Introduction
Thyroid cancer is the most prevalent endocrine
cancer worldwide with rapidly rising morbidity in the past decade
owing to the wide use of diagnostic imaging, especially in women
(1,2).
In 2015, thyroid cancer had the most rapidly increasing incidence
rate for women in China (3).
Papillary thyroid cancer (PTC) is a major type of thyroid
malignancy, accounting for 85% of all thyroid cancer cases
(4). Although the mortality rate of
PTC is relatively low, with a 5-year survival rate over 95%, the
clinical outcomes for severe PTC patients are unfavorable, and the
recurrence rate within 3 years was reported to be up to 15.6% in
patients who underwent postoperative radioactive iodine (RAI,
131I) treatment (3,5). A primary
reason for the unsatisfactory outcome is the failure to respond to
131I ablation treatment after surgical resection caused
by the impaired ability of 131I aggregation of thyroid
follicular cells (6). Thus,
discovering additional molecular mechanisms in 131I
resistance and developing potential targets for TC prevention and
treatment is of great importance.
Noncoding RNAs, mainly long noncoding RNAs (lncRNAs)
and microRNAs (miRs) are well known for their roles in diverse
processes through regulation of gene expression (7). Recently, it has been well established
that lncRNAs may serve as competing endogenous RNAs (ceRNAs) of
miRs by binding to the mRNA response element in miRs, thus
regulating gene expression (8). The
ceRNA network has been documented to be involved in
radioresistance. For instance, lncRNA nuclear-enriched autosomal
transcript 1 (NEAT1) has been suggested to act as a ceRNA of
miR-101-3p to promote 131I resistance of PTC cells via
inactivation of the phosphoinositide 3-kinase/protein kinase B
(PI3K/Akt) signaling pathway (5).
Among lncRNAs, the small nucleolar RNA host gene (SNHG7) has
recently aroused great interest for its tumor promoting roles in
several cancer types through binding with miRs (9,10), however
its role in PTC remains unclear. miR-9-5p is a well-known tumor
inhibitor in multiple cancer types (11,12),
including PTC (13). Notably, a
previous study suggested that SNHG7 acts as a sponge for miR-9
(14). The present study further
identified the target relationship between miR-9-5p and
dipeptidyl-peptidase 4 (DPP4). Importantly, upregulation of DPP4
has been suggested to promote PTC cell growth and metastasis
(15). Moreover, downregulation of
the PI3K/Akt signaling pathway has been documented to participate
in the suppression of PTC growth (16). Overall, this study was designed to
identify the interaction among SNHG7, miR-9-5p and DPP4 and to
evaluate their roles in the growth and radioresistance of TC
cells.
Materials and methods
Ethics statement
The present study was approved and supervised by the
Clinical Ethics Committee of the Second Affiliated Hospital of
Nanchang University (Nanchang, China). Signed informed consent was
obtained from each eligible participant. Animal studies were
conducted according to the principles and procedures approved by
the Committee on the Ethics of Animal Experiments of the Second
Affiliated Hospital of Nanchang University. All experimental
procedures were conducted in line with the ethical guidelines for
the study of experimental pain in conscious animals.
Clinical sample collection
PTC and paracancerous tissues were collected from 50
PTC patients who were admitted to the Second Affiliated Hospital of
Nanchang University from January 2017 to January 2018. Another 50
PTC patients were recruited, and each of them was treated with 10.0
mCi/131I once a month for a total of 6 months. Then, the
patients were categorized into responders (n=28) and nonresponders
(n=22) to irradiation in accordance with the Response Evaluation
Criteria in Solid Tumors version 1.1 (17). We used Arraystar Human lncRNA
Microarrray version 2.0 (8660 K; Arraystar) sequencing to analyze
the differential expression of lncRNAs between PTC tissues and
adjacent tissues. To validate the diagnostic reliability of the
lncRNAs in PTC, a receiver operating characteristic (ROC) curve was
constructed, and the area under the ROC curve (AUC) of SNHG7 was
analyzed.
Microarray analysis
The lncRNAs with differential expression in PTC and
paracancerous tissues were predicted and analyzed. The
transcriptome data were obtained on an Illumina HiSeq RNA-Seq
platform [Illumina HiSeq (Illumina, Inc.), Life Technologies SOLiD
(Thermo Fisher Scientific, Inc.), and Roche 454 (Roche
Diagnostics)] for 6 PTC tissues and 6 paracancerous tissues.
According to The Cancer Genome Atlas (TCGA) (http://ualcan.path.uab.edu/analysis.html) research
network, the RNA'Seq PTC dataset contained 60,483 mRNAs that
included 7,589 lncRNAs, as described in the NCBI (https://www.ncbi.nlm.nih.gov/) and Ensembl databases
(http://www.ensembl.org/). Next, the differential
expression of the lncRNAs was assessed using the R language package
DESeq (http://www.bioconductor.org/packages/release/bioc/html/limma.html)
(18) (criteria set at P<0.05 and
|log2 FC| >2). lncRNAs whose expression fold changes
were <1 in over 10% of the samples were excluded. Additionally,
the expression of each lncRNA was log2-transformed for
the subsequent analysis. Then the online bioinformatics analysis
website UALCAN (http://ualcan.path.uab.edu/index.html) (19) was used to predict and screen lncRNA
differentially expressed in patients with PTC.
Cell culture and
131I-resistant cell line construction
The TC cell lines TPC-1 (BNCC337912) and B-CPAP
(BNCC338685) were purchased from BeNa Culture Collection (Beijing,
China). The B-CPAP cell line was identified correctly by STR, and
it was not cross-contaminated or misidentified. TPC-1 cells were
incubated in 90% Roswell Park Memorial Institute (RPMI)-1640 medium
with 10% fetal bovine serum (FBS), while B-CPAP cells were cultured
in 90% F-12K medium containing 10% FBS, and they were all incubated
at 37°C with 5% CO2. All reagents were purchased from
Gibco; Thermo Fisher Scientific, Inc. To construct
131I-resistant TPC-1 and 131I-resistant
B-CPAP cell lines, the cell medium was mixed with a median lethal
dose of 131I. After 24 h of treatment, the cell
viability was evaluated using a
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay (Promega Corporation) by evaluating the half-maximal
inhibitory concentration of 131I (IC50).
Cell transfection
miR-9-5p mimic (50 nM), miR-control, empty vector
(pcDNA3.1), overexpression (Oe) SNHG7 vector, Oe-SNHG5 + miR-9-5p,
and miR-9-5p + DPP4 were transfected into parent TPC-1 and B-CPAP
cells, while Scramble (50 nM), small interfering RNA (siRNA) to
SNHG7 (si-SNHG7), miR-9-5p inhibitor (200 nM), si-SNHG7 + miR-9-5p
inhibitor and miR-9-5p inhibitor + si-DPP4 (50 nM, Shanghai
GenePharma Co., Ltd.) were transfected into res-TPC-1/B-CPAP cells
using 2 µl Lipofectamine™ 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). Cells were cultured for 37°C 24 h, and reverse
transcription quantitative polymerase chain reaction (RT-qPCR) was
used to verify the transfection efficiency. The sequences were as
follows: si-SNHG7-1 sense, 5′-AAUAAUCCGUUUUUACUCCCC-3′ and
antisense, 5′-GGAGUAAAAACGGAUUAUUUA-3′; si-SNHG7-2 sense,
5′-CGGAUUAUUUAGUCUUCAACA-3′ and antisense,
5′-UUGAAGACUAAAUAAUCCGUU-3′; si-SNHG7-3 sense,
5′-GAGACAGUAUCAAGAAAGAUA-3′ and antisense,
5′-UCUUUCUUGAUACUGUCUCUU-3′; Scramble sense,
5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense,
5′-ACGUGACACGUUCGGAGAATT-3′; si-DPP4, 5′-CCCTATGAAACCGCTGGAAAT-3′;
si-DPP4 negative control, 5″TTCTCCGAACGTGTCACGT′3′.
miR-9-5p mimic, 5′-CGAGCTCTGTGTGTGTGTGTGTGTG-3′;
miR-9-5p inhibitor:
5′-TTCCGCGGCCGCTATGGCCGACGTCGACGGGAATGGGGAAAGGGAA-3′.
miR-9-5p mimic negative control,
5′-UUCUCCGAACGUGUCACGUTT-3′; miR-9-5p inhibitor negative control,
5′-CAGUACUUUUGUGUAGUACAA-3′.
RT-qPCR
Total RNA of cells was extracted according to the
instructions of a TRIzol kit (Invitrogen; Thermo Fisher Scientific,
Inc.). The purity of total RNA was evaluated using an ultraviolet
spectrophotometer, and the absorbance values at 260 nm (A260) and
280 nm (A280) were measured. RNA samples with A260/A280 no less
than 1.70 were qualified for subsequent studies. Single-strand cDNA
templates were produced by reverse transcription strictly in
accordance to the instructions of a Revert Aid First Strand cDNA
Synthesis Kit (Thermo Fisher Scientific, Inc.). qRCR was performed
using SYBR Premix Ex Taq (Takara Bio, Inc.) on an MX3000P qPCR
system (Stratagene; Agilent Technologies, Inc.). Then, PCR
amplification was performed with the following conditions:
Pre-denaturation at 95°C for 60 sec, denaturation at 95°C for 20
sec, annealing at 58°C for 30 sec and extension at 74°C for 30 sec.
Each experiment was performed in triplicate. The PCR primer
sequences and the internal references U6 and
glyceraldehyde-3-phosphate dehydrogenase (GAPDH) are presented in
Table I. The relative expression was
calculated using 2−ΔΔCq (20).
 | Table I.Primer sequences for RT-qPCR. |
Table I.
Primer sequences for RT-qPCR.
| Primer | Sequence
(5′-3′) |
|---|
| SNHG7 | F:
GTGACTTCGCCTGTGATGGA |
|
| R:
GGCCTCTATCTGTACCTTTATTC |
| LINC01977 | F:
CCCCTTTCCCCAGGGTACTA |
|
| R:
CGTTAGACAGCCTCTTGGGG |
| RP11′363E7.4 | F:
GGCACTTTTCAGAACATC |
|
| R:
TGTCGTGTATCACAGCAT |
| RP3′483K16.4 | F:
CCTCGTGCACTCTGGGGTA |
|
| R:
GTGGAACTGCTGTGCCAATG |
| RUNDC3A'AS1 | F:
CAGTCTCTGTGCGTTGAAGC |
|
| R:
GGAGTACACATTGACGGCCA |
| AC093609.1 | F:
CGAGTCGGGTTCTGATCCAC |
|
| R:
GGATGCTGCTTTCCACCCAT |
| CTD-2008L17.2 | F:
AGGGGCCTTCCAGATTAAGG |
|
| R:
CGAGTCGGGTTCTGATCCAC |
| HAGLROS | F:
GTCACCCTTAAATACCGCTCT |
|
| R:
CTTCCTCCCACACAAATACTCC |
| UNC5B'AS1 | F:
CTGGAGGATGAAGGATCGGG |
|
| R:
GGCTAGGGGGAAATGTCGG |
| LINC01354 | F:
TGCGTTCAGTAAAACGGGCA |
|
| R:
TGTGGGAAATGCAGGGTTCT |
| miR-9-5p | F:
CCTGGGAGTATGTCGATCTATTG |
|
| R:
TGGTGTCGTGGAGTCG |
| DPP4 | F:
AAGTGGCGTGTTCAAGTGTG |
|
| R:
GGCTTTGGAGATCTGAGCTG |
| GADPH | F:
CGGACCAATACGACCAA |
|
| R:
AGCCACATCGCTCAGACACC |
Flow cytometry
Cells (2×106) were washed with
phosphate-buffered saline (PBS) and fixed in Hank's buffered salt
solution (product code H1025; Solarbio Life Sciences) (137 mmol/l
NaCl, 0.25 mmol/l Na2HPO4, 5.4 mmol/l KCl,
0.44 mmol/l KH2PO4, 1.0 mmol/l
MgSO4, 1.3 mmol/l CaCl2 and 4.2 mmol/l
NaHCO3) supplemented with cold 80% ethanol for 30 min.
Then, the cells underwent centrifugation at 167.7 × g for 10 min at
4°C and two PBS washes. Next, the cells were stained with 50 mg/ml
propidium iodide [PI; Hangzhou MultiSciences (Lianke) Biotech Co.,
Ltd.] containing 0.1 mg/ml RNase A and 0.6% NP-40 (Thermo Fisher
Scientific, Inc.) at 4°C under dark conditions for 30 min,
subjected to centrifugation at 167.7 × g for 10 min at 4°C, and
washed with PBS. For the apoptosis assay, cells were stained with
fluorescein isothiocyanate (FITC)-Annexin V and PI based on the
instructions of an Annexin V-FITC/PI kit (BD Biosciences) and then
incubated on ice for 15 min without light exposure. Flow cytometry
was conducted using a FACSCalibur flow cytometer (BD Biosciences),
and FlowJo 10.0.4 software (FlowJo LLC) was used for data analysis.
Each procedure was performed 3 times.
5-Ethynyl-2′-deoxyuridine (EdU)
labeling assay
An EdU assay was performed to measure DNA synthesis.
The inhibition rate of cell proliferation was assessed using a
Click-iT EdU Imaging Kit (Thermo Fisher Scientific, Inc.) following
a previously reported procedure (21).
MTT assay
An MTT assay was conducted to assess the
proliferation of transfected cells. Cells were sorted into 96-well
plates at a density of 3×103 cells/well for 3 days,
following exposure to 1.0 mCi/well 131I or 0.45 mCi/well
131I. Then, the cells were cultured in 10 µl MTT-8
solution (5 mg/ml, Sigma-Aldrich; Merck KGaA) at 37°C for 4 h.
After 2 h of incubation at 37°C, 150 µl dimethyl sulfoxide
(Sigma-Aldrich; Merck KGaA) was added into each well to dissolve
the formazan crystals. Absorbance at 570 nm was read using a
microplate reader (Bio-Rad Laboratories, Inc.), and growth curves
were generated for each group of cells.
Colony formation assay
Cells were resuspended in RPMI-1640 culture medium
in a Luria-Bertani culture plate (cat. no. D0110; Beijing
Nobleryder Science and Technology Co., Ltd.) with each plate
containing 10 cm medium and 500 cells and cultured in a 37°C
incubator with 5% CO2 for 2 weeks. Then, the TPC-1 cells
were exposed to 1.0 mCi/well 131I, while the B-CPAP
cells were exposed to 0.45 mCi/well 131I. Following 20
min of 4% paraformaldehyde fixation at room temperature, the cells
were stained with 0.1% crystal violet for 10 min. Then, the plates
were air dried, and colonies with at least 50 cells were evaluated
under a light microscope (magnification, ×100). The relative colony
rate was calculated as follows: Relative colony rate=cell colonies
in the experimental group/cell colonies in the control group.
Cell cycle assays
Cells (2×106) were harvested after
transfection and then resuspended and fixed in 75% cold ethanol at
4°C overnight, followed by staining with 100 µl of PI/RNase (BD
Biosciences) at 37°C for 30 min without light exposure. Flow
cytometry (Beckman Coulter, Inc.) was used to measure cell
populations in the G0/G1, S and G2/M phases. The results were
analyzed with cell ModFit LT V5.0 software (Verity Software House,
Inc.).
Fluorescence in situ hybridization
(FISH)
The subcellular localization of SNHG7 was predicted
in accordance with the available subcellular localization data at
LncATLAS (http://lncatlas.crg.eu/) (22). Next, FISH was conducted to further
confirm the subcellular localization of SNHG7 according to the
instructions of Ribo™ lncRNA FISH Probe Mix (Green) (Guangzhou
RiboBio Co., Ltd.). TPC-1 and B-CPAP cells (3.5×104
cells/well) were mounted onto slides and fixed in 4% formaldehyde
at 4°C for 10 min. Slides were pretreated with protease K (2 µg/ml;
product code P9460; Solarbio Life Sciences), glycine and acetic
anhydride, which was followed by prehybridization at 42°C for 1 h.
Next, the slides were subjected to hybridization using probes
against SNHG7 (250 µl; 300 ng/ml) at 42°C overnight. Then, the
slides were stained with PBS-0.1% Tween-diluted
4′,6-diamidino-2-phenyl indole (DAPI; product code C0065; Solarbio
Life Sciences) at room temperature for 6 min. Images were acquired
with a fluorescence microscope (magnification, ×1,000) (Eclipse Ti
microscope; Nikon Instruments), with five fields randomly selected
from each slide.
Fractionation of nuclear/cytoplasmic
RNA
The nuclear and cytoplasmic RNA was separated
according to the instructions of a PARIS™ Kit (Life Technologies,
Inc.; Thermo Fisher Scientific, Inc.). In brief, TPC-1 and B-CPAP
cells were resuspended in 500 µl cell fractionation buffer for 5–10
min. Then, the RNA fractions were centrifuged at 500 × g at 4°C for
5 min for separation. The supernatant (cytoplasmic fraction) was
collected into a 2-ml sterile enzyme-free tube and centrifuged,
while the pellet (nuclear fraction) was resuspended in 500 µl cell
disruption buffer and centrifuged. Next, the fractions were washed
with 500 µl 2X lysis/binding solution and centrifuged. Each
fraction was then mixed with 500 µl absolute ethanol, transferred
into a filter cartridge and rinsed with wash solution. After
elution, nuclear and cytoplasmic RNA was collected. SNHG7
expression was detected using RT-qPCR, with U6 as the internal
control for nuclear RNA expression and GAPDH for cytoplasmic RNA
expression. The primers are presented in Table I.
Western blot analysis
Cells (2×106) were lysed in cold
radioimmunoprecipitation buffer (product code R0010; Solarbio Life
Sciences) supplemented with 1 mM phenylmethylsulfonyl fluoride
(product code P8340; Solarbio Life Sciences). The bicinchoninic
acid method was used to quantify the extracted protein. Equal
volumes of proteins (50 µg) were run on 10% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (Bio-Rad Laboratories,
Inc.) and transferred to polyvinylidene fluoride membranes
(Amersham Pharmacia; GE Healthcare). After being sealed in 5% skim
milk at room temperature for 1 h, western blots were probed with
antibodies against Bax (1:1,000; product code ab32503), Bcl-2
(1:1,000; product code ab32124), DPP4 (1:1,000, product code
ab114033), Akt (1:500; product code ab8805), p-Akt (1:10,000,
product code ab81283), PI3K (1:1,000, product code ab191606),
p-PI3K (1:1,000, product code ab182651) and β-actin (1:5,000;
product code ab179467) (all purchased from Abcam,) at 4°C
overnight, followed by incubation with the secondary antibody
horseradish peroxidase-labeled goat anti-rabbit immunoglobulin G
(IgG; 1:2,000; cat. no. A0208, Beyotime Institute of Biotechnology)
at 4°C for 2 h. β-actin was used as a loading control for
normalization. Immunoblots were visualized with enhanced
chemiluminescence (Amersham Pharmacia; GE Healthcare) and analyzed
using ImageJ v1.48u software (National Institutes of Health).
Dual-luciferase reporter assay
starBase (http://starbase.sysu.edu.cn/) (23) and TargetScan (http://www.targetscan.org/vert_72/) (23) were used to predict the target gene of
miR-9-5p. The 3′-untranslated region (3′-UTR) fragment of DPP4 mRNA
was amplified and cloned into the PmeI and XbaI sites
of the pmirCytomegalovirus vector (pmirCMV; Promega Corparation) to
construct DPP4-wild-type (WT) and DPP4-mutant (MUT) plasmids.
Similarly, pmirCMV plasmids containing lncRNA SNHG7-WT and lncRNA
SNHG7-MUT were also constructed in a similar way. Next, the
verified plasmids along with either miR-9-5p mimic or mimic
negative control were cotransfected into 293T cells. Every assay
was repeated 3 times. Cells were collected 48 h later, and then the
luciferase activities were detected using the Dual-CMV Luciferase
Assay System (Promega Corpation) and a MicroLumatPlus LB96V
luminometer (Berthold Technologies GmbH). The relative luciferase
activity was evaluated as the ratio of the firefly luciferase
activity to the Renilla luciferase activity.
RNA pull-down assay
Cell lysates were treated with RNase-free DNase I
(Sigma-Aldrich; Merck KGaA) and cultured with a mixture of 1 µg
biotin-labeled miR-9-5p RNA fragments and streptavidin-coated
magnetic beads (Sigma-Aldrich; Merck KGaA) in a 4°C incubator for 3
h. The RNA was isolated from the obtained RNA-protein complexes,
and the interacting proteins were subjected to western blot
analysis.
Xenograft tumors in nude mice
Female nude mice (N=36; BALB/C nu/nu; ~18–22 g;
Beijing Vital River Laboratory Animal Technology Co., Ltd.) had
free access to water and food. The mice were raised at 20–22°C,
with humidity 50–60% and a 12-h light/dark cycle. TPC-1 cells
transfected with the overexpression plasmid (Oe)-SNHG7,
cotransfected with Oe-SNHG7 and miR-9-5p, or transfected with empty
vector, and 131I resistant (RAI-res-TPC-1) cells
transfected with si-SNHG7 or scramble siRNA were subcutaneously
injected into mice at a dose of 3×106 cells per mouse.
From the day of transplantation to the end of the experiment, the
health status and behavior of nude mice were observed every day.
Next, the tumor volume (V) was recorded every 3 days and calculated
as follows: V (mm3)=(axb2)/2, in which ‘a’
refers to the largest diameter while ‘b’ refers to the
perpendicular diameter. The mice were randomly allocated into 6
groups (6 mice per group) and treated with 2.0 mCi/100 g
131I once the tumor volume reached approximately 70
mm3. The tumor volume was recorded for up to 35 days.
According to the method introduced in the literature (24), the nude mice were euthanized by
intraperitoneal injection of pentobarbital (800 mg/kg), and the
physical signs of the nude mice were monitored strictly according
to the operation. The following experiment was carried out after
the complete cessation of the heartbeat of the animals. After
subcutaneous injection, all animals bore only one tumor.
Immunohistochemical staining
Sections (5 µm) from xenograft tumors were stained
using anti-DPP4 (1:50; product code ab114033; Abcam), anti-p-Akt
(1:10,000; ab81283; Abcam) and=anti-Ki67 (1: 200, ab16667; Abcam)
at 4°C overnight and then incubated with anti-IgG secondary
antibody (1:1,000; ab6721; Abcam) for 30 min. Staining results were
visualized with 3,3-diaminobenzidine (cat. no. DA1010; Solarbio).
Five fields were randomly selected and observed under an inverted
microscope (Nikon Instruments) at a magnification of ×200.
Statistical analysis
Data were analyzed with SPSS 21.0 software (IBM
Corp.). Each experiment was performed 3 times. The results are
presented as the mean ± standard deviation. Differences in multiple
groups were analyzed using one-way or two-way analysis of variance
(ANOVA). Tukey's multiple comparison test was utilized for pairwise
comparisons after ANOVA. The correlation analysis between each
group pair was performed with Pearson's correlation coefficient
test. ROC curves were generated, and the area under the ROC curve
for lncRNA SNHG7 was analyzed. The P-value was obtained by a
two-tailed test, and P<0.05 was considered to indicate a
statistically significant difference.
Results
SNHG7 is highly expressed in PTC
patients and correlated with 131I resistance
Initially, we analyzed the differentially expressed
lncRNAs in PTC and paracancerous tissues, and it was revealed that
a total of 169 lncRNAs were differentially expressed. Among them,
145 lncRNAs were upregulated while the remaining 24 were
downregulated in PTC tissues (Fig.
1A). To validate the accuracy of the transcriptome data, the
top 10 differentially expressed lncRNAs (Table II) were subjected to RT-qPCR in 50
pairs of PTC and paracancerous tissues, and the results revealed
the same trend as the transcriptome sequencing results (all
P<0.05) (Fig. 1B). Among these
lncRNAs, SNHG7 has been demonstrated to increase tumor growth in
numerous cancer types (25,26). According to the bioinformation
analysis software UALCAN, SNHG7 expression was higher in PTC
patients (P<0.05) (Fig. 1C).
Moreover, it was determined that high SNHG7 expression was
positively associated with 131I resistance in PTC
patients (P<0.05) (Fig. 1D). SNHG7
expression revealed diagnostic potential in the prediction of
131I resistance in PTC patients according to the ROC
curve and AUC of SNHG7 (area under the ROC curve: 0.708;
sensitivity: 66.7% and specificity: 75.9%) (P<0.05) (Fig. 1E).
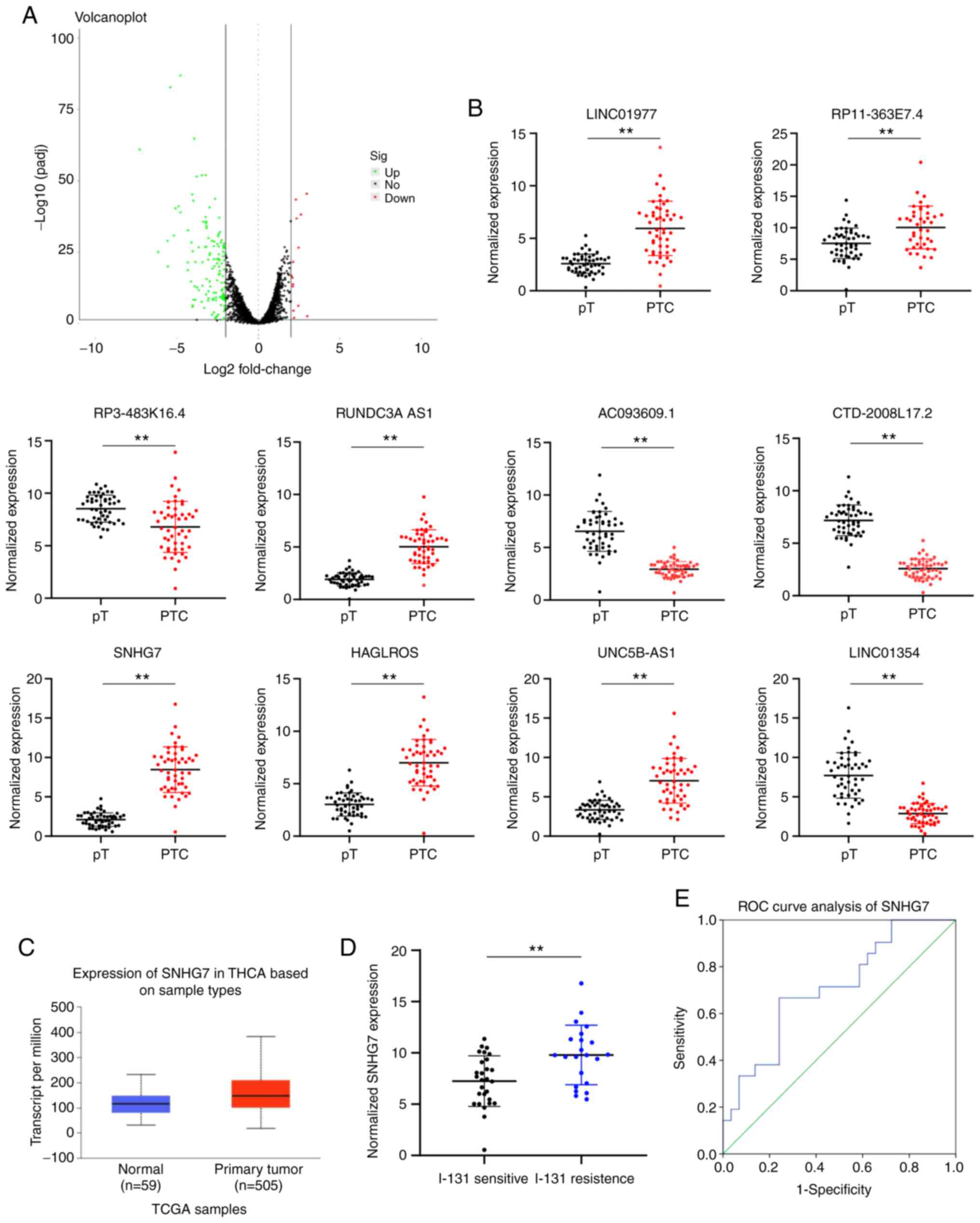 | Figure 1.SNHG7 is highly expressed in PTC
patients and correlated with 131I resistance. (A)
Volcano plot of differentially expressed lncRNAs in PTC and
paracancerous tissues, in which the green dots indicate upregulated
lncRNAs while the red dots indicate downregulated lncRNAs, and the
black dots represent lncRNAs with expression of |log2 FC| <2.
(B) Top 10 differentially expressed lncRNAs in PTC and
paracancerous tissues identified using RT-qPCR. (C) SNHG7
expression in PTC patients and normal people evaluated via the
bioinformatics software UALCAN. (D) The SNHG7 level in responders
(131I-sensitive patients) and nonresponders
(131I-resistant patients) measured using RT-qPCR. (E)
ROC analysis of SNHG7 levels (P<0.05, area under the ROC curve:
0.708; sensitivity: 66.7% and specificity: 75.9%). In B, C and D,
data were analyzed using unpaired t-tests. **P<0.01. SNHG7,
small nucleolar RNA host gene 7; PTC, thyroid cancer;
131I, radioactive iodine; lncRNA, long noncoding RNA;
RT-qPCR, reverse transcription quantitative polymerase chain
reaction; ROC, receiver operator characteristic. |
 | Table II.Characteristics of the top 10
lncRNAs. |
Table II.
Characteristics of the top 10
lncRNAs.
| Ensemble | Gene | Dysregulation | Fold change | P-value |
|---|
|
ENSG00000262772.1 | SNHG7 | Up | 15.19 |
1.15×10−68 |
|
ENSG00000260912.1 | LINC01977 | Up | 4.03 |
3.75×10−27 |
|
ENSG00000271367.1 | RP11′363E7.4 | Up | 7.86 |
5.67×10−49 |
|
ENSG00000233542.1 | RP3′483K16.4 | Down | 11.02 |
1.55×10−55 |
|
ENSG00000267750.4 | RUNDC3A'AS1 | Up | 4.38 |
5.34×10−30 |
|
ENSG00000230587.1 | AC093609.1 | Down | 4.96 |
9.37×10−47 |
|
ENSG00000206129.3 | CTD-2008L17.2 | Down | 6.15 |
2.35×10−41 |
|
ENSG00000226363.3 | HAGLROS | Up | 14.37 |
4.78×10−24 |
|
ENSG00000237512.5 | UNC5B'AS1 | Up | 6.06 |
1.12×10−33 |
|
ENSG00000231768.1 | LINC01354 | Down | 4.07 |
4.87×10−19 |
131I treatment has a poor
effect on 131I-resistant PTC cell lines
To further confirm the role of SNHG7 in the
131I resistance and growth of PTC cells, the
131I-resistant cell lines RAI-res-TPC-1 and
RAI-res-B-CPAP were constructed by treating the WT PTC cell lines
with a median lethal dose of 131I for a total of 8
generations of cells, and the results revealed that the
IC50 of 131I presented a generation-dependent
uptrend. In detail, the IC50 of 131I in the
first generation of TPC-1 cells was 1.0 mCi/well, while it was 1.9
mCi/well in the 8th generation of RAI-res-TPC-1 cells. The
IC50 of 131I in the first generation of
B-CPAP cells was 0.45 mCi/well, while it was 1.05 mCi/well in the
8th generation of RAI-res-B-CPAP cells (all P<0.05) (Fig. 2A and B). It can be observed from the
aforementioned data that the 131I resistance of TPC-1
cells was lower than that of B-CPAP cells. Then, SNHG7 expression
was detected in TPC-1 cells and B-CPAP cells and it was revealed
that SNHG7 expression in TPC-1 cells was higher than that in B-CPAP
cells (Fig. 2C). The MTT assay, flow
cytometry and western blot analysis demonstrated that the
131I-resistant cell lines presented higher cell
viabilities, less apoptosis (reduced Bax level and increased Bcl-2
level) and higher SNHG7 expression than the parent cells (all
P<0.05) (Fig. 2D-H).
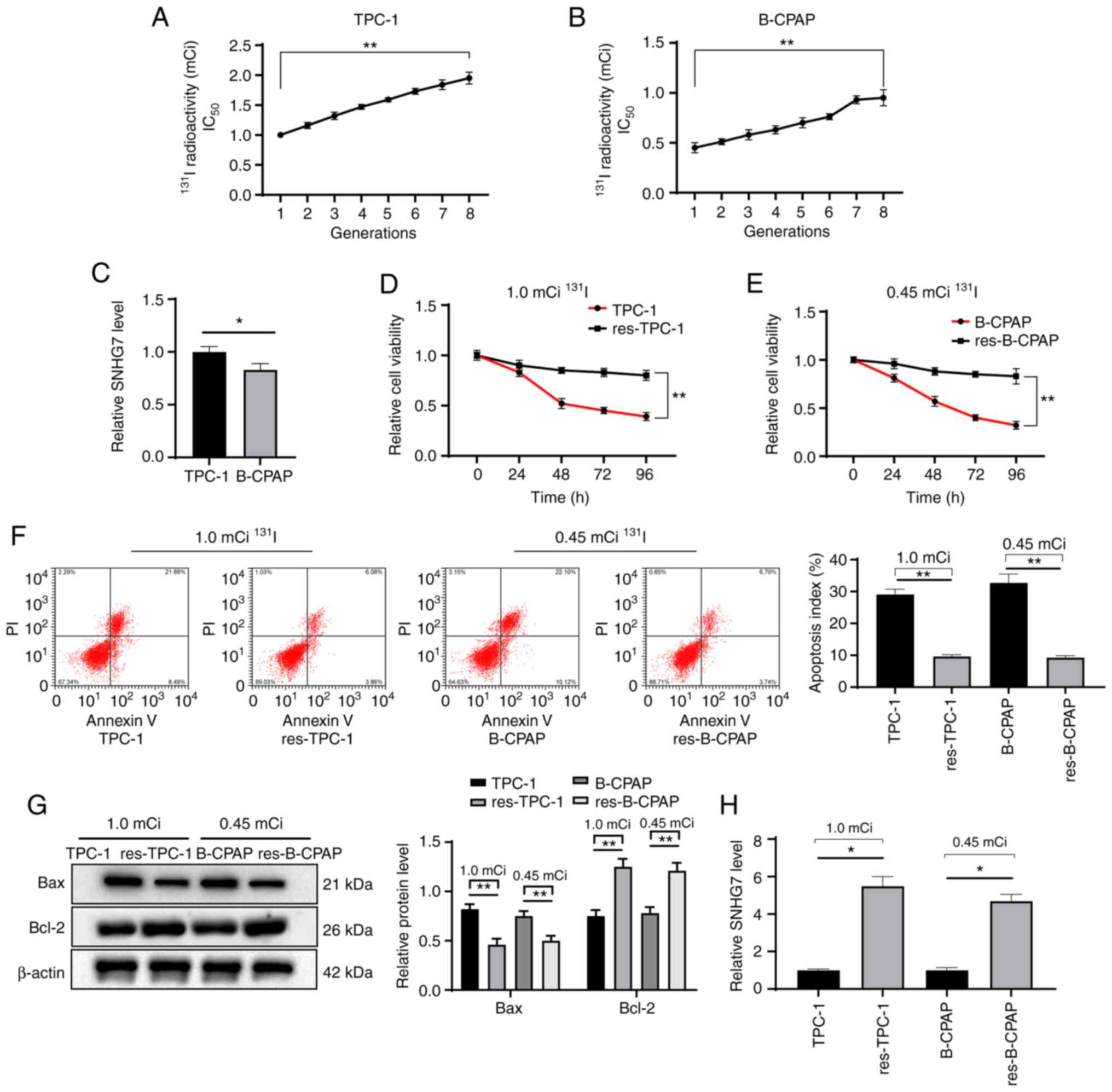 | Figure 2.131I treatment has a poor
effect on 131I-resistant PTC cell lines. (A and B)
Continuous treatment with the median-lethal dose of 131I
in (A) TPC-1 and (B) B-CPAP cell lines. The 1st generation of TPC-1
or B-CPAP was set as the normal cell line, while the 8th generation
was the (A) RAI-res-TPC-1 cell line or (B) RAI-res-B-CPAP cell
line. (C) SNHG7 expression in TPC-1 and B-CPAP cells determined
using RT-qPCR. (D and E) The viability of (D) TPC-1 and
RAI-res-TPC-1 cells and (E) B-CPAP and RAI-res-B-CPAP cell lines
after 131I treatment assessed with MTT assays. (F and G)
Apoptosis of TPC-1, RAI-res-TPC-1, B-CPAP and RAI-res-B-CPAP cell
lines treated with 131I assessed with (F) flow cytometry
and (G) western blot analysis. (H) SNHG7 expression in TPC-1 and
B-CPAP cells determined using RT-qPCR. Replicates=3, data were
calculated from one representative experiment and expressed as the
means ± standard deviation; in A and B, data were analyzed using
one-way ANOVA; in D, E, F, G and H, data were analyzed using
two-way ANOVA, and data in C were analyzed by the t-test;
*P<0.05, **P<0.01. 131I, radioactive iodine;
RAI-res, radioactive iodine (131I)-resistant; PTC,
thyroid cancer; MTT,
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide;
RT-qPCR, reverse transcription quantitative polymerase chain
reaction; ANOVA, analysis of variance. |
Downregulated SNHG7 inhibits the
growth and 131I resistance of PTC cells in vitro
To determine the roles of SNHG7 in the growth and
131I resistance of PTC cells, RAI-res-TPC-1 and
RAI-res-B-CPAP cells were treated with si-SNHG7 and the parent
TPC-1 and B-CPAP cells were treated with Oe-SNHG7 (P<0.05)
(Fig. 3A). The results revealed that
SNHG7 overexpression promoted the growth of parent PTC cells,
demonstrated by fewer cells in the G0/G1 phase and more cells in
the S phase (P<0.05) (Fig. 3B-D).
Next, PTC-1 and B-CPAP cells were subjected to 1.0 mCi/well or 0.45
mCi/well 131I treatment. Then, it was revealed that
silencing of SNHG7 enhanced 131I sensitivity in PTC
cells, and overexpressing SNHG7 promoted 131I resistance
in parent PTC cells and reduced cell apoptosis after
131I treatment and increased the formation of colonies
(P<0.05) (Fig. 3E-G).
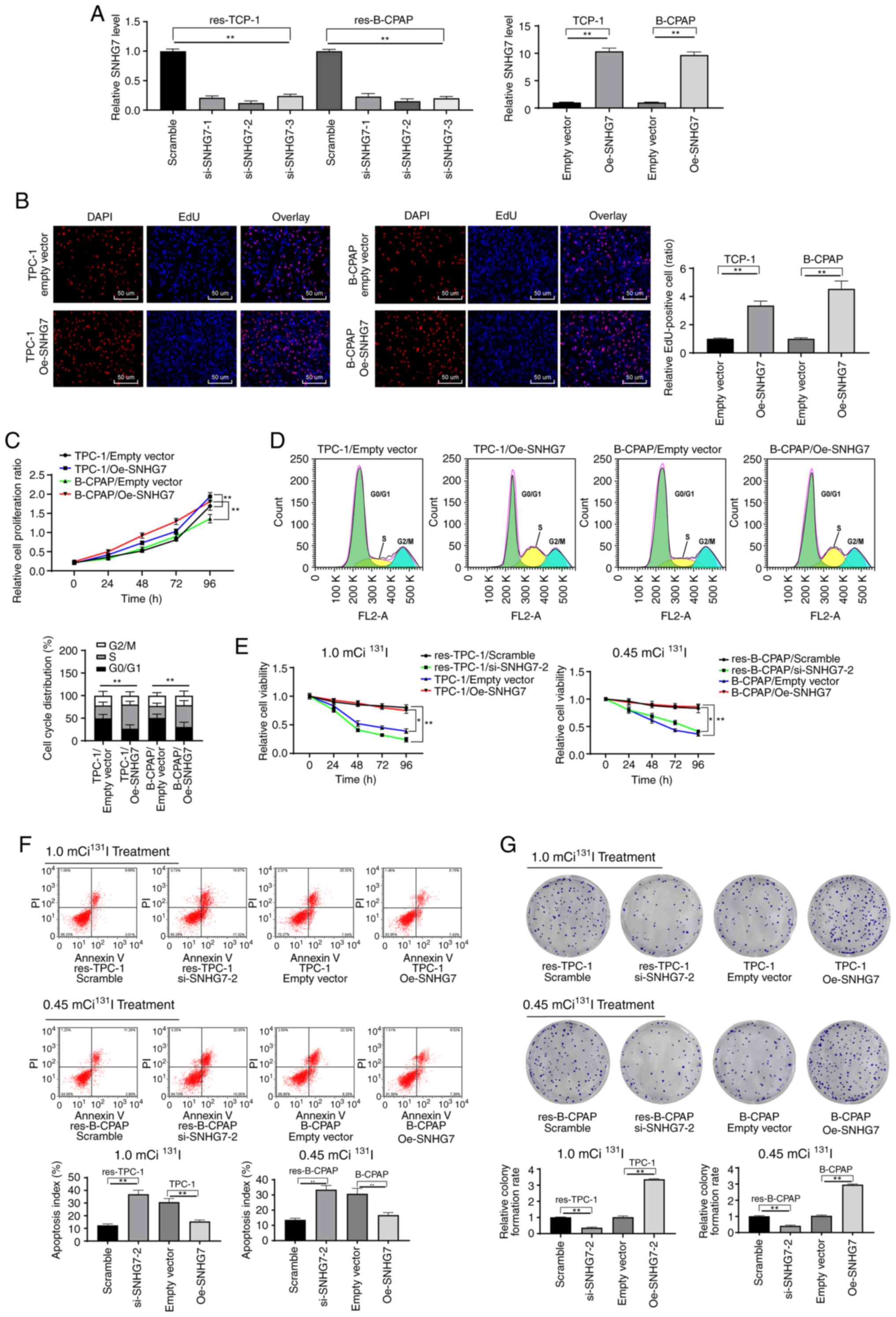 | Figure 3.Downregulated SNHG7 inhibits the
growth and 131I resistance of PTC cells. Three siRNAs
targeting SNHG7 were transfected into RAI-res-TPC-1 and
RAI-res-B-CPAP cells (si-SNHG7-1, si-SNHG7-2 and si-SNHG7-3), and
an expression vector containing SNHG7 was transfected into parent
TPC-1 and B-CPAP cells (Oe-SNHG7 group), with scramble siRNA (mock
group) and empty vector (empty vector group) as the negative
controls. (A) siRNA and expression vector transfection efficiency
validated using RT-qPCR. (B and C) Growth, proliferation and the
cell cycle of parent TPC-1 and B-CPAP cells transfected with
Oe-SNHG7 detected using (B) EdU, (C) MTT and (D) cell cycle assays.
(E) Proliferation of 131I-sensitive and
131I-resistant TPC-1 and B-CPAP cell lines after 96 h of
131I treatment assessed using MTT assays. (F) Apoptosis
and (G) colony formation of TPC-1, RAI-res-TPC-1, B-CPAP and
RAI-res-B-CPAP cell lines treated with 131I for 12 h
measured by flow cytometry. TPC-1 cells were treated with 1.0 mCi
131I, and B-CPA cells were treated with 0.45 mCi
131I. All experiments were repeated three times, and
one-way ANOVA was used to determine statistical significance in A,
B, F and G; two-way ANOVA was used to determine statistical
significance in C, D and E; n=50, *P<0.05, **P<0.01; SNHG7,
small nucleolar RNA host gene 7; PTC, thyroid cancer;
131I, radioactive iodine; RAI-res, radioactive iodine
(131I)-resistant; Oe, overexpression; si, small
interfering; EdU, 5-ethynyl-2′-deoxyuridine; MTT,
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide;
ANOVA, analysis of variance. |
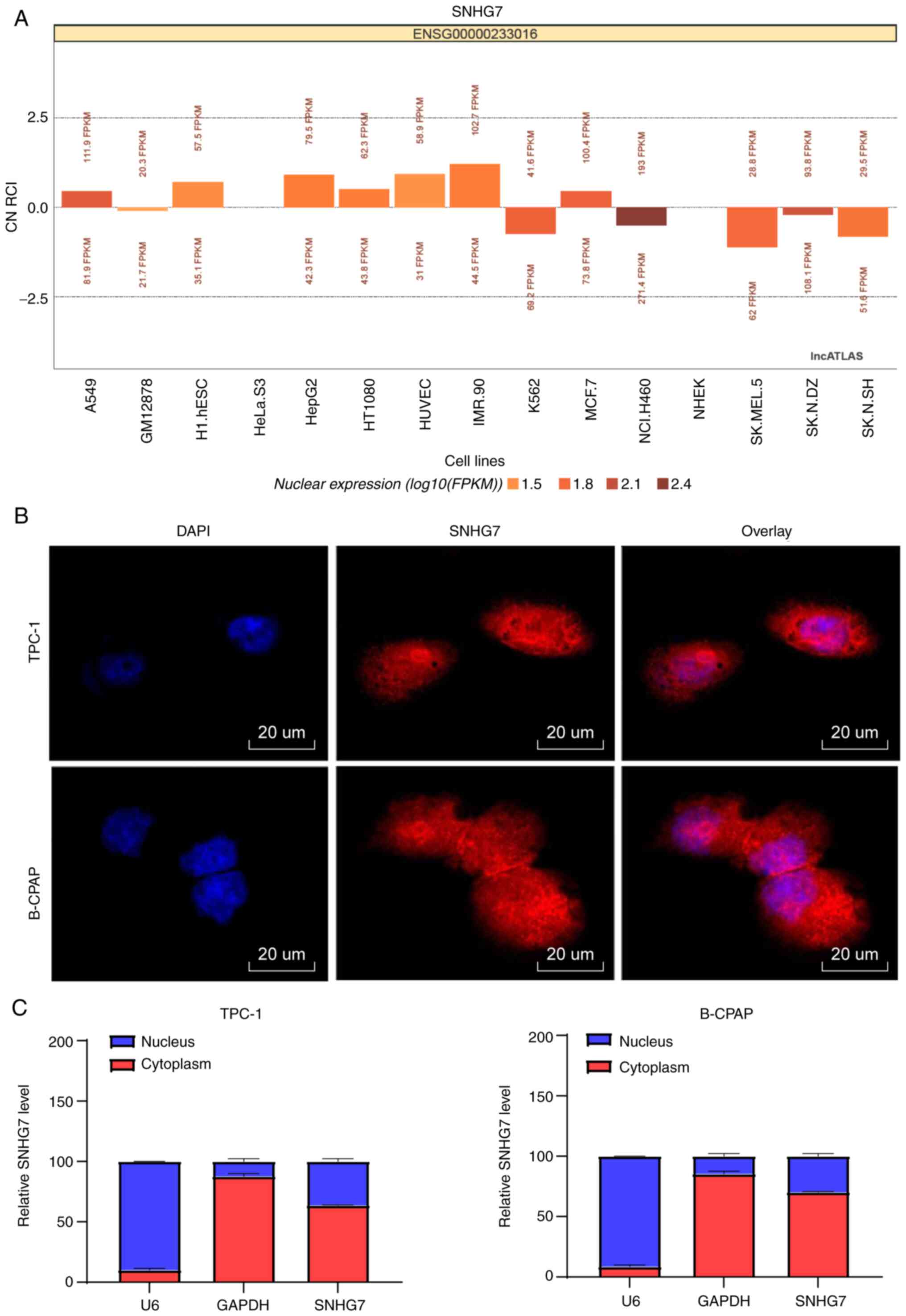
SNHG7 is mainly located in the
cytoplasm
According to the LncATLAS database, SNHG7 was
predicted to mainly localize in the cytoplasm (Fig. 4A). Next, FISH results further
demonstrated that SNHG7 was mainly located in the cytoplasm of
TPC-1 and B-CPAP cells, since the cytoplasm was stained red by
probes targeting SNHG7 and the nucleus was stained blue by DAPI
(Fig. 4B). Moreover, total nuclear
and cytoplasmic RNA in TPC-1 and B-CPAP cells was separated, and
SNHG7 expression in the cytoplasm was higher than that in the
nucleus (P<0.05) (Fig. 4C).
SNHG7 functions as a ceRNA of miR-9-5p
to upregulate DPP4 expression
The radioresistance-promoting effect of SNHG7 on PTC
cells prompted us to determine the underlying mechanism. First, the
miRs that could bind to SNHG7 were predicted via starBase. Among
them, miR-9-5p has been documented to reverse cell resistance to
multiple chemotherapeutics in chronic myelogenous leukemia
(27) and to promote the sensitivity
of glioma to chemotherapy (28). We
hypothesized that SNHG7 may bind to miR-9-5p to reduce the
131I sensitivity of PTC cells. To test this hypothesis,
a CMV-based luciferase reporter plasmid containing the binding site
of the miR-9-5p mimic or miR-negative control and also SNHG7-WT and
SNHG7-MUT were designed in accordance with the starBase prediction,
and it was determined that miR-9-5p directly bound to SNHG7
(Fig. 5A-B). This binding
relationship was further confirmed by the RNA pull-down assay (all
P<0.05) (Fig. 5C). In addition,
RT-qPCR revealed a negative correlation between miR-9-5p and SNHG7
expression in 50 PTC patients (Fig.
5D), and miR-9-5p expression in PTC cells treated with Oe-SNHG7
was further inhibited, while that in cells treated with si-SNHG7
was enhanced (all P<0.05) (Fig.
5E).
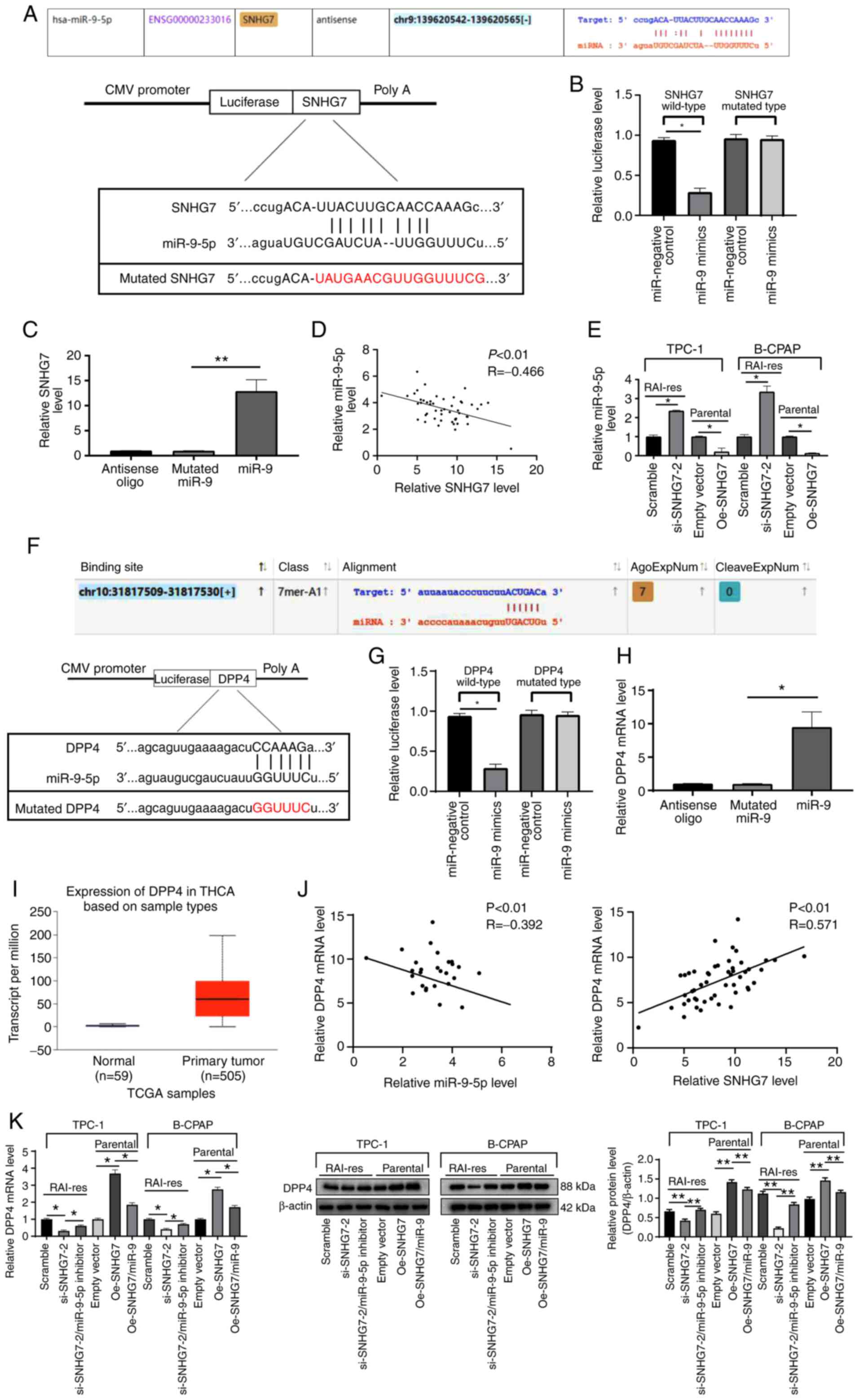 | Figure 5.SNHG7 functions as a ceRNA of
miR-9-5p to upregulate DPP4 expression. (A) The SNHG7 transcript
contains putative miRNA recognition sites complementary to
miR-9-5p. (B and C) Target relationship between SNHG7 and miR-9-5p
identified using (B) dual luciferase reporter and (C) RNA pull-down
assays. (D) Plot analysis of SNHG7 and miR-9-5p expression in 50
PTC patients. (E) miR-9-5p expression after Oe-SNHG7or si-SNHG7
treatment measured using RT-qPCR. (F) Binding relationship between
miR-9-5p and DPP4 predicted via starBase. (G and H) Target
relationship between miR-9-5p and DPP4 identified using (G)
dual-luciferase reporter and (H) RNA pull-down assays. (I) SNHG7
expression in PTC and paracancerous tissues evaluated on UACAN. (J)
Plot analysis of miR-9-5p and DPP4 expression in 50 PTC patients.
(K) Relative expression of DPP4 mRNA and protein levels after
Oe-SNHG7 or si-SNHG7 treatment measured using RT-qPCR and western
blot analysis. si-SNHG7-treated RAI-res-TCP-1 and RAI-res-B-CPAP
cells were transfected with miR-9-5p inhibitor, while
Oe-SNHG7-treated parental TPC-1 and B-CPAP cells were transfected
with miR-9-5p mimic. miR-9-5p expression was normalized to U6
expression, while DPP4 mRNA expression was normalized to GAPDH
expression. In B, C, E, G, H and K, each experiment was repeated
three times, and one-way ANOVA and Tukey's multiple comparison test
were used to determine statistical significance, while in D and J,
Pearson's correlation coefficient test was utilized; n=50.
*P<0.05, **P<0.01. SNHG7, small nucleolar RNA host gene 7;
ceRNA, competing endogenous RNA; miRNA, microRNA; DPP4,
dipeptidyl-peptidase 4; PTC, thyroid cancer; Oe, overexpression;
si, small interfering; RT-qPCR, reverse transcription quantitative
polymerase chain reaction; GAPDH, glyceraldehyde-3-phosphate
dehydrogenase; ANOVA, analysis of variance; miR-9,
microRNA-9-5p. |
Following the result that SNHG7 could target
miR-9-5p, we further predicted the target genes of miR-9-5p through
starBase and TargetScan. Among the target genes, DPP4 has been
documented to increase the growth and metastasis of PTC cells
(15). starBase prediction revealed
that miR-9-5p could bind to a specific site in the 3′-UTR of DPP4
(P<0.05) (Fig. 5F). Similarly, the
dual luciferase reporter assay and RNA pull-down assay were
performed, and the results further demonstrated that miR-9-5p could
directly bind to DPP4 (all P<0.05) (Fig. 5G and H). The online bioinformatic
analysis website UACAN (http://ualcan.path.uab.edu/index.html) (19) indicated that DPP4 expression was
higher in PTC patients than in healthy individuals (P<0.05)
(Fig. 5I). Moreover, the mRNA
expression of DPP4 in PTC patients was negatively associated with
miR-9-5p expression and positively associated with SNHG7 (all
P<0.05) (Fig. 5J). Oe-SNHG7 (or
si-SNHG7) led to enhanced DPP4 levels (or decreased), respectively,
and miR-9-5p inhibitor reversed the inhibition of si-SNHG7 on DPP4
expression (all P<0.05) (Fig. 5K).
The transfection effect of miR-9-5p inhibitor is presented in
Fig. S1.
Overexpressed miR-9-5p reduces
SNHG7-induced PTC cell proliferation and 131I
resistance
To further identify the roles of miR-9 in the growth
and 131I resistance of PTC cells, miR-9-5p mimic was
transfected into parental TPC-1 and B-CPAP cells and into
RAI-res-TPC-1 and RAI-res-TPC cells, and cotransfected miR-9-5p
mimic and Oe-SNHG7 into parental TPC-1 and B-CPAP cells, and the
transfection effect was evaluated using RT-qPCR (P<0.05)
(Fig. 6A). The results revealed that
overexpression of miR-9-5p inhibited TPC-1 and B-CPAP cell
proliferation and reduced the Oe-SNHG7-induced 131I
resistance of TPC-1 and B-CPAP cells (all P<0.05) (Fig. 6B-F).
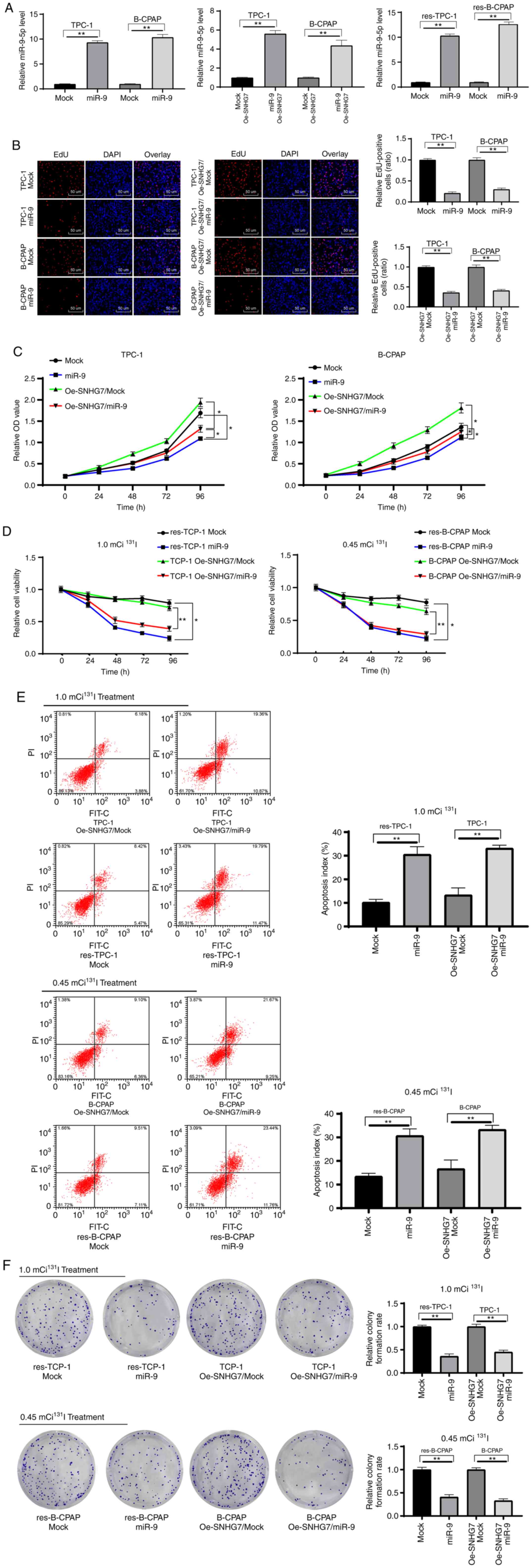 | Figure 6.Overexpression of miR-9-5p reduces
SNHG7-induced PTC cell proliferation and 131I
resistance. Parental TPC-1 and B-CPAP cells, RAI-res-TPC-1 and
RAI-res-TPC cells, and Oe-SNHG7-treated parental TPC-1 and B-CPAP
cells were transfected with miR-9-5p mimic, and miR-negative
control mimic (mock group) was used as the negative control. (A)
Transfection efficiency of miR-9-5p evaluated using RT-qPCR. (B and
C) The effects of Oe-SNHG7 on parental TPC-1 and B-CPAP cells
measured using (B) EdU assay and (C) MTT assay. (D) Proliferation
of TPC-1 and B-CPAP cell lines treated for 96 h with
131I (1.0 mCi/well) and 131I (0.45 mCi/well),
respectively. (E) Apoptosis and (F) colony formation assays for
TPC-1, B-CPAP, RAI-res-TPC-1 and RAI-res-B-CPAP cells subjected to
12 h of 131I treatment measured by flow cytometry. TPC-1
cells were treated with 1.0 mCi 131I, and B-CPAP cells
were treated with 0.45 mCi 131I. Replicates=3; one-way
ANOVA and Tukey's multiple comparison test were used for data
analysis. *P<0.05, **P<0.01. SNHG7, small nucleolar RNA host
gene 7; 131I, radioactive iodine; RAI-res, radioactive
iodine (131I)-resistant; RT-qPCR, transcription
quantitative polymerase chain reaction; Oe, overexpression; EdU,
5-ethynyl-2′-deoxyuridine; MTT,
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide;
ANOVA, analysis of variance; miR-9, microRNA-9-5p. |
SNHG7/miR-9-5p/DPP4 ceRNA network
promotes PI3K/Akt activation
Liu et al reported that inhibition of the
PI3K/Akt signaling pathway induced by silencing SNHG7 can reduce
131I resistance of PTC cells (5). Thus, PI3K/Akt-related protein levels in
TPC-1 and B-CPAP cells after SNHG7 interference were assessed. In
TPC-1 and B-CPAP cells, compared with the empty vector group, after
overexpression of SNHG7, the PI3K/Akt signaling pathway was
activated, while overexpression of miR-9-5p reversed these results.
Compared with miR-9-5p overexpression alone, overexpression of
miR-9-5p and DPP4 activated the PI3K/Akt signaling pathway. In
res-TPC-1 and res-B-CPAP cells, the PI3K/Akt signaling pathway was
inhibited after SNHG7 expression was inhibited compared with the
Scramble group, while the results were reversed after miR-9-5p
overexpression. Compared with the inhibition of miR-9-5p expression
alone, inhibition of miR-9-5p and DPP4 blocked the PI3K/Akt
signaling pathway (all P<0.01) (Fig.
7).
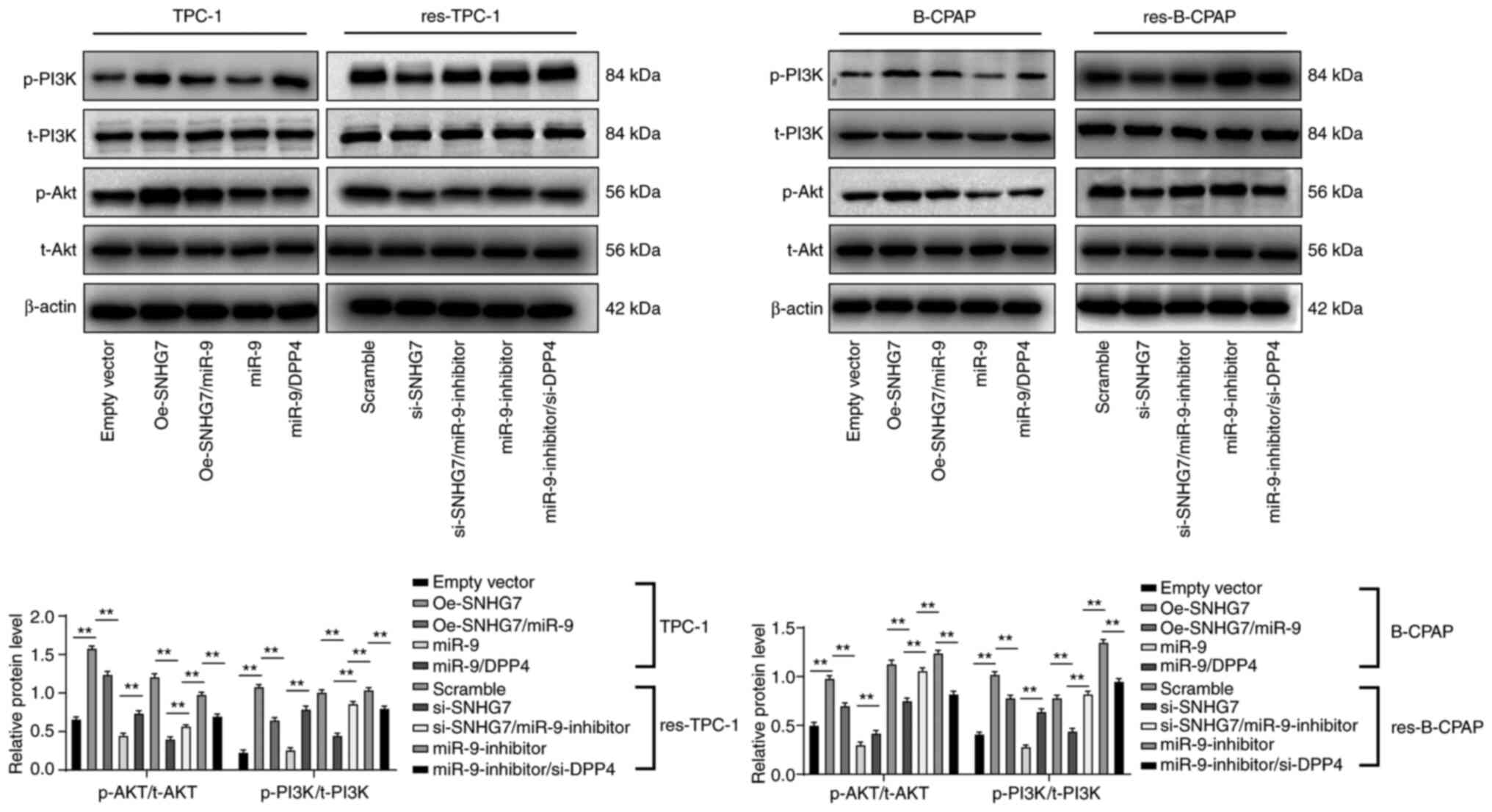 | Figure 7.SNHG7 promotes PTC cell resistance
and proliferation by activating the PI3K/Akt signaling pathway.
TPC-1/B-CPAP cells were transfected with Oe-SNHG7, miR-9-5p mimic,
Oe-SNHG7 + miR-9-5p mimic (Oe-SNHG7/miR-9), or miR-9-5p mimic +
pcDNA-DPP4 (miR-9/DPP4); while res-TPC-1/B-CPAP cells were
transfected with si-SNHG7, si-SNHG7 + miR-9-inhibitor,
miR-9-inhibitor, or miR-9-inhibitor + si-DPP4. Scramble siRNA (mock
group) and empty vector (empty vector group) served as negative
controls. Western blot analysis was performed to determine the
activation of the PI3K/Akt signaling pathway in TPC-1 and B-CPAP
cells. For PI3K activation, the phosphorylation site was Y-607,
while the Akt phosphorylation site was T-308. Replicates=3; two-way
ANOVA and Tukey's multiple comparison test were used for data
analysis. **P<0.01. SNHG7, small nucleolar RNA host gene 7;
siRNA, small interfering RNA; PI3K/Akt, phosphatidyl inositol
3-kinase/protein kinase B; 131I, radioactive iodine;
RAI-res, radioactive iodine (131I)-resistant; miR-9,
microRNA-9-5p. |
Downregulated SNHG7 inhibits the
growth and 131I resistance of PTC cells in vivo
To further investigate the roles of SNHG7 in PTC
cell growth in vivo, xenograft tumors in nude mice were
generated from Oe-SNHG7-treated or miR-9 + Oe-SNHG7-cotreated TPC-1
cells and si-SNHG7-treated RAI-res-TPC-1 cells. No nude mice died
in any group during the experiment. The results revealed that
Oe-SNHG7 promoted TPC-1 growth in vivo and 131I
resistance in TPC-1 cells, while si-SNHG7 led to markedly reduced
tumor volume and weight (all P<0.05) (Fig. 8A). Moreover, the immunohistochemical
results indicated that silencing of SNHG7 led to significantly
reduced levels of pAkt, DPP4 and Ki67 (all P<0.05) (Fig. 8B).
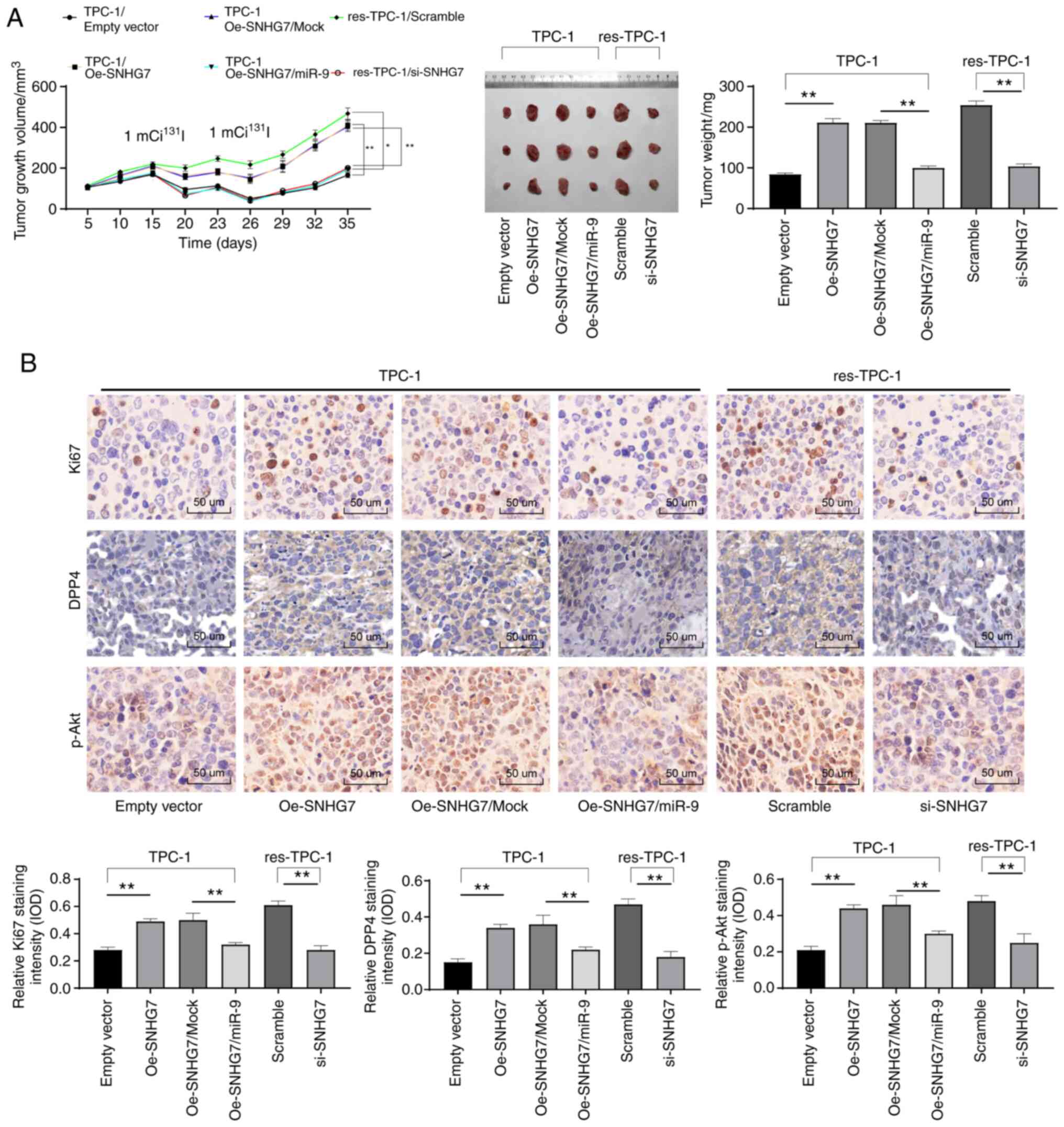 | Figure 8.Downregulated SNHG7 inhibits the
growth and 131I resistance of PTC cells in vivo.
TPC-1 cells transfected with Oe-SNHG7 or Oe + miR-9 or scramble
siRNA or RAI-res-TPC-1 cells transfected with si-SNHG7 were
transfected into BALB/c nude mice (n=6 in each group). From the
first day to the 20th day, tumor growth was measured every 5 days,
and 20 days later, tumor growth was monitored every 3 days.
131I treatments were conducted with 2.0 mCi/100 g on
days 15 and 25 after cell injection. (A) Measurement of tumor
volumes and weights in each group of nude mice; tumor sections were
collected and stained with anti-DPP4, anti-pAkt and anti-Ki67
antibodies. (B) Relative levels of DPP4, pAkt and Ki67 in each
group of tumor cells identified using immunohistochemistry. In A,
two-way analysis was used, while in B, one-way analysis was used
for data analysis. *P<0.05, **P<0.01. SNHG7, small nucleolar
RNA host gene 7; PTC, thyroid cancer; Oe, overexpressed; siRNA,
small interfering RNA; 131I, radioactive iodine;
RAI-res, radioactive iodine (131I)-resistant; DPP4,
dipeptidyl-peptidase 4, pAkt, phosphorylated-protein kinase B;
ANOVA, analysis of variance; miR-9, microRNA-9-5p. |
Discussion
131I remains a major therapeutic option
for differentiated thyroid carcinoma, including PTC, however a
great number of PTC patients have reduced iodine uptake thus
leading to RAI resistance, which is the main obstacle in PTC
treatment and a major cause of poor prognosis (29). lncRNA/miR ceRNA networks have recently
gained attention for their roles in regulating gene expression and
radiotherapy sensitivity (8,30). The present study was performed to
determine the roles of SNHG7 and miR-9-5p in the growth and
131I resistance of PTC cells. The results demonstrated
that SNHG7 could serve as a sponge for miR-9-5p to enhance DPP4
expression, and this ceRNA network could induce the growth and
131I resistance of PTC cells via PI3K/Akt
activation.
The initial finding of the study was that SNHG7 was
aberrantly highly expressed in PTC cells, and overexpressing SNHG7
promoted PTC cell proliferation, while silencing of SNHG7 enhanced
the radiosensitivity of 131I-resistant PTC cells. High
SNHG7 expression has been revealed in several tumor types, such as
nasopharyngeal carcinoma (31),
hepatocellular carcinoma (32) and,
importantly, thyroid carcinoma (33),
with knockdown of SNHG7 leading to inhibited proliferation and
metastasis of cancer cells. In addition, downregulation of SNHG7
has been documented to reduce cisplatin resistance in NSCLC cells
via PI3K/Akt inhibition (34).
Similarly, high expression of SNHG7 has been suggested to be
related to an advanced hypopharyngeal cancer stage and higher drug
resistance (35). In the present
study, it was reported that SNHG7 could also lead to
radioresistance in PTC cells.
The promoting role of SNHG7 in PTC cell growth
prompted us to further identify the molecules involved in this
event. First, we determined the sublocalization of SNHG7 and
determined that it was mainly located in the cytoplasm in PTC
cells, which provided insights about the potential factors that
were involved in SNHG7-induced cell growth. Notably, the present
study revealed that SNHG7 could directly bind with miR-9-5p, which
was consistent with a previous study (14). In addition, overexpression of miR-9-5p
reversed SNHG7-induced growth and radioresistance of PTC cells in
the present study. miR-9-5p has been well documented as a tumor
inhibitor in several cancer types, such as human gastric cancer
(36) and pancreatic cancer (11), and downregulation of miR-9-5p affected
by ceRNA networks has been reported to lead to the pathogenesis of
numerous cancers (36,37). Notably, miR-9-5p has also been
identified as an inhibitor of PTC, whose downregulation led to
enhanced proliferation and resistance to apoptosis of PTC cells
(13). In addition, miR-9 has been
suggested to enhance the sensitivity of glioma to chemotherapy
(28) and to markedly improve the
therapeutic efficiency of radiotherapy in NSCLC cells (38). Moreover, the present study further
determined that miR-9-5p negatively targeted DPP4, and accordingly,
DPP4 expression was positively correlated with SNHG7 expression in
PTC patients. DPP4 serves as a tumor biomarker since it is
inversely associated with survival in most cancer types (39). Furthermore, it has been documented
that upregulation of DPP4 is involved in the growth and development
of PTC cells (15). Herein, it was
demonstrated that SNHG7 promoted the growth and 131I
resistance of PTC cells through the SNHG7/miR-9-5p/DPP4 ceRNA
network. In addition, the present study determined that this
network promoted PI3K/Akt axis activation. As aforementioned, SNHG7
has been demonstrated to lead to cisplatin resistance through
PI3K/Akt activation (34), and a
similar trend has also been found in colorectal cancer (40). Moreover, it has also been revealed
that overexpression of miR-9-5p inhibits the PI3K/Akt signaling
pathway (41). Notably, as
aforementioned, PI3K/Akt activation has been implicated in
NEAT1-induced 131I resistance in PTC cells (5). Similarly, inhibition of PI3K/Akt
signaling has been documented to enhance the radiosensitivity of
glioma cells (42). In the present
study, it was concluded that the SNHG7/miR-9-5p/DPP4 network could
promote the growth and 131I resistance in PTC cells via
PI3K/Akt activation.
In summary, the present study demonstrated that
SNHG7 could act as a sponge for miR-9-5p to enhance DPP4 expression
and further lead to PI3K/Akt activation, thus promoting the growth
and radioresistance of PTC cells. However, this research is still
at the preclinical stage, and the mechanism of action is not
totally elucidated. Thus, more studies in this area are required to
validate our findings and to develop clinical strategies or novel
therapeutic options for TC treatment.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Natural
Science Foundation of China (grant no. 81760320).
Availability of data and materials
All the data generated or analyzed during this study
are included in this published article.
Authors' contributions
WC is the guarantor of integrity of the entire study
and contributed to the concepts of this study. JY and MZ
contributed to the experimental studies and design of this study.
RX and TZ contributed to the data and statistical analysis. SZ
performed the clinical studies. CX drafted the manuscript and
performed the experiments. WC contributed to the critical review of
the manuscript for important intellectual content. All authors read
and approved the final manuscript.
Ethical approval and consent to
participate
The present study was approved and supervised by the
Clinical Ethics Committee of the Second Affiliated Hospital of
Nanchang University. Signed informed consent was obtained from each
eligible participant. Animal studies were conducted according to
the principles and procedures approved by the Committee on the
Ethics of Animal Experiments of Second Affiliated Hospital of
Nanchang University. All experimental procedures were conducted in
line with the ethical guidelines for the study of experimental pain
in conscious animals.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ding C, Yu H, Shi C, Shi T, Qin H and Cui
Y: MiR-let-7e inhibits invasion and magration and regulates HMGB1
expression in thyroid cancer. Biomed Pharmacother. 110:528–536.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li H, Zhao L, Zhang Z, Zhang H, Ding C and
Su Z: Roles of microRNA let-7b in thyroid cancer by regulating
HMGA2. Tumour Biol. 39:10104283177192742017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Huang Y, Yu S, Cao S, Yin Y, Hong S, Guan
H, Li Y and Xiao H: MicroRNA-222 promotes invasion and metastasis
of papillary thyroid cancer through targeting protein phosphatase 2
regulatory subunit b alpha expression. Thyroid. 28:1162–1173. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gezer E, Selek A, Tarkun I, Canturk Z and
Çetinarslan B: Papillary thyroid cancer presenting as a primary
renal tumor with multiple pulmonary and bone metastases: A case
report. J Med Case Rep. 13:952019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liu C, Feng Z, Chen T, Lv J, Liu P, Jia L,
Zhu J, Chen F, Yang C and Deng Z: Downregulation of NEAT1 reverses
the radioactive iodine resistance of thyroid cancer cell via
miR-101-3p/FN1/PI3K-AKT signaling pathway. Cell Cycle. 18:167–203.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Xiang C, Zhang ML, Zhao QZ, Xie QP, Yan
HC, Yu X, Wang P and Wang Y: lncRNA-SLC6A9-5:2: A potent sensitizer
in 131I-resistant thyroid cancer with PARP-1 induction.
Oncotarget. 8:22954–22967. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wan ZY, Song F, Sun Z, Chen YF, Zhang WL,
Samartzis D, Ma CJ, Che L, Liu X, Ali MA, et al: Aberrantly
expressed long noncoding RNAs in human intervertebral disc
degeneration: A microarray related study. Arthritis Res Ther.
16:4652014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen Y, Shen Z, Zhi Y, Zhou H, Zhang K,
Wang T, Feng B, Chen Y, Song H, Wang R and Chu X: Long non-coding
RNA ROR promotes radioresistance in hepatocelluar carcinoma cells
by acting as a ceRNA for microRNA-145 to regulate RAD18 expression.
Arch Biochem Biophys. 645:117–125. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gao YT and Zhou YC: Long non-coding RNA
(lncRNA) small nucleolar RNA host gene 7 (SNHG7) promotes breast
cancer progression by sponging miRNA-381. Eur Rev Med Pharmacol
Sci. 23:6588–6595. 2019.PubMed/NCBI
|
|
10
|
Yao X, Liu C, Liu C, Xi W, Sun S and Gao
Z: lncRNA SNHG7 sponges miR-425 to promote proliferation,
migration, and invasion of hepatic carcinoma cells via
Wnt/β-catenin/EMT signalling pathway. Cell Biochem Funct.
37:525–533. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang J, Wang B, Ren H and Chen W: miR-9-5p
inhibits pancreatic cancer cell proliferation, invasion and
glutamine metabolism by targeting GOT1. Biochem Biophys Res Commun.
509:241–248. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shi D, Wang H, Ding M, Yang M, Li C, Yang
W and Chen L: MicroRNA-26a-5p inhibits proliferation, invasion and
metastasis by repressing the expression of Wnt5a in thyroid cancer.
Onco Targets Ther. 12:6605–6616. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Guo F, Hou X and Sun Q: MicroRNA-9-5p
functions as a tumor suppressor in papillary thyroid cancer via
targeting BRAF. Oncol Lett. 16:6815–6821. 2018.PubMed/NCBI
|
|
14
|
Chen Z, Liu Z, Shen L and Jiang H: Long
non-coding RNA SNHG7 promotes the fracture repair through negative
modulation of miR-9. Am J Transl Res. 11:974–982. 2019.PubMed/NCBI
|
|
15
|
Wang Y, Han J, Lv Y and Zhang G: miR-29a
inhibits proliferation, invasion, and migration of papillary
thyroid cancer by targeting DPP4. Onco Targets Ther. 12:4225–4233.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li C, Zhou Y, Cai Y, Shui C, Liu W, Wang
X, Jiang J, Zeng D, Gui C and Sun R: Parthenolide inhibits the
proliferation of MDA-T32 thyroid cancer cells in vitro and in mouse
tumor xenografts and activates autophagy and apoptosis by
downregulation of the mammalian target of rapamycin (mTOR)/PI3K/AKT
Signaling Pathway. Med Sci Monit. 25:5054–5061. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Eisenhauer EA, Therasse P, Bogaerts J,
Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S,
Mooney M, et al: New response evaluation criteria in solid tumours:
Revised RECIST guideline (version 1.1). Eur J Cancer. 45:228–247.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chandrashekar DS, Bashel B, Balasubramanya
SA, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BV and Varambally
S: UALCAN: A portal for facilitating tumor subgroup gene expression
and survival analyses. Neoplasia. 19:649–658. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chehrehasa F, Meedeniya AC, Dwyer P,
Abrahamsen G and Mackay-Sim A: EdU, a new thymidine analogue for
labelling proliferating cells in the nervous system. J Neurosci
Methods. 177:122–130. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mas-Ponte D, Carlevaro-Fita J, Palumbo E,
Hermoso Pulido T, Guigo R and Johnson R: LncATLAS database for
subcellular localization of long noncoding RNAs. RNA. 23:1080–1087.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:e050052015. View Article : Google Scholar
|
|
24
|
Zatroch KK, Knight CG, Reimer JN and Pang
DS: Refinement of intraperitoneal injection of sodium pentobarbital
for euthanasia in laboratory rats (Rattus Norvegicus). BMC Vet Res.
13:602017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Deng Y, Zhao F, Zhang Z, Sun F and Wang M:
Long noncoding RNA SNHG7 promotes the tumor growth and
epithelial-to-mesenchymal transition via regulation of miR-34a
signals in osteosarcoma. Cancer Biother Radiopharm. 33:365–372.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shan Y, Ma J, Pan Y, Hu J, Liu B and Jia
L: lncRNA SNHG7 sponges miR-216b to promote proliferation and liver
metastasis of colorectal cancer through upregulating GALNT1. Cell
Death Dis. 9:7222018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li Y, Zhao L, Li N, Miao Y, Zhou H and Jia
L: miR-9 regulates the multidrug resistance of chronic myelogenous
leukemia by targeting ABCB1. Oncol Rep. 37:2193–2200. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li Q, Chang Y, Mu L and Song Y: MicroRNA-9
enhances chemotherapy sensitivity of glioma to TMZ by suppressing
TOPO II via the NF-κB signaling pathway. Oncol Lett. 17:4819–4826.
2019.PubMed/NCBI
|
|
29
|
Amit M, Na'ara S, Francis D, Matanis W,
Zolotov S, Eisenhaber B, Eisenhaber F, Weiler Sagie M, Malkin L,
Billan S, et al: Post-translational regulation of radioactive
iodine therapy response in thyroid cancer. J Natl Cancer Inst. Dec
1–2017.(Epub ahead of print). doi: 10.1093/jnci/djx092. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hu X, Ding D, Zhang J and Cui J: Knockdown
of lncRNA HOTAIR sensitizes breast cancer cells to ionizing
radiation through activating miR-218. Biosci Rep.
39:BSR201810382019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang L, Xu T, Cui X, Han M, Zhou LH, Wei
ZX, Xu ZJ and Jiang Y: Downregulation of lncRNA SNHG7 inhibits
proliferation and invasion of nasopharyngeal carcinoma cells
through repressing ROCK1. Eur Rev Med Pharmacol Sci. 23:6186–6193.
2019.PubMed/NCBI
|
|
32
|
Sun BZ, Ji DG, Feng ZX and Wang Y: Long
noncoding RNA SNHG7 represses the expression of RBM5 to strengthen
metastasis of hepatocellular carcinoma. Eur Rev Med Pharmacol Sci.
23:5699–5704. 2019.PubMed/NCBI
|
|
33
|
Wang YH, Huo BL, Li C, Ma G and Cao W:
Knockdown of long noncoding RNA SNHG7 inhibits the proliferation
and promotes apoptosis of thyroid cancer cells by downregulating
BDNF. Eur Rev Med Pharmacol Sci. 23:4815–4821. 2019.PubMed/NCBI
|
|
34
|
Chen K, Abuduwufuer A, Zhang H, Luo L,
Suotesiyali M and Zou Y: SNHG7 mediates cisplatin-resistance in
non-small cell lung cancer by activating PI3K/AKT pathway. Eur Rev
Med Pharmacol Sci. 23:6935–6943. 2019.PubMed/NCBI
|
|
35
|
Wu P, Tang Y, Fang X, Xie C, Zeng J, Wang
W and Zhao S: Metformin suppresses hypopharyngeal cancer growth by
epigenetically silencing long Non-coding RNA SNHG7 in FaDu Cells.
Front Pharmacol. 10:1432019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Fan Y, Shi Y, Lin Z, Huang X, Li J, Huang
W, Shen D, Zhuang G and Liu W: miR-9-5p suppresses malignant
biological behaviors of human gastric cancer cells by negative
regulation of TNFAIP8L3. Dig Dis Sci. 64:2823–2829. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Xie CH, Cao YM, Huang Y, Shi QW, Guo JH,
Fan ZW, Li JG, Chen BW and Wu BY: Long non-coding RNA TUG1
contributes to tumorigenesis of human osteosarcoma by sponging
miR-9-5p and regulating POU2F1 expression. Tumour Biol.
37:15031–15041. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wei W, Dong Z, Gao H, Zhang YY, Shao LH,
Jin LL, Lv YH, Zhao G, Shen YN and Jin SZ: MicroRNA-9 enhanced
radiosensitivity and its mechanism of DNA methylation in non-small
cell lung cancer. Gene. 710:178–185. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Javidroozi M, Zucker S and Chen WT: Plasma
seprase and DPP4 levels as markers of disease and prognosis in
cancer. Dis Markers. 32:309–320. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li Y, Zeng C, Hu J, Pan Y, Shan Y, Liu B
and Jia L: Long non-coding RNA-SNHG7 acts as a target of miR-34a to
increase GALNT7 level and regulate PI3K/Akt/mTOR pathway in
colorectal cancer progression. J Hematol Oncol. 11:892018.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yi J and Gao ZF: MicroRNA-9-5p promotes
angiogenesis but inhibits apoptosis and inflammation of high
glucose-induced injury in human umbilical vascular endothelial
cells by targeting CXCR4. Int J Biol Macromol. 130:1–9. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Gwak HS, Kim TH, Jo GH, Kim YJ, Kwak HJ,
Kim JH, Yin J, Yoo H, Lee SH and Park JB: Silencing of microRNA-21
confers radio-sensitivity through inhibition of the PI3K/AKT
pathway and enhancing autophagy in malignant glioma cell lines.
PLoS One. 7:e474492012. View Article : Google Scholar : PubMed/NCBI
|