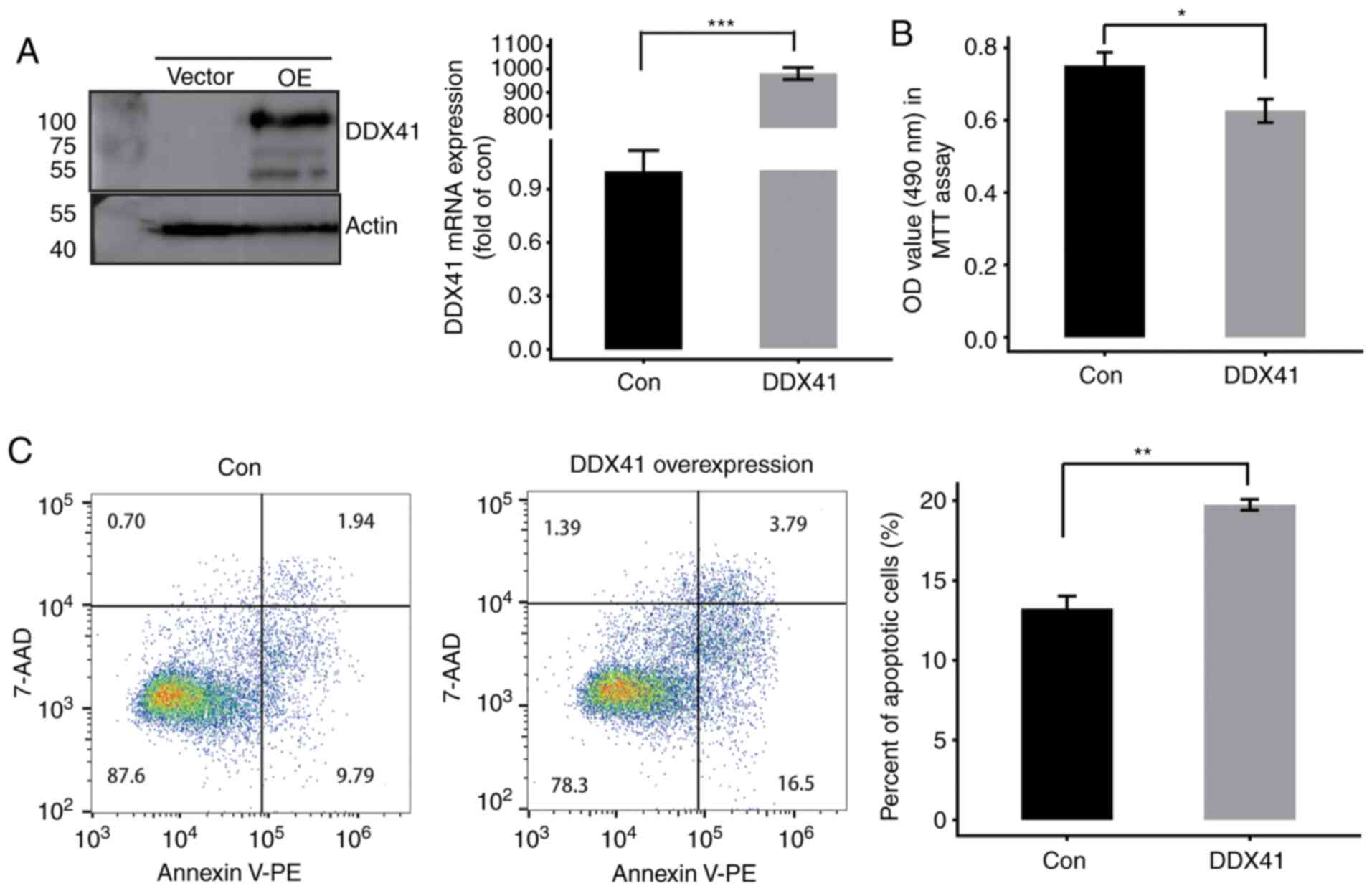
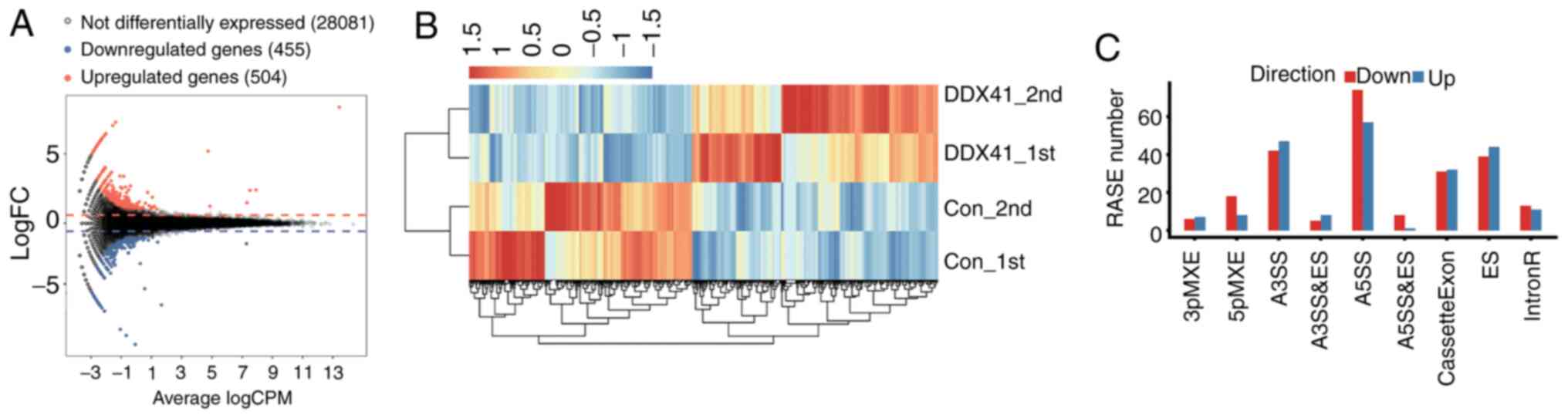
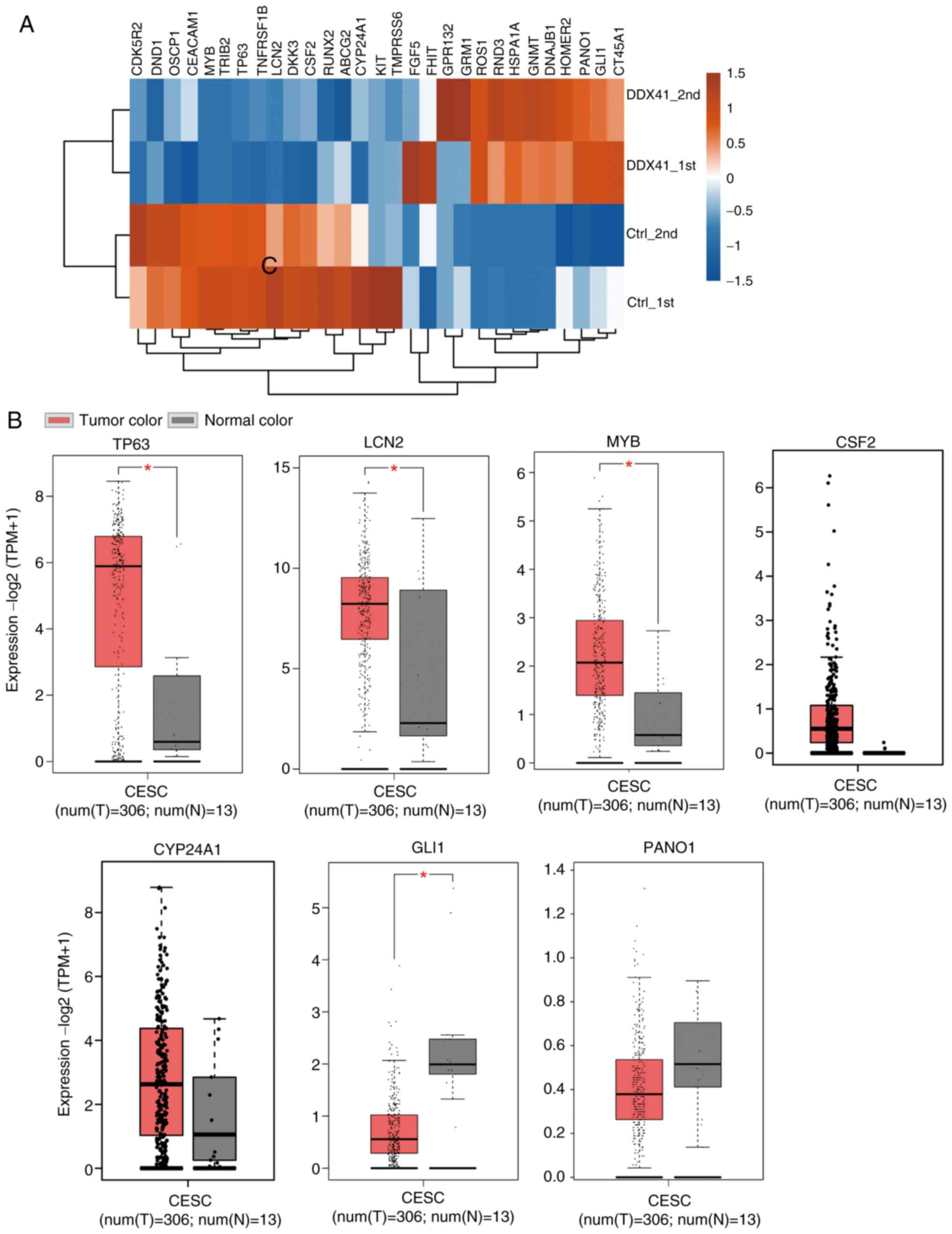
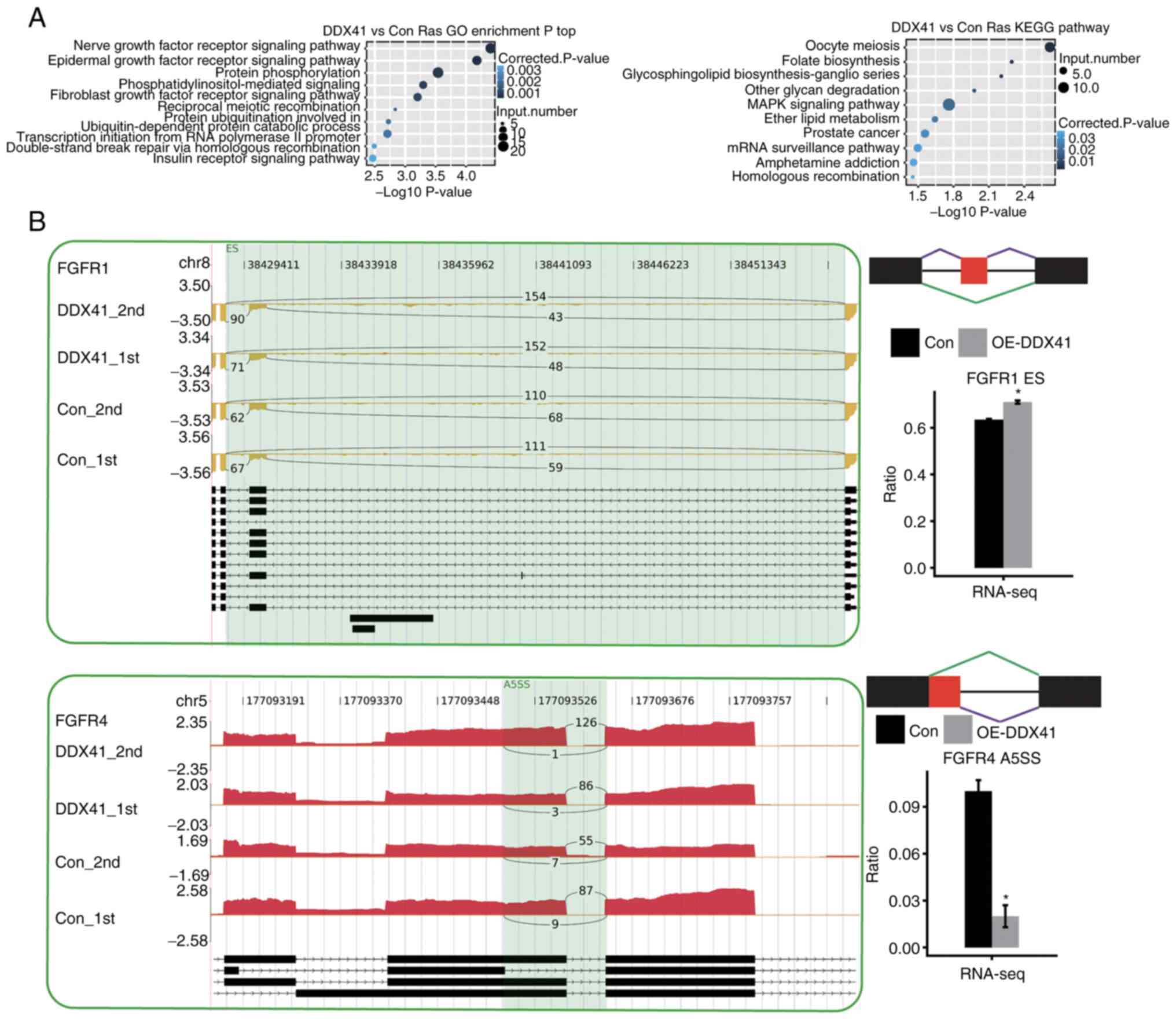
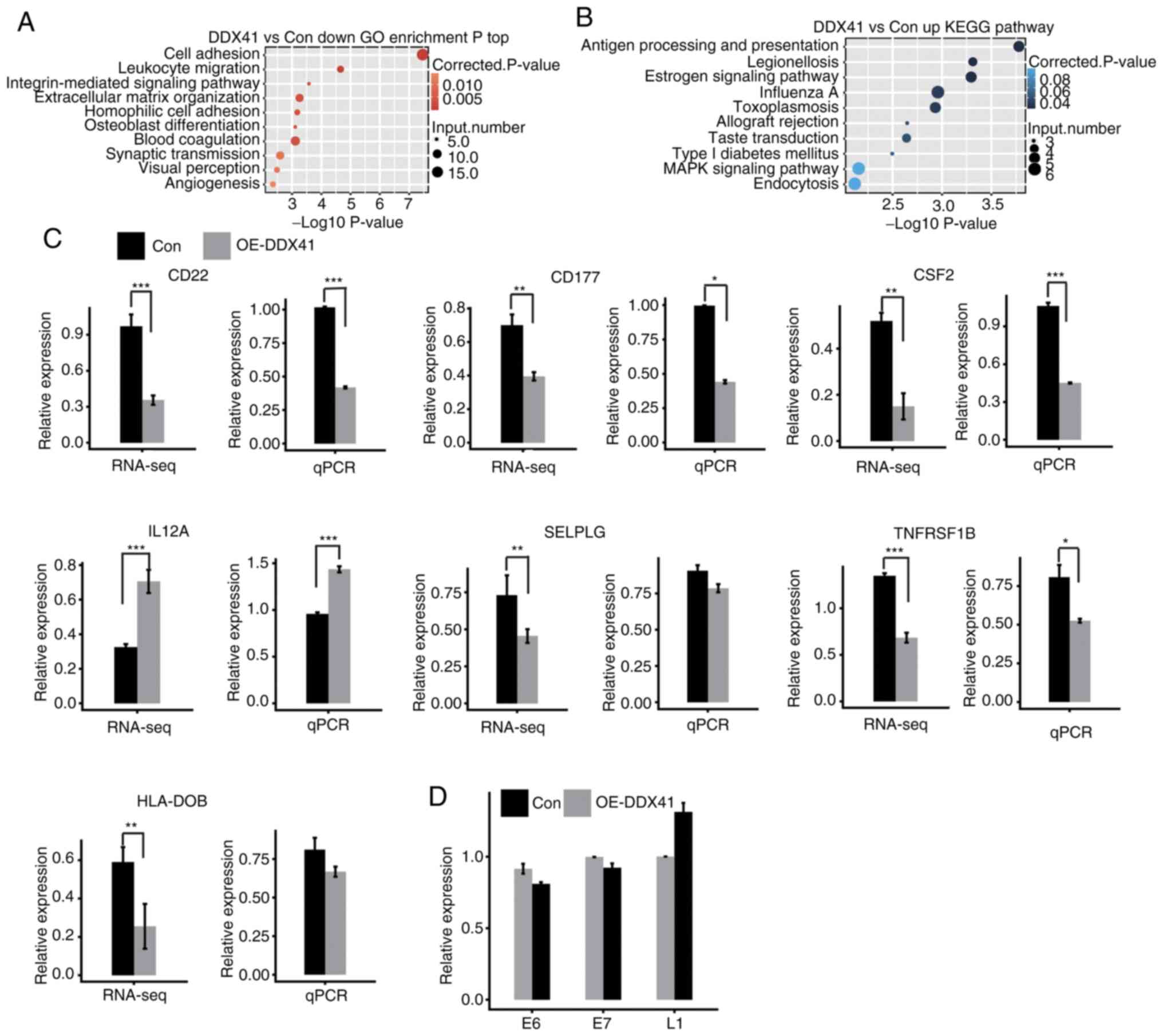
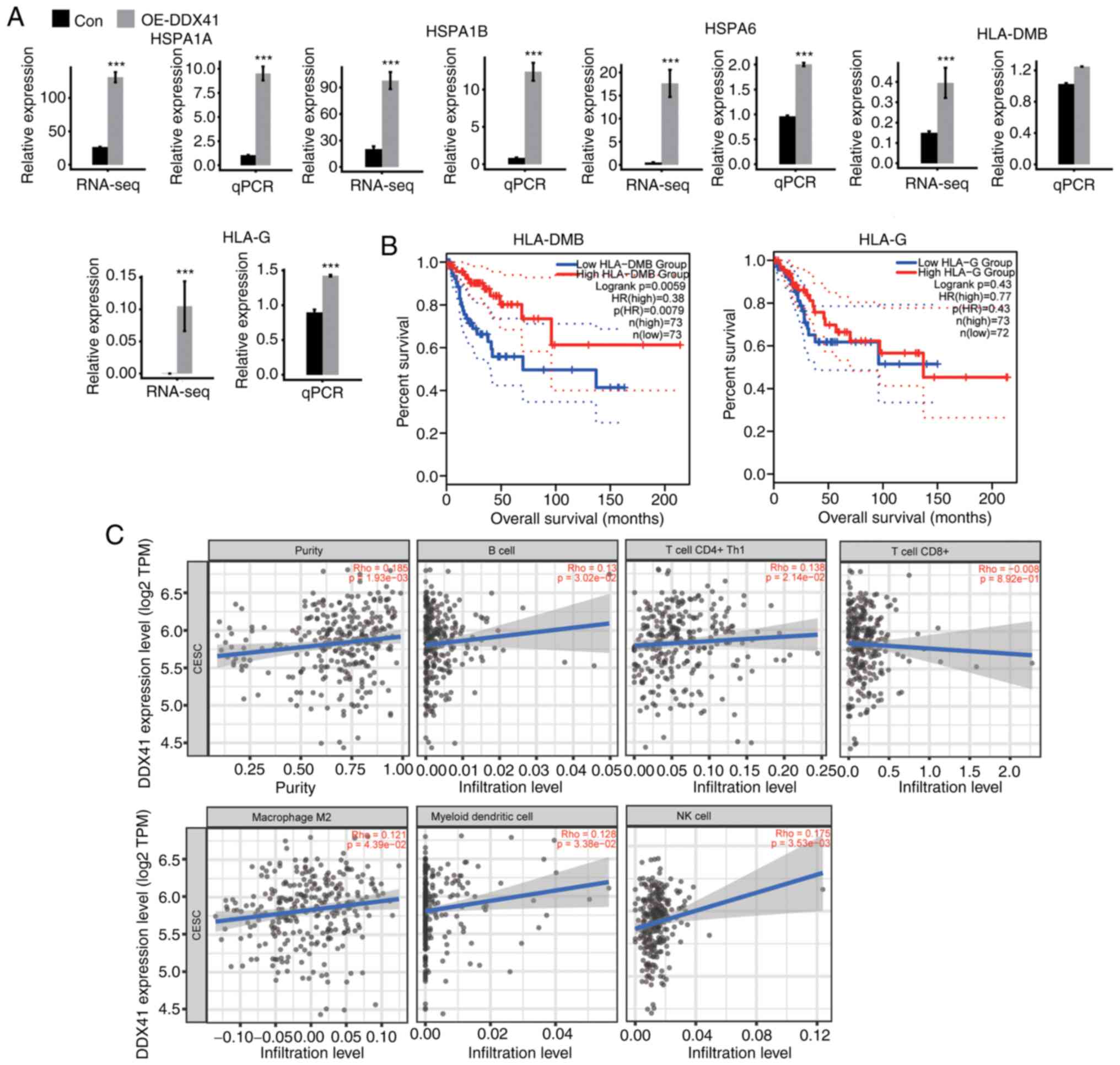
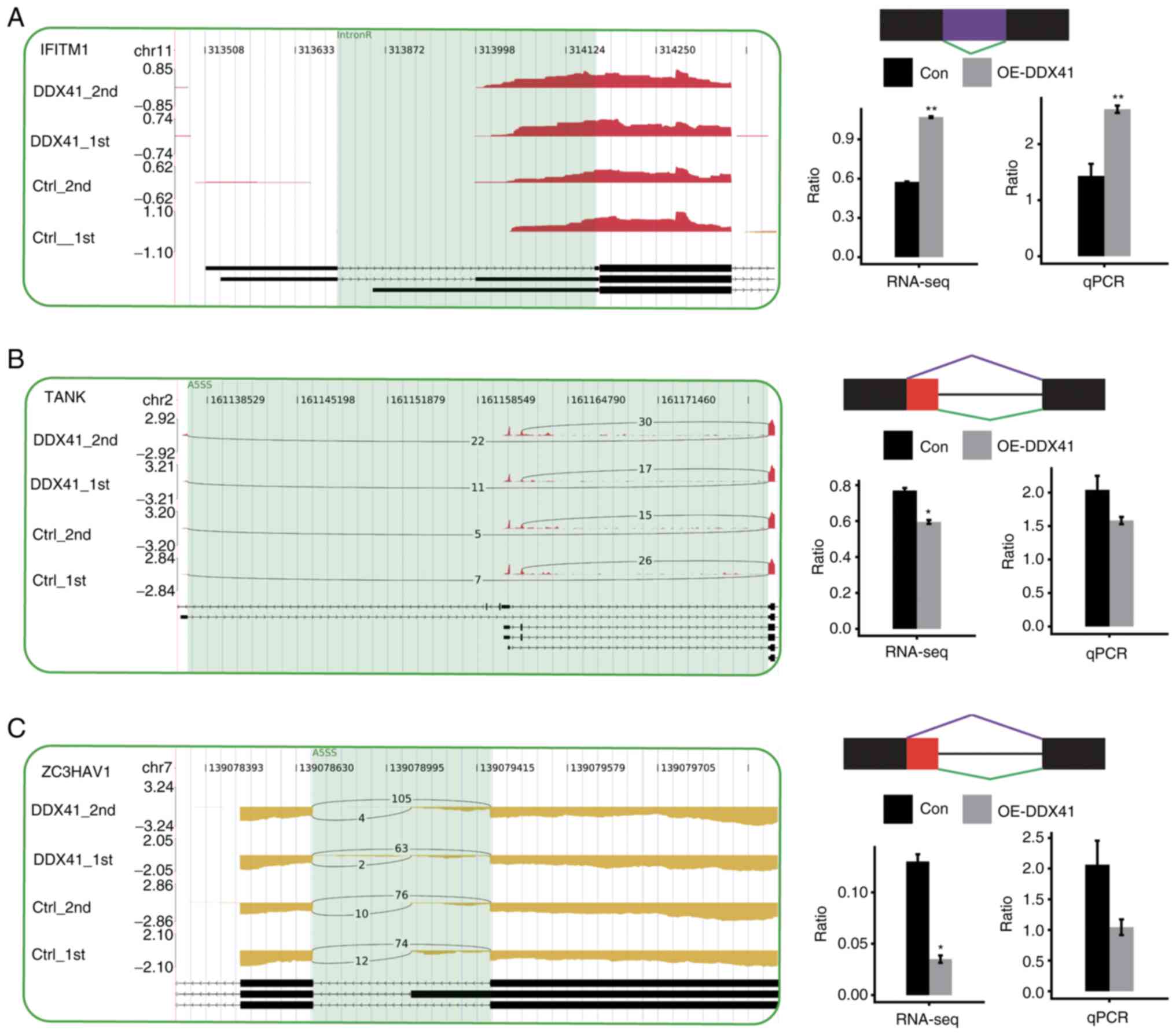
|
1
|
Jankowsky E: RNA helicases. 511. 1st
edition. Academic Press; 2012
|
|
2
|
Tanner NK and Linder P: DExD/H Box RNA
helicases: From generic motors to specific dissociation functions.
Mol Cell. 8:251–262. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bourgeois CF, Mortreux F and Auboeuf D:
The multiple functions of RNA helicases as drivers and regulators
of gene expression. Nat Rev Mol Cell Biol. 17:426–438. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rocak S and Linder P: DEAD-Box proteins:
The driving forces behind RNA metabolism. Nat Rev Mol Cell Biol.
5:232–241. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jankowsky E: RNA helicases at work:
Binding and rearranging. Trends Biochem Sci. 36:19–29. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fuller-Pace FV: DExD/H box RNA helicases:
Multifunctional proteins with important roles in transcriptional
regulation. Nucleic Acids Res. 34:4206–4215. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cruciat CM, Dolde C, de Groot RE, Ohkawara
B, Reinhard C, Korswagen HC and Niehrs C: RNA helicase DDX3 is a
regulatory subunit of casein kinase 1 in wnt-β-catenin signaling.
Science. 339:1436–1441. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bleichert F and Baserga SJ: The long
unwinding road of RNA helicases. Mol Cell. 27:339–352. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mosallanejad K, Sekine Y,
Ishikura-Kinoshita S, Kumagai K, Nagano T, Matsuzawa A, Takeda K,
Naguro I and Ichijo H: The DEAH-Box RNA helicase DHX15 activates
NF-κB and MAPK signaling downstream of MAVS during antiviral
responses. Sci Signal. 7:ra402014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fuller-Pace FV: DEAD box RNA helicase
functions in cancer. RNA Biol. 10:121–132. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Francis R and Jerry P: Perturbations of
RNA helicases in cancer. Wiley Interdiscip Rev RNA. 4:333–349.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Abdelhaleem M: Do human RNA helicases have
a role in cancer? Biochim Biophys Acta. 1704:37–46. 2004.PubMed/NCBI
|
|
13
|
Cai W, Chen ZX, Rane G, Singh SS, Choo Z,
Wang C, Yuan Y, Tan TZ, Arfuso F, Yap CT, et al: Wanted DEAD/H or
alive: Helicases winding up in cancers. J Natl Cancer Inst.
25:1092017.
|
|
14
|
Chlon TM, Stepanchick E, Choi K, Zheng Y,
Hueneman K, Davis A and Starczynowski DT: The inherited MDS gene
DDX41 is required for ribosome biogenesis and cell viability.
Blood. 134 (Suppl 1):S7732019. View Article : Google Scholar
|
|
15
|
Ding L, Ley TJ, Larson DE, Miller CA,
Koboldt DC, Welch JS, Ritchey JK, Young MA, Lamprecht T, McLellan
MD, et al: Clonal evolution in relapsed acute myeloid leukaemia
revealed by whole-genome sequencing. Nature. 481:506–510. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Polprasert C, Schulze I, Sekeres MA,
Makishima H, Przychodzen B, Hosono N, Singh J, Padgett RA, Gu X,
Phillips JG, et al: Inherited and somatic defects in DDX41 in
myeloid neoplasms. Cancer Cell. 27:658–670. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Venugopal P, Cheah JJC, Eshraghi L,
Shahrin NH, Homan C, Feng J, Schreiber AW, Fine M, Phillips K,
Poplawski N, et al: An integrative genomic approach to examine
mutations and biological pathways associated with hematological
malignancy development in DDX41 mutated families. Blood. 134 (Suppl
1):S26862019. View Article : Google Scholar
|
|
18
|
Cheah JJC, Hahn CN, Hiwase DK, Scott HS
and Brown AL: Myeloid neoplasms with germline DDX41 mutation. Int J
Hematol. 106:163–174. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Peters D, Radine C, Reese A, Budach W,
Sohn D and Jänicke RU: The DEAD-box RNA helicase DDX41 is a novel
repressor of p21 WAF1/CIP1 mRNA translation. J Biol
Chem. 292:8331–8341. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Stavrou S, Aguilera AN, Blouch K and Ross
SR: DDX41 recognizes RNA/DNA retroviral reverse transcripts and is
critical for in vivo control of murine leukemia virus infection.
mBio. 9:e00923–18. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Duan Y, Zeng J, Fan S, Liao Y, Feng M,
Wang L, Zhang Y and Li Q: Herpes simplex virus type 1-encoded
miR-H2-3p manipulates cytosolic DNA-stimulated antiviral innate
immune response by targeting DDX41. Viruses. 15:7562019. View Article : Google Scholar
|
|
22
|
Zhang Z, Yuan B, Bao M, Lu N, Kim T and
Liu YJ: The helicase DDX41 senses intracellular DNA mediated by the
adaptor STING in dendritic cells. Nat Immunol. 12:959–965. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Parvatiyar K, Zhang Z, Teles RM, Ouyang S,
Jiang Y, Iyer SS, Zaver SA, Schenk M, Zeng S, Zhong W, et al: The
helicase DDX41 recognizes the bacterial secondary messengers cyclic
di-GMP and cyclic di-AMP to activate a type I interferon immune
response. Nat Immunol. 13:1155–1161. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nakamura T, Miyabe H, Hyodo M, Sato Y,
Hayakawa Y and Harashima H: Liposomes loaded with a STING pathway
ligand, cyclic di-GMP, enhance cancer immunotherapy against
metastatic melanoma. J Control Release. 216:149–157. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Miyabe H, Hyodo M, Nakamura T, Sato Y,
Hayakawa Y and Harashima H: A new adjuvant delivery system ‘cyclic
di-GMP/YSK05 liposome’ for cancer immunotherapy. J Control Release.
184:20–27. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cordin O and Beggs JD: RNA helicases in
splicing. RNA Biol. 10:83–95. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jurica MS, Licklider LJ, Gygi SR,
Grigorieff N and Moore MJ: Purification and characterization of
native spliceosomes suitable for three-dimensional structural
analysis. RNA. 8:426–439. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bessonov S, Anokhina M, Will CL, Urlaub H
and Lührmann R: Isolation of an active step I spliceosome and
composition of its RNP core. Nature. 452:846–850. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Nam DK, Lee S, Zhou G, Cao X, Wang C,
Clark T, Chen J, Rowley JD and Wang SM: Oligo(dT) primer generates
a high frequency of truncated cDNAs through internal poly(A)
priming during reverse transcription. Proc Natl Acad Sci USA.
99:6152–6156. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Robinson MD, McCarthy DJ and Smyth GK:
EdgeR: A bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xie C, Mao X, Huang J, Ding Y, Wu J, Dong
S, Kong L, Gao G, Li CY and Wei L: KOBAS 2.0: A web server for
annotation and identification of enriched pathways and diseases.
Nucleic Acids Res. 39((Web Server Issue)): W316–W322. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Xia H, Chen D, Wu Q, Wu G, Zhou Y, Zhang Y
and Zhang L: CELF1 preferentially binds to exon-intron boundary and
regulates alternative splicing in HeLa cells. Biochim Biophys Acta
Gene Regul Mech. 1860:911–921. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Pryke A, Mostaghim S and Nazemi A: Heatmap
visualization of population based multi objective algorithms.
Evolutionary multi-criterion optimization. Obayashi S, Deb K,
Poloni C, Hiroyasu T and Murata T: Springer Berlin Heidelberg;
Berlin, Heidelberg: pp. 361–375. 2007, View Article : Google Scholar
|
|
35
|
Katz Y, Wang ET, Silterra J, Schwartz S,
Wong B, Thorvaldsdóttir H, Robinson JT, Mesirov JP, Airoldi EM and
Burge CB: Quantitative visualization of alternative exon expression
from RNA-seq data. Bioinformatics. 31:2400–2402. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kim D, Pertea G, Trapnell C, Pimentel H,
Kelley R and Salzberg SL: TopHat2: Accurate alignment of
transcriptomes in the presence of insertions, deletions and gene
fusions. Genome Biol. 14:R362013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhao M, Kim P, Mitra R, Zhao J and Zhao Z:
TSGene 2.0: An updated literature-based knowledgebase for tumor
suppressor genes. Nucleic Acids Res. 44(D1): D1023–D1031. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Adey A, Burton JN, Kitzman JO, Hiatt JB,
Lewis AP, Martin BK, Qiu R, Lee C and Shendure J: The
haplotype-resolved genome and epigenome of the aneuploid HeLa
cancer cell line. Nature. 500:207–211. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Landry JJ, Pyl PT, Rausch T, Zichner T,
Tekkedil MM, Stütz AM, Jauch A, Aiyar RS, Pau G, Delhomme N, et al:
The genomic and transcriptomic landscape of a HeLa cell line. G3
(Bethesda). 7:1213–1224. 2013. View Article : Google Scholar
|
|
40
|
Li Y, Qi H, Li X, Hou X, Lu X and Xiao X:
A novel dithiocarbamate derivative induces cell apoptosis through
p53-dependent intrinsic pathway and suppresses the expression of
the E6 oncogene of human papillomavirus 18 in HeLa cells.
Apoptosis. 20:787–795. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Jensen PE: Recent advances in antigen
processing and presentation. Nat Immunol. 8:1041–1048. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Murata S, Takahama Y, Kasahara M and
Tanaka K: The immunoproteasome and thymoproteasome: Functions,
evolution and human disease. Nat Immunol. 19:923–931. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Rock KL, Reits E and Neefjes J: Present
yourself! by MHC class I and MHC class II molecules. Trends
Immunol. 37:724–737. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Becht E, Giraldo NA, Lacroix L, Buttard B,
Elarouci N, Petitprez F, Selves J, Laurent-Puig P, Sautès-Fridman
C, Fridman WH and de Reyniès A: Estimating the population abundance
of tissue-infiltrating immune and stromal cell populations using
gene expression. Genome Biol. 17:2182016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q,
Li B and Liu XS: TIMER2.0 for analysis of tumor-infiltrating immune
cells. Nucleic Acids Res. 48((W1)): W509–W514. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Flood BA, Higgs EF, Li S, Luke JJ and
Gajewski TF: STING pathway agonism as a cancer therapeutic. Immunol
Rev. 290:24–38. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yoneyama-Hirozane M, Kondo M, Matsumoto
SI, Morikawa-Oki A, Morishita D, Nakanishi A, Kawamoto T and
Nakayama M: High-throughput screening to identify inhibitors of
DEAD box helicase DDX41. SLAS Discov. 22:1084–1092. 2017.PubMed/NCBI
|
|
48
|
Polprasert C, Schulze I, Sekeres MA,
Makishima H, Przychodzen BP, Hosono N, Singh J, Padgett RA, Gu X,
Jankowsky E, et al: DDX41 is a tumor suppressor gene associated
with inherited and acquired mutations. Blood. 124:1252014.
View Article : Google Scholar
|
|
49
|
Nekulova M, Holcakova J, Coates P,
Vojtesek BJC and Letters MB: The role of P63 in cancer, stem cells
and cancer stem cells. Cell Mol Biol Lett. 16:296–327. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Graziano V and De Laurenzi V: Role of p63
in cancer development. Biochim Biophys Acta. 1816:57–66.
2011.PubMed/NCBI
|
|
51
|
Wang TY, Chen BF, Yang YC, Chen H, Wang Y,
Cviko A, Quade BJ, Sun D, Yang A, McKeon FD and Crum CP: Histologic
and immunophenotypic classification of cervical carcinomas by
expression of the p53 homologue p63: A study of 250 cases. Hum
Pathol. 32:479–486. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
McCluggage WG: Immunohistochemistry as a
diagnostic aid in cervical pathology. Pathology. 39:97–111. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Houghton O and McCluggage WG: The
expression and diagnostic utility of p63 in the female genital
tract. Adv Anat Pathol. 16:316–321. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Saritha VN, Veena VS, Jagathnath KM,
Somanathan T and Sujathan K: Significance of DNA replication
licensing proteins (MCM2, MCM5 and CDC6), p16 and p63 as markers of
premalignant lesions of the uterine cervix: Its usefulness to
predict malignant potential. Asian Pac J Cancer Prev. 27:141–148.
2018.
|
|
55
|
Pattabiraman D and Gonda T: Role and
potential for therapeutic targeting of MYB in leukemia. Leukemia.
27:269–277. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Ramsay RG and Gonda TJ: MYB function in
normal and cancer cells. Nat Rev Cancer. 8:523–534. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Drier Y, Cotton MJ, Williamson KE,
Gillespie SM, Ryan RJ, Kluk MJ, Carey CD, Rodig SJ, Sholl LM,
Afrogheh AH, et al: An oncogenic MYB feedback loop drives alternate
cell fates in adenoid cystic carcinoma. Nat Genet. 48:265–272.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Sarvaiya PJ, Schwartz JR, Hernandez CP,
Rodriguez PC and Vedeckis WV: Role of c-myb in the survival of pre
B-cell acute lymphoblastic leukemia and leukemogenesis. Am J
Hematol. 87:969–976. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Biroccio A, Benassi B, D'Agnano I,
D'Angelo C, Buglioni S, Mottolese M, Ricciotti A, Citro G,
Cosimelli M, Ramsay RG, et al: C-Myb and bcl-x overexpression
predicts poor prognosis in colorectal cancer: Clinical and
experimental findings. Am J Pathol. 158:1289–1299. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Knopfová L, Biglieri E, Volodko N, Masařík
M, Hermanová M, Garzón JF, Dúcka M, Kučírková T, Souček K, Šmarda
J, et al: Transcription factor c-myb inhibits breast cancer lung
metastasis by suppression of tumor cell seeding. Oncogene.
37:1020–1030. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Ma M, Zhao R, Yang X, Zhao L, Liu L, Zhang
C, Wang X and Shan B: Low expression of Mda-7/IL-24 and high
expression of C-myb in tumour tissues are predictors of poor
prognosis for burkitt lymphoma patients. Hematology. 23:448–455.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Werner S, Duan DS, de Vries C, Peters KG,
Johnson DE and Williams LT: Differential splicing in the
extracellular region of fibroblast growth factor receptor 1
generates receptor variants with different ligand-binding
specificities. Mol Cell Biol. 12:82–88. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Johnson DE, Lu J, Chen H, Werner S and
Williams LT: The human fibroblast growth factor receptor genes: A
common structural arrangement underlies the mechanisms for
generating receptor forms that differ in their third immunoglobulin
domain. Mol Cell Biol. 11:4627–4634. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Chellaiah AT, McEwen DG, Werner S, Xu J
and Ornitz M: Fibroblast growth factor receptor (FGFR) 3.
Alternative splicing in immunoglobulin-like domain III creates a
receptor highly specific for acidic FGF/FGF-1. J Biol Chem.
269:11620–11627. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Vainikka S, Partanen J, Bellosta P,
Coulier F, Birnbaum D, Basilico C, Jaye M and Alitalo K: Fibroblast
growth factor receptor-4 shows novel features in genomic structure,
ligand binding and signal transduction. EMBO J. 12:4273–4280. 1992.
View Article : Google Scholar
|
|
66
|
Tomlinson DC and Knowles MA: Altered
splicing of FGFR1 is associated with high tumor grade and stage and
leads to increased sensitivity to FGF1 in bladder cancer. Am J
Pathol. 177:2379–2386. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Tang S, Hao Y, Yuan Y, Liu R and Chen Q:
Role of fibroblast growth factor receptor 4 in cancer. Cancer Sci.
109:3024–3031. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Touat M, Ileana E, Postel-Vinay S, André F
and Soria JC: Targeting FGFR signaling in cancer. Clin Cancer Res.
21:2684–2694. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Babina IS and Turner NC: Advances and
challenges in targeting FGFR signalling in cancer. Nat Rev Cancer.
17:318–332. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Trembleau S, Penna G, Bosi E, Mortara A,
Gately MK and Adorini L: Interleukin 12 administration induces T
helper type 1 cells and accelerates autoimmune diabetes in NOD
mice. J Exp Med. 181:817–821. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Poe JC, Fujimoto Y, Hasegawa M, Haas KM,
Miller AS, Sanford IG, Bock CB, Fujimoto M and Tedder TF: CD22
regulates B lymphocyte function in vivo through both
ligand-dependent and ligand-independent mechanisms. Nat Immunol.
5:1078–1087. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
72
|
Milani V, Noessner E, Ghose S, Kuppner M,
Ahrens B, Scharner A, Gastpar R and Issels RD: Heat shock protein
70: Role in antigen presentation and immune stimulation. Int J
Hyperthermia. 18:563–575. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Castelli C, Ciupitu AM, Rini F, Rivoltini
L, Mazzocchi A, Kiessling R and Parmiani G: Human heat shock
protein 70 peptide complexes specifically activate antimelanoma T
cells. Cancer Res. 61:222–227. 2001.PubMed/NCBI
|
|
74
|
Noessner E, Gastpar R, Milani V, Brandl A,
Hutzler PJ, Kuppner MC, Roos M, Kremmer E, Asea A, Calderwood SK
and Issels RD: Tumor-Derived heat shock protein 70 peptide
complexes are cross-presented by human dendritic cells. J Immunol.
169:5424–5432. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Banchereau J and Palucka AK: Dendritic
cells as therapeutic vaccines against cancer. Nat Rev Immunol.
5:296–306. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Noessner E: Thermal stress-related
modulation of tumor cell physiology and immune responses. Cancer
Immunol Immunother. 55:289–291. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Albakova Z, Armeev GA, Kanevskiy LM,
Kovalenko EI and Sapozhnikov AM: HSP70 multi-functionality in
cancer. Cells. 9:5872020. View Article : Google Scholar
|
|
78
|
Lee KG, Susana SY, Kui L, Chih-Cheng Voon
D, Mauduit M, Bist P, Bi X, Pereira NA, Liu C, Sukumaran B, et al:
Bruton's tyrosine kinase phosphorylates DDX41 and activates its
binding of dsDNA and STING to initiate type 1 interferon response.
Cell Rep. 10:1055–1065. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Ma JX, Li JY, Fan DD, Feng W, Lin AF,
Xiang LX and Shao JZ: Identification of DEAD-box RNA helicase DDX41
as a trafficking protein that involves in multiple innate immune
signaling pathways in a zebrafish model. Front Immunol. 9:13272018.
View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Woo SR, Corrales L and Gajewski TF: The
STING pathway and the T cell-inflamed tumor microenvironment.
Trends Immunol. 36:250–256. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Barber GN: STING-Dependent cytosolic DNA
sensing pathways. Trends Immunol. 35:88–93. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Barber GN: STING: Infection, inflammation
and cancer. Nat Rev Immunol. 15:760–770. 2015. View Article : Google Scholar : PubMed/NCBI
|