Introduction
Glioblastoma multiforme (GBM), an incurable primary
brain tumor with a poor prognosis, is characterized by various
genetic alterations, such as mutation of isocitrate dehydrogenase
(IDH)1/2, amplification of epidermal growth factor receptor (EGFR)
and dysregulation of multiple signaling pathways, such as the
PI3K/AKT/mTOR, Wnt/β-catenin and NF/κB pathways (1,2). The
high degree of heterogeneity based on this complicated molecular
network in GBM is responsible for its poor treatment response,
which is influenced by chemo- and radiotherapy resistance (3). Despite current progress in
high-throughput data containing more significant subtype genes
identifications based on The Cancer Genome Atlas (TCGA) (1,4,5), the
precise molecular mechanism remains poorly understood. Previously,
non-coding RNAs, including microRNAs (miRNAs or miRs) have been
implicated in GBM and have been shown to form extensive complex
crosstalk networks to participate in GBM initiation and development
(6,7); therefore, further identification of
relevant complex signaling networks is urgently required to provide
insights into combinational targeted therapy and treatment
resistance.
miRNAs regulate gene expression by primarily binding
to the 3′-untranslated region of target mRNAs, leading to
translational inhibition or mRNA destabilization, which is
extensively implicated in numerous cancers (8). Evidence suggests that miR-422a
downregulation is closely associated with poor prognosis and
unfavorable clinicopathologic parameters. For example, the miR-422a
expression level is negatively associated with pathological grade,
recurrence and metastasis in hepatic cell carcinoma (9), and has also been reported to predict
lymphatic metastasis with high diagnostic accuracy in lung cancer
(10). Additionally, serum miR-422a
is regarded as a biomarker for the early diagnosis of colorectal
adenocarcinoma (11). Furthermore,
miR-422a, in combination with multiple key target genes, such as
forkhead box Q1 (FOXQ1) and S-phase kinase associated protein 2,
has been extensively identified to act as a crucial tumor
suppressor and prognostic factor during cancer progression and
development in solid malignancies, including in nasopharyngeal
carcinoma (12) and in
retinoblastoma (13). The decreased
expression of miR-422a and the identification of certain targets,
such as PIK3CA, have been reported in glioma (14); however, the molecular mechanism of
the miR-422a-mediated inhibitory effect on the glioma malignant
phenotype is poorly understood and requires further
investigation.
Ribophorin II (RPN2), which belongs to the key part
of the oligosaccharyltransferase complex, is responsible for the
N-glycosylation of multiple proteins, and its glycosylation
alterations have been confirmed to be associated with GBM malignant
progression (15–17). Accumulating evidence has
demonstrated that aberrant RPN2 overexpression is frequently
associated with multiple clinical parameters, such as lymphatic
metastasis, pathological grade and poor prognosis in a variety of
tumors, including osteosarcoma, non-small cell lung cancer,
advanced gastric cancer and colorectal cancer (16,18–20).
However, to the best of our knowledge, there is no report that
indicates a relationship between miR-422a and RPN2 in glioma.
The present study initially investigated the
miR-422a downregulation in GBM tissues and cell lines, and verified
its negative relationship with the World Health Organization (WHO)
grade. Additionally, overexpression of miR-422a markedly suppressed
cell proliferation and invasion, and promoted apoptosis and cell
cycle arrest at the G0/G1 phase. Mechanistically, miR-422a
inhibited the Wnt/β-catenin signaling pathway, as indicated by
TOP/FOP luciferase and western blot assays. Furthermore, it was
demonstrated that RPN2 was a direct functional target of miR-422a
and plays significant roles in miR-422a-mediated inhibitory effects
on Wnt/β-catenin signaling, as well as the malignant phenotype.
Therefore, the present study explored a novel potential axis
involving miR-422a/RPN2/β-catenin, which represents a novel
therapeutic target for GBM.
Materials and methods
Patients and samples
A total of 39 glioma samples undergoing craniotomy
for tumor were obtained from 15 male and 24 female patients (age
range, 9–76 years; mean age, 42.4±16.2 years) who were diagnosed by
surgeons and pathologists at Tianjin Huan Hu Hospital (Tianjin,
China) between January 2010 and June 2018. Among the 10 low-grade
gliomas, 6 were WHO grade I (pilocytic astrocytoma) and 4 were WHO
grade II (diffuse astrocytoma). All 29 GBM samples were WHO grade
IV. None of the patients had received any radiotherapy,
chemotherapy or any other anticancer treatments prior to surgery.
Five normal adult brain tissue samples were collected from 4 males
and 1 female patient (age range, 58–74 years; mean age, 66±6.3
years) undergoing post-trauma surgery for severe traumatic brain
injury. All the collected tissues were frozen immediately in liquid
nitrogen and stored at −80°C. This study was approved by the
Institutional Review Board of Tianjin Huan Hu Hospital, and written
informed consent was obtained from all participants or the parents
of patients below the legal age. Gene expression data from the
mRNAseq_325 dataset and the relevant clinicopathological variables
dataset were obtained from the Chinese Glioma Genome Atlas (CGGA)
database (http://www.cgga.org.cn/), which is a web
application for data storage and analysis to explore brain tumor
datasets of >2,000 samples from Chinese cohorts (21). In addition, the ‘Survival Map’
module from the online databases Gene Expression Profiling
Interactive Analysis 2 (GEPIA2; http://gepia.cancer-pku.cn/index.html), which is an
interactive website that includes information for 514 low-grade and
162 high-grade glioma tissues obtained from TCGA and GTEx projects
(22), was used to evaluate the
relationships between RPN2 expression and overall survival and
disease free survival. Moreover, UALCAN (http://ualcan.path.uab.edu) (23), which is a comprehensive interactive
database containing data of 513 low-grade gliomas (248 grade II and
265 grade III) from The Cancer Genome Atlas, was also utilized to
analyze RPN2 expression level in gliomas with different grades, and
the association with the patient prognosis.
Cell culture and transfection
The human glioblastoma cell lines U87, U251 and A172
and the low-grade glioma cell line H4 were obtained from the Peking
Union Medical College Cell Library. Human LN308, LN229 and T98G
glioblastoma cells were obtained from the China Academia Sinica
Cell Repository. The SNB19 cell line was purchased from iCell
Bioscience, Inc. U87 and SNB 19 cell lines were authenticated using
short tandem repeat profiling analysis. The U87 cell line
(glioblastoma of unknown origin) used in the present study was of
the American Type Culture Collection type. The cells were
maintained in Dulbecco's modified Eagle's medium (Gibco; Thermo
Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum
(Gibco; Thermo Fisher Scientific, Inc.) and 1%
penicillin/streptomycin (Invitrogen; Thermo Fisher Scientific,
Inc.). All cell lines were incubated at 37°C with 5%
CO2.
The oligonucleotide sequences of the human miR-422a
mimic and miRNA-negative control (miR-NC), and RPN2 overexpression
plasmid and empty vector were purchased from Shanghai GenePharma
Co., Ltd. The human miR-422a sequence was
5′-ACUGGACUUAGGGUCAGAAGGC-3′, and the scrambled sequence was
5′-UUGUACUACACAAAAGUACUG-3′. Prior to transfection, U87 and LN229
cells were incubated in a 6-well plate at a density of
2×105 cells/well and then co-transfected with 200 pmol
miR-422a mimic or miR-NC and RPN2 overexpression plasmid or empty
vector (2 µg) using X-tremeGENE transfection reagent (Roche
Diagnostics) according to the manufacturer's protocol. Subsequent
experiments were performed after 72 h.
For RPN2-knockdown, the shRNA lentiviral vector
targeting RPN2 (shRPN2; 5′-GGATCGCCCTTTCACAAAT-3′) and lentiviral
vector negative control (sh-NC; 5′-TTCTCCGAACGTGTCACGT-3′) were
also purchased from Shanghai GenePharma Co., Ltd. LN229 cells were
infected at a multiplicity of infection of 10 in the presence of 5
µg/ml polybrene (Shanghai GenePharma Co., Ltd.), following the
manufacturer's instructions. Western blotting was performed to
identify the knockdown efficiency after 72 h.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was isolated from glioma tissues and cells
using TRIzol reagent (Thermo Fisher Scientific, Inc.), reverse
transcribed to cDNA using PrimeScript RT Master mix (Takara Bio,
Inc.), according to the manufacturer's instructions and
subsequently qPCR was performed using TB Green Premix EXTaq II kit
(Takara Bio, Inc.) for analysis of the mRNA expression of RPN2,
TCF4, c-myc and cyclinD1. The amplification conditions were 95°C
for 5 sec, followed by 40 cycles at 60°C for 30 sec and 50°C for 30
sec. To analyze miRNA-422a expression, stem-loop RT was performed
with an miScript PCR starter kit (Qiagen GmbH) according to the
manufacturer's instructions. qPCR was performed using miScript SYBR
Premix Green PCR kit (Qiagen GmbH) and Roche LC480 quantitative
Real-Time PCR system (Roche Diagnostics). The amplification
conditions were 95°C for 15 min, followed by 40 cycles at 94°C for
15 sec and 55°C for 30 sec. GAPDH or U6 levels were selected as
internal controls for mRNA and miRNA expression, respectively, and
fold changes were calculated using the 2−∆∆Cq method
(24). Data were analyzed from
three independent experiments and are presented as the mean ±
standard deviation. The primer sequences for the detection of mRNAs
and miR-422a expression were as follows: RPN2 forward,
5′-CTCTGACGCCCACTCACTAC-3′ and reverse,
5′-AATAGAGATCTTTGCATCTGGCAC-3′; TCF4 forward,
5′-CAAATAGAGGAAGCGGGGC-3′ and reverse, 5′-TGCTGAGAGAGATGGAGGAGA-3′;
cyclinD1 forward, 5′-AACTACCTGGACCGCTTCCT-3′ and reverse,
5′-CCACTTGAGCTTGTTCACCA-3′; c-myc forward,
5′-TTCGGGTAGTGGAAAACCAG-3′ and reverse, 5′-CAGCAGCTCGAATTTCTTCC-3′;
GAPDH forward, 5′-CATGAGAAGATGACAACAGCCT-3′ and reverse,
5′-AGTCCTTCCACGATACCAAAGT-3′; miR-422a forward,
5′-GGGTCAGAAGGCCTGAGTCT-3′ and reverse, 5′-CAAAGCTTGGCTCAGGGACA-3′;
and U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse,
5′-AACGCTTCACGAATTTGCGT-3′.
Dual-luciferase reporter assay
The PicTar (http://www.pictar.org/), miRmap (http://miRNAMap.mbc.nctu.edu.tw/.) and miRanda
(http://www.microrna.org/microrna/home.do) databases
were applied to identify targets of miR-422a, and the seed sequence
of miR-422a was identified to matched the 3′-untranslated region
(3′UTR) of the RPN2 gene. The 3′UTR of RPN2 containing the miR-422a
binding site and corresponding mutant site were inserted into the
pMIR-REPORT vector (Promega Corporation). LN229 and U87 cells
(1×104/well) were cultured in 96-well plates, and
co-transfected with wild-type or mutant luciferase reporters and
the miR-422a mimic or miR-NC using the X-tremeGENE transfection
reagent (Roche Diagnostics). To evaluate the β-catenin/CF-4
transcriptional activity, TOP-FLASH and FOP-FLASH luciferase
reporter constructs were used. TOP-FLASH (with repeats of the TCF
binding site) or FOP-FLASH (with repeats of the mutant TCF binding
site) plasmids (EMD Millipore) were transfected into LN229 and U87
cells transfected with miR-422a mimic. Furthermore, RPN2
overexpression plasmid transfection was performed 24 h after miRNA
transfection. Following a further 48 h incubation, luciferase
activity was measured using the Dual-Luciferase Reporter assay
system (Promega Corporation), and Renilla luciferase
activity was used as an internal control.
RNA immunoprecipitation (RIP)
assay
The Magna RIP RNA-Binding Protein
Immunoprecipitation kit (EMD Millipore) was used to perform RIP
assays according to the manufacturer's instructions. LN229 and U87
cells transfected with miR-422 mimic or miR-NC for 48 h were
collected by centrifugation at 150 × g for 5 min at 4°C and
incubated overnight at 4°C with RIP buffer containing protein A/G
magnetic beads coated with anti-Ago2 or anti-IgG antibody (catalog
no. 03-110; EMD Millipore; 1:5,000) as a negative control.
Following overnight incubation at overnight at 4°C with rotation,
and following washing, RNase-free DNase I and proteinase K (EMD
Millipore) were used to purify the RNA in the immunoprecipitated
complex, and RT-qPCR was then performed to detect the RPN2
expression level, as described previously.
Viability assay
Cell viability was analyzed by Cell Counting Kit-8
(CCK-8; Dojindo Molecular Technologies, Inc.) according to the
manufacturer's instructions. Following transfection with miR-422a
mimic or co-transfection with miR-422a mimic and RPN2 plasmid,
LN229 and U87 cells were incubated for 24, 48, 72 and 96 h. CCK-8
solution (10 µl) was then added to each well, and the absorbance at
490 nm was measured after incubation for 2 h to estimate the number
of viable cells.
Cell cycle analysis
Cell cycle analysis was performed by flow cytometry,
and transfected and control LN229 and U87 cells in the log phase of
growth were harvested by trypsinization, washed with PBS, fixed
with 75% ethanol overnight at 4°C and then incubated with RNase at
37°C for 30 min. A total of 1×104 nuclei were assessed
with a FACS Calibur Flow Cytometer (Becton, Dickinson and Company),
and DNA histograms were analyzed using Modfit software (Modfit LT
4.0; Becton, Dickinson and Company). Experiments were performed in
triplicate.
Transwell assay
Transwell membranes coated with Matrigel (BD
Biosciences) were placed in an incubator for 30 min at 37°C, which
were used to quantify glioma cell invasion. Transfected cells were
plated at 5×104 per well in the upper chamber in
serum-free medium. DMEM supplemented with 10% FBS (600 µl) were
used as a chemoattractant and placed in the bottom chamber. After
24 h of incubation, the filters were gently removed, and the medium
was removed from the upper chamber. The cells that had migrated
through the Matrigel into the pores of the inserted filter were
fixed with 100% methanol, stained with hematoxylin for 20 min at
room temperature, and mounted. The number of cells that invaded
through the Matrigel was counted in three randomly selected visual
fields from the central and peripheral portions of the filter using
an inverted microscope (magnification, ×200).
Apoptosis assay
Apoptosis was quantified following transfection by
assessing Annexin V labelling and caspase 3/7 activity. For the
Annexin V assay, an annexin V-FITC-labeled apoptosis detection kit
(Abcam) was used according to the manufacturer's protocol. Caspase
3/7 activity was analyzed using Caspase-Glo 3/7 reagent (catalog
no. G8091; Promega Corporation). Briefly, caspase-Glo 3/7 reagent
(100 µl) was added to a white-walled 96-well plate, which was
gently mixed using a plate shaker at 50 × g for 30 sec. Following
incubation at room temperature for 1–2 h, the luminescence of each
sample was measured in a plate-reading luminometer.
Western blotting
The total protein extraction of different
transfected LN229 and U87 cells was performed using ExKine Total
Protein Extraction kit (Abbkine), and nuclear protein extraction
was performed using DUALXtract Cytoplasmic and Nuclear Protein
Extraction kit (Dualsystems Biotech AG), according to
manufacturer's protocol. Protein concentrations were determined
using a BCA Protein assay kit (Thermo Fisher Scientific, Inc.).
Equal amounts of protein (30 µg/lane) were separated by 10%
SDS-PAGE and subsequently transferred to PVDF membranes (EMD
Millipore). After blocking in 5% skimmed milk for 1 h, the
membranes were incubated with diluted primary antibodies at 4°C
overnight. After washing with TBST (1% Tween-20), the membranes
were incubated with horseradish peroxidase-conjugated secondary
antibodies (1:5,000; cat. nos. ab222772 and ab222759; Abcam) for 1
h at room temperature, and the immune-reactive bands were
visualized using ECL Western Blot Detection reagents (EMD
Millipore). The expression levels were normalized to the levels of
β-actin or GAPDH of the total protein and fibrillarin of nuclear
protein. The protein band intensities were quantified using ImageJ
software (version 1.48; National Institutes of Health). The primary
antibodies used were as follows: RPN2 (1:200; catalog no. ab244399;
Abcam), β-catenin (1:5,000; catalog no. ab32572; Abcam), cyclinD1
(1:10,000; catalog no. ab134175; Abcam), TCF4 (1:10,000; catalog
no. ab76151; Abcam), c-myc (1:1,000; ab39688; Abcam), β-actin
(1:5,000; catalog no. ab6276; Abcam), fibrillarin (1:5,000; catalog
no. ab4566; Abcam) and GAPDH (1:1,000; catalog no. 5174; Cell
Signaling Technology, Inc.).
Immunohistochemistry (IHC)
analysis
For IHC assays, detailed protocols were performed as
previously described (25).
Briefly, the animal tumor samples were fixed with 10% formalin for
48 h at room temperature, embedded with paraffin, and sliced into
4-µm sections, followed by dewaxing. Subsequently, these sections
were incubated with 3% hydrogen peroxide for 20 min to block
endogenous peroxidase. Following antigen retrieval, the slides were
blocked with 10% normal goat serum (Beijing Solarbio Science &
Technology Co., Ltd.) for 30 min and then incubated at 4°C
overnight with appropriate primary antibody. After washing with
PBS, the slides were incubated with biotinylated secondary antibody
(cat. no. Sp-9001; 1:1,000; OriGene Technologies, Inc.) for 1 h at
room temperature. A DAB Horseradish Peroxidase Color Development
kit (Beyotime Institute of Biotechnology) was used for color
development, and neutral gum (Sinopharm Chemical Reagent Co., Ltd.)
was used for sealing. Subsequently, five fields were randomly
selected and imaged using the Olympus X71 inverted microscope
(Olympus Corporation). The images were assessed using Image-Pro
Plus 6.0 (Media Cybernetics, Inc.). The antibodies used for IHC
were as follows: RPN2 (1:200; cat. no. ab244399; Abcam), β-catenin
(1:500; cat. no. ab32572; Abcam), c-myc (1:100; cat. no. ab39688;
Abcam), caspase 3 (1:100; cat. no. 9662; Cell Signaling Technology,
Inc.) and PCNA (1:4,000; cat. no. 2586; Cell Signaling Technology,
Inc.).
Xenograft model with nude mice
All animal protocols were performed with the
approval of the Animal Care and Used Committee of Tianjin Huanhu
Hospital (Tianjin, China). A total of 10 female BALB/c nude mice at
5 weeks of age (weight, 18–21 g) were purchased from the Animal
Center of the Cancer Institute, Chinese Academy of Medical Science.
Animal welfare was taken seriously and all the mice were housed
with filtered air, a 12-h light/dark cycle, regulated temperature
(25±2°C) and humidity (50±10%), and with ad libitum access
to sterilized food and water. A total of 1×107 U87 GBM
cells in 100 µl PBS were subcutaneously injected into the left
flank region of nude mice. When the tumor volume reached 100
mm3, mice were randomly divided into two groups (5 mice
per group) and subsequently treated with oligonucleotides (10 µg
miR-422a mimic or miR-NC) with a mixture of 10 µl Lipofectamine
2000 (Invitrogen; Thermo Fisher Scientific, Inc.) through local
injection of the xenograft tumor at multiple sites, respectively.
The treatment was performed once every 3 days for 27 days. The
tumor volume was monitored with a caliper, using the formula:
Volume=length × width2/2. The health and behavior of
mice were monitored daily. Humane endpoints used in the study to
verify when to euthanize the mice included: i) Tumors, 2.0 cm
diameter; ii) weight loss, ≥20%; iii) tumor ulceration, iv)
abnormal posture; and v) quick weight loss assessed by observations
and weighing. At the termination of the experiment, no mice reached
these criteria. The mice were anesthetized by injecting 1%
pentobarbital sodium (100 mg/kg body weight) and sacrificed by
cervical vertebra dislocation, when the diameter of the largest
tumor was 1.93 cm. If there was no heartbeat for 30 sec accompanied
by no response to the toe pinch reflex, mice were considered to be
euthanized. Finally, tumor tissues were removed, tumor weight was
calculated, and RNA extraction and IHC assays were performed.
Statistical analysis
All statistical analyses were performed using
GraphPad software version 6.0 (GraphPad Software, Inc.) or SPSS
23.0 (IBM Corp.). Data are presented as the mean ± standard
deviation of at least three independent experiments. An unpaired
Student's t-test and one-way ANOVA were performed to analyze
significant differences between two groups or multiple groups,
respectively. Additionally, Tukey's test was used for multiple
comparisons following ANOVA. Fisher's exact test was used to
analyze the associations between miR-422a and clinicopathological
characteristics of patients with glioma. The correlation between
miR-422a and c-myc expression in 29 GBM patients was assessed using
Pearson's correlation analysis. The survival curves from online
databases were plotted using the Kaplan-Meier method and compared
using the log-rank test. P<0.05 was considered to indicate a
statically significant difference.
Results
miR-422a is significantly
downregulated in GBM specimens and cell lines
To investigate the expression level of miR-422a in
glioma cell lines and samples, total RNA was isolated from eight
glioma cell lines, GBM specimens and normal brain tissues, and the
levels of miR-422a were evaluated by RT-qPCR. The results
demonstrated that miR-422a was significantly downregulated in
glioma tissues compared with normal brain tissues, and miR-422a
expression was significantly lower in WHO grade IV GBM compared
with in low-grade glioma samples (WHO grades I–II) (Fig. 1A). The clinicopathological
characteristics of 39 glioma specimens are summarized in Table I, and no significant associations
were identified between miR-422a expression and other
clinicopathological variables, including gender and age, although
it was significantly association with histology. Additionally,
miR-422a was also significantly lower in the seven GBM cell lines
compared with the low-grade cell line H4 (Fig. 1B). Furthermore, miR-422a expression
was analyzed in CGGA, which revealed that the miR-422a expression
level was associated with the histopathological subtypes,
specifically, the expression in subtypes O and A (WHO II) were
significantly higher compared with in the subtypes of AOA, AO and
AA (WHO III) and GBM (WHO IV) (Fig.
1C). Similarly, a high expression of miR-422a was significantly
associated with a lower WHO grade (Fig.
1D). These data indicate that miR-422a may act as a key tumor
suppressor in glioma progression.
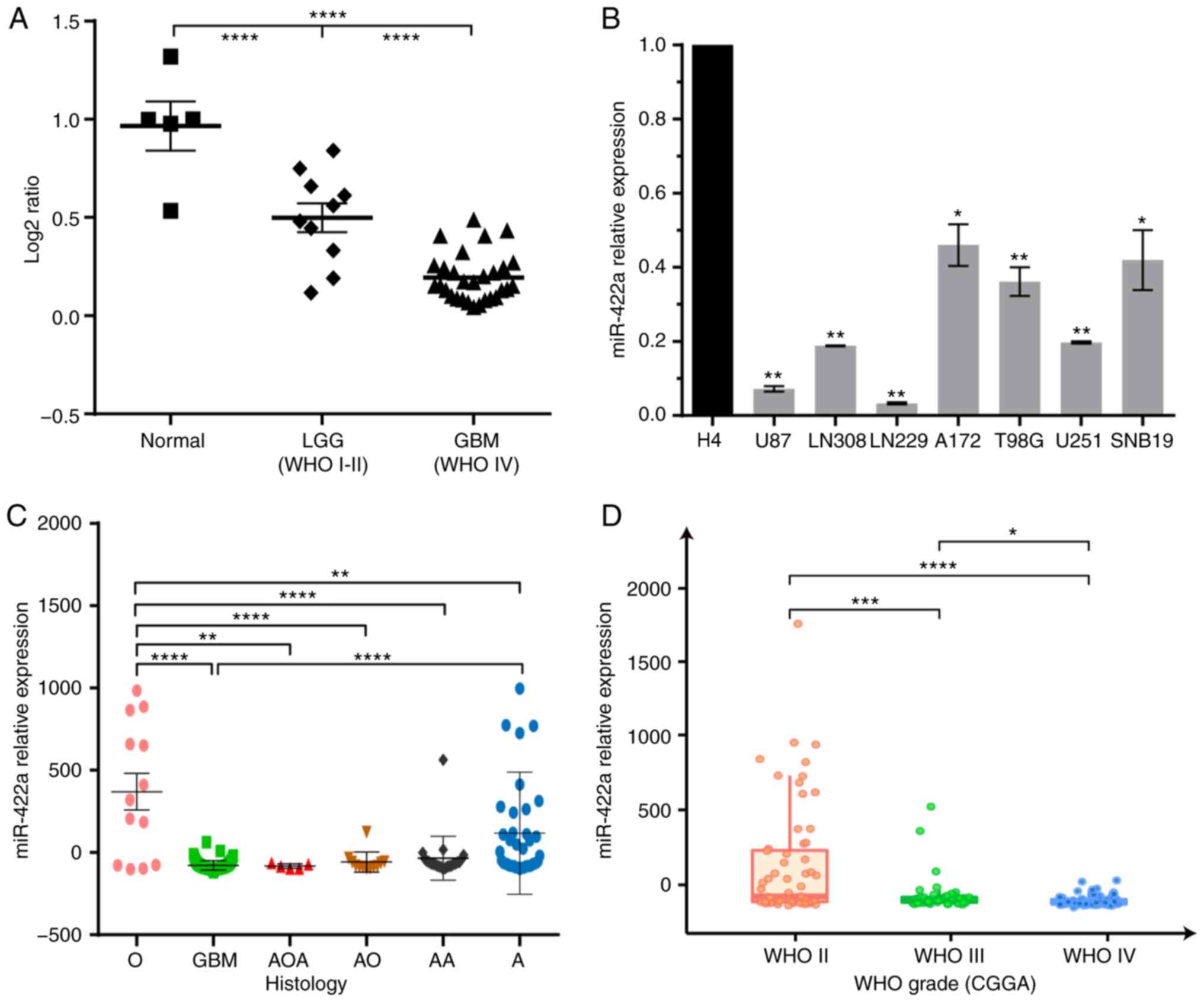 | Figure 1.miR-422a is significantly
downregulated in GBM clinical samples and cell lines. (A) RT-qPCR
was used to evaluate the miR-422a expression in 10 LGG (WHO grade
I–II) and 29 GBM (WHO grade IV) resected specimens compared with 5
normal brain tissues. (B) RT-qPCR analysis of the low-grade H4 cell
line and GBM cell lines (U87, LN308, LN229, A172, T98G, U251 and
SNB19). *P<0.05, **P<0.01 vs. H4 cells. (C) The association
between miR-422a and histology in gliomas according to the CGGA
database. (D) miR-422a expression was negatively associated with
the WHO grade of glioma samples from the CGGA database. *P<0.05,
**P<0.01, ***P<0.001, ****P<0.0001. miR-422a,
microRNA-422a; GBM, glioblastoma multiforme; RT-qPCR, reverse
transcription-quantitative PCR; WHO, World Health Organization;
CGGA, Chinese Glioma Genome Atlas; LGG, low-grade glioma. |
 | Table I.Associations between miR-422a and
clinicopathological characteristics of patients with glioma. |
Table I.
Associations between miR-422a and
clinicopathological characteristics of patients with glioma.
|
|
| miR-422a
expression |
|
|---|
|
|
|
|
|
|---|
| Variable | No. of cases | Low, n | High n | P-value |
|---|
| Sex |
|
|
| 0.1532 |
|
Female | 15 | 7 | 8 |
|
|
Male | 24 | 5 | 19 |
|
| Age, years |
|
|
| >0.999 |
|
<42 | 20 | 13 | 7 |
|
|
≥42 | 19 | 13 | 6 |
|
| Histology |
|
|
| 0.0015 |
|
Astrocytoma | 10 | 2 | 8 |
|
|
Glioblastoma | 29 | 23 | 6 |
|
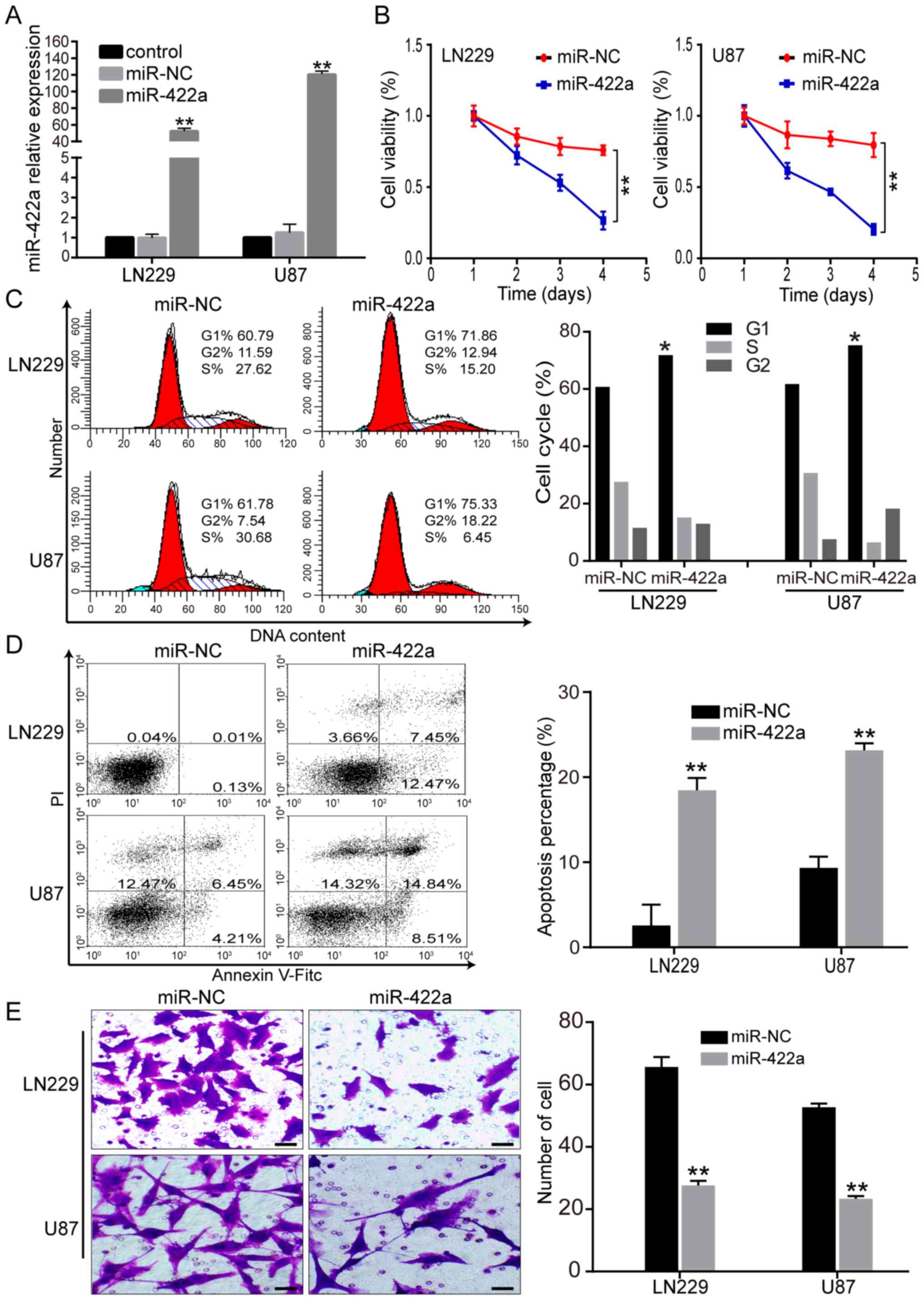
Ectopic miR-422a expression suppresses
the GBM malignant phenotype
Based on the low expression of miR-422a in GBM, the
present study then investigated how the malignant phenotype of GBM
was mediated by miR-422a. LN229 and U87 cells were transfected with
miR-422a mimic or miR-NC, and RT-qPCR was performed to verify
miR-422a overexpression (Fig. 2A).
The CCK-8 assay revealed a significant decrease in cell viability
for miR-422a mimic-transfected GBM cells compared with the miR-NC
group (Fig. 2B). Flow cytometry
analysis demonstrated a significant increase in G1 cell cycle
arrest in miR-422a mimic-transfected LN229 and U87 cells (Fig. 2C). Additionally, in vitro
Annexin-V and Transwell assays revealed that miR-422a
overexpression significantly enhanced tumor cell apoptosis and
significantly reduced the number of invasive LN229 and U87 cells
(Fig. 3D and E). These results
suggest that miR-422a overexpression suppressed GBM cell
proliferation and invasion, and promoted G1 cell cycle arrest and
apoptosis, indicating a crucial role in the progression of GBM.
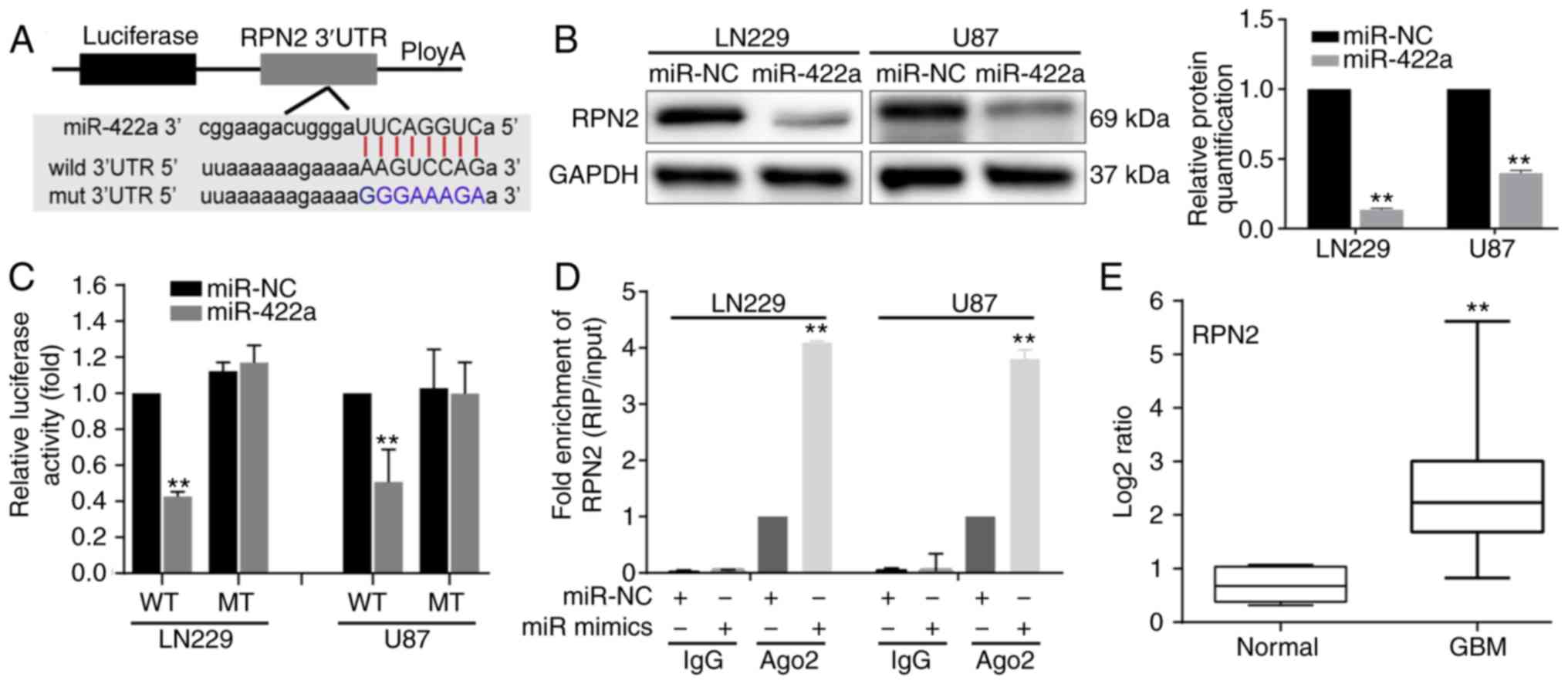
RPN2 is a direct target of
miR-422a
To verify the downstream effectors of miR-422a,
PITA, miRmap and miRanda databases were used. Based on these
databases, it was identified that the ‘seed sequence’ of miR-422a
matched the 3′UTR of the RPN2 mRNA (Fig. 3A). To determine whether RPN2
expression was modulated by miR-422a, western blotting was
performed to detect RPN2 protein expression in LN229 and U87 cells
transfected with miR-422a mimic, and the results revealed that
miR-422a overexpression significantly reduced the RPN2 expression
level (Fig. 3B). To confirm that
RPN2 was a direct target of miR-422a, luciferase reporter
constructs carrying the RPN2 3′UTR with wild-type or mutant
miR-422a binding sites were constructed and co-transfected with
miR-422a mimic and miR-NC. In comparison to miR-NC, miR-422a mimic
significantly decreased the luciferase activities (Fig. 3C). However, the miR-NC and miR-422a
mimic did not affect the luciferase activity in mutant constructs.
Furthermore, a RIP assay was performed using an Ago2 antibody to
investigate the direct association between miR-422a and RPN2. The
results indicated that RPN2 was enriched in Ago2-coated beads
compared with the IgG control group and that miR-422a mimic
resulted in a marked upregulation of RPN2 level in the Ago2
immunoprecipitation complex in LN229 and U87 cells (Fig. 3D). In addition, RPN2 expression
level was detected in five normal and 29 GBM specimens, and RPN2
was significantly upregulated in the GBM samples compared with the
normal samples (Fig. 3E). These
data indicate that miR-422a directly regulates RPN2 expression by
binding to the 3′UTR of RPN2.
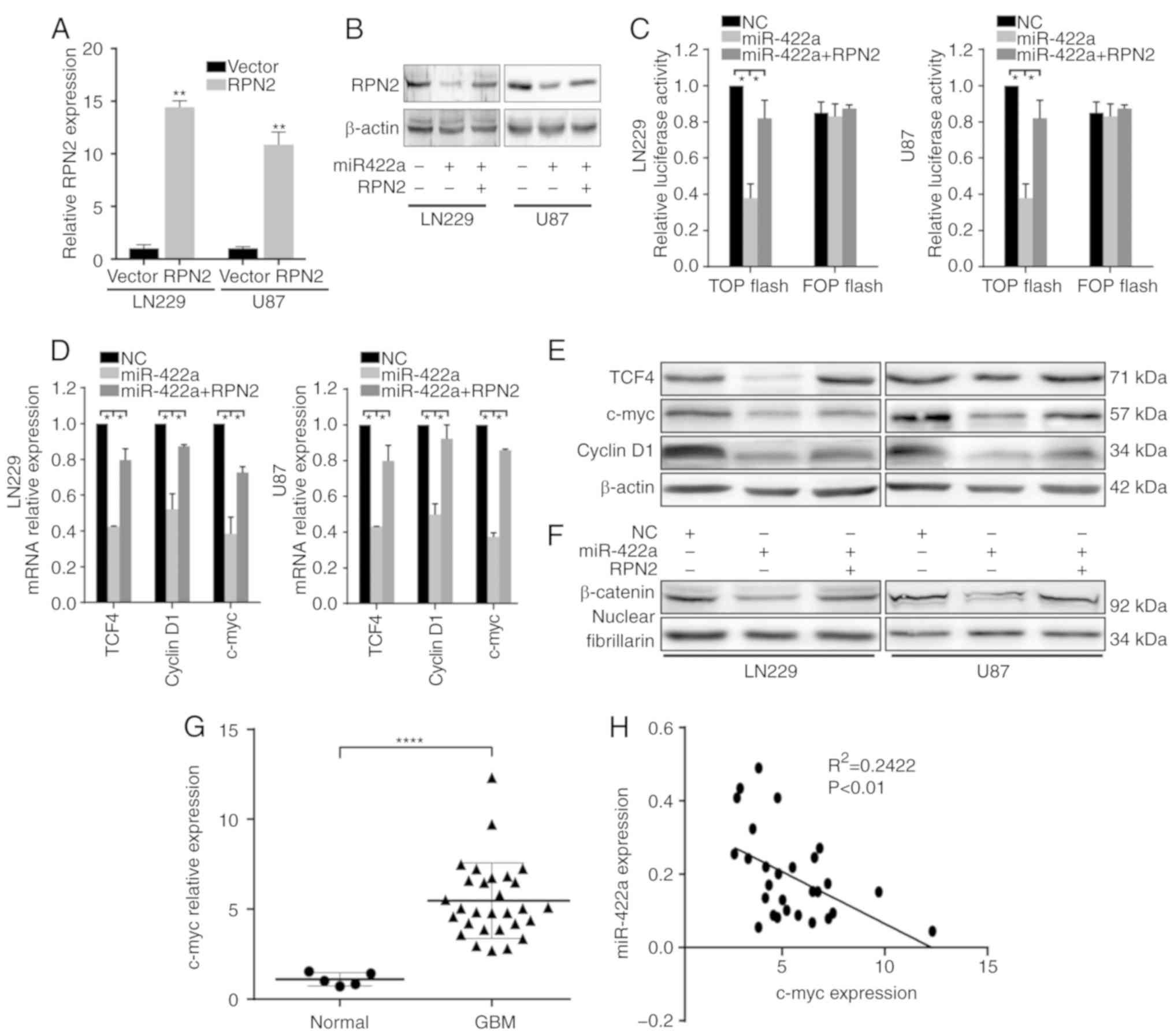
miR-422a suppresses the Wnt/β-catenin
signaling pathway, at least partially through RPN2
Our previous studies confirmed that RPN2 promotes
activation of the Wnt/β-catenin signaling pathway (data not shown).
In combination with the fact that RPN2 is a direct functional
target of miR-422a, the present study further investigated whether
miR-422a also regulates the Wnt/β-catenin signaling pathway via
RPN2. RT-qPCR demonstrated successful overexpression of RPN2 in
LN229 and U87 cells following transfection with RPN2 overexpression
plasmid (Fig. 4A), and results of
the western blot analysis of RPN2 expression in LN229 and U87 cells
transfected with miR-422a mimic or miR-422a and RPN2 overexpression
plasmid are shown in Fig. 4B,
indicating that the transfection of RPN2 overexpression plasmid can
markedly recover RPN2 protein level in LN229 and U87 cells
transfected with miR-422a mimic.
A TOP/FOP assay was used to analyze regulation of
β-catenin/TCF4 transcriptional activity by miR-422a, and the
results demonstrated that miR-422a significantly suppressed TOP
luciferase activity, with no apparent change in FOP activity
(Fig. 4C). Furthermore, ectopic
miR-422a expression could inhibit TCF4, c-myc and cyclinD1
expression, and β-catenin nuclear translocation, as verified by
RT-qPCR and western blotting (Fig.
4D-F). However, overexpression of RPN2 following the
transfection with miR-422a reversed the effects mediated by
miR-422a (Fig. 4C-F). To further
verify the relationship between miR-422a and Wnt signaling, c-myc,
a critical downstream factor of the Wnt/β-catenin pathway, was
detected by RT-qPCR, which revealed that it was significantly
higher in GBM tissue samples compared with normal samples (Fig. 4G). Additionally, Pearson's
correlation analysis demonstrated that miR-422a was negatively
correlated with the expression of c-myc in 29 GBM patient samples
(Fig. 4H). These data demonstrate
that miR-422a can attenuate the Wnt/β-catenin signaling pathway, at
least partially through RPN2 in LN229 and U87 cells.
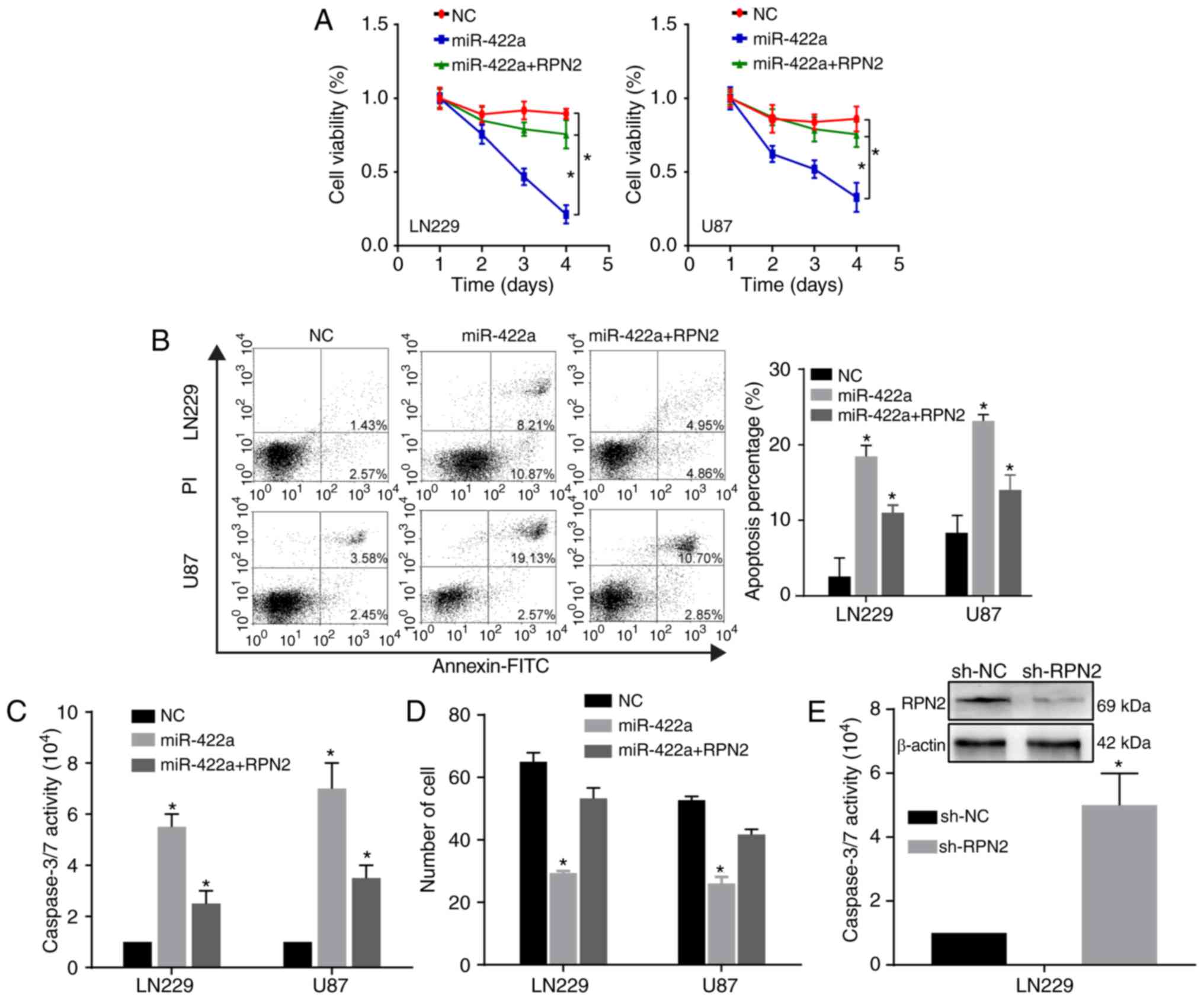
RPN2 is required for the biological
effects of miR-422a on the GBM malignant phenotype
To further verify the functional association between
miR-422a and its target RPN2, the role of RPN2 in the
miR-422a-mediated effect on the proliferation, invasion and
apoptosis of GBM cells was investigated. When RPN2 was ectopically
overexpressed in LN229 and U87 cells transfected with miR-422a
mimic, the inhibition of proliferation and invasion, and the
promotion of apoptosis induced by miR-422a was partially reversed
(Fig. 5A-D). Furthermore, the
knockdown of RPN2 by shRPN2 significantly enhanced the caspase-3/7
activity (Fig. 5E), which was
consistent with the effect mediated by miR-422a overexpression.
These results indicate that miR-422 inhibits the GBM malignant
phenotype, partially through the oncogene RPN2.
miR-422a inhibits tumor growth in
vivo
Based on the in vitro experimental findings,
the effects of miR-422a on tumor growth and Wnt/β-catenin signaling
were further examined in vivo. U87 cells were implanted into
the left flanks of nude mice by subcutaneous injection. miR-422a
mimic and miR-NC were injected in multiple sites of the tumor every
3 days. The study was terminated on day 27, and the tumors were
excised and further analyzed. The tumor growth curve was generated
from data obtained every 3 days, which demonstrated that the
overexpression of miR-422a significantly reduced the tumor volume
(Fig. 6A). Additionally, a
significant decrease in tumor weight was observed for the miR-422a
mimic-treated tumors compared with the miR-NC-treated tumors
(Fig. 6B). Evidence that miR-422a
mimic significantly increased miR-422a expression in tumor tissues
is presented in Fig. 6C.
Furthermore, the IHC assay demonstrated that miR-422a
overexpression significantly inhibited RPN2, β-catenin, c-myc and
PCNA expression, and increased the caspase3 expression level
(Fig. 6D), which was consistent
with the in vitro results.
 | Figure 6.miR-422a inhibits GBM xenograft tumor
growth. (A) U87 cells were subcutaneously injected into nude mice.
When tumors were established uniformly in the two groups, miR-422a
was administered by multisite injection every 3 days. Tumor volumes
were evaluated every 3 days during treatment. *P<0.05. (B) At
the termination of the experiment, tumor weights were measured.
*P<0.05. (C) Reverse transcription-quantitative PCR was
performed to examine miR-422a expression in resected tumor tissues.
**P<0.01 vs. miR-NC. (D) IHC assays were performed to assess
RPN2, β-catenin, c-myc, PCNA and caspase3 expression in xenograft
tumor sections. Scale bar, 50 µm. *P<0.05 vs. miR-NC. IHC,
immunohistochemistry; miR-422a, microRNA-422a; NC, negative
control; RPN2, ribophorin II. |
Increased RPN2 expression is
associated with poor prognosis in human glioma
Based on the association between miR-422a and RPN2,
RPN2 expression and its correlation with prognosis in the CGGA
dataset (CGGA Mseq325) and the UALCAN and GEPIA public databases
was investigated. The CGGA data revealed that the mRNA level of
RPN2 was significantly higher in high-grade gliomas compared with
the low-grade glioma (Fig. 7A).
Retrospective analysis of the clinical outcome of these patients
revealed that low expression of RPN2 was significantly associated
with a longer overall survival, in both primary and recurrent
gliomas (Fig. 7B and C). Further
analysis of RPN2 expression and relevant clinical characteristics
indicated that RPN2 expression was significantly associated with
IDH1 gene mutation status, age and 1p/19q codeletion status
(Fig. S1A-C). Furthermore, the
low-grade glioma data analysis of the UALCAN database demonstrated
that RPN2 expression in grade 3 was significantly higher than that
in grade 2 (Fig. 7D), and the
higher RPN2 expression was significantly associated with a poorer
survival (Fig. 7E). Moreover, the
data obtained from the ‘Survival Map’ module of GEPIA demonstrated
that the high RPN2 expression group had a poorer overall survival
and disease free survival compared with the low RPN2 expression
group of patients with low-grade glioma (n=514) and gliomas,
including low-grade and high-grade glioma (n=676) (Fig. 7F-I). Hence, these data indicate that
RPN2 predicts poor prognosis and is associated with the progression
of gliomas.
Discussion
Accumulating data have demonstrated that miR-422a,
which serves as a key anticancer gene, exerts a crucial influence
on the initiation, development and drug resistance of several
tumors. He et al (26)
reported that miR-422a downregulation contributes to malignancy by
targeting pyruvate dehydrogenase kinase 2 in gastric cancer. Zhang
et al (9) demonstrated that
miR-422a suppresses tumor growth and metastasis in hepatocellular
carcinoma. Additionally, miR-422a inhibits osteosarcoma cell cycle
arrest and induces apoptosis by directly targeting BCL2L2 and KRAS
(27). Furthermore, a study of
glioma analyzing a TCGA dataset indicated that miR-422a expression
predicts the neural subtype of GBM and patient outcome (28). In combination with the public
database analysis, this evidence prompted us to further explore the
expression of miR-422a in clinical tissues of GBM with different
grades and GBM cell lines. Moreover, the role of miR-422a in the
malignant phenotype of GBM and the underlying antitumor mechanism
was also investigated.
Although previous studies revealed that miR-422a
markedly suppressed glioma cell proliferation, migration and
invasion by targeting PIK3CA, insulin-like growth factor 1 (IGF1)
and IGF1 receptor (IGF1R), which are new prognostic biomarkers for
human glioblastoma (14,29,30),
the detailed mechanism of the miR-422a-mediated inhibitory effect
on tumor growth remains poorly understood. The present study first
confirmed the downregulation of miR-422a, which was negatively
associated with the WHO grade, and identified its antitumor
function. Mechanistically, by analyzing PITA, miRmap and miRdanda
public databases, it was identified that RPN2 had highly conserved
binding sites for miR-422a in its 3′UTR region. Experimental
evidence from a luciferase reporter assay, western blotting and RIP
assay verified that RPN2 was a direct target of miR-422a. These
data also demonstrated that miR-422a suppressed GBM growth at least
partially through RPN2. Furthermore, the present study investigated
the role of RPN2 in the miR-422a-mediated regulation of GBM
biological behaviors and Wnt/β-catenin signaling pathway.
Collectively, the findings revealed a novel
miR-422a/RPN2/Wnt/β-catenin axis implicated in GBM development and
progression, deepening the understanding of the etiology of
glioma.
Extensive studies have verified that RPN2
upregulation is involved in the progression of various
malignancies, and that RPN2-mediated glycosylation of P-gp (encoded
by MDR1) is responsible for drug resistance in a number of cancers,
including breast cancer, ovarian cancer, gastric cancer and
esophageal squamous cell carcinoma (31–34).
Additionally, it has been reported that the P-gp gene is closely
associated with poor prognosis and temozolomide (TMZ) resistance in
high-grade gliomas (35,36). Accordingly, whether miR-422a
modulates TMZ chemosensitivity via RPN2 may be worth investigating.
However, to the best of our knowledge, there have been no reports
regarding the expression and function of RPN2 in gliomas. Our
previous unpublished study systematically evaluated RPN2
upregulation in glioma specimens and GBM cell lines and
investigated the tumor-promoting mechanism (data not shown).
Furthermore, in the present study, survival analysis from online
public databases indicated that high RPN2 expression is associated
with poor prognosis. A limitation of the current study is that a
log-rank test may not applicable for survival plots where
late-stage crossover is present, although it is the most common way
to analyze survival differences. However, the survival time of
gliomas is generally similar to the normal distribution, few
patients have a very short or long survival time. Hence, the
late-stage crossover observed for patients with very long survival
may not be significant. Indeed, further verification of these data
is necessary to determine the prognostic significance of RPN2 using
a weighted test, such as Renyi or Cramér-von Mises.
Wnt/β-catenin signaling is one of the most important
oncogenic pathways in GBM and represents a promising target for
glioma treatment (37,38). β-catenin in the nucleus interacts
with the TCF/LEF family to regulate the transcription of multiple
genes, including c-myc and cyclinD1, while GSK-3β promotes its
degradation contributing to Wnt signal inactivation (37–39).
Takahashi et al (40)
confirmed that RPN2 antagonizes GSK3β through physical interactions
and subsequently suppresses heat shock protein-containing HSP70 and
HSP90, which are essential for mtp53 stabilization, enhanced
initiation, metastasis and cancer stem cell property acquisition in
breast cancer. Our previous experimental data also verified that
the knockdown of RPN2 inhibited the Wnt/β-catenin pathway in glioma
(data not shown); hence, it was hypothesized that miR-422a
suppresses glioma growth through the Wnt/β-catenin signaling
pathway via RPN2. Indeed, the present results revealed that
miR-422a markedly disrupted the Wnt/β-catenin pathway, whereas
restoration of RPN2 in LN229 and U87 cells with miR-422a
overexpression could in part abrogate the inhibitory effect of
miR-422a. Therefore, to the best of our knowledge, the present
study demonstrated for the first time the relationship between
miR-422a, RPN2 and the Wnt pathway in glioma.
The miRNA/mRNA/signaling pathway is a complicated
regulatory network that is a critical molecular mechanism for GBM
recurrence, TMZ resistance and EMT (41–43).
miR-422a negatively modulates the EGFR/MEK/ERK signaling pathway by
targeting the mediator complex subunit (Med19) (44). Additionally, miR-422a serves as a
tumor suppressor through the SULF2-mediated TGF-β/SMAD signaling
pathway in non-small cell lung cancer (45). Furthermore, miR-422a inhibits the
PI3K/AKT pathway by directly targeting PIK3CA and AKT1 in glioma
and colorectal cancer, respectively (14,46).
Combined with the present data on Wnt signaling pathway regulation
by miR-422a, the data suggested that miR-422a may be a crucial
anticancer gene that acts through multiple signaling pathways. In
addition, Huang et al (47)
reported that RPN2 promotes metastasis and suppresses autophagy via
STAT3 and NF-κB signaling pathways in hepatocellular carcinoma.
These reports reflect the complexity of the signal transduction
pathways mediated by miR-422a that are implicated in cancer
progression, including glioma. However, the specific regulatory
mechanism between miR-422a and other pathways, such as NF/KB and
STAT3, is worthy of further study.
Mounting evidence has demonstrated that competing
endogenous RNA (ceRNA) networks play significant roles in tumor
biology. A variety of miRNAs are regulated by other non-coding
RNAs, such as circular RNAs (circRNAs) and long non-coding RNAs
(lncRNAs) (6,48). For instance, Hong et al
(12) reported that circCRIM1
competitively sponges miR-422a to block the inhibitory effect of
miR-422a on FOXQ1 and subsequently contributes to nasopharyngeal
carcinoma cell metastasis, EMT and docetaxel chemoresistance. Zhou
et al (49), demonstrated
that lncRNAD63785 acts as a ceRNA of miR-422a and enhances
chemoresistance by retarding miR-422a-dependent suppression of
MEF2D. However, more circRNAs or lncRNAs sponging miR-422a have
been identified and validated (50,51),
which may be of significance in understanding the in-depth
molecular mechanism of glioma initiation and progression, and
combined treatment responses.
In conclusion, the present study confirmed that
ectopic miR-422a expression suppresses GBM tumorigenesis and
promotes apoptosis by regulating the Wnt/β-catenin signaling
pathway, and that RPN2 plays significant roles in the
miR-422a-mediated effect on tumor growth and Wnt pathway regulation
as a direct functional target of miR-422a. These results revealed a
novel miR-422a/RPN2/Wnt/β-catenin axis in glioma, representing a
potential candidate target for GBM therapy.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The study was supported by the Foundation of Tianjin
Science and Technology Committee (grant nos. 14JCZDJC35600 and
12ZCDZSY17700), the National Key Technology Support Program (grant
nos. 2014BAI04B00 and 2015BAI03B05) and the National Natural
Science Fund of China (grant no. 81671169).
Availability of data and materials
The datasets used and/or analysed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
JS, ZC and JW conceived and designed the
experiments. JS, JX, LM and CW performed the in vitro
experiments. JS and QW analyzed the data and prepared the figures.
WF, JX, XZ and FT collected specimens and performed the animal
experiments. JS and ZC drafted the manuscript. JS, JX, ZC, QW and
JW interpreted the data, reviewed and revised the manuscript. All
authors have read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was reviewed and approved by
Institutional Review Board of Tianjin Huanhu Hospital (Tianjin,
China; approval no. CK19-190318), and written informed consent was
obtained from all participating patients. All experimental
procedures involving the use of animals were approved by the Animal
Ethical and Welfare Committee of Tianjin Huanhu Hospital of Nankai
University (Tianjin, China; approval no. SYXK2019-001).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Aldape K, Zadeh G, Mansouri S,
Reifenberger G and von Deimling A: Glioblastoma: Pathology,
molecular mechanisms and markers. Acta Neuropathol. 129:829–848.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kanu OO, Hughes B, Di C, Lin N, Fu J,
Bigner DD, Yan H and Adamson C: Glioblastoma multiforme
oncogenomics and signaling pathways. Clin Med Oncol. 3:39–52.
2009.PubMed/NCBI
|
|
3
|
Cloughesy TF, Cavenee WK and Mischel PS:
Glioblastoma: From molecular pathology to targeted treatment. Annu
Rev Pathol. 9:1–25. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Carlsson SK, Brothers SP and Wahlestedt C:
Emerging treatment strategies for glioblastoma multiforme. EMBO Mol
Med. 6:1359–1370. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lapointe S, Perry A and Butowski NA:
Primary brain tumours in adults. Lancet. 392:432–446. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chan JJ and Tay Y: Noncoding RNA:RNA
regulatory networks in cancer. Int J Mol Sci. 19:13102018.
View Article : Google Scholar
|
|
7
|
Floyd D and Purow B: Micro-masters of
glioblastoma biology and therapy: Increasingly recognized roles for
microRNAs. Neuro Oncol. 16:622–627. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Slack FJ and Chinnaiyan AM: The role of
Non-coding RNAs in oncology. Cell. 179:1033–1055. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang J, Yang Y, Yang T, Yuan S, Wang R,
Pan Z, Yang Y, Huang G, Gu F, Jiang B, et al: Double-negative
feedback loop between microRNA-422a and forkhead box (FOX)G1/Q1/E1
regulates hepatocellular carcinoma tumor growth and metastasis.
Hepatology. 61:561–573. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wu L, Hu B, Zhao B, Liu Y, Yang Y, Zhang L
and Chen J: Circulating microRNA-422a is associated with lymphatic
metastasis in lung cancer. Oncotarget. 8:42173–42188. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zheng G, Du L, Yang X, Zhang X, Wang L,
Yang Y, Li J and Wang C: Serum microRNA panel as biomarkers for
early diagnosis of colorectal adenocarcinoma. Br J Cancer.
111:1985–1992. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hong X, Liu N, Liang Y, He Q, Yang X, Lei
Y, Zhang P, Zhao Y, He S, Wang Y, et al: Circular RNA CRIM1
functions as a ceRNA to promote nasopharyngeal carcinoma metastasis
and docetaxel chemoresistance through upregulating FOXQ1. Mol
Cancer. 19:332020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Du S, Wang S, Zhang F and Lv Y: SKP2,
positively regulated by circ_ODC1/miR-422a axis, promotes the
proliferation of retinoblastoma. J Cell Biochem. 121:322–331. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liang H, Wang R, Jin Y, Li J and Zhang S:
MiR-422a acts as a tumor suppressor in glioblastoma by targeting
PIK3CA. Am J Cancer Res. 6:1695–1707. 2016.PubMed/NCBI
|
|
15
|
Lemjabbar-Alaoui H, McKinney A, Yang YW,
Tran VM and Phillips JJ: Glycosylation alterations in lung and
brain cancer. Adv Cancer Res. 126:305–344. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ono M, Tsuda H, Kobayashi T, Takeshita F,
Takahashi RU, Tamura K, Akashi-Tanaka S, Moriya T, Yamasaki T,
Kinoshita T, et al: The expression and clinical significance of
ribophorin II (RPN2) in human breast cancer. Pathol Int.
65:301–308. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Veillon L, Fakih C, Abou-El-Hassan H,
Kobeissy F and Mechref Y: Glycosylation changes in brain cancer.
ACS Chem Neurosci. 9:51–72. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fujimoto D, Goi T, Koneri K and Hirono Y:
RPN2 is effective biomarker to predict the outcome of combined
chemotherapy docetaxel and cisplatin for advanced gastric cancer.
Oncotarget. 9:15208–15218. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fujiwara T, Takahashi RU, Kosaka N, Nezu
Y, Kawai A, Ozaki T and Ochiya T: RPN2 gene confers osteosarcoma
cell malignant phenotypes and determines clinical prognosis. Mol
Ther Nucleic Acids. 3:e1892014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang J, Yan B, Spath SS, Qun H, Cornelius
S, Guan D, Shao J, Hagiwara K, Van Waes C, Chen Z, et al:
Integrated transcriptional profiling and genomic analyses reveal
RPN2 and HMGB1 as promising biomarkers in colorectal cancer. Cell
Biosci. 5:532015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhao Z, Meng F, Wang W, Wang Z, Zhang C
and Jiang T: Comprehensive RNA-seq transcriptomic profiling in the
malignant progression of gliomas. Sci Data. 4:1700242017.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tang Z, Kang B, Li C, Chen T and Zhang Z:
GEPIA2: An enhanced web server for large-scale expression profiling
and interactive analysis. Nucleic Acids Res. 47((W1)): W556–W560.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sun J, Jia Z, Li B, Zhang A, Wang G, Pu P,
Chen Z, Wang Z and Yang W: MiR-19 regulates the proliferation and
invasion of glioma by RUNX3 via β-catenin/Tcf-4 signaling.
Oncotarget. 8:110785–110796. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
He Z, Li Z, Zhang X, Yin K, Wang W, Xu Z,
Li B, Zhang L, Xu J, Sun G, et al: MiR-422a regulates cellular
metabolism and malignancy by targeting pyruvate dehydrogenase
kinase 2 in gastric cancer. Cell Death Dis. 9:5052018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang H, He QY, Wang GC, Tong DK, Wang RK,
Ding WB, Li C, Wei Q, Ding C, Liu PZ, et al: miR-422a inhibits
osteosarcoma proliferation by targeting BCL2L2 and KRAS. Biosci
Rep. 38:BSR201703392018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li R, Gao K, Luo H, Wang X, Shi Y, Dong Q,
Luan W and You Y: Identification of intrinsic subtype-specific
prognostic microRNAs in primary glioblastoma. J Exp Clin Cancer
Res. 33:92014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Maris C, D'Haene N, Trepant AL, Le Mercier
M, Sauvage S, Allard J, Rorive S, Demetter P, Decaestecker C and
Salmon I: IGF-IR: A new prognostic biomarker for human
glioblastoma. Br J Cancer. 113:729–737. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang H, Tang C, Na M, Ma W, Jiang Z, Gu Y,
Ma G, Ge H, Shen H and Lin Z: miR-422a inhibits glioma
proliferation and invasion by targeting IGF1 and IGF1R. Oncol Res.
25:187–194. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
De Souza R, Zahedi P, Badame RM, Allen C
and Piquette- Miller M: Chemotherapy dosing schedule influences
drug resistance development in ovarian cancer. Mol Cancer Ther.
10:1289–1299. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Honma K, Iwao-Koizumi K, Takeshita F,
Yamamoto Y, Yoshida T, Nishio K, Nagahara S, Kato K and Ochiya T:
RPN2 gene confers docetaxel resistance in breast cancer. Nat Med.
14:939–948. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kurashige J, Watanabe M, Iwatsuki M,
Kinoshita K, Saito S, Nagai Y, Ishimoto T, Baba Y, Mimori K and
Baba H: RPN2 expression predicts response to docetaxel in
oesophageal squamous cell carcinoma. Br J Cancer. 107:1233–1238.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang H, Jiang H, Zhang H, Liu J, Hu X and
Chen L: Ribophorin II potentiates P-glycoprotein- and
ABCG2-mediated multidrug resistance via activating ERK pathway in
gastric cancer. Int J Biol Macromol. 128:574–582. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Koshkin PA, Chistiakov DA, Nikitin AG,
Konovalov AN, Potapov AA, Usachev DY, Pitskhelauri DI, Kobyakov GL,
Shishkina LV and Chekhonin VP: Analysis of expression of microRNAs
and genes involved in the control of key signaling mechanisms that
support or inhibit development of brain tumors of different grades.
Clin Chim Acta. 430:55–62. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Stavrovskaya AA, Shushanov SS and
Rybalkina EY: Problems of glioblastoma multiforme drug resistance.
Biochemistry (Mosc). 81:91–100. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang K, Zhang J, Han L, Pu P and Kang C:
Wnt/beta-catenin signaling in glioma. J Neuroimmune Pharmacol.
7:740–749. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
He L, Zhou H, Zeng Z, Yao H, Jiang W and
Qu H: Wnt/β-catenin signaling cascade: A promising target for
glioma therapy. J Cell Physiol. 234:2217–2228. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lim JC, Kania KD, Wijesuriya H, Chawla S,
Sethi JK, Pulaski L, Romero IA, Couraud PO, Weksler BB, Hladky SB
and Barrand MA: Activation of beta-catenin signalling by GSK-3
inhibition increases p-glycoprotein expression in brain endothelial
cells. J Neurochem. 106:1855–1865. 2008.PubMed/NCBI
|
|
40
|
Takahashi RU, Takeshita F, Honma K, Ono M,
Kato K and Ochiya T: Ribophorin II regulates breast tumor
initiation and metastasis through the functional suppression of
GSK3β. Sci Rep. 3:24742013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Low SY, Ho YK, Too HP, Yap CT and Ng WH:
MicroRNA as potential modulators in chemoresistant high-grade
gliomas. J Clin Neurosci. 21:395–400. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Novakova J, Slaby O, Vyzula R and Michalek
J: MicroRNA involvement in glioblastoma pathogenesis. Biochem
Biophys Res Commun. 386:1–5. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Xin S, Huang K and Zhu XG: Non-coding
RNAs: Regulators of glioma cell epithelial-mesenchymal
transformation. Pathol Res Pract. 215:1525392019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhang X, Gao D, Fang K, Guo Z and Li L:
Med19 is targeted by miR-101-3p/miR-422a and promotes breast cancer
progression by regulating the EGFR/MEK/ERK signaling pathway.
Cancer Lett. 444:105–115. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Li WQ, Zhang JP, Wang YY, Li XZ and Sun L:
MicroRNA-422a functions as a tumor suppressor in non-small cell
lung cancer through SULF2-mediated TGF-β/SMAD signaling pathway.
Cell Cycle. 18:1727–1744. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wei WT, Nian XX, Wang SY, Jiao HL, Wang
YX, Xiao ZY, Yang RW, Ding YQ, Ye YP and Liao WT: miR-422a inhibits
cell proliferation in colorectal cancer by targeting AKT1 and
MAPK1. Cancer Cell Int. 17:912017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Huang L, Jian Z, Gao Y, Zhou P, Zhang G,
Jiang B and Lv Y: RPN2 promotes metastasis of hepatocellular
carcinoma cell and inhibits autophagy via STAT3 and NF-KB pathways.
Aging (Albany NY). 11:6674–6690. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Dragomir M, Mafra ACP, Dias SMG, Vasilescu
C and Calin GA: Using microRNA networks to understand cancer. Int J
Mol Sci. 19:18712018. View Article : Google Scholar
|
|
49
|
Zhou Z, Lin Z, He Y, Pang X, Wang Y,
Ponnusamy M, Ao X, Shan P, Tariq MA, Li P and Wang J: The long
noncoding RNA D63785 regulates chemotherapy sensitivity in human
gastric cancer by targeting miR-422a. Mol Ther Nucleic Acids.
12:405–419. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wang R, Zhang S, Chen X, Li N, Li J, Jia
R, Pan Y and Liang H: CircNT5E Acts as a Sponge of miR-422a to
promote glioblastoma tumorigenesis. Cancer Res. 78:4812–4825. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wei F, Yang L, Jiang D, Pan M, Tang G,
Huang M and Zhang J: Long noncoding RNA DUXAP8 contributes to the
progression of hepatocellular carcinoma via regulating
miR-422a/PDK2 axis. Cancer Med. 9:2480–2490. 2020. View Article : Google Scholar : PubMed/NCBI
|