Introduction
Prostate cancer (PCa) is the second leading cause of
cancer-associated mortality in men in America and Europe (1). In China, the morbidity and mortality
rates of PCa have been markedly increasing for the last two decades
(2). The majority of newly
diagnosed PCa patients in China are at an advanced or metastatic
stage that can only be treated by androgen deprivation therapy
(ADT) instead of radical therapy options such as radical
prostatectomy and radical radiotherapy (3). However, the efficacy of ADT eventually
decreases and cancer will inevitably develop into
castration-resistant prostate cancer (CRPC) (4).
Currently, CRPC can be treated by chemotherapy,
immunotherapy, and new androgen receptor blockade agents (5). For example, when CRPC patients were
treated with abiraterone and enzalutamide, the overall survival
period was extended for 15.8 and 18.4 months, respectively
(6). However, the efficacy was
primarily dependent on the AR-V7 expression status in the cancer
tissue or the circulating cancer cell (7). For instance, several chemotherapeutic
agents such as methotrexate, 5-fluorouracil, cyclophosphamide,
doxorubicin, and docetaxel, were heavily investigated for the
treatment of CPRC before the new androgen receptor blockade agents
were used. The results indicated that docetaxel is the most
effective agent for CRPC compared with other chemotherapeutic
agents (8). Therefore,
docetaxel-based chemotherapy is still widely used for the treatment
of CRPC (5). The majority of
patients exhibit limited disease control and overall survival
(9). Most of these patients have
become refractory to second-line chemotherapy such as cabazitaxel
(10). The overall efficacy of all
palliative methods is far from satisfactory.
The mechanism by which CRPC patients display
different therapeutic efficacy to docetaxel remains unexplored to
this day. It suggested that increased cancer stem cell population
(11), dysregulated transcription
factors (12), anti-apoptosis
factors (13), miRNAs (14), as well as testicular nuclear
receptor 4 (TR4) (15) are
associated with chemoresistance in CRPC.
MicroRNAs (miRNAs) are small sets of non-coding RNAs
that are 21–23 nucleotides in length, which regulate target genes
by the degradation of mRNA or by suppressing mRNA translation.
miRNAs regulate the initiation, progression, and chemosensitivity
of different types of cancers, including prostate cancer. For
example, miR-200a has been revealed to modulate chemoresistance by
targeting TP53INP1 and YAP1 in breast cancer (16). miR-429 sensitized pancreatic cancer
cells to gemcitabine through the negative regulation of PDCD4
(17). miRNAs such as miR-200c
(18), miR-205 (18), miR-21 (19), and miR-34 (20) have been reported to modulate
chemosensitivity in PCa.
Although the transcription of miRNA is non-coding,
it is regulated by transcription factors (TFs). It has been
revealed that miRNAs and TFs can cooperate to regulate gene
expression and biological processes (21). Increasing evidence has indicated
that the aberrant regulation of miRNAs by TFs can cause various
diseases, including cancer. For example, Li et al reported
that HIF1A suppresses the expression of miR-34a in colorectal
cancer cells, which in turn promotes the expression of PPP1R11,
thereby activating the epithelial-mesenchymal transition (22). Chang et al revealed that
miR-137 exerts its pro-metastatic function by targeting TFAP2C in
non-small cell lung cancer cells (23). A great amount of recent research has
investigated how miRNAs act to regulate their target genes
(miRNA-gene regulation) and the role they play in various types of
cancer (24). However, studies on
how miRNAs are regulated by TFs (TF-miRNA regulation) are quite
limited.
TR4 is a transcriptional factor that modulates
various molecular signals in multiple malignant tumors including
PCa (25). A previous study has
revealed that TR4 can affect the chemosensitivity of PCa
stem/progenitor cells by regulating octamer-binding transcription
factor 4 (OCT4) expression (15).
In addition, OCT4 is also regulated by miRNAs to exert its function
to affect chemosensitivity (25).
The present study focused on whether any ‘intermediary miRNA’
exists between TR4 and OCT4 to form the TR4/miRNA/OCT4 axis than
can regulate chemosensitivity in PCa.
Materials and methods
Bioinformatics methods
Text mining was used to screen
chemoresistance-related miRNAs in PCa. Four online tools including
TargetScan (http://targetscan.org), miRDB
(http://mirdb.org), miRanda (http://microrna.org), and miRTarBase (http://mirtarbase.mbc.nctu.edu.tw) predicted
microRNAs which targeted the human OCT4 gene. Results were merged
from each database into one dataset. The JASPAR database
(http://jaspar.genereg.net) was utilized
to predict the putative TR4 transcription factor binding site of
the promoter region of microRNA.
Cell culture
The C4–2 human PCa cell line (from ATCC) was
provided by Dr Jer-Tsong Hsieh of the University of Texas
Southwestern Medical Center (Dallas, USA). The PC3 and DU-145 human
PCa cell lines were purchased from the Cell Bank of Type Culture
Collection of the Chinese Academy of Sciences (CBTCCCAS; Shanghai,
China). All of the cell lines were verified by STR analysis. PC3
and C4-2 cells were maintained in RPMI-1640 medium containing
penicillin (25 U/ml), streptomycin (25 g/ml), 1% L-glutamine and
10% fetal bovine serum (all from Gibco; Thermo Fisher Scientific,
Inc.), at 37°C with 5% CO2 in a humidified atmosphere.
The DU-145 cells were maintained in DMEM medium (Gibco; Thermo
Fisher Scientific, Inc.) containing penicillin (25 U/ml),
streptomycin (25 g/ml), 1% L-glutamine, and 10% FBS, at 37°C with
5% CO2 in a humidified atmosphere. After three to four
(3–4) passages, the cells grew well and were
ready to use in the experiments.
Plasmids and lentivirus
A second-generation lentiviral vector and packing
system was used. The TR4 short hairpin (sh)RNA sequence
(5′-cgggagaaaccaagcaattg-3′) was cloned into pLKO.1 puro plasmid
(Addgene, Inc.). In order to overexpress TR4, TR4 cDNA was cloned
into the PWPI vector (Addgene, Inc.). Lentivirus packaging and
purification were the same as previously described (15). The concentration of the lentiviral
vectors was 1 µg/µl. The 293T cell line (ATCC) was used to generate
the packaged lentivirus. For transfection, 20 µg lentiviral plasmid
(1 µg/µl) was used, and the ratio of the lentiviral
plasmid:packaging vector:envelope was was 2:1:1. The duration of
transfection was 48 h. Then conditioned medium containing viral
particles was collected, cellular debris was removed, and viral
particles were concentrated. Lentivirus expressing or inhibiting
hsa-miR-145 or non-targeting negative control was purchased from
Shanghai GeneChem Co., Ltd. Primers for cloning were obtained from
Sangon Biotech Co., Ltd. Human genomic DNA (Promega Corporation)
was used as the template DNA. Lentivirus multiplicity of infection
(MOI) for the C-2, PC-3, and DU-145 cell lines was 20. The duration
of transduction into cells of interest was 24 h and the time
interval between transduction and subsequent experimentation was 24
h.
RNA transfection
Chemically synthesized human miR-145 mimic and
inhibitor and their corresponding non-targeting negative controls
were purchased from Shanghai Genechem Co., Ltd. The sequences were
as follows: miR-145-5p mimics sense, 5′-GUCCAGUUUUCCCAGGAAUCCCU-3′
and antisense, 5′-GGAUUCCUGGGAAAACUGGACUU-3′; mimics negative
control sense, 5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense,
5′-ACGUGACACGUUCGGAGAATT-3′; miR-145-5p inhibitor,
5′-AGGGAUUCCUGGGAAAACUGGAC-3′; inhibitor negative control,
5′-CAGUACUUUUGUGUAGUACAA-3′. According to the manufacturer's
instructions for transient transfection, cells at 50% confluence
were transfected with miRNA mimics (final concertration 50 nmol/l)
or inhibitors (final concertration 100 nmol/l) using Lipofectamine
3000 (Invitrogen; Thermo Fisher Scientific, Inc.) at room
temperature. The duration of transfection was 10 min. The cells
were then incubated under 37°C for 24 h before subsequent
experimentations.
RNA extraction and reverse
transcription-quantitative (RT-q)PCR
For both cells and tissues, total RNAs were isolated
using Trizol reagent (Invitrogen; Thermo Fisher Scientific, Inc.).
Total RNA (2 µg) was subjected to reverse transcription using
Superscript III transcriptase (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions.
Quantitative real-time PCR (qPCR) was conducted using a Bio-Rad
CFX96 system with SYBR-Green (Invitrogen; Thermo Fisher Scientific,
Inc.) to determine the mRNA expression level of the target gene.
Thermocycling conditions were as follows: Samples were initially
held at 95°C for 3 min, then processed through 40 cycles at 95°C
for 15 sec and 60°C for 45 sec. The 2−ΔΔCq method was
used for quantification (26). The
expression levels of specific genes were normalized by being
compared to the expression of GAPDH mRNA. The sequences of the
primers were as follows: OCT4 forward, 5′-CTGGGTTGATCCTCGGACCT-3′
and reverse, 5′-CCATCGGAGTTGCTCTCCA-3′; survivin forward,
5′-AGGACCACCGCATCTCTACAT-3′ and reverse,
5′-AAGTCTGGCTCGTTCTCAGTG-3′; and GAPDH forward,
5′-GGAGCGAGATCCCTCCAAAAT-3′ and reverse,
5′-GGCTGTTGTCATACTTCTCATGG-3′.
For miRNA detection, a PureLink miRNA isolation kit
(Invitrogen; Thermo Fisher Scientific, Inc.) was used to isolate
the miRNAs. Small RNAs (50 ng) were processed for poly-A addition
and cDNA synthesis, as previously described. Quantitative real-time
PCR was then conducted using a Bio-Rad CFX96 system with FAM/FITC
to determine the expression level of the miRNAs of interest.
Expression levels were normalized against U6 small nuclear RNA.
Western blotting
Cells were washed with PBS and lysed in an RIPA
buffer (Sigma-Aldrich; Merck KGaA). Protein concentration was
determined using the bicinchoninic acid (BCA) method of protein
determination with a Microplate Manager V6 (Bio-Rad Laboratories,
Inc.). Proteins (20 µg) were separated on 8–10% SDS-PAGE gel and
then transferred to PVDF membranes (EMD Millipore). Membranes were
blocked in 5% non-fat milk in PBST for 1 h at room temperature, and
then incubated with diluted primary antibodies against GAPDH
(dilution 1:1,000; cat. no. sc-166574; Santa Cruz Biotechnology,
Inc.), TR4 (dilution 1:500; product code ab109513; Abcam), OCT4
(dilution 1:500; product no. 2750; Cell Signaling Technology, Inc.)
overnight at 4°C. The blots were then incubated with HRP-conjugated
secondary antibody (goat anti mouse; 1:10,000; cat. no. A16078; or
goat anti rabbit; 1:10,000; cat. no. A16110; both from Thermo
Fisher Scientific, Inc.) for 1 h at room temperature, washed, and
developed in an ECL system (Bio-Rad Laboratories, Inc.).
MTT assay
Cells were infected with lentivirus for 48 h and
then seeded in a 96-well plate at a concentration of
1×104 cells/well. Docetaxel was added at various
concentrations (0, 2, 5, 10, 15, 20, 30, 40 µg/ml) to the wells 24
h later. After 48 h of incubation, cells from each well were
treated with MTT (0.5 mg/ml) for 4 h at 37°C. The absorbance at 570
nm was determined using a microplate reader (Model 550; Bio-Rad
Laboratories, Inc.).
Chromatin immunoprecipitation
assay
Regarding chromatin immunoprecipitation (ChIP),
cultured C4-2 prostate cancer cells (2×107) were
harvested and treated with 1% formaldehyde at room temperature for
10 min to cross-linked DNA. After sonication, cross-linked
chromatin was precleared with A/G-agarose protein beads and then
immunoprecipitated using anti-TR4-specific antibody (product code
ab109513; Abcam) or Anti-Histone H3 (acetyl K27) antibody (product
code ab4729) or IgG (product code ab172730) overnight at 4°C. IgG
was used as the negative control and Anti-Histone H3 as the
positive control. Supernatants from the no-antibody-added samples
were used to measure total input chromatin. The chromatins were
incubated overnight at 65°C to reverse cross-links. DNA was then
treated with 20 µg/ml RNase A (37°C, 30 min), purified using gel
extraction columns (OMEGA), and resuspended in 50 ml TE buffer.
Then, the TR4 occupancy on chromatin was assessed by PCR with
locus-specific primers.
Luciferase reporter assay
The full-length sequence of the human miR-145
promoter was cloned into a pGL3-basic luciferase reporter vector
(Promega Corporation). Site-directed mutagenesis of the TR4 binding
site in the promoter was achieved with the Quick-Change mutagenesis
(Stratagene; Agilent Technologies, Inc.) according to the
manufacturer's protocols.
The 3′UTR of the human OCT4 gene with predicted
miRNA-responsive elements was cloned into the pGL3-promoter vector
(Promega Corporation) downstream of the firefly luciferase ORF.
Site-directed mutagenesis of miRNA-responsive elements in 3′UTR was
achieved with the Quick-Change mutagenesis according to the
manufacturer's protocols.
Cells were placed in 24-well plates. The plasmids or
chemical miRNA mimics were transfected with Lipofectamine 3000
(Thermo Fisher Scientific, Inc.) according to the manufacturer's
instructions. PRL-TK was used as an internal control. Cell lysates
were prepared with Passive Lysis Buffer (Promega Corporation) 24 h
after transfection. Luciferase activity was measured using the
Dual-Luciferase Reporter Assay (Promega Corporation) according to
the manufacturer's manual. Firefly luciferase activity was
normalized by comparing to Renilla luciferase activity.
In vivo mouse model
Animal experiments were performed according to a
protocol approved by the Ethics Committee of the Second Affiliated
Hospital of Soochow University (Suzhou, China) and followed the
ARRIVE Guidelines Checklist (27).
Forty 8-week-old BALB/c-Foxn1nu male mice with a body
weight of 18–19 g were purchased from the Model Animal Research
Center of Nanjing University (Nanjing, China). They were housed
under standard conditions with a 12-h light/dark cycle at 23±3°C,
55±5% relative humidity, and access to food and water ad
libitum. All the mice received anesthesia by intraperitoneal
injection of pentobarbital 50 mg/kg. Then PC3 human PCa cells
(5×106/mouse) mixed with Matrigel (BD Biosciences) were
concurrently injected into the right axillary space of each mouse.
Tumor sizes were measured every four days with a caliper. The mice
were randomly divided into four groups 21 days after tumor cell
inoculation, specifically, a negative control group (NC), shTR4
group (shTR4), shmiR-145 group (sh145), and a shTR4+shmiR-145
(shTR4+145) group. Lentivirus was injected into the tumor of each
mouse. After 48 h, 5 mg/kg docetaxel (Sigma Aldrich; Merck KGaA)
was injected intraperitoneally twice a week for three weeks. Then
the mice were sacrificed by cervical dislocation and tumors were
removed for further characterization.
The duration of the experiment was 51 days. During
the experiment, mice health and behaviour were monitored daily. The
humane endpoint for mice was a tumor size >17 mm in diameter of
any mouse. All 40 mice were euthanized by cervical dislocation. No
mouse was found dead before euthanasia. Animal welfare
considerations were taken including anaesthetics (intraperitoneal
injection of pentobarbital 50 mg/kg) to minimize suffering and
distress.
Immunohistochemistry (IHC)
Tumor tissues were fixed in 4% neutral buffered
paraformaldehyde (room temperature, 12 h) and embedded in paraffin.
Immunohistochemical staining was performed on 4-µm sections of
paraffin blocks. After blocking with 3% hydrogen peroxide for 10
min at room temperature, the slides were then incubated overnight
at 4°C with primary antibody. The primary antibodies, OCT4 (diluted
1:800; product no. 2750; Cell Signaling Technology, Inc.) and
survivin (diluted 1:500; product code ab76424; Abcam) were used for
staining. The slides were then incubated with the biotinylated
goat-anti-rabbit secondary antibody (diluted 1:500; product no.
BA-1000; Vector Laboratories, Inc.) for 30 min at room temperature,
and visualized by VECTASTAIN ABC peroxidase system and peroxidase
substrate DAB kit (Vector Laboratories, Inc.) according to the
manufacturer's instructions. The slides were analyzed using an
Olympus BX51 fluorescence microscope (Olympus Corporation).
Statistical analysis
Values were expressed as the mean ± standard
deviation. The unpaired t-test was used for comparing two groups
and ANOVA followed by the Tukey's test was used for multiple
comparisons using SPSS 19.0 software (IBM Corp.). P<0.05
(two-sided) indicated a statistically significant difference.
Results
miR-145 mediates PCa
chemoresistance
In our previous study, it was revealed that TR4 can
mediate the chemoresistance of PCa by upregulating the expression
of OCT4 at the transcriptional level (15). However, more evidence revealed that
miRNAs and TFs may cooperate to control gene expression via
TF-miRNA-target axes. For example, TR4 has been revealed to promote
prostate cancer metastasis through the TR4/miR-373/TGFβR2 signaling
axis (28). This prompted us to
investigate the existence of a TR4/miRNA/OCT4 axis that regulates
chemoresistance of PCa.
In total, 38 miRNAs that are related to
chemoresistance in PCa by text mining were collected. The
intersection was calculated between these 38 miRNAs and another ten
key miRNAs in CRPC previously identified (14) since chemoresistance occurs in the
CRPC stage. It was revealed that four miRNAs, mainly let-7b,
miR-125b, miR-197 and miR-145 played essential roles in the
chemoresistance of CRPC (Fig. 1).
Then, four online databases (TargetScan, miRDB, miRanda,
miRTarBase) were used to predict whether any of these four miRNAs
can target OCT4. Unfortunately, only miR-145 was predicted to
target OCT4 in one database (miRTarBase). Consequently, miR-145 was
the only miRNA needed for further validatation (Fig. 1).
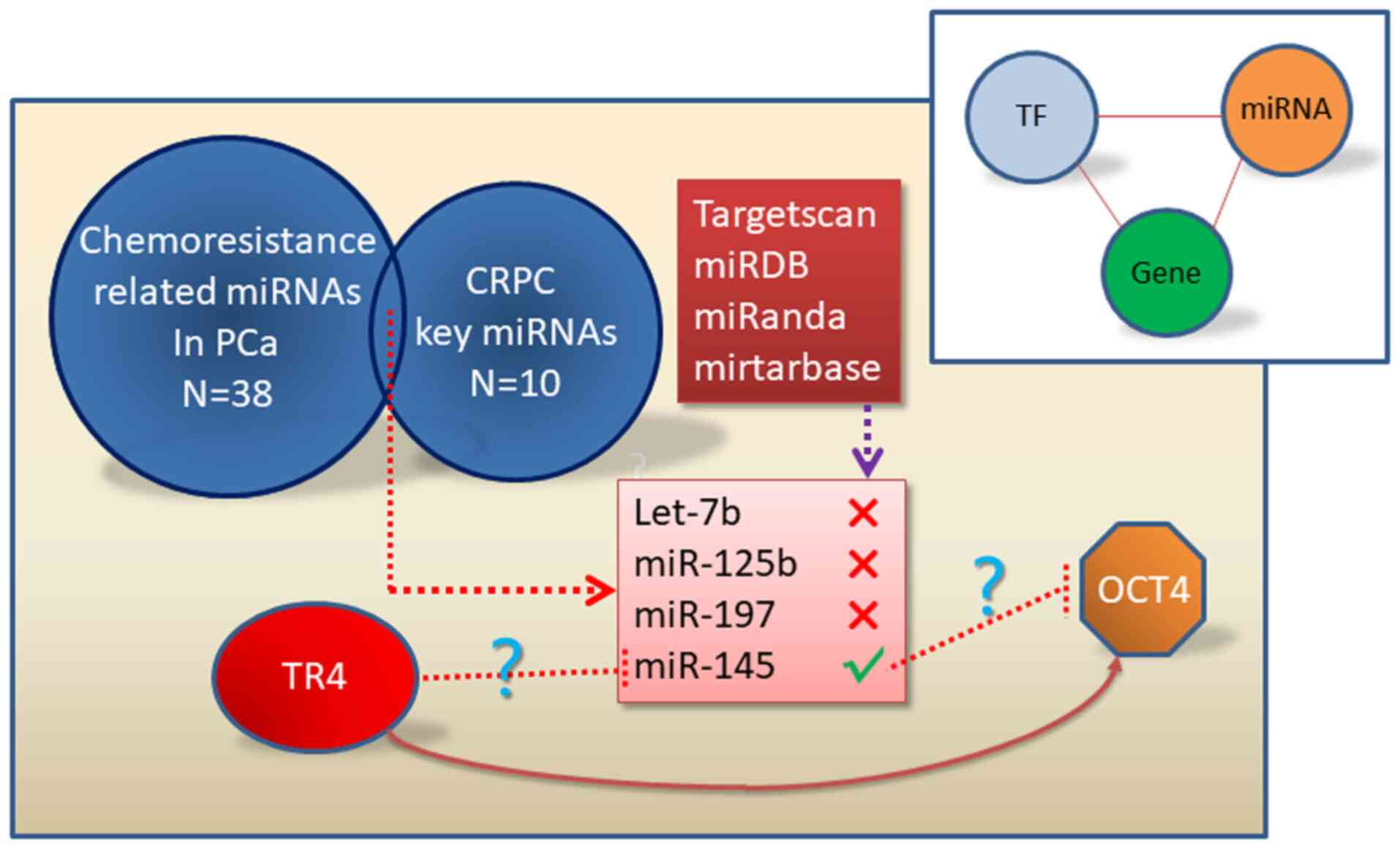 | Figure 1.miR-145 is predicted to join the
TR4/OCT4 signaling to mediate PCa chemoresistance. A total of 38
chemoresistance-related miRNAs in PCa were listed by text mining.
The intersection between the 38 miRNAs and the 10 key miRNAs in
CRPC previously identified by our research group was calculated.
Four miRNAs (let-7b, miR-125b, miR-197, and miR-145) were revealed
to play key roles in the chemoresistance in CRPC. Four online
databases (TargetScan, miRDB, miRanda, miRTarBase) were used and
predicted miR-145 as the only miRNA to target OCT4. TR4, testicular
nuclear receptor 4; OCT4, octamer-binding transcription factor 4;
PCa, prostate cancer; miR/miRNAs, microRNAs; CRPC,
castration-resistant prostate cancer. |
TR4 suppresses miR-145 but promotes
OCT4 expression in PCa cell lines
It was first evaluated whether TR4 can affect the
expression of miR-145. PCa PC3 cells were infected with either a
TR4 knockdown (shTR4) or negative control (scr) lentivirus, and
then the miR-145 expression levels were determined by qPCR. OCT4
mRNA and protein expression levels were also determined by qPCR and
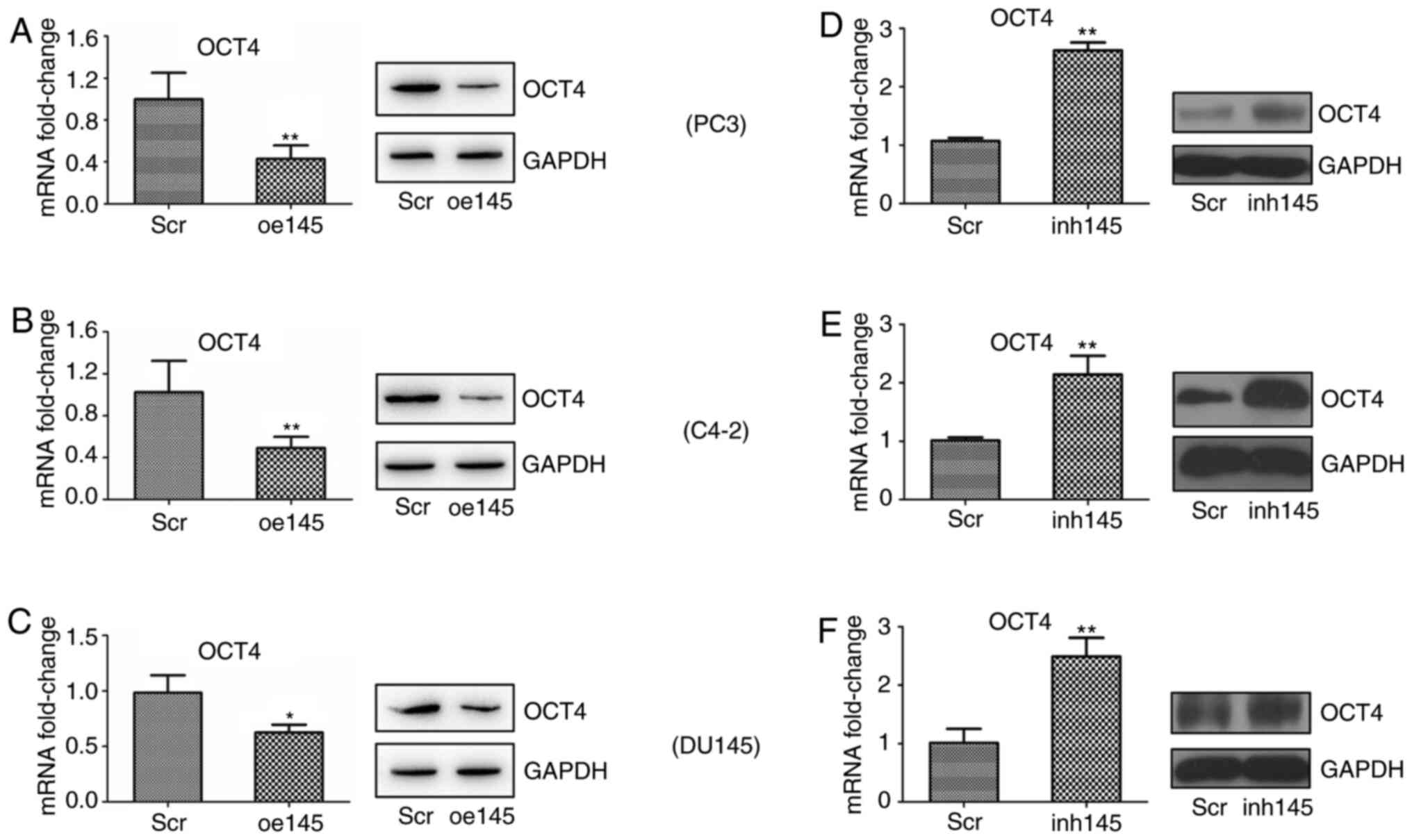
western blot analyses. As revealed in Fig. 2A, the knocking down of TR4
significantly increased the expression of miR-145 while decreasing
OCT4 mRNA and protein expression in PC3 cells.
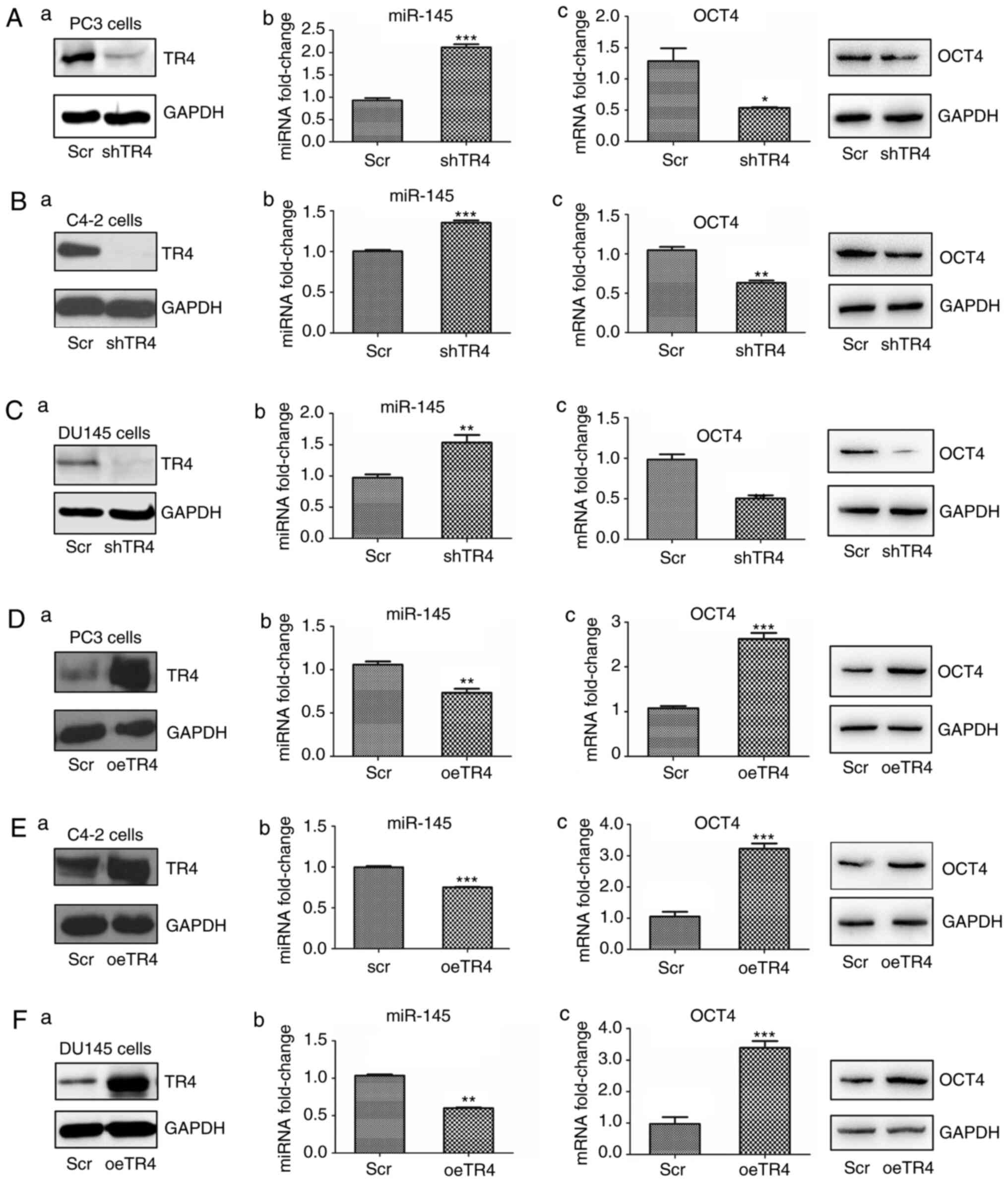 | Figure 2.TR4 suppresses miR-145 but promotes
OCT4 expression in PCa cell lines. TR4 was (A-a to C-a) inhibited
or (D-a to F-a) overexpressed in three PCa cell lines (PC3, C4-2,
DU145). (A-b to F-b) The expression of miR-145 was determined by
qPCR. (A-c to F-c) OCT4 mRNA and protein expression levels were
determined by qPCR and western blotting analyses, respectively.
*P<0.05, **P<0.01 and ***P<0.001. TR4, testicular nuclear
receptor 4; miR, microRNA; OCT4, octamer-binding transcription
factor 4; PCa, prostate cancer; qPCR, quantitative PCR; scr,
scambled; oe, overexpressed. |
Similarly, the other two PCa cell lines, C4-2, and
DU-145, were used for validatation. As anticipated, upon inhibition
of TR4, miR-145 increased and OCT4 decreased in both C4-2 (Fig. 2B) and DU-145 cells (Fig. 2C).
To further confirm the TR4 negative regulation on
miR-145, TR4 was overexpressed in all three cell lines using
lentivirus. The results revealed that miR-145 expression decreased
and OCT4 increased at both mRNA and protein levels (Fig. 2D-F).
The results (Fig.
2A-F) revealed that TR4 suppressed miR-145 but promoted OCT4
expression in PCa.
miR-145 negatively regulates OCT4
expression in PCa cell lines
Since the TR4/miR-145 regulation was confirmed, the
following goal was to assess the miR-145/OCT4 signal. miR-145 has
been reported to directly target OCT4 in several cancer types
(29,30). However, in PCa, the miR-145/OCT4
signal was reported only once in one cell line (PC3), and the
detailed mechanism was not discussed (31). Consequently, it was neccessary to
confirm the effect of miR-145 on OCT4 expression in more PCa cell
lines.
Experiments were performed on the PC3 PCa cell line
and chemically synthesized miR-145 mimic or negative control was
transfected into PC3 cells. OCT4 expression was evaluated using
qPCR and western blotting. As revealed in Fig. 3A, when miR-145 was overexpressed,
OCT4 expression decreased significantly both at the mRNA and
protein levels.
In the other two cell lines, C4-2 and DU-145,
miR-145 mimics were transfected and similar results were obtained
(Fig. 3B and C).
For further confirmation, the miR-145 inhibitor was
transfected into the three cell lines. The results revealed that
the expression of OCT4 significantly increased in all three cell
lines at the mRNA and protein level when miR-145 was inhibited
(Fig. 3D-F).
Collectively, the results (Fig. 3A-F) revealed that miR-145 suppressed
OCT4 expression in PCa cells.
TR4-mediated chemoresistance in PCa is
attenuated by miR-145
Although all of the data revealed the regulatory
axis of the TR4/miR-145/OCT4 signal, further experimention was
conducted to confirm that miR-145 had a real function in
TR4/OCT4-mediated PCa chemoresistance.
First, TR4 expression was inhibited using a
lentivirus in PC3 cells and the cell survival was assessed after
treatment with docetaxel. The results revealed that the sensitivity
to docetaxel of the shTR4 group was significantly increased
compared with the control group (Fig.
4A). Then, miR-145 was inhibited using a lentivirus and it was
reveled that PC3 cells lacking miR-145 were more resistant to
docetaxel compared with the control group (Fig. 4B). Next, the chemosensitivity to
docetaxel between the control group and the TR4/miR-145
double-knockdown group was compared; the results revealed that
there was no significant difference in chemosensitivity to
docetaxel between these two groups (Fig. 4C).
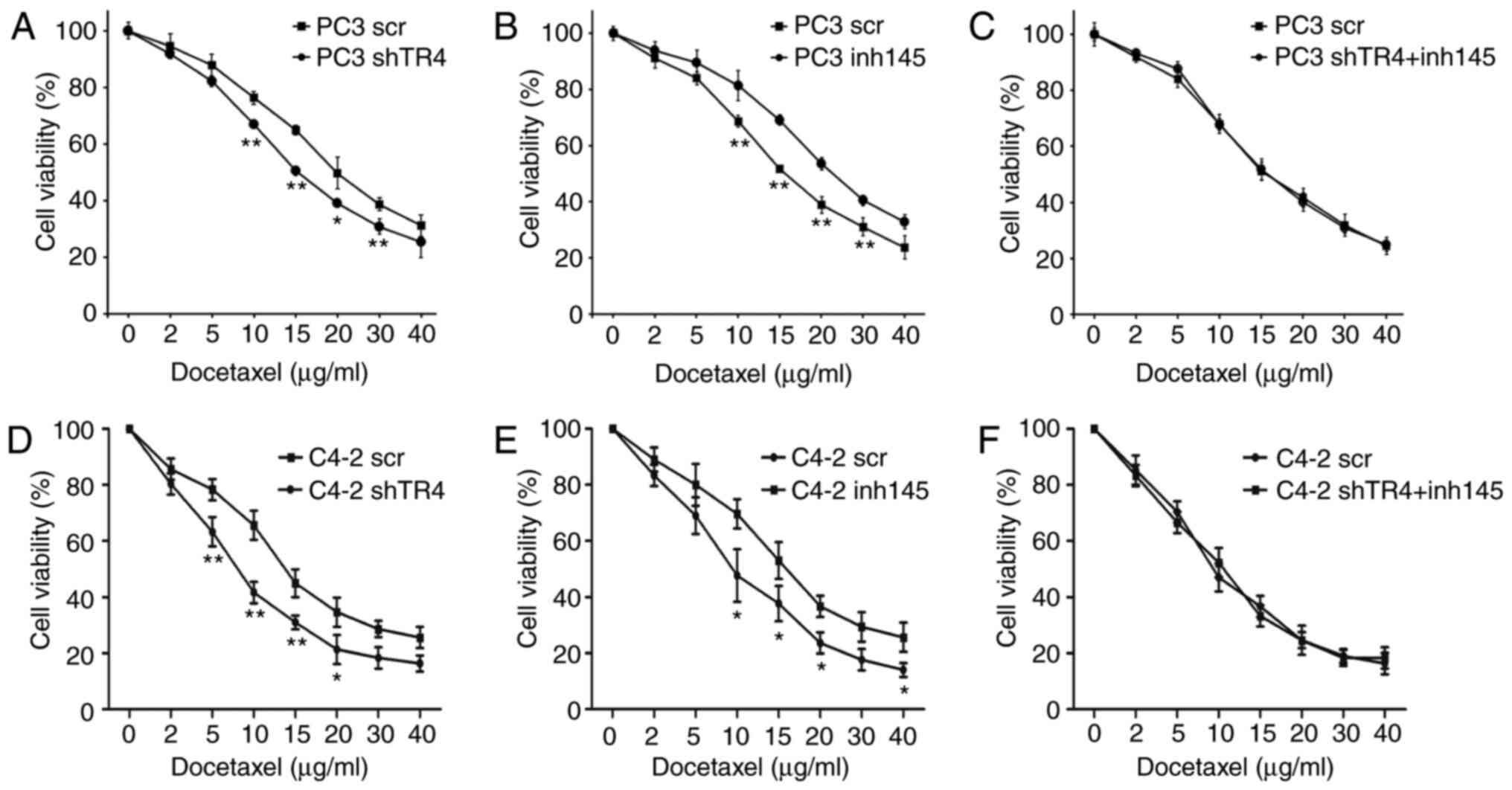 | Figure 4.TR4-mediated chemoresistance in PCa
is attenuated by miR-145. PC3 or C4-2 cells were divided into three
groups: (A and D) TR4-silenced group, (B and E) miR-145 inhibitor
group, and (C and F) TR4 and miR-145 double-inhibition group. Cells
were treated with a specific concentration (0, 2, 5, 10, 15, 20,
30, 40 µg/ml) of docetaxel. Cell survival after the treatment of
docetaxel was recorded. *P<0.05 and **P<0.01. TR4, testicular
nuclear receptor 4; PCa, prostate cancer; miR, microRNA; scr,
scrambled; sh, short hairpin; inh, inhibitor. |
One more cell line (C4-2 line) was used to repeat
the three tests. As anticipated, TR4 shRNA increased C4-2
sensitivity to docetaxel (Fig. 4D),
while the inhibition of miR-145 caused more chemoresistance to C4-2
cells (Fig. 4E). Notably, in C4-2
cells, TR4/miR-145 double-knockdown cells also had a similar
sensitivity to docetaxel when compared to the control group
(Fig. 4F).
The results (Fig.
4A-F) revealed that miR-145 could attenuate TR4 mediated
chemoresistance in PCa.
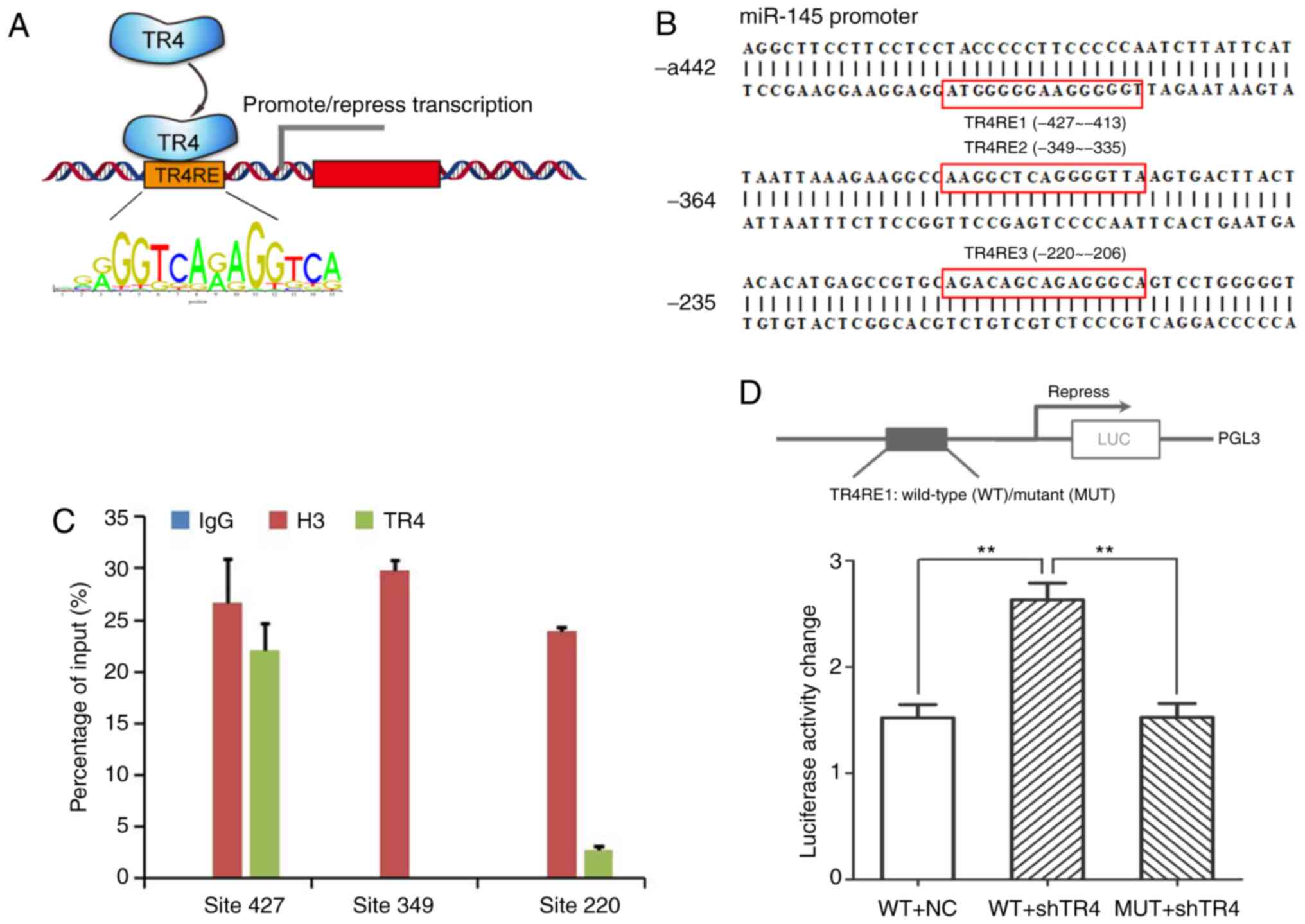
TR4 inhibits the expression of miR-145
at the transcriptional level
TR4 is a transcription factor that can regulate a
large number of molecules in PCa and other diseases at the
transcriptional level (25)
(Fig. 5A). Given this reason, it
was decided to further investigate whether TR4 could bind to the
promoter region of miR-145 to regulate it.
The online tool JASPAR was used to predict the
putative TR4 response elements (TR4REs) for the promoter of
miR-145. As revealed in Fig. 5B,
three putative TR4REs were identified, including TR4RE1 (~-427 to
−413), TR4RE2 (~-349 to −335), and TR4RE3 (~-220 to −206).
Then, a ChIP assay was performed to verify whether
TR4 binds to any of the three putative TR4REs. As revealed in
Fig. 5C, IgG served as the negative
control and anti-histone H3 as the positive control; the TR4
protein could only robustly bind to the first predicted TR4
response element (TR4RE1, ~-427 to −413).
Then, the full promoter DNA sequence of miR-145 was
cloned into the PGL-3 primary luciferase vector and another
promoter with a mutant in the TR4RE1 region. The luciferase assays
were then performed (Fig. 5D); upon
the inhibition of TR4, the activity of miR-145 promoter
significantly increased compared with the control group. As
anticipated, when the wild-type promoter was substituted with the
mutant promoter, TR4 inhibition no longer had any effect on
luciferase activity.
Collectively, the results (Fig. 5A-D) revealed that TR4
transcriptionally supressed miR-145 via binding to the TR4 response
element (~-427 to −413) in the promoter region of miR-145.
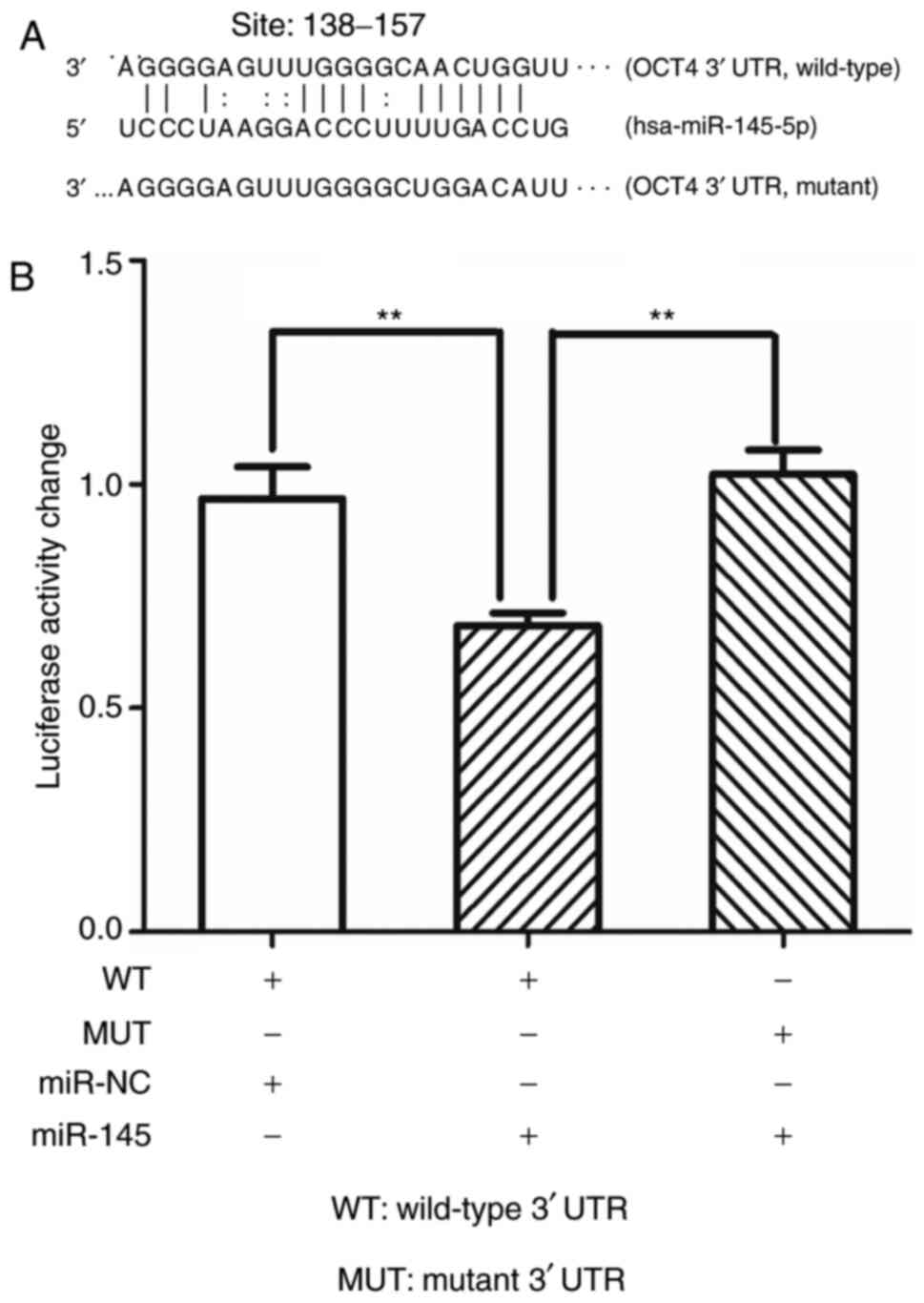
miR-145 targets OCT4 by directly
targeting its 3′untranslated (3′UTR) region
The mechanism for the regulation of miR-145 on OCT4
has been previously reported in other cancers but not in PCa
(29,30). To further confirm the present
findings, it was attempted to reveal the mechanism in C4-2 PCa
cells.
As revealed in Fig.
6A, the 3′UTR of OCT4 was predicted to have putative miRNA
response elements (MRE) of miR-145 (site: ~138–157). Thus, the full
3′UTR sequence of OCT4 was cloned into the PGL-3
promoter-luciferase vector downstream to the firefly luciferase. In
addition, the 3′UTR was cloned with a mutant in the seed region.
Luciferase assays were then performed, and as revealed in Fig. 6B, upon transfection of miR-145,
luciferase activity significantly decreased compared with the
control group. As anticipated, when the wild-type 3′UTR was
substituted with the mutant one, miR-145 no longer had any effect
on luciferase activity.
Collectively, the results (Fig. 6A and B) revealed that miR-145
targeted OCT4 by directly targeting its 3′UTR.
TR4 causes chemoresistance to PCa via
miR-145/OCT4 signals in vivo
As the in vitro data revealed that TR4
regulates the chemosensitivity of PCa via the miR-145/OCT4 signal,
the present study investigated whether the same effects were
observed in vivo.
A total of 40 male nude mice were used in the
experiment. For each mouse, 5×106 cells were
subcutaneously injected into the right axillary space. The mice
were then divided into four groups 21 days after tumor cell
inoculation: A control group (NC), a TR4-inhibiting group (shTR4),
a miR-145-inhibiting group (sh145), and a TR4/miR-145
double-inhibiting group (shTR4+145). Lentivirus was injected into
the tumor of each mouse. After 48 h, 5 mg/kg docetaxel was injected
intraperitoneally at a frequency of twice a week for 3 weeks. Tumor
sizes were measured every 4 days and the mice were then sacrificed.
Then, the tumors were collected and the tumor volumes were
calculated.
The results demonstrated that tumors in the shTR4
group were smaller than those in the control group (Fig. 7A-C). Tumors in the sh145 group were
had a larger tumor size than those in the control group (Fig. 7A-C). Notably, tumors in the
TR4/miR-145 double-inhibiting group had a similar size compared to
those in the control group (Fig.
7A-C).
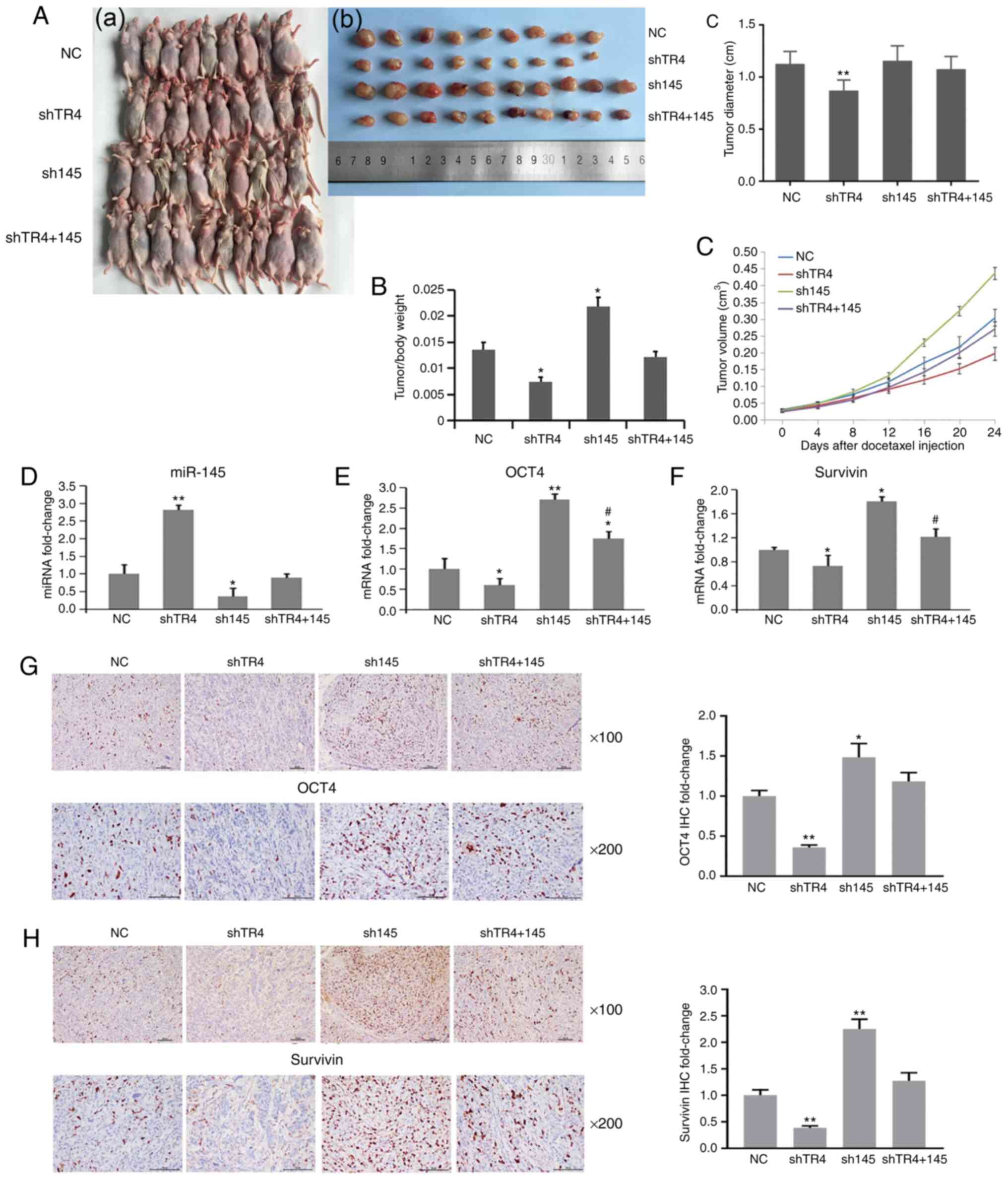 | Figure 7.TR4 causes chemoresistance to PCa via
miR-145/OCT4 signals in vivo. Forty male nude mice were used
in the experiment. For each mouse, 5×106 cells were
subcutaneously injected into the right axillary space. Then the
mice were divided into four groups 21 days after tumor cell
inoculation: The control group (NC), the TR4-inhibiting group
(shTR4), the miR-145-inhibiting group (sh145), and the TR4/miR-145
double-inhibiting group (shTR4 and 145). Lentivirus was injected
into the tumor of each mouse. After 48 h, 5 mg/kg docetaxel was
injected intraperitoneally at a frequency of twice a week for three
weeks. Mice were sacrificed and (A-a to A-c) tumors were collected,
and (B) tumor/body weight ratio was recorded. (C) The tumor volume
was measured every four days after docetaxel injection. Expression
of (D) miR-145, (E) OCT4, and (F) survivin of tumor tissue from
each group was determined by qPCR. Expression of (G) OCT4 and (H)
survivin protein in tumor tissues from each group was determined by
IHC staining. *P<0.05 and **P<0.01. #P<0.05,
shTR4+145 group compared with shTR4 group. TR4, testicular nuclear
receptor 4; PCa, prostate cancer; miR, microRNA; OCT4,
octamer-binding transcription factor 4; NC, negative control; sh,
short hairpin. |
The expression of miR-145, OCT4, and survivin was
then determined using qPCR. Survivin is the downstream gene of OCT4
(32) and is an antiapoptotic
factor in PCa (33). As revealed in
Fig. 7D-F, miR-145 was increased
while OCT4 and survivin were decreased in the shTR4 group. In the
sh145 group, OCT4 (Fig. 7E) and
survivin (Fig. 7F) mRNA were
significantly increased. Notably, the shTR4 effect on OCT4 and
survivin was attenuated by concurrent inhibition of miR-145
(shTR4+145 group compared with shTR4 group) (Fig. 7E and F).
Similarly, when immunohistochemical staining was
used to analyze OCT4 and survivin protein expression, it was
revealed that the same expression pattern of both OCT4 (Fig. 7G) and survivin (Fig. 7H) occurred in the four groups as
compared with the qPCR results.
Collectively, these data indicated that TR4
transcriptionally inhibited the expression of miR-145 and
consequently promoted the expression of OCT4, causing
chemoresistance of PCa in vitro and in vivo. miR-145
could reverse the TR4 effect on chemosensitivity in PCa. The in
vitro and in vivo data obtained in the present study
confirmed the TR4/miR-145/OCT4 signaling in PCa
chemoresistance.
Discussion
The mechanism by which CRPC patients display
different therapeutic efficacy to docetaxel needs to be further
explored. In our previous study, it was identified that TR4 may
promote chemoresistance of PCa via increasing the expression of
OCT4 (15). In the present study,
the purpose was to identify an ‘intermediary miRNA’ between TR4 and
OCT4 to regulate the chemosensitivity in PCa. miR-145 was predicted
as the key miRNA by using integrative bioinformatics analysis.
Findings revealed that TR4 could inhibit miR-145 expression while
increasing OCT4 expression and, consequently, causing
chemoresistance to docetaxel in vitro and in vivo and
miR-145 was able to reverse the effects. Furthermore, findings
demonstrated that TR4 enhanced the chemoresistance of PCa at least
partially via the axis of TR4/miR-145/OCT4.
The role of nuclear receptors in prostate cancer has
received increasing attention. For example, recent study has
revealed that RORγ mediated resistance of prostate cancer to
doxorubicin chemotherapy (34).
Nuclear receptor HNF4α funtioned as a tumor suppressor in prostate
cancer via its induction of p21-driven cellular senescence
(35). In addition, TR4 is a
nuclear receptor and has been revealed to play an important role in
tumors, especially prostate cancer (28).
TR4 nuclear receptor plays a crucial role in the
development and progression of various types of cancer including
liver (36), breast (37), lung (38), and renal cancer (39). For example, TR4 promoted miR-32-5p
to suppress clear cell renal cell carcinoma metastasis via altering
the miR-32-5p/TR4/HGF/Met signaling (40). Shen et al revealed that TR4
enhanced cisplatin chemosensitivity via altering ATF3 expression in
hepatic cell carcinoma (36). TR4
is especially important in PCa because TR4 regulates PCa initiation
and progression. TR4 has been revealed to inhibit tumor initiation
(41) but promote metastasis
(42), radiation resistance
(43), and chemoresistance
(44) via several distinct signals.
High TR4 expression in PCa tissues was revealed to be correlated
with high Gleason scores and is usually correlated with a poor
prognosis (42). In the present
study, it was revealed that TR4 could be the cause of
chemoresistance to docetaxel in PCa. This result is consistent with
a study by Chen et al (44)
and our previous study, which revealed that TR4 promoted PCa stem
cell-mediated chemoresistance (15). Thus, it was concluded by the
findings in the present study that TR4 is a strong drive factor to
promote PCa progression from various aspects of tumor
behaviors.
Several studies have indicated that human miR-145 is
involved in the progression of numerous cancers acting as a tumor
suppressor (45–48). With regard to prostate cancer,
miR-145 is expressed at a low level, therefore, the restoration of
miR-145 could inhibit the proliferation and invasion restoring
radiation sensitivity via targeting multiple genes (49,50).
Additionally, our previous study identified miR-145 as one of the
key miRNAs involved in CRPC (14).
However, no previous published data focused on the effect of
miR-145 on the chemosensitivity of PCa. In the present study, it
was revealed that miR-145 can re-sensitize PCa to docetaxel via
targeting OCT4. The present findings provided more insight into the
function of miR-145 in PCa.
Furthermore, the study linked the TR4 transcription
factor and miR-145 together. Other transcription factors may also
have this TF/miRNA regulation pattern. In lung adenocarcinoma,
thyroid transcription factor-1 has been reported to regulate
miR-532-5p consequently inducing apoptosis (51). Nanog has been reported to
transcriptionally suppress miR-200 family in colon cancer cells and
induce epithelial-mesenchymal transition (52). ChIP-seq data also revealed that TR4
could bind to the upstream locus of multiple miRNAs (53). The existence of this TF/miRNA
regulation can modulate the biological function in cells in a
precised manner.
A previous study reported that miR-145 targeted AR
and inhibited prostate cancer progression (54). In addition, AR could regulate the
activity of OCT4 (55). The present
study revealed that miR-145 could directly target the 3′UTR of OCT4
to regulate OCT4 in both AR-positive cell line C4-2 and AR-negative
cell lines PC3 and DU145. Therefore, it was speculated that miR-145
may regulate OCT4 directly, as well as indirectly through the
AR/OCT4 pathway, and thus the completion of the regulatory goal is
ensured through multiple pathways.
There is increasing interest in the study of
TF/miRNA/gene axes in different diseases. For instance, Qiu et
al revealed that TR4 could promote PCa metastasis via
TR4/miR-373/TGFβR2 signaling (28).
Wang et al revealed that TR4 promoted clear cell renal cell
carcinoma vasculogenic mimicry formation and metastasis via
altering the TR4/miR490-3p/vimentin signal (39). All of these studies have added
evidence to the TF/miRNA/gene axis hypothesis.
For further investigation, web-based TF/miRNA/gene
prediction tools have been developed such as TransmiR (56). These prediction tools are
constructed with specific algorithms and ChIP-Seq based data.
However, numerous TFs are not included in the databases and
ChIP-Seq data are obtained from a minimal number of cell lines,
thus limiting the use of these database tools. In the present
study, online TF/miRNA/gene prediction tools were not used; instead
bioinformatics methods were used including text mining and
traditional miRNA/mRNA interaction databases. The researchers
consider that with the decreasing cost for ChIP-Seq, it is possible
to perform more ChIP-seq experiments based on PCa cells to better
study PCa.
The researchers admit that bioinformatics methods
have limitations. As was observed, only one miRNA was identified in
the present study indicating that some other relevant miRNAs may
have been missed. In addition, the possibility of adapting this
method to future studies is unknown yet.
The present study revealed that TR4 inhibited
miR-145 promoting the chemoresistance to docetaxel. This prompted
our research group to hypothesize that the expression of TR4 and
miR-145 may be used in combination to decide whether individual
patients could accept docetaxel treatment. On the other hand, TR4
is an orphan receptor that lacks ligands, thus it may not be easy
to target. Theoretically, the researchers can treat PCa with
miR-145, since small RNA-delivering techniques are approaching
clinical use at present.
In conclusion, the present study associated the
expression of the transcription factor TR4 to miR-145 in PCa.
Ectopic TR4 inhibited miR-145, thus increasing the expression of
OCT4 and consequently causing chemoresistance to docetaxel. These
findings provide insight into the interplay between TR4 and miR-145
in PCa chemosensitivity and open up new perspectives concerning the
function and regulatory axes of transcription factors.
Acknowledgments
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81472776 and
81773221; to DY and JZ), the Natural Science Foundation of Jiangsu
Province (grant nos. BK20161222 and BK20191170; to JZ and LX). It
was also funded by Suzhou Science and Technology Planned Projects
(grant nos. SYS201629, SS201857 and SYS2018062; to JZ and LX), and
the Key Young Talents of Medicine in Jiangsu (grant no.
QNRC2016875; to JZ)
Availability of data and materials
The researchers declare that the raw data,
certificates of the cell lines and other materials can be provided,
if requested.
Authors' contributions
JZ and PQ performed the experiments and statistical
analyses, and created the figures. CC and GD assisted with the
interpretation of data and reviewed the manuscript. JZ, LX and DY
conceived the study and wrote the manuscript. All authors read and
approved the manuscript and agree to be accountable for all aspects
of the research in ensuring that the accuracy or integrity of any
part of the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
Animal experiments were performed according to a
protocol approved by the Ethics Committee of the Second Affiliated
Hospital of Soochow University (Suzhou, China) and followed the
ARRIVE Guidelines Checklist.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Cronin KA, Lake AJ, Scott S, Sherman RL,
Noone AM, Howlader N, Henley SJ, Anderson RN, Firth AU, Ma J, et
al: Annual report to the nation on the status of cancer, part I:
National cancer statistics. Cancer. 124:2785–2800. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhao F, Shen J, Yuan Z, Yu X, Jiang P,
Zhong B, Xiang J, Ren G, Xie L and Yan S: Trends in treatment for
prostate cancer in china: Preliminary patterns of care study in a
single institution. J Cancer. 9:1797–1803. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Huang Y, Jiang X, Liang X and Jiang G:
Molecular and cellular mechanisms of castration resistant prostate
cancer. Oncol Lett. 15:6063–6076. 2018.PubMed/NCBI
|
|
5
|
Mottet N, Bellmunt J, Bolla M, Joniau S,
Mason M, Matveev V, Schmid HP, Van der Kwast T, Wiegel T, Zattoni F
and Heidenreich A: EAU Guidelines on prostate cancer. Part II:
Treatment of advanced, relapsing, and castration-resistant prostate
cancer. Eur Urol. 59:572–583. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Summers N, Vanderpuye-Orgle J, Reinhart M,
Gallagher M and Sartor O: Efficacy and safety of post-docetaxel
therapies in metastatic castrate-resistant prostate cancer: A
systematic review of the literature. Curr Med Res Opin.
33:1995–2008. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Seitz AK, Thoene S, Bietenbeck A, Nawroth
R, Tauber R, Thalgott M, Schmid S, Secci R, Retz M, Gschwend JE, et
al: AR-V7 in peripheral whole blood of patients with
castration-resistant prostate cancer: Association with
treatment-specific outcome under abiraterone and enzalutamide. Eur
Urol. 72:828–834. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Berthold DR, Pond GR, Roessner M, de Wit
R, Eisenberger M and Tannock AI; TAX-327 investigators, : Treatment
of hormone-refractory prostate cancer with docetaxel or
mitoxantrone: Relationships between prostate-specific antigen,
pain, and quality of life response and survival in the TAX-327
Study. Clin Cancer Res. 14:2763–2767. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhao Y, Huang H, Chen C, Liu H, Liu H, Su
F, Bi J, Lam TB, Li J, Lin T and Huang J: Efficacy and safety of
different interventions in castration resistant prostate cancer
progressing after docetaxel-based chemotherapy: Bayesian network
analysis of randomized controlled trials. J Cancer. 9:690–701.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hongo H, Kosaka T and Oya M: Analysis of
cabazitaxel-resistant mechanism in human castration-resistant
prostate cancer. Cancer Sci. 109:2937–2945. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yun EJ, Lo UG and Hsieh JT: The evolving
landscape of prostate cancer stem cell: Therapeutic implications
and future challenges. Asian J Urol. 3:203–210. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Faivre EJ, Wilcox D, Lin X, Hessler P,
Torrent M, He W, Uziel T, Albert DH, McDaniel K, Kati W and Shen Y:
Exploitation of castration-resistant prostate cancer transcription
factor dependencies by the novel BET inhibitor ABBV-075. Mol Cancer
Res. 15:35–44. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhu J, Sun C, Wang L, Xu M, Zang Y, Zhou
Y, Liu X, Tao W, Xue B, Shan Y and Yang D: Targeting survivin using
a combination of miR-494 and survivin shRNA has synergistic effects
on the suppression of prostate cancer growth. Mol Med Rep.
13:1602–1610. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhu J, Wang S, Zhang W, Qiu J, Shan Y,
Yang D and Shen B: Screening key microRNAs for castration-resistant
prostate cancer based on miRNA/mRNA functional synergistic network.
Oncotarget. 6:43819–43830. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang DR, Ding XF, Luo J, Shan YX, Wang R,
Lin SJ, Li G, Huang CK, Zhu J, Chen Y, et al: Increased
chemosensitivity via targeting testicular nuclear receptor 4
(TR4)-Oct4-Interleukin 1 receptor antagonist (IL1Ra) axis in
prostate cancer CD133+ Stem/progenitor cells to battle prostate
cancer. J Biol Chem. 288:16476–16483. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yu SJ, Yang L, Hong Q, Kuang XY, Di GH and
Shao ZM: MicroRNA-200a confers chemoresistance by antagonizing
TP53INP1 and YAP1 in human breast cancer. BMC Cancer. 18:742018.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yu G, Jia B, Cheng Y, Zhou L, Qian B, Liu
Z and Wang Y: MicroRNA-429 sensitizes pancreatic cancer cells to
gemcitabine through regulation of PDCD4. Am J Transl Res.
9:5048–5055. 2017.PubMed/NCBI
|
|
18
|
Puhr M, Hoefer J, Schäfer G, Erb HH, Oh
SJ, Klocker H, Heidegger I, Neuwirt H and Culig Z:
Epithelial-to-mesenchymal transition leads to docetaxel resistance
in prostate cancer and is mediated by reduced expression of
miR-200c and miR-205. Am J Pathol. 181:2188–2201. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang HL, Yang LF, Zhu Y, Yao XD, Zhang
SL, Dai B, Zhu YP, Shen YJ, Shi GH and Ye DW: Serum miRNA-21:
Elevated levels in patients with metastatic hormone-refractory
prostate cancer and potential predictive factor for the efficacy of
docetaxel-based chemotherapy. Prostate. 71:326–331. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Rokhlin OW, Scheinker VS, Taghiyev AF,
Bumcrot D, Glover RA and Cohen MB: MicroRNA-34 mediates
AR-dependent p53-induced apoptosis in prostate cancer. Cancer Biol
Ther. 7:1288–1296. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gov E and Arga KY: Interactive cooperation
and hierarchical operation of microRNA and transcription factor
crosstalk in human transcriptional regulatory network. IET Syst
Biol. 10:219–228. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li H, Rokavec M, Jiang L, Horst D and
Hermeking H: Antagonistic Effects of p53 and HIF1A on microRNA-34a
regulation of PPP1R11 and STAT3 and Hypoxia-induced Epithelial to
mesenchymal transition in colorectal cancer cells.
Gastroenterology. 153:505–520. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chang TH, Tsai MF, Gow CH, Wu SG, Liu YN,
Chang YL, Yu SL, Tsai HC, Lin SW, Chen YW, et al: Upregulation of
microRNA-137 expression by Slug promotes tumor invasion and
metastasis of non-small cell lung cancer cells through suppression
of TFAP2C. Cancer Lett. 402:190–202. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mcmanus MT: MicroRNAs and cancer. Semin
Cancer Biol. 13:253–258. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gao L, Yang Y, Xu H, Liu R, Li D, Hong H,
Qin M and Wang Y: MiR-335 functions as a tumor suppressor in
pancreatic cancer by targeting OCT4. Tumour Biol. 35:8309–8318.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kilkenny C, Browne WJ, Cuthill IC, Emerson
M and Altman DG: Improving bioscience research reporting: The
ARRIVE guidelines for reporting animal research. PLoS Biol.
8:e10004122010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Qiu X, Zhu J, Sun Y, Fan K, Yang DR, Li G,
Yang G and Chang C: TR4 nuclear receptor increases prostate cancer
invasion via decreasing the miR-373-3p expression to alter
TGFβR2/p-Smad3 signals. Oncotarget. 6:15397–15409. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sachdeva M, Zhu S, Wu F, Wu H, Walia V,
Kumar S, Elble R, Watabe K and Mo YY: p53 represses c-Myc through
induction of the tumor suppressor miR-145. Proc Natl Acad Sci USA.
106:3207–3212. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yin R, Zhang S, Wu Y, Fan X, Jiang F,
Zhang Z, Feng D, Guo X and Xu L: microRNA-145 suppresses lung
adenocarcinoma-initiating cell proliferation by targeting OCT4.
Oncol Rep. 25:1747–1754. 2011.PubMed/NCBI
|
|
31
|
Ren D, Wang M, Guo W, Zhao X, Tu X, Huang
S, Zou X and Peng X: Wild-type p53 suppresses the
epithelial-mesenchymal transition and stemness in PC-3 prostate
cancer cells by modulating miR-145. Int J Oncol. 42:1473–1481.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cao L, Li C, Shen S, Yan Y, Ji W, Wang J,
Qian H, Jiang X, Li Z, Wu M, et al: OCT4 increases BIRC5 and CCND1
expression and promotes cancer progression in hepatocellular
carcinoma. BMC Cancer. 13:822013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang M, Latham DE, Delaney MA and
Chakravarti A: Survivin mediates resistance to antiandrogen therapy
in prostate cancer. Oncogene. 24:2474–2482. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gao M, Guo L, Wang H, Huang J, Han F,
Xiang S and Wang J: Orphan nuclear receptor RORgamma confers
doxorubicin resistance in prostate cancer. Cell Biol Int.
44:2170–2176. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang Z, Li Y, Wu D, Yu S, Wang Y and Chan
FL: Correction: Nuclear receptor HNF4α performs a tumor suppressor
function in prostate cancer via its induction of p21-driven
cellular senescence. Oncogene. 39:62632020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shen J, Lin H, Li G, Jin RA, Shi L, Chen
M, Chang C and Cai X: TR4 nuclear receptor enhances the cisplatin
chemo-sensitivity via altering the ATF3 expression to better
suppress HCC cell growth. Oncotarget. 7:32088–32099. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Shyr CR, Hu YC, Kim E and Chang C:
Modulation of estrogen receptor-mediated transactivation by orphan
receptor TR4 in MCF-7 cells. J Biol Chem. 277:14622–14628. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang L, Zhang J, Ma Y, Chen J, Dong B,
Zhao W, Wang X, Zheng Q, Fang F and Yang Y: Testicular orphan
receptor 4 (TR4) is a marker for metastasis and poor prognosis in
non-small cell lung cancer that drives the EMT phenotype. Lung
Cancer. 89:320–328. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Bai J, Yeh S, Qiu X, Hu L, Zeng J, Cai Y,
Zuo L, Li G, Yang G and Chang C: TR4 nuclear receptor promotes
clear cell renal cell carcinoma (ccRCC) vasculogenic mimicry (VM)
formation and metastasis via altering the miR490-3p/vimentin
signals. Oncogene. 37:5901–5912. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang M, Sun Y, Xu J, Lu J, Wang K, Yang
DR, Yang G, Li G and Chang C: Preclinical studies using miR-32-5p
to suppress clear cell renal cell carcinoma metastasis via altering
the miR-32-5p/TR4/HGF/Met signaling. Int J Cancer. 143:100–112.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lin SJ, Lee SO, Lee YF, Miyamoto H, Yang
DR, Li G and Chang C: TR4 nuclear receptor functions as a tumor
suppressor for prostate tumorigenesis via modulation of DNA
damage/repair system. Carcinogenesis. 35:1399–1406. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ding X, Yang DR, Lee SO, Chen YL, Xia L,
Lin SJ, Yu S, Niu YJ, Li G and Chang C: TR4 nuclear receptor
promotes prostate cancer metastasis via upregulation of CCL2/CCR2
signaling. Int J Cancer. 136:955–964. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yu S, Wang M, Ding X, Xia L, Chen B, Chen
Y, Zhang Z, Niu Y, Li G and Chang C: Testicular orphan nuclear
receptor 4 is associated with the radio-sensitivity of prostate
cancer. Prostate. 75:1632–1642. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Chen B, Yu S, Ding X, Jing C, Xia L, Wang
M, Matro E, Rehman F, Niu Y, Li G and Chang C: The role of
testicular nuclear receptor 4 in chemo-resistance of docetaxel in
castration-resistant prostate cancer. Cancer Gene Ther. 21:411–415.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Li S, Wu X, Xu Y, Wu S, Li Z, Chen R,
Huang N, Zhu Z and Xu X: miR-145 suppresses colorectal cancer cell
migration and invasion by targeting an ETS-related gene. Oncol Rep.
36:1917–1926. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Xu Q, Liu LZ, Qian X, Chen Q, Jiang Y, Li
D, Lai L and Jiang BH: miR-145 directly targets p70S6K1 in cancer
cells to inhibit tumor growth and angiogenesis. Nuclc Acids Res.
40:761–774. 2012. View Article : Google Scholar
|
|
47
|
Cho WC, Chow AS and Au JS: miR-145
inhibits cell proliferation of human lung adenocarcinoma by
targeting EGFR and NUDT1. RNA Biol. 8:125–131. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Chiyomaru T, Enokida H, Tatarano S,
Kawahara K, Uchida Y, Nishiyama K, Fujimura L, Kikkawa N, Seki N
and Nakagawa M: miR-145 and miR-133a function as tumour suppressors
and directly regulate FSCN1 expression in bladder cancer. Br J
Cancer. 102:883–891. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Fuse M, Nohata N, Kojima S, Sakamoto S,
Chiyomaru T, Kawakami K, Enokida H, Nakagawa M, Naya Y, Ichikawa T
and Seki N: Restoration of miR-145 expression suppresses cell
proliferation, migration and invasion in prostate cancer by
targeting FSCN1. Int J Oncol. 38:1093–1101. 2011.PubMed/NCBI
|
|
50
|
Gong P, Zhang T, He D and Hsieh JT:
MicroRNA-145 modulates tumor sensitivity to radiation in prostate
cancer. Radiat Res. 184:630–638. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Griesing S, Kajino T, Tai MC, Liu Z,
Nakatochi M, Shimada Y, Suzuki M and Takahashi T: Thyroid
transcription factor-1-regulated microRNA-532-5p targets KRAS and
MKL2 oncogenes and induces apoptosis in lung adenocarcinoma cells.
Cancer Sci. 108:1394–1404. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Pan Q, Meng L, Ye J, Wei X, Shang Y, Tian
Y, He Y, Peng Z, Chen L, Chen W, et al: Transcriptional repression
of miR-200 family members by Nanog in colon cancer cells induces
epithelial-mesenchymal transition (EMT). Cancer Lett. 392:26–38.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
O'Geen H, Lin YH, Xu X, Echipare L,
Komashko VM, He D, Frietze S, Tanabe O, Shi L, Sartor MA, et al:
Genome-wide binding of the orphan nuclear receptor TR4 suggests its
general role in fundamental biological processes. BMC Genomics.
11:6892010. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Larne O, Hagman Z, Lilja H, Bjartell A,
Edsjö A and Ceder Y: miR-145 suppress the androgen receptor in
prostate cancer cells and correlates to prostate cancer prognosis.
Carcinogenesis. 36:858–866. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Li R, Azzollini D, Shen R, Thorup J,
Clasen-Linde E, Cortes D and Hutson JM: Postnatal germ cell
development during first 18 months of life in testes from boys with
non-syndromic cryptorchidism and complete or partial androgen
insensitivity syndrome. J Pediatr Surg. 54:1654–1659. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Wang J, Lu M, Qiu C and Cui Q: TransmiR: A
transcription factor-microRNA regulation database. Nucleic Acids
Res. 38((Database Issue)): D119–D122. 2010. View Article : Google Scholar : PubMed/NCBI
|