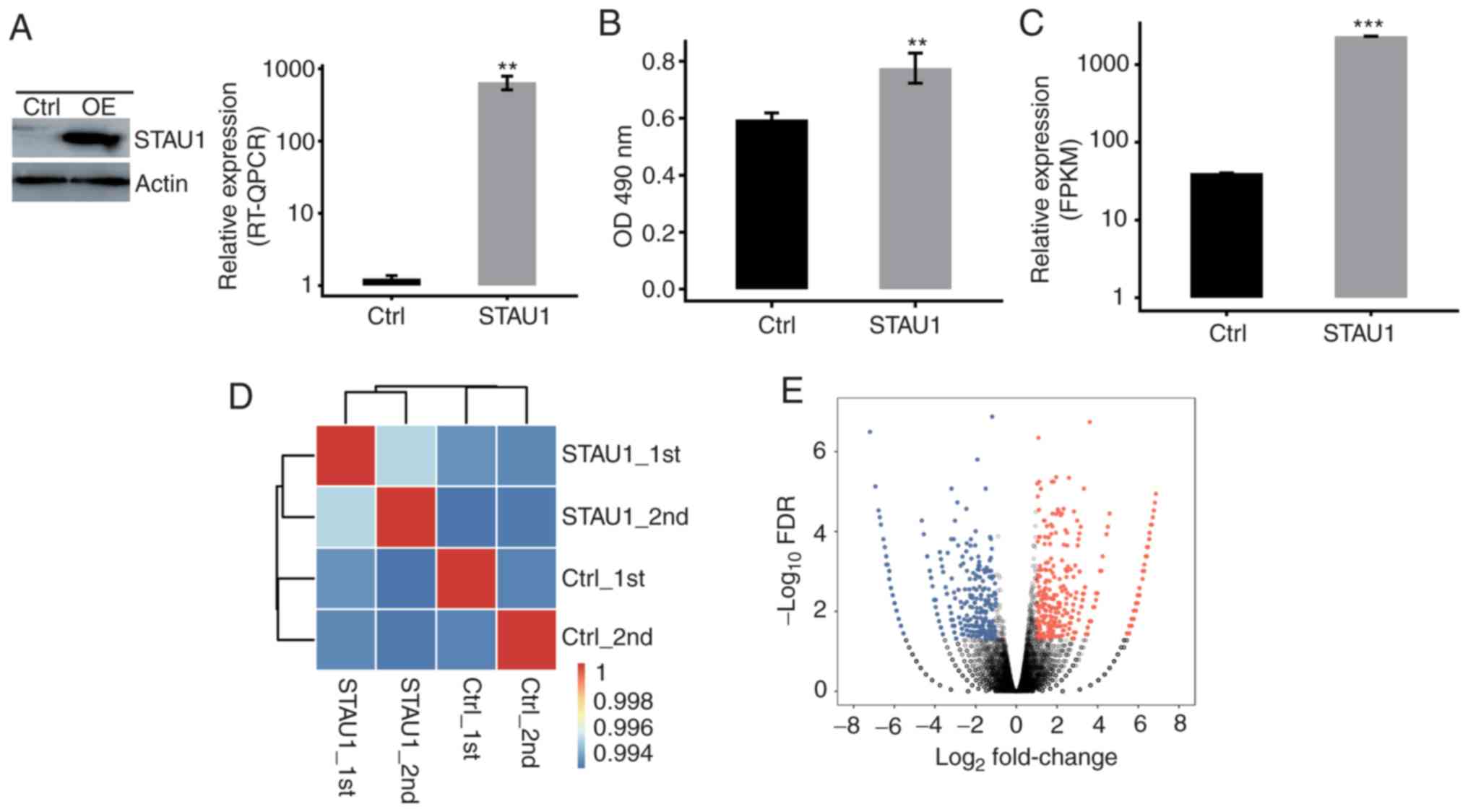
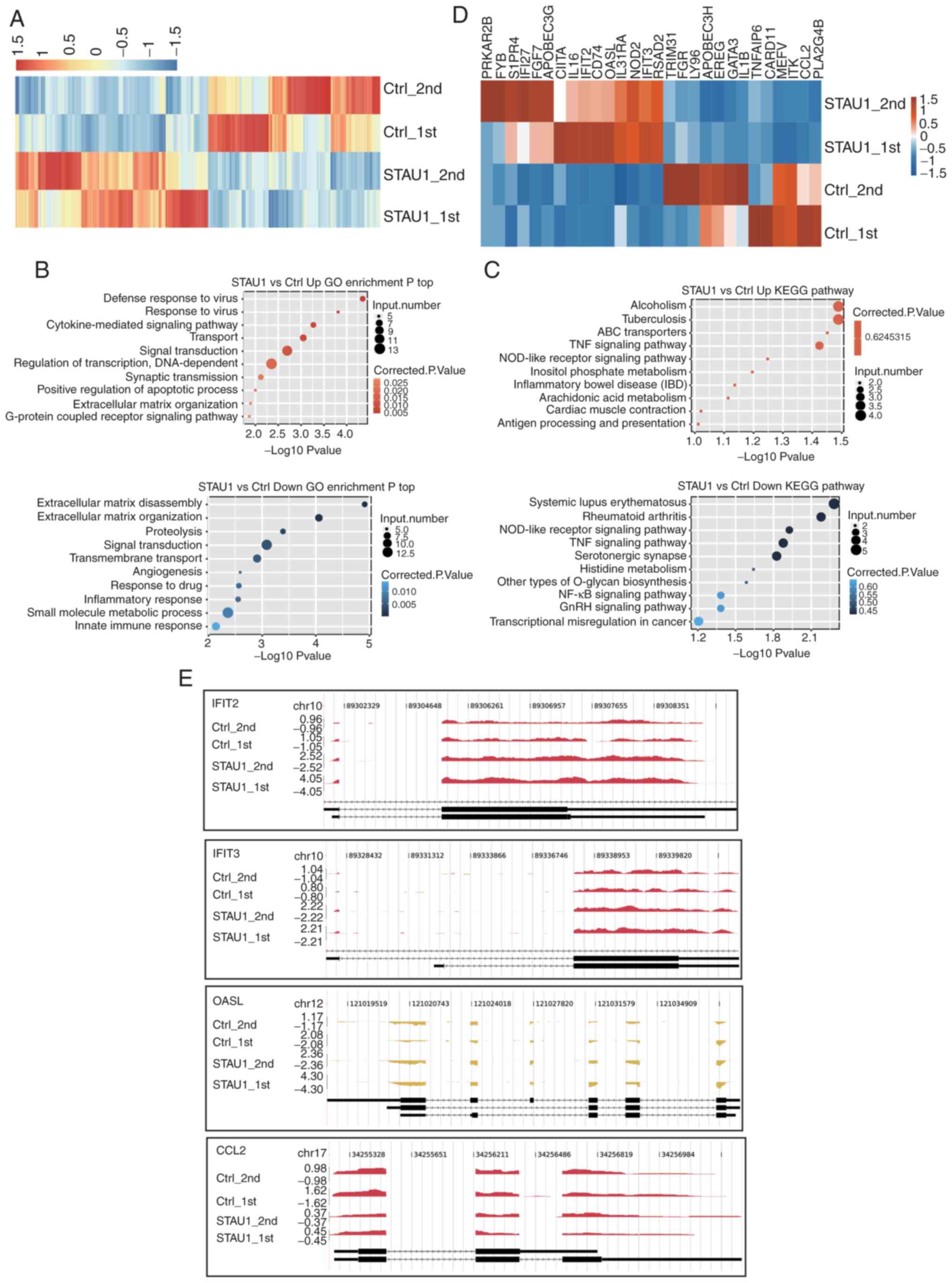
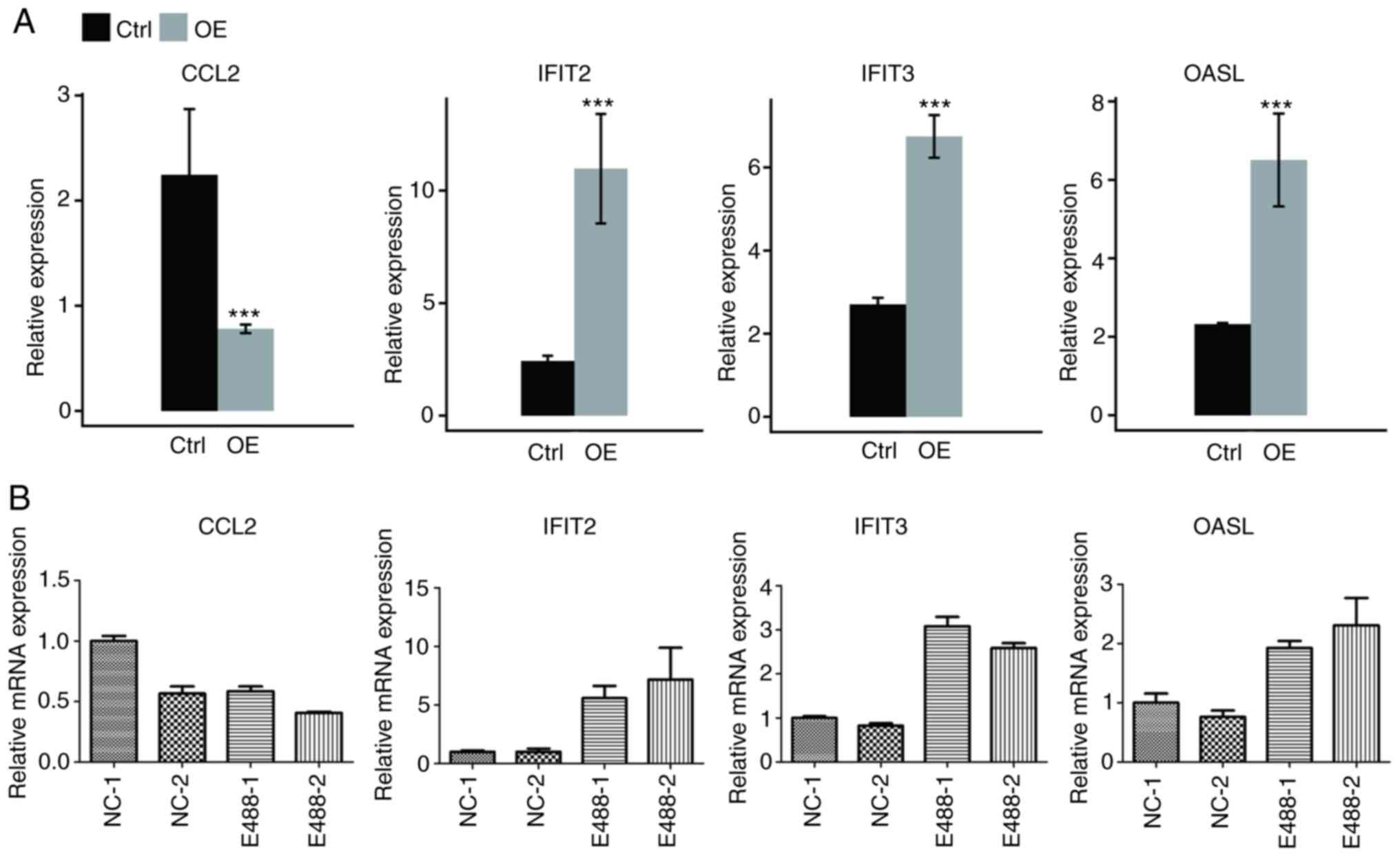
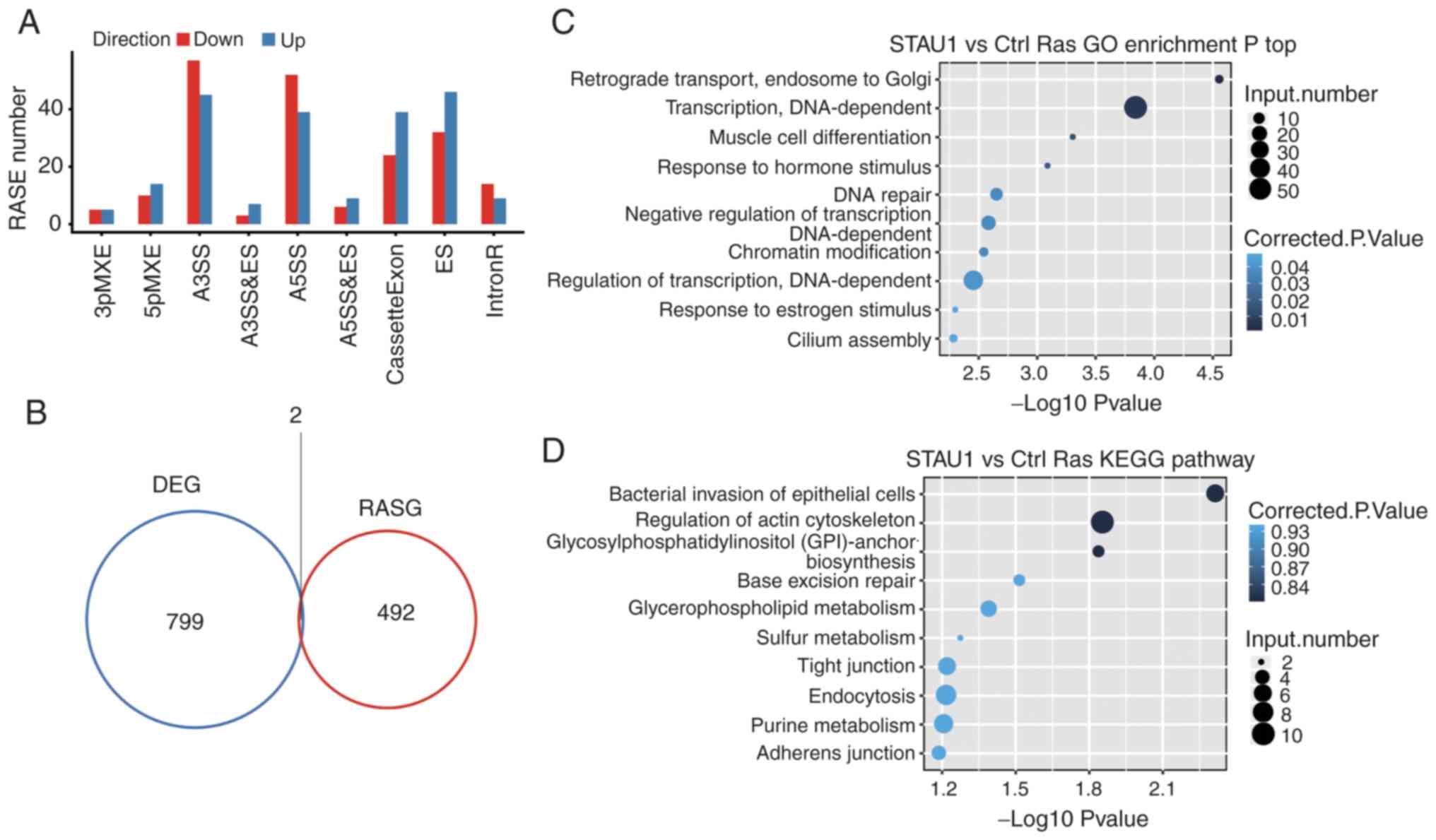
|
1
|
Castello A, Fischer B, Hentze MW and
Preiss T: RNA-binding proteins in mendelian disease. Trends Genet.
29:318–327. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lunde BM, Moore C and Varani G:
RNA-binding proteins: Modular design for efficient function. Nat
Rev Mol Cell Biol. 8:479–490. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Thandapani P, O'Connor TR, Bailey TL and
Richard S: Defining the RGG/RG motif. Mol Cell. 50:613–623. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hentze MW, Castello A, Schwarzl T and
Preiss T: A brave new world of RNA-binding proteins. Nat Rev Mol
Cell Biol. 19:327–341. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Furic L, Maher-Laporte M and
DesGroseillers L: A genome-wide approach identifies distinct but
overlapping subsets of cellular mRNAs associated with staufen1- and
staufen2-containing ribonucleoprotein complexes. RNA. 14:324–335.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bondy-Chorney E, Crawford Parks TE,
Ravel-Chapuis A, Klinck R, Rocheleau L, Pelchat M, Chabot B, Jasmin
BJ and Côté J: Staufen1 regulates multiple alternative splicing
events either positively or negatively in DM1 Indicating its role
as a disease modifier. PLoS Genet. 12:e10058272016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gong C and Maquat LE: lncRNAs
transactivate STAU1-mediated mRNA decay by duplexing with 3′UTRs
via Alu elements. Nature. 470:284–288. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kretz M, Siprashvili Z, Chu C, Webster DE,
Zehnder A, Qu K, Lee CS, Flockhart RJ, Groff AF, Chow J, et al:
Control of somatic tissue differentiation by the long non-coding
RNA TINCR. Nature. 493:231–235. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kretz M: TINCR, staufen1, and cellular
differentiation. RNA Biol. 10:1597–1601. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Elbarbary RA, Li W, Tian B and Maquat LE:
STAU1 binding 3′ UTR IRAlus complements nuclear retention to
protect cells from PKR-mediated translational shutdown. Genes Dev.
27:1495–1510. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Giorgi C, Yeo GW, Stone ME, Katz DB, Burge
C, Turrigiano G and Moore MJ: The EJC factor eIF4AIII modulates
synaptic strength and neuronal protein expression. Cell.
130:179–191. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Oh Y, Park J, Kim JI, Chang MY, Lee SH,
Cho YH and Hwang J: Lin28B and miR-142-3p regulate neuronal
differentiation by modulating staufen1 expression. Cell Death
Differ. 25:432–443. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
de Lucas S, Peredo J, Marion RM, Sanchez C
and Ortin J: Human staufen1 protein interacts with influenza virus
ribonucleoproteins and is required for efficient virus
multiplication. J Virol. 84:7603–7612. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lee JH, Oh JY, Pascua PN, Kim EG, Choi YK
and Kim HK: Impairment of the staufen1-NS1 interaction reduces
influenza viral replication. Biochem Biophys Res Commun.
414:153–158. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rao S, Hassine S, Monette A, Amorim R,
DesGroseillers L and Mouland AJ: HIV-1 requires staufen1 to
dissociate stress granules and to produce infectious viral
particles. RNA. 25:727–736. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xue Y, Ouyang K, Huang J, Zhou Y, Ouyang
H, Li H, Wang G, Wu Q, Wei C, Bi Y, et al: Direct conversion of
fibroblasts to neurons by reprogramming PTB-regulated microRNA
circuits. Cell. 152:82–96. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gentry JJ, Casaccia-Bonnefil P and Carter
BD: Nerve growth factor activation of nuclear factor kappaB through
its p75 receptor is an anti-apoptotic signal in RN22 schwannoma
cells. J Biol Chem. 275:7558–7565. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nykjaer A, Lee R, Teng KK, Jansen P,
Madsen P, Nielsen MS, Jacobsen C, Kliemannel M, Schwarz E, Willnow
TE, et al: Sortilin is essential for proNGF-induced neuronal cell
death. Nature. 427:843–848. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rabizadeh S, Rabizadeh S, Ye X, Wang JJ
and Bredesen DE: Neurotrophin dependence mediated by p75NTR:
Contrast between rescue by BDNF and NGF. Cell Death Differ.
6:1222–1227. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Notterpek L: Neurotrophins in myelination:
A new role for a puzzling receptor. Trends Neurosci. 26:232–234.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fu C, Donovan WP, Shikapwashya-Hasser O,
Ye X and Cole RH: Hot fusion: An efficient method to clone multiple
DNA fragments as well as inverted repeats without ligase. PLoS One.
9:e1153182014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
McGinn S and Gut IG: DNA
sequencing-spanning the generations. N Biotechnol. 30:366–372.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kim D, Pertea G, Trapnell C, Pimentel H,
Kelley R and Salzberg SL: TopHat2: Accurate alignment of
transcriptomes in the presence of insertions, deletions and gene
fusions. Genome Biol. 14:R362013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Trapnell C, Williams BA, Pertea G,
Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ and Pachter
L: Transcript assembly and quantification by RNA-Seq reveals
unannotated transcripts and isoform switching during cell
differentiation. Nat Biotechnol. 28:511–515. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Robinson MD, McCarthy DJ and Smyth GK:
EdgeR: A bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jin L, Li G, Yu D, Huang W, Cheng C, Liao
S, Wu Q and Zhang Y: Transcriptome analysis reveals the complexity
of alternative splicing regulation in the fungus verticillium
dahliae. BMC Genomics. 18:1302017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xia H, Chen D, Wu Q, Wu G, Zhou Y, Zhang Y
and Zhang L: CELF1 preferentially binds to exon-intron boundary and
regulates alternative splicing in HeLa cells. Biochim Biophys Acta
Gene Regul Mech. 1860:911–921. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xie C, Mao X, Huang J, Ding Y, Wu J, Dong
S, Kong L, Gao G, Li CY and Wei L: KOBAS 2.0: A web server for
annotation and identification of enriched pathways and diseases.
Nucleic Acids Res. 39((Web Server issue)): W316–W322. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Erlich Y, Mitra PP, delaBastide M,
McCombie WR and Hannon GJ: Alta-cyclic: A self-optimizing base
caller for next-generation sequencing. Nat Methods. 5:679–682.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lebeau G, DesGroseillers L, Sossin W and
Lacaille JC: mRNA binding protein staufen 1-dependent regulation of
pyramidal cell spine morphology via NMDA receptor-mediated synaptic
plasticity. Mol Brain. 4:222011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lebeau G, Maher-Laporte M, Topolnik L,
Laurent CE, Sossin W, Desgroseillers L and Lacaille JC: Staufen1
regulation of protein synthesis-dependent long-term potentiation
and synaptic function in hippocampal pyramidal cells. Mol Cell
Biol. 28:2896–2907. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Vessey JP, Macchi P, Stein JM, Mikl M,
Hawker KN, Vogelsang P, Wieczorek K, Vendra G, Riefler J, Tübing F,
et al: A loss of function allele for murine staufen1 leads to
impairment of dendritic staufen1-RNP delivery and dendritic spine
morphogenesis. Proc Natl Acad Sci USA. 105:16374–16379. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang Z, Gerstein M and Snyder M: RNA-Seq:
A revolutionary tool for transcriptomics. Nat Rev Genet. 10:57–63.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Underwood JG, Uzilov AV, Katzman S,
Onodera CS, Mainzer JE, Mathews DH, Lowe TM, Salama SR and Haussler
D: FragSeq: Transcriptome-wide RNA structure probing using
high-throughput sequencing. Nat Methods. 7:995–1001. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Palanisamy N, Ateeq B, Kalyana-Sundaram S,
Pflueger D, Ramnarayanan K, Shankar S, Han B, Cao Q, Cao X, Suleman
K, et al: Rearrangements of the RAF kinase pathway in prostate
cancer, gastric cancer and melanoma. Nat Med. 16:793–798. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chu CT, Ji J, Dagda RK, Jiang JF, Tyurina
YY, Kapralov AA, Tyurin VA, Yanamala N, Shrivastava IH,
Mohammadyani D, et al: Cardiolipin externalization to the outer
mitochondrial membrane acts as an elimination signal for mitophagy
in neuronal cells. Nat Cell Biol. 15:1197–1205. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Fensterl V, Wetzel JL, Ramachandran S,
Ogino T, Stohlman SA, Bergmann CC, Diamond MS, Virgin HW and Sen
GC: Interferon-induced Ifit2/ISG54 protects mice from lethal VSV
neuropathogenesis. PLoS Pathog. 8:e10027122012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Stawowczyk M, Van Scoy S, Kumar KP and
Reich NC: The interferon stimulated gene 54 promotes apoptosis. J
Biol Chem. 286:7257–7266. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang B, Liu X, Chen W and Chen L: IFIT5
potentiates anti-viral response through enhancing innate immune
signaling pathways. Acta Biochim Biophys Sin (Shanghai).
45:867–874. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Siegfried A, Berchtold S, Manncke B,
Deuschle E, Reber J, Ott T, Weber M, Kalinke U, Hofer MJ, Hatesuer
B, et al: IFIT2 is an effector protein of type I IFN-mediated
amplification of lipopolysaccharide (LPS)-induced TNF-α secretion
and LPS-induced endotoxin shock. J Immunol. 191:3913–3921. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Berchtold S, Manncke B, Klenk J, Geisel J,
Autenrieth IB and Bohn E: Forced IFIT-2 expression represses LPS
induced TNF-alpha expression at posttranscriptional levels. BMC
Immunol. 9:752008. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Niess H, Camaj P, Mair R, Renner A, Zhao
Y, Jäckel C, Nelson PJ, Jauch KW and Bruns CJ: Overexpression of
IFN-induced protein with tetratricopeptide repeats 3 (IFIT3) in
pancreatic cancer: Cellular ‘pseudoinflammation’ contributing to an
aggressive phenotype. Oncotarget. 6:3306–3318. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Liu XY, Chen W, Wei B, Shan YF and Wang C:
IFN-induced TPR protein IFIT3 potentiates antiviral signaling by
bridging MAVS and TBK1. J Immunol. 187:2559–2568. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Melchjorsen J, Kristiansen H, Christiansen
R, Rintahaka J, Matikainen S, Paludan SR and Hartmann R:
Differential regulation of the OASL and OAS1 genes in response to
viral infections. J Interferon Cytokine Res. 29:199–207. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhu J, Ghosh A and Sarkar SN: OASL-a new
player in controlling antiviral innate immunity. Curr Opin Virol.
12:15–19. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Sun L, Li Y, Jia X, Wang Q, Li Y, Hu M,
Tian L, Yang J, Xing W, Zhang W, et al: Neuroprotection by IFN-ү
via astrocyte-secreted IL-6 in acute neuroinflammation. Oncotarget.
8:40065–40078. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ellis A and Bennett DL: Neuroinflammation
and the generation of neuropathic pain. Br J Anaesth. 111:26–37.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Kawasaki Y, Zhang L, Cheng JK and Ji RR:
Cytokine mechanisms of central sensitization: Distinct and
overlapping role of interleukin-1beta, interleukin-6, and tumor
necrosis factor-alpha in regulating synaptic and neuronal activity
in the superficial spinal cord. J Neurosci. 28:5189–5194. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Leung L and Cahill CM: TNF-alpha and
neuropathic pain-a review. J Neuroinflammation. 7:272010.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Deshmane SL, Kremlev S, Amini S and Sawaya
BE: Monocyte chemoattractant protein-1 (MCP-1): An overview. J
Interferon Cytokine Res. 29:313–326. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Dansereau MA, Gosselin RD, Pohl M, Pommier
B, Mechighel P, Mauborgne A, Rostene W, Kitabgi P, Beaudet N,
Sarret P and Melik-Parsadaniantz S: Spinal CCL2 pronociceptive
action is no longer effective in CCR2 receptor antagonist-treated
rats. J Neurochem. 106:757–769. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Thacker MA, Clark AK, Bishop T, Grist J,
Yip PK, Moon LD, Thompson SW, Marchand F and McMahon SB: CCL2 is a
key mediator of microglia activation in neuropathic pain states.
Eur J Pain. 13:263–272. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Gao YJ, Zhang L, Samad OA, Suter MR,
Yasuhiko K, Xu ZZ, Park JY, Lind AL, Ma Q and Ji RR: JNK-induced
MCP-1 production in spinal cord astrocytes contributes to central
sensitization and neuropathic pain. J Neurosci. 29:4096–4108. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zhuang ZY, Kawasaki Y, Tan PH, Wen YR,
Huang J and Ji RR: Role of the CX3CR1/p38 MAPK pathway in spinal
microglia for the development of neuropathic pain following nerve
injury-induced cleavage of fractalkine. Brain Behav Immun.
21:642–651. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zhang H, Boyette-Davis JA, Kosturakis AK,
Li Y, Yoon SY, Walters ET and Dougherty PM: Induction of monocyte
chemoattractant protein-1 (MCP-1) and its receptor CCR2 in primary
sensory neurons contributes to paclitaxel-induced peripheral
neuropathy. J Pain. 14:1031–1044. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Rougerie P and Delon J: Rho GTPases:
Masters of T lymphocyte migration and activation. Immunol Lett.
142:1–13. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Sato K, Sugiyama T, Nagase T, Kitade Y and
Ueda H: Threonine 680 phosphorylation of FLJ00018/PLEKHG2, a Rho
family-specific guanine nucleotide exchange factor, by epidermal
growth factor receptor signaling regulates cell morphology of
Neuro-2a cells. J Biol Chem. 289:10045–10056. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Shimizu Y, Dobashi K, Iizuka K, Horie T,
Suzuki K, Tukagoshi H, Nakazawa T, Nakazato Y and Mori M:
Contribution of small GTPase Rho and its target protein rock in a
murine model of lung fibrosis. Am J Respir Crit Care Med.
163:210–217. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Sato K, Kimura M, Sugiyama K, Nishikawa M,
Okano Y, Nagaoka H, Nagase T, Kitade Y and Ueda H: Four-and-a-half
LIM domains 1 (FHL1) protein interacts with the rho guanine
nucleotide exchange factor PLEKHG2/FLJ00018 and regulates cell
morphogenesis. J Biol Chem. 291:25227–25238. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Sun X, Zhou Z, Fink DJ and Mata M: HspB1
silences translation of PDZ-RhoGEF by enhancing miR-20a and miR-128
expression to promote neurite extension. Mol Cell Neurosci.
57:111–119. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Hofer F, Fields S, Schneider C and Martin
GS: Activated ras interacts with the ral guanine nucleotide
dissociation stimulator. Proc Natl Acad Sci USA. 91:11089–11093.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Boriack-Sjodin PA, Margarit SM, Bar-Sagi D
and Kuriyan J: The structural basis of the activation of ras by
sos. Nature. 394:337–343. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
64
|
Mitra S, Cheng KW and Mills GB: Rab
GTPases implicated in inherited and acquired disorders. Semin Cell
Dev Biol. 22:57–68. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Bhattacharya M, Anborgh PH, Babwah AV,
Dale LB, Dobransky T, Benovic JL, Feldman RD, Verdi JM, Rylett RJ
and Ferguson SS: Beta-arrestins regulate a Ral-GDS Ral effector
pathway that mediates cytoskeletal reorganization. Nat Cell Biol.
4:547–555. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
66
|
Hao Y, Wong R and Feig LA: RalGDS couples
growth factor signaling to Akt activation. Mol Cell Biol.
28:2851–2859. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Yin J, Pollock C, Tracy K, Chock M, Martin
P, Oberst M and Kelly K: Activation of the RalGEF/Ral pathway
promotes prostate cancer metastasis to bone. Mol Cell Biol.
27:7538–7550. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Rondaij MG, Bierings R, van Agtmaal EL,
Gijzen KA, Sellink E, Kragt A, Ferguson SS, Mertens K, Hannah MJ,
van Mourik JA, et al: Guanine exchange factor RalGDS mediates
exocytosis of weibel-palade bodies from endothelial cells. Blood.
112:56–63. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Anteby EY, Natanson-Yaron S, Hamani Y,
Sciaki Y, Goldman-Wohl D, Greenfield C, Ariel I and Yagel S:
Fibroblast growth factor-10 and fibroblast growth factor receptors
1–4: Expression and peptide localization in human decidua and
placenta. Eur J Obstet Gynecol Reprod Biol. 119:27–35. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Kang K and Song MR: Diverse FGF receptor
signaling controls astrocyte specification and proliferation.
Biochem Biophys Res Commun. 395:324–329. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Goetz R and Mohammadi M: Exploring
mechanisms of FGF signalling through the lens of structural
biology. Nat Rev Mol Cell Biol. 14:166–180. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Jaye M, Schlessinger J and Dionne CA:
Fibroblast growth factor receptor tyrosine kinases: Molecular
analysis and signal transduction. Biochim Biophys Acta.
1135:185–199. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Barnes GL, Kostenuik PJ, Gerstenfeld LC
and Einhorn TA: Growth factor regulation of fracture repair. J Bone
Miner Res. 14:1805–1815. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Grothe C and Nikkhah G: The role of basic
fibroblast growth factor in peripheral nerve regeneration. Anat
Embryol (Berl). 204:171–177. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Pringle NP, Yu WP, Howell M, Colvin JS,
Ornitz DM and Richardson WD: Fgfr3 expression by astrocytes and
their precursors: Evidence that astrocytes and oligodendrocytes
originate in distinct neuroepithelial domains. Development.
130:93–102. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Beenken A and Mohammadi M: The FGF family:
Biology, pathophysiology and therapy. Nat Rev Drug Discov.
8:235–253. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Bono F, De Smet F, Herbert C, De Bock K,
Georgiadou M, Fons P, Tjwa M, Alcouffe C, Ny A, Bianciotto M, et
al: Inhibition of tumor angiogenesis and growth by a small-molecule
multi-FGF receptor blocker with allosteric properties. Cancer Cell.
23:477–488. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Murphy PR and Knee RS: Basic fibroblast
growth factor binding and processing by human glioma cells. Mol
Cell Endocrinol. 114:193–203. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Chang F, Steelman LS, Lee JT, Shelton JG,
Navolanic PM, Blalock WL, Franklin RA and McCubrey JA: Signal
transduction mediated by the Ras/Raf/MEK/ERK pathway from cytokine
receptors to transcription factors: Potential targeting for
therapeutic intervention. Leukemia. 17:1263–1293. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Fredriksson L, Li H and Eriksson U: The
PDGF family: Four gene products form five dimeric isoforms.
Cytokine Growth Factor Rev. 15:197–204. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Reigstad LJ, Varhaug JE and Lillehaug JR:
Structural and functional specificities of PDGF-C and PDGF-D, the
novel members of the platelet-derived growth factors family. FEBS
J. 272:5723–5741. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Kohler N and Lipton A: Platelets as a
source of fibroblast growth-promoting activity. Exp Cell Res.
87:297–301. 1974. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Westermark B and Wasteson A: A platelet
factor stimulating human normal glial cells. Exp Cell Res.
98:170–174. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Ross R, Glomset J, Kariya B and Harker L:
A platelet-dependent serum factor that stimulates the proliferation
of arterial smooth muscle cells in vitro. Proc Natl Acad Sci USA.
71:1207–1210. 1974. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Williams BP, Park JK, Alberta JA,
Muhlebach SG, Hwang GY, Roberts TM and Stiles CD: A PDGF-regulated
immediate early gene response initiates neuronal differentiation in
ventricular zone progenitor cells. Neuron. 18:553–562. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Pietz K, Odin P, Funa K and Lindvall O:
Protective effect of platelet-derived growth factor against
6-hydroxydopamine-induced lesion of rat dopaminergic neurons in
culture. Neurosci Lett. 204:101–104. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Smits A, Kato M, Westermark B, Nister M,
Heldin CH and Funa K: Neurotrophic activity of platelet-derived
growth factor (PDGF): Rat neuronal cells possess functional PDGF
beta-type receptors and respond to PDGF. Proc Natl Acad Sci USA.
88:8159–8163. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Erlandsson A, Enarsson M and
Forsberg-Nilsson K: Immature neurons from CNS stem cells
proliferate in response to platelet-derived growth factor. J
Neurosci. 21:3483–3491. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Peng F, Dhillon N, Callen S, Yao H,
Bokhari S, Zhu X, Baydoun HH and Buch S: Platelet-derived growth
factor protects neurons against gp120-mediated toxicity. J
Neurovirol. 14:62–72. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Yao H, Duan M, Yang L and Buch S:
Platelet-derived growth factor-BB restores human immunodeficiency
virus Tat-cocaine-mediated impairment of neurogenesis: Role of
TRPC1 channels. J Neurosci. 32:9835–9847. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
de la Pena JB and Campbell ZT: RNA-binding
proteins as targets for pain therapeutics. Neurobiol Pain. 4:2–7.
2018. View Article : Google Scholar : PubMed/NCBI
|