Introduction
Prostate cancer was the second leading cause of
cancer incidence (13.5% of approximately 9.5 million new cases) and
the fifth leading cause of mortality (6.7% of approximately 5.4
million deaths) globally in 2018 for males according to Globocan
(1). Advanced or metastatic
prostate cancer patients usually respond well to initial androgen
deprivation therapy (ADT). However, ADT does not prevent
progression of prostate cancer despite the maintenance of low
levels of testosterone over an extended period of time. Disease at
this stage is termed castration-resistant prostate cancer (CRPC).
Several systemic agents have been approved for the treatment of
CRPC. However, despite the significant development of treatment
options, CRPC remains as a lethal disease (2).
Genomic aberrations are common in prostate cancer
cells. Various oncogenes and tumor suppressor genes are related to
prostate cancer (3–8). PTEN, a tumor suppressor gene (9,10),
regulates androgen receptor (AR) signaling (11) in prostate cancer. A change in AR
signaling is associated with the acquisition of castration
resistance in prostate cancer (12). However, the mechanism of prostate
cancer progression is still not completely understood.
Since the 1990s, the Hippo signaling pathway has
been revealed as a tumor suppressor signaling pathway.
Yes-associated protein (YAP) plays a central role in the Hippo
pathway and has been revealed to regulate cell proliferation,
migration, and invasion in various cancers including prostate
cancer (13). High expression of
YAP has been revealed to be associated with the differentiation and
extra-prostatic extension of prostate cancer (14,15).
YAP expression has also been revealed to be associated with
castration-resistant growth of prostate cancer cells as well as
proliferation of androgen-independent human prostate cancer cells
(13,16). It is also known that YAP is
activated by mechano-transduction via the hardness of the ECM
(17).
YAP was revealed to bind to certain Rho
GTPase-activating proteins (ARHGAPs), resulting in cytoskeletal
rearrangement and the promotion of cell migration by altering the
dynamics of F-actin/G-actin turnover in gastric cancer (18,19).
Furthermore, some ARHGAPs are regarded as effectors of YAP
(17,18). Thus far, the function of Rho
GTPase-activating protein 29 (ARHGAP29) has been unclear in
prostate cancer. Therefore, YAP and ARHGAP29 were examined, to
investigate the role of ARHGAP29 in prostate cancer.
The aim of this study was to elucidate the role of
ARHGAP29 by in vitro analysis and determine whether its
protein expression is associated with prostate cancer
prognosis.
Materials and methods
Patients
In total, 133 patients who underwent radical
prostatectomy at Yamaguchi University Hospital from November 2000
to September 2016 were enrolled in the present study. All patients
were diagnosed pathologically with prostate cancer. Detailed
patient characteristics are presented in Table I. The present study was approved by
the Institutional Ethics Committee of the Graduate School of
Medicine of Yamaguchi University and written informed consent was
obtained from all individuals enrolled in the study.
 | Table I.Characteristics of 133 patients who
underwent radical prostatectomy. |
Table I.
Characteristics of 133 patients who
underwent radical prostatectomy.
|
Characteristics | n (%) |
|---|
| Age in years,
median (range) | 67 (54–76) |
| Initial PSA, median
(range) | 8.69 ng/ml |
|
| (3.53–354
ng/ml) |
| Clinical T
category |
|
|
≤T1c | 30 (23) |
|
T2a | 21 (16) |
|
T2b | 60 (45) |
|
T2c | 17 (13) |
|
≥T3 | 5 (4) |
| D'Amico risk
classification |
|
|
Low | 15 (11) |
|
Intermediate | 55 (41) |
|
High | 63 (47) |
| Gleason score |
|
| ≤6 | 33 (25) |
| 7 | 65 (49) |
| ≥8 | 35 (26) |
| Preoperative
ADT |
|
|
Yes | 35 (25) |
| No | 98 (75) |
Immunohistochemistry
Formalin-fixed and paraffin-embedded tissue
specimens were subjected to H&E staining and
immunohistochemical (IHC) staining. For each sample, 3-µm-thick
sections were deparaffinized in xylene, dehydrated in ethanol, and
incubated in a 0.3% hydrogen peroxide solution in methanol for 10
min at room temperature. The sections were then microwaved in a
0.01 M citrate-buffered solution (pH 6.0) for 15 min and covered in
blocking solution (IMMUNO SHOT; Cosmo Bio Co., Ltd.) for 30 min at
room temperature. Then, a primary antibody [anti-ARHGAP29 (1:200
dilution; cat. no. HPA026534; Atlas Antibodies) or anti-YAP (1:200
dilution; product no. 14074; Cell Signaling Technology, Inc.)] was
incubated according to the manufacturers' instructions overnight at
4°C, followed by incubation with the respective secondary antibody
(N-Histofine Simple Stain MAX PO MULTI; cat. no. 414152F; Nichirei
Biosciences, Inc.) for 30 min at room temperature. To evaluate IHC
staining, the H-score was used in the present study. Briefly,
>500 tumor cells were counted in five different fields of vision
in each section (×100, magnification), and the H-score was
calculated by multiplying the percentage of positive cells by the
intensity (strongly stained, 3×; moderately stained, 2×; weakly
stained, 1×), yielding a possible range of 0–300 (20–22).
Two independent examiners (KS and HM) judged the scores and the
mean score was set to the representative score. Cut-off of the
H-score was determined by receiver operating characteristic (ROC)
curve.
Cell lines
Four primary prostate cancer cell lines (22Rv1, ATCC
no. CRL-2505; LNCaP, ATCC no. CRL-1740; DU145, ATCC no. HTB-81;
PC-3, ATCC no. CRL-1435) were purchased from the American Type
Culture Collection. Cells were cultured in RPMI-1640 and DMEM (Life
Technologies; Thermo Fisher Scientific, Inc.) supplemented with 10%
fetal bovine serum (Biological Industries) and maintained in
humidified incubators with 5% CO2 at 37°C.
siRNA knockdown of ARHGAP29
si-ARHGAP29 and control siRNAs were obtained from
Life Technologies; Thermo Fisher Scientific, Inc. siRNA sequences
were as follows: ARHGAP29-#1 siRNA sense,
5′-GCAUAGGUGUUGUUGAUCAtt-3′ and antisense,
5′-UGAUCAACAACACCUAUGCta-3′; ARHGAP29-#2 siRNA sense,
5′-GACCAAGGCUAAAACGAAUtt-3′ and antisense,
5′-AUUCGUUUUAGCCUUGGUCtc-3′. The PC-3 cell line was transiently
transfected with siRNA using Lipofectamine RNAi MAX (Thermo Fisher
Scientific, Inc.), according to the manufacturer's instructions.
After transfection, cells were incubated at 37°C in a
CO2 incubator for 48 h. Quantitative evaluations of mRNA
and protein expression were performed by western blotting and
RT-qPCR, respectively.
Plasmid construction and
transfection
A mammalian expression of HA tagged ARHGAP29
(#104154) was purchased from Addgene, Inc. Cells were seeded on
culture dishes at density of 1×105/well in a 6-well
plate, and the pcDNA3.1 empty vector plasmid (mock) (Thermo Fisher
Scientific, Inc.) or 2 µg of ARHGAP29 expressing plasmid were
transfected using X-tremeGENE HP DNA transfection Reagent
(Sigma-Aldrich; Merck KGaA) for 48 h, according to the
manufacturer's instructions.
Regarding the DU145 cell line, plasmid transfection
was performed via electroporesis system using an Amaxa cell line
Nucleofector Kit L (cat. no. VACA-1005; Lonza Group, Ltd.)
according to the manufacturer's instructions. Prior to
electroporation, 1×106 DU145 cells were centrifuged at
90 × g for 5 min, resuspended in 100 µl of Nucleofector solution
and mixed with 2 µg of pmaxGFP or 2 µg of ARHGAP29 plasmid. The
aforementioned cells were transferred to cuvettes and immediately
electroporated based on the DU145 program (Nucleofector Program
A-023) using Nucleofector 2b Device. After electroporation, cells
were incubated in the cuvette at room temperature for 10 min and
then 500 µl of pre-warmed RPMI-1640 medium supplemented with 10%
FBS were added to the cuvette. Cells were transferred to a 6-well
plate and incubated at 37°C 5% CO2 overnight. The day
after electroporation, cells were centrifuged and the medium was
replaced by RPMI-1640 supplemented with 10% FBS and incubated for
48 h and then performed subsequent experiments were performed.
Reverse transcription quantitative PCR
(RT-qPCR)
We created cDNA by reverse transcription of mRNAs
extracted from each prostate cancer cell line (22Rv1, LNCaP, DU145
and PC-3), using iScript Advanced cDNA Synthesis Kit for RT-qPCR
(cat. no. 1725037; Bio-Rad Laboratories, Inc.). Quantitative
real-time RT-PCR was performed in triplicate with an Applied
Biosystems StepOnePlus using TaqMan universal PCR master mix
(Applied Biosystems; Thermo Fisher Scientific, Inc.), according to
the manufacturer's protocols. The TaqMan probes and primers were
purchased from Applied Biosystems. Human GAPDH (assay ID: 02786624)
was used as an endogenous control. Levels of ARHGAP29 (assay ID:
00191351) and MMP-2 (assay ID: 01548727) RNA expression were
determined using StepOnePlus software (version 2.2.2; Applied
Biosystems; Thermo Fisher Scientific, Inc.). The miRNA expression
levels were determined using the 2−ΔΔCq method (23). The cycling conditions consisted of
an initial denaturation at 95°C for 30 sec and PCR at 40 cycles at
95°C for 5 sec and 60°C for 30 sec.
Gene expression analysis by qPCR
Total RNA was isolated from cells (PC-3-si-NC and
si-ARHGAP29), and an RT2 Profiler PCR Array (Qiagen
RT2 Profiler PCR Array Human Cell Motility; cat. no.
PAHS-128Z, product no. 330231) was used to examine the expression
patterns of genes involved in human cell motility, according to the
manufacturer's instructions. We analyzed the gene expression levels
and produced a heatmap using the web-based software ‘RT2 Profiler
PCR Array’ Data Analysis version 3.5 (Qiagen, Inc.).
Western blotting
Cells samples were lysed in RIPA buffer (cat. no.
89900; Thermo Fisher Scientific, Inc.) supplemented with 1%
protease inhibitors (cOmplete™, Mini, cat. no. 04693124001;
Sigma-Aldrich) for total protein extraction. We quantified the
concentration of total proteins using BCA. Each lysate sample (30
µg protein) was separated by 4–20% SDS-PAGE (Mini-PROTEAN TGX
Stain-Free Gels, cat. no. 4568095; Bio-Rad Laboratories), and then
electro-transferred to a PVDF membrane. After blocking in 5% dry
non-fat milk or 5% BSA for 1 h at room temperature, the membranes
were incubated with a primary antibody overnight at 4°C. After
washing in TBS with 0.05% Tween-20 (TBST), the membranes were
incubated with an HRP-conjugated secondary antibody for 1 h at room
temperature. After washing with TBST, signals were detected using
an ECL detection system (ChemiDoc™ XRS+; Bio-Rad Laboratories,
Inc). Primary antibodies were as follows: anti-ARHGAP29 (product
code ab85853, 1:2,000 dilution), anti-AR (product code ab133273,
1:1,000 dilution), anti-F-actin (product code ab205, 1:500
dilution) and anti-MMP-2 (product code ab97779, 1:1,000 dilution)
from Abcam and anti-YAP (cat. no. 14074S; 1:1,000 dilution),
anti-phosphorylated YAP (cat. no. 13008S; 1:1,000 dilution),
anti-GAPDH (cat. no. 5174S; 1:1,000 dilution), anti-Cofilin (cat.
no. 5175T; 1:1,000 dilution) and anti-phospho-Cofilin (cat. no.
3313T; 1:1,000 dilution) were obtained from Cell Signaling
Technology Inc. Secondary antibodies were as follows: goat
anti-rabbit IgG H&L (HRP) (product code ab6721; 1:10,000
dilution) and goat anti-mouse IgG H&L (HRP) (product code
ab6789; 1:10,000 dilution) from Abcam. GAPDH was used for protein
normalization. We performed densitometry using the public domain
free software ImageJ (version 1.51; National Institutes of
Health).
Cell viability and invasion
assays
Cell viability was assessed using an MTS assay
(CellTiter 96 AQueous One Solution Cell Proliferation
Assay; Promega Corporation). After the cells were seeded at density
of 5×103/well in a 96-well plate, cell viability was
measured at 24, 48, and 72 h at an OD of 490 nm. Data are expressed
as the mean ± SD of three independent experiments. Cell invasion
assays were performed using a CytoSelect 24-well cell invasion
assay kit (Cell BioLabs, Inc.). The CytoSelect™ Cell Invasion Assay
Kit contains polycarbonate membrane inserts (8-µm pore size) in a
24-well plate. The upper surface of the insert membrane is coated
with a uniform layer of dried basement membrane matrix solution.
This basement membrane layer serves as a barrier to discriminate
invasive cells from non-invasive cells. A cell suspension
containing 0.5–1.0×106 cells/ml was placed in upper
chamber in serum-free media. A total of 500 µl of media containing
10% fetal bovine serum was added to the lower well of the invasion
plate. After 48 h of incubation at 37°C with 5% CO2,
cells invaded through the basement membrane layer and clung to the
bottom of the insert membrane. Non-invasive cells remained in the
upper chamber. After removal of non-invasive cells, invasive cells
were stained for 10 min at room temperature using Cell Stain
Solution (Part no. 11002; CytoSelect 24-well cell invasion assay
kit) and then quantified. Each insert was transferred to an empty
well, 200 µl of Extraction Solution (Part no. 11003; CytoSelect
24-well cell invasion assay kit) was added per well and then
incubation followed for 10 min on an orbital shaker. Subsequently
100 µl from each sample was transferred to a 96-well microtiter
plate and the OD 560 nm of each sample was measured on a plate
reader, according to the manufacturer's instructions.
Database
The Cancer Genome Atlas (TCGA) accessed from the
data portal of the National Cancer Institute Home Page (http://cancergenome.nih.gov/) was used for comparison
with our data.
Statistical analysis
Categorical variables were compared by the
Chi-squared test. Continuous variables were analyzed using the
unpaired Student's t-test when comparing two groups. One-way ANOVA
followed by Tukey-Kramer test were used when comparing more than
two groups. Survival analysis was estimated by the Kaplan-Meier
method and compared by the log-rank test. A Cox proportional
hazards regression model was used in the multivariable analysis to
identify risk factors for disease progression. Statistical analysis
was performed using JMP software (Pro.13; SAS Institute). P-values
were two-sided, and statistical significance was defined as
P<0.05 in all tests. Regarding protein expression, bivariate
analysis was performed and a ROC curve was constructed using JMP
software to set the cutoff value and determine the high/low
expression of proteins (24).
Results
AR, YAP, ARHGAP29, and F-actin
expression in prostate cancer cell lines
RT-qPCR and western blotting were performed to
clarify whether there was a difference in the expression of
ARHGAP29 between prostate cancer cell lines depending on AR and YAP
expression. AR was expressed in 22Rv1 and LNCaP cell lines. YAP was
expressed in all four prostate cancer cell lines (Fig. 1A and B). The expression level of YAP
was higher in DU-145 and PC-3 cells than in LNCaP and 22Rv1 cells.
ARHGAP29 protein expression was higher in PC-3 cells than in the
other cell lines. F-actin was the most weakly expressed in PC-3
cells compared with the other cell lines (Fig. 1C).
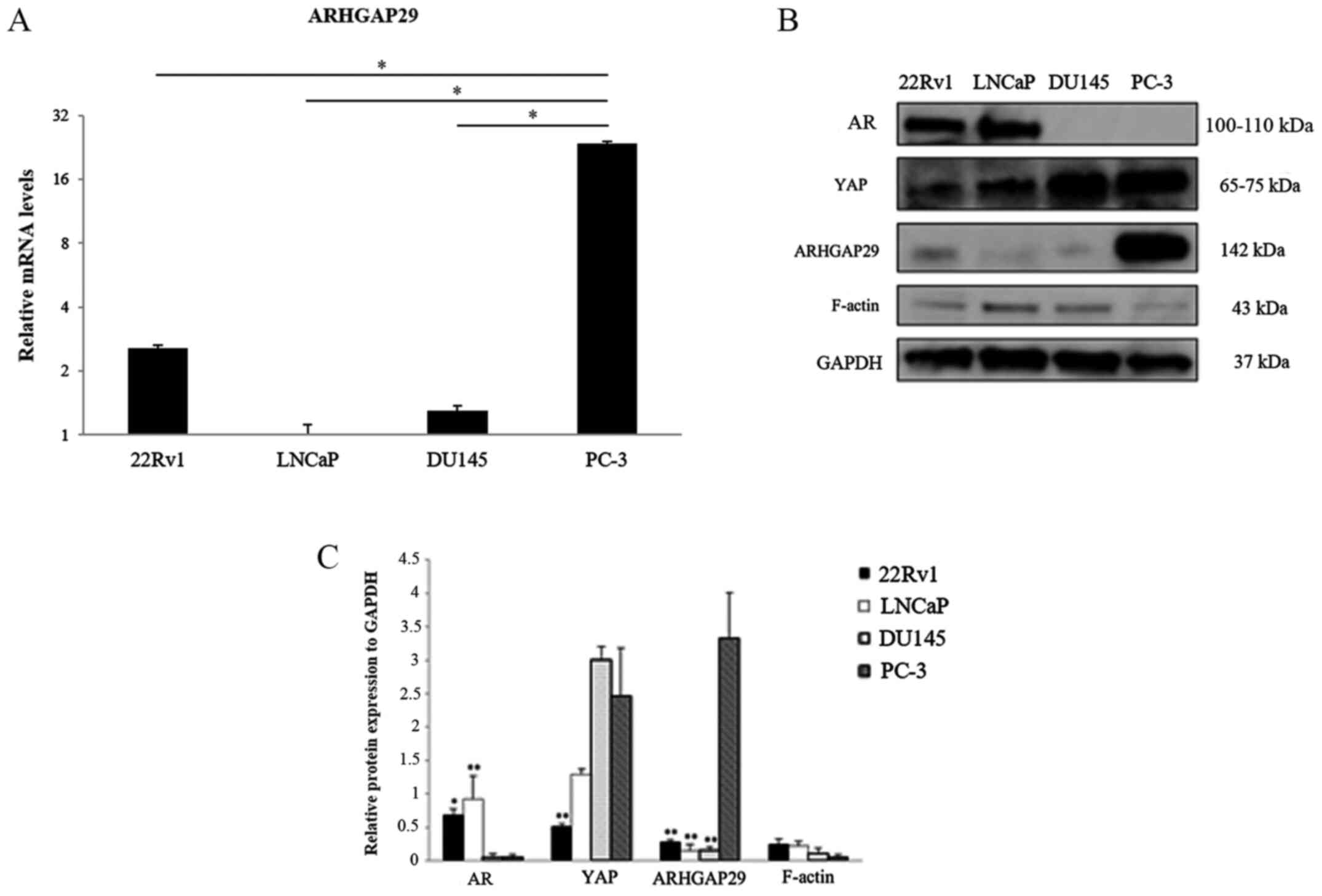 | Figure 1.AR, YAP, ARHGAP29, and F-actin
expression in prostate cancer cell lines. (A) mRNA levels of
ARHGAP29 in prostate cancer cell lines (22Rv1, LNCaP, DU145 and
PC-3). Experiments were performed in triplicate. The vertical axis
of the graph is presented on a logarithmic scale. The results are
expressed as the mean ± SD. *P<0.01 compared with PC-3 cells.
(B) Western blotting of the expression of various proteins in
prostate cancer cell lines. (C) Densitometric analysis of B
(relative protein expression to GAPDH). There was an inverse
association between ARHGAP29 and F-actin expression, but no clear
association between the expression of ARHGAP29 and that of other
proteins. The results are expressed as the mean ± SD (at least
three independent experiments). *P<0.05, **P<0.01 compared
with PC-3 cells. AR, androgen receptor; YAP, yes-associated
protein; ARHGAP29, Rho GTPase-activating protein 29. |
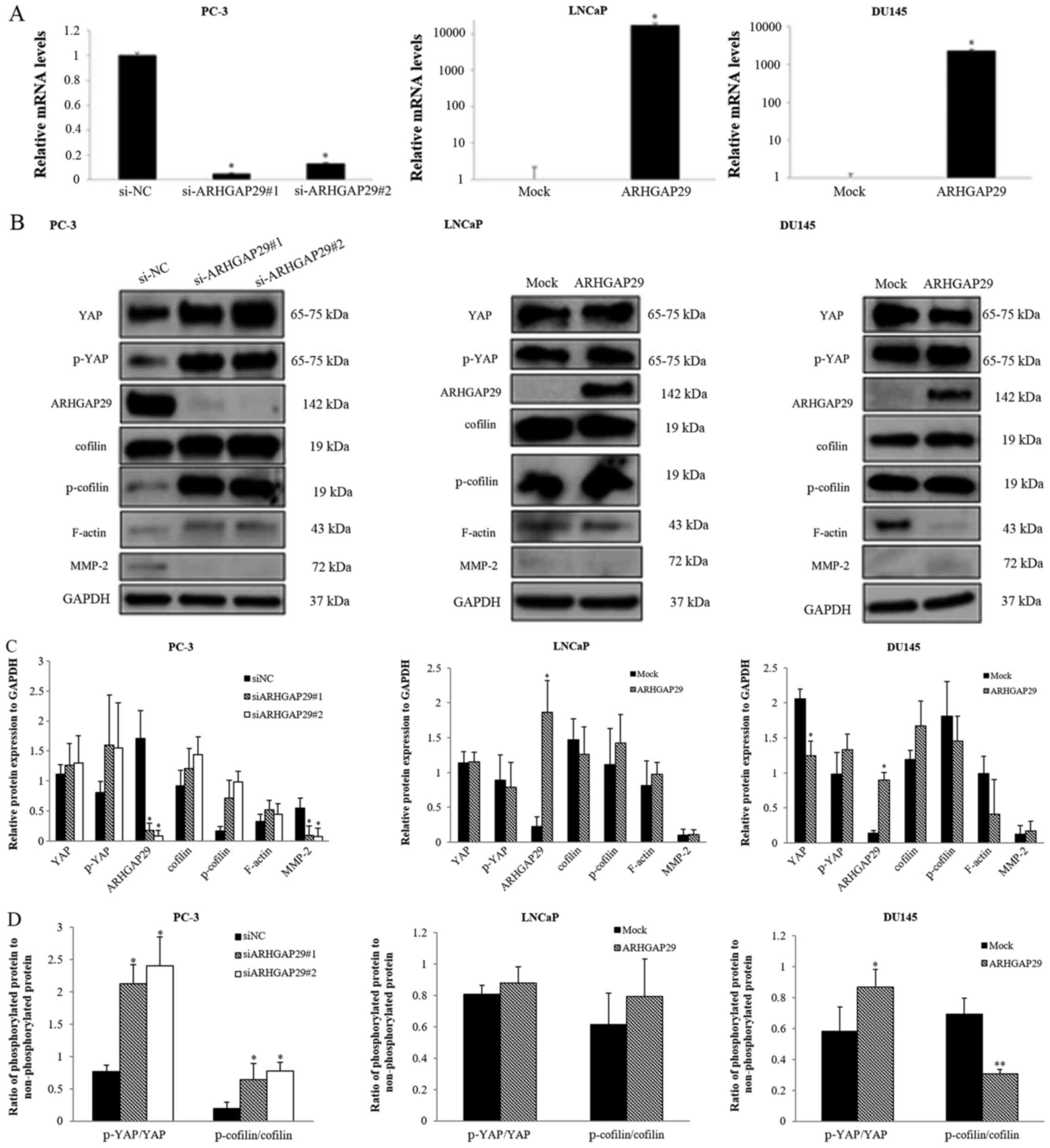
Effect of downregulation or
upregulation of ARHGAP29 in prostate cancer cell lines (PC-3, LNCaP
and DU145)
After downregulation (PC-3 cells) or upregulation
(LNCaP and DU145 cells) of ARHGAP29 in prostate cancer cell lines
(Fig. 2A and B), the expression of
several proteins was examined. Based on a recent study (18), the RhoA-LIMK-cofilin signaling
pathway has been revealed to be affected by ARHGAP29 in a gastric
cancer cell line. Therefore, certain related genes (cofilin,
p-cofilin and F-actin) were analyzed by western blotting (Fig. 2B).
After almost complete knockdown of ARHGAP in PC-3
cells, phosphorylated cofilin and F-actin were increased and
cofilin expression was unchanged. In contrast, after overexpression
of ARHGAP29 in DU145 cells, F-actin was slightly decreased but
phosphorylated cofilin was not altered. YAP and phosphorylated YAP
were slightly recovered without significant differences in
si-ARHGAP29 PC-3 transfectants compared with the si-NC control.
Conversely, YAP was decreased after overexpression of ARHGAP29 in
DU145 cells. Expression of these proteins relative to that of the
housekeeping gene GAPDH and the ratio of phosphorylated protein to
non-phosphorylated protein (YAP and cofilin) are presented in
Fig. 2C and D.
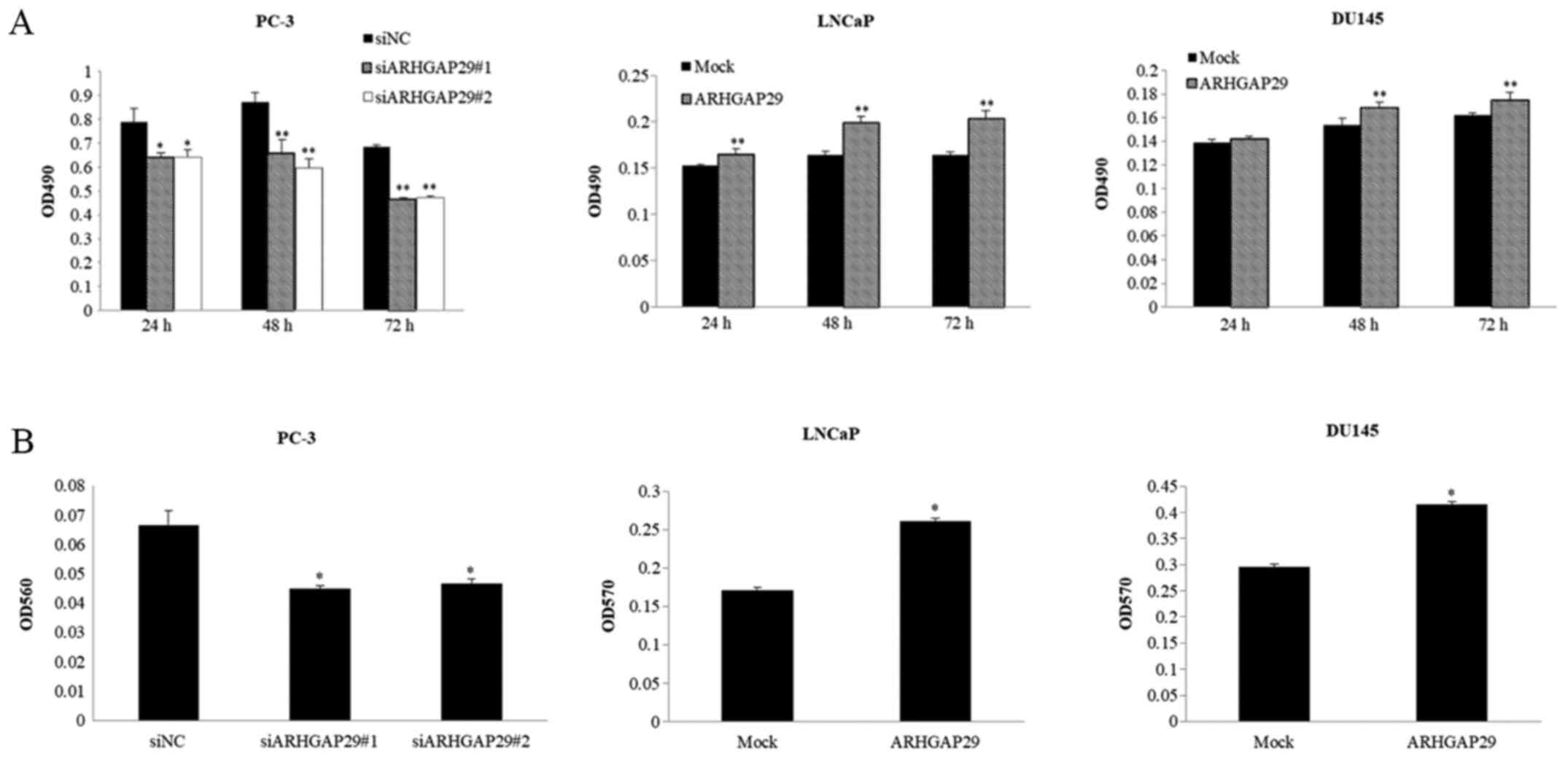
Functional analyses by MTS and cell invasion assays
were also performed in these three cell lines (Fig. 3). Cell viability and invasion were
significantly decreased after downregulation of ARHGAP29 in PC-3
cells (Fig. 3A and B). After
upregulation of ARHGAP29 in LNCaP and DU145 cells, cell viability
and invasion were significantly increased (Fig. 3A and B).
Identification of cell
motility-related genes after knock down of ARHGAP29
Based on the functional analyses, ARHGAP29 may be
involved in cell proliferation or invasion. To determine new
therapeutic targets or genes related to ARHGAP29 in prostate cancer
cells, Qiagen RT2 Profiler PCR Array Human Cell Motility
was used.
The pre-designed array included 84 genes related to
cell motility (Fig. 4A). Data
analysis was performed using the web-based software ‘RT2 Profiler
PCR Array’ Data Analysis version 3.5 as aforementioned. A heatmap
is presented in Fig. 4B. When the
boundary was set at 3, one gene (STAT3) was upregulated and
numerous genes (including CSF1, ACTN3 and HGF) were downregulated
after knocking down ARHGAP29 in PC3 cells (Fig. 4C). Regulation of some proteins, such
as HGF, RHO, CAPN1, was validated by western blotting. However,
there was no difference in the expression of these proteins between
si-NC and si-ARHGAP29 cells (data not shown). Among the
downregulated genes of the 84 genes in the array, active MMP2
expression was significantly decreased at mRNA and protein levels
after knockdown of ARHGAP29 in PC3 cells (Figs. 2B and C and 4D).
Association between the expression
level of ARHGAP29 and prognosis in prostate cancer patients
The expression level of ARHGAP29 was evaluated by
IHC in 133 prostate cancer patients who had undergone radical
prostatectomy. Representative images of YAP and ARHGAP29 staining
in prostate cancer specimens (negative and positive) are presented
Fig. 5A.
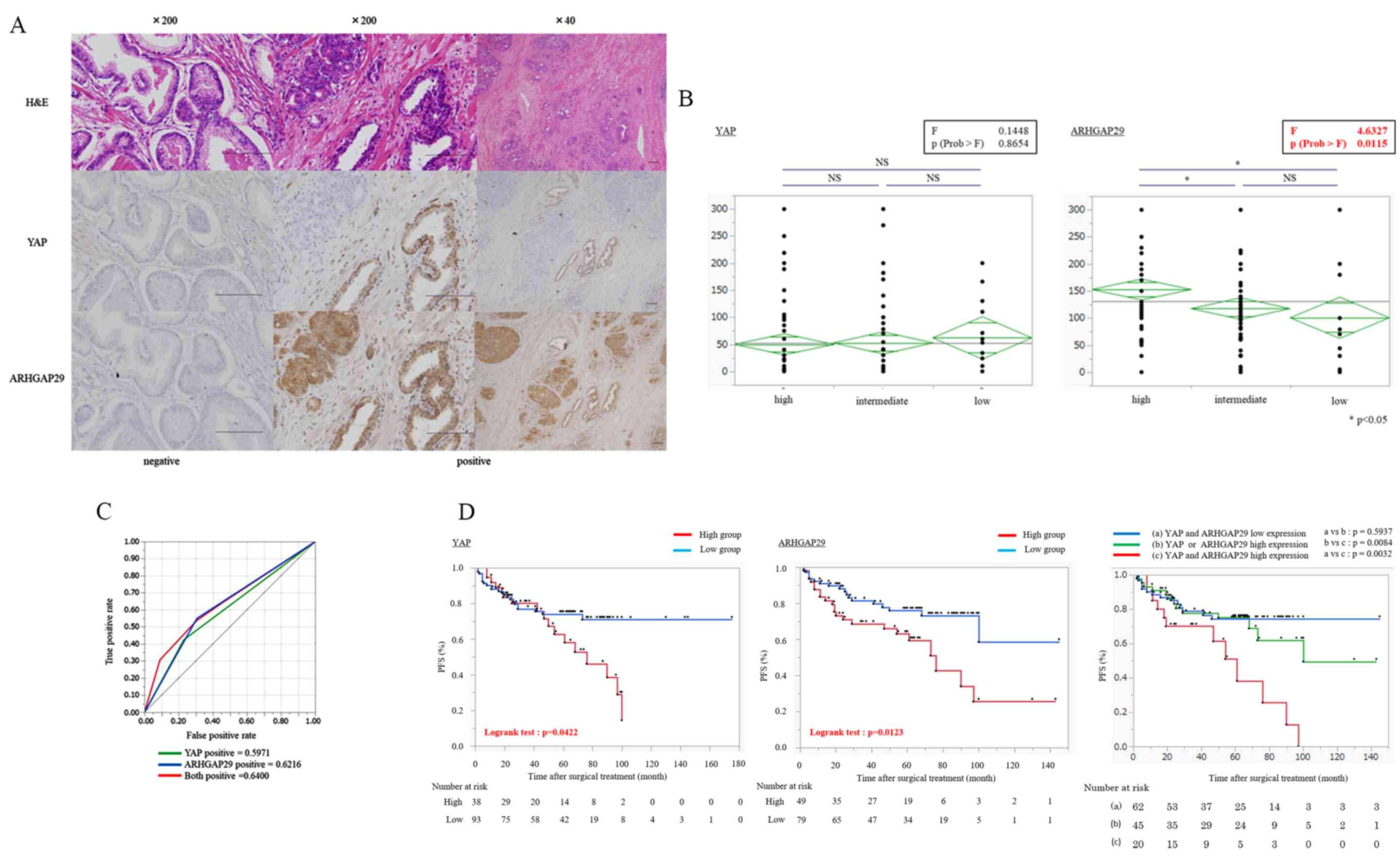 | Figure 5.Association between the expression
levels of YAP and ARHGAP29 and the prognosis of prostate cancer
patients. (A) Histology (H&E staining) and IHC staining of YAP
or ARHGAP29. Protein expression patterns were different between YAP
and ARHGAP29. YAP was heterogeneously stained. Scale bars represent
200 µM. (B) Association of the D'Amico risk classification with the
expression of YAP and ARHGAP29. When comparing three groups,
one-way ANOVA (inside the black frame) followed by Tukey-Kramer
test were used. *P<0.05. YAP was unrelated to the risk
classification, but ARHGAP29 was significantly associated with the
risk classification. The diamond indicates the mean (long
horizontal line) and 95% confidence interval of the H-score. (C)
ROC curve of YAP, ARHGAP29 and both (AUC: 0.5971, 0.6216 and
0.6400, respectively). Both proteins had low AUC scores as
prognostic markers. (D) Kaplan-Meier plot of biochemical PFS
stratified by the expression levels of YAP, ARHGAP29 and both. For
each protein, high expression was associated with a poor prognosis
of prostate cancer patients. Furthermore, the group with high
expression of both YAP and ARHGAP29 had the worst prognosis. PFS
was compared by a log-rank test. P<0.05 was considered to
indicate a statistically significant difference. YAP,
yes-associated protein; ARHGAP29, Rho GTPase-activating protein 29;
ROC, receiver operating characteristic; AUC, area under the curve;
PFS, progression-free survival; NS, not significant. |
YAP expression was high in the nucleus of basal
cells and the cytoplasm of luminal cells, but ARHGAP29 expression
was high in the cytoplasm of both cells. Notably, YAP expression
was unrelated to the Gleason score. The characteristics of the
prostate cancer patients are presented in Table I. ARHGAP29 expression was
significantly associated with the risk classification of prostate
cancer (Fig. 5B). Both YAP and
ARHGAP29 had low area under the curve (AUC) scores as prognostic
markers, but there was a significant difference between the
expression of these proteins and biochemical progression-free
survival (b-PFS: P=0.0422, and P=0.0123, respectively) (Fig. 5C and D). In addition, high
expression of both proteins was significantly associated with poor
prognoses (Fig. 5D). In TCGA
database, YAP did not exhibit a tendency for a poor prognosis in
patients with high expression. In contrast, ARHGAP29 exhibited a
tendency for a poor prognosis in patients with high expression in
TCGA (Fig. S1A and B). Moreover,
the prognostic significance of clinicopathological parameters,
including prostate specific antigen (PSA), the D'Amico risk
classification, Gleason score, and pathological T category, and the
expression levels of YAP and ARHGAP29 were evaluated in prostate
cancer patients (Table II). As a
result, high ARHGAP29 expression was a significant independent risk
factor related to b-PFS in multivariate analysis (HR=2.27;
P<0.05; data not shown).
 | Table II.Univariate and multivariate analyses
of prognostic factors associated with biochemical recurrence-free
survival of prostate cancer patients. |
Table II.
Univariate and multivariate analyses
of prognostic factors associated with biochemical recurrence-free
survival of prostate cancer patients.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
|
| HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Initial PSA*
(ng/ml) |
| <20
vs. ≥20 | 0.53
(0.30–1.00) | 0.049 | 0.37
(0.15–0.88) | 0.025 |
| D'Amico risk
classification |
| Low,
intermediate vs. high | 0.73
(0.43–1.24) | 0.248 |
|
|
| Gleason score |
| <8
vs. ≥8 | 0.84
(0.47–1.57) | 0.567 |
|
|
| Pathological T
category |
|
<pT2c vs. ≥pT2c | 0.60
(0.30–1.10) | 0.099 |
|
|
| Expression of
YAP |
| Low vs.
high | 0.53
(0.28–1.01) | 0.052 |
|
|
| Expression of
ARHGAP29 |
| Low vs.
high | 0.46
(0.24–086) | 0.015 | 0.44
(0.19–0.95) | 0.037 |
Discussion
YAP has been revealed as an oncogenic protein in
several cancers, such as gastric, breast, hepatocellular,
pancreatic, and lung cancers as well as melanoma (25–31).
Similar to other cancers, YAP regulates cell migration and invasion
in prostate cancer (13). Several
ARHGAPs, which enhance Rho GTPase activity in almost all basic
cellular processes, are oncogenic or tumor suppressor proteins
(32). For example, ARHGAP5 and
ARHGAP42 have been revealed to be oncogenic proteins in
nasopharyngeal cancer (33,34), whereas ARHGAP24 has been
demonstrated to be a tumor suppressor protein in lung, breast, and
colorectal cancers (35–38). Numerous studies have demonstrated a
close association of ARHGAPs with several malignancies.
Recently, other ARHGAPs such as ARHGAP18 and
ARHGAP29 (17,18) were identified as transcriptional
targets of YAP, and ARHGAP29 was reported as a prognostic marker
for gastric cancer. Since there have been no studies on ARHGAP29 in
prostate cancer, in the present study, ARHGAP29 was examined to
investigate how it affects progression or metastasis of prostate
cancer and whether ARHGAP29 may be a prognostic marker for prostate
cancer. Initially, protein expression in four prostate cancer cell
lines (22Rv1, LNCaP, DU145 and PC-3) was assessed. Among these cell
lines, PC-3 and DU145 did not express AR, but highly expressed YAP.
In contrast, YAP expression was low in AR-expressing cell lines
(22Rv1 and LNCaP). PC-3 cells highly expressed ARHGAP29 compared
with the other three cell lines. AR-null PC-3 cells are derived
from bone metastasis (39,40). After complete knockdown of ARHGAP29
in PC-3 cells, their proliferation and invasion were significantly
decreased. In contrast, cell proliferation and invasion were
increased after upregulation of ARHGAP29 in LNCaP and DU145 cells.
In the present study, we did not investigate a direct interaction
between AR and ARHGAP29. However, the present results indicated
that ARHGAP29 regulates cell proliferation and invasion in prostate
cancer cells. Recently, Qiao et al demonstrated that
ARHGAP29 suppressed the RhoA-cofilin pathway and destabilized
F-actin, which caused cytoskeletal rearrangement and promoted
migration (18). In the present
study, certain proteins in the RhoA-cofilin pathway were analyzed
in PC-3, LNCaP and DU145 cells. Specifically, phosphorylated
cofilin and F-actin were recovered when ARHGAP29 was completely
knocked down in PC-3 cells. Moreover, the relative protein level of
phosphorylated cofilin to cofilin was increased. These data were
consistent with the results from a recent study on a gastric cancer
cell line (18). F-actin was
slightly decreased in DU145 cells following upregulation of
ARHGAP29. In PC-3 cells, ARHGAP29 may be associated with cell
migration by suppressing the RhoA-cofilin pathway similar to a
previous study (18). However,
cofilin and p-cofilin expression were not altered in LNCaP and
DU145 cells. Specifically, the results of LNCaP and Du145 cells
were not demonstrated as the reverse of the observations made in
PC-3 ARHGAP29-knockdown cells. These results may be explained by
the fact that each cell line has a different genotype/phenotype as
revealed in a previous study (for instance only PC-3 cells do not
express α-catenin) (40).
Upregulation of ARHGAP29 may lead to decrease of F-actin via
another pathway in LNCaP and DU145 cells, however, to demonstrate
this, further experiments are required.
Apart from the Rho-A-cofilin pathway, to identify
new targets or cancer pathways related to ARHGAP29, a pre-designed
array (Human Cell Motility), based on the functional analysis data
in the present study, was used. Among the 84 genes in the array,
expression of several genes was altered after knocking down
ARHGAP29 in PC-3 cells. Among the genes, expression of MMP-2 was
validated by RT-qPCR and western blotting. Among the matrix
metalloproteinase (MMP) family, which degrade the ECM, MMP-2, also
known as gelatinase A, is reported to be correlated with the
invasion and metastasis of cancer cells as well as angiogenesis in
numerous human cancer tissues (41,42).
Moreover, Zhang et al indicated a role of YAP in gastric
cancer and revealed that LATS1 inhibited the growth and metastasis
of gastric cancer cells by restraining nuclear transfer of YAP and
downregulating MMP-2 expression concurrently (43). This suggests that the YAP pathway,
which regulates the progression of cancer cells, is associated with
MMP-2. In previous studies on prostate cancer development, it has
been similarly demonstrated that MMP-2 is associated with invasion,
metastasis, and a poor prognosis (44–46).
It is theorized that ARHGAP29 may activate cell motility to
upregulate MMP-2. In the present, we did not establish a direct
interaction between ARGAP29 and MMP-2. Therefore, further
experiments are required to support this theory.
Next, IHC staining was performed to investigate the
clinical role of YAP and ARHGAP29 protein expression in prostate
cancer patients. High expression levels of ARHGAP29 were related to
the D'Amico risk classification, which is the risk classification
of prostate cancer, and prognosis of prostate cancer patients (PSA
PFS). In the present study, prostate cancer patients with high YAP
or ARHGAP29 expression had a significantly poor prognosis. These
differences between the TCGA database and our data may be due to
different characteristics of the cohort including racial bias.
Based on our data, ARHGAP29 may be a prognostic marker and
therapeutic target. To confirm the present results, large-scale
data analysis using Japanese samples is required in the future.
A limitation of the present study is the lack of
co-localization studies of AR, YAP and ARHGAP29 in human prostate
specimens as well as lack of ARHGAP29 rescue experiments in
prostate cancer cells.
In conclusion, it was demonstrated that ARHGAP29 may
be associated with prostate cancer cell growth and invasion as well
as a clinically poor prognosis of prostate cancer patients.
Therefore, ARHGAP29 may serve as a new biomarker or novel
therapeutic target in prostate cancer. In a future study, the
investigation of the relationship between YAP and the ARHGAP29
pathway is required to elucidate the underlying mechanism in
prostate cancer.
Supplementary Material
Supporting Data
Acknowledgements
We thank Kiyomi Fujita and Takao Kitagawa for
technical assistance.
Funding
The present study was funded by a Grant-in-Aid for
Scientific Research (C) (KAKENHI-PROJECT-16K11008).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HMatsum, HH, MFS and HMatsuy conceived and designed
the study. All authors advised the work. KS, HH and KU performed
experiments. KS, MS and HH prepared the figures and drafted the
original manuscript. JM, NF, YK, RI, YY, SY and TS contributed to
the analysis or interpretation of the data. HMatsum, HH and HMatsuy
reviewed and revised the manuscript. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Institutional
Ethics Committee of the Graduate School of Medicine of Yamaguchi
University and written informed consent was obtained from all
individuals enrolled in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pagliuca M, Buonerba C, Fizazi K and Di
Lorenzo G: The evolving systemic treatment landscape for patients
with advanced prostate cancer. Drugs. 79:381–400. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xu J, Zheng SL, Komiya A, Mychaleckyj JC,
Isaacs SD, Hu JJ, Sterling D, Lange EM, Hawkins GA, Turner A, et
al: Germline mutations and sequence variants of the macrophage
scavenger receptor 1 gene are associated with prostate cancer risk.
Nat Genet. 32:321–325. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jefferies MT, Cox AC, Shorning BY, Meniel
V, Griffiths D, Kynaston HG, Smalley MJ and Clarke AR: PTEN loss
and activation of K-RAS and β-catenin cooperate to accelerate
prostate tumourigenesis. J Pathol. 243:442–456. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Suzuki H, Freije D, Nusskern DR, Okami K,
Cairns P, Sidransky D, Isaacs WB and Bova GS: Interfocal
heterogeneity of PTEN/MMAC1 gene alterations in multiple metastatic
prostate cancer tissues. Cancer Res. 58:204–209. 1998.PubMed/NCBI
|
|
6
|
Suzuki H, Komiya A, Aida S, Ito H, Yatani
R and Shimazaki J: Detection of human papillomavirus DNA and p53
gene mutations in human prostate cancer. Prostate. 28:318–324.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Grignon DJ, Caplan R, Sarkar FH, Lawton
CA, Hammond EH, Pilepich MV, Forman JD, Mesic J, Fu KK, Abrams RA,
et al: p53 status and prognosis of locally advanced prostatic
adenocarcinoma: A study based on RTOG 8610. J Natl Cancer Inst.
89:158–165. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhou Z, Flesken-Nikitin A, Corney DC, Wang
W, Goodrich DW, Roy-Burman P and Nikitin AY: Synergy of p53 and Rb
deficiency in a conditional mouse model for metastatic prostate
cancer. Cancer Res. 66:7889–7898. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dong JT, Li CL, Sipe TW and Frierson HF
Jr: Mutations of PTEN/MMAC1 in primary prostate cancers from
Chinese patients. Clin Cancer Res. 7:304–308. 2001.PubMed/NCBI
|
|
10
|
Wang SI, Parsons R and Ittmann M:
Homozygous deletion of the PTEN tumor suppressor gene in a subset
of prostate adenocarcinomas. Clin Cancer Res. 4:811–815.
1998.PubMed/NCBI
|
|
11
|
Lin HK, Hu YC, Lee DK and Chang C:
Regulation of androgen receptor signaling by PTEN (phosphatase and
tensin homolog deleted on chromosome 10) tumor suppressor through
distinct mechanisms in prostate cancer cells. Mol Endocrinol.
18:2409–2423. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nelson PS: Molecular states underlying
androgen receptor activation: A framework for therapeutics
targeting androgen signaling in prostate cancer. J Clin Oncol.
30:644–646. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang L, Yang S, Chen X, Stauffer S, Yu F,
Lele SM, Fu K, Datta K, Palermo N, Chen Y, et al: The hippo pathway
effector YAP regulates motility, invasion, and castration-resistant
growth of prostate cancer cells. Mol Cell Biol. 35:1350–1362. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Noh MG, Kim SS, Hwang EC, Kwon DD and Choi
C: Yes-associated protein expression is correlated to the
differentiation of prostate adenocarcinoma. J Pathol Transl Med.
51:365–373. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Collak FK, Demir U, Ozkanli S, Kurum E and
Zerk PE: Increased expression of YAP1 in prostate cancer correlates
with extraprostatic extension. Cancer Biol Med. 14:405–413. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jin X, Zhao W, Zhou P and Niu T: YAP
knockdown inhibits proliferation and induces apoptosis of human
prostate cancer DU145 cells. Mol Med Rep. 17:3783–3788.
2018.PubMed/NCBI
|
|
17
|
Porazinski S, Wang H, Asaoka Y, Behrndt M,
Miyamoto T, Morita H, Hata S, Sasaki T, Krens SFG, Osada Y, et al:
YAP is essential for tissue tension to ensure vertebrate 3D body
shape. Nature. 521:217–221. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Qiao Y, Chen J, Lim YB, Finch-Edmondson
ML, Seshachalam VP, Qin L, Jiang T, Low BC, Singh H, Lim CT, et al:
YAP regulates actin dynamics through ARHGAP29 and promotes
metastasis. Cell Rep. 19:1495–1502. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dupont S, Morsut L, Aragona M, Enzo E,
Giulitti S, Cordenonsi M, Zanconato, Le Digabel J, Forcato M,
Bicciato S, et al: Role of YAP/TAZ in mechanotransduction. Nature.
474:179–183. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Goulding H, Pinder S, Cannon P, Pearson D,
Nicholson R, Snead D, Bell J, Elston CW, Robertson JF, Blamey RW,
et al: A new immunohistochemical antibody for the assessment of
estrogen receptor status on routine formalin-fixed tissue samples.
Hum Pathol. 26:291–294. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ishibashi H, Suzuki T, Suzuki S, Moriya T,
Kaneko C, Takizawa T, Sunamori M, Handa M, Kondo T and Sasano H:
Sex steroid hormone receptors in human thymoma. J Clin Endocrinol
Metab. 88:2309–2317. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pirker R, Pereira JR, von Pawel J,
Krzakowski M, Ramlau R, Park K, de Marinis F, Eberhardt WE,
Paz-Ares L, Störkel S, et al: EGFR expression as a predictor of
survival for first-line chemotherapy plus cetuximab in patients
with advanced non-small-cell lung cancer: Analysis of data from the
phase 3 FLEX study. Lancet Oncol. 13:33–42. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kawakami T, Takeuchi S, Arimura Y and Soma
Y: Elevated antilysosomal-associated membrane protein-2 antibody
levels in patients with adult Henoch-Schönlein purpura. Br J
Dermatol. 166:1206–1212. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pan Z, Tian Y, Zhang B, Zhang X, Shi H,
Liang Z, Wu P, Li R, You B, Yang L, et al: YAP signaling in gastric
cancer-derived mesenchymal stem cells is critical for its promoting
role in cancer progression. Int J Oncol. 51:1055–1066. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen D, Sun Y, Wei Y, Zhang P, Rezaeian
AH, Teruya-Feldstein J, Gupta S, Liang H, Lin HK, Hung MC, et al:
LIFR is a breast cancer metastasis suppressor upstream of the
Hippo-YAP pathway and a prognostic marker. Nat Med. 18:1511–1517.
2012. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lamar JM, Stern P, Liu H, Schindler JW,
Jiang ZG and Hynes RO: The Hippo pathway target, YAP, promotes
metastasis through its TEAD-interaction domain. Proc Natl Acad Sci
USA. 109:E2441–E2450. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xu MZ, Chan SW, Liu AM, Wong KF, Fan ST,
Chen J, Poon RT, Zender L, Lowe SW, Hong W, et al: AXL receptor
kinase is a mediator of YAP-dependent oncogenic functions in
hepatocellular carcinoma. Oncogene. 30:1229–1240. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang M, Zhao Y, Zhang Y, Wang D, Gu S,
Feng W, Peng W, Gong A and Xu M: LncRNA UCA1 promotes migration and
invasion in pancreatic cancer cells via the Hippo pathway. Biochim
Biophys Acta Mol Basis Dis. 1864:1770–1782. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hsu PC, Miao J, Huang Z, Yang YL, Xu Z,
You J, Dai Y, Yeh CC, Chan G, Liu S, et al: Inhibition of
yes-associated protein suppresses brain metastasis of human lung
adenocarcinoma in a murine model. J Cell Mol Med. 22:3073–3085.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jin D, Guo J, Wang D, Wu Y, Wang X, Gao Y,
Shao C, Xu X and Tan S: The antineoplastic drug metformin
downregulates YAP by interfering with IRF-1 binding to the YAP
promoter in NSCLC. EBioMedicine. 37:188–204. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tcherkezian J and Lamarche-Vane N: Current
knowledge of the large RhoGAP family of proteins. Biol Cell.
99:67–86. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fang Y, Zhu X, Wang J, Li N, Li D, Sakib
N, Sha Z and Song W: MiR-744 functions as a proto-oncogene in
nasopharyngeal carcinoma progression and metastasis via
transcriptional control of ARHGAP5. Oncotarget. 6:13164–13175.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hu Q, Lin X, Ding L, Zeng Y, Pang D,
Ouyang N, Xiang Y and Yao H: ARHGAP42 promotes cell migration and
invasion involving PI3K/Akt signaling pathway in nasopharyngeal
carcinoma. Cancer Med. 7:3862–3874. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang L, Shen S, Wang M, Ding F, Xiao H, Li
G and Hu F: Rho GTPase activating protein 24 (ARHGAP24) silencing
promotes lung cancer cell migration and invasion by activating
β-catenin signaling. Med Sci Monit. 25:21–31. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Dai X, Geng F, Dai J, Li M and Liu M: Rho
GTPase activating Protein 24 (ARHGAP24) regulates the anti-cancer
activity of sorafenib against breast cancer MDA-MB-231 cells via
the signal transducer and activator of transcription 3 (STAT3)
signaling pathway. Med Sci Monit. 24:8669–8677. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Uehara S, Saito K, Asami H and Ohta Y:
Role of ARHGAP24 in ADP ribosylation factor 6 (ARF6)-dependent
pseudopod formation in human breast carcinoma cells. Anticancer
Res. 37:4837–4844. 2017.PubMed/NCBI
|
|
38
|
Zhang S, Sui L, Zhuang J, He S, Song Y, Ye
Y and Xia W: ARHGAP24 regulates cell ability and apoptosis of
colorectal cancer cells via the regulation of P53. Oncol Lett.
16:3517–3524. 2018.PubMed/NCBI
|
|
39
|
Kaighn ME, Narayan KS, Ohnuki Y, Lechner
JF and Jones LW: Establishment and characterization of a human
prostatic carcinoma cell line (PC-3). Invest Urol. 17:16–23.
1979.PubMed/NCBI
|
|
40
|
Mitchell S, Abel P, Ware M, Stamp G and
Lalani E: Phenotypic and genotypic characterization of commonly
used human prostatic cell lines. BJU Int. 85:932–944. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li Y, Song T, Chen Z, Wang Y, Zhang J and
Wang X: Pancreatic stellate cells activation and matrix
metallopeptidase 2 expression correlate with lymph node metastasis
in pancreatic carcinoma. Am J Med Sci. 357:16–22. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Maekawa R, Maki H, Yoshida H, Hojo K,
Tanaka H, Wada T, Uchida N, Takeda Y, Kasai H, Okamoto H, et al:
Correlation of antiangiogenic and antitumor efficacy of N-biphenyl
sulfonyl-phenylalanine hydroxiamic acid (BPHA), an orally-active,
selective matrix metalloproteinase inhibitor. Cancer Res.
59:1231–1235. 1999.PubMed/NCBI
|
|
43
|
Zhang J, Wang G, Chu SJ, Zhu JS, Zhang R,
Lu WW, Xia LQ, Lu YM, Da W and Sun Q: Loss of large tumor
suppressor 1 promotes growth and metastasis of gastric cancer cells
through upregulation of the YAP signaling. Oncotarget.
7:16180–16193. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Trudel D, Fradet Y, Meyer F, Harel F and
Têtu B: Significance of MMP-2 expression in prostate cancer: An
immunohistochemical study. Cancer Res. 63:8511–8515.
2003.PubMed/NCBI
|
|
45
|
Chen PC, Tang CH, Lin LW, Tsai CH, Chu CY,
Lin TH and Huang YL: Thrombospondin-2 promotes prostate cancer bone
metastasis by the up-regulation of matrix metalloproteinase-2
through down-regulating miR-376c expression. J Hematol Oncol.
10:332017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ito Y, Ishiguro H, Kobayashi N, Hasumi H,
Watanabe M, Yao M and Uemura H: Adipocyte-derived monocyte
chemotactic protein-1 (MCP-1) promotes prostate cancer progression
through the induction of MMP-2 activity. Prostate. 75:1009–1019.
2015. View Article : Google Scholar : PubMed/NCBI
|