Introduction
The incidence of human papillomavirus-positive
(HPV+) tonsillar and base of tongue squamous cell
carcinomas (TSCC/BOTSCC), the major subsites of oropharyngeal
squamous cell cancer (OPSCC), (but not other OPSCC subsites) is
still increasing in most western countries (1–4).
Together they account for an increasing proportion of head and neck
squamous cell carcinomas (HNSCC), especially in countries where
smoking, a main HNSCC risk factor is decreasing (4,5).
Patients with HPV+ TSCC/BOTSCC usually
respond well to treatment and have better long-term survival
compared to HPV− TSCC/BOTSCC and HNSCC in general,
irrespective if they receive radiotherapy alone, or
radio-chemotherapy (i.e. 80 vs. 40–50% 5-year overall survival)
(1,5–9).
However, over recent decades, treatment for HNSCC, as well as for
HPV+ TSCC/BOTSCC, has gradually been intensified with
radiotherapy and induction or concomitant chemotherapy, and the
numbers of side effects have increased (5). Most likely this intensified therapy
could be de-escalated for most patients with HPV+
TSCC/BOTSCC, but to facilitate this, greater knowledge of the
responsiveness of these tumours to different treatment strategies
is required, both through clinical and preclinical studies
(10).
Previous in vitro studies have investigated
response to ionizing irradiation (IR) in a range of HNSCC cell
lines. These studies describe differences in sensitivity to IR
correlating with HPV status (11–17).
Some of these reports suggest an association between a greater
intrinsic cellular radio-sensitivity and increased survival in
HPV+ HNSCC cell lines (11–13).
However, in these studies the numbers of HPV+ OPSCC cell
lines have been limited, making it difficult to assess the
generalizability of these observations (11–17).
There is a clear need to assess the correlation between HPV status
and radiosensitivity in a greater number of cell lines, but
HPV+ OPSCC cell lines are relatively scarce and are
notoriously difficult to establish.
In 2014, to our knowledge, there are eight published
HPV+ HNSCC cell lines: UPCI-SCC-90, UPCI-SCC-154,
UPCI-SCC-152, UM-SCC-104, UM-SCC-47, 93-VU-147T, UD-SCC-2 and
UT-SCC-45 (11–18). In 2018, we established and
characterised two new HPV+ cell lines CU-OP-2 and
CU-OP-3 (18). In this report, two
additional cell lines CU-OP-17 and CU-OP-20 have been derived and
characterised. We have used these 4 novel lines, and 6 established
OPSCC lines to investigate the effects of IR on surviving fraction
(SF), cell cycle distribution, and global transcription patterns,
similar to studies performed by others (11–13).
In addition, circos plots were established, to identify possible
changes in HPV integration sites.
Materials and methods
Patients, cell lines, and sample
collection
Five HPV+ and five HPV− OPSCC
cell lines were included in this study.
To derive novel lines, TSCC biopsies were obtained,
during the diagnostic procedure (where ultimately diagnosis is
finalised by multidisciplinary meetings) from patients, prior to
treatment at Cardiff and Vale University Health Board, in
concordance with Ethical and NHS R&D approval (reference number
13/WA/0002), by written consent. The derivation of HPV+
CU-OP-2 and CU-OP-3 has been described previously (18). CU-OP-17 and CU-OP-20, described here
for the first time, were established using the same processes
(18), and were shown below to be
HPV− and HPV+ respectively. The identity of
all CU-OP cell lines included in this report was established by
short tandem repeats (STR) as described previously for CU-OP-2 and
CU-OP-3 (18), and by Public Health
England (Promega, Southampton, UK) for CU-OP-17 and CU-OP-20. The
characteristics of the patients from which CU-OP-17 and CU-OP-20
were derived and their tumours are shown in Table I. For CU-OP-2 and CU-OP-3 these
details have been published previously (18). All CU-OP cell lines were grown on 60
Gy irradiated J2 3T3 feeder layers (a kind gift from Dr Sally
Roberts, University of Birmingham, UK) in GMEM medium with 10% FBS,
and further details have been described previously (18).
 | Table I.CU-OP-17 and CU-OP-20 cell lines and
the corresponding patient characteristics. |
Table I.
CU-OP-17 and CU-OP-20 cell lines and
the corresponding patient characteristics.
| Study number | Sex (M/F) | Age (years) | Smoking
statusa | Site | TNM
stageb | P16 IHC | P53
statusc | HPV type |
Treatmentd | Recurrence | Cell line
population doublingse |
|---|
| CU-OP-17 | M | 68 | Y (100
pack/yr) | Tonsil | T4aN1M0 | Y | WT | Negative | Induction chemo,
then CRT, then neck dissection | None | 125 |
| CU-OP-20 | M | 55 | Never | Tonsil | T2N2aM0 | Y | WT | 16 | Neck dissection
followed by CRT to primary | None | 120 |
HPV+ UM-SCC-47 and HPV−
UM-SCC-6, UM-SCC-19, UM-SCC-74a and UM-SCC-4 were obtained from
Professor Thomas Carey at the University of Michigan USA.
HPV+ UPCI-SCC-90 was purchased from Deutsche Sammlung
von Mikoorganismen und Zellkulturen (DSMZ), Leibniz, Germany. All
these cell lines are described in the data base https://web.expasy.org/cellosaurus/. These cell
lines were all cultured in DMEM (Sigma-Aldrich; Merck KGaA), with
1% L-glutamine, 100 U/ml of penicillin, and 100 µg/ml streptomycin
and 10% foetal bovine serum (Sigma-Aldrich; Merck KGaA).
Human epithelial keratinocytes (HEKn) were purchased
from Thermo Fisher Scientific, Inc. and grown as described
previously in EpiLife media (18).
All cell lines were tested for absence of
mycoplasma, by standardised PCR, using the Venor®GeM
detection kit (Minerva Biolabs), which is specific to the highly
conserved 16s rRNA coding region, thereby detecting a wide range of
mycoplasma species.
HPV PCR
The presence of HPV in CU-OP-17 and CU-OP-20 was
confirmed by a bead-based multiplex assay for 27 HPV types as
described in detail previously (19). This PCR uses
BSG5+/G6+ primers targeting the L1 region, as
well as primers targeting the HPV16 E6 region, and includes the
analysis of all high-risk HPV types using a PCR-based bead-based
multiplex-assay on a MagPix instrument (Luminex Corp.) as
previously described (19). A mean
fluorescence intensity (MFI) value above 1.5× background + 8 for
specific HPV primers were considered as HPV-positive as previously
described (19).
Clonogenic assay and analysis
Cells were cultured at low cell densities, more
specifically in ranges of 2,500-12,500 cells per plate on 6-cm
culture dishes (VWR) and incubated at 37°C with 5% CO2
for 24 h. Cell density per cell line, was defined before the
initiation of clonogenic assays. Cell density, was defined by
individual cell line density tests (using ranges of cells between
1,000-20,000 cells/well lasting for ~10-15 days depending on the
cell line. After 24 h the cells were irradiated with 0–6 Gy
(Gammacell-1000 MDS Nordion; a caesium-137 source) and the media
were changed after 7 days. The assays were stopped 10–15 days later
(depending on the growth of the cell lines) and the cells stained
with crystal violet, and the colonies counted using a Colony
counter (ColonyDoc-It Imaging Station, UVP).
Data analysis and statistical
evaluation of clonogenic assay
All experiments were performed in triplicates. The
plating efficiency (PE) and survival fraction (SF) of each cell
line per IR dose was calculated. Cells were classified as
radiosensitive SF <0.40 or radio-resistant SF >0.40 as
described before (20). The mean
value was calculated and standard deviations (SD) are indicated as
error bars. Statistical evaluation was performed using GraphPad
Prism (GraphPad Version 7, GraphPad Software), by using a two-way
ANOVA with a Sidak post-test. Strength of significance is indicated
as follows: *P<0.05, **P<0.01, ***P<0.01 and
****P<0.001). ns stands for not significant.
Flow cytometry
Sample preparation and collection
Cells were cultured without feeder cells in 6-cm
culture dishes (VWR) and treated with 0–6 Gy 24 h after seeding.
Untreated cells were collected 24 h after seeding, while irradiated
cells were collected 8, 24 and 48 h after treatment. Approximately
500,000 cells/cell line were fixed in 1 ml of 70% ethanol in
fluorescence-activated cell sorting (FACS) tubes and stored at
−20°C for at least 1 h. Cells were then washed with PBS, incubated
with 100 µl of RNase A (10 µg/ml) (Sigma-Aldrich; Merck KGaA) for
45 min at 37°C, centrifuged at 270 × g, and resuspended in 200 µl
propidium iodide (PI) solution (50 µg/ml) (Sigma-Aldrich; Merck
KGaA) and incubated for 15 min at 37°C.
Cell cycle measurement and statistical
analysis
The PI-stained cells were analysed using a BD Accuri
C6 (BD Biosciences) low-pressure flow cytometer (absorbance 488
nm). Data were extracted as FCS files and the cell cycle
distribution was analysed with FlowJo analysis software (version
10; FlowJo LLC), using the cell cycle tool, based on the Watson
pragmatic algorithm (21). For each
cell cycle phase the mean value was calculated based on a total of
three experimental runs. Each IR dose and time point were compared
to the non-treated sample ‘time zero’, to quantify changes in cell
cycle distribution after IR treatments. Excel 2016 was used to
generate 100% stacked bar charts. Statistical analysis was
performed in GraphPad Prism 7 (GraphPad Software, Inc.) using a
two-way ANOVA with a Sidak post-test to the panel of cell lines
(treatment dose and time points). Strength of significance is
indicated as follows: *P<0.05, **P<0.01, ***P<0.01 and
****P<0.001). ns stands for not significant.
mRNA sequencing
Experimental set up
Cells (untreated and 2 Gy) were collected 24 h after
treatment and RNA was extracted with QiaAMP Mini kit (Qiagen)
according to the manufacturer's instructions.
Library preparation
Library preparation and validation for mRNA
sequencing was performed through a commercial service/collaboration
with Wales Gene Park (Cardiff University, UK). Library preparation,
including depletion of ribosomal RNA was carried out using the
Illumina® TruSeq® Stranded Total RNA with
Ribo-Zero Gold™ kit (Illumina Inc.) according to the instructions
of the manufacturer.
Sequencing and data analysis
A 75-base paired-end dual index read format was used
on the Illumina® HiSeq2500 in high-output mode by the
Wales Gene Park (Cardiff University, UK). Sequencing data were
analysed by the bioinformatics service by Dr Peter Giles at the
Wales Gene Park (Cardiff University, UK). Trimmed reads were mapped
against a combined human sequence genome hg19 and an HPV16 genome
reference sequence NC_001526 using STAR (Alex Dobin, Git Hub) to
generate circos plots. For both exons and transcripts, gene
expression counts were calculated, using Subread feature Counts
Version 1.5.1 (22). Data were
visualised using The Integrative Genomics Viewer (IGV) downloaded
from the Broad Institute (23) and
Geneview software (written by Dr Peter Giles of the Wales Gene
Park).
Differential gene expression
The DEseq2 analysis tool was used to identify
differentially expressed genes (statistical analysis of count
matrices for systematic changes between conditions) (24). For multiple testing and false
discovery issues, the generated P-values were corrected using the
FDR method (25). The data are
shown in heatmaps in 3 colour patterns, where green represents
underexpressed (compared to median); black represents values
approximately median expression; and red represents overexpressed
values.
Human viral fusion transcript plots
(circos plots)
Circos plots were used to visualise and identify
human (hg19) and viral (HPV-16) mRNA fusion transcripts (26).
Results
Derivation of novel OPSSC cell lines
CU-OP-17 and CU-OP-20
Explant cultures using fresh primary biopsies from
two patients with TSCC were attempted as described previously
(18). In addition, and the
identity of the cell lines CU-OP-17 and CU-OP-20 to the tumour
biopsies was confirmed by STR. Details of these two patients and
their tumours are shown in Table I.
The HPV status of the biopsy and resulting cell line was assessed
by PCR-based bead-based multiplex-assay on a MagPix instrument and
the obtained MFIs calculated as described above: CU-OP-20 tested
positive for HPV-16; CU-OP-17 was HPV−. TP53 status
assessed by RNA sequencing was considered as being wild-type in
both cell lines.
Surviving fraction (SF) of
HPV+ and HPV−
OPSCC cell lines after ionising
irradiation (IR)
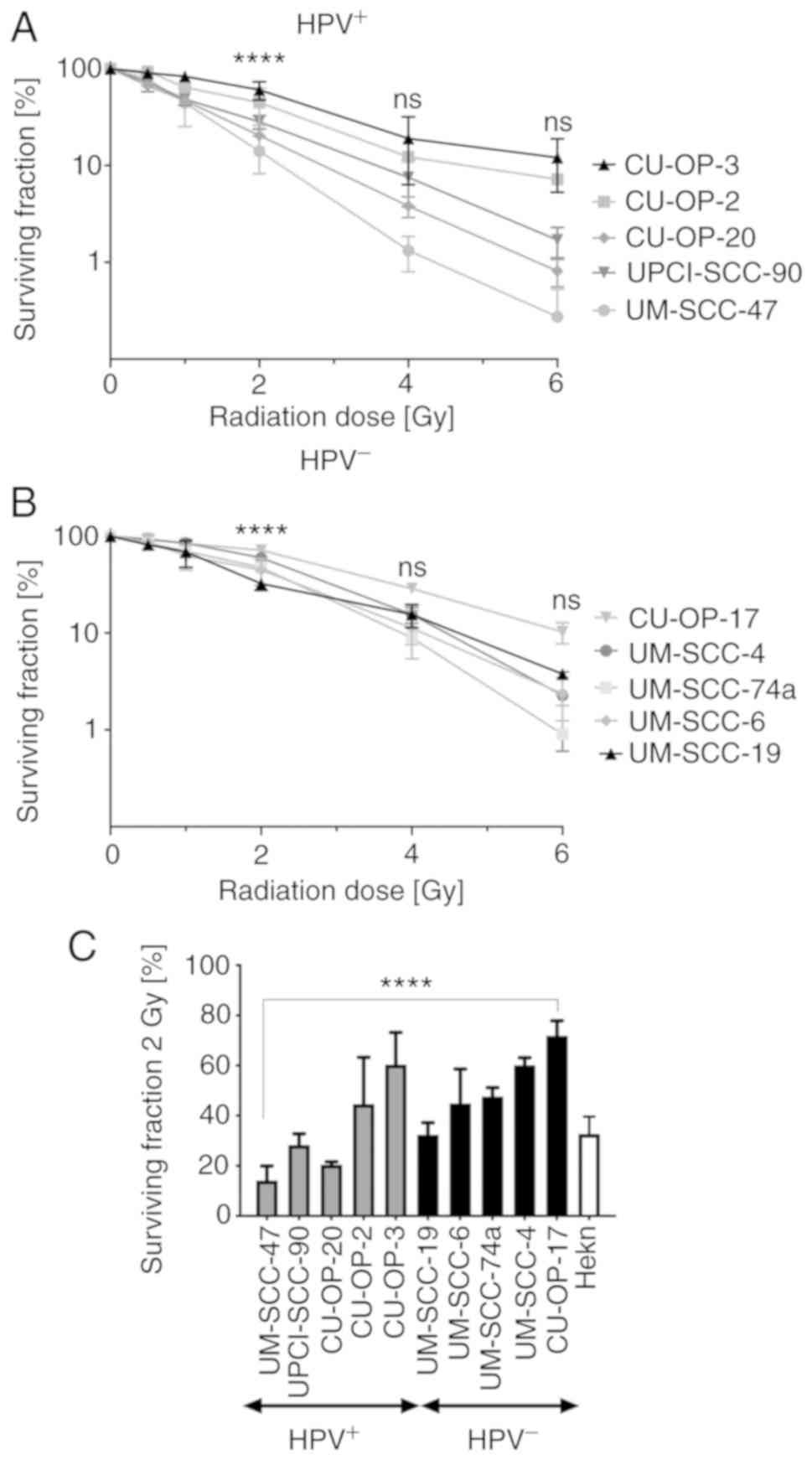
The sensitivity of the panel of 10 OPSCC cell lines
to IR was assessed using clonogenic survival assays. The SFs of all
OPSCC cell lines to 0.5–6 Gy respectively are shown in Fig. 1, and Tables II and III, respectively.
 | Table II.Surviving fraction (SF) of
HPV+ cell lines after treatment with 0.5–6 Gy. |
Table II.
Surviving fraction (SF) of
HPV+ cell lines after treatment with 0.5–6 Gy.
|
| Surviving fraction
(%) |
|---|
|
|
|
|---|
|
| HPV+
cell lines sensitive to IR | HPV+
cell lines resistant to IR |
|---|
| Treatment dose
(Gy) | UMSCC-47 | UPCI-SCC-90 | CU-OP-20 | CU-OP-2 | CU-OP-3 |
|---|
| 0.5 | 78.4 | 72.5 | 66.8 | 93.6 | 90.5 |
| 1 | 44.4 | 48.5 | 46.9 | 64.1 | 83.3 |
| 2a | 14.1 | 28.3 | 20.4 | 44.7 | 60.4 |
| 4 | 1.3 | 7.5 | 3.8 | 12.2 | 19.1 |
| 6 | 0.3 | 1.7 | 0.8 | 7.2 | 12.1 |
 | Table III.Surviving fraction (SF) of
HPV− OPSCC cell lines and HEKn after treatment with
0.5–6 Gy. |
Table III.
Surviving fraction (SF) of
HPV− OPSCC cell lines and HEKn after treatment with
0.5–6 Gy.
|
| Surviving fraction
(%) |
|
|---|
|
|
|
|
|---|
|
| HPV−
cell line sensitive to IR | HPV−
cell lines resistant to IR | Non-cancer cell
line response to IR |
|---|
|
|
|
|
|
|---|
| Treatment dose
(Gy) | UMSCC-19 | UMSCC-6 | UMSCC-74a | UMSCC-4 | CU-OP-17 | HEKn |
|---|
| 0.5 | 81.7 | 87.4 | 85.3 | 92.3 | 94.3 | 79.8 |
| 1 | 69.1 | 62.9 | 70.1 | 85.1 | 84.2 | 70.6 |
| 2a | 32.4 | 44.9 | 47.7 | 60.1 | 71.8 | 32.7 |
| 4 | 15.5 | 11.3 | 8.9 | 15.6 | 28.8 | 17.6 |
| 6 | 3.8 | 2.4 | 0.9 | 2.3 | 10.3 | 1.6 |
Special focus was put on an indicated clinically
relevant dose of 2 Gy (20), which
identified three HPV+ OPSCC cell lines UM-SCC-47
(SF=14.1) UPCI-SCC-90 (SF=28.3) and CU-OP-20 (SF=20.4) and only one
HPV− OPSCC cell line UM-SSC-19 (SF=32.4) as
radiosensitive, Tables II and
III, respectively. All other cell
lines were by definition radioresistant (20). Furthermore, for 2 Gy,
HPV+ OPSCC cell lines tended to be more sensitive and
showed a wider variability in SF values (14.1–60.4, mean 33.6) than
the HPV− OPSCC cell lines (32.4–71.8, 51.4) P=0.0754
(Mann-Whitney one-tailed t-test as conducted in (13) (Fig.
1, Tables II and III, respectively).
There were significant differences in SF within the
HPV+ and HPV− groups after IR with 2 Gy.
HPV+ UM-SCC-47 and CU-OP-20 were more radiosensitive to
2 Gy compared to CU-OP-2 and CU-OP-3, and UPCI-SSC-90 was more
radiosensitive than CU-OP-3 (for all at least P<0.05) (Fig. 1A). Among the HPV− OPSCC
cell lines, UM-SCC-19 was more radiosensitive than CU-OP-17
(P<0.0001) (Fig. 1B). Fig. 1C shows the SF values in more detail
after treatment with 2 Gy for all OPSCC cell lines as well as HEKn.
Fig. 1C demonstrates that
HPV+ OPSCC cell lines have a variable sensitivity to IR,
as well as an evident overlap with HPV− OPSCC cell
lines, but still tend to be more radiosensitive than the
HPV− OPSCC cell lines.
Effects of IR on cell cycle
distribution in HPV+ and HPV− OPSCC cell
lines
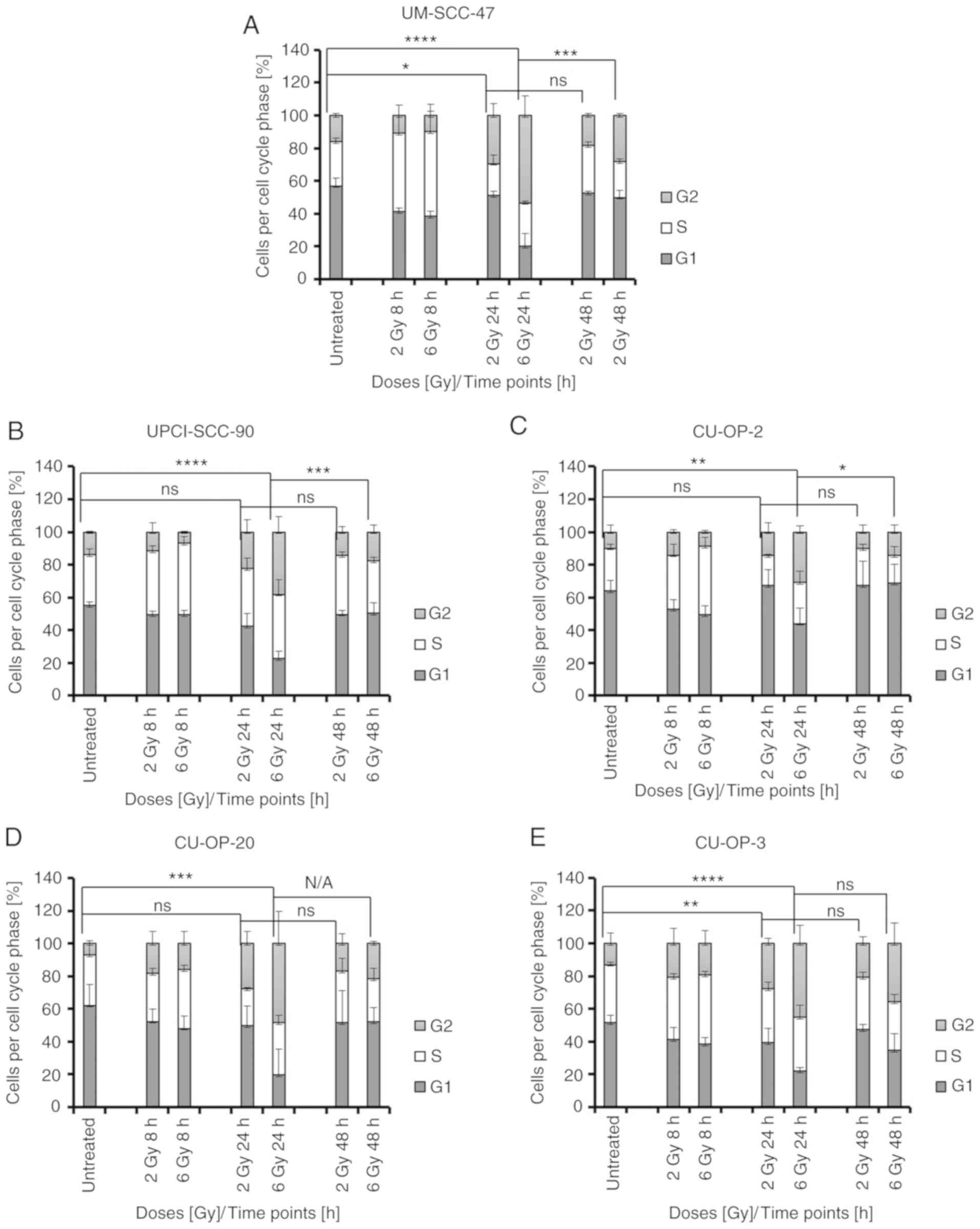
To gain insight into the mechanisms underlying the
differences in sensitivity to IR, the cell lines were treated with
IR and the effects on cell cycle distribution were assessed by flow
cytometry. IR induced cell cycle effects primarily on
HPV+ OPSCC as compared to HPV− OPSCC and
mainly in the proportion of cells in G2, and the data are presented
in detail below.
Changes in G2 arrest in
HPV+ OPSCC cell lines
No significant changes were observed in cell cycle
distribution 8 h after 2–6 Gy for any cell line. However, after 24
h (2 Gy), a significant increase in cells in G2 phase was observed
for two HPV+ OPSCC cell lines: UM-SCC-47 (the most
radiosensitive line, P=0.0135) and CU-OP-3 (the most radioresistant
line, P=0.0075) (Fig. 2A and E).
For the 6 Gy treatment after 24 h, all HPV+ OPSCC cell
lines showed a significant increase in G2 arrest (UM-SCC-47,
P<0.0001; UPCI-SCC-90, P<0.0001; CU-OP-20, P=0.0004; CU-OP-2,
P=0.0019; and CU-OP-3, P<0.0001) (Fig. 2). After 48 h post 6 Gy treatment the
proportion of cells in G2 was generally reduced. In UM-SCC-47,
UPCI-SSC-90 and CU-OP-2, the reduction in G2 fraction was
significant (P<0.05) (Fig.
2A-C).
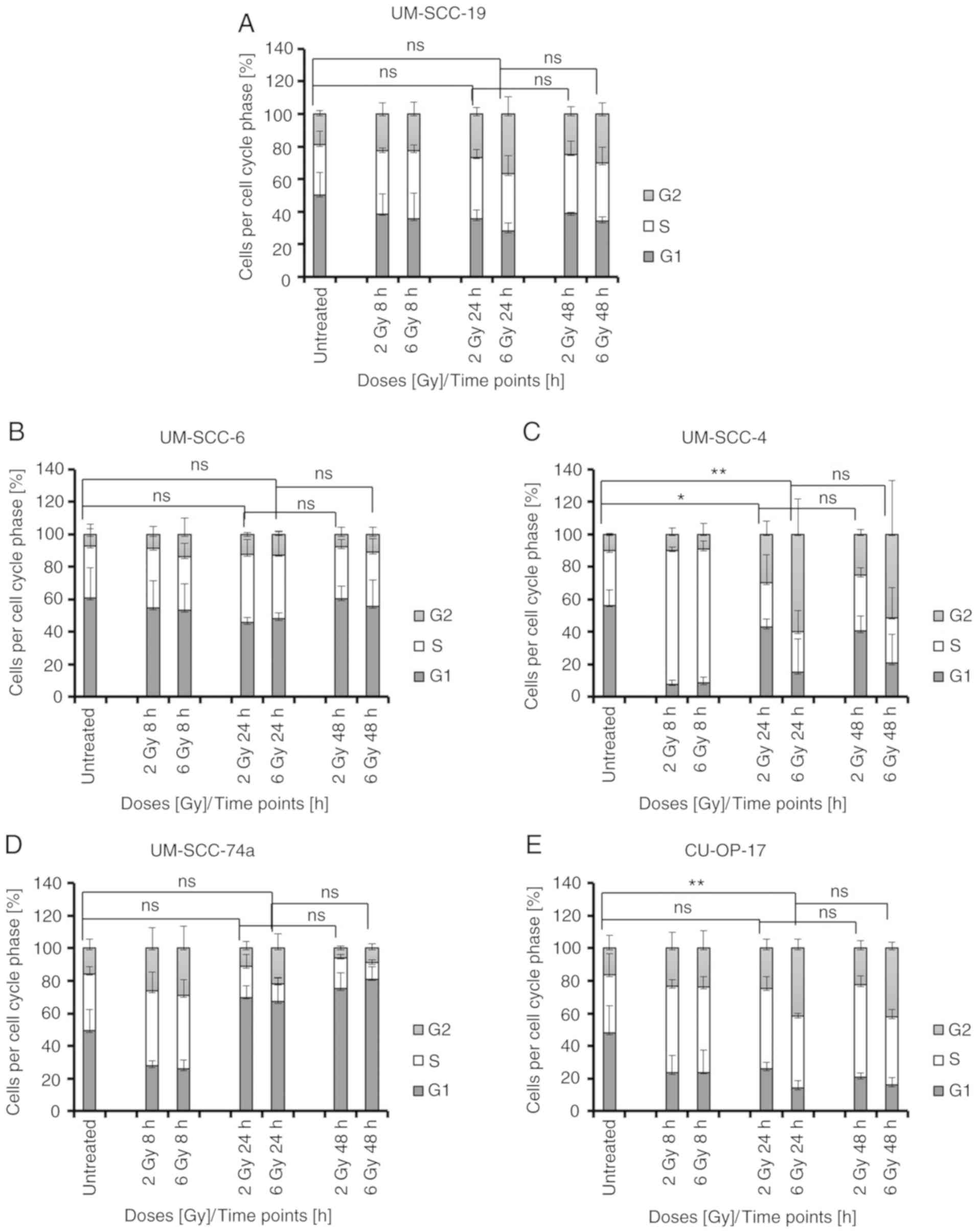
Changes in G2 arrest in
HPV−
OPSCC cell lines
Compared to untreated controls, at 2 Gy after 24 h,
a significant increase in cells in G2 was observed in one
HPV− OPSCC cell line, UM-SCC-4, P=0.0222 (Fig. 3C). With 6 Gy after 24 h, an increase
in G2 arrest was only observed in two lines, and these were the two
most IR resistant ones of the five HPV− cell lines:
UM-SCC-4, P=0.0027 and CU-OP-17, P=0.0052 (Fig. 3C and E). It was notable that in
contrast to the HPV+ lines, neither UM-SCC-4, nor
CU-OP-17 showed a reduction in G2 fraction after 48 h.
Changes in G1 in HPV+ and
HPV−
OPSCC cell lines
Roughly, 50–60% of HPV+ and
HPV− OPSCC cells were initially in G1, and few changes
occurred 8–48 h after IR (Figs. 2
and 3). Compared to controls, with
2 Gy after 8 h, decreases in the proportion of cells in G1 were
noted in HPV+ UM-SCC-47, P=0.0031; HPV−
UM-SCC-4, P<0.0001; and HPV− UM-SCC-74a, P=0.0168;
(Figs. 2A and 3C and D), and this was the case after 24 h
for CU-OP-17, P=0.0402; (Fig. 3E),
while instead an increase in G1 was seen at 24 h in UM-SCC-74a,
P=0.0037; (Fig. 3D). At 6 Gy, after
24 h, a decrease in G1 could be seen for HPV+ UM-SCC-47,
P<0.0001; and UPCI-SCC-90, P<0.0001; (Fig. 2A and B) and HPV−
UM-SCC-19, P=0.0312; and UM-SCC-4, P=0.0305 (Fig. 3A and C).
Changes in S-phase in HPV+ and
HPV−
OPSCC cell lines
Compared to untreated controls some changes in the
proportion of cells in S phase occurred early (8 h), but not later
(24–48 h) after IR in both HPV+ and HPV−
OPSCC cell lines (Figs. 2 and
3). Compared to controls, with 2 Gy
after 8 h an increase in the proportion of cells in S-phase was
observed in HPV+ UM-SCC-47; P=0.0002; (Fig. 2A) and HPV− UM-SCC-4,
P<0.0001; (Fig. 3C). Compared to
controls, with 6 Gy after 8 h, HPV+ UM-SCC-47, P=0.0004;
UPCI-SCC-90, P=0.0123; CU-OP-2; P=0.0144; (Fig. 2A-C) and HPV− UM-SCC-4,
P=0.0061; (Fig. 3C) showed
increases in the proportion of cells in S-phase.
No changes in the cell cycle after IR
of HEKn
No significant changes were observed in the cell
cycle of HEKn between the non-treated samples and the 2 Gy and 6 Gy
samples (8, 24 and 48 h) (Fig.
S1). One representative flow cytometry plot of each cell line
is presented in Figs. S2 and
S3.
Influence of IR on HPV
integration
RNA sequencing was used to identify the presence on
human: Viral fusion transcripts before and after treatment with 2
Gy IR. This data was visualised using circos plots, which
demonstrated fusion transcripts as described previously for
UM-SCC-47, UPCI-SCC-90, CU-OP-2 and CU-OP-3 (18). The main integration sites were:
UM-SCC-47-chromosome 3 to TP63 from E6/E7; UPCI-SCC-90-C9orf156
(chromosome 9 open reading frame 156 and 200 bp before FOXE1 from
E7; CU-OP-2-chromosome 10 to E2 (YME1L1 gene at introns 6 and 7);
CU-OP-3-chromosome 20 at the gene CEBPB (CCAT/enhancer binding
protein β) (Fig. S4) (18). Novel minor HPV integration sites
were identified for CU-OP-20, but a high incidence of fusion
transcripts above 5 mapped reads, as described previously (18) were not disclosed (Fig. S4). Treatment with 2 Gy did not
change the main integration sites of UM-SCC-47, UPCI-SCC-90,
CU-OP-2 and CU-OP-3 (Fig. S4).
However, the frequency of additional minor chromosomal integration
sites per chromosome increased after 2 Gy IR, i.e. for the
radiosensitive lines: UM-SCC-47, from 3 to 16; UPCI-SCC-90, from 17
to 20; and CU-OP-20; from 7 to 20; as well as for the
radioresistant lines: CU-OP-3, from 6 to 21 and CU-OP-2, from 1 to
10 (Fig. S4A and B,
respectively).
Changes in gene expression in
HPV+ and HPV−
OPSCC cell lines after IR with 2
Gy
The RNA-sequencing data was also examined to
determine whether there were consistent differences in response to
IR between the HPV+ and HPV− cell lines. An
unsupervised analysis comparing treated and untreated cells was
initially performed. After correction for multiple testing, 519
transcripts or genes were indicated as significant (data not
shown). This indicated 2 Gy had a significant effect on
transcription of multiple genes. The top 19 genes with significant
differences after treatment of the OPSCC lines with 2 Gy are
presented in Table SI. None of the
genes however, showed an obvious link to DNA repair, DNA damage or
stress mechanisms, while transcription of the HPV oncogenes, E6 and
E7, was noted in all HPV+ cell lines (data not
shown).
Discussion
The present study investigated responses to ionising
irradiation (IR) in five human papillomavirus-positive
(HPV+) and five HPV− oropharyngeal squamous
cell carcinoma (OPSCC) cell lines. Clonogenic survival assays, flow
cytometry and RNA sequencing were used to assess survival, cell
cycle distribution and HPV integration. We also report derivation
and characterisation of two novel OPSCC cell lines. HPV+
OPSCC cell lines showed greater variation in radiosensitivity than
was apparent among the HPV− cell lines, and we observed
a tendency for greater sensitivity to IR in HPV+ lines,
although the correlation between radiosensitivity and HPV status
was not perfect. HPV+ cells more frequently demonstrated
an increase in G2 arrest following IR as compared to the
HPV− OPSCC cell lines. It was interesting and
potentially clinically relevant to note that radiation treatment
resulted in a marked increase in novel HPV: Human fusion
transcripts, which may suggest that radiation could facilitate
integration of HPV DNA into novel genomic sites. However, due to
the limited number of available cell lines and without further
studies, a definite conclusion cannot be made after the current
study. RNA sequencing did not indicate major changes in
transcription of genes associated with DNA repair, DNA damage or
stress mechanisms.
Following irradiation of HPV+ cell lines
(UM-SCC-47, UPCI-SCC-90, CU-OP-2, CU-OP-3, and CU-OP-20) and the
HPV− cell lines (UM-SCC-6, UM-SCC-4, UM-SCC-19,
UM-SCC-74a and CU-OP-17), our observations show some important
differences from earlier reports (11–13).
These data are based on an expanded panel of cell lines, since
three more HPV+ and two HPV− OPSCC cell lines
(HPV+ CU-OP-2, CU-OP-3, CU-OP-20 and HPV−
CU-OP-17 and UM-SCC-74a) were added to the previously relatively
limited panel of IR tested OPSCC cell lines (11–18).
Notably, in this report we did not find that HPV+ OPSCC
cell lines were consistently and significantly more sensitive to IR
relative to HPV− OPSCC cells (11–13),
although this trend was evident in a limited number of lines. We
suggest that it would be more accurate to state that
HPV+ OPSCC cell lines show wider variation in
radio-sensitivity than is observed among the HPV− OPSCC
cell lines.
The wider variability in sensitivity to IR in the
HPV+ group is in line with one previous report, where
the authors also therefore suggest caution in de-intensification of
therapy via dose reduction (13).
An example of this in the present study was that the
HPV+ CU-OP-2 and CU-OP-3 cell lines were relatively
radioresistant as compared to the other HPV+ cell
lines.
Of note, the HPV+ CU-OP-20, which
according to our circos plots lacked major integrated HPV sites,
was among the most radiosensitive HPV+ OPSCC cell line.
The data are of course very limited, but would be consistent with
suggestions that HPV+ OPSCC with episomal HPV DNA may
have an even better prognosis (27,28).
Similar to other publications, we found that upon IR
the proportion of HPV+ cell lines often exhibited an
increase in G2 arrest as compared to HPV− OPSCC lines,
while changes in the proportion of cells in G1 and S-phase were not
as apparent for any of the OPSCC cell lines (11–13).
In two of the previous studies it was hypothesised that due to
HPV16 E6-mediated degradation of p53, a greater G2 arrest should be
expected in the HPV+ OPSCC cell lines (12,13).
Notably, an increase in G2 arrest was the case for the
HPV+ OPSCC cell lines in this study, irrespective as to
whether the cell lines were radiosensitive or radioresistant. The
data would still be in line with the hypothesis above indicating
that it is due to HPV16 E6-mediated degradation of p53 rather than
a correlation to radiosensitivity per se (12,13).
This would be consistent with the observation that the most
radiosensitive line, UM-SCC-47, and the most radioresistant CU-OP-3
were the two cell lines with the highest proportion of G2 arrest.
To our knowledge this has not been demonstrated before. In fact, in
an earlier report the numbers of cells arrested in G2 correlated to
the radiosensitivity of the HPV+ OPSCC cell lines
(13). The differences in the data
reported between the present study and the study by Rieckmann et
al (13) reflects the
difficulties in drawing generalised conclusions on limited numbers
of HPV+ OPSCC cell lines.
In this study, we also demonstrated an increase in
HPV minor integration sites upon IR. We suggest that this could be
due to an increase in DNA lesions following IR, and that this
promotes integration of HPV DNA (possibly previously episomal) into
new genomic locations.
mRNA sequencing IR treated and untreated cells also
allowed investigation of changes in transcription of genes
correlated to DNA repair, cell cycle arrest and apoptosis. Our data
did not indicate an increase in transcription of specific DNA
repair genes after IR, which could be interpreted as consistent
with the suggestion that the IR sensitivity of HPV+
OPSCC cell lines could be due to impaired capability to repair
double-stranded DNA breaks (13).
However, it may equally reflect the limitation of an experiment
involving a single dose at a single time point. Other factors may
better explain the better prognosis of HPV+ OPSCC
patients. One report suggests e.g. that IR increases the levels of
MHC class I in HPV+ OPSCC, thereby allowing for better
immune recognition, while another study implies the necessity of an
immune system to benefit from IR or chemotherapy (29,30).
The role of CD47 has also been investigated, showing that this
protein is downregulated upon IR, and that this in turn enhances
immune recognition of HPV+ OPSCC (31).
There are limitations inherent in our study; only
10 OPSCC cell lines were assessed. Nonetheless this is a larger
sample than investigated in several previous studies. By including
five OPSCC cell lines previously not tested with IR, we could show
a tendency, but not a significantly increased sensitivity to IR in
HPV+ as compared to HPV− OPSCC cell lines,
and this may challenge a previous dogma (12,13).
Nevertheless, caution is still warranted to draw any major
conclusions, since the numbers of cell lines in each category were
limited.
Furthermore as already mentioned, another
limitation was the special focus on one irradiation dose 2 Gy.
However, we could confirm that upon IR, the proportion of cells in
G2 increased much more in HPV+ OPSCC cell lines as
compared to HPV− OPSCC, but in this study the increase
in G2 was not correlated to radiosensitivity as reported previously
(13). Whether this is due to
unique features of the CU-OP-3 cell line, or whether caution should
be exercised in regarding G2 arrest as a measure of
radiosensitivity, remains to be established.
To summarise, in this report five HPV+
and five HPV− OPSCCs cell lines, including five lines
not previously assessed, were examined for radiosensitivity.
HPV+ OPSCC lines demonstrated a wide range of
sensitivity to IR, and importantly not all cell lines were
radiosensitive, although they still tended to be more sensitive to
irradiation than HPV− OPSCC lines. Furthermore, upon IR,
HPV+ OPSCCs more often increased the proportion of cells
in G2 arrest as compared to HPV− OPSCC cell lines, but
the increases in G2 arrest were not correlated to radiosensitivity.
Lastly, upon treatment with 2 Gy some increases in minor HPV
integration sites were noted and changes in gene expression were
demonstrated, but not in genes primarily associated with DNA
repair.
To conclude, our data suggest that HPV+
OPSCC cell lines may possibly vary more in radiosensitivity than
previously anticipated, and despite they are generally more
sensitive than HPV− OPSCC cell lines, individual
variations may exist within both the HPV+ and the
HPV− OPSCC groups. Furthermore, in spite of the fact
that 10 cell lines were tested, the data are limited and additional
studies are warranted.
Supplementary Material
Supporting Data
Acknowledgements
We are very thankful for the support provided by
Cancer Research Wales as well as the patients who took part in this
study. Tina Dalianis and Nicole Wild are acknowledged for carefully
reading the manuscript and providing valuable suggestions, and
Ramona Ursu for technical help with the bead based multiplex
assay.
Funding
Grant funding from Cancer Research Wales (grant no.
CRW14-506688) supported this study. The funders had no role in
study design, data collection and analysis, decision to publish, or
preparation of the manuscript.
Availability of data and materials
RNA-seq data discussed in this publication is
available in the NCBIs Gene Expression Omnibus (GEO) and is
accessible through GEO series accession number GSE153966
(https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE153966).
The remaining datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SH performed the majority of the experiments,
interpreted the data, calculated the statistics and contributed to
the writing of the manuscript. EP and JJ collaborated with SH and
performed some experiments. PG contributed by analyzing and
providing the raw data. DO, AAH and ME contributed by identifying
and consenting patients and by providing the biopsy samples for the
establishment of the cell lines. NP and SM assisted all the
authors, with the planning of some experiments, performed
combinational analyses, and assisted in final interpretation and
presentation of the data. NP and SH were involved in the work from
its initiation and bringing into completion and were involved in
writing and finalizing the work and the manuscript. All authors
critically read and approved the manuscript.
Ethics approval and consent to
participate
The study with regard to patient material was
performed according to permission from the Cardiff and Vale
University Health board, in concordance with Ethical and NHS
R&D approval (reference no. 13/WA/0002).
Patient consent for publication
Not applicable.
Competing interests
Authors report no competing interests to
disclose.
References
|
1
|
Evans M, Newcombe R, Fiander A, Powell J,
Rolles M, Thavaraj S, Robinson M and Powell N: Human
papillomavirus-associated oropharyngeal cancer: An observational
study of diagnosis, prevalence and prognosis in a UK population.
BMC Cancer. 13:2202013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Haeggblom L, Attoff T, Yu J, Holzhauser S,
Vlastos A, Mirzae L, Ährlund-Richter A, Munck-Wikland E, Marklund
L, Hammarstedt-Nordenvall L, et al: Changes in incidence and
prevalence of human papillomavirus in tonsillar and base of tongue
cancer during 2000–2016 in the Stockholm region and Sweden. Head
Neck. 41:1583–1590. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Näsman A, Nordfors C, Holzhauser S,
Vlastos A, Tertipis N, Hammar U, Hammarstedt-Nordenvall L, Marklund
L, Munck-Wikland E, Ramqvist T, et al: Incidence of human
papillomavirus positive tonsillar and base of tongue carcinoma: A
stabilisation of an epidemic of viral induced carcinoma? Eur J
Cancer. 51:55–61. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chaturvedi AK, Engels EA, Pfeiffer RM,
Hernandez BY, Xiao W, Kim E, Jiang B, Goodman MT, Sibug-Saber M,
Cozen W, et al: Human papillomavirus and rising oropharyngeal
cancer incidence in the United States. J Clin Oncol. 29:4294–4301.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Näsman A, Du J and Dalianis T: A global
epidemic increase of an HPV-induced tonsil and tongue base
cancer-potential benefit from a pan-gender use of HPV vaccine. J
Intern Med. 287:134–152. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mellin H, Friesland S, Lewensohn R,
Dalianis T and Munck-Wikland E: Human papillomavirus (HPV) DNA in
tonsillar cancer: Clinical correlates, risk of relapse, and
survival. Int J Cancer. 89:300–304. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Schache AG, Powell NG, Cuschieri KS,
Robinson M, Leary S, Mehanna H, Rapozo D, Long A, Cubie H, Junor E,
et al: HPV-related oropharynx cancer in the United Kingdom: An
evolution in the understanding of disease etiology. Cancer Res.
76:6598–6606. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Schache AG, Simcock R, Gilbert DC and Shaw
RJ: Changing face of HPV related cancer in the UK. BMJ.
343:d66752011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bersani C, Mints M, Tertipis N, Haeggblom
L, Sivars L, Ährlund-Richter A, Vlastos A, Smedberg C, Grün N,
Munck-Wikland E, et al: A model using concomitant markers for
predicting outcome in human papillomavirus positive oropharyngeal
cancer. Oral Oncol. 68:53–59. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Owadally W, Hurt C, Timmins H, Parsons E,
Townsend S, Patterson J, Hutcheson K, Powell N, Beasley M,
Palaniappan N, et al: PATHOS: A phase II/III trial of
risk-stratified, reduced intensity adjuvant treatment in patients
undergoing transoral surgery for human papillomavirus (HPV)
positive oropharyngeal cancer. BMC Cancer. 15:6022015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Arenz A, Ziemann F, Mayer C, Wittig A,
Dreffke K, Preising S, Wagner S, Klussmann JP, Engenhart-Cabillic R
and Wittekindt C: Increased radiosensitivity of HPV-positive head
and neck cancer cell lines due to cell cycle dysregulation and
induction of apoptosis. Strahlenther Onkol. 190:839–846. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kimple RJ, Smith MA, Blitzer GC, Torres
AD, Martin JA, Yang RZ, Peet CR, Lorenz LD, Nickel KP, Klingelhutz
AJ, et al: Enhanced radiation sensitivity in HPV-positive head and
neck cancer. Cancer Res. 73:4791–4800. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rieckmann T, Tribius S, Grob TJ, Meyer F,
Busch CJ, Petersen C, Dikomey E and Kriegs M: HNSCC cell lines
positive for HPV and p16 possess higher cellular radiosensitivity
due to an impaired DSB repair capacity. Radiother Oncol.
107:242–246. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Vlashi E, Chen AM, Boyrie S, Yu G, Nguyen
A, Brower PA, Hess CB and Pajonk F: Radiation-induced
dedifferentiation of head and neck cancer cells into cancer stem
cells depends on human papillomavirus status. Int J Radiat Oncol
Biol Phys. 94:1198–1206. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang L, Wang X, Li Y, Han S, Zhu J, Wang
X, Molkentine DP, Blanchard P, Yang Y, Zhang R, et al: Human
papillomavirus status and the relative biological effectiveness of
proton radiotherapy in head and neck cancer cells. Head Neck.
39:708–715. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Reid P, Wilson P, Li Y, Marcu LG,
Staudacher AH, Brown MP and Bezak E: Experimental investigation of
radiobiology in head and neck cancer cell lines as a function of
HPV status, by MTT assay. Sci Rep. 8:77442018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Reid P, Wilson P, Li Y, Marcu LG,
Staudacher AH, Brown MP and Bezak E: In vitro investigation of head
and neck cancer stem cell proportions and their changes following
X-ray irradiation as a function of HPV status. PLoS One.
12:e01861862017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pirotte EF, Holzhauser S, Owens D, Quine
S, Al-Hussaini A, Christian AD, Giles PJ, Man ST, Evans M and
Powell NG: Sensitivity to inhibition of DNA repair by olaparib in
novel oropharyngeal cancer cell lines infected with human
papillomavirus. PLoS One. 13:e02079342018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nordfors C, Vlastos A, Du J,
Ahrlund-Richter A, Tertipis N, Grün N, Romanitan M, Haeggblom L,
Roosaar A, Dahllöf G, et al: Human papillomavirus prevalence is
high in oral samples of patients with tonsillar and base of tongue
cancer. Oral Oncol. 50:491–497. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
West CM, Davidson SE, Roberts SA and
Hunter RD: Intrinsic radiosensitivity and prediction of patient
response to radiotherapy for carcinoma of the cervix. Br J Cancer.
68:819–823. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Watson JV, Chambers SH and Smith PJ: A
pragmatic approach to the analysis of DNA histograms with a
definable G1 peak. Cytometry. 8:1–8. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liao Y, Smyth GK and Shi W: FeatureCounts:
An efficient general purpose program for assigning sequence reads
to genomic features. Bioinformatics. 30:923–930. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Robinson JT, Thorvaldsdottir H, Winckler
W, Guttman M, Lander ES, Getz G and Mesirov JP: Integrative
genomics viewer. Nat Biotechnol. 29:24–26. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15:5502014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Benjamini Y and Hochberg Y: Controlling
the false fiscovery rate: A practical and powerful approach to
multiple testing. J R Stat Soc B. 57:289–300. 1995.
|
|
26
|
Krzywinski M, Schein J, Birol I, Connors
J, Gascoyne R, Horsman D, Jones SJ and Marra MA: Circos: An
information aesthetic for comparative genomics. Genome Res.
19:1639–1645. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nulton TJ, Kim NK, DiNardo LJ, Morgan IM
and Windle B: Patients with integrated HPV16 in head and neck
cancer show poor survival. Oral Oncol. 80:52–55. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
McBride AA and Warburton A: The role of
integration in oncogenic progression of HPV-associated cancers.
PLoS Pathog. 13:e10062112017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Haeggblom L, Nordfors C, Tertipis N,
Bersani C, Ramqvist T, Näsman A and Dalianis T: Effects of
irradiation on human leukocyte antigen class I expression in human
papillomavirus positive and negative base of tongue and mobile
tongue squamous cell carcinoma cell lines. Int J Oncol.
50:1423–1430. 2017. View Article : Google Scholar
|
|
30
|
Spanos WC, Nowicki P, Lee DW, Hoover A,
Hostager B, Gupta A, Anderson ME and Lee JH: Immune response during
therapy with cisplatin or radiation for human
papillomavirus-related head and neck cancer. Arch Otolaryngol Head
Neck Surg. 135:1137–1146. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Vermeer DW, Spanos WC, Vermeer PD, Bruns
AM, Lee KM and Lee JH: Radiation-induced loss of cell surface CD47
enhances immune-mediated clearance of human papillomavirus-positive
cancer. Int J Cancer. 133:120–129. 2013. View Article : Google Scholar : PubMed/NCBI
|