Introduction
Ovarian cancer is one of the most lethal types of
gynecological cancer. Although surgery and chemotherapy can improve
survival, the 5-year survival rate remained low at ~50% in the USA
in 2015 (1). Epithelial cancer is
the most common type of malignant ovarian tumor, followed by
malignant germ cell tumor (2–4).
Ovarian cancer is difficult to detect during the early stages due
to the small size of the ovary, which is located deep in the pelvic
cavity, and patients may not exhibit symptoms or may only vaguely
show symptoms when tumor cells invade or spread to other parts of
the body (5). Patients with ovarian
cancer typically have a poor prognosis due to a lack of signs or
screening tests that would facilitate early detection, resulting in
a considerable proportion of cases being diagnosed at an advanced
stage (6). During
epithelial-mesenchymal transition (EMT), epithelial cells can
repress their epithelial morphology and acquire motile and invasive
properties during tumor progression (7). Surgery combined with chemotherapy is
the primary treatment modality for patients with ovarian cancer;
however, among patients who undergo surgery for removal of an
ovarian epithelial tumor, only ~30% of the tumors are confined to
the ovary, and the majority of patients have tumors that have
spread to pelvic and abdominal organs, resulting in a low success
rate of surgical excision (8).
Furthermore, the beneficial effects of maintenance chemotherapy
have not been clearly demonstrated (9). Therefore, determining the molecular
mechanisms underlying metastasis of ovarian cancer may highlight
potential novel therapeutic targets.
Keratin 7 (KRT7) is a member of the keratin gene
family, which is subdivided into type I (KRT9-KRT22) and type II
(KRT1-KRT8) (10). Type II
cytokeratin is composed of alkaline or neutral proteins and is
specifically expressed in simple epithelial cells in the lumen of
the internal organs and in the ducts and blood vessels of glands
(11). KRT7 is abnormally expressed
in various types of cancer, such as esophageal squamous cell,
cervical and colorectal cancers (12–15).
KRT7 also participates in cervical cancer invasion, metastasis and
promotes the malignant progression of cancer (16). Several studies have reported an
association between KRT7 dysregulation and clinical pathology or
prognosis (15,17); however, to the best of our
knowledge, the molecular mechanisms involving KRT7 in promoting
malignant progression have not been determined. KLK4-7 serve key
roles in regulating the expression of their associated genes and
proteins in ovarian cancer (18).
As a downstream effector, KRT7 may be an important therapeutic
target for treatment of ovarian cancer. However, the expression and
function of KRT7 in ovarian cancer has not been determined, to the
best of our knowledge.
The present study validates the role of KRT7 in
ovarian cancer progression. By using TCGA database and in
vitro and in vivo experiments, the crucial role of KRT7
in the metastasis and proliferation of ovarian cancer cells has
been confirmed. This indicates the role of KRT7 in ovarian cancer
and may provide a potential target for cancer therapy.
Materials and methods
Cell culture
In the present study, seven ovarian cancer cell
lines (HEY, 59M, COV504, COV413A, DOV13, COV318 and OVCAR433),
obtained from Nanjing KeyGen Biotech Co., Ltd., were cultured and
maintained. Ovarian cancer cells were cultured in RPMI-1640 medium
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS
(Thermo Fisher Scientific, Inc.) and 1% penicillin-streptomycin
solution (Beyotime Institute of Biotechnology) in a 5%
CO2 humidified incubator at 37°C.
Plasmid construction and cell
transfection
pcDNA3.1-KRT7, pcDNA3.1-TGFβ1 and short hairpin
psi-H1-shKRT7 plasmids were synthesized by GeneCopoeia Co., Ltd.
Plasmids were transfected into HEY and OVCAR433 cells (1 µg
plasmid/105 cells) using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.). pcDNA3.1 and psi-H1
empty vectors were used as the control. After 24 or 48 h, total RNA
and protein were extracted, respectively, and then analyzed using
reverse transcription-quantitative PCR (RT-qPCR) and western
blotting to determine the transfection efficiency. For stable
KRT7-knockdown cells, OVCAR433 cells were transfected with the
shKRT7 plasmid (1 µg plasmid/105 cells) using
Lipofectamine® 2000, and cultured with puromycin at a
final concentration of 5 µg/ml for 3 weeks to obtain a stable cell
line. The KRT7 shRNA sequences were as follows: shKRT7-top,
5′-AATTCGGAATACCCGGAATGAGATTTCGAAAAATCTCATTCCGGGTATTCCG-3′; and
shKRT7-bot,
5′-GATCCGGAATACCCGGAATGAGATTTTTCGAAATCTCATTCCGGGTATTCCG-3′.
RNA extraction and reverse
transcription-quantitative PCR
Total RNA in cells was extracted using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. cDNA was
synthesized using HiScript QRT SuperMix from the qPCR kit (Vazyme
Biotech Co., Ltd.) according to the manufacturer's protocol. qPCR
was performed using TransStart Top Green qPCR Super Mix (TransGen
Biotech Co., Ltd.) on an ABI Step One Plus (Applied Biosystem;
Thermo Fisher Scientific, Inc.). The thermocycling conditions of
qPCR were as follows: 94°C for 4 min, 30 cycles of 94°C for 45 sec,
56°C for 30 sec and 72°C for 30 sec, and 72°C for 10 min. The
sequences of the primers for amplification of KRT7 were: KRT7
forward, 5′-CGAGGATATTGCCAACCGCAG−3′ and reverse,
5′-CCTCAATCTCAGCCTGGAGCC−3′; and GAPDH forward,
5′-GGAGTCCACTGGCGTCTT−3′ and reverse, 5′-AGTCCTTCCACGATACCAA-3′ and
were synthesized by GeneCopoeia, Inc. Data were quantified using
the 2−ΔΔCq method (19)
and normalized to GAPDH expression in each respective sample.
Western blotting
Total cellular proteins were lysed on ice using RIPA
lysis buffer [consisting of 50 mM Tris (pH 7.4), 150 mM NaCl, 1%
NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 100X PMSF and 100X PMSF
protease inhibitor cocktail]. After the protein concentration was
measured by the BCA method, 100 µg protein was loaded per lane on a
10% SDS gel, resolved using SDS-PAGE and transferred to a PVDF
membrane. The PVDF membrane was blocked using TBS with 0.1%
Tween-20 buffer containing 5% skimmed milk at room temperature for
2 h, incubated with primary antibodies at room temperature for 2 h,
and then incubated with HRP-labeled goat anti-rabbit IgG (catalog
no. S0001; Affinity Biosciences; 1:3,000) and HRP-labeled goat
anti-mouse IgG (catalog no. S0002; Affinity Biosciences; 1:5,000)
antibodies at room temperature for 2 h. Finally, proteins were
detected using the BeyoECL Plus kit (catalog no. P0018S; Beyotime
Institute of Biotechnology). The relative density of the bands was
quantified using ImageJ software (version 1.52; National Institutes
of Health). The following antibodies were used: Anti-KRT7 (catalog
no. AF0195; Affinity Biosciences; 1:500), anti-GAPDH (catalog no.
AF7021; Affinity Biosciences; 1:2,000), anti-E-cadherin (catalog
no. AF0131; Affinity Biosciences; 1:1,000), anti-N-cadherin
(catalog no. AF4039; Affinity Biosciences; 1:1,000), anti-vimentin
(catalog no. AF7013; Affinity Biosciences; 1:1,000), anti-Snail
(catalog no. AF6032; Affinity Biosciences; 1:500), anti-matrix
metalloproteinase (MMP)2 (catalog no. AF0577; Affinity Biosciences;
1:1000), anti-MMP9 (catalog no. AF5228; Affinity Biosciences;
1:5,000), anti-fibronectin (FN) (catalog no. AF5335; Affinity
Biosciences; 1:200), anti-integrin-β1 (catalog no. AF5379; Affinity
Biosciences; 1:500), anti-FAK (catalog no. AF6397; Affinity
Biosciences; 1:1,000), anti-phosphorylated (p)-FAK (catalog no.
AF3398; Affinity Biosciences; 1:500), anti-Smad2/3 (catalog no.
AF6367; Affinity Biosciences; 1:500), anti-phosphorylated Smad2/3
(catalog no. AF3367; Affinity Biosciences; 1:500) and anti-TGF-β1
(catalog no. AF1027; Affinity Biosciences; 1:1,000).
Transwell migration assay
Transwell experiments were performed using a CoStar
Transwell chamber (8-µm pore size; BD Biosciences). HEY and
OVCAR433 cells were seeded in the upper chamber at a concentration
of 1×109 cells/well in 300 µl serum-free medium, and the
lower chamber was filled with 700 µl medium supplemented with 10%
FBS to induce cell migration. The cells were incubated for 24 h at
37°C with 5% CO2, and the cells on the upper surface of
the membrane were gently removed using a cotton swab. Cells that
had migrated to the lower surface of the membrane were fixed with
4% paraformaldehyde and stained with crystal violet at room
temperature. Images of the cells were obtained using a light
microscope (magnification, ×100).
Wound healing assay
HEY and OVCAR433 cells were seeded in 24-well plates
in serum-free medium at a final concentration of 5×105
cells per well. After 24 h, the cell layer was scraped with a 10-µl
pipette tip to form a straight wound. After removing suspended
cells with PBS, cells were grown for 48 h. Pictures of the wound
area were taken under a light microscope (magnification, ×100) at
0, 24 and 48 h. The difference between the wound area at different
time points and 0 h was used to calculate the cell migration rate.
Each experiment was performed in triplicate.
Cell proliferation assay
HEY and OVCAR433 cell proliferation was examined
using a Cell Counting Kit-8 (CCK-8) assay (catalog no. C0037;
Beyotime Institute of Biotechnology) according to the
manufacturer's protocol. For the assay, ~3×103
transfected cells were seeded in 96-well plates and cultured for
24, 48 and 72 h. Subsequently, 10 µl CCK-8 solution was added to
each well and incubated for 1 h. The absorbance at 450 nm was
measured at 0, 24 and 48 h using a microplate reader.
Colony formation assay
HEY and OVCAR433 cells were harvested, seeded on
six-well plates (1×103 cells/well) and cultured for 14
days at 37°C in a humidified incubator with 5% CO2. The
medium was changed every 3 days. After 14 days, cells were fixed
with 4% paraformaldehyde and stained using crystal violet at room
temperature. Clone numbers were counted by eye after photographs
were obtained.
Analysis of clinical data of patients
with ovarian cancer
The representative images of the immunohistochemical
assay were obtained from The Human Protein Atlas (http://www.proteinatlas.org; normal patient id: 2344,
tumor patient ID: 1115) (20). TCGA
data for transcriptional analysis were obtained from the UALCAN
database (ualcan.path.uab.edu/index.html) (21). The clinical stage and pathological
grade information of patients with ovarian cancer were obtained
from TCGA database (https://www.cancer.gov/tcga). The correlation
analyisis between KRT7 and TGF-β1, integrin-β1 (ITGB1), MMP2 and
SMAD2 expression was performed using ChIPBase version 2.0
(http://rna.sysu.edu.cn/chipbase/index.php).
Differentially expressed gene analysis of KRT7 in TCGA ovarian
cancer samples was performed using DECenter (Sanger Box; http://soft.sangerbox.com/). The significantly
upregulated genes [log(fold-change)| ≥1.0] were analyzed by Gene
Ontology (GO) enrichment using Metascape (metascape.org/) (22).
In vivo experiments
A total of 12 5-week-old female BALB/c mice (~16 g)
were purchased from Charles River Laboratories, Inc., and mice were
randomly divided into two groups. One group was injected with
OVCAR433/shNC cells and the other group was injected with
OVCAR433/shKRT7 cells. All animal experiments were performed in
accordance with ethical standards of the Institutional Animal Use
and Care Committee of the Affiliated Hospital of Zunyi Medical
University, and ethical approval (approval no. 2019-11) was
obtained from the Institutional Animal Use and Care Committee of
the Affiliated Hospital of Zunyi Medical University prior to the
commencement of the study. The animals were kept in a constant
temperature environment of 26-28°C and kept in light for 10 h a
day. All drinking water and food were sterilized and provided ad
libitum. OVCAR433/shNC and OVCAR433/shKRT7 cells
(2×106 cells) were subcutaneously injected into nude
mice (n=6 per group). Tumor volumes were monitored every 3 days and
animal health was continuously monitored throughout the study. The
tumor volume was calculated as follows: Volume = (length ×
width2)/2. A total of 1 month after injection, all
animals were euthanized through intravenous injection of sodium
pentobarbital at a final concentration of 100 mg/kg. The mice were
checked for >5 min and death was confirmed by observing lack of
respiration and cardiac output. Subsequently, the solid tumors were
harvested from the mice by surgery. All tissues were fixed in 4%
formalin and embedded in paraffin for immunohistochemical
staining.
Embedded tumor tissues were cut into sections (4-µm
thickness) and treated with 3% hydrogen peroxide for 10 min to
inhibit endogenous peroxidase activity. Subsequently, 5% bovine
serum albumin (Shanghai Shenggong Biology Engineering Technology
Service, Ltd.) was used to block non-specific binding at 37°C for
30 min. Tissue sections were treated with primary antibodies
against E-cadherin (catalog no. AF0131; Affinity Biosciences;
1:200) and vimentin (catalog no. AF7013; Affinity Biosciences;
1:200) and incubated overnight in a humidified chamber at 4°C.
Sections were visualized using 3,3-diaminobenzidine at room
temperature for 5 min and counterstained with hematoxylin for 1 min
at room temperature. Then pictures were examined under a light
microscope (magnification, ×100). The expression levels of
E-cadherin and Vimentin protein in the tumor were scored based on
the staining intensity and the percentage of positively stained
cells. Six fields were randomly selected for each slice. The
scoring system used was as follows: Staining intensity × number of
cells with positive scores. Staining intensity: 0, no positive
cells; 1, yellow staining; 2, light brown staining; and 3, dark
brown staining. Percentage of cells with positive scores: 1,
<25%; 2, 25-50%; 3, 51-75%; 4, >75%).
Statistical analysis
All statistical analyses were performed using SPSS
version 19.0 (IBM Corp.). Significant differences between two
groups were compared using an unpaired Student's t-test.
Comparisons of KRT7 expression levels between ovarian cancer
tissues and adjacent normal tissues were analyzed using a paired
Student's t-test. Comparisons among three or more groups were
conducted using ANOVA with post hoc Student-Newman-Keuls test. The
correlation between KRT7 expression and clinicopathological factors
was estimated using Pearson's correlation analysis. The
relationship between the survival rate and KRT7 expression was
analyzed using the Kaplan-Meier method. P<0.05 was considered to
indicate a statistically significant difference.
Results
KRT7 expression is upregulated in
ovarian cancer tissues
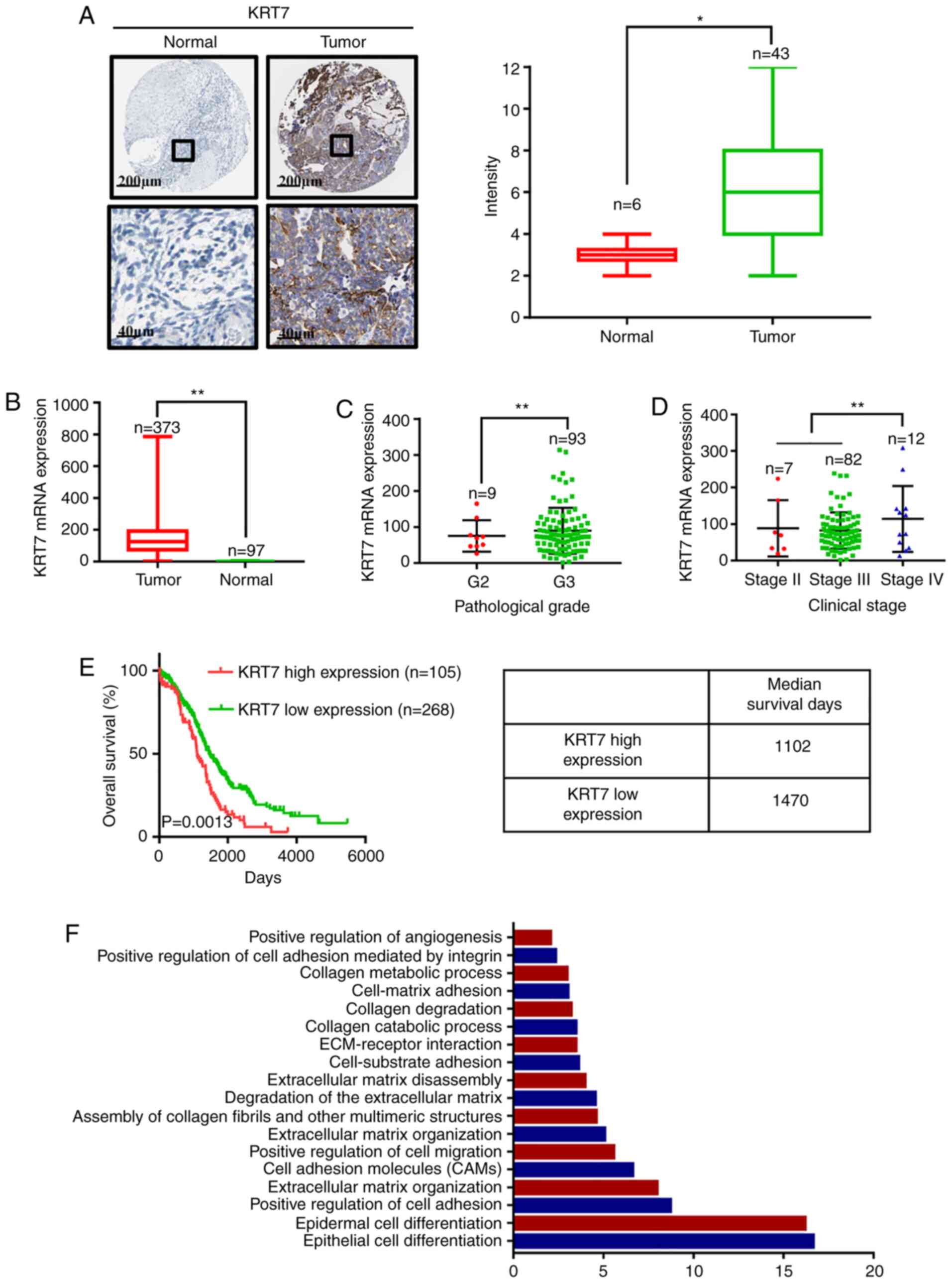
To investigate the clinical significance of KRT7 in
ovarian cancer, the expression levels of KRT7 in ovarian cancer
tissues and adjacent normal tissues was determined using the data
obtained from the Human Protein Atlas. The expression of KRT7
protein was analyzed by immunohistochemical staining in tumor
tissues and corresponding normal (non-tumor) tissues. The
representative images and final scoring results are shown in
Fig. 1A. There was a significant
increase in KRT7 staining in cancer tissues compared with adjacent
normal tissues. Analysis of data obtained from TCGA demonstrated
that KRT7 mRNA expression was higher in tumor tissues compared with
normal tissues (Fig. 1B). By
analyzing TCGA data, KRT7 expression levels in grade 3 samples were
found to be higher than that in grade 2 samples (Fig. 1C). In addition, KRT7 expression was
observed to be the highest in patients with clinical stage IV
ovarian cancer (Fig. 1D).
KRT7 expression levels are associated
with the survival of patients with ovarian cancer
To further elucidate the role of KRT7 in the
survival of patients with ovarian cancer, data obtained from TCGA
were used to determine the relationship between KRT7 mRNA
expression and survival in patients with ovarian cancer.
Kaplan-Meier analysis showed that the median survival of patients
with high and low KRT7 expression levels was 1,102 and 1,470 days,
respectively, indicating a poorer prognosis in patients with high
KRT7 expression (Fig. 1E). After
stratifying ovarian cancer patient data from TCGA based on KRT7
expression levels, the differentially expressed genes between the
high- and low-expression groups were determined. GO enrichment
analysis results showed that KRT7 is associated with ‘positive
regulation of cell migration’ and ‘positive regulation of
angiogenesis’ (Fig. 1F). Therefore,
upregulated KRT7 expression may promote the malignant progression
of ovarian cancer in patients.
KRT7 expression levels in human
ovarian cancer cell lines
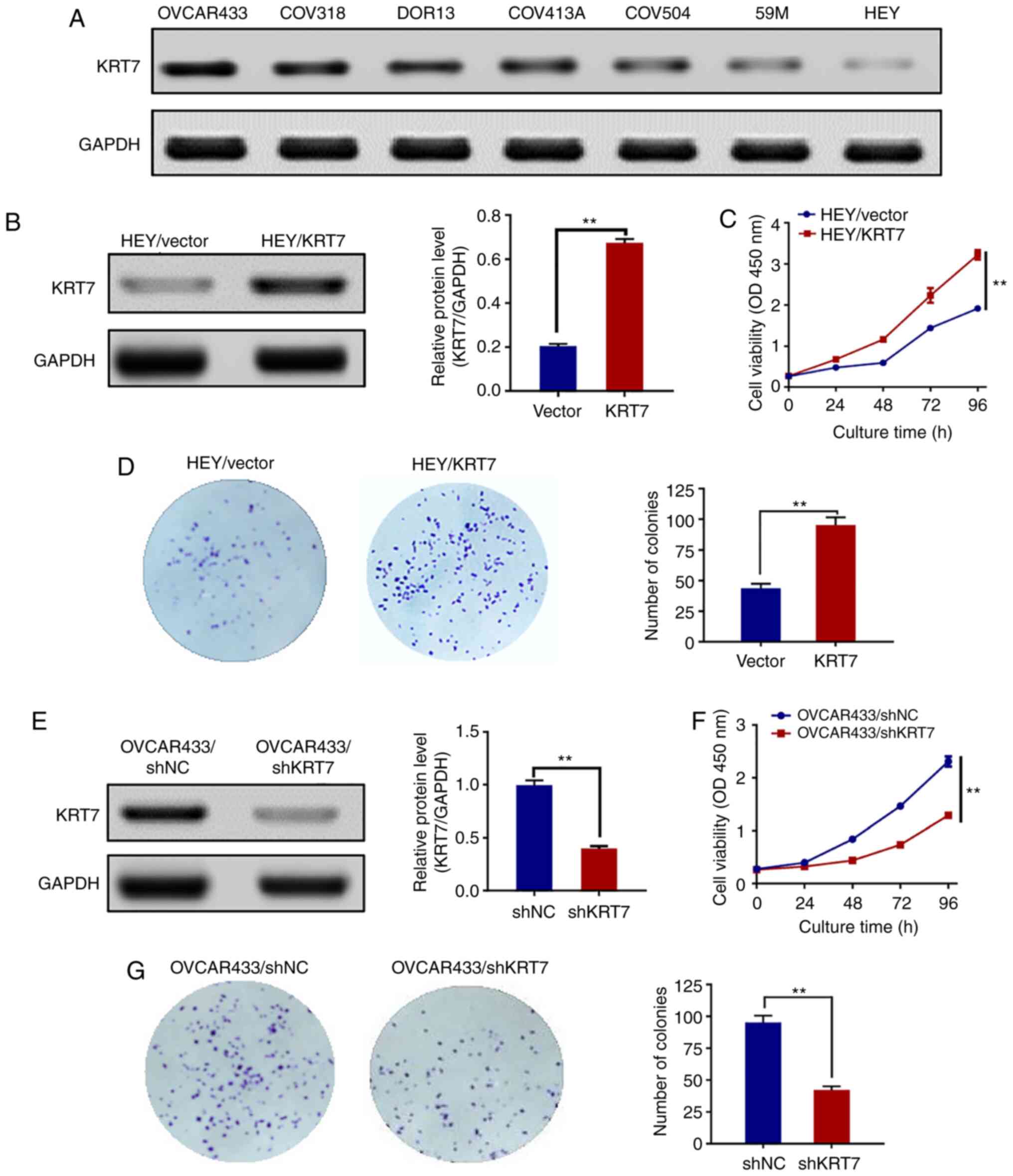
To examine the expression levels of KRT7 in human
ovarian cancer cell lines, seven cell lines (HEY, 59M, COV504,
COV413A, DOR13, COV318 and OVCAR433) were analyzed by western
blotting. KRT7 protein was expressed in all seven human ovarian
cancer cell lines. The expression of the KRT7 protein was the
highest and lowest in the OVCAR433 and HEY cell lines, respectively
(Fig. 2A). Therefore, OVCAR433 and
HEY cells were used for the subsequent experiments.
KRT7 overexpression affects ovarian
cancer cell proliferation
To investigate whether KRT7 affected ovarian cancer
cell proliferation, HEY cells were transfected with pcDNA3.1/KRT7
to enhance KRT7 expression, and HEY cells transfected with pcDNA3.0
blank vector were used as a control. KRT7 overexpression in HEY
cells was confirmed by western blotting (Fig. 2B). Thereafter, the proliferation of
pcDNA3.0-KRT7 and pcDNA3.0 (blank vector) HEY cells was assayed
using the CCK-8 assay. KRT7 overexpression significantly increased
HEY cell proliferation (Fig. 2C)
and resulted in increased colony formation compared with the
control cells (Fig. 2D). KRT7
expression in OVCAR433 cells was then knocked down using shRNA
(Fig. 2E). The proliferation and
colony formation abilities of OVCAR433 cells transfected with
si-KRT7 were decreased compared with the control group (Fig. 2F and G). Results of colony formation
suggest that KRT7 can promote the proliferation of ovarian cancer
cells and may affect the stemness of the cells.
KRT7 affects the migration of ovarian
cancer cells
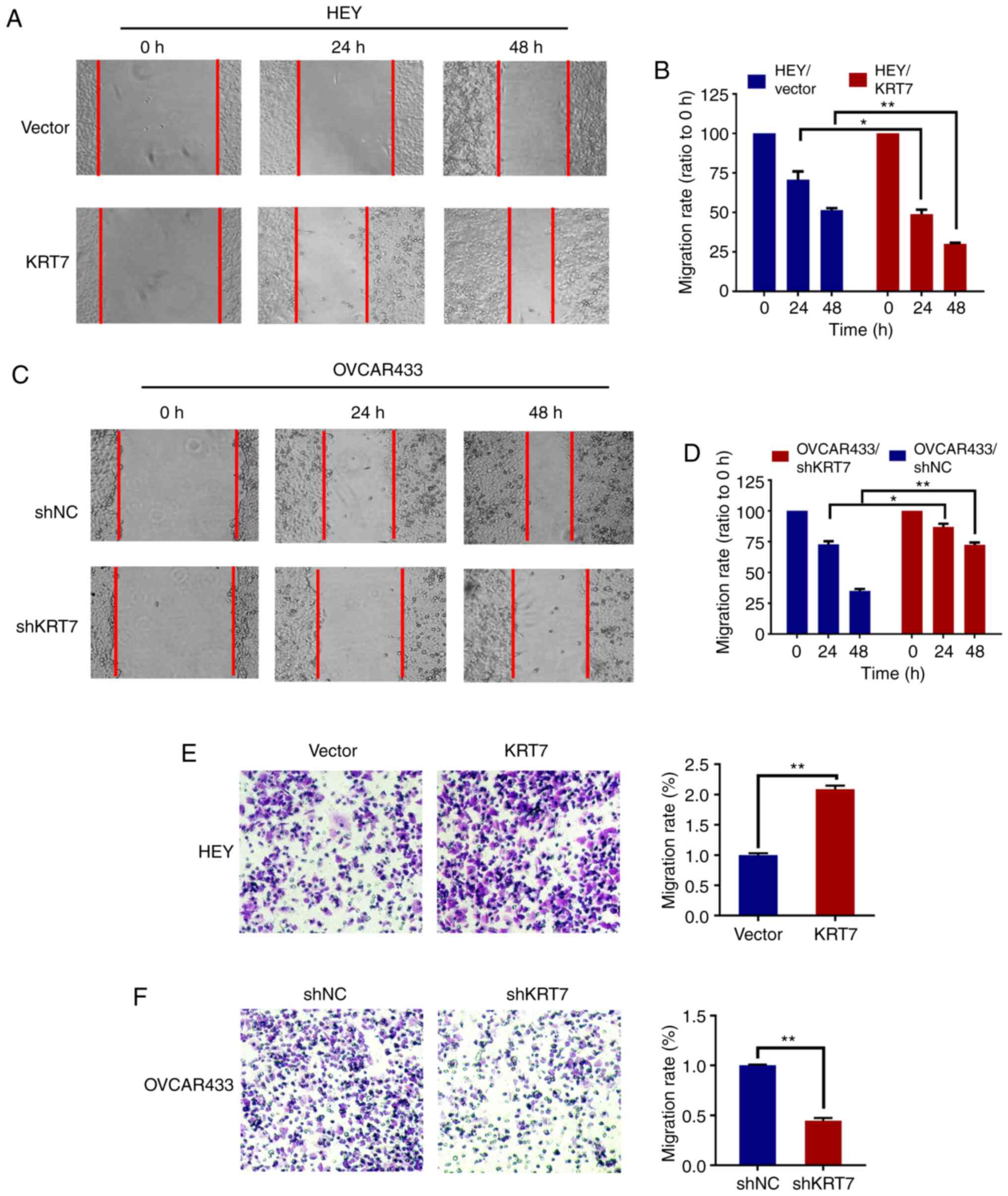
To evaluate the effect of KRT7 on ovarian cell
migration, a wound healing assay with HEY and OVCAR433 cells was
used. Compared with the vector group, the cell migration rate was
significantly increased following KRT7 overexpression in HEY cells
(Fig. 3A and B). In KRT7 -knockdown
OVCAR433 cells, the migration rate decreased significantly
(Fig. 3C and D). The effect of KRT7
on ovarian cell migration was verified using a Transwell assay. The
results demonstrated that KRT7 overexpression significantly
increased the number of migrating HEY cells compared with the
vector group (Fig. 3E). By
contrast, KRT7 knockdown significantly reduced the number of
invading OVCAR433 cells (Fig.
3F).
KRT7 overexpression induces EMT of
ovarian cancer cells
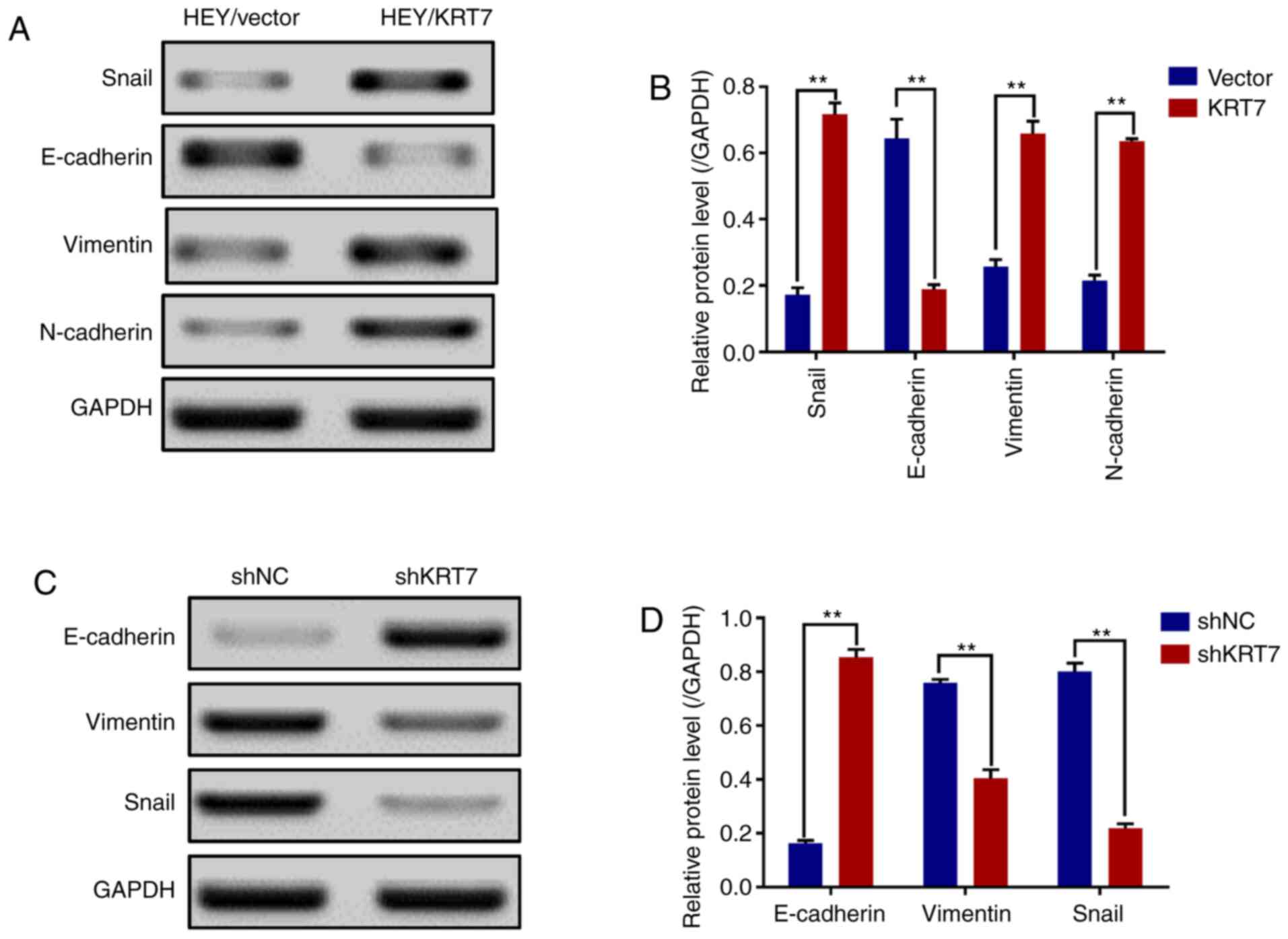
Before migration and invasion of epithelial cells
can occur, there are genotypic and morphological changes which
occur; collectively referred to as EMT. Snail, vimentin, N-cadherin
and E-cadherin are EMT markers (23). The results demonstrated that the
expression levels of Snail, vimentin and N-cadherin were
significantly increased, and that of E-cadherin was significantly
decreased in HEY cell lines overexpressing KRT7 (Fig. 4A and B). By contrast, KRT7 knockdown
in OVCAR433 cells significantly increased E-cadherin expression and
significantly decreased vimentin and Snail expression (Fig. 4C and D). In summary, these data
suggest that KRT7 regulates EMT, resulting in the increased
migratory capacity of ovarian cancer cells in vitro.
KRT7 overexpression increases
cell-matrix adhesion through integrin-β1-FAK signaling
Increasing cell-matrix adhesion is an important step
in tumor cell metastasis (24). The
role of KRT7 in regulating cell-matrix adhesion in ovarian cancer
cells was investigated by detecting the expression of FN,
integrin-β1, FAK and other genes involved in proliferation and
migration. Western blotting revealed that the expression levels of
PCNA, FN, MMP9, integrin-β1 and p-FAK/FAK (Tyr397) were increased,
but TIMP-1 was decreased in HEY cells overexpressing KRT7 (Fig. 5A-D). By contrast, KRT7 knockdown
decreased MMP2 and MMP9 expression in OVCAR433 cells (Fig. 5E). Thus, the adhesion-promoting
effect of KRT7 overexpression was likely mediated through
integrin-β1-FAK signaling.
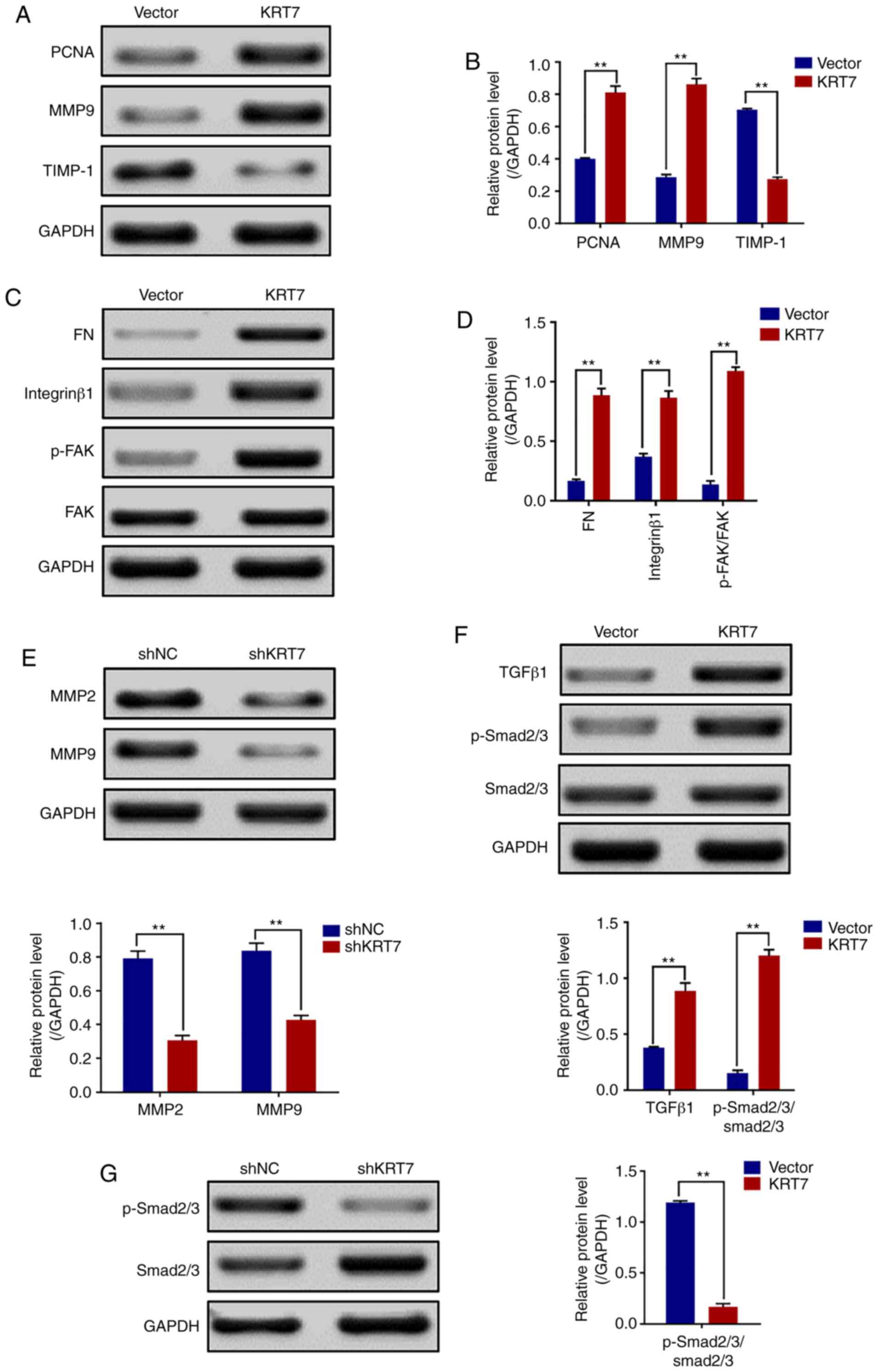 | Figure 5.KRT7 expression affects the
integrin-β1-FAK signaling and TGF-β signaling pathways. (A and B)
Expression of proliferation- and migration-associated genes (PCNA,
MMP9 and TIMP-1) were evaluated using western blotting in HEY
cells. (C and D) Western blotting of proteins involved in
integrin-β1-FAK signaling pathway in the KRT7-overexpressing HEY
cells. (E) Expression of MMPs after knockdown of KRT7 in OVCAR433
cells. (F and G) Expression of the TGF-β signaling pathway-related
proteins was evaluated by western blotting in KRT7-overexpressing
HEY cells and KRT7-knockdown OVCAR433 cells. All experiments were
performed at least three times. Results are presented as the mean ±
standard deviation. **P<0.01. FAK, focal adhesion kinase; PCNA,
proliferating cell nuclear antigen; FN, fibronectin; TIMP-1, TIMP
metallopeptidase inhibitor 1; p-, phosphorylated; MMP, matrix
metalloproteinase; KRT7, keratin 7; sh, short hairpin RNA; NC,
negative control. |
KRT7-induced EMT and cell migration
are mediated by TGF-β signaling
Several pathways regulate cancer cell EMT and
migration. One of the key mechanisms by which TGF-β promotes cell
migration, invasion and metastasis is through induction of EMT
(25). Thus, whether KRT7-induced
EMT was mediated by the TGF-β signaling pathway was determined.
TGF-β1, Smad2/3 and p-Smad2/3, which are important downstream
regulators of the TGF-β signaling pathway, were detected in the
present study. The results showed that the levels of
p-Smad2/3/Smad2/3 in KRT7-overexpressing HEY cells was
significantly increased (Fig. 5F).
The levels of p-Smad2/3/Smad2/3 in KRT7 knockdown OVCAR433 cells
was decreased (Fig. 5G). These data
suggest that KRT7 can partially promote cellular EMT and migration
by activating the TGF-β/Smad2/3 pathway. To verify the
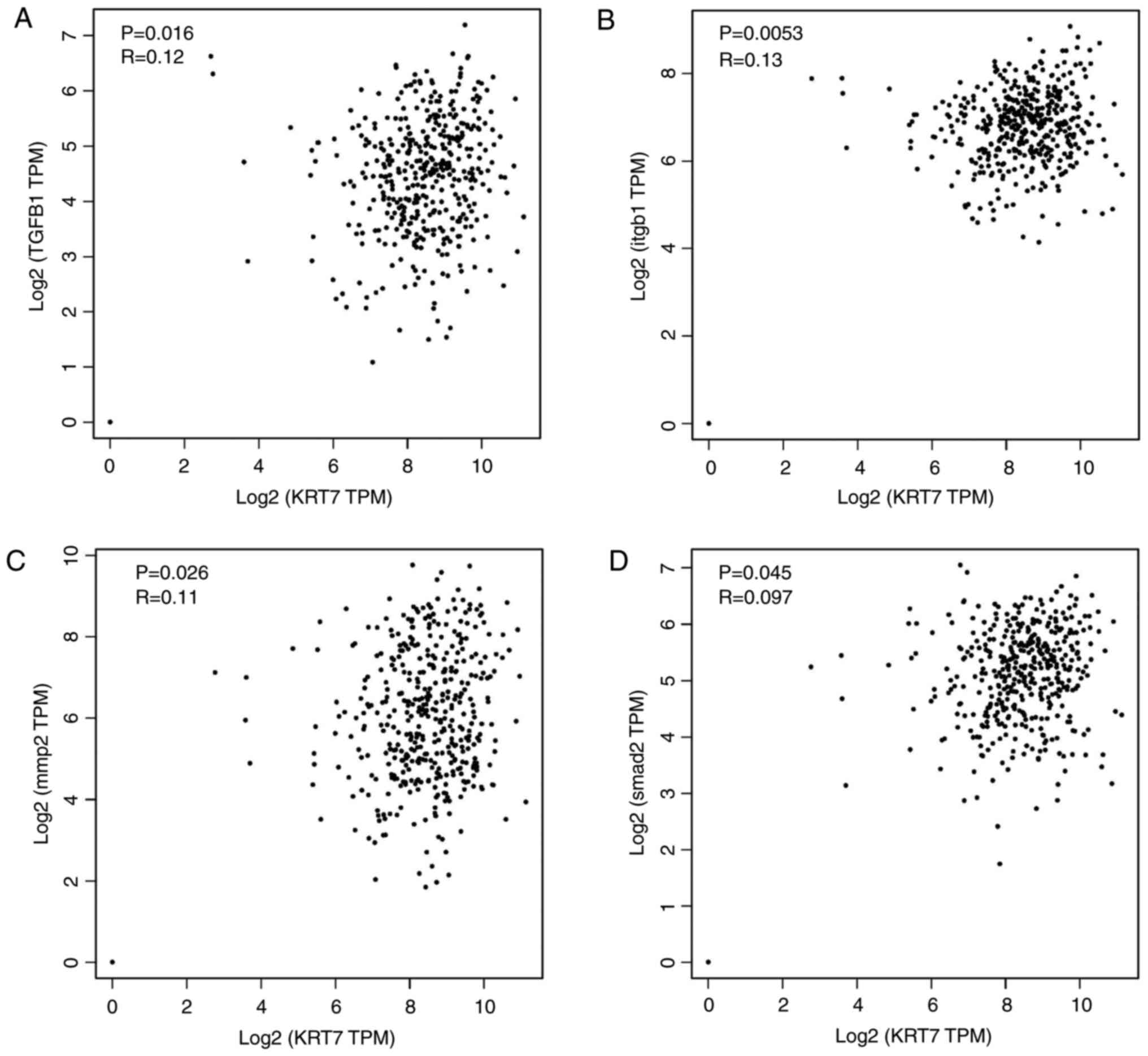
aforementioned molecular mechanism, ChIPBase was used to analyze
the correlation of KRT7 protein expression with TGF-β1, ITGB1, MMP2
and SMAD2 protein expression in patients with ovarian cancer. The
results demonstrated that KRT7 expression levels were positively
correlated with the expression of TGF-β1, ITG-β1, MMP2 and SMAD2
(Fig. 6A-D), verifying the role of
KRT7 in EMT and cell migration through the TGF-β pathway.
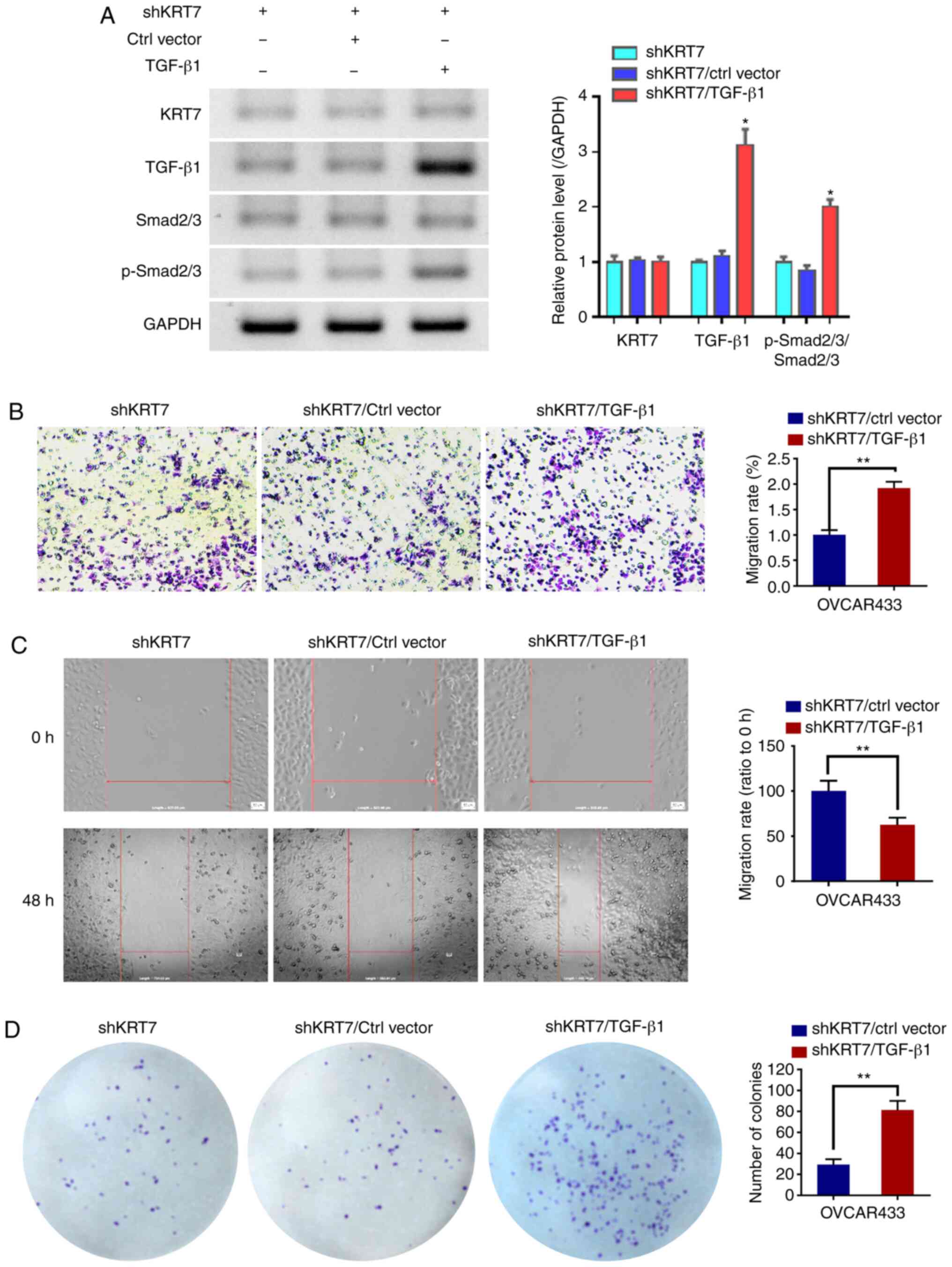
TGF-β1 counteracts the inhibition of
KRT7 knockdown on OVCAR433 cells
To verify whether the function of KRT7 in ovarian
cancer cells was dependent on the TGF-β1 pathway, TGF-β1 was
overexpressed in the KRT7-knockdown OVCAR433 cells. Western
blotting was used to detect TGF-β1 and p-Smad2/3. The results
demonstrated that TGF-β1 could increase the ratio of
p-Smad2/3/Smad2/3. Following confirmation of overexpression
(Fig. 7A), the results of the
Transwell and would healing experiments showed that the
overexpression of TGF-β1 restored the inhibitory effects of KRT7
knockdown (Fig. 7B and C). In
addition, colony formation assays also showed similar results
(Fig. 7D). These results suggest
that the role of KRT7 in ovarian cancer cells is partially
dependent on the TGF-β1 pathway.
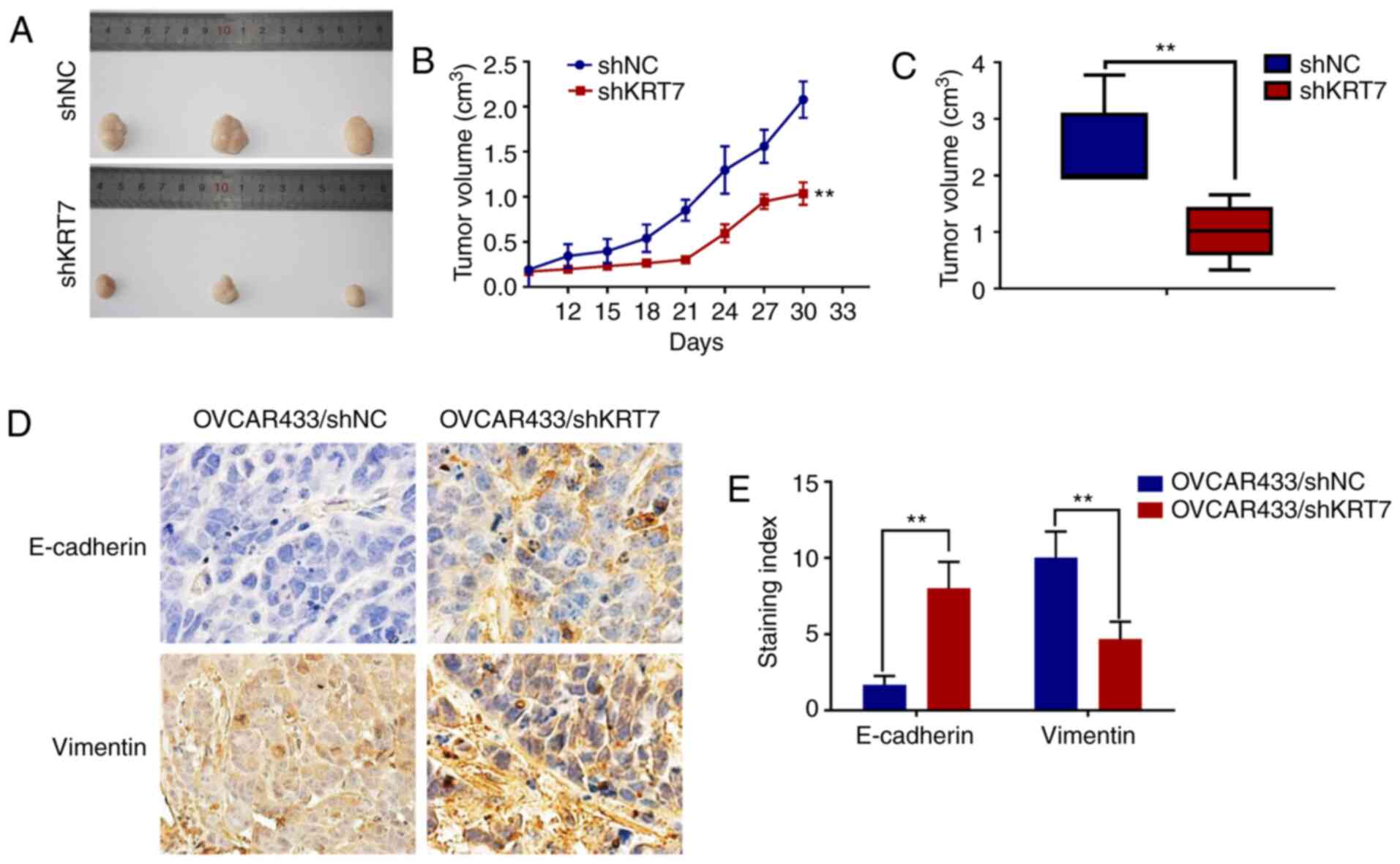
KRT7 knockdown inhibits tumor growth
in vivo
Tumor-bearing mice were established using
KRT7-knockdown OVCAR433 stable cells. Tumor growth in the KRT7
knockdown group was significantly inhibited compared with the
control group (Fig. 8A-C).
Immunohistochemical staining of E-cadherin and vimentin in the
tumors showed significantly higher expression of E-cadherin and
significantly lower expression of vimentin in the KRT7 knockdown
group compared with the control group (Fig. 8D and E). These data suggest that
KRT7 knockdown may inhibit tumor growth in vivo.
Discussion
Although KRT7 has been reported to be overexpressed
in several types of malignant tumors, including gastric cancer and
ovarian cancer, and is associated cell proliferation, EMT and
stemness (26,27). The abnormal expression and possible
roles of KRT7 in ovarian cancer have not been reported previously,
to the best of our knowledge. In the present study, KRT7 expression
in ovarian cancer cells was significantly higher compared with the
normal tissues. The upregulated expression of KRT7 resulted in the
migration of cancer cells and in the alteration in the expression
levels of a series of EMT-related genes, which decreased the
survival time of patients. The present study also demonstrated that
the abnormal expression of KRT7 regulated changes in extracellular
matrix (ECM) and collagen-associated genes, resulting in cancer
cell migration.
The ECM provides biophysical and biochemical signals
that regulate cell proliferation, differentiation, migration and
invasion. It is a dynamic environment that constantly influences
cell response (28). ECM and its
modifications are key factors involved in determining metastatic
tumor formation (29). In several
types of solid cancer, the increased expression of matrix proteins
is associated with increased mortality (30). In addition, changes in matrix
hardness and fibrosis, and excessive deposition of fibrous ECM
components (primarily collagen) are associated with tumor
progression (31). Changes in the
structure and hardness of ECM result in changes in cellular
mechanical transduction, and the associated molecular pathways are
considered potential targets for treatment of cancer (32). The key role of ECM in tumor
migration and metastasis is the expression of ECM receptor
(integrin) on the surface of tumor cells and the ability of tumor
cells to degrade the ECM through MMPs (33). Therefore, the increase in
integrin-β1 and MMP9 expression is associated with cancer cell
migration. MMP-9 is the primary target of colonial migration, which
is associated with a high invasive capacity of cancer cells
(34). FAK is a downstream target
of integrin and an important signaling molecule regulating the cell
response (35). In the present
study, the expression levels of FAK and integrin-β1 were
upregulated when KRT7 was overexpressed, suggesting that KRT7 can
regulate the degradation of the cancer cell ECM, thereby enhancing
the invasive capacity of cells.
TGF-β signal transduction is closely associated with
cancer and serves a dual role in tumorigenesis (36). Generally, TGF-β inhibits cell
proliferation and stimulates normal cell differentiation; thus, it
can be used as a tumor suppressor (37). However, in advanced stage cancer,
TGF-β promotes tumor progression and metastasis, thus acting as a
carcinogen (37). TGF-β-induced EMT
supports tumor invasion and transmission by releasing tumor cells
into the surrounding environment and promoting their movement
(38). In several types of cancer,
TGF-β-induced EMT transcription regulates E-cadherin, Snail,
N-cadherin and vimentin. It also induces Sox4 expression and
promotes mesenchymal transition, facilitating tumor progression and
cancer cell invasion (39–41). In the present study, KRT7 was shown
to be involved in EMT through TGF-β signaling, which in turn
resulted in increased ovarian cancer cell migration. E-cadherin
protein expression was decreased in KRT7-overexpressing ovarian
cancer cells, whereas expression of MMP9, FN and TGF-β signaling
(TGF-β1 and p-Smad2/3) pathway-associated proteins was increased.
Therefore, KRT7 enhanced EMT through the TGF-β/Smad2/3 signaling
pathway and promoted the progression of cancer.
In conclusion, the present study demonstrated that
upregulated expression of KRT7 was associated with ovarian cancer
cell migration and EMT pathways, thereby promoting ovarian cancer
progression, such as tumor invasion depth, and leading to decreased
survival. Therefore, KRT7 may serve as a potential target for
treatment of ovarian cancer.
Acknowledgements
The results shown here are in whole or part based
upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga.
Funding
This study was supported by the Science and
Technology Foundation of Guizhou Provincial Science and Technology
Department (grant no. JLKZ201120).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
BY conceived and designed the study. QA performed
all the experiments. TL, MYW, YJY, ZDZ and ZJL analyzed the data.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
All animal experiments were performed in accordance
with ethical standards of the Institutional Animal Use and Care
Committee of the Affiliated Hospital of Zunyi Medical University,
and ethical approval (approval no. 2019-11) was obtained prior to
the commencement of the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Coukos G, Tanyi J and Kandalaft LE:
Opportunities in immunotherapy of ovarian cancer. Ann Oncol. 27
(Suppl 1):i11–i15. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Smith RA, Andrews KS, Brooks D, Fedewa SA,
Manassaram-Baptiste D, Saslow D, Brawley OW and Wender RC: Cancer
screening in the United States, 2017: A review of current American
Cancer Society guidelines and current issues in cancer screening.
CA Cancer J Clin. 67:100–121. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
US Preventive Services Task Force, ;
Grossman DC, Curry SJ, Owens DK, Barry MJ, Davidson KW, Doubeni CA,
Epling JW Jr, Kemper AR, Krist AH, et al: Screening for ovarian
cancer: US preventive services task force recommendation statement.
JAMA. 319:588–594. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Corrado G, Salutari V, Palluzzi E,
Distefano MG, Scambia G and Ferrandina G: Optimizing treatment in
recurrent epithelial ovarian cancer. Expert Rev Anticancer Ther.
17:1147–1158. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bristow RE and Chi DS: Platinum-based
neoadjuvant chemotherapy and interval surgical cytoreduction for
advanced ovarian cancer: A meta-analysis. Gynecol Oncol.
103:1070–1076. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Derynck R and Weinberg RA: EMT and cancer:
More than meets the eye. Dev Cell. 49:313–316. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Feng W, Dean DC, Hornicek FJ, Shi H and
Duan Z: Exosomes promote pre-metastatic niche formation in ovarian
cancer. Mol Cancer. 18:1242019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Javan Maasomi Z, Pilehvar Soltanahmadi Y,
Dadashpour M, Alipour S, Abolhasani S and Zarghami N: Synergistic
anticancer effects of silibinin and chrysin in T47D breast cancer
cells. Asian Pac J Cancer Prev. 18:1283–1287. 2017.PubMed/NCBI
|
|
10
|
Sandilands A, Smith FJ, Lunny DP, Campbell
LE, Davidson KM, MacCallum SF, Corden LD, Christie L, Fleming S,
Lane EB and McLean WH: Generation and characterisation of keratin 7
(K7) knockout mice. PLoS One. 8:e644042013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Woods RSR, Keegan H, White C, Tewari P,
Toner M, Kennedy S, O'Regan EM, Martin CM, Timon CVI and O'Leary
JJ: Cytokeratin 7 in oropharyngeal squamous cell carcinoma: A
junctional biomarker for human papillomavirus-related tumors.
Cancer Epidemiol Biomarkers Prev. 26:702–710. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hosoya A, Kwak S, Kim EJ, Lunny DP, Lane
EB, Cho SW and Jung HS: Immunohistochemical localization of
cytokeratins in the junctional region of ectoderm and endoderm.
Anat Rec (Hoboken). 293:1864–1872. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Karantza V: Keratins in health and cancer:
More than mere epithelial cell markers. Oncogene. 30:127–138. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Oue N, Noguchi T, Anami K, Kitano S,
Sakamoto N, Sentani K, Uraoka N, Aoyagi K, Yoshida T, Sasaki H and
Yasui W: Cytokeratin 7 is a predictive marker for survival in
patients with esophageal squamous cell carcinoma. Ann Surg Oncol.
19:1902–1910. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Harbaum L, Pollheimer MJ, Kornprat P,
Lindtner RA, Schlemmer A, Rehak P and Langner C: Keratin 7
expression in colorectal cancer-freak of nature or significant
finding? Histopathology. 59:225–234. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lambaudie E, Chereau E, Pouget N,
Thomassin J, Minsat M, Charafe-Jauffret E, Jacquemier J and
Houvenaeghel G: Cytokeratin 7 as a predictive factor for response
to concommitant radiochemotherapy for locally advanced cervical
cancer: A preliminary study. Anticancer Res. 34:177–181.
2014.PubMed/NCBI
|
|
17
|
Kuroda H, Imai Y, Yamagishi H, Ueda Y,
Kuroso K, Oishi Y, Ohashi H, Yamashita A, Yashiro Y and Fukushima
H: Aberrant keratin 7 and 20 expression in triple-negative
carcinoma of the breast. Ann Diagn Pathol. 20:36–39. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang P, Magdolen V, Seidl C, Dorn J,
Drecoll E, Kotzsch M, Yang F, Schmitt M, Schilling O, Rockstroh A,
et al: Kallikrein-related peptidases 4, 5, 6 and 7 regulate
tumour-associated factors in serous ovarian cancer. Br J Cancer.
119:1–9. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Uhlén M, Fagerberg L, Hallström BM,
Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C,
Sjöstedt E, Asplund A, et al: Proteomics. Tissue-based map of the
human proteome. Science. 347:12604192015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chandrashekar DS, Bashel B, Balasubramanya
SA, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BV and Varambally
S: UALCAN: A portal for facilitating tumor subgroup gene expression
and survival analyses. Neoplasia. 19:649–658. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhou Y, Zhou B, Pache L, Chang M,
Khodabakhshi AH, Tanaseichuk O, Benner C and Chanda SK: Metascape
provides a biologist-oriented resource for the analysis of
systems-level datasets. Nat Commun. 10:15232019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pastushenko I and Blanpain C: EMT
transition states during tumor progression and metastasis. Trends
Cell Biol. 29:212–226. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kai F, Drain AP and Weaver VM: The
extracellular matrix modulates the metastatic journey. Dev Cell.
49:332–346. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cantelli G, Orgaz JL, Rodriguez-Hernandez
I, Karagiannis P, Maiques O, Matias-Guiu X, Nestle FO, Marti RM,
Karagiannis SN and Sanz-Moreno V: TGF-β-induced transcription
sustains amoeboid melanoma migration and dissemination. Curr Biol.
25:2899–2914. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Huang B, Song JH, Cheng Y, Abraham JM,
Ibrahim S, Sun Z, Ke X and Meltzer SJ: Long non-coding antisense
RNA KRT7-AS-as is activated in gastric cancers and supports cancer
cell progression by increasing krt7 expression. Oncogene.
35:4927–4936. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tajima Y, Ito K, Umino A, Wilkinson AC,
Nakauchi H and Yamazaki S: Continuous cell supply from
Krt7-expressing hematopoietic stem cells during native
hematopoiesis revealed by targeted in vivo gene transfer method.
Sci Rep. 7:406842017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lu P, Weaver VM and Werb Z: The
extracellular matrix: A dynamic niche in cancer progression. J Cell
Biol. 196:395–406. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Filipe EC, Chitty JL and Cox TR: Charting
the unexplored extracellular matrix in cancer. Int J Exp Pathol.
99:58–76. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Alexander J and Cukierman E: Stromal
dynamic reciprocity in cancer: Intricacies of fibroblastic-ECM
interactions. Curr Opin Cell Biol. 42:80–93. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Byron A, Humphries JD and Humphries MJ:
Defining the extracellular matrix using proteomics. Int J Exp
Pathol. 94:75–92. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pickup MW, Mouw JK and Weaver VM: The
extracellular matrix modulates the hallmarks of cancer. EMBO Rep.
15:1243–1253. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Piwko-Czuchra A, Koegel H, Meyer H, Bauer
M, Werner S, Brakebusch C and Fässler R: Beta1 integrin-mediated
adhesion signalling is essential for epidermal progenitor cell
expansion. PLoS One. 4:e54882009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Khan Z and Marshall JF: The role of
integrins in TGFβ activation in the tumour stroma. Cell Tissue Res.
365:657–673. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Golubovskaya V: Targeting FAK in human
cancer: From finding to first clinical trials. Front Biosci
(Landmark Ed). 19:687–706. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
36
|
Peixoto P, Etcheverry A, Aubry M, Missey
A, Lachat C, Perrard J, Hendrick E, Delage-Mourroux R, Mosser J,
Borg C, et al: EMT is associated with an epigenetic signature of
ECM remodeling genes. Cell Death Dis. 10:2052019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cicchini C, Laudadio I, Citarella F,
Corazzari M, Steindler C, Conigliaro A, Fantoni A, Amicone L and
Tripodi M: TGFbeta-induced EMT requires focal adhesion kinase (FAK)
signaling. Exp Cell Res. 314:143–152. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Attisano L and Wrana JL: Signal
transduction by the TGF-beta superfamily. Science. 296:1646–1647.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Pardali E, Goumans MJ and Ten Dijke P:
Signaling by members of the TGF-beta family in vascular
morphogenesis and disease. Trends Cell Biol. 20:556–567. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zavadil J and Böttinger EP: TGF-beta and
epithelial-to-mesenchymal transitions. Oncogene. 24:5764–5774.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hajra KM, Chen DY and Fearon ER: The SLUG
zinc-finger protein represses E-cadherin in breast cancer. Cancer
Res. 62:1613–1618. 2002.PubMed/NCBI
|