1
inhibited the migration, invasion and proliferation of GC
cells.
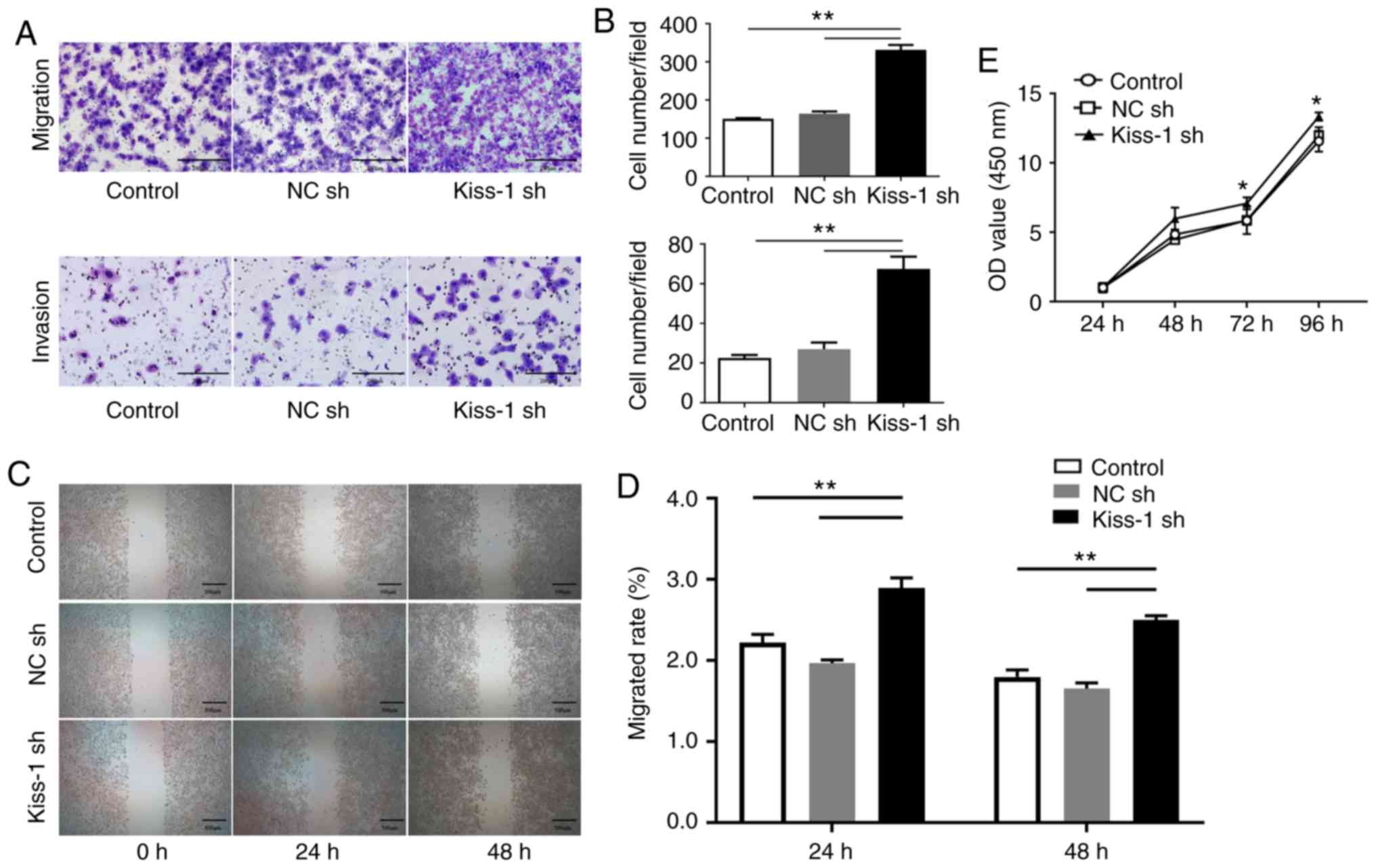
Effects of Kiss-1 knockdown on
migration, invasion and proliferation of HGC-27 cells
Transwell migration, matrigel invasion and wound
healing assays were performed following transfection of HGC-27
cells with Kiss-1 sh in order to assess migration and
invasion in GC cells devoid of Kiss-1. The number of cells
migrating through the chamber in Kiss-1 sh cells was
significantly higher than that in the NC sh and control groups
(Fig. 5, P<0.05). Similarly, the
number of cells invading through the chamber in the Kiss-1
sh group was significantly higher than that noted in the NC sh and
control groups (P<0.05). The scratch healing ability of
Kiss-1 sh-transfected HGC-27 cells was significantly higher
than that of the NC sh and control groups (P<0.05). In addition,
the OD values were significantly increased at 48, 72, and 96 h in
the sh group (Fig. 5E). It was
deduced that knockdown of Kiss-1 enhanced cell migration,
invasion and proliferation.
Discussion
GC is the fifth most common cancer in the world and
the second most lethal cancer (1).
The vast majority of GC fatalities are caused by complications
caused by tumor cell metastasis. The signaling pathways involved in
the initial control of the tumor cells are activated and the
primary tumor cells migrate into adjacent tissues (12). Following contact of the tumor cells
with blood and lymphatic vessels, the basement membrane and the
endothelial wall are penetrated and the cells are dispersed through
the lumen of blood vessels to reach distant organs, facilitating
the progression of metastasis (13,14).
Therefore, investigation of the expression of tumor metastasis
suppressor genes that interfere with specific points in these steps
and block the metastatic cascade is critical for early diagnosis,
treatment and improved clinical outcomes of GC patients. The
Kiss-1 gene was initially reported as a novel metastasis
suppressor gene in human melanoma and breast cancer cells (15,16).
The translation product of Kiss-1 is a protein containing
145 amino acids, which is further cleaved into Kisspeptin-10, −13,
−14 and −54 proteins (17,18). Kiss-1 proteins bind
specifically to GPR54 (AXOR12 or hOT7T175) and induce the release
of the secondary messenger inositol trisphosphate (IP3) and of the
diglycerides, which play a role in cell proliferation,
differentiation and apoptosis (5,6).
However, the expression levels of Kiss-1 and
its pathogenesis in GC remain unclear. To investigate the mechanism
of action of Kiss-1 in GC, the association of promoter
methylation with the clinicopathological data was investigated with
regard to GC progression. In the present study, hypermethylation of
Kiss-1 was present in GC tissues compared with the
corresponding levels noted in adjacent tissues, indicating that
methylation of Kiss-1 may contribute to the progression of
GC. Statistical analysis of the levels of the Kiss-1
promoter methylation and the pathological parameters of the GC
patients indicated association among lymph node metastasis, TNM
III+IV stage and the higher methylation positive rate of the
Kiss-1 promoter. This demonstrated that the complexity of
tumor pathogenesis was, not only a reflection of genetic change by
mutation or deletion, but also a reflection of epigenetic
alterations, such as DNA methylation. In addition to the deletion
and mutation of the associated genes, aberrant changes in DNA
methylation were considered as the third mechanism leading to
anti-oncogenic inactivation (19,20),
which played an essential role in tumor development. A previous
study suggested that hypermethylation of Kiss-1 occurred
frequently in colorectal cancer (CRC) (21). Moreover, it has been confirmed that
Kiss-1 methylation is associated with tumor differentiation,
depth of invasion, distant lymph node metastasis and predictive
recurrence (8,21,22).
These data indicated that Kiss-1 methylation may be
associated with invasion, metastasis and poor prognosis in GC.
Results of those studies are similar to those presented in this
study.
Kiss-1 mRNA and protein expression levels are
substantially downregulated in GC tissues compared with the
corresponding levels noted in non-tumor tissues. This is consistent
with the findings of Kostakis et al demonstrating that
Kiss-1 expression in adjacent gastric mucosa was
considerably higher than that noted in malignant mucosa (23). Subsequently, we performed
statistical analysis of the pathological features of the GC tissues
and confirmed that low protein expression levels of Kiss-1
were significantly associated with poor histological grade, lymph
node metastasis and TNM III+IV stage. This is consistent with other
studies reporting that Kiss-1 exhibited low expression
levels in CRC and that it may be considered a putative metastasis
suppressor in human CRC (24–26).
It was hypothesized that Kiss-1 promoter methylation
resulted in loss of Kiss-1 expression and metastasis of GC.
Previous findings have demonstrated that Kiss-1
hypermethylation is associated with loss of transcription and
protein expression in CRC (8).
Furthermore, promoter CpG island methylation has been shown to
reduce the expression levels of related tumor suppressor genes and
is considered the main tumor suppressor-inactivation mechanism in
GC (27). Therefore, it was
essential to examine whether Kiss-1 expression in GC tissues
is also directly affected by the methylation levels of its
promoter. Based on results of the present study, we sugget that the
Kiss-1 protein plays a role in inhibiting tumor metastasis
during the development of GC, further confirming that the
Kiss-1 gene is a metastasis suppressor gene in GC and that
the downregulation of its expression exhibits considerable
significance for clinical development of individualized treatment
and disease prognosis.
The biological function of Kiss-1 on AGS and
HGC-27 cells was further examined. The data indicated that
overexpression of the Kiss-1 gene significantly inhibited
migration and invasion of AGS in the GC cell lines used. Its
decreased expression was able to promote migration and invasion of
HGC-27 cells. These results are similar to those reported by Lee
and Kim demonstrating that Kiss-1 may inhibit the invasion of
NUGC-3 and MKN-28 GC cells (28).
Chen et al reported that Kiss-1 overexpression
significantly decreased the invasiveness of CRC cells (29). Previous reports have also suggested
that the reduction of Kiss-1 expression promotes cell
migration and invasion in pancreatic (30), ovarian (31), prostate (32) and endometrial cancer (33) as well as in nasopharyngeal carcinoma
(34). Considering the importance
of migration and invasion as two key processes required for tumor
progression and metastasis, the results demonstrated the
therapeutic potential of Kiss-1 by reducing tumor cell
metastatic activity.
The data from the proliferation experiments
indicated that the overexpression of Kiss-1 exhibited an
inhibitory effect in AGS cells, whereas its knockdown exhibited a
promoting effect in the proliferation of HGC-27 cells. These
effects appeared 48 h following treatment mainly because
Kiss-1 required a longer time to exert its effect on
proliferation. Notably, the role of Kiss-1 in the
proliferation of various tumors has been well established. Chen
et al (29) demonstrated
that silencing of the Kiss-1 gene did not influence
proliferation of HCT-116 CRC cells, whereas its overexpression
resulted in the opposite effects. Knockdown of Kiss-1 in
HT115 and HRT18 CRC cells did not have an effect on their
proliferation (25). Certain
inconsistent proliferation results may be associated with the
characteristics of different tumor cells. It has been reported that
Kiss-1 inhibits growth of matrix-independent tumors but not
of matrix-dependent tumors (34).
Therefore, the regulation of Kiss-1 in different tumor
phenotypes is more complex than expected and requires further
investigation. Some previous studies have only examined
Kiss-1 expression in GC tissues or part of its biological
role in GC cells (23,28). By contrast, our research
systematically investigated the role of Kiss-1 in GC,
including mRNA expression, protein expression, methylation status,
and clinicopathological data in GC tissues, as well as biological
functions after upregulation and downregulation of Kiss-1 in
GC cells. We hypothesized that Kiss-1 promoter methylation
would lead to loss of Kiss-1 expression, thereby promoting
the metastasis of GC. This makes our research content more diverse
and the results clearer.
In conclusion, the experiments demonstrated that
Kiss-1 may be considered a tumor metastasis suppressor gene
closely associated with the development of GC. Kiss-1 was
able to inhibit migration and invasion of GC to a certain extent.
Consequently, Kiss-1 can be used as a new target for
clinical treatment, which may not only eliminate local disease, but
also inhibit the systemic spread of GC cells. Additional future
studies should be performed to confirm these findings.
Acknowledgements
We would like to thank Professor Quanhai Li and Dr
Xia Jiang for experimental suggestions and writing guidance.
Funding
This study was supported by the Hebei Province Key
Research and Development Project (grant no. 18277741D) and other
Hebei Province Projects (grant nos. 1387, SGH201501, A201802017,
LNB201809, G2019035, ZD20140126 and XH201805).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WY designed the study. CL, MX and DL performed the
experiments. LY, SH and BT analyzed the data. CL and LY wrote the
manuscript together. WY helped to revise the manuscript. All
authors read and approved the final manuscript and agree to be
accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
The study was approved by the Institutional Ethical
Review Committee of the First Hospital of Hebei Medical University
and adhered to the principles of the Declaration of Helsinki.
Informed consent was obtained from each patient before collection
of tissues.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Sun K, Zheng R, Zeng H, Zhang S,
Xia C, Yang Z, Li H, Zou X and He J: Cancer incidence and mortality
in China, 2014. Chin J Cancer Res. 30:1–12. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Samant RS, Seraj MJ, Saunders MM, Sakamaki
TS, Shevde LA, Harms JF, Leonard TO, Goldberg SF, Budgeon L, Meehan
WJ, et al: Analysis of mechanisms underlying BRMS1 suppression of
metastasis. Clin Exp Metastasis. 18:683–693. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lee JH, Miele ME, Hicks DJ, Phillips KK,
Trent JJM, Weissman BE and Welch DR: KiSS-1, a novel human
malignant melanoma metastasis-suppressor gene. J Natl Cancer Inst.
88:1731–1737. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Clements MK, McDonald TP, Wang R, Xie G,
O'Dowd BF, George SR, Austin CP and Liu Q: FMRFamide-related
neuropeptides are agonists of the orphan G-protein-coupled receptor
GPR54. Biochem Biophys Res Commun. 284:1189–1193. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Stafford LJ, Xia C, Ma W, Cai Y and Liu M:
Identification and characterization of mouse metastasis-suppressor
KiSS1 and its G-protein-coupled receptor. Cancer Res. 62:5399–5404.
2002.PubMed/NCBI
|
|
7
|
Zhang Y, Huang Z, Zhu Z, Zheng X, Liu J,
Han Z, Ma X and Zhang Y: Upregulated UHRF1 promotes bladder cancer
cell invasion by epigenetic silencing of KiSS1. PLoS One.
9:e1042522014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Moya P, Esteban S, Fernandez-Suarez A,
Maestro M, Morente M and Sánchez-Carbayo M: KiSS-1 methylation and
protein expression patterns contribute to diagnostic and prognostic
assessments in tissue specimens for colorectal cancer. Tumour Biol.
34:471–479. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Teng Y, Mei Y, Hawthorn L and Cowell JK:
WASF3 regulates miR-200 inactivation by ZEB1 through suppression of
KISS1 leading to increased invasiveness in breast cancer cells.
Oncogene. 33:203–211. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yamashita S, Tsujino Y, Moriguchi K,
Tatematsu M and Ushijima T: Chemical genomic screening for
methylation-silenced genes in gastric cancer cell lines using
5-aza-2′-deoxycytidine treatment and oligonucleotide microarray.
Cancer Sci. 97:64–71. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wolf K, Wu YI, Liu Y, Geiger J, Tam E,
Overall C, Stack MS and Friedl P: Multi-step pericellular
proteolysis controls the transition from individual to collective
cancer cell invasion. Nat Cell Biol. 9:893–904. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hou JM, Krebs M, Ward T, Sloane R, Priest
L, Hughes A, Clack G, Ranson M, Blackhall F and Dive C: Circulating
tumor cells as a window on metastasis biology in lung cancer. Am J
Pathol. 178:989–996. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kienast Y, von Baumgarten L, Fuhrmann M,
Klinkert WEF, Goldbrunner R, Herms J and Winkler F: Real-time
imaging reveals the single steps of brain metastasis formation. Nat
Med. 16:116–122. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lee JH and Welch DR: Suppression of
metastasis in human breast carcinoma MDA-MB-435 cells after
transfection with the metastasis suppressor gene, KiSS-1. Cancer
Res. 57:2384–2387. 1997.PubMed/NCBI
|
|
16
|
Lee JH and Welch DR: Identification of
highly expressed genes in metastasis-suppressed chromosome 6/human
malignant melanoma hybrid cells using subtractive hybridization and
differential display. Int J Cancer. 71:1035–1044. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kotani M, Detheux M, Vandenbogaerde A,
Communi D, Vanderwinden JM, Poul EL, Brézillon S, Tyldesley R,
Suarez-Huerta N, Vandeput F, et al: The metastasis suppressor gene
KiSS-1 encodes kisspeptins, the natural ligands of the orphan G
protein-coupled receptor GPR54. J Biol Chem. 276:34631–34636. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ohtaki T, Shintani Y, Honda S, Matsumoto
H, Hori A, Kanehashi K, Terao Y, Kumano S, Takatsu Y, Masuda Y, et
al: Metastasis suppressor gene KiSS-1 encodes peptide ligand of a
G-protein-coupled receptor. Nature. 411:613–617. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fang JY and Lu YY: Effects of histone
acetylation and DNA methylation on p21( WAF1) regulation. World J
Gastroenterol. 8:400–405. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kumar R and Xi Y: MicroRNA, epigenetic
machinery and lung cancer. Thora Cancer. 2:35–44. 2011. View Article : Google Scholar
|
|
21
|
Chen SQ, Chen ZH, Lin SY, Dai QB, Fu LX
and Chen RQ: KISS1 methylation and expression as predictors of
disease progression in colorectal cancer patients. World J
Gastroenterol. 20:10071–10081. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cebrian V, Fierro M, Orenes-Piñero E, Grau
L, Moya P, Ecke T, Alvarez M, Gil M, Algaba F, Bellmunt J, et al:
KISS1 methylation and expression as tumor stratification biomarkers
and clinical outcome prognosticators for bladder cancer patients.
Am J Pathol. 179:540–546. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kostakis ID, Agrogiannis G, Vaiopoulos AG,
Mylona E, Patsouris E, Kouraklis G and Koutsilieris M: KISS1 and
KISS1R expression in gastric cancer. J BUON. 23:79–84.
2018.PubMed/NCBI
|
|
24
|
Kostakis ID, Agrogiannis G, Vaiopoulos AG,
Mylona E, Patsouris E, Kouraklis G and Koutsilieris M: A
clinicopathological analysis of KISS1 and KISS1R expression in
colorectal cancer. APMIS. 123:629–637. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ji K, Ye L, Ruge F, Hargest R, Mason MD
and Jiang WG: Implication of metastasis suppressor gene, Kiss-1 and
its receptor Kiss-1R in colorectal cancer. BMC Cancer. 14:7232014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Huo X, Zhang L and Li T: Analysis of the
association of the expression of KiSS-1 in colorectal cancer
tissues with the pathology and prognosis. Oncol Lett. 15:3056–3060.
2018.PubMed/NCBI
|
|
27
|
Guan Z, Zhang J, Song S and Dai D:
Promoter methylation and expression of TIMP3 gene in gastric
cancer. Diagn Pathol. 8:1102013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lee KH and Kim JR: Kiss-1 suppresses MMP-9
expression by activating p38 MAP kinase in human stomach cancer.
Oncol Res. 18:107–116. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen S, Chen W, Zhang X, Lin S and Chen Z:
Overexpression of KiSS-1 reduces colorectal cancer cell invasion by
downregulating MMP-9 via blocking PI3K/Akt/NF-κB signal pathway.
Int J Oncol. 48:1391–1398. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang CH, Qiao C, Wang RC and Zhou WP:
KiSS-1-mediated suppression of the invasive ability of human
pancreatic carcinoma cells is not dependent on the level of KiSS-1
receptor GPR54. Mol Med Rep. 13:123–129. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jiang Y, Berk M, Singh LS, Tan H, Yin L,
Powell CT and Xu Y: KiSS1 suppresses metastasis in human ovarian
cancer via inhibition of protein kinase C alpha. Clin Exp
Metastasis. 22:369–376. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang H, Jones J, Turner T, He QP, Hardy S,
Grizzle WE, Welch DR and Yates C: Clinical and biological
significance of KISS1 expression in prostate cancer. Am J Pathol.
180:1170–1178. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kang HS, Baba T, Mandai M, Matsumura N,
Hamanishi J, Kharma B, Kondoh E, Yoshioka Y, Oishi S, Fujii N, et
al: GPR54 is a target for suppression of metastasis in endometrial
cancer. Mol Cancer Ther. 10:580–590. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang GM, Liu JF, Zhang L, Sun Q, Zhou Y,
Xu HB, Zhang YJ, Cai F, Cheng ZN, Xiang P and Jiang H: Inhibition
of KiSS-1 on metastasis of nasopharyngeal carcinoma implant tumor
in nude mice. Am J Transl Res. 11:904–910. 2019.PubMed/NCBI
|