Introduction
Osteosarcoma is the most common malignant bone tumor
affecting children and adolescents worldwide, and it has features
of local aggressive growth and a high metastatic potential
(1). It occurs more frequently in
males than in females with an incidence ratio of 1.43:1 (2,3). The
metaphysis of the lower long bones is typically the primary site of
osteosarcoma, and metastases are present in approximately 20% of
cases (4,5). Distant metastases of osteosarcoma,
such as lung metastases, are difficult to control and usually have
a poor prognosis (6). The etiology
of osteosarcoma is complex and linearly associated with increasing
doses of radiation (7). Certain
rare heritable cancer predisposition syndromes are also associated
with an increased risk of osteosarcoma (8). The majority of patients with
osteosarcoma suffer pain in the affected region, partially from
localized swelling, motion limitation and pathological fractures
(4,9). Currently, chemotherapeutic agents,
such as cisplatin, methotrexate, doxorubicin, etoposide and
ifosfamide, are usually administered before or after surgery to
prevent the tumor from spreading throughout the body (5,10).
However, their outcomes are not satisfactory and the 5-year overall
survival rate of affecting patients is estimated at approximately
25% (11). Patients with distant
metastases have a poorer prognosis, with a 5-year survival rate of
only 20% (6). Therefore, novel
treatment strategies are urgently required to improve the outcome
and quality of life of patients with osteosarcoma.
Tea has a history with dates back centuries, and has
become the most popular beverage in Asia, particularly in China. By
using fresh or fermented leaves of Camellia sinensis O.
Kuntze, tea drinks have unique flavors and potential health
benefits, such as anti-fatigue, anti-hyperlipidemia, and
anti-hypercholesterolemia activities (12–14).
Epidemiological surveys showed that consuming more than 10 cups of
tea per day could prevent cancer occurrence, indicating the
anti-cancer potential of tea (15,16).
As a representative bioactive component of tea, theabrownin (TB)
has been reported to possess anticancer activity against lung
carcinoma and osteosarcoma in previous studies by the authors
(17,18). TB is a reddish-brown pigment derived
from polyphenol components by oxidation, and it has the advantages
of high water-solubility and low toxicity over other pigments. The
authors have previously demonstrated that TB exerts potent
pro-apoptotic effects against U2OS osteosarcoma cells by triggering
DNA damage through the p53 signaling pathway, whereas it exhibited
no toxicity on normal tissue in vivo or on normal cells
[bone marrow-derived mesenchymal stem cells (BMSCs)] (17). These findings indicate that TB may
be a promising candidate for osteosarcoma therapy.
Considering that both U2OS cells and BMSCs are p53
wild-type cells, the activation of the p53 signaling pathway may
not be the only mechanism of TB, since it cannot explain the
diverse effectiveness of TB on U2OS cells and normal cells.
Therefore, it was hypothesized that TB may have other mechanisms of
action in addition to its p53-mediated mechanisms. To examine this
hypothesis, the present study employed xenograft zebrafish samples
to conduct RNA sequencing and performed molecular experiments for
further verification and investigation. To date, only the
p53-mediated mechanism has been reported to be associated with the
anticancer effects of TB. Thus, the present study presents a novel
report on the anti-osteosarcoma mechanisms of TB.
Materials and methods
Chemicals and reagents
TB (>90% purity) was purchased from Theabio Co.,
Ltd. Fetal bovine serum (FBS) was purchased from CellMax.
Phosphate-buffered saline (PBS) and McCoys 5A medium were purchased
from Gibco; Thermo Fisher Scientific, Inc. CCK-8, phosphatase
inhibitor cocktail, All-in-One cDNA Synthesis SuperMix and 2X
SYBR-Green qPCR Master Mix were purchased from Bimake. RNAiso Plus
was purchased from Takara Biotechnology Co., Ltd. Crystal violet
(0.1%) was purchased from Sigma-Aldrich; Merck KGaA. Nocodazole was
purchased from Selleck Chemicals. Propidium iodide/RNase staining
buffer was purchased from BD Biosciences. Acti-stain™ 535
phalloidins (cat. no. PHDR1) was purchased from Cytoskeleton, Inc.
Tris-buffered saline, Tween-20, Triton X-100, paraformaldehyde and
bovine serum albumin (BSA) were purchased from Sangon
Biotechnology, Inc. ProLong Diamond Antifade Mountant with DAPI,
the Pierce BCA protein assay kit and RIPA lysis buffer were
purchased from Thermo Fisher Scientific, Inc. Polyvinylidene
fluoride membranes and Immobilon Western Chemiluminescent HRP
substrate were purchased from Merck KGaA. Proteinase inhibitor
cocktail was purchased from Roche Diagnostics. Transwell chamber
systems and Matrigel were purchased from Corning, Inc. Recombinant
human TNF-α (cat. no. 300-01A) was purchased from PeproTech Inc.
Anti-tubulin antibody-microtubule marker (cat. no. ab195883) was
purchased from Abcam. β-actin (cat. no. A3854) was purchased from
Sigma Chemical Co. All the primary antibodies (Vimentin (cat. no.
5741), Slug (cat. no. 9585), Snail (cat. no. 3879), Claudin-1 (cat.
no. 13255), ZEB1 (cat. no. 3396), E-Cadherin (cat. no. 3195),
N-Cadherin (cat. no. 13116), IKKα (cat. no. 11930), IKKβ (cat. no.
8943), IκBα (cat. no. 4814), NF-κB (cat. no. 8242), phospho-IKKα/β
(Ser176/180) (cat. no. 2697), phospho-IκBα (Ser32) (cat. no. 2859),
Histone-H3 (cat. no. 4499) used for western blot analysis were
purchased from Cell Signaling Technology, Inc. Fluorescein
(FITC)-conjugated AffiniPure Donkey Anti-Rabbit IgG (H+L) (cat. no.
711-095-152) and Peroxidase AffiniPure Goat Anti-Mouse IgG (H+L)
(cat. no. 115-035-062) was purchased from Jackson ImmunoResearch
Laboratories, Inc.
Xenograft experiment and RNA
sequencing (RNA-seq)
The xenograft experiment on larval zebrafishes was
conducted in a previous study by the authors (17). Briefly, a xenograft model of
osteosarcoma was established by the microinjection of U2OS cells to
larval zebrafishes at the age of 3 days followed by the orally
administration of TB at 2.13 to 21.3 µg/ml. Following 24 h of
treatment, all fish at the age of <5 days were anesthetized by
freezing at 0°C for 10 min to observed and detect tumor growth,
followed by sacrificing using liquid nitrogen. The whole larval
zebrafishes were used for RNA extraction as follows: i) The fish
bodies in each group [model group and TB-treated group (2.13
µg/ml)] were collected into an Eppendorf tube and mixed with 0.5 ml
of TRIzol reagent for cell lysis; ii) the mixture was homogenized
and supplemented with 0.5 ml chloroform, followed by centrifugation
at 12,000 × g for 10 min at 4°C to separate the RNA and protein;
and iii) the separated aqueous phase was collected and supplemented
with the same amount of isopropanol, followed by centrifugation at
12,000 × g for 10 min at 4°C to deposit the RNA. The concentration
of total RNA was quantified using a NanoDrop 2000 spectrophotometer
(Thermo Fisher Scientific, Inc.). For cDNA library preparation, 3
µg total RNA was captured by Dynabeads Oligo (Life Technologies;
Thermo Fisher Scientific, Inc.) and sheared to fragments of ~200
bp. Reverse transcription was performed using the Superscript III
cDNA Synthesis Kit (Life Technologies; Thermo Fisher Scientific,
Inc.). The cDNA was end-repaired, A-tailed and ligated to Illumina
sequencing adapters and amplified by PCR. Library preparation was
performed using the TruSeq RNA LT V2 Sample Prep Kit (Illumina).
Sequencing was performed on the Illumina XTen Sequencing System
(Illumina). The raw reads were processed by removing the adaptors,
sequences with uncertain bases, low-quality sequence, and sequences
of <50 bp to generate clean reads. The clean reads from the
Fastq files were mapped to human reference genome using Spliced
Transcripts Alignment to Reference (STAR) software (version
2.7.1a). Differential expression analysis of the different groups
was performed with biological replicates using DESeq software
(version 1.30.0). A P-value of 0.05 was set as the threshold for
significant differential expression.
Cell line and culture
The human osteosarcoma cell line, U2OS, without
mycoplasma infection was identified and provided from Shanghai Cell
Bank of Chinese Academy of Sciences. The cells were cultured in
McCoys 5A medium containing 10% FBS at 37°C in humidified
atmosphere containing 5% CO2. The medium was changed
daily and the cells at logarithmic growth phase were prepared for
the subsequent experiments.
Reverse transcription-quantitative PCR
(RT-qPCR)
For verifying the results of RNA sequencing, qPCR
assay was performed. Total RNA was extracted from the U2OS cells
with and without TB treatment using RNAiso Plus (Takara
Biotechnology Co., Ltd.), followed by cDNA synthesis using an
All-in-One cDNA Synthesis SuperMix. Subsequently, the 2X SYBR-Green
qPCR Master Mix kit was used for transcript quantification with
specific primers. All the reactions were set up according to the
manufacturers instructions. At the end of each reaction, a melting
curve analysis was performed. Data were normalized to the
expression of ACTIN and presented using the 2−Δ∆Cq
method (Table I) (19).
 | Table I.Primer sequences used for
RT-qPCR. |
Table I.
Primer sequences used for
RT-qPCR.
| Gene | Forward primer | Reverse primer |
|---|
| ACTIN |
5′-ATAGCACAGCCTGGATAGCAACGTAC-3′ |
5′-CACCTTCTACAATGAGCTGCGTGTG-3′ |
| MYH2 |
5′-CTGAGGGAGGAGCGACTCT-3′ |
5′-CTCGGGCTTATACACAGGCA-3′ |
| MYH7 |
5′-ACTGCCGAGACCGAGTATG-3′ |
5′-GCGATCCTTGAGGTTGTAGAGC-3′ |
| TUBA1B |
5′-GTACCGTGGTGACGTGGTTC-3′ |
5′-CTTGGCATACATCAGGTCAA-3′ |
| TUBB2B |
5′-ATCAGCAAGATCCGGGAAGAG-3′ |
5′-CCGTGTCTGACACCTTGGGT-3′ |
Cell viability assay
The viability of the U2OS cells was determined by
CCK-8 assay. The cells were seeded on 96-well plates at a density
of 1×104 cells/well in 200 µl McCoys 5A medium contained
10% FBS. Following adherence for 24 h, the cells were respectively
treated with TB at 0, 10, 20, 30, 40, 50, 60, 70, 80, 90 and 100
µg/ml for 24, 48 and 72 h. Following treatment, CCK-8 was added and
incubated with the cells at 37°C for 2 h. The optical density was
measured at 450 nm using Synergy H1 microplate reader (BioTek
Instruments, Inc.) and calculated as follows: Inhibitory rate
(%)=[1-(TB-treat OD-blank OD)/(untreated OD-blank OD)] ×100%.
Clone formation assay
The U2OS cells were seeded into 6-well plates in
triplicate (1,000 cells/well) for 48 h adherence and then treated
with TB at 0, 5 and 10 µg/ml for 3 h. Following treatment, the
cells were washed with PBS and the medium was replaced with fresh
medium. In the following 8 days, the cells were treated TB for a
further 3 times. The cell colonies were fixed with paraformaldehyde
(4%) at 4°C for 20 min and stained with 0.1% crystal violet at room
temperature for 20 min (Sigma-Aldrich; Merck KGaA).
Cell cycle analysis
The U2OS cells were seeded in 10 cm dishes
(1.5×106 cells/dish) for 24 h adherence and were
synchronized by 10 µM Nocodazole for 8 h. The cells were then
treated with TB at 12.5, 25 and 50 µg/ml for 12, 24 and 48 h.
Subsequently, the cells were washed with PBS and fixed with 70%
ice-cold ethanol overnight at −20°C. Fixed cells were washed in PBS
and stained with Propidium iodide/RNase Staining Buffer for 15 min
at room temperature, followed by analysis using a BD Accuri™ C6
flow cytometer (BD Biosciences).
Immunofluorescence
The cell cytoskeleton of U2OS cells treated with TB
was observed using Anti-Tubulin antibody-Microtubule Marker and
Acti-stain™ Fluorescent Phalloidins. Briefly, the cells
(4×104) were fixed with 4% paraformaldehyde for 15 min
at room temperature and then permeabilized with PBS containing 0.1%
Triton X-100 at 4°C for 10 min. The cells were blocked with 1% BSA
in tris-buffered saline-Tween 20 (TBST) solution at 4°C for 1 h,
and were then incubated with the anti-tubulin antibody (1:100, v/v)
and anti-NF-κB antibody (1:400, v/v) at 4°C overnight.
Subsequently, Fluorescein (FITC)-conjugated AffiniPure Donkey
Anti-Rabbit IgG (H+L) (1:2,000, v/v) was added to the cells
incubated with anti-NF-κB antibody at 4°C in dark for 2 h. The
cells were incubated with Acti-stain™ 535 Phalloidin (100 nM) at
room temperature for 30 min. Finally, the cells on coverslip slides
were stained with ProLong Diamond Antifade Mountant with DAPI at
room temperature overnight and observed under a fluorescence
microscope (Zeiss AG). TNF-α (10 ng/ml) was used to stimulate the
microfilament and microtubule formation of U2OS cells for 12 h as a
model, and confocal laser scanning microscopy was employed to
observe the effects of TB. The cells incubated with Phalloidin were
observed under a Leica TCS SP5 confocal laser scanning microscope
(Leica Microsystems GmbH) at settled excited wavelength (DAPI 405
nm, FITC 488 nm) and consistent detecting wave band (DAPI 420–470
nm, FITC 503–550 nm).
Cell migration and invasion assay
Cell migration assay and invasion assay were
performed using a Transwell chamber system without and with
Matrigel, respectively (Corning Costar, Inc.). The cells were
starved in serum-free medium for 12 h and seeded into the upper
chambers at 5×104 cells/well in serum-free medium.
Complete medium containing TB was added to the lower chambers and
incubated with the upper chambers at 37°C for 12 h without Matrigel
and for 24 h with Matrigel. The migrated cells were then fixed with
4% paraformaldehyde and stained with 0.1% crystal violet at room
temperature for 20 min for counting under an optical microscope
(Zeiss AG).
Wound healing assay
The cells were seeded into 6-well plates
(5×105 cells/well) and cultured for 24 h, followed by ab
artificial scratch being made in a cross form using a micropipette
tip. The cells were then cultured with fresh medium containing 0.5%
FBS and TB at 10, 20 and 40 µg/ml. The cells were observed and
imaged at 5 different time points (0, 12, 24 and 48 h) under an
inverted microscope (Zeiss AG). The wound area was calculated using
Image J 1.47 software. Each experiment was conducted in
triplicate.
Western blot analysis
Total proteins were extracted from the TB-treated
U2OS cells (1.5×106) using RIPA lysis buffer with
proteinase inhibitor cocktail and phosphatase inhibitor cocktail
for 30 min on ice. The proteins were quantified using the Pierce
BCA Protein Assay kit, separated by electrophoresis and transferred
onto polyvinylidene fluoride membranes (30 µg protein per lane).
The membranes were blocked with 5% non-fat milk or 5% BSA for 2 h
at 4°C, followed by overnight incubation at 4°C with primary
antibodies (Vimentin, Slug, Snail, Claudin-1, ZEB1, E-Cadherin,
N-Cadherin, IKKα, IKKβ, IκBα, NF-κB, phospho-IKKα/β, phospho-IκBα
and Histone-H3) at dilution rate of 1:20,000 (v/v). The membranes
were then washed 3 times with TBST for 5 min and incubated with
Peroxidase AffiniPure Goat Anti-Mouse IgG (H+L) (1:20,000, v/v) at
4°C for 2 h. Target proteins were visualized using Immobilon
Western Chemiluminescent HRP Substrate and detected using X-ray
film and scanned. The densitometry of each blot was tested by using
ImageJ software (version 1.8.0).
Statistical analysis
Data are expressed as the means ± SD and analyzed by
a normal distribution test, followed by one-way ANOVA coupled with
Dunnetts multiple comparisons test. All analyses were performed
using GraphPad Prism 8.0. A value of P<0.05 was considered to
indicate a statistically significant difference.
Results
RNA-seq-based transcriptional action
of TB
The tumor-inhibitory effect of TB on U2OS cells in
zebrafish has been previously reported (17). In the present study, by using RNA
samples from model group and TB (2.13 µg/ml)-treated group, RNA-seq
analysis was performed by Gene Ontology analysis. The results
revealed that TB regulated a number of biological processes, in
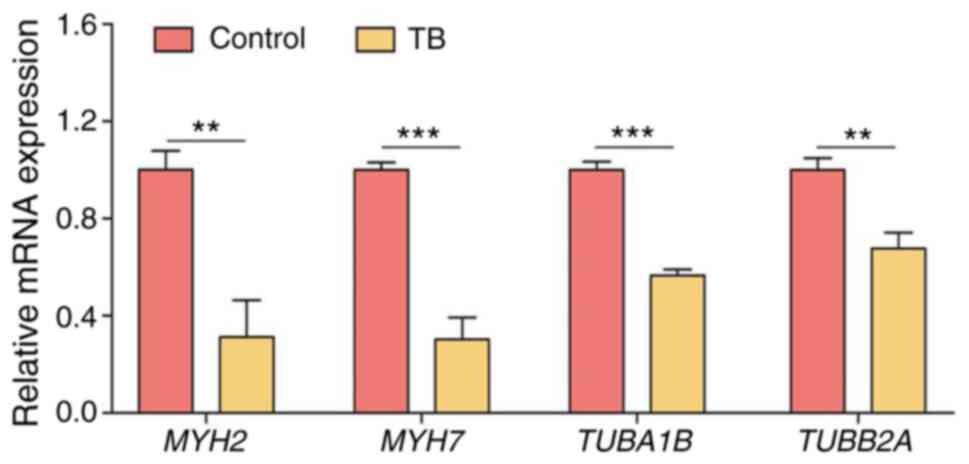
which the microtubule bundle formation was inhibited (Fig. S1). As shown in Table II, a number of genes involved in
the microtubule bundle formation of U2OS cells, including MYH2,
MYH7, TUBA1B and TUBB2A, were significantly
downregulated by treatment with TB, indicating the inhibition of
microtubule bundle formation by TB. To verify this result, RT-qPCR
was performed and a similar result was obtained, in that the
expression levels of all these genes were significantly
downregulated following treatment with TB (each P<0.01)
(Fig. 1).
 | Table II.Gene expression in zebrafish differed
significantly between the model group and TB-treated group (model
vs. TB) in RNA sequencing analysis. |
Table II.
Gene expression in zebrafish differed
significantly between the model group and TB-treated group (model
vs. TB) in RNA sequencing analysis.
| Gene name | log2 (fold
change) | P-value | Adjusted
P-value | Direction to
model |
|---|
| MYH2 | −1.868 | 3.24E-116 | 1.86E-114 | Down |
| MYH7 | −1.046 | 1.08E-109 | 5.57E-108 | Down |
| TUBA1B | −1.234 | 7.82E-17 | 8.78E-16 | Down |
| TUBB2A | −4.728 | 3.41E-29 | 6.30E-28 | Down |
Anti-colony formation and cell
cycle-arresting effects of TB
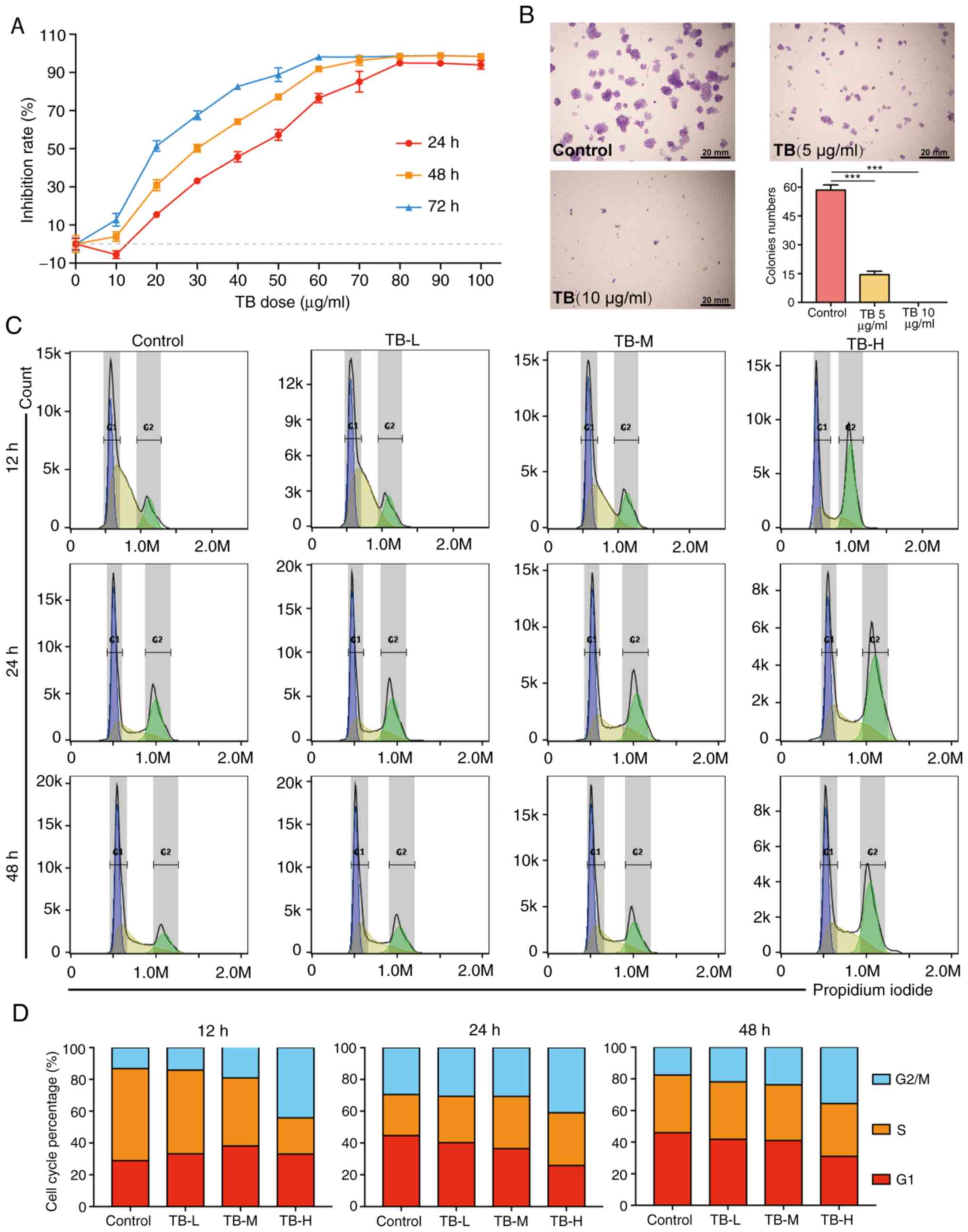
As shown in Fig. 2A,
the viability of the U2OS cells was significantly inhibited by TB
and the inhibitory rate was increased with the increasing
concentrations of TB from 10 to 100 µg/ml, and with the increasing
treatment duration from 24 to 72 h. It was found that TB exerted
anti-proliferative effects in a dose-dependent and time-dependent
manner. As shown in Fig. 2B, the
results of colony formation assay revealed that TB at 5 µg/ml
significantly decreased the U2OS cell colony numbers (>50
cells/colony; P<0.001), and there was almost no colony formation
observed following treatment with TB at 10 µg/ml. As shown in
Fig. 2C and D, TB blocked the cell
cycle progression of U2OS cells at the G2/M phase from
12 to 48 h. With the increasing concentration of TB, the
G2/M phase cell percentage progressively increased when
compared with the control.
Cytoskeleton-inhibitory effects of
TB
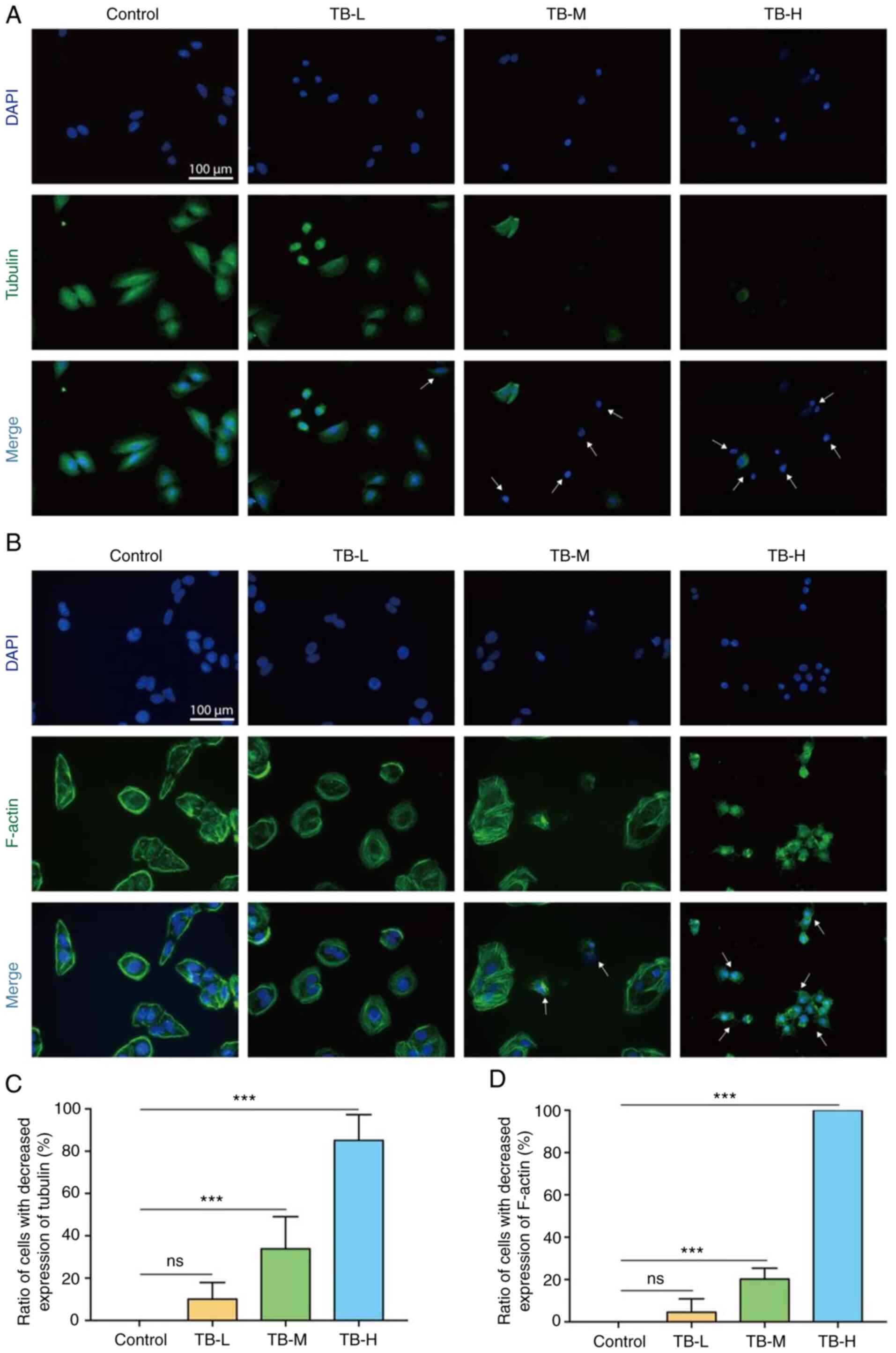
According to the result of RNA-seq analysis, the
inhibition of microtubule bundle formation was the primary effects
through which TB affected U2OS cells and zebrafish. To confirm this
effect, immunofluorescence using antibodies against cytoskeleton
proteins was performed. As shown in Fig. 3A and B, the cellular expression of
Tubulin and F-actin was markedly decreased, with the visible loss
of microfilament and microtubule bundles induced by TB at 12.5 to
50 µg/ml, indicating that TB exerted cytoskeleton-inhibitory
effects on the U2OS cells. This effect was significantly exerted by
TB at 25 (TB-L) and 50 µg/ml (TB-H) (each P<0.001) (Fig. 3C and D).
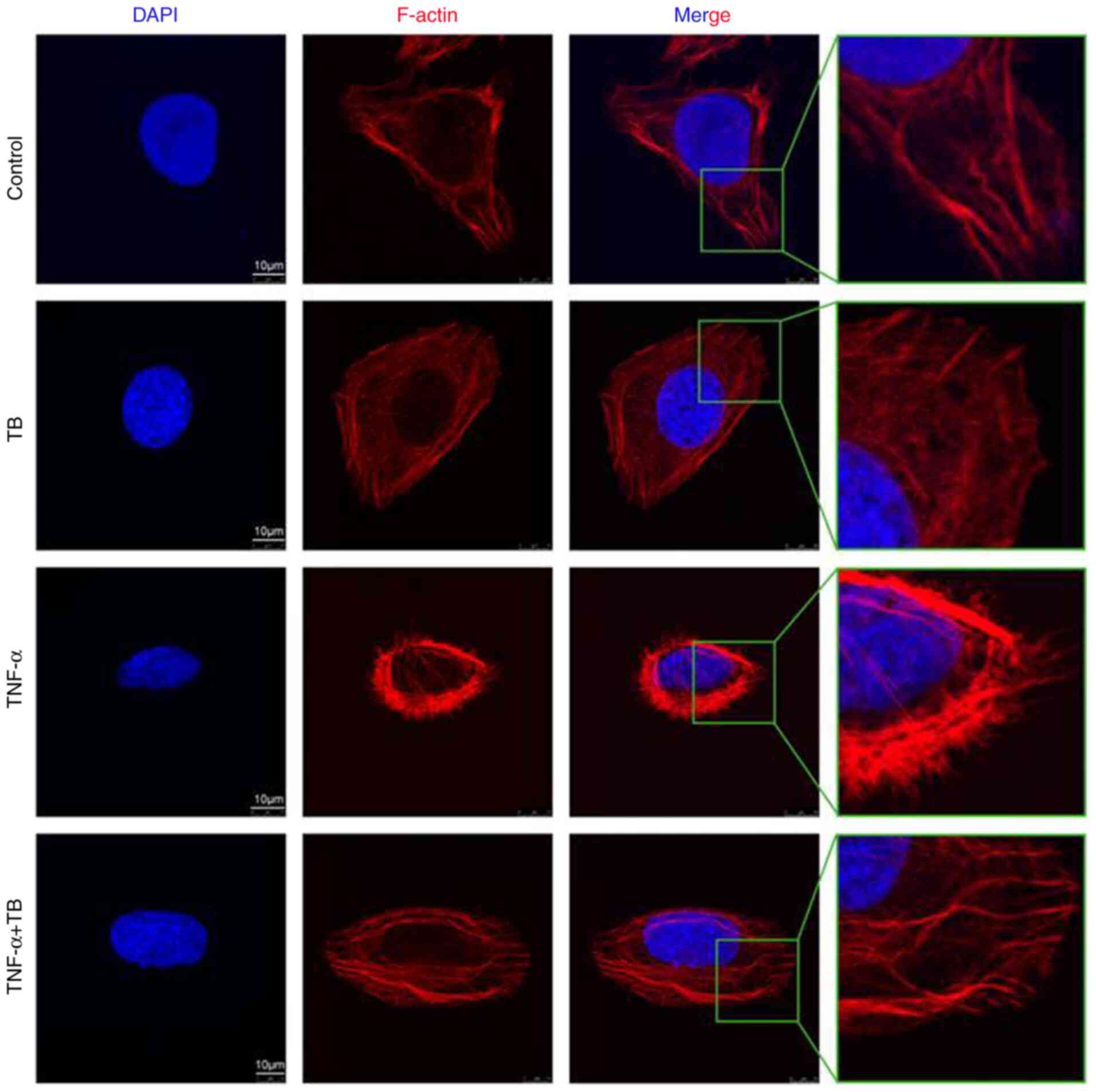
To further confirm the cytoskeleton-inhibitory
effects of TB, A TNF-α-induced microfilament- and
microtubule-formation model was applied. As shown in Fig. 4, the fluorescence intensity of
F-actin and the formation of microfilaments were evidently
increased with TNF-α pre-treatment, and the phenotype was reversed
to normal by treatment with TB, verifying the inhibitory effects of
TB on the cytoskeleton.
Anti-migratory and anti-invasive
effects of TB
The integrities of microfilament and microtubule
formation are crucial for the functions of cell migration and
invasion, which may be disrupted by TB in U2OS cells. To determine
whether TB exerts inhibitory effects on the migration and invasion
of the U2OS cells, Transwell and wound healing assays were
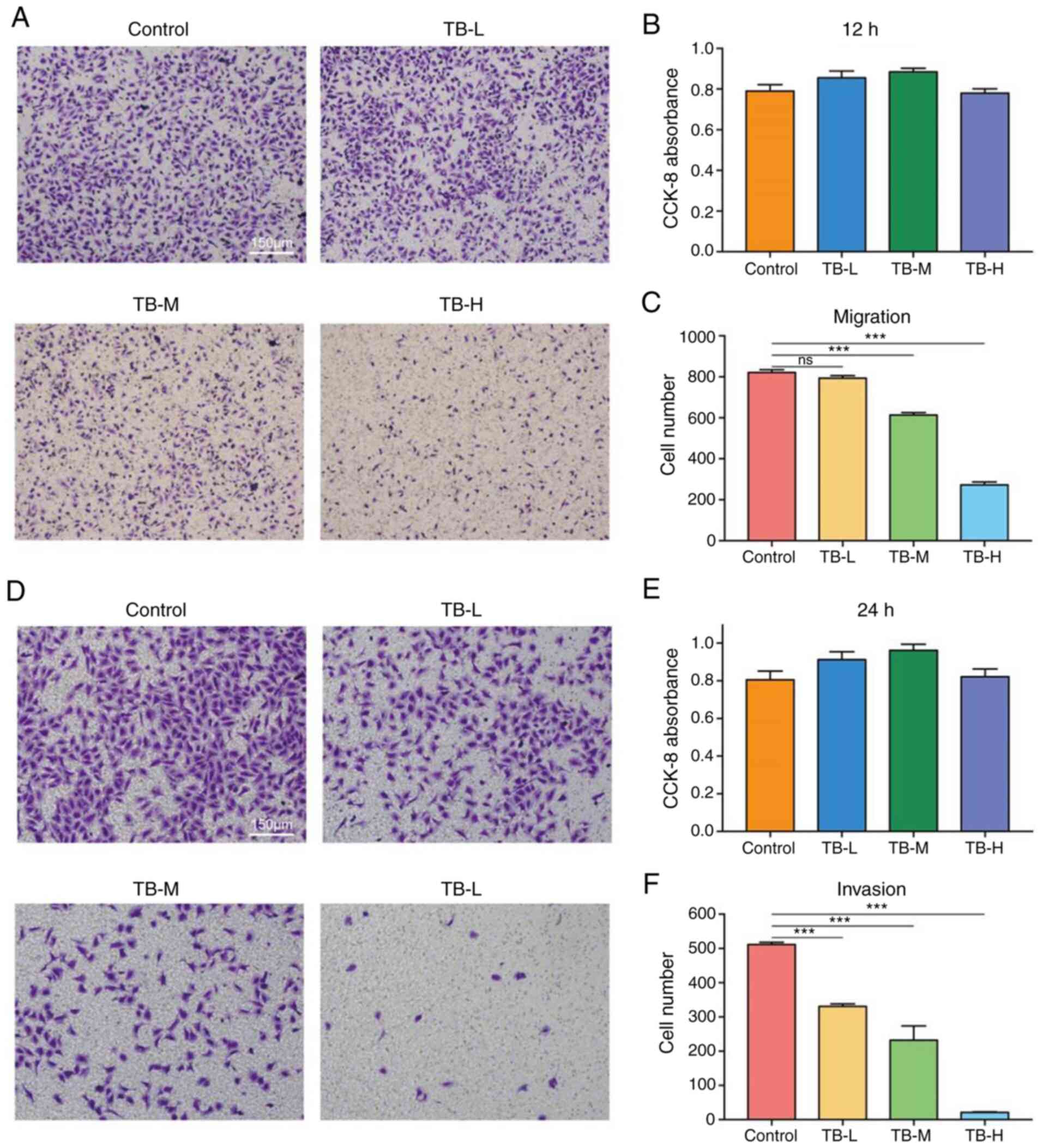
performed. As shown in Fig. 5, TB
significantly inhibited the migration of U2OS cells following 12 h
of treatment at the middle and high doses, and significantly
inhibited U2OS cell invasion following treatment for 24 h treatment
at the low, middle and high doses, with significant decreases
observed in the numbers of migrated and invaded cells (each
P<0.001). Moreover, the results of CCK-8 assays revealed that TB
at these concentrations had no significant effect on the viability
of the U2OS cells, indicating that the anti-migratory and
anti-invasive effects of TB were independent on its
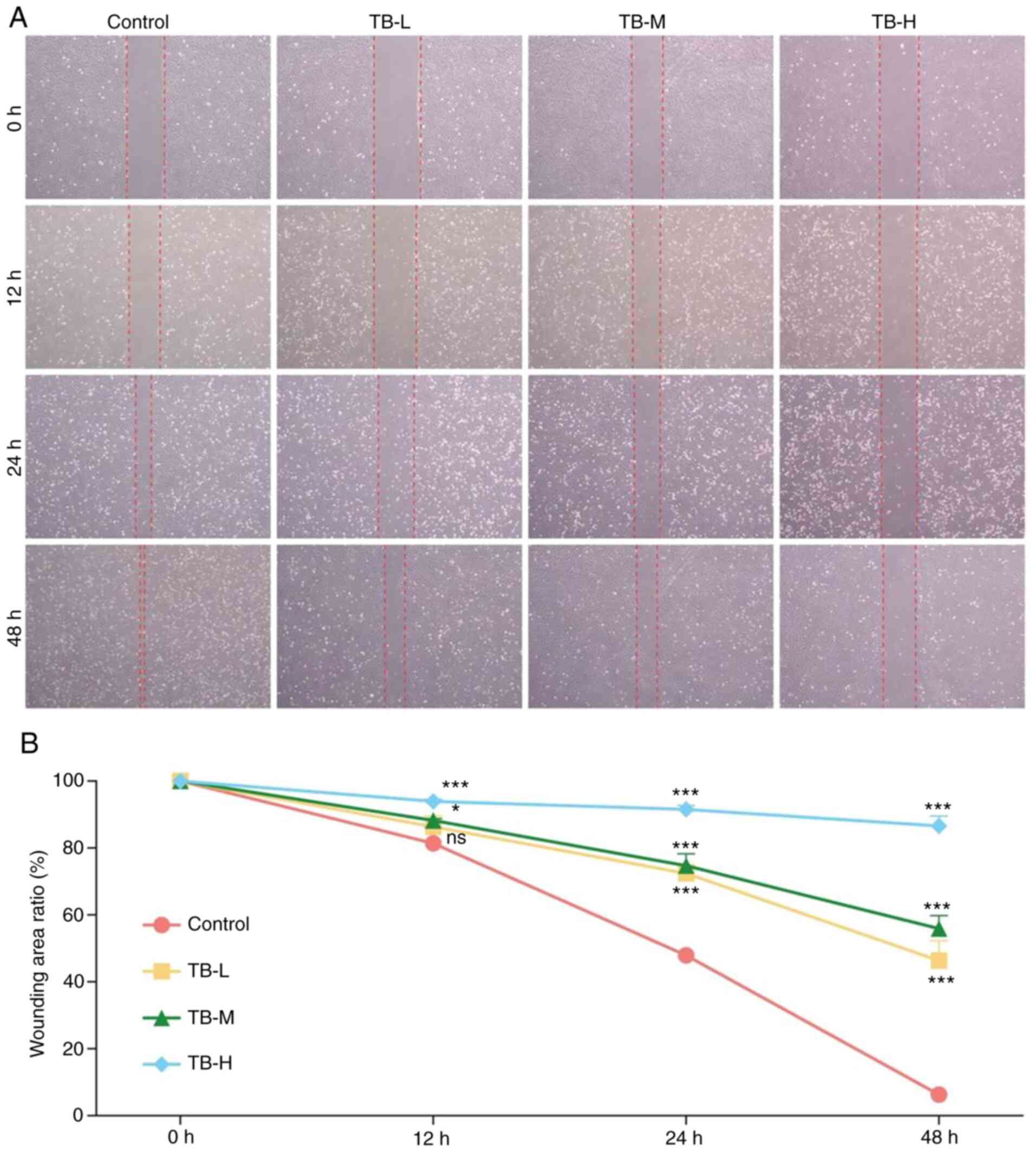
anti-proliferative effects. As shown in Fig. 6, TB significantly inhibited the
wound healing ability of the U2OS cells following 12 h of treatment
at the middle and high doses, and following 24 and 48 h of
treatment at the low, middle and high doses (each P<0.05). These
results indicated that TB inhibited U2OS cell migration through the
blockage of wound closure in a dose-dependent manner.
Molecular mechanisms of TB
To clarify the underlying mechanisms of the
anti-migratory and anti-invasive effects of TB, western blot
analysis was conducted on the proteins involved in
epithelial-mesenchymal transition (EMT) and the related pathways.
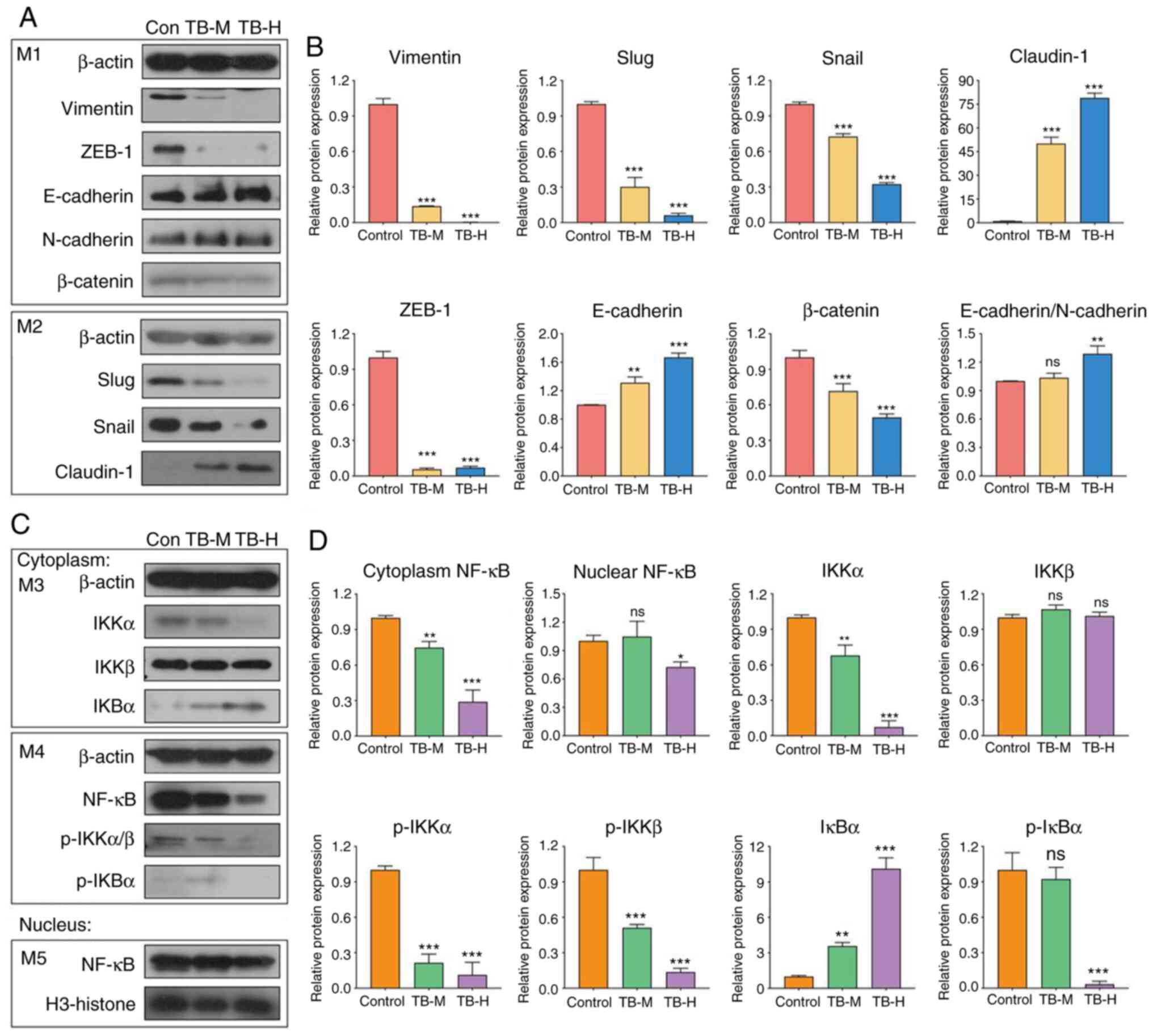
As shown in Fig. 7A and B, the
expression levels of vimentin, Slug, Snail-1, ZEB-1 and β-catenin
were significantly downregulated, while those of Claudin-1 and
E-cadherin were significantly upregulated by TB at 25 and 50 µg/ml,
indicating that the anti-migratory and anti-invasive effects of TB
were mediated by the reversal of EMT. Moreover, the levels of key
molecules in the NF-κB pathway, including cytoplasmic NF-κB,
nuclear NF-κB, IKKα, phospho-IKKα (p-IKKα), phospho-IKKβ (p-IKKβ),
and phospho-IκBα (p-IκBα), were all significantly downregulated by
TB (Fig. 7C and D), indicating that
NF-κB pathway may participate in the mechanisms of TB. As shown in
Fig. S2, the immunofluorescent
data verified that TB inhibited NF-κB in U2OS cells. The
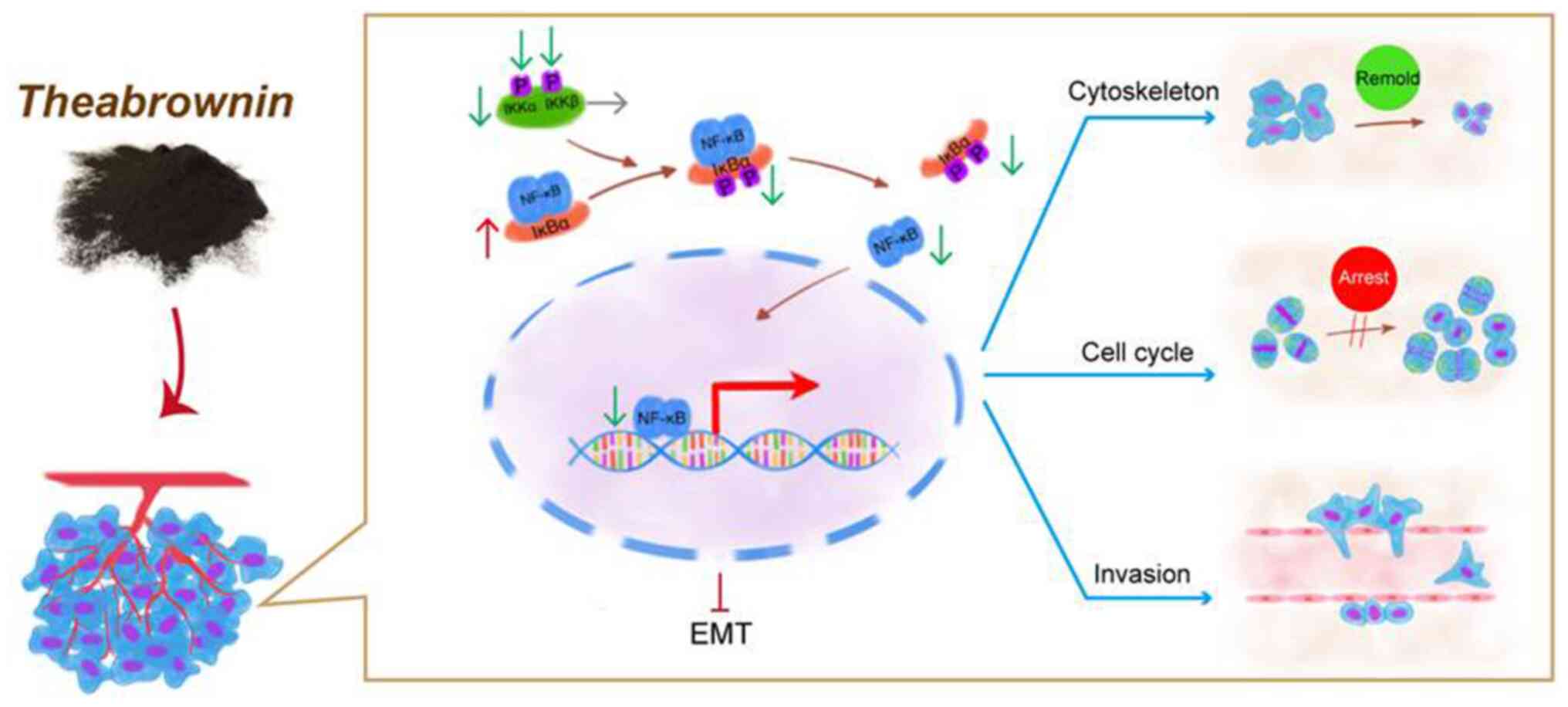
above-mentioned results revealed that the mechanisms of TB were
mediated by the reversal of EMT and associated with the inhibition
of NF-κB signaling (Fig. 8).
Discussion
In a previous study, the authors demonstrated that
the pro-apoptotic and tumor-inhibitory effects and p53 signaling
pathway-dependent mechanism of TB on osteosarcoma, providing a
promising candidate pro-apoptotic anti-osteosarcoma agent (17). Considering the high metastatic
potential of osteosarcoma, both pro-apoptotic and anti-metastatic
effects are important and needed for anti-osteosarcoma therapy.
However, to the best of our knowledge, there are no available drugs
that effectively inhibit the metastases of osteosarcoma. For the
first time, to the best of our knowledge, the present study
evaluated the anti-metastatic potential of TB by exploring its
inhibitory effects on the metastasis-associated activities of
osteosarcoma cells and the related mechanisms. Biologically, it was
found that TB significantly inhibited the colony formation, cell
cycle transition, microtubule and microfilament formation,
migration and invasion of U2OS osteosarcoma cells, indicating its
anti-migratory and anti-invasive effects. On a molecular level, it
was revealed that the anti-migratory and anti-invasive mechanisms
of TB was mediated by the reversal of EMT and associated with the
inhibition of NF-κB signaling (Fig.
8). The innovative contribution of the present study is the
discovery of the cytoskeleton-specific anti-migratory and
anti-invasive effects and mechanisms of TB, which provide a
promising candidate for both pro-apoptotic and anti-metastatic
purposes for the treatment of osteosarcoma.
EMT was originally described in the early 1980s as a
biological process through which epithelial cells lose their
epithelial characteristics and acquire mesenchymal characteristics
(20). During EMT, epithelial cells
lose their junctions and apical-basal polarity, reorganize their
cytoskeleton, undergo a change in cell shape and alter their gene
expression, and increase the motility and invasiveness of
individual cells (21,22). EMT is activated during wound
healing, fibrosis and cancer progression (23,24). A
number of studies have indicated that EMT plays a vital role in the
metastatic initiation of several types of cancer, such as
osteosarcoma, by enabling tumour cells to gain invasive properties
and metastatic growth characteristics (25). The major events in EMT are the loss
of E-cadherin, the translocation of β-catenin from the cell
membrane to the nucleus, and the activation of vimentin, induced by
transcription factors, such as Snail-1, Slug and ZEB-1 (26,27).
E-cadherin is a 120 kDa Ca2+-dependent transmembrane
glycoprotein present on the surface of epithelial cells and
mediates cell-cell adhesion at adherens junctions through
homophilic binding, thus maintaining epithelial cellular adhesion
and integrity to support the tissue architecture (28). The cytoplasmic domain of E-cadherin
interacts with α, β and γ catenins to act on the actin cytoskeleton
in cells (29). E-cadherin
switching is indispensable during EMT, and the loss or suppression
of E-cadherin expression leads to cancer development and
progression, as well as metastasis (30–32).
Moreover, as a major cytoskeletal protein in the large intermediate
filament, vimentin is important for the structural integrity of
tumour cells and is often overexpressed at the end stages of
progression in EMT, representing the highly proliferative and
invasive characteristics of tumour cells (33). Conversely, the knockdown of vimentin
with antisense oligonucleotides reduces cell motility (34). In the present study, the expression
of E-cadherin and E-cadherin/N-cadherin was upregulated, and that
of vimentin and β-catenin was downregulated by TB, indicating that
the mechanisms of TB are mediated by the suppression of EMT. Snail
proteins, including Snail-1 and Slug (also known as Snail-2), are
zinc finger-containing transcription factors that act as repressors
of E-cadherin by binding to the E-box (5′-CACCTG-3′) in the
E-cadherin promoter (35). Snail
and Slug are overexpressed in osteosarcoma and induce EMT by
downregulating E-cadherin, leading to an increase in cell
migration, invasion and tumour progression of osteosarcoma
(35–37). The ZEB family (ZEB-1 and ZEB-2) is
another group of zinc finger transcription factors targeting the
E-box and E-box-like DNA sequences in the target gene promoters
(38). ZEB proteins induce EMT by
downregulating E-cadherin, which is overexpressed, and plays
critical roles in cell proliferation, migration and progression of
osteosarcoma (39,40). In the present study, TB
downregulated the expression of Snail-1, Slug and ZEB-1, indicating
that Snails/ZEB-E-cadherin/β-catenin are the targets of TB in
osteosarcoma treatment.
Emerging evidence has indicated that the NF-κB
signaling pathway plays central roles in the control of cell
growth, apoptosis, stress response, and several other physiological
processes (41,42). It protects tumour cells from
apoptosis, while also supporting cell invasion, metastasis, and
angiogenesis of tumour cells (43,44).
In addition, the activation of the NF-κB pathway has been confirmed
as an essential step towards EMT in tumor cells. NF-κB directly
leads to EMT in tumor cells by activating the transcription of
SNAIL and ZEB-1, resulting in E-cadherin
suppression and vimentin overexpression (33,45,46).
The underlying mechanisms of the association between NF-κB and EMT
have been clarified, in which the phosphorylation of IκB increases
the abundance of nuclear-localized NF-κB to promote the expression
of SNAIL and ZEB-1 (47). Therefore, the NF-κB signaling
pathway is essential for tumor invasion and metastasis via EMT
(48). Conversely, the inhibition
of the NF-κB pathway can induce a 10-fold reduction in the
metastasis of cancer due to the blockage of EMT (33,49).
In the present study, TB inhibited the NF-κB pathway by
downregulating NF-κB, IKKα, p-IKKα, p-IKKβ and p-IκBα in U2OS
cells, resulting in EMT blockade with the downregulation of
vimentin, Slug, Snail-1, ZEB-1 and β-catenin (Fig. 7). The cytoplasm and nuclear NF-κB
data indicated that TB not only blocked the transition of NF-κB
from the cytoplasm to the nucleus, but also inhibited the total
expression of NF-κB. The cell immunofluorescent data provided
evidence of this conclusion (Fig.
S2). The results indicated that the NF-κB pathway may
participate in the EMT-mediated anti-osteosarcoma mechanisms of TB
(Fig. 8). A key question remains
however, as to what connects TB and NF-κB. Most likely, receptors
on the osteosarcoma cell membrane, such as CXCR4, endothelin-1
receptors (ETA), adenosine A3 receptor (A3AR), Toll-like
receptor 4 (TLR4) and estrogen receptor β (ERβ), are the answer.
CXCR4 is an SDF-1 receptor overexpressed in human osteosarcoma
cells that mediates the migration of osteosarcoma cells by inducing
IκB kinase (IKKα/β) phosphorylation, IκB phosphorylation, p65
phosphorylation and κB-luciferase activity in the NF-κB pathway
(50). As an ET-1-specific
receptor, ETA controls the invasive ability of
osteosarcoma cells through matrix metalloproteinases (MMPs; MMP-2
and MMP-9) and NF-κB transcription factors, and targeting this
receptor may prove to be a significant treatment strategy for
osteosarcoma (51). A3AR expression
is lower in human osteosarcoma tissues than in normal tissues, and
it acts as a suppressor of osteosarcoma migration and progression
by inhibiting the PKA/Akt/NF-κB axis (52). TLR4 is an upstream receptor of the
MAPK/NF-κB pathway in human osteosarcoma cells, and the inhibition
of TLR4 expression exerts suppressive effets on osteosarcoma
(53). ERβ has been found to exert
evident anti-tumor effects on osteosarcoma cells by regulating the
integrin, PI3K/Akt and NF-κB signal pathways, and the viability,
migration and invasion of U2OS cells can be significantly inhibited
by ERβ agonists (54). These
receptors may be potential targets of TB for the modulation of the
NF-κB pathway in U2OS cells, warranting further investigation.
In conclusion, to the best of our knowledge, the
present study is the first to demonstrate the inhibitory effects of
TB on the cytoskeleton-dependent cell cycle, migration and invasion
of human osteosarcoma cells. The mechanisms of TB were found to be
mediated by the suppression of EMT, which may be associated with
the NF-κB pathway. Therefore, TB can be regarded as an
anti-metastatic agent for the treatment of osteosarcoma.
Considering that only one osteosarcoma cell line was used in the
present study, further studies are required to determine the
anti-metastatic effects of TB on other tumor cell lines. In
particular, in vivo studies are also required in the future
to verify these effects of TB. Taken together, the findings of the
present study provide novel evidence of the anti-osteosarcoma
effects of TB and contribute to the development of natural
product-derived anti-metastatic agents for osteosarcoma
therapy.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81774331 and
81873049), the Zhejiang Provincial Natural Science Foundation of
China (grant nos. LY18H270016, LY18H270004 and LY17H270001), the
Medical Health Science and Technology Project of Zhejiang
Provincial Health Commission (grant no. 2012ZA045); and the
Zhejiang Provincial Key Construction University Superiority
Characteristic Discipline (Traditional Chinese Pharmacology)
Opening Foundation of China (grant no. ZYX2018006).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article or are available from the
corresponding author on reasonable request.
Authors' contributions
WJ conducted the main work of the cellular and
molecular experiments and contributed to the revision of this
manuscript. CG, LZ, XY and MG contributed to the cellular and
molecular experiments. JZ provided the TB and improved the
experimental design of the study. JC and XD were involved in the
conception of the study. QY was involved in the conception of the
study and provided funding to support to the study. LS designed the
study, drafted and revised the manuscript, and provided funding
support to the study. All listed authors approved the manuscript
for publication, and agree to be accountable for all aspects of the
study.
Ethics approval and consent to
participate
The zebrafish larvae used in the present study were
<5 days post-fertilization and thus were not free-feeding and
thus do not count as a protected species. Thus, ethics approval was
not required for the zebrafish experiments in the present study, as
the larvae were sacrificed at 4 days post-fertilization.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
TB
|
theabrowin
|
|
RNA-seq
|
RNA sequencing
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
|
EMT
|
epithelial-mesenchymal transition
|
|
p-IKKα
|
phospho-IKKα
|
|
p-IKKβ
|
phospho-IKKβ
|
|
BMSCs
|
bone marrow-derived mesenchymal stem
cells
|
|
p-IκBα
|
phospho-IκBα
|
|
FBS
|
fetal bovine serum
|
|
PBS
|
phosphate-buffered saline
|
|
CCK-8
|
Cell Counting Kit-8
|
|
BSA
|
bovine serum albumin
|
|
OD
|
optical density
|
|
TBST
|
tris-buffered saline-Tween-20
|
References
|
1
|
Mirabello L, Troisi RJ and Savage SA:
Osteosarcoma incidence and survival rates from 1973 to 2004: Data
from the surveillance, epidemiology, and end results program.
Cancer. 115:1531–1543. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mirabello L, Troisi RJ and Savage SA:
International osteosarcoma incidence patterns in children and
adolescents, middle ages and elderly persons. Int J Cancer.
125:229–234. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Valery PC, Laversanne M and Bray F: Bone
cancer incidence by morphological subtype: A global assessment.
Cancer Causes Control. 26:1127–1139. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bielack SS, Kempf-Bielack B, Delling G,
Exner GU, Flege S, Helmke K, Kotz R, Salzer-Kuntschik M, Werner M,
Winkelmann W, et al: Prognostic factors in high-grade osteosarcoma
of the extremities or trunk: An analysis of 1,702 patients treated
on neoadjuvant cooperative osteosarcoma study group protocols. J
Clin Oncol. 20:776–790. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Whelan JS, Bielack SS, Marina N, Smeland
S, Jovic G, Hook JM, Krailo M, Anninga J, Butterfass-Bahloul T,
Böhling T, et al: EURAMOS-1, an international randomised study for
osteosarcoma: Results from pre-randomisation treatment. Ann Oncol.
26:407–414. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Guo J, Reddick WE, Glass JO, Ji Q, Billups
CA, Wu J, Hoffer FA, Kaste SC, Jenkins JJ, Ortega Flores XC, et al:
Dynamic contrast-enhanced magnetic resonance imaging as a
prognostic factor in predicting event-free and overall survival in
pediatric patients with osteosarcoma. Cancer. 118:3776–3785. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Le Vu B, de Vathaire F, Shamsaldin A,
Hawkins MM, Grimaud E, Hardiman C, Diallo I, Vassal G, Bessa E,
Campbell S, et al: Radiation dose, chemotherapy and risk of
osteosarcoma after solid tumours during childhood. Int J Cancer.
77:370–377. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Calvert GT, Randall RL, Jones KB,
Cannon-Albright L, Lessnick S and Schiffman JD: At-risk populations
for osteosarcoma: The syndromes and beyond. Sarcoma.
2012:1523822012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Klein MJ and Siegal GP: Osteosarcoma:
Anatomic and histologic variants. Am J Clin Pathol. 125:555–581.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Meyers PA, Schwartz CL, Krailo M,
Kleinerman ES, Betcher D, Bernstein ML, Conrad E, Ferguson W,
Gebhardt M, Goorin AM, et al: Osteosarcoma: A randomized,
prospective trial of the addition of ifosfamide and/or muramyl
tripeptide to cisplatin, doxorubicin, and high-dose methotrexate. J
Clin Oncol. 23:2004–2011. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kempf-Bielack B, Bielack SS, Jurgens H,
Branscheid D, Berdel WE, Exner GU, Göbel U, Helmke K, Jundt G,
Kabisch H, et al: Osteosarcoma relapse after combined modality
therapy: An analysis of unselected patients in the cooperative
osteosarcoma study group (COSS). J Clin Oncol. 23:559–568. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jensen GS, Beaman JL, He Y, Guo Z and Sun
H: Reduction of body fat and improved lipid profile associated with
daily consumption of a Puer tea extract in a hyperlipidemic
population: A randomized placebo-controlled trial. Clin Interv
Aging. 11:367–376. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang M, Wang C and Chen H: Green, oolong
and black tea extracts modulate lipid metabolism in hyperlipidemia
rats fed high-sucrose diet. J Nutr Biochem. 12:14–20. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wu LY, Juan CC, Hwang LS, Hsu YP, Ho PH
and Ho LT: Green tea supplementation ameliorates insulin resistance
and increases glucose transporter IV content in a fructose-fed rat
model. Eur J Nutr. 43:116–124. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Imai K, Suga K and Nakachi K:
Cancer-preventive effects of drinking green tea among a Japanese
population. Prev Med. 26:769–775. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nakachi K, Suemasu K, Suga K, Takeo T,
Imai K and Higashi Y: Influence of drinking green tea on breast
cancer malignancy among Japanese patients. Jpn J Cancer Res.
89:254–261. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jin W, Zhou L, Yan B, Yan L, Liu F, Tong
P, Yu W, Dong X, Xie L, Zhang J, et al: Theabrownin triggers DNA
damage to suppress human osteosarcoma U2OS cells by activating p53
signalling pathway. J Cell Mol Med. 22:4423–4436. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhou L, Wu F, Jin W, Yan B, Chen X, He Y,
Yang W, Du W, Zhang Q, Guo Y, et al: Theabrownin inhibits cell
cycle progression and tumor growth of lung carcinoma through
c-myc-Related mechanism. Front Pharmacol. 8:752017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hay ED: An overview of
epithelio-mesenchymal transformation. Acta Anat (Basel). 154:8–20.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Thiery JP and Sleeman JP: Complex networks
orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell
Biol. 7:131–142. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Thiery JP, Acloque H, Huang RY and Nieto
MA: Epithelial-mesenchymal transitions in development and disease.
Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lamouille S, Xu J and Derynck R: Molecular
mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell
Biol. 15:178–196. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chapman HA: Epithelial-mesenchymal
interactions in pulmonary fibrosis. Annu Rev Physiol. 73:413–435.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang G, Yuan J and Li K: EMT transcription
factors: Implication in osteosarcoma. Med Oncol. 30:6972013.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Peinado H, Olmeda D and Cano A: Snail, Zeb
and bHLH factors in tumour progression: An alliance against the
epithelial phenotype? Nat Rev Cancer. 7:415–428. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
De Craene B and Berx G: Regulatory
networks defining EMT during cancer initiation and progression. Nat
Rev Cancer. 13:97–110. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Damsky CH, Richa J, Solter D, Knudsen K
and Buck CA: Identification and purification of a cell surface
glycoprotein mediating intercellular adhesion in embryonic and
adult tissue. Cell. 34:455–466. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kobielak A and Fuchs E: Alpha-catenin: At
the junction of intercellular adhesion and actin dynamics. Nat Rev
Mol Cell Biol. 5:614–625. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Oka H, Shiozaki H, Kobayashi K, Inoue M,
Tahara H, Kobayashi T, Takatsuka Y, Matsuyoshi N, Hirano S,
Takeichi M, et al: Expression of E-cadherin cell adhesion molecules
in human breast cancer tissues and its relationship to metastasis.
Cancer Res. 53:1696–1701. 1993.PubMed/NCBI
|
|
31
|
Gumbiner BM: Cell adhesion: The molecular
basis of tissue architecture and morphogenesis. Cell. 84:345–357.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hirohashi S: Inactivation of the
E-cadherin-mediated cell adhesion system in human cancers. Am J
Pathol. 153:333–339. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Min C, Eddy SF, Sherr DH and Sonenshein
GE: NF-kappaB and epithelial to mesenchymal transition of cancer. J
Cell Biochem. 104:733–744. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hendrix MJ, Seftor EA, Chu YW, Trevor KT
and Seftor RE: Role of intermediate filaments in migration,
invasion and metastasis. Cancer Metastasis Rev. 15:507–525. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cano A, Perez-Moreno MA, Rodrigo I,
Locascio A, Blanco MJ, del Barrio MG, Portillo F and Nieto MA: The
transcription factor snail controls epithelial-mesenchymal
transitions by repressing E-cadherin expression. Nat Cell Biol.
2:76–83. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yang H, Zhang Y, Zhou Z, Jiang X and Shen
A: Snail-1 regulates VDR signaling and inhibits
1,25(OH)-D3 action in osteosarcoma. Eur J Pharmacol.
670:341–346. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sharili AS, Allen S, Smith K, Hargreaves
J, Price J and McGonnell I: Expression of Snail2 in long bone
osteosarcomas correlates with tumour malignancy. Tumour Biol.
32:515–526. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Gheldof A, Hulpiau P, van Roy F, De Craene
B and Berx G: Evolutionary functional analysis and molecular
regulation of the ZEB transcription factors. Cell Mol Life Sci.
69:2527–2541. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Huang Y, Yang Y, Gao R, Yang X, Yan X,
Wang C, Jiang S and Yu L: RLIM interacts with Smurf2 and promotes
TGF-β induced U2OS cell migration. Biochem Biophys Res Commun.
414:181–185. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Arima Y, Inoue Y, Shibata T, Hayashi H,
Nagano O, Saya H and Taya Y: Rb depletion results in deregulation
of E-cadherin and induction of cellular phenotypic changes that are
characteristic of the epithelial-to-mesenchymal transition. Cancer
Res. 68:5104–5112. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Barnes PJ and Karin M: Nuclear
factor-kappaB: A pivotal transcription factor in chronic
inflammatory diseases. N Engl J Med. 336:1066–1071. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Karin M and Greten FR: NF-kappaB: Linking
inflammation and immunity to cancer development and progression.
Nat Rev Immunol. 5:749–759. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Huang S, Pettaway CA, Uehara H, Bucana CD
and Fidler IJ: Blockade of NF-kappaB activity in human prostate
cancer cells is associated with suppression of angiogenesis,
invasion, and metastasis. Oncogene. 20:4188–4197. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Helbig G, Christopherson KW II,
Bhat-Nakshatri P, Kumar S, Kishimoto H, Miller KD, Broxmeyer HE and
Nakshatri H: NF-kappaB promotes breast cancer cell migration and
metastasis by inducing the expression of the chemokine receptor
CXCR4. J Biol Chem. 278:21631–21638. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Radisky DC and Bissell MJ: NF-kappaB links
oestrogen receptor signalling and EMT. Nat Cell Biol. 9:361–363.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Chua HL, Bhat-Nakshatri P, Clare SE,
Morimiya A, Badve S and Nakshatri H: NF-kappaB represses E-cadherin
expression and enhances epithelial to mesenchymal transition of
mammary epithelial cells: Potential involvement of ZEB-1 and ZEB-2.
Oncogene. 26:711–724. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Medici D and Nawshad A: Type I collagen
promotes epithelial-mesenchymal transition through ILK-dependent
activation of NF-kappaB and LEF-1. Matrix Biol. 29:161–165. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wang X, Belguise K, Kersual N, Kirsch KH,
Mineva ND, Galtier F, Chalbos D and Sonenshein GE: Oestrogen
signalling inhibits invasive phenotype by repressing RelB and its
target BCL2. Nat Cell Biol. 9:470–478. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Sarkar FH, Li Y, Wang Z and Kong D:
NF-kappaB signaling pathway and its therapeutic implications in
human diseases. Int Rev Immunol. 27:293–319. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Huang CY, Lee CY, Chen MY, Yang WH, Chen
YH, Chang CH, Hsu HC, Fong YC and Tang CH: Stromal cell-derived
factor-1/CXCR4 enhanced motility of human osteosarcoma cells
involves MEK1/2, ERK and NF-kappaB-dependent pathways. J Cell
Physiol. 221:204–212. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Felx M, Guyot MC, Isler M, Turcotte RE,
Doyon J, Khatib AM, Leclerc S, Moreau A and Moldovan F:
Endothelin-1 (ET-1) promotes MMP-2 and MMP-9 induction involving
the transcription factor NF-kappaB in human osteosarcoma. Clin Sci
(Lond). 110:645–654. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Iyer SV, Ranjan A, Elias HK, Parrales A,
Sasaki H, Roy BC, Umar S, Tawfik OW and Iwakuma T: Genome-wide RNAi
screening identifies TMIGD3 isoform1 as a suppressor of NF-κB and
osteosarcoma progression. Nat Commun. 7:135612016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhou J, Liu Q, Qian R, Liu S, Hu W and Liu
Z: Paeonol antagonizes oncogenesis of osteosarcoma by inhibiting
the function of TLR4/MAPK/NF-κB pathway. Acta Histochem.
122:1514552020. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Yang M, Liu B, Jin L, Tao H and Yang Z:
Estrogen receptor β exhibited anti-tumor effects on osteosarcoma
cells by regulating integrin, IAP, NF-kB/BCL-2 and PI3K/Akt signal
pathway. J Bone Oncol. 9:15–20. 2017. View Article : Google Scholar : PubMed/NCBI
|