Introduction
Esophageal cancer (EC) is a common human malignant
tumor type with high incidence and mortality and the sixth leading
cause of cancer-associated deaths worldwide (1–3). While
esophageal adenocarcinoma (EAC) is the main histologic subtype in
Western countries, esophageal squamous cell carcinoma (ESCC)
remains the predominant form in Asian countries, accounting for
more than 90% of EC (4). The
development of early diagnostic and adjuvant treatment methods has
provided a benefit for certain ESCC patients in recent decades.
However, the poor prognosis of patients remains a major clinical
problem (5). Recurrence and the
ability to form metastasis are crucial characteristics of malignant
compared with benign tumors, and are accountable for the mortality
of a considerable proportion of affected patients. Tumor metastasis
is multistep and complex biological process, including the spread
of tumor cells from their primary location and disseminating
through the bloodstream to eventually colonize in situ or in
distant organs (6). Identification
of effective regulators in every key regulatory step of malignant
progression is anticipated to provide improvement for the diagnosis
and treatment of ESCC.
Sex-determining region Y-box 12 (SOX12) is
well-known as a member of a transcription factor superfamily
belonging to the high-mobility group, and has been revealed to
perform key functions in embryonic development and the
determination of cell fate (7,8).
Previous studies have also revealed that aberrant expression of
SOX12 affects the biological behaviors of tumor cells and has a
potential prognostic value in various type of tumors, such as liver
(9), breast (10), and lung cancer (11), as well as renal carcinoma (12) and acute myeloid leukemia (13). However, the potential roles and
specific regulatory mechanisms of SOX12 in ESCC remain unknown.
The purpose of this study was to investigate the
role of SOX12 in the biological behavior of ESCC cells and its
predictive value for clinical prognosis
Materials and methods
Patients and collection of tissue
samples
Esophageal squamous cell carcinoma tissue and paired
adjacent non-cancerous tissues (n=56) were collected from patients
who were diagnosed with primary ESCC and received radical
esophageal surgery without preoperative radiotherapy, chemotherapy
or biotherapy from August 2012 to September 2013 at Changhai
hospital, Second Military Medical University (Shanghai, China). The
median age of patients at the time of admission was 58 years, range
43–72 years. In total, 15 patients were women, and 41 were men.
Total RNA from 56 pairs of tissue samples were extracted for
RT-qPCR detection. Total protein from 6 pairs of tissue samples
were extracted for western blot detection. All tissues (tumor
tissue and paired para-cancerous tissue) were embedded in paraffin
immediately after surgery. Each sample was confirmed by
pathological diagnosis and was followed up until March 2018. The
use of esophageal cancer tissues and clinical data was approved by
the Biomedical Ethics Committee of the Second Military Medical
University (Shanghai, China). Informed consent from each patient
before surgery was obtained.
Immunohistochemistry (IHC)
IHC was used to evaluate the expression level of
SOX12 in paraffin-embedded tissues. All specimens had been fixed in
10% buffered formalin and embedded in paraffin. The embedded
tissues were cut into 4-µm-thick serial sections and stored at 4°C.
After deparaffinization, rehydration and antigen retrieval, each
slide was blocked with 10% (w/v) normal goat serum (product no. SP
KIT-B1; Fuzhou Maixin Biotech Co., Ltd.) at room temperature for 1
h and then incubated with anti-SOX12 primary antibody (dilution,
1:100; cat. no. SAB4502835; Sigma-Aldrich; Merck KGaA) at 4°C
overnight, followed by incubation with secondary antibodies
(EliVision plus; product no. KIT-9901; Fuzhou Maixin Biotech Co.,
Ltd.) at room temperature for 1 h and diaminobenzidine dye (product
no. DAB-0031 (20X); Fuzhou Maixin Biotech Co., Ltd.) at room
temperature for 1–9 min. Counterstaining of the nucleus was
performed with hematoxylin (product no. CTS-1099; Fuzhou Maixin
Biotech Co., Ltd.) at room temperature for 10 min. Negative
controls were prepared by replacing the primary antibody with PBS.
Images were obtained with light microscope at a magnification of
×200. All tissues were reviewed by experienced pathologists. For
each patient sample, the number and proportion of immunopositive
cells was determined in 10 randomly selected high-power fields and
immunoreactive scoring was performed. A score of >4 was
considered as high expression.
Cell lines culture and vectors
The human ESCC cell lines Eca109, TE1 and the human
normal esophageal epithelial cell line HEEC (used as control) were
obtained from the Cell Bank of the Shanghai Institutes for
Biological Sciences. The cells were maintained in DMEM medium
supplemented with 10% FBS (HyClone; Cytiva) at 37°C in a 5%
CO2 incubator. The plasmids expressing short hairpin
(sh)RNAs targeting SOX12 or scrambled shRNA (shRNA1:
5′-CATGGCGGATTACCCGGACTA; shRNA2: 5′-TCCGCAGTCTTACGAGGAGTC;
scrambled shRNA: 5′-GAGCTCTCTCTGCACATTCTT;) were purchased from
Guangzhou RiboBio Co., Ltd. (product nos. stB0007967A-1-5,
stB0007967B-1–5 and stB0007967C-1-5). Cell culture was performed in
six well plates transfected with a total of 4 µg shRNA using
Lipofectamine® 2000 transfection reagent (Invitrogen;
Thermo Fisher Scientific, Inc.), used according to the
manufacturer's protocol, that was added in each well when the cell
density reached 50–60%, and culture was continued at 37°C for 48–72
h according to the experimental requirements.
Recombinant human SOX12 protein (cat. no.
H00006666-Q01; Abnova) and WP1066 (cat. no. HY-15312; MCE), a
potent inhibitor of the JAK2/STAT3 signaling pathway were purchased
and used according to the manufacturer's instructions. Recombinant
human SOX12 protein (10 µl) was co-cultured with shRNA1 in the
colony formation, Transwell and cellular immunofluorescence assays.
WP1066 (5 µl), an inhibitor of the JAK2/STAT3 signaling pathway,
was added to the cell culture medium in the colony formation,
Transwell and western blotting assays. DMSO was used as a solvent
control.
Colony formation assay
After transfection with the indicated plasmids,
Eca109 and TE1 cells were collected and re-suspended in 1.5 ml
complete culture medium supplemented with 0.45% low-melting point
agarose (Invitrogen; Thermo Fisher Scientific, Inc.). The cells
(1×103) were inoculated in 35-mm tissue culture dishes
containing 1.5 ml culture medium and agarose (0.75%) on the bottom
layer. The dishes were cultured for two weeks. Cell colonies were
fixed with 4% formaldehyde solution for 15 min and stained with
0.005% crystal violet for 10–30 min at room temperature. The number
of colonies with more than 10 cells were counted by light
microscope at a magnification of ×10. Experimental groups are
representative of at least three independent experiments.
Migration and invasion assays
The migratory and invasive capacities of Eca109 and
TE1 were assessed by using Transwell assays. A normal Transwell
chamber was used for the migration assay, and a Transwell chamber
whose membrane was pre-coated with Matrigel® (cat. no.
356231; Corning; BD Bisociences) was used for the invasion assay.
For each group, 5×104 cells in 200 µl serum-free medium
were added to each upper chamber, while 500 µl DMEM containing 20%
FBS was added to the lower chambers. After incubation for 48 h,
cells were fixed with 4% formaldehyde solution for 15 min and
stained with 0.005% crystal violet for 10 min at room temperature
and images were obtained by light microscope (magnification, ×200).
Experimental groups are representative of at least three
independent experiments.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA extraction from tissues and cell lines was
performed with TRIzol™ reagent (cat. no. 15596026; Invitrogen;
Thermo Fisher Scientific, Inc.) and then reverse-transcribed into
complementary DNA immediately with the PrimeScript™ RT Reagent kit
(cat. no. RR037B; Takara Bio, Inc.) according to the manufacturer's
instructions. Fast SYBR™ Green Master Mix (cat. no. 4385612; Thermo
Fisher Scientific;) was added to the PCR reaction mixture according
to the manufacturers' protocols. The thermocycling conditions used
for qPCR were as follows: 95°C for 2 min; 45 cycles at 95°C for 5
sec, 55°C for 20 sec and 72°C, 30 sec. A melting curve was
established under the following reaction conditions: 95°C for 15
sec, 60°C for 1 min and 95°C for 15 sec. The experiment was
performed in triplicate. GAPDH was used as internal control. The
2−ΔΔCq method (14) was
conducted to calculate the relative expression of the genes. The
primer sequences used were as follows: SOX12 (NM_006943.3) forward,
5′-AGCACCCGTGTGACTCTTTCC-3′ and reverse,
5′-AGCAGAACCAAGCCCTGTCTC-3′; GAPDH (NM_001256799.1) forward,
5′-CACCCACTCCTCCACCTTTG-3′ and reverse,
5′-CCACCACCCTGTTGCTGTAG-3′.
Western blot analysis
Total protein from tissues and cell lines was
extracted by using radioimmunoprecipitation assay lysis buffer
(product no. P0013B; Beyotime Institute of Biotechnology). The
total protein concentration was determined using the BCA method. A
total of 30 µg of protein was separated by SDS-PAGE (10%) and
transferred to a PVDF membrane. Non-specific binding sites were
blocked by incubating with TBST (0.1% Tween 20) containing 5% (w/v)
non-fat dried milk for 1 h at room temperature. The antibodies used
were as follows: Anti-SOX12 primary antibody (product no.
SAB1412152; dilution, 1:1,000; Sigma-Aldrich; Merck KGaA), anti-JAK
primary antibody (product code ab108596; dilution, 1:5,000),
anti-phosphorylated (p)-JAK at Tyr1007+1008 primary antibody
(product code ab32101; dilution, 1:2,000), anti-STAT3 primary
antibody (product code ab68153; dilution, 1:1,000), anti-p-STAT3
(phosphorylated at Tyr705) antibody (product code ab76315;
dilution, 1:5,000), anti-GAPDH antibody (product code ab9482;
dilution, 1:5,000; all from Abcam) and goat anti-rabbit
immunoglobulin G H&L (cat. no. ab205718; dilution, 1:5,000;
Abcam). Signals were visualized by ECL chemiluminescence (cat. no.
34095, Thermo Fisher Scientific, Inc.). The results were evaluated
with Quantity One software (v4.6.6; Bio-Rad Laboratories,
Inc.).
Cellular immunofluorescence
ESCC cell lines Eca109 and TE1 were seeded onto
coverslips in six-well cell culture plates at 2×104
cells/well with complete medium and transfected with different
shRNAs as aforementioned. After culture for 48 h, the cells were
rinsed three times with PBS carefully and then fixed with 4%
formaldehyde solution for 15 min at room temperature. Cells were
then permeabilized with Triton X-100 solution (1% v/v) for 10 min
and blocked with non-immune goat serum for 1 h at room temperature.
Cells were incubated with various antibodies as follows: SOX12
(product no. HPA055052; 1:100 dilution; Sigma-Aldrich; Merck KGaA),
and the aforementioned antibodies p-JAK (phosphorylated at
Tyr1007+1008; 1:100 dilution) and p-STAT3 (phosphorylated at
Tyr705; dilution, 1:100) at 4°C overnight. Cells were then rinsed
with PBS and incubated with cyanine 3-labeled (product no. A0516,
1:500 dilution) and Alexa 488-labeled (product no. A0423; 1:500
dilution) secondary antibodies (Beyotime Beyotime Institute of
Biotechnology) for 1 h at room temperature, and stained with DAPI
(1 µg/ml; Sigma-Aldrich; Merck KGaA) for 5 min at room temperature.
Images were obtained using a fluorescence microscope
(magnification, ×200).
Statistical analysis
Data analyses were calculated by SPSS 18.0
statistical software (SPSS, Inc.) and values are expressed as the
mean ± standard deviation. ANOVA and Dunnett's test were used for
comparing the mean of multiple experimental groups and that of the
control group. The chi-square test and Spearman's rank correlation
were used to analyze the correlation between gene expression and
clinical pathological data. Survival analysis was performed using
the Kaplan-Meier method and significant differences in overall
survival (OS) and disease-free survival (DFS) was determined with
the log-rank test. Univariate and multivariate analyses were
performed using Cox's proportional hazard model. Differences
between multiple groups were analyzed using one-way analysis of
variance. P<0.05 was considered to indicate a statistically
significant difference. The experimental data were representative
of three independent experiments.
Results
SOX12 is aberrantly upregulated in
ESCC tissues and cell lines
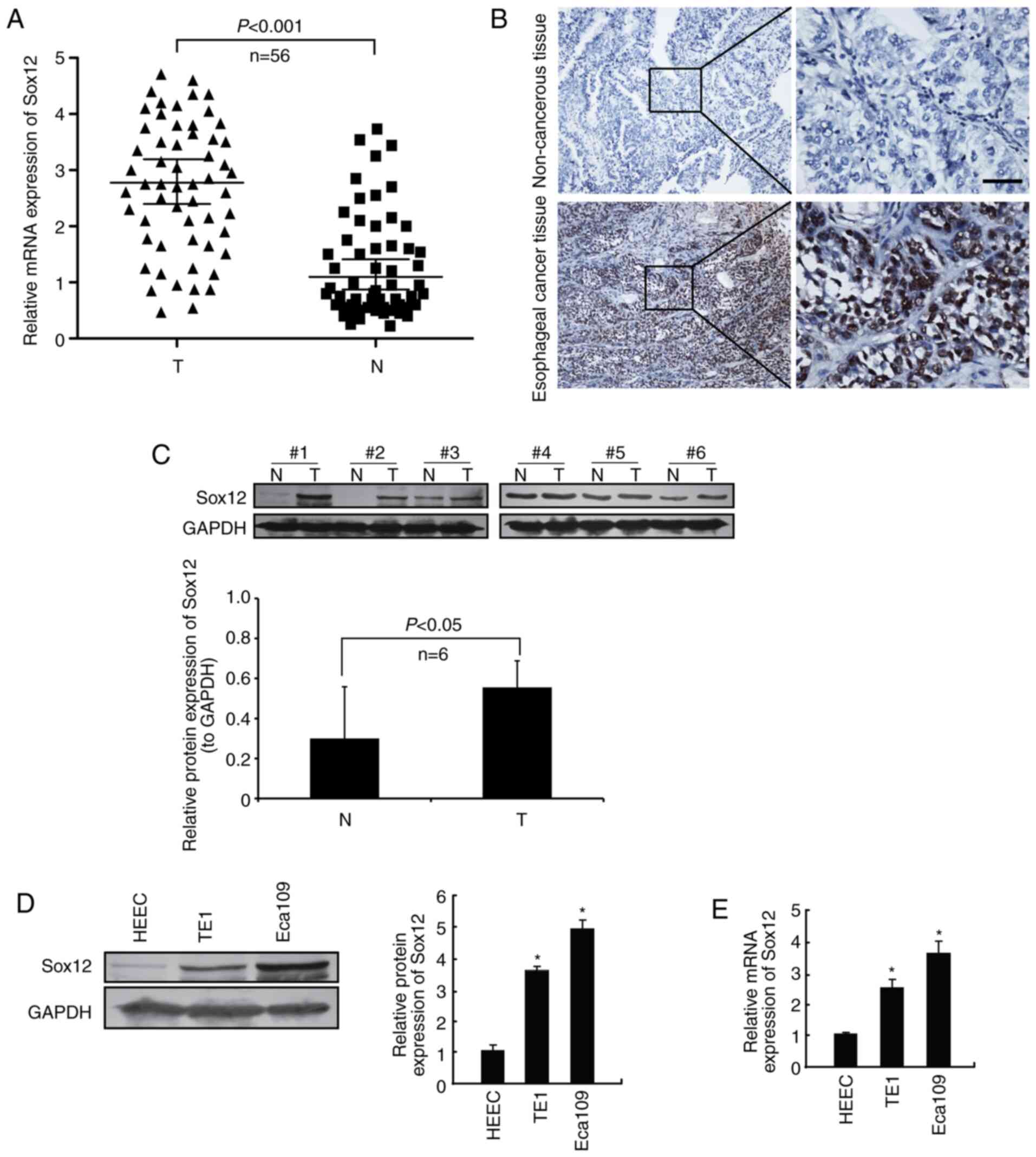
In the present study, the protein level of SOX12 in
56 ESCC samples and adjacent non-cancerous tissues was examined by
IHC. The results indicated that SOX12 was upregulated in ESCC
tissues compared with adjacent non-cancerous tissues in 38 out of
56 cases (67.9%; Fig. 1A and B).
Then 6 pairs of fresh tissue samples were collected and total
proteins were extracted. The protein level of SOX12 in ESCC tissues
and adjacent non-cancerous tissues was detected by western blot
analysis. The results revealed that the protein levels of SOX12
were markedly upregulated in ESCC tissue samples compared with
those in the corresponding non-cancerous tissues (Fig. 1C). In addition, the protein and mRNA
expression of SOX12 in ESCC cell lines was evaluated by western
blot and RT-qPCR analysis, respectively. Human normal esophageal
epithelial cell line (HEEC) was used as a control. As presented in
Fig. 1D and E, compared with that
in the HEEC human normal esophageal epithelial cell line, SOX12 was
significantly overexpressed in both Eca109 and TE1. Collectively,
these results indicated that SOX12 was aberrantly overexpressed in
ESCC cell lines and tissue samples.
Knockdown of SOX12 inhibits the colony
formation, migration and invasion of ESCC cell lines
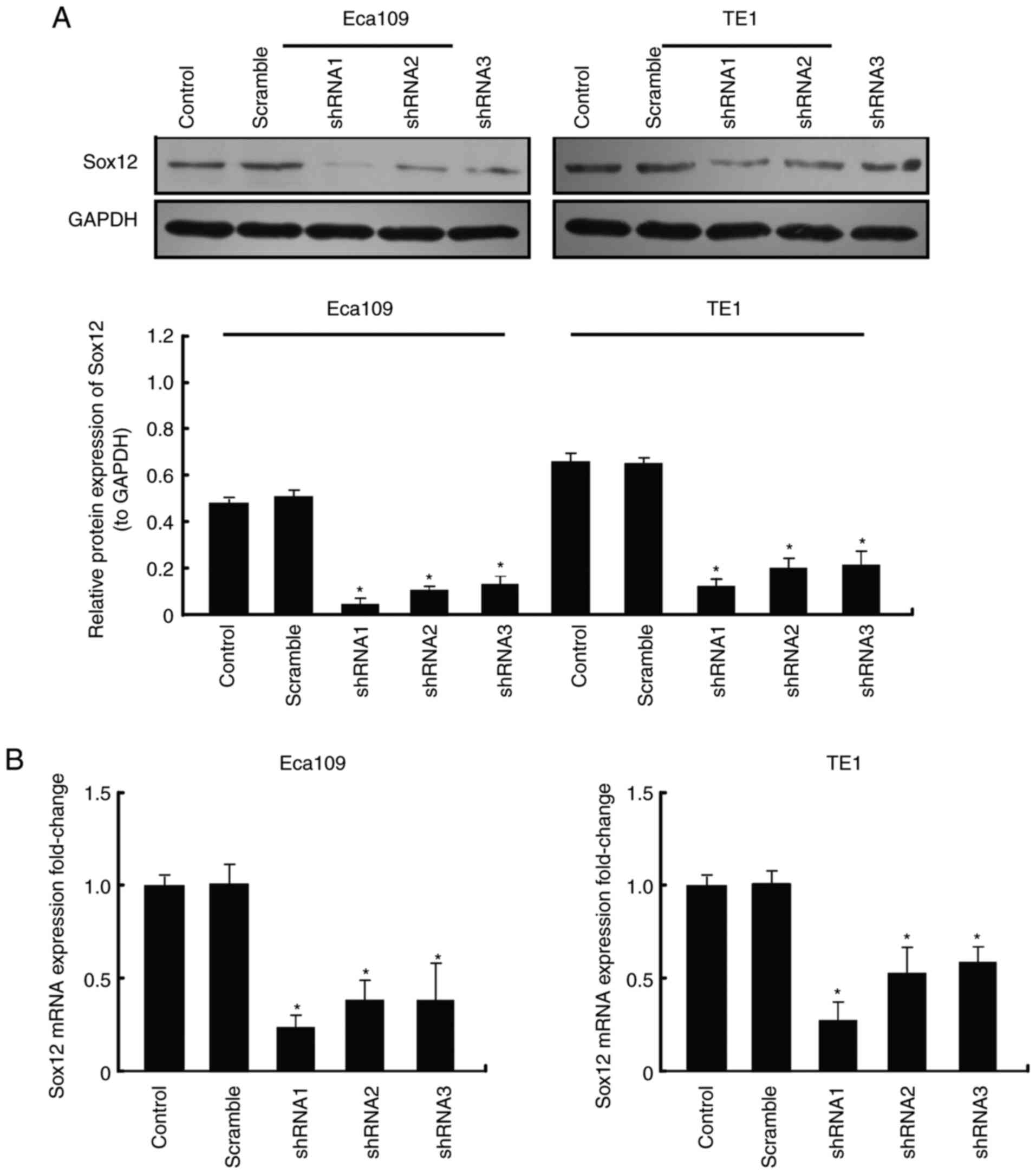
To explore the function of SOX12 which regulates the
biological characteristics of ESCC cells, three vectors carrying
shRNA targeting SOX12, shRNA1, shRNA2 and shRNA3, were constructed
and respectively transfected into Eca109 and TE1 cells. Two of the
vectors, shRNA1 and shRNA2, were more effective at knocking down
the expression of SOX12 (Fig. 2),
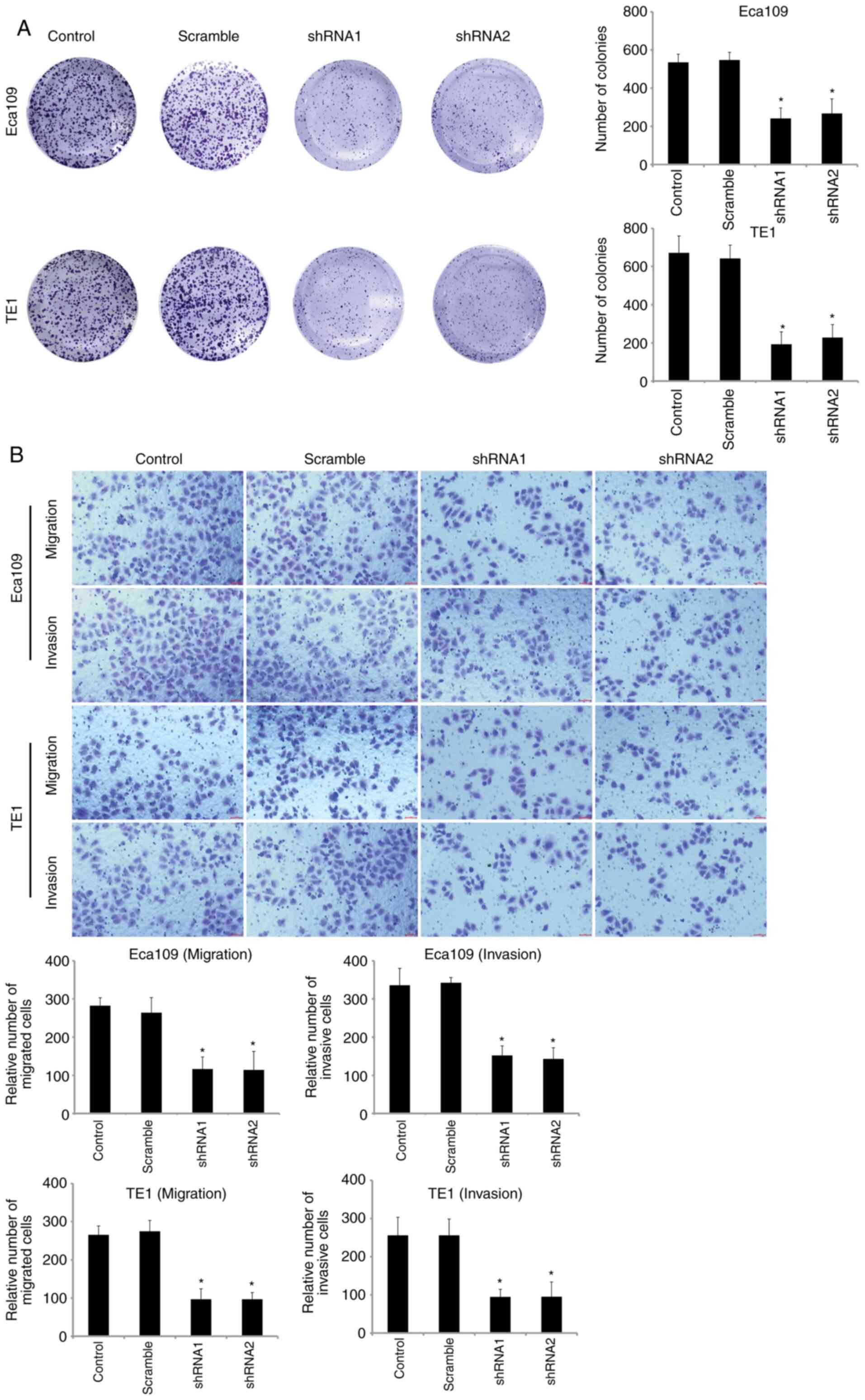
as determined by RT-qPCR and western blot analysis. Colony
formation and Transwell assays were then performed to examine the
neoplastic capacity and motility of SOX12-silenced Eca109 and TE1
cells. The results revealed that downregulation of SOX12 could
inhibit the viability and motility capacities of ESCC cells in
vitro (Fig. 3). When
recombinant SOX12 (10 µM) was co-cultured in transfected cells, the
proliferation and motility of ESCC cells could be restored
(Fig. S1A and B). It was therefore
revealed that overexpression of SOX12 was closely related with the
malignant biological behavior of ESCC cells.
Knockdown of SOX12 expression
suppresses the JAK2/STAT3 signaling pathway in ESCC cells
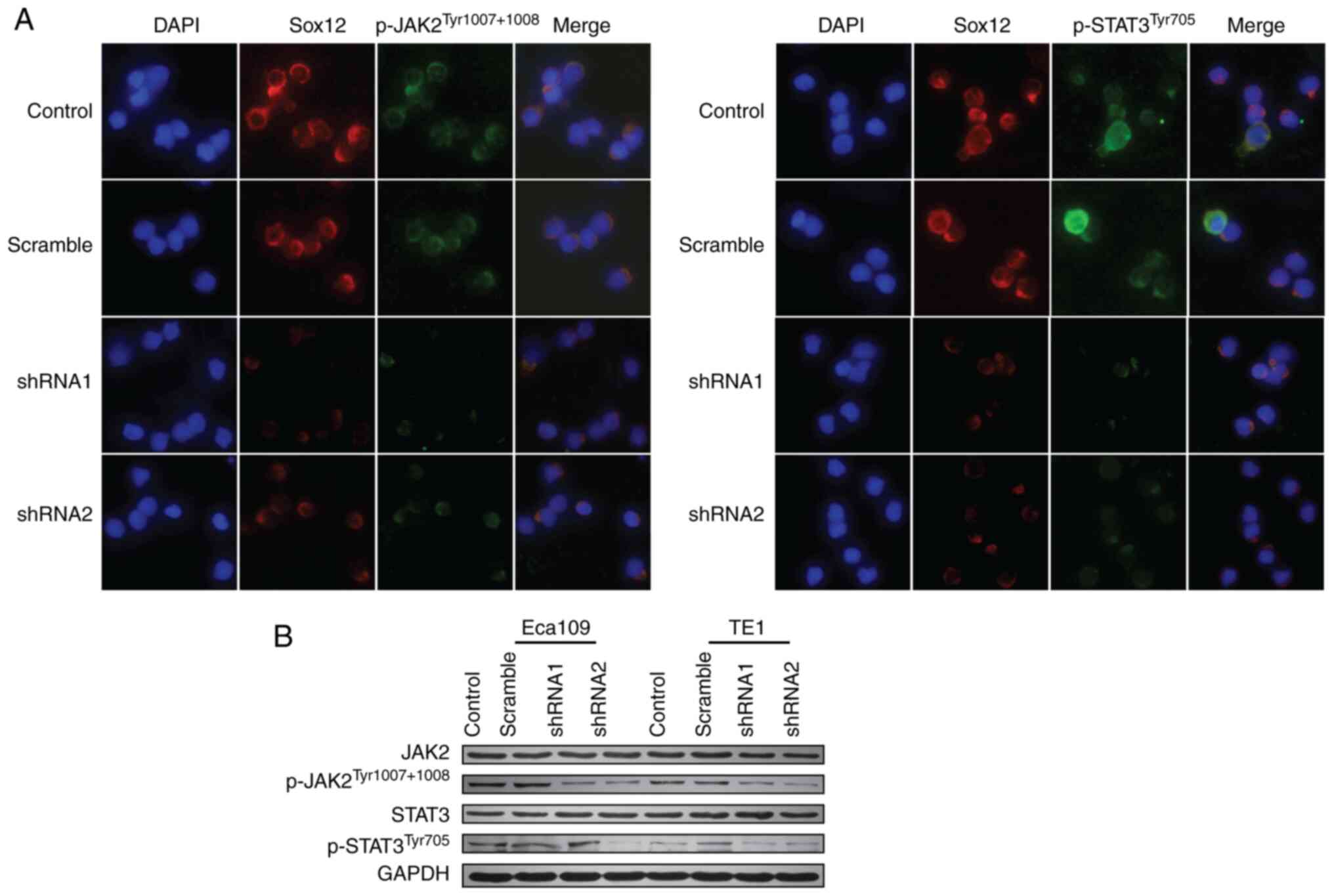
Mutational activation of the JAK2/STAT3 signaling
pathway is responsible for the malignant transformation and
progression of various types of tumors (15). In this study, the protein expression
levels of JAK2/STAT3 signaling proteins were measured by
immunofluorescence and western blot assay. The results revealed
that knockdown of SOX12 in ESCC cells decreased the levels of
p-JAK2Tyr1007+1008 and p-STAT3Tyr705
(Fig. 4), which could be reversed
by co-culture with recombinant SOX12 (10 µM) (Fig. S1C). In addition, WP1066 (5 µM)
could inhibit colony formation and motility of ESCC cells in
vitro (Fig. S2). Thus, it may
be inferred that SOX12 promotes the malignant biological behavior
of ESCC cells, including proliferation, invasion and migration, at
least in part through activation of JAK2/STAT3 signaling
pathway.
Increased expression of SOX12 predicts
poor prognosis for ESCC patients
The correlation between SOX12 and
clinicopathological factors of patients with ESCC was then
evaluated. Statistical analysis indicated no significant difference
in any of the clinicopathological characteristics of ESCC patients,
including sex, age, degree of differentiation, tumor invasion and
lymph node metastasis, between patients with high and low
expression of SOX12 in their ESCC tissues (Table I). To explore the effect of SOX12
expression (high vs. low) on the clinical prognosis of ESCC
patients, Kaplan-Meier survival analysis was used. As presented in
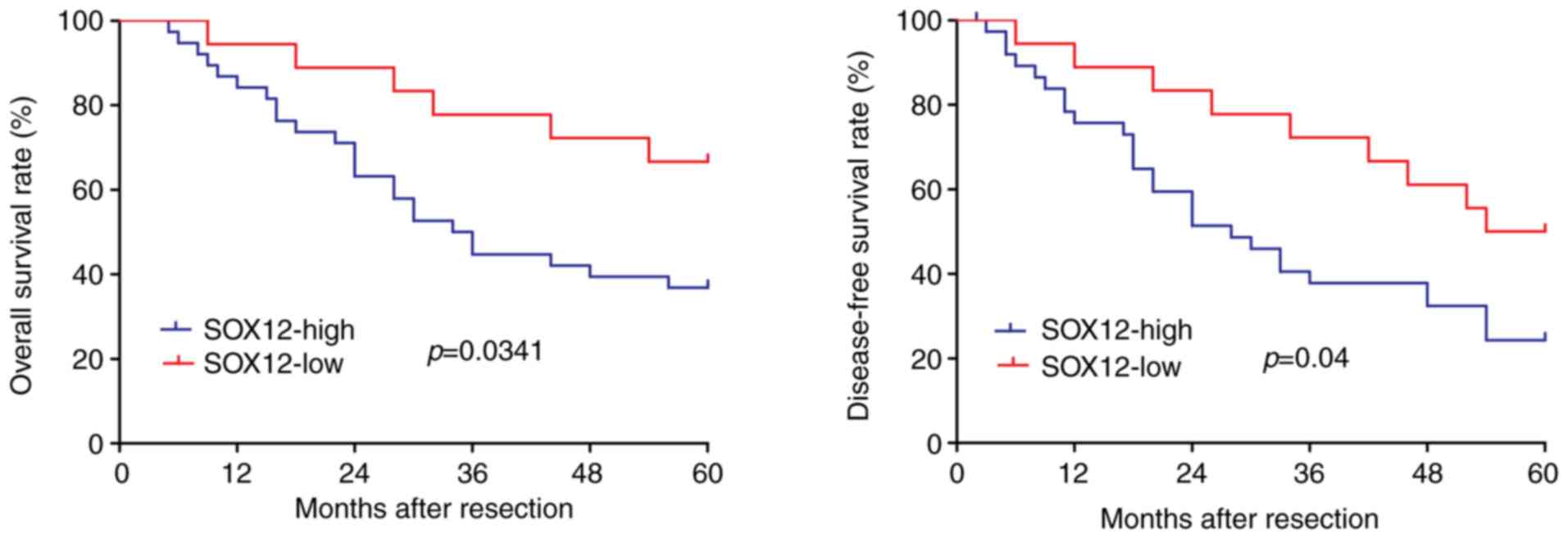
Fig. 5, high expression of SOX12
was associated with lower OS (P=0.0341) and DFS (P=0.04) compared
with low expression of SOX12. Accordingly, overexpression of SOX12
was closely associated to the poor prognosis and survival of
patients.
 | Table I.Association between SOX12 expression
and ESCC clinicopathological features. |
Table I.
Association between SOX12 expression
and ESCC clinicopathological features.
|
|
| SOX12 expression |
|
|
|---|
|
|
|
|
|
|
|---|
| Variables | n | High | Low | χ2 | P-value |
|---|
| Sex |
|
|
| 0.282 | 0.596 |
| Male | 41 | 27 | 14 |
|
|
|
Female | 15 | 11 | 4 |
|
|
| Age (years) |
|
|
| 0.136 | 0.712 |
| ≤60 | 26 | 17 | 9 |
|
|
|
>60 | 30 | 21 | 9 |
|
|
| Differentiation |
|
|
| 2.994 | 0.224 |
| Grade
1 | 17 | 9 | 8 |
|
|
| Grade
2 | 32 | 23 | 9 |
|
|
| Grade
3 | 7 | 6 | 1 |
|
|
| p-T |
|
|
| 1.309 | 0.253 |
|
T1/2 | 19 | 11 | 8 |
|
|
| T3 | 37 | 27 | 10 |
|
|
| p-N |
|
|
| 0.083 | 0.773 |
| N0 | 17 | 12 | 5 |
|
|
|
N1/2 | 39 | 26 | 13 |
|
|
Analysis of the clinical data by univariate logistic
regression using Cox's proportional hazards model indicated that
SOX12 is a significant prognostic factor for ESCC patients
(P=0.016; Table II). Multivariate
logistic regression analysis revealed that SOX12 is an independent
prognostic factor for ESCC patients (P=0.044; Table II). The results of clinical
analysis revealed that overexpression of SOX12 may affect the
malignant characteristics of ESCC and poor prognosis.
 | Table II.Univariate and Multivariate Cox's
proportional hazard models (n=56). |
Table II.
Univariate and Multivariate Cox's
proportional hazard models (n=56).
| Term | Risk ratio | 95% Confidence
interval | P-value |
|---|
| Univariate |
|
|
|
|
Grade | 1.372 | 0.742–2.594 |
0.229 |
|
p-T | 1.775 | 0.825–3.767 |
0.106 |
|
P-N | 1.546 | 0.797–3.141 |
0.157 |
|
SOX12 | 2.242 | 1.161–4.673 |
0.016 |
| Multivariate |
|
|
|
|
Grade | 1.423 | 0.765–2.903 |
0.257 |
|
p-T | 1.814 | 0.894–3.885 |
0.142 |
|
P-N | 1.606 | 0.807–3.302 |
0.183 |
|
SOX12 | 2.105 | 1.073–4.353 |
0.044 |
Discussion
ESCC is the main cause of cancer-related deaths in
China (16). Radical resection
remains the first choice of treatment for ESCC (17). Postoperative recurrence and
metastasis are the key factors limiting the long-term prognosis of
patients (18). Elucidation of the
potential mechanisms of tumor progression may enhance the current
understanding of ESCC and lead to the identification of valuable
biomarkers for diagnosis and targets for treatment.
A previous study has indicated that SOX12, a
multifunctional nuclear transcription factor, serves as a crucial
role in embryonic development and cell-fate determination (8). A recent study indicated that SOX12 may
functionally contribute to maintain stem-like characteristics of
hepatocellular carcinoma (HCC) cells (19). Furthermore, SOX12 was reported to be
responsible for metastasis via activation of the
epithelial-mesenchymal transition process in HCC (20). However, at present, the
understanding of the function of SOX12 in ESCC remains limited.
In the present study, SOX12 was revealed to be
significantly upregulated in ESCC cell lines and tissues. These
results revealed that SOX12 could acts as an oncogene and its
upregulation may participate in the initial tumorigenesis as well
as the malignant development of human cancer, including HCC
(9), breast cancer (10), lung cancer (11), renal cancer (12) and acute myeloid leukemia (13).
The role of SOX12 in ESCC cells was then explored
using knockdown experiments. The present findings indicated that
downregulation of SOX12 significantly inhibited the proliferation
and motility of ESCC cells in vitro. Co-culture of
recombinant SOX12 in transfected cells could recover the
proliferation and motility abilities of ESCC cells. These results
indicated that SOX12 has a significant effect on the malignant
biological behavior of ESCC cells.
Furthermore, clinical data revealed that high
expression of SOX12 in ESCC tissues was closely associated to the
poor prognosis of patients. Overexpression of SOX12 indicated
shorter OS time (P=0.0341) and DFS time (P=0.04). The expression of
SOX12 in esophageal cancer vs. adjacent non-cancerous tissues was
higher in 67.9% of cases. Univariate logistic regression analysis
using Cox's proportional hazards model revealed that SOX12 could be
a significant prognostic factor for ESCC patients (P=0.016).
Furthermore, multivariate logistic regression analysis indicated
that SOX12 was an independent prognostic factor in ESCC patients
(P=0.044). However, there were no significant differences in the
clinicopathological characteristics between patients with high and
low expression of SOX12. This result may be due to two reasons.
First, the small size of the cohort. Second, post-translational
modification (such as methylation modification) may be involved and
play a key role. In a subsequent study, the clinical sample size
will be expanded, and further investigation will be performed to
verify the results. In conclusion, these results indicated that
SOX12 was associated with malignant transformation of ESCC and poor
prognosis.
As a multifunctional nuclear transcription factor,
SOX12 is able to participate in the regulation of multiple
signaling pathways, including WNT/T-cell factor (21). In our pre-experiment (data not
shown), several signaling pathways such as PI3K/AKT, MAPK/ERK,
Wnt/β-catenin and JAK2/STAT3 had been detected. Among all the
signaling pathway proteins we detected, JAK2/STAT3 showed the most
significant difference. This pathway can regulate several cellular
behaviors by activating receptors or intracellular kinases to the
nucleus to regulate gene transcription (22). In recent studies, the JAK2/STAT3
signaling pathway has been revealed to play a crucial role in
proliferation, motility and stemness in cancer cells. USP9X
positively regulated the JAK2/STAT3 pathway to promote the
malignant progression of liver cancer cells (15). miR-375 inhibited the stemness of
breast cancer cells by blocking the JAK2/STAT3 signaling (23). In the present study, knockdown of
SOX12 in ESCC cells decreased the levels of
p-JAK2Tyr1007+1008 and p-STAT3Tyr705, which
could be reversed by co-culture with recombinant SOX12 (10 µM).
WP1066 (5 µM), a novel potent inhibitor of the JAK2/STAT3 signaling
pathway (24), could inhibit colony
formation and motility of ESCC cells in vitro. The present
results indicated that SOX12 could increase the colony formation
rate, mobility of ESCC cells through activation of the JAK2/STAT3
signaling pathway which has been intensely investigated in various
cancer types (25). Mutational
activation of the JAK2/STAT3 is responsible for the malignant
transformation and progression of several types of tumor (26–29).
The present findings indicated that SOX12 could serve a crucial
function in maintaining the malignant biological phenotype of ESCC
cells via activating the JAK2/STAT3 signaling pathway. In a
subsequent study, we will further explore the exact mechanism
between SOX12 and ESCC, since more in vitro and in
vivo experimental evidence is required to be investigated.
Collectively, the present findings revealed that
SOX12 could serve as an oncogenic factor in ESCC. Aberrantly high
levels of SOX12 were indicated to be associated with malignant
biological behavior exhibited in ESCC cells and were revealed to be
an independent unfavorable prognostic factor for patients with
ESCC. SOX12 may become a novel prognostic biomarker and candidate
for the targeted therapy of ESCC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
Not applicable.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
CL and MZ collected, analyzed and interpreted the
data as well as were major contributors in writing the manuscript.
JZ provided the majority of statistical analysis as well as
provided the figures and tables for the manuscript. QL and BoS
collected a large amount of data for the dataset. HC and BiS
oversaw the analysis of the dataset, provided guidance in creating
the dataset and manuscript, and were major contributors in writing
the manuscript. All authors have read and approved the manuscript
in its current state.
Ethics approval and consent to
participate
The present study was approved by the Biomedical
Ethics Committee of the Second Military Medical University
(Shanghai, China). Written informed consent was obtained from all
patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kamangar F, Dores GM and Anderson WF:
Patterns of cancer incidence, mortality, and prevalence across five
continents: Defining priorities to reduce cancer disparities in
different geographic regions of the world. J Clin Oncol.
24:2137–2150. 2006. View Article : Google Scholar
|
|
2
|
Wen SW, Zhang YF, Li Y, Liu ZX, Lv HL, Li
ZH, Xu YZ, Zhu YG and Tian ZQ: Association of miR-21 with
esophageal cancer prognosis: A meta-analysis. Genet Mol Res.
14:6578–6582. 2015. View Article : Google Scholar
|
|
3
|
Zhao K, Chen BJ, Chen ZG, Zhang YJ, Xu D
and Liu Q: Effect of miR-503 down-regulation on growth and invasion
of esophagus carcinoma and related immune function. Med Sci Monit.
21:3564–3569. 2015. View Article : Google Scholar
|
|
4
|
Domper Arnal MJ, Ferrández Arenas Á and
Lanas Arbeloa Á: Esophageal cancer: Risk factors, screening and
endoscopic treatment in Western and Eastern countries. World J
Gastroenterol. 21:7933–7943. 2015. View Article : Google Scholar
|
|
5
|
Xu Y, Yu X, Chen Q and Mao W: Neoadjuvant
versus adjuvant treatment: Which one is better for resectable
esophageal squamous cell carcinoma? World J Surg Oncol. 10:1732012.
View Article : Google Scholar
|
|
6
|
Han TS, Hur K, Xu G, Choi B, Okugawa Y,
Toiyama Y, Oshima H, Oshima M, Lee HJ, Kim VN, et al: MicroRNA-29c
mediates initiation of gastric carcinogenesis by directly targeting
ITGB1. Gut. 64:203–214. 2015. View Article : Google Scholar
|
|
7
|
Dy P, Penzo-Mendez A, Wang H, Pedraza CE,
Macklin WB and Lefebvre V: The three SoxC proteins-Sox4, Sox11 and
Sox12-exhibit overlapping expression patterns and molecular
properties. Nucleic Acids Res. 36:3101–3117. 2008. View Article : Google Scholar
|
|
8
|
Hoser M, Potzner MR, Koch JM, Bosl MR,
Wegner M and Sock E: Sox12 deletion in the mouse reveals
nonreciprocal redundancy with the related Sox4 and Sox11
transcription factors. Mol Cell Biol. 28:4675–4687. 2008.
View Article : Google Scholar
|
|
9
|
Huang W, Chen Z, Shang X, Tian D, Wang D,
Wu K, Fan D and Xia L: Sox12, a direct target of FoxQ1, promotes
hepatocellular carcinoma metastasis through up-regulating Twist1
and FGFBP1. Hepatology. 61:1920–1933. 2015. View Article : Google Scholar
|
|
10
|
Ding H, Quan H, Yan W and Han J: Silencing
of SOX12 by shRNA suppresses migration, invasion and proliferation
of breast cancer cells. Biosci Rep. 36:e003892016. View Article : Google Scholar
|
|
11
|
Wang L, Hu F, Shen S, Xiao H, Li G, Wang M
and Mei J: Knockdown of SOX12 expression inhibits the proliferation
and metastasis of lung cancer cells. Am J Transl Res. 9:4003–4014.
2017.
|
|
12
|
Gu W, Wang B, Wan F, Wu J, Lu X, Wang H,
Zhu Y, Zhang H, Shi G, Dai B and Ye D: SOX2 and SOX12 are
predictive of prognosis in patients with clear cell renal cell
carcinoma. Oncol Lett. 15:4564–4570. 2018.
|
|
13
|
Wan H, Cai J, Chen F, Zhu J, Zhong J and
Zhong H: SOX12: A novel potential target for acute myeloid
leukaemia. Br J Haematol. 176:421–430. 2017. View Article : Google Scholar
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
15
|
Song X, Yang W, Wu C, Han YM and Lu Y:
USP9X promotes the proliferation, invasion and metastasis of liver
cancer cells through regulating the JAK2/STAT3 signaling. Oncol
Lett. 20:2897–2905. 2020. View Article : Google Scholar
|
|
16
|
Chen W, Zheng R, Zhang S, Zeng H, Fan Y,
Qiao Y and Zhou Q: Esophageal cancer incidence and mortality in
China, 2010. Thoracic Cancer. 5:343–348. 2014. View Article : Google Scholar
|
|
17
|
Wang X, Lu Q, Fei X, Zhao Y, Shi B, Li C
and Chen H: Expression and Prognostic Value of Id-4 in patients
with esophageal squamous cell carcinoma. Onco Targets Ther.
13:1225–1234. 2020. View Article : Google Scholar
|
|
18
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar
|
|
19
|
Zou S, Wang C, Liu J, Wang Q, Zhang D, Zhu
S, Xu S, Kang M and He S: Sox12 is a cancer stem-like cell marker
in hepatocellular carcinoma. Mol Cells. 40:847–854. 2017.
|
|
20
|
Jiang T, Guan LY, Ye YS, Liu HY and Li R:
MiR-874 inhibits metastasis and epithelial-mesenchymal transition
in hepatocellular carcinoma by targeting SOX12. Am J Cancer
Res. 7:1310–1321. 2017.
|
|
21
|
Sinner D, Kordich JJ, Spence JR, Opoka R,
Rankin S, Lin SC, Jonatan D, Zorn AM and Wells JM: Sox17 and Sox4
differentially regulate beta-catenin/T-cell factor activity and
proliferation of colon carcinoma cells. Mol Cell Biol.
27:7802–7815. 2007. View Article : Google Scholar
|
|
22
|
Wu R, Liu Y, Zhao Y, Bi Z, Yao Y, Liu Q,
Wang F, Wang Y and Wang X: m6A methylation controls pluripotency of
porcine induced pluripotent stem cells by targeting
SOCS3/JAK2/STAT3 pathway in a YTHDF1/YTHDF2-orchestrated manner.
Cell Death Dis. 10:1712019. View Article : Google Scholar
|
|
23
|
Zhao Q, Liu Y, Wang T, Yang Y, Ni H, Liu
H, Guo Q, Xi T and Zheng L: MiR-375 inhibits the stemness of breast
cancer cells by blocking the JAK2/STAT3 signaling. Eur J Pharmacol.
884:1733592020. View Article : Google Scholar
|
|
24
|
Verstovsek S, Manshouri T, Quintás-Cardama
A, Harris D, Cortes J, Giles FJ, Kantarjian H, Priebe W and Estrov
Z: WP1066, a novel JAK2 inhibitor, suppresses proliferation and
induces apoptosis in erythroid human cells carrying the JAK2 V617F
mutation. Clin Cancer Res. 14:788–96. 2008. View Article : Google Scholar
|
|
25
|
Bollrath J and Greten FR: IKK/NF-kappaB
and STAT3 pathways: Central signalling hubs in
inflammation-mediated tumour promotion and metastasis. EMBO Rep.
10:1314–1319. 2009. View Article : Google Scholar
|
|
26
|
Yang Y, Zhou H, Liu W, Wu J, Yue X, Wang
J, Quan L, Liu H, Guo L, Wang Z, et al: Ganoderic acid A exerts
antitumor activity against MDA-MB-231 human breast cancer cells by
inhibiting the Janus kinase 2/signal transducer and activator of
transcription 3 signaling pathway. Oncol Lett. 16:6515–6521.
2018.
|
|
27
|
Zhou X, Yan T, Huang C, Xu Z, Wang L,
Jiang E, Wang H, Chen Y, Liu K, Shao Z and Shang Z: Melanoma
cell-secreted exosomal miR-155-5p induce proangiogenic switch of
cancer-associated fibroblasts via SOCS1/JAK2/STAT3 signaling
pathway. J Exp Clin Cancer Res. 37:2422018. View Article : Google Scholar
|
|
28
|
Chang R, Song L, Xu Y, Wu Y, Dai C, Wang
X, Sun X, Hou Y, Li W, Zhan X and Zhan L: Loss of Wwox drives
metastasis in triple-negative breast cancer by JAK2/STAT3 axis. Nat
Commun. 9:34862018. View Article : Google Scholar
|
|
29
|
Zhang W, Qiao B and Fan J: Overexpression
of miR-4443 promotes the resistance of non-small cell lung cancer
cells to epirubicin by targeting INPP4A and regulating the
activation of JAK2/STAT3 pathway. Pharmazie. 73:386–392. 2018.
|