Introduction
Esophageal cancer (EC) is the 6th leading cause of
cancer-related mortality worldwide and the incidence is 45,600
cases each year (1). The etiology
of EC varies with histological type and population. Moreover, the
histopathology of EC is composed of ESCC and esophageal
adenocarcinoma (EA), and ESCC accounts for ~90% of the EC incident
each year. ESCC is a complex disease, and metastasis and invasion
significantly promote the development and progression of ESCC.
Moreover, epithelial-to-mesenchymal transition (EMT) contributes to
the metastasis and invasion of ESCC cells by disassembling the
cell-cell junctions, increasing cell motility and conferring
invasive properties (2). Therefore,
identifying the key inducers and understanding the molecular
mechanisms underlying EMT in ESCC is crucial.
MicroRNAs (miRNAs/miRs) are a class of endogenous
non-coding RNAs, ~18-25 nucleotides in length, which are found in
eukaryotes (3). miRNAs regulate
gene expression by combining with the 3′-untranslated region
(3′-UTR) of their target mRNAs to then induce translational
repression and/or mRNAs degradation (4). Previous studies have shown that miRNAs
are implicated in numerous important physiological and pathological
processes, such as stem cell differentiation (5,6), cell
proliferation and apoptosis (7,8),
immune responses (9), viral
infection (10) and tumorigenesis
(11,12). miR-590, a newly identified miRNA,
was reported to act as an anti-oncogene in osteosarcoma (13), non-small cell lung cancer (14,15)
and hepatocellular carcinoma (16,17).
However, the clinical significance and biological mechanism of
action of miR-590, as well as its association with EMT in ESCC
progression remain elusive.
The present study aimed to investigate the
association between the expression levels of miR-590 and
low-density lipoprotein receptor-related protein 6 (LRP6) in ESCC
tissues and cell lines. The effects of miR-590 and LRP6 on the
migration, invasion and EMT process in ESCC cells were also
examined in order to identify a novel regulatory mechanism
involving the miR-590/LRP6/EMT axis, which may be of therapeutic
value in the treatment of ESCC.
Materials and methods
Tissue specimens
A total of 28 paired human ESCC and corresponding
adjacent non-tumor tissues (age range, 43–77 years; mean age,
62.79±7.09 years; males, 20; females, 8) were obtained from the
Zhengzhou Central Hospital between July 2017 and June 2018. The
inclusion criteria included patients who diagnosed pathologically
with ESCC who had not undergone radiotherapy or chemotherapy prior
to surgery. The exclusion criteria included patients who had
undergone radiotherapy and chemotherapy before operation or whose
tissues did not meet the pathology requirements, which were
independently determined by two pathologists. All patients signed
an informed consent form for having their data collected and
analyzed for research purposes. The clinical specimens were
promptly snap-frozen in liquid nitrogen (−196°C) after resection
and then maintained at −80°C for further use. The research protocol
was approved by the Ethics Review Committee of Zhengzhou
University.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR) analysis
Total RNA was extracted from tissue samples and
cultured cells using TRIzol® reagent (Thermo Fisher
Scientific, Inc.). Subsequently, a Reverse Transcriptase kit
(Thermo Fisher Scientific, Inc.; cat. no. AB1454LDB) was used to
reverse transcribe RNA into cDNA at 50°C for 15 min. qPCR was
performed using Applied Biosystems™ SYBR™ Green (Thermo Fisher
Scientific, Inc.) on the ABI Real-Time PCR System (Thermo Fisher
Scientific, Inc.) following the manufacturer's protocols. The
relative expression was calculated using the 2−ΔΔCq
method (18). The expression of
LRP6 was normalized to β-actin expression, and the expression of
miR-590 was normalized to U6. The PCR cycle parameters for miR-590
and U6 were as follows: Initial denaturation at 95°C for 3 min,
followed by 40 cycles at 95°C for 15 sec and 60°C for 30 sec. The
RT-qPCR parameters for LRP6 and β-actin were as follows: Initial
denaturation at 95°C for 10 min, followed by 40 cycles at 95°C for
15 sec and 60°C for 1 min. The primer sequences used included:
miR-590 forward, 5′-TAATTTTATGTATAAGCTAGT-3′ and reverse,
5′-TGGTGTCGTGGAGTCG-3′; U6 forward, 5′-TCCGATCGTGAAGCGTTC-3′ and
reverse, 5′-GTGCAGGGTCCGAGGT-3′; LRP6 forward,
5′-TGTCAGCGAAGAAGCCATTAAA-3′ and reverse,
5′-TCTAAGGCAATAGCTCTGGGT-3′; and β-actin forward,
5′-GCACCACACCTTCTACAATG-3′ and reverse,
5′-TGCTTGCTGATCCACATCTG-3′.
Cell culture and transfection
The human esophageal cell line HET-1A and five ESCC
cell lines (EC-109, KYSE30, EC-9706, TE-1 and KYSE150) were
obtained from Peking Union Medical College Cell Bank. Cells were
cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific,
Inc.), containing 10% FBS (Hyclone; Cytiva) and 1% penicillin and
streptomycin with 5% CO2 at 37°C. EC-109 and EC-9706
cells were transfected with 40 nM small interfering RNA (si)-LRP6
fragments or miR-590 mimics. After 48 h, transfected cells were
used for further experiments. To assess miR-590 overexpression or
knockdown of LRP6, mimics negative controls (NC) and si-NC,
respectively, were used as corresponding controls. miRNA mimics and
siRNAs were obtained from Shanghai GenePharma Co., Ltd. Cell
transfection was conducted using Lipofectamine® 3000
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. The sequences were as follows: miR-590
mimics forward, 5′-UAAUUUUAUGUAUAAGCUAGU-3′ and reverse,
5′-UAGCUUAUACAUAAAAUUAUU-3′; miR-590 mimics NC forward,
5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse,
5′-ACGUGACACGUUCGGAGAATT-3′; si-LRP6 forward,
5′-UUGCAUAAAGCAACAAAGGGG-3′ and reverse,
5′-CCUUUGUUGCUUUAUGCAAAC-3′; and si-NC forward,
5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse,
5′-ACGUGACACGUUCGGAGAATT-3′.
Bioinformatics predictions and
luciferase assay
Putative binding sites of miR-590 and LRP6 were
predicted using miRDB 4.0 (http://www.mirdb.org/), TargetScan 7.1 (http://www.targetscan.org/vert_71/) and miRbase
21.0 (http://www.mirbase.org/).
Wild-type (WT) LRP6 3′-UTR fragments were amplified
with Phusion® High-Fidelity DNA polymerase (New England
Biolabs, Inc.) from human genomic DNA using PCR. The PCR
thermocycling conditions were as follows: Initial denaturation for
3 min at 94°C, followed by 35 cycles at 94°C for 30 sec, 58°C for
30 sec and 72°C for 45 sec. The sequences of LRP6 for PCR were as
follows: forward, 5′-CCGCTCGAGCTCCTCTGACTGCCTCCAAC-3′ and reverse,
5′-GCTCTAGATTATGGCACAAGCAGCAA3′. The LRP6 3′-UTR (including WT and
mutant fragments) were inserted into pmirGLO reporter vectors
(Promega Corporation). When EC-109 and EC-9706 cells had grown to
80% confluence, they were co-transfected into 40 nM miR-590 mimics
(or control miRNAs) and luciferase plasmids using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). After transfection for 36 h, firefly and
Renilla luciferase activities were detected using the
Dual-Luciferase Reporter Assay system (Promega Corporation).
Transwell assay
For invasion assays, the upper chamber of a 24-well
Transwell insert (size, 8-µm pores; Merck KGaA) was pre-coated with
100 µl Matrigel (pre-coating at 4°C for overnight).
3×104 cells in serum-free medium (200 µl) were added to
the upper chamber, while 500 µl culture medium was added to the
lower chamber. The loaded device was then placed into an incubator
for 24 h at 37°C. The invading cells were fixed with 4%
paraformaldehyde for 20 min at room temperature, stained with 0.1%
crystal violet for 30 min at room temperature and counted under
inverted fluorescent microscope (Olympus Corporation) from five
randomly selected fields (magnification, ×200). For the migration
assay, the experimental procedures were similar, but no Matrigel
was added to the upper chamber.
Western blotting
After transfected cells were lysed using RIPA lysis
buffer (Beyotime Institute of Biotechnology) and determined using a
bicinchoninic acid protein assay kit (Beyotime Institute of
Biotechnology), an equal amount of protein (40 µg per lane) in each
group was separated via SDS-PAGE (10%) and transferred onto PVDF
membranes. After blocking at room temperature for 2 h with 10%
skimmed milk, primary antibodies were incubated with the PVDF
membranes overnight at 4°C. The primary antibodies used (all from
Abcam) were as follows: Anti-N-cadherin (1:500; Mouse; cat. no.
ab98952), anti-vimentin (1:800; Mouse; cat. no. ab20346),
anti-E-cadherin (1:500; Mouse; cat. no. ab219332) and anti-β-actin
(1:30,000; Mouse; cat. no. ab49900). The primary antibodies were
discarded, and the membranes were washed with PBS/0.1% (v/v)
Tween-20 (three times; 10 min/time). Subsequently, the membranes
were incubated with a horseradish peroxidase-conjugated secondary
goat anti-mouse IgG antibody (1:4,000; cat. no. ab205719; Abcam)
for 2 h at room temperature. Then, the membranes were washed with
PBS/0.1% (v/v) Tween-20 (three times; 10 min/time) and incubated
with chemiluminescence (ECL) solution (Beyotime Institute of
Biotechnology.) for 1 min at room temperature on a Bio-Rad ChemiDoc
MP system (Bio-Rad Laboratories, Inc.) and exposed in a dark room.
ImageJ software version 1.46 (National Institutes of Health) was
used to measure the density of the protein bands.
Statistical analysis
SPSS 21.0 software (IBM Corp.) was used to analyze
data. Data are presented as the mean ± standard deviation. Each
experiment was repeated ≥3 times. Student's t-test was used to
analyze the differences between two groups, and one-way ANOVA
followed by Tukey's post hoc test was applied to analyze the
differences among ≥3 groups. A paired t-test was used to analyze
the statistical differences between tumors and adjacent non-tumor
samples from the same individual. Fisher's exact test was used to
assess the association between the expression of LRP6 or miR-590
and clinicopathological features. The correlation of LRP6 and
miR-590 in tissue samples was evaluated using Pearson's correlation
coefficient. P<0.05 was considered to indicate a statistically
significant difference.
Results
Expression of miR-590 is low, while
that of LRP6 is high in human ESCC tissues and cell lines
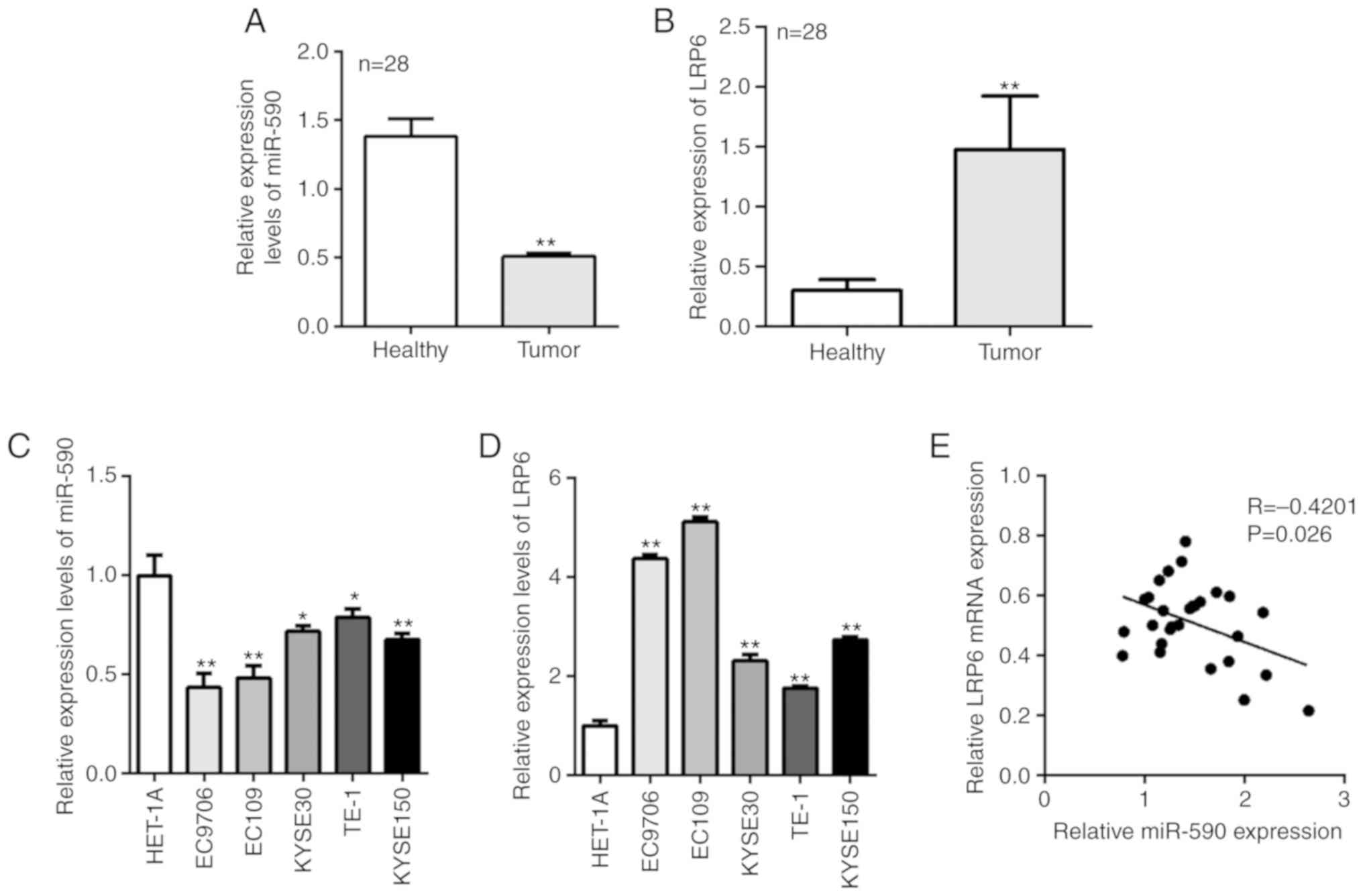
The RT-qPCR results demonstrated that miR-590 was
significantly downregulated and LRP6 was upregulated in tumor
tissues compared with healthy tissues (Fig. 1A and B). It was then examined
whether the expression pattern of miR-590 and LRP6 was the same in
ESCC cell lines; miR-590 was also significantly downregulated and
LRP6 was upregulated in ESCC cell lines, compare with HET-1A cells
(Fig. 1C and D). Pearson's
correlation analysis identified that there was a negative
correlation between miR-590 and LRP6 expression in ESCC tissues
(Fig. 1E). Furthermore, TNM stage
and lymph node metastasis were the two clinicopathological
characteristics that were significantly associated with miR-590 and
LRP6 expression levels in the clinical samples (Tables I and II).
 | Table I.Association of miR-590 expression
with clinicopathological characteristics in patients with
esophageal squamous cell carcinoma. |
Table I.
Association of miR-590 expression
with clinicopathological characteristics in patients with
esophageal squamous cell carcinoma.
|
|
| miR-590
expression |
|
|
|---|
|
|
|
|
|
|
|---|
| Variables | Cases | Low | High | P-value |
|---|
| Sex |
|
|
| 0.651 |
|
Male | 20 | 5 | 15 |
|
|
Female | 8 | 3 | 5 |
|
| Age, years |
|
|
| 1.000 |
|
<60 | 7 | 2 | 5 |
|
|
≥60 | 21 | 6 | 15 |
|
| Lymph node
metastasis |
|
|
| 0.030a |
| No | 11 | 6 | 5 |
|
|
Yes | 17 | 2 | 15 |
|
| TNM stage |
|
|
| 0.004a |
|
I/II | 12 | 7 | 5 |
|
|
III/IV | 16 | 1 | 15 |
|
| Distant
metastasis |
|
|
| 0.284 |
| No | 25 | 8 | 17 |
|
|
Yes | 3 | 2 | 1 |
|
 | Table II.Association of LRP6 expression with
clinicopathological characteristics in patients with esophageal
squamous cell carcinoma. |
Table II.
Association of LRP6 expression with
clinicopathological characteristics in patients with esophageal
squamous cell carcinoma.
|
|
| LRP6
expression |
|
|---|
|
|
|
|
|
|---|
| Variables | Cases | Low | High | P-value |
|---|
| Sex |
|
|
| 0.705 |
|
Male | 15 | 6 | 9 |
|
|
Female | 13 | 4 | 9 |
|
| Age, years |
|
|
| 0.236 |
|
<60 | 10 | 7 | 3 |
|
|
≥60 | 18 | 7 | 11 |
|
| Lymph node
metastasis |
|
|
| 0.019a |
| No | 12 | 8 | 4 |
|
|
Yes | 16 | 3 | 13 |
|
| TNM stage |
|
|
| 0.044a |
|
I/II | 12 | 6 | 6 |
|
|
III/IV | 16 | 2 | 14 |
|
| Distant
metastasis |
|
|
| 0.311 |
| No | 24 | 10 | 14 |
|
|
Yes | 4 | 3 | 1 |
|
Overexpression of miR-590 inhibits
migration and invasion in EC-109 and EC-9706 cells
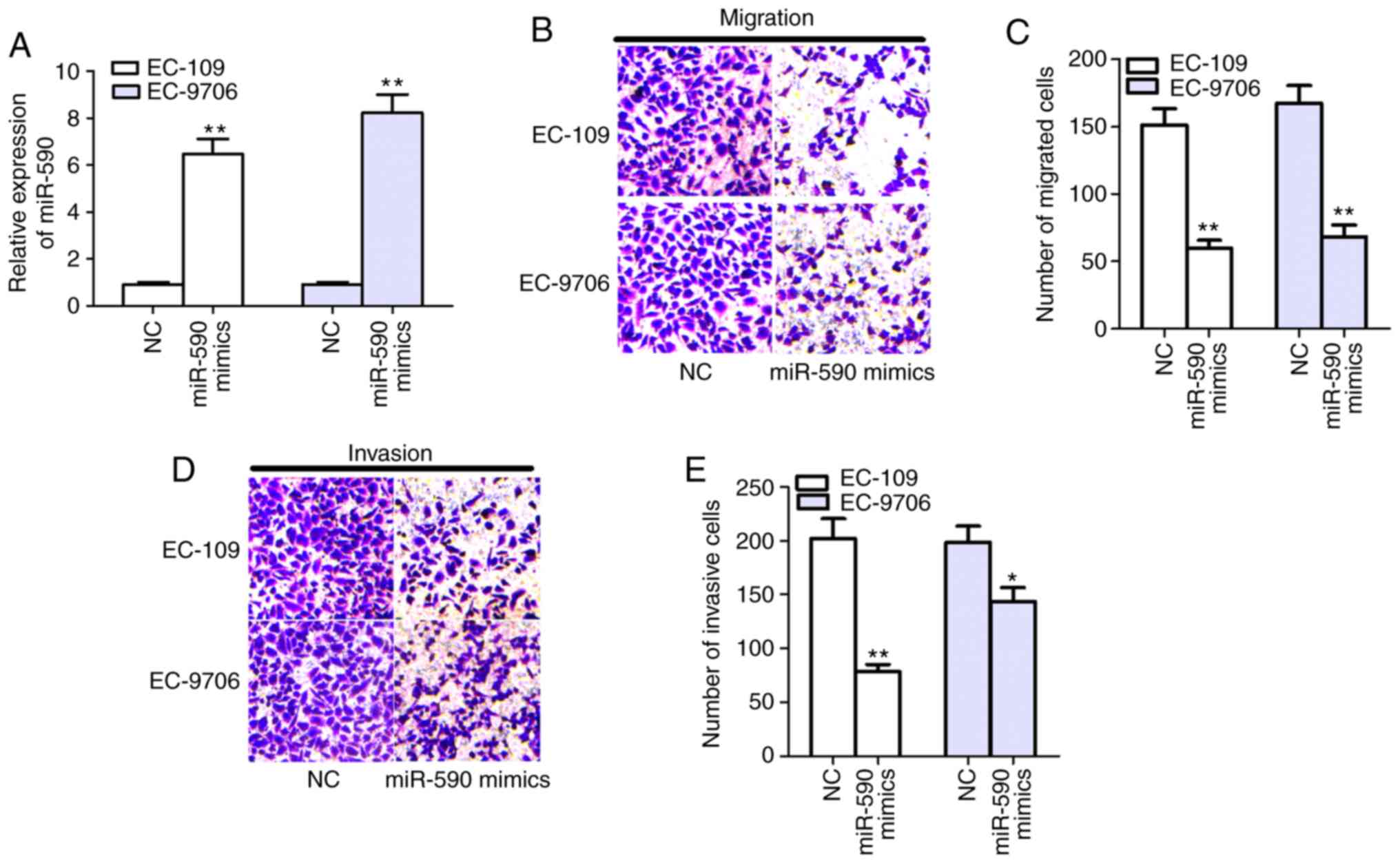
Following transfection of miR-590 mimics, its
expression was significantly increased in EC-109 and EC-9706 cells
(Fig. 2A). Moreover, the Transwell
assay results demonstrated that cell migratory and invasive
activities were significantly reduced in EC-109 and EC-9706 cells
when transfected with miR-590 mimics after 24 h in culture,
compared with NC (Fig. 2B-E).
Overexpression of miR-590 inhibits EMT
of EC-109 and EC-9706 cells
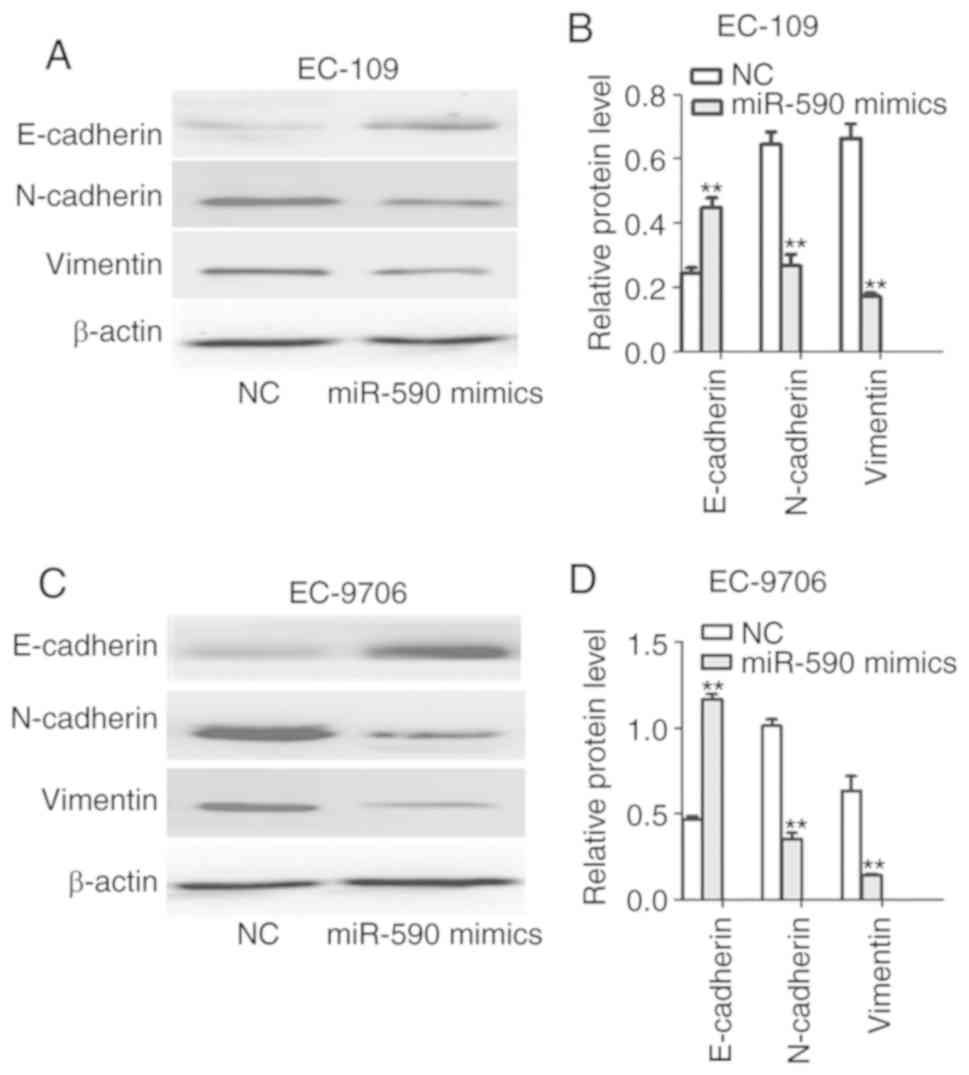
Subsequently, the expression levels of EMT-related
molecular markers were assessed. It was found that miR-590
overexpression led to downregulation of N-cadherin and vimentin,
and upregulation of E-cadherin expression in EC-109 and EC-9706
cells compared with the NC group (Fig.
3A-D), indicating that miR-590 inhibited the EMT process in
ESCC. Together with the aforementioned results, it may be inferred
that miR-590 inhibits the invasive phenotype of ESCC cells.
Knockdown of LRP6 inhibits migration
and invasion of EC-109 and EC-9706 cells
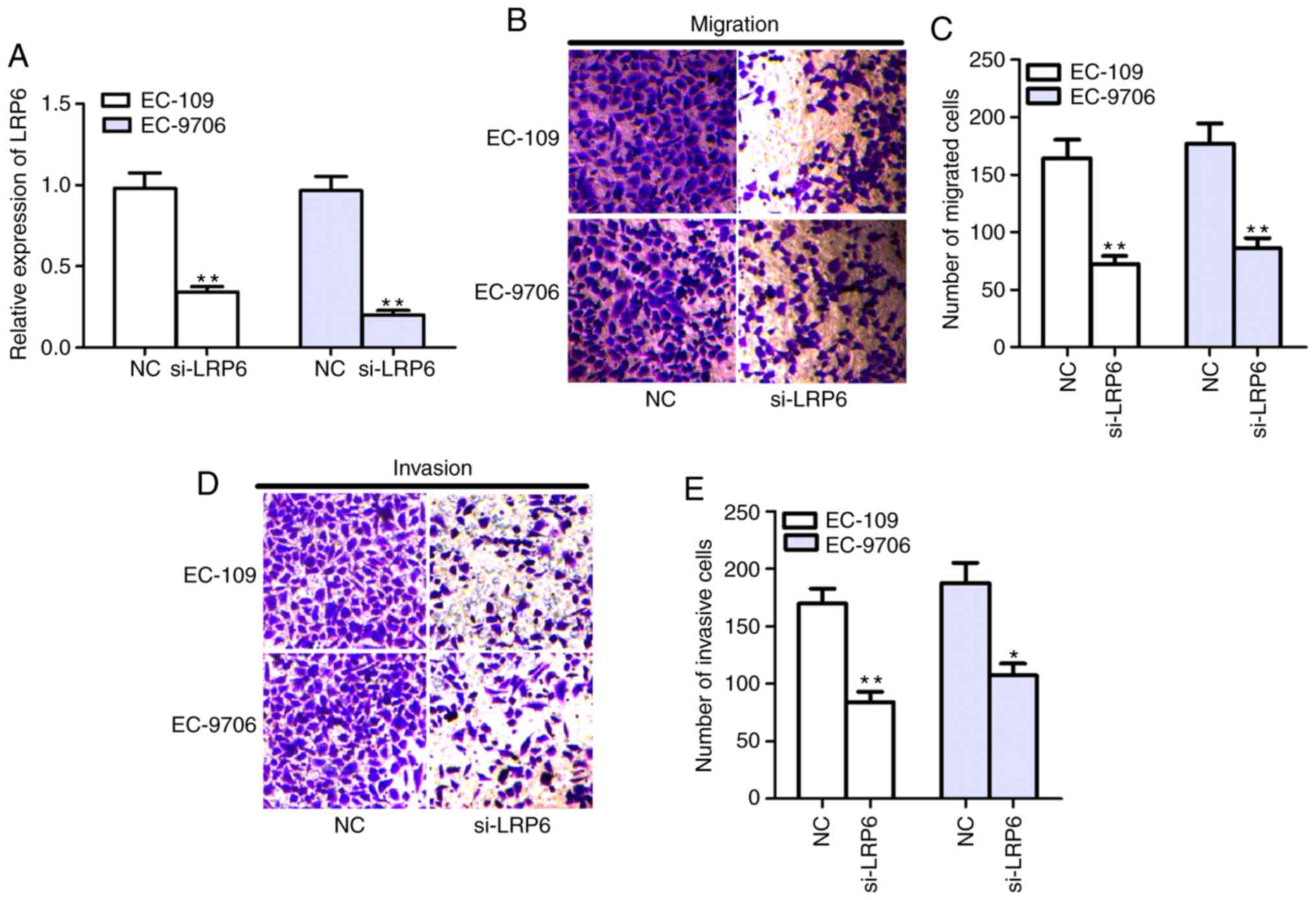
To evaluate the effects of LRP6 on ESCC cells,
siRNAs targeted LRP6 and NC were designed and transfected into
EC-109 and EC-9706 cells. After transfection of si-LRP6, LRP6
expression was significantly decreased in EC-109 and EC-9706 cells
(Fig. 4A). In addition, Transwell
assay results identified that cell migratory and invasive abilities
were significantly reduced in EC-109 and EC-9706 cells following
transfection with si-LRP6 (Fig.
4B-E), which indicated that downregulation of LRP6 exerted the
same effect as overexpression of miR-590.
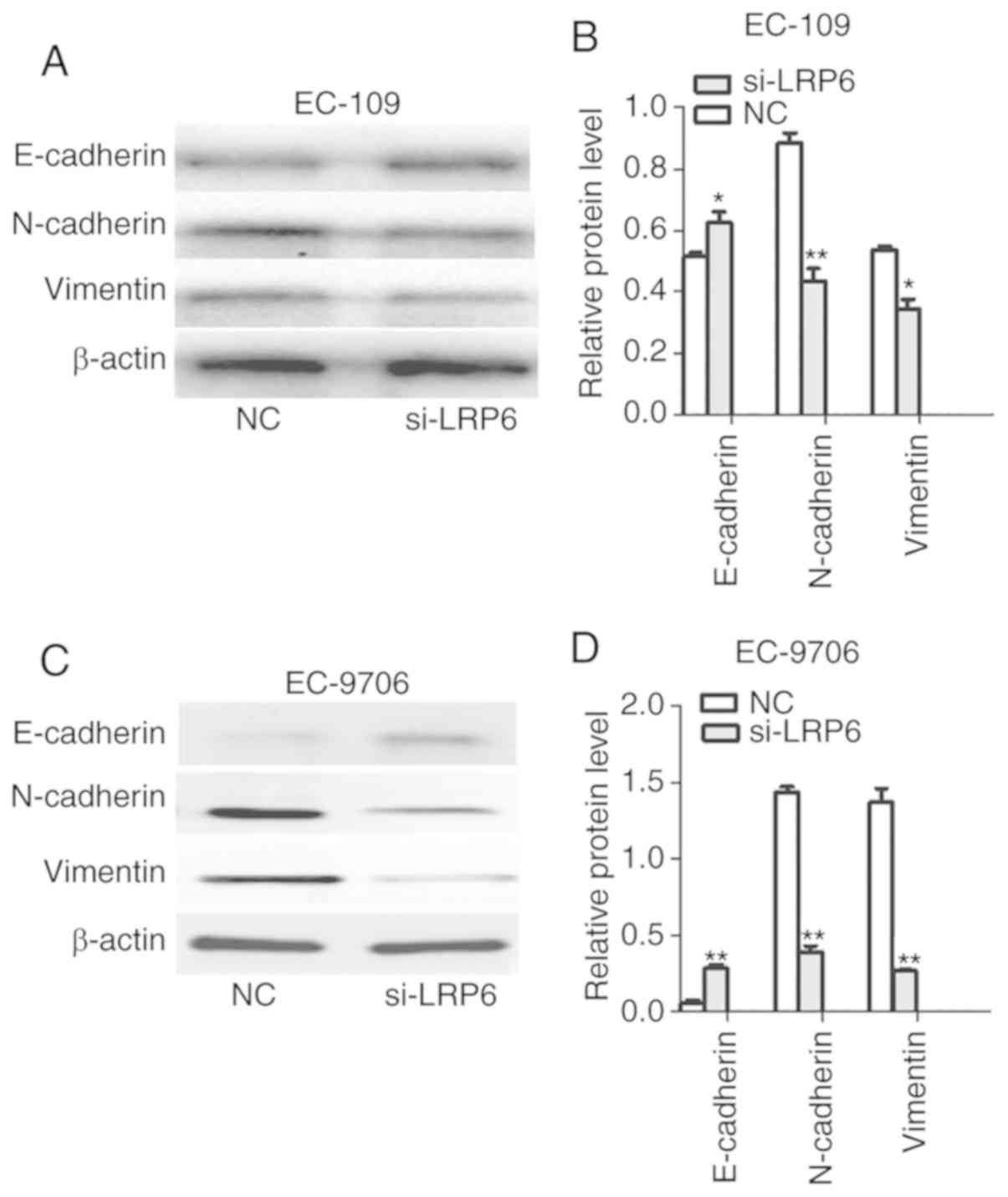
Knockdown of LRP6 inhibits EMT of
EC-109 and EC-9706 cells
Since LRP6 silencing inhibited cell invasion and
migration, it was then investigated whether these effects were
mediated via inhibition of the EMT process. The protein expression
levels of N-cadherin and vimentin were significantly downregulated,
while the expression of E-cadherin was upregulated after LRP6
silencing (Fig. 5A-D). These
results suggested that downregulation of LRP6 suppresses EMT in
ESCC.
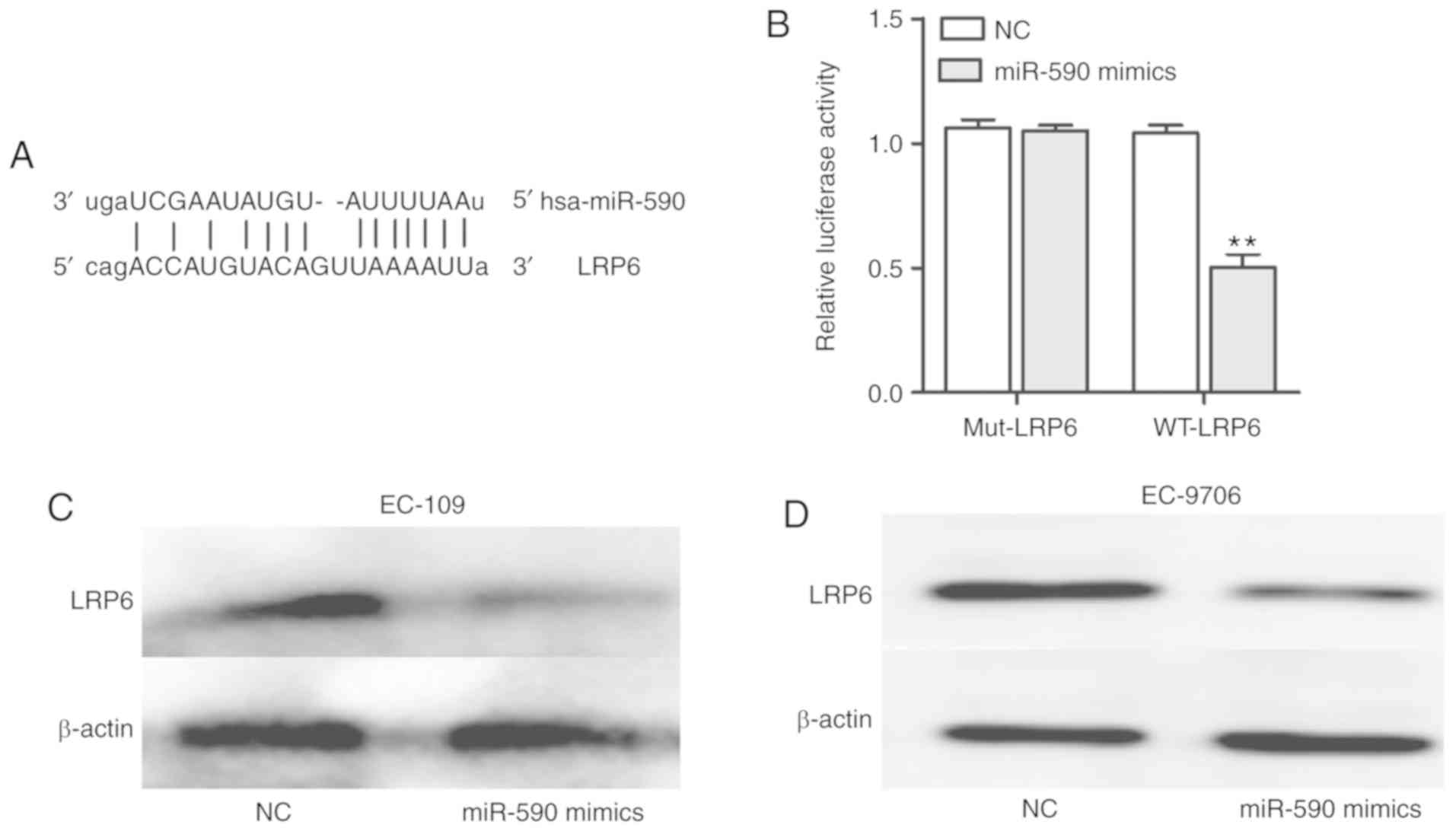
LRP6 is a potential target of
miR-590
miRNAs can regulate their targets to exert their
biological effects (19,20). For example, in glioma, miR-320a
inhibits cell invasion and migration by targeting aquaporin 4
(21), and in liver cancer, miR-498
inhibits cell proliferation and metastasis by targeting zinc finger
E-box binding homeobox 2 (22).
Therefore, the association between miR-590 and LRP6 was examined,
and it was identified that LRP6 served as a potential target of
miR-590 (Fig. 6A), which was
further assessed with a luciferase reporter assay. The luciferase
results demonstrated the interactions of miR-590 with LRP6
(Fig. 6B). In addition, western
blotting was used to demonstrate that miR-590 suppressed the
protein expression of LRP6 in EC-109 and EC-9706 cells (Fig. 6C and D). Collectively, the results
suggested that LRP6 may serve as a potential target of miR-590.
Discussion
Over the past few years, previous studies have
reported that non-coding RNAs, including miRNAs and long non-coding
RNAs, serve important roles in cancer (23–25).
For example, miR-543 was observed to promote ESCC cell migration,
invasion and EMT via targeting phospholipase A2 Group IVA (26). It has also been shown that miR-145
can promote the proliferation and metastasis of ESCC cells by
targeting SMAD5 (27). Moreover,
miR-590 was revealed to inhibit the invasion and metastasis of
triple-negative breast cancer (28). Similar anti-tumor effects have been
reported in osteosarcoma (13) and
tongue SCC (29). The present study
investigated miR-590 expression, which was found it to be
significantly decreased in ESCC tissues and cell lines.
LRP6, a member of the low-density lipoprotein
receptor family, has been reported to be closely associated with
the invasion, metastasis and EMT of cancer cells (30,31).
Furthermore, LRP6 is a key receptor protein in the Wnt signaling
pathway, and activates Wnt signaling by binding to the Wnt ligand
protein (32). In the present
study, LRP6 expression was found to be upregulated, which was
consistent with other studies reporting that LRP6 expression was
also increased in different tumor types, such as human triple
negative breast cancer (31) and
prostate cancer (33). It is well
known that miRNAs may exert their biological roles via the
regulation of their target mRNAs (19–22).
Therefore, the current study investigated the association between
miR-590 and LRP6 by analyzing their expression in 28 pairs of ESCC
tissues, and subsequently identified a weak negative correlation
between the two factors.
The functional roles of miR-590 and LRP6 in ESCC
cells were also examined in the present study. Following
transfection of miR-590 mimics into EC-109 and EC-9706 cells,
Transwell assays were performed, the results of which demonstrated
that miR-590 suppressed cell migration and invasion. EMT is a key
mechanism involved in cancer metastasis, which includes the process
of polarized epithelial cells transforming into motile stromal
cells (34,35). EMT is also considered to be one of
the most important mechanisms for promoting tumor migration and
invasion. For instance, it has been shown that a number of key
EMT-related factors, such as E-cadherin, N-cadherin and vimentin,
serve important roles in tumor invasion and metastasis (36). Moreover, the findings of the present
study suggested that miR-590 reduced the expression levels of
N-cadherin and vimentin, while enhancing E-cadherin expression.
This indicated that miR-590 not only inhibited cell migration and
invasion, but also suppressed the EMT process in ESCC cells.
However, the mechanism underlying the role of miR-590 in EMT
remains unknown. Therefore, the function of LRP6 in ESCC cells was
assessed, which identified that LRP6 knockdown exerted the same
effects as miR-590 overexpression.
The Wnt signaling pathway is often abnormally
activated and is the key signal transduction pathway that induces
EMT (37,38). In addition, the Wnt signaling
pathway can coordinate with or antagonize other signaling pathways
to regulate the proliferation, migration, invasion and EMT of tumor
cells (39,40). LRP6 is a key receptor protein in the
Wnt signaling pathway (41), which
may explain why LRP6 knockdown inhibited the migration, invasion
and EMT of ESCC cells. In order to evaluate whether the changes
caused by miR-590 were mediated by LRP6 in ESCC cells, biological
analysis and luciferase experiments were performed, and LRP6 was
identified as one of potential targets of miR-590. Western blotting
results demonstrated that the expression of LRP6 was reduced
following overexpression miR-590. Therefore, these findings may
explain how the overexpression miR-590 can inhibit the migration,
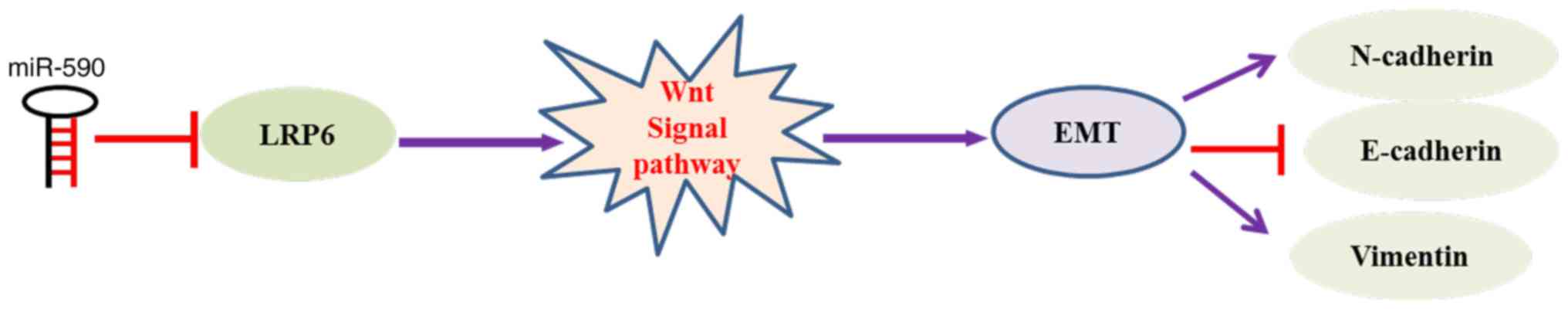
invasion and EMT in ESCC cells. It was suggested that miR-590
targeting of LRP6 leads to inactivation of the Wnt signaling
pathway, thus suppressing EMT and invasion of ESCC cells (Fig. 7). Therefore, miR-590 appears to act
as a tumor inhibitor by targeting LRP6 and restricting cell
migration, invasion and EMT in ESCC. However, there were certain
limitations to the present study, such as the limited number of
tissue samples. The focus of future studies will be to further
elucidate the roles of miR-590 and LRP6 in ESCC tissues, and
identify additional potential targets and signaling pathways of
miR-590, to determine whether these may interact with LRP6 to
affect cell migration, invasion and EMT in ESCC.
In conclusion, the findings of the present study
demonstrated that miR-590 acted as an anti-oncogene in ESCC, and
inhibited ESCC cell migration, invasion and EMT by targeting LRP6.
These findings suggested that the miR-590/LRP6/EMT axis may be a
novel potential therapeutic target for ESCC.
Acknowledgements
Not applicable.
Funding
This study was funded by the National Natural
Science Foundation of China (grant nos. 81171992 and 31570899), the
Natural Science Foundation of Henan (grant nos. 182102310328 and
162300410279), the Education Department of Henan Province (grant
nos. 18B310022 and 19A320037) and the Medical Science Research
Project of Henan Province (grant nos. 2018020765 and
2018020760).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
HG performed all of the experiments and draft the
manuscript. JL revised the manuscript. PL and LZ performed the
statistical analysis. JZ and WC designed the study. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the Zhengzhou University and all the participants
signed informed consent. All patients provided informed consent to
undergo the procedures and for having their data collected and
analyzed for research purposes.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Abnet CC, Arnold M and Wei WQ:
Epidemiology of esophageal squamous cell carcinoma.
Gastroenterology. 154:360–373. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kong D, Li Y, Wang Z and Sarkar FH: Cancer
stem cells and epithelial-to-mesenchymal transition
(EMT)-phenotypic cells: Are they cousins or twins? Cancers (Basel).
3:716–729. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J,
Lee J, Provost P, Radmark O, Kim S and Kim VN: The nuclear RNase
III Drosha initiates microRNA processing. Nature. 425:415–419.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lai EC: Micro RNAs are complementary to
3′UTR sequence motifs that mediate negative post-transcriptional
regulation. Nat Genet. 30:363–364. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shim J and Nam JW: The expression and
functional roles of microRNAs in stem cell differentiation. BMB
Rep. 49:3–10. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gu C, Xu Y, Zhang S, Guan H, Song S, Wang
X, Wang Y, Li Y and Zhao G: miR-27a attenuates adipogenesis and
promotes osteogenesis in steroid-induced rat BMSCs by targeting
PPARγ and GREM1. Sci Rep. 6:384912016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang Y, Zhang S, Xu Y, Zhang Y, Guan H, Li
X, Li Y and Wang Y: Upregulation of miR-192 inhibits cell growth
and invasion and induces cell apoptosis by targeting TCF7 in human
osteosarcoma. Tumour Biol. 37:15211–15220. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yi Y, Chen J, Jiao C, Zhong J, Song Z, Yu
X, Lu X and Lin B: Upregulated miR-193a-3p as an oncogene in
esophageal squamous cell carcinoma regulating cellular
proliferation, migration and apoptosis. Oncol Lett. 12:4779–4784.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yan L, Hu F, Yan X, Wei Y, Ma W, Wang Y,
Lu S and Wang Z: Inhibition of microRNA-155 ameliorates
experimental autoimmune myocarditis by modulating Th17/Treg immune
response. J Mol Med (Berl). 94:1063–1079. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fan N and Wang J: MicroRNA 34a contributes
to virus-mediated apoptosis through binding to its target gene Bax
in influenza A virus infection. Biomed Pharmacother. 83:1464–1470.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhao X, Xu Y, Sun X, Ma Y, Zhang Y and
Wang Y, Guan H, Jia Z, Li Y and Wang Y: miR-17-5p promotes
proliferation and epithelial-mesenchymal transition in human
osteosarcoma cells by targeting SRC kinase signaling inhibitor 1. J
Cell Biochem. 120:5495–5504. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Svoronos AA, Engelman DM and Slack FJ:
OncomiR or tumor suppressor? The duplicity of MicroRNAs in cancer.
Cancer Res. 76:3666–3670. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang WT, Qi Q, Zhao P, Li CY, Yin XY and
Yan RB: miR-590-3p is a novel microRNA which suppresses
osteosarcoma progression by targeting SOX9. Biomed Pharmacother.
107:1763–1769. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xu BB, Gu ZF, Ma M, Wang JY and Wang HN:
MicroRNA-590-5p suppresses the proliferation and invasion of
non-small cell lung cancer by regulating GAB1. Eur Rev Med
Pharmacol Sci. 22:5954–5963. 2018.PubMed/NCBI
|
|
15
|
Wang FF, Wang S, Xue WH and Cheng JL:
microRNA-590 suppresses the tumorigenesis and invasiveness of
non-small cell lung cancer cells by targeting ADAM9. Mol Cell
Biochem. 423:29–37. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ge X and Gong L: MiR-590-3p suppresses
hepatocellular carcinoma growth by targeting TEAD1. Tumour Biol.
39:10104283176959472017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shan X, Miao Y, Fan R, Qian H, Chen P, Liu
H, Yan X, Li J and Zhou F: MiR-590-5P inhibits growth of HepG2
cells via decrease of S100A10 expression and Inhibition of the Wnt
pathway. Int J Mol Sci. 14:8556–8569. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hong Z, Hong C, Ma B, Wang Q, Zhang X, Li
L, Wang C and Chen D: MicroRNA-126-3p inhibits the proliferation,
migration, invasion, and angiogenesis of triple negative breast
cancer cells by targeting RGS3. Oncol Rep. 42:1569–1579.
2019.PubMed/NCBI
|
|
20
|
Alcantara KMM and Garcia RL: MicroRNA-92a
promotes cell proliferation, migration and survival by directly
targeting the tumor suppressor gene NF2 in colorectal and lung
cancer cells. Oncol Rep. 41:2103–2116. 2019.PubMed/NCBI
|
|
21
|
Xiong W, Ran J, Jiang R, Guo P, Shi X, Li
H, Lv X, Li J and Chen D: miRNA-320a inhibits glioma cell invasion
and migration by directly targeting aquaporin 4. Oncol Rep.
39:1939–1947. 2018.PubMed/NCBI
|
|
22
|
Zhang X, Xu X, Ge G, Zang X, Shao M, Zou
S, Zhang Y, Mao Z, Zhang J, Mao F, et al: miR-498 inhibits the
growth and metastasis of liver cancer by targeting ZEB2. Oncol Rep.
41:1638–1648. 2019.PubMed/NCBI
|
|
23
|
Guan H, Mei Y, Mi Y, Li C, Sun X, Zhao X,
Liu J, Cao W, Li Y and Wang Y: Downregulation of lncRNA
ANRIL suppresses growth and metastasis in human osteosarcoma
cells. Onco Targets Ther. 11:4893–4899. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Guan H, Shang G, Cui Y, Liu J, Sun X, Cao
W, Wang Y and Li Y: Long noncoding RNA APTR contributes to
osteosarcoma progression through repression of miR-132-3p and
upregulation of yes-associated protein 1. J Cell Physiol.
2234:8998–9007. 2019. View Article : Google Scholar
|
|
25
|
Chen Z, Lin J, Wu S, Xu C, Chen F and
Huang Z: Up-regulated miR-548k promotes esophageal squamous cell
carcinoma progression via targeting long noncoding RNA-LET. Exp
Cell Res. 362:90–101. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhao H, Diao C, Wang X, Xie Y, Liu Y, Gao
X, Han J and Li S: MiR-543 promotes migration, invasion and
epithelial-mesenchymal transition of esophageal cancer cells by
targeting phospholipase A2 Group IVA. Cell Physiol Biochem.
48:1595–1604. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang Q, Gan H, Song W, Chai D and Wu S:
MicroRNA-145 promotes esophageal cancer cells proliferation and
metastasis by targeting SMAD5. Scand J Gastroenterol. 53:769–776.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yan M, Ye L, Feng X, Shi R, Sun Z, Li Z
and Liu T: MicroRNA-590-3p inhibits invasion and metastasis in
triple-negative breast cancer by targeting slug. Am J Cancer Res.
10:965–974. 2020.PubMed/NCBI
|
|
29
|
Xu Y, Han T, Zhu Y and Chen Q: miR-590-5P
inhibits the progression of tongue squamous cell carcinoma by
targeting FasL. Int J Clin Exp Pathol. 10:11880–11887.
2017.PubMed/NCBI
|
|
30
|
Yao Q, An Y, Hou W, Cao YN, Yao MF, Ma NN,
Hou L, Zhang H, Liu HJ and Zhang B: LRP6 promotes invasion and
metastasis of colorectal cancer through cytoskeleton dynamics.
Oncotarget. 8:109632–109645. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ma J, Lu W, Chen D, Xu B and Li Y: Role of
Wnt co-receptor LRP6 in triple negative breast cancer cell
migration and invasion. J Cell Biochem. 118:2968–2976. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hua Y, Yang Y, Li Q, He X, Zhu W, Wang J
and Gan X: Oligomerization of Frizzled and LRP5/6 protein initiates
intracellular signaling for the canonical WNT/β-catenin pathway. J
Biol Chem. 293:19710–19724. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lu W and Li Y: Salinomycin suppresses LRP6
expression and inhibits both Wnt/β-catenin and mTORC1 signaling in
breast and prostate cancer cells. J Cell Biochem. 115:1799–807.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Pastushenko I and Blanpain C: EMT
transition states during tumor progression and metastasis. Trends
Cell Biol. 29:212–226. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yilmaz M and Christofori G: EMT, the
cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev.
28:15–33. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Paolillo M and Schinelli S: Extracellular
matrix alterations in metastatic processes. Int J Mol Sci.
20:49472019. View Article : Google Scholar
|
|
37
|
Arend RC, Londoño-Joshi AI, Straughn JM Jr
and Buchsbaum DJ: The Wnt/β-catenin pathway in ovarian cancer: A
review. Gynecol Oncol. 131:772–779. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Gonzalez DM and Medici D: Signaling
mechanisms of the epithelial-mesenchymal transition. Sci Signal.
7:re82014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang J, Tian XJ and Xing J: Signal
transduction pathways of EMT induced by TGF-β, SHH, and WNT and
their crosstalks. J Clin Med. 5:412016. View Article : Google Scholar
|
|
40
|
Liu X, Yun F, Shi L, Li ZH, Luo NR and Jia
YF: Roles of signaling pathways in the epithelial-mesenchymal
transition in cancer. Asian Pac J Cancer Prev. 16:6201–6206. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Dong Y, Zhang Y, Kang W, Wang G, Chen H,
Higashimori A, Nakatsu G, Go M, Tong JH, Zheng S, et al: VSTM2A
suppresses colorectal cancer and antagonizes Wnt signaling receptor
LRP6. Theranostics. 9:6517–6531. 2019. View Article : Google Scholar : PubMed/NCBI
|