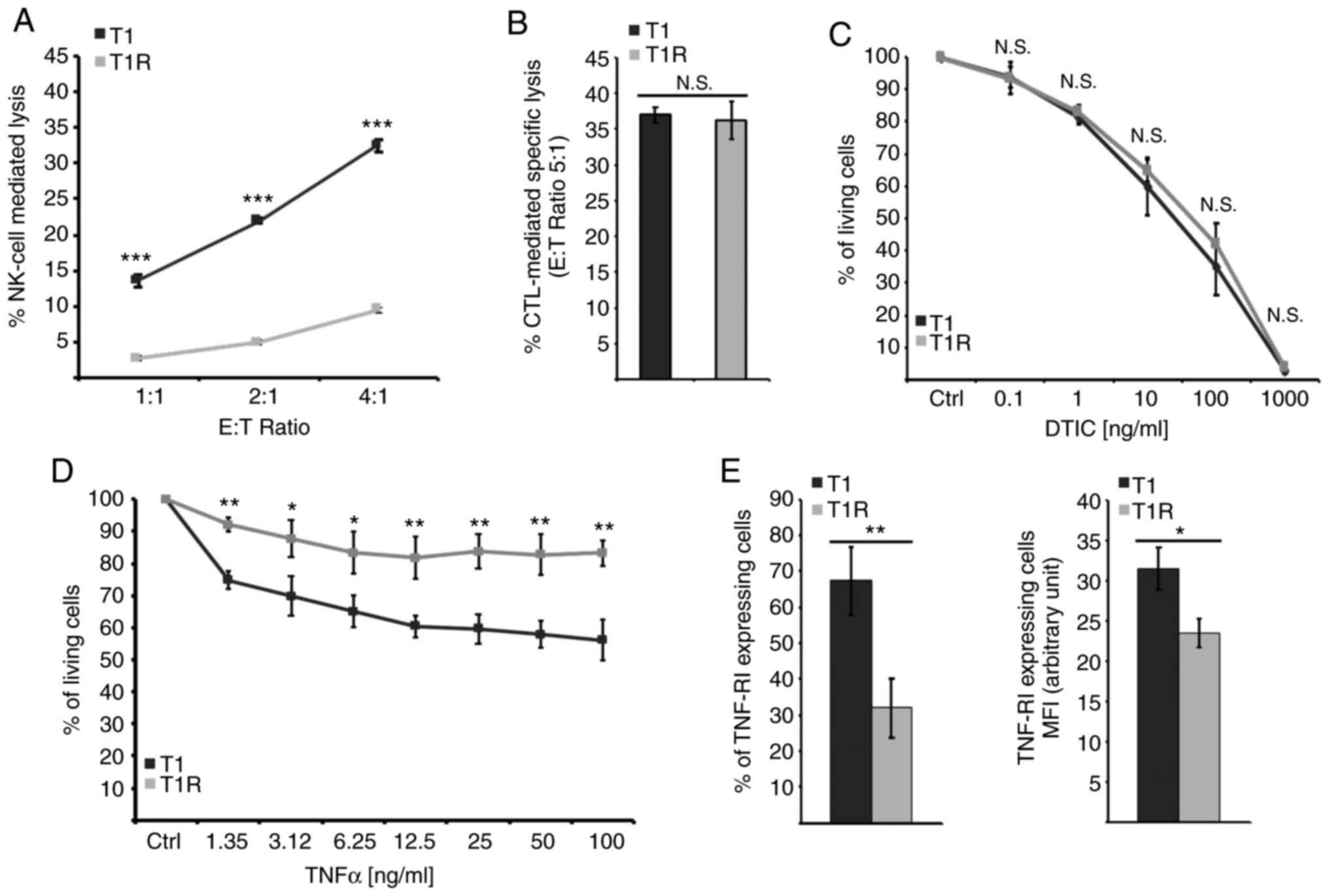
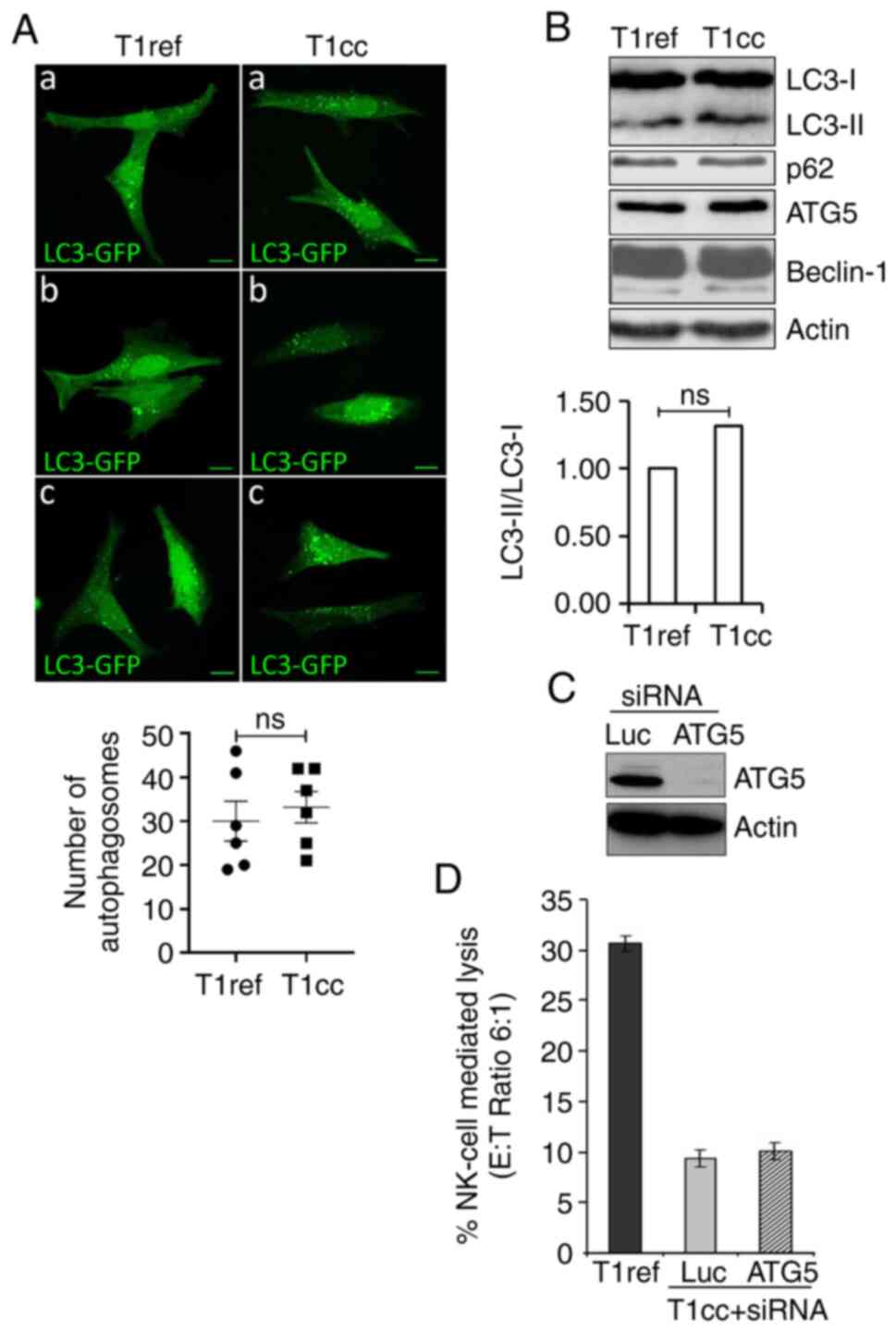
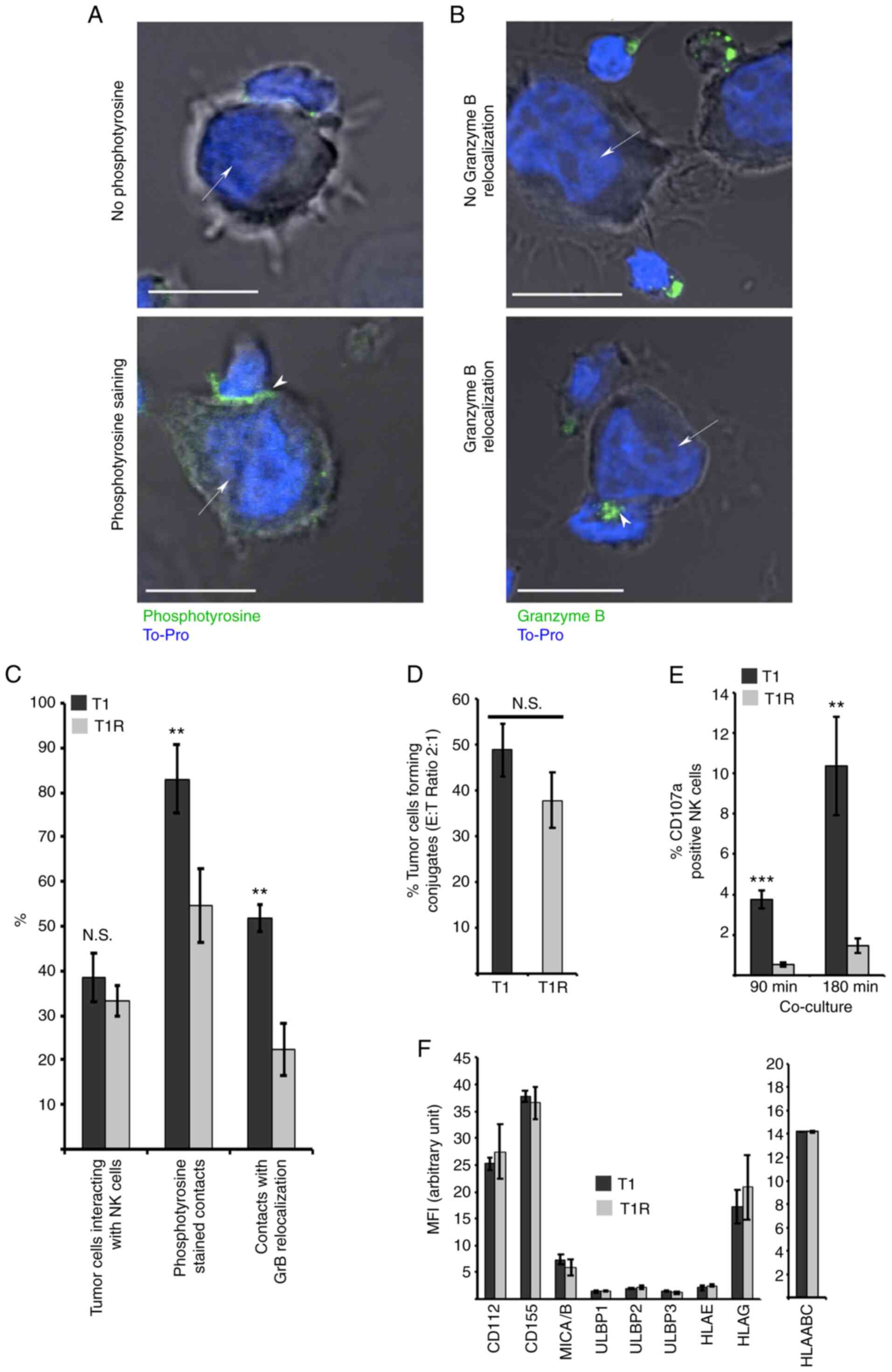
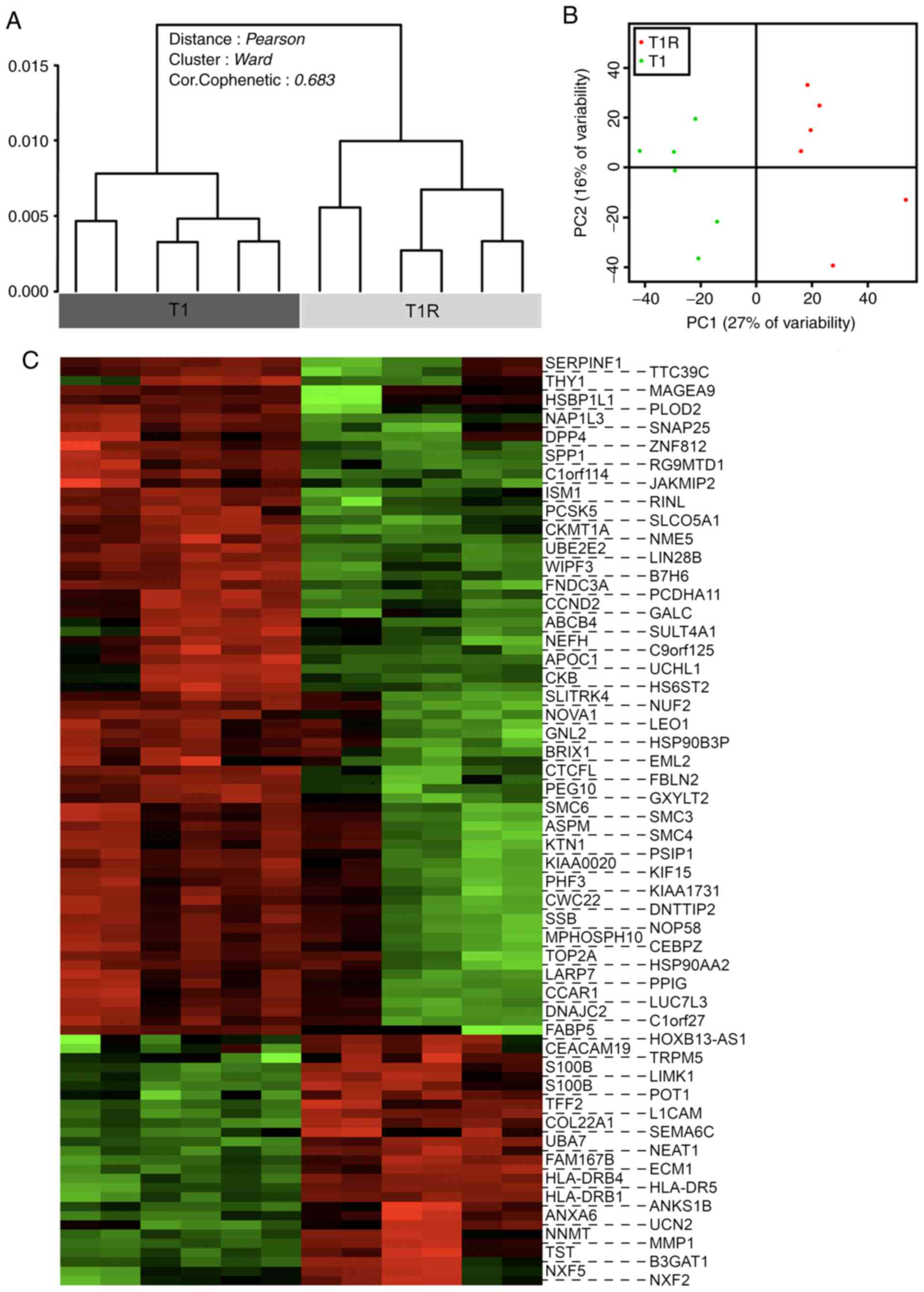
|
1
|
Shankaran V, Ikeda H, Bruce AT, White JM,
Swanson PE, Old LJ and Schreiber RD: Pillars article: IFNγ and
lymphocytes prevent primary tumour development and shape tumour
immunogenicity. Nature. 2001. 410: 1107-1111. J Immunol.
201:827–831. 2018.PubMed/NCBI
|
|
2
|
Dunn GP, Old LJ and Schreiber RD: The
three Es of cancer immunoediting. Annu Rev Immunol. 22:329–360.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Vesely MD, Kershaw MH, Schreiber RD and
Smyth MJ: Natural innate and adaptive immunity to cancer. Annu Rev
Immunol. 29:235–271. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dunn GP, Bruce AT, Ikeda H, Old LJ and
Schreiber RD: Cancer immunoediting: From immunosurveillance to
tumor escape. Nat Immunol. 3:991–998. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dunn GP, Old LJ and Schreiber RD: The
immunobiology of cancer immunosurveillance and immunoediting.
Immunity. 21:137–148. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Abouzahr S, Bismuth G, Gaudin C, Caroll O,
Van Endert P, Jalil A, Dausset J, Vergnon I, Richon C, Kauffmann A,
et al: Identification of target actin content and polymerization
status as a mechanism of tumor resistance after cytolytic T
lymphocyte pressure. Proc Natl Acad Sci USA. 103:1428–1433. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Khong HT and Restifo NP: Natural selection
of tumor variants in the generation of ‘tumor escape’ phenotypes.
Nat Immunol. 3:999–1005. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Trinchieri G: Biology of natural killer
cells. Adv Immunol. 47:187–376. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Vivier E, Tomasello E, Baratin M, Walzer T
and Ugolini S: Functions of natural killer cells. Nature
Immunology. 9:503–510. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Coca S, Perez-Piqueras J, Martinez D,
Colmenarejo A, Saez MA, Vallejo C, Martos JA and Moreno M: The
prognostic significance of intratumoral natural killer cells in
patients with colorectal carcinoma. Cancer. 79:2320–2328. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ishigami S, Natsugoe S, Tokuda K, Nakajo
A, Che X, Iwashige H, Aridome K, Hokita S and Aikou T: Prognostic
value of intratumoral natural killer cells in gastric carcinoma.
Cancer. 88:577–583. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Villegas FR, Coca S, Villarrubia VG,
Jiménez R, Chillón MJ, Jareño J, Zuil M and Callol L: Prognostic
significance of tumor infiltrating natural killer cells subset CD57
in patients with squamous cell lung cancer. Lung Cancer. 35:23–28.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Martín-Fontecha A, Thomsen LL, Brett S,
Gerard C, Lipp M, Lanzavecchia A and Sallusto F: Induced
recruitment of NK cells to lymph nodes provides IFN-γ for T(H)1
priming. Nat Immunol. 5:1260–1265. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Morandi B, Mortara L, Chiossone L, Accolla
RS, Mingari MC, Moretta L, Moretta A and Ferlazzo G: Dendritic cell
editing by activated natural killer cells results in a more
protective cancer-specific immune response. PLoS One. 7:e391702012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Moretta L, Ferlazzo G, Bottino C, Vitale
M, Pende D, Mingari MC and Moretta A: Effector and regulatory
events during natural killer-dendritic cell interactions. Immunol
Rev. 214:219–228. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kijima M, Saio M, Oyang G-F, Suwa T,
Miyauchi R, Kojima Y, Imai H, Nakagawa J, Nonaka K, Umemura N, et
al: Natural killer cells play a role in MHC class I in vivo
induction in tumor cells that are MHC negative in vitro. Int
J Oncol. 26:679–684. 2005.PubMed/NCBI
|
|
17
|
Farag SS, Fehniger TA, Ruggeri L, Velardi
A and Caligiuri MA: Natural killer cell receptors: New biology and
insights into the graft-versus-leukemia effect. Blood.
100:1935–1947. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ferlazzo G and Moretta L: Dendritic cell
editing by natural killer cells. Crit Rev Oncog. 19:67–75. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dunn GP, Bruce AT, Sheehan KCF, Shankaran
V, Uppaluri R, Bui JD, Diamond MS, Koebel CM, Arthur C, White JM
and Schreiber RD: A critical function for type I interferons in
cancer immunoediting. Nat Immunol. 6:722–729. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Smyth MJ, Dunn GP and Schreiber RD: Cancer
immunosurveillance and immunoediting: The roles of immunity in
suppressing tumor development and shaping tumor immunogenicity. Adv
Immunol. 90:1–50. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Smyth MJ, Swann J, Cretney E, Zerafa N,
Yokoyama WM and Hayakawa Y: NKG2D function protects the host from
tumor initiation. J Exp Med. 202:583–588. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Street SE, Hayakawa Y, Zhan Y, Lew AM,
MacGregor D, Jamieson AM, Diefenbach A, Yagita H, Godfrey DI and
Smyth MJ: Innate immune surveillance of spontaneous B cell
lymphomas by natural killer cells and gammadelta T cells. J Exp
Med. 199:879–884. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Crowe NY, Smyth MJ and Godfrey DI: A
critical role for natural killer T cells in immunosurveillance of
methylcholanthrene-induced sarcomas. J Exp Med. 196:119–127. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Takeda K, Smyth MJ, Cretney E, Hayakawa Y,
Kayagaki N, Yagita H and Okumura K: Critical role for tumor
necrosis factor-related apoptosis-inducing ligand in immune
surveillance against tumor development. J Exp Med. 195:161–169.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Smyth MJ, Crowe NY and Godfrey DI: NK
cells and NKT cells collaborate in host protection from
methylcholanthrene-induced fibrosarcoma. Int Immunol. 13:459–463.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ben Safta T, Ziani L, Favre L, Lamendour
L, Gros G, Mami-Chouaib F, Martinvalet D, Chouaib S and Thiery J:
Granzyme B-activated p53 interacts with Bcl-2 to promote cytotoxic
lymphocyte-mediated apoptosis. J Immunol. 194:418–428. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Noman MZ, Janji B, Kaminska B, Van Moer K,
Pierson S, Przanowski P, Buart S, Berchem G, Romero P, Mami-Chouaib
F and Chouaib S: Blocking hypoxia-induced autophagy in tumors
restores cytotoxic T-cell activity and promotes regression. Cancer
Res. 71:5976–5986. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Janji B, Viry E, Moussay E, Paggetti J,
Arakelian T, Mgrditchian T, Messai Y, Noman MZ, Van Moer K, Hasmim
M, et al: The multifaceted role of autophagy in tumor evasion from
immune surveillance. Oncotarget. 7:17591–17607. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Dufour E, Carcelain G, Gaudin C, Flament
C, Avril MF and Faure F: Diversity of the cytotoxic
melanoma-specific immune response: Some CTL clones recognize
autologous fresh tumor cells and not tumor cell lines. J Immunol.
158:3787–3795. 1997.PubMed/NCBI
|
|
30
|
Smyth GK: Linear models and empirical
bayes methods for assessing differential expression in microarray
experiments. Stat Appl Genet Mol Biol. 3:Article32004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Le Maux Chansac B, Missé D, Richon C,
Vergnon I, Kubin M, Soria JC, Moretta A, Chouaib S and Mami-Chouaib
F: Potentiation of NK cell-mediated cytotoxicity in human lung
adenocarcinoma: Role of NKG2D-dependent pathway. Int Immunol.
20:801–810. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Weichold FF, Jiang YZ, Dunn DE, Bloom M,
Malkovska V, Hensel NF and Barrett AJ: Regulation of a
graft-versus-leukemia effect by major histocompatibility complex
class II molecules on leukemia cells: HLA-DR1 expression renders
K562 cell tumors resistant to adoptively transferred lymphocytes in
severe combined immunodeficiency mice/nonobese diabetic mice.
Blood. 90:4553–4558. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wan SM, Tie J, Zhang YF, Guo J, Yang LQ,
Wang J, Xia SH, Yang SM, Wang RQ and Fang DC: Silencing of the
hPOT1 gene by RNA inference promotes apoptosis and inhibits
proliferation and aggressive phenotype of gastric cancer cells,
likely through up-regulating PinX1 expression. J Clin Pathol.
64:1051–1057. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bellucci R, Nguyen HN, Martin A, Heinrichs
S, Schinzel AC, Hahn WC and Ritz J: Tyrosine kinase pathways
modulate tumor susceptibility to natural killer cells. J Clin
Invest. 122:2369–2383. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hai J, Zhu CQ, Bandarchi B, Wang YH, Navab
R, Shepherd FA, Jurisica I and Tsao MS: L1 cell adhesion molecule
promotes tumorigenicity and metastatic potential in non-small cell
lung cancer. Clin Cancer Res. 18:1914–1924. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Meier F, Busch S, Gast D, Göppert A,
Altevogt P, Maczey E, Riedle S, Garbe C and Schittek B: The
adhesion molecule L1 (CD171) promotes melanoma progression. Int J
Cancer. 119:549–555. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Stoeck A, Gast D, Sanderson MP, Issa Y,
Gutwein P and Altevogt P: L1-CAM in a membrane-bound or soluble
form augments protection from apoptosis in ovarian carcinoma cells.
Gynecol Oncol. 104:461–469. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Peric B, Zagar I, Novakovic S, Zgajnar J
and Hocevar M: Role of serum S100B and PET-CT in follow-up of
patients with cutaneous melanoma. BMC Cancer. 11:3282011.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Xiong GP, Zhang JX, Gu SP, Wu YB and Liu
JF: Overexpression of ECM1 contributes to migration and invasion in
cholangiocarcinoma cell. Neoplasma. 59:409–415. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Stern N, Markel G, Arnon TI, Gruda R, Wong
H, Gray-Owen SD and Mandelboim O: Carcinoembryonic antigen (CEA)
inhibits NK killing via interaction with CEA-related cell adhesion
molecule 1. J Immunol. 174:6692–6701. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang WJ, Li QQ, Xu JD, Cao XX, Li HX, Tang
F, Chen Q, Yang JM, Xu ZD and Liu XP: Over-expression of ubiquitin
carboxy terminal hydrolase-L1 induces apoptosis in breast cancer
cells. Int J Oncol. 33:1037–1045. 2008.PubMed/NCBI
|
|
42
|
Wu QJ, Gong CY, Luo ST, Zhang DM, Zhang S,
Shi HS, Lu L, Yan HX, He SS, Li DD, et al: AAV-mediated human PEDF
inhibits tumor growth and metastasis in murine colorectal
peritoneal carcinomatosis model. BMC Cancer. 12:1292012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Iguchi-Manaka A, Kai H, Yamashita Y,
Shibata K, Tahara-Hanaoka S, Honda S, Yasui T, Kikutani H, Shibuya
K and Shibuya A: Accelerated tumor growth in mice deficient in
DNAM-1 receptor. J Exp Med. 205:2959–2964. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Guillerey C and Smyth MJ: NK cells and
cancer immunoediting. Curr Top Microbiol Immunol. 395:115–145.
2016.PubMed/NCBI
|
|
45
|
Ferlazzo G, Tsang ML, Moretta L, Melioli
G, Steinman RM and Münz C: Human dendritic cells activate resting
natural killer (NK) cells and are recognized via the NKp30 receptor
by activated NK cells. J Exp Med. 195:343–351. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Brandt CS, Baratin M, Yi EC, Kennedy J,
Gao Z, Fox B, Haldeman B, Ostrander CD, Kaifu T, Chabannon C, et
al: The B7 family member B7-H6 is a tumor cell ligand for the
activating natural killer cell receptor NKp30 in humans. J Exp Med.
206:1495–1503. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Balsamo M, Vermi W, Parodi M, Pietra G,
Manzini C, Queirolo P, Lonardi S, Augugliaro R, Moretta A,
Facchetti F, et al: Melanoma cells become resistant to
NK-cell-mediated killing when exposed to NK-cell numbers compatible
with NK-cell infiltration in the tumor: Innate immunity. Eur J
Immunol. 42:1833–1842. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Fiegler N, Textor S, Arnold A, Rölle A,
Oehme I, Breuhahn K, Moldenhauer G, Witzens-Harig M and Cerwenka A:
Downregulation of the activating NKp30 ligand B7-H6 by HDAC
inhibitors impairs tumor cell recognition by NK cells. Blood.
122:684–693. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Kaifu T, Escalière B, Gastinel LN, Vivier
E and Baratin M: B7-H6/NKp30 interaction: A mechanism of alerting
NK cells against tumors. Cell Mol Life Sci. 68:3531–3539. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Lakshmikanth T, Burke S, Ali TH, Kimpfler
S, Ursini F, Ruggeri L, Capanni M, Umansky V, Paschen A, Sucker A,
et al: NCRs and DNAM-1 mediate NK cell recognition and lysis of
human and mouse melanoma cell lines in vitro and in vivo. J Clin
Invest. 119:1251–1263. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Fang L, Lonsdorf AS and Hwang ST:
Immunotherapy for advanced melanoma. J Invest Dermatol.
128:2596–2605. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Sutlu T and Alici E: Natural killer
cell-based immunotherapy in cancer: Current insights and future
prospects. J Intern Med. 266:154–181. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Ljunggren HG and Malmberg KJ: Prospects
for the use of NK cells in immunotherapy of human cancer. Nat Rev
Immunol. 7:329–339. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Pachynski RK, Zabel BA, Kohrt HE, Tejeda
NM, Monnier J, Swanson CD, Holzer AK, Gentles AJ, Sperinde GV,
Edalati A, et al: The chemoattractant chemerin suppresses melanoma
by recruiting natural killer cell antitumor defenses. J Exp Med.
209:1427–1435. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Albertsson PA, Basse PH, Hokland M,
Goldfarb RH, Nagelkerke JF, Nannmark U and Kuppen PJK: NK cells and
the tumour microenvironment: Implications for NK-cell function and
anti-tumour activity. Trends Immunol. 24:603–609. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Kim MH, Kitson RP, Albertsson P, Nannmark
U, Basse PH, Kuppen PJ, Hokland ME and Goldfarb RH: Secreted and
membrane-associated matrix metalloproteinases of IL-2-activated NK
cells and their inhibitors. J Immunol. 164:5883–5889. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Balsamo M, Scordamaglia F, Pietra G,
Manzini C, Cantoni C, Boitano M, Queirolo P, Vermi W, Facchetti F,
Moretta A, et al: Melanoma-associated fibroblasts modulate NK cell
phenotype and antitumor cytotoxicity. Proc Natl Acad Sci USA.
106:20847–20852. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Drake CG, Jaffee E and Pardoll DM:
Mechanisms of immune evasion by tumors. Adv Immunol. 90:51–81.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Rabinovich GA, Gabrilovich D and Sotomayor
EM: Immunosuppressive strategies that are mediated by tumor cells.
Annu Rev Immunol. 25:267–296. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Pietra G, Manzini C, Rivara S, Vitale M,
Cantoni C, Petretto A, Balsamo M, Conte R, Benelli R, Minghelli S,
et al: Melanoma cells inhibit natural killer cell function by
modulating the expression of activating receptors and cytolytic
activity. Cancer Res. 72:1407–1415. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Kaiser BK, Yim D, Chow IT, Gonzalez S, Dai
Z, Mann HH, Strong RK, Groh V and Spies T:
Disulphide-isomerase-enabled shedding of tumour-associated NKG2D
ligands. Nature. 447:482–486. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Soriani A, Zingoni A, Cerboni C, Iannitto
ML, Ricciardi MR, Di Gialleonardo V, Cippitelli M, Fionda C,
Petrucci MT, Guarini A, et al: ATM-ATR-dependent up-regulation of
DNAM-1 and NKG2D ligands on multiple myeloma cells by therapeutic
agents results in enhanced NK-cell susceptibility and is associated
with a senescent phenotype. Blood. 113:3503–3511. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Valés-Gómez M, Chisholm SE, Cassady-Cain
RL, Roda-Navarro P and Reyburn HT: Selective induction of
expression of a ligand for the NKG2D receptor by proteasome
inhibitors. Cancer Res. 68:1546–1554. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Waldhauer I, Goehlsdorf D, Gieseke F,
Weinschenk T, Wittenbrink M, Ludwig A, Stevanovic S, Rammensee H-G
and Steinle A: Tumor-associated MICA is shed by ADAM proteases.
Cancer Res. 68:6368–6376. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Burke S, Lakshmikanth T, Colucci F and
Carbone E: New views on natural killer cell-based immunotherapy for
melanoma treatment. Trends Immunol. 31:339–345. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Veuillen C, Aurran-Schleinitz T,
Castellano R, Rey J, Mallet F, Orlanducci F, Pouyet L, Just-Landi
S, Coso D, Ivanov V, et al: Primary B-CLL resistance to NK cell
cytotoxicity can be overcome in vitro and in vivo by priming NK
cells and monoclonal antibody therapy. J Clin Immunol. 32:632–646.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Chouaib S, Pittari G, Nanbakhsh A, El
Ayoubi H, Amsellem S, Bourhis JH and Spanholtz J: Improving the
outcome of leukemia by natural killer cell-based immunotherapeutic
strategies. Front Immunol. 5:952014. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Terme M, Ullrich E, Delahaye NF, Chaput N
and Zitvogel L: Natural killer cell-directed therapies: Moving from
unexpected results to successful strategies. Nat Immunol.
9:486–494. 2008. View
Article : Google Scholar : PubMed/NCBI
|