Introduction
Hepatocellular carcinoma (HCC) is a frequently
occurring malignant tumor, with more than half a million new
patients being diagnosed each year worldwide (1). Importantly, owing to high metastasis and
limited treatment options, patients with HCC usually suffer from a
poor prognosis (2). Hepatitis virus
infection may be partially responsible for the occurrence of HCC
(3); however, the exact underlying
molecular mechanism is complex and not entirely clear. Thus,
further study of the pathogenesis of HCC is pivotal for providing
more therapeutic options and improving the survival of patients
with HCC.
Long non-coding RNAs (lncRNAs), which are >200
nucleotides in length, are involved in multiple biological
activities and pathological processes by regulating a range of
genes at the transcriptional or posttranscriptional level (4,5). Recently,
numerous studies have attempted to determine the impact of lncRNAs
in carcinogenesis. Liao et al (6) reported that the lcnRNA LGMN is
significantly upregulated in glioblastoma tissues, and that
increased LGMN expression facilitates glioblastoma growth and
metastasis. lncMat2B was confirmed to promote cisplatin resistance
by facilitating DNA damage repair in breast cancer cells (7). Elevated lncRNA H19 expression was
reported to be associated with the metastasis of colorectal cancer
(CRC) via upregulation of heterogeneous nuclear ribonucleoprotein
A2B1 and Raf-1 expression (8). lncRNA
LOC284454 is a stable chromatin-associated lncRNA and seems to be
differentially regulated in different types of cancer. For example,
LOC284454 expression is markedly downregulated in various types of
human cancer, including prostate, breast and kidney cancer
(9). However, in nasopharyngeal
carcinoma (NPC), oral cancer and thyroid cancer, LOC284454
expression is significantly upregulated (10). A recent study revealed that increased
LOC284454 expression indicates a poor prognosis in NPC and
facilitates NPC cell migration and invasion (11). However, the role of LOC284454 in HCC
has not been investigated.
E-cadherin serves a critical role in regulating
cellular adhesion and represses metastasis of human cancers by
blocking epithelial-mesenchymal transition (EMT), which is
critically responsible for tumor metastasis (12,13). Our
previous study suggested that autophagic degradation of E-cadherin
induces EMT and promotes HCC metastasis (14). Consistent with this finding, Zhou
et al (15) observed that
decreased E-cadherin expression in HCC caused by overexpression of
plant homeodomain finger protein 8 promotes HCC metastasis.
The RNA-binding protein (RBP) enhancer of zeste
homolog 2 (EZH2) acts as a histone inhibitor and significantly
contributes to the regulation of its target genes by influencing
automethylation (16). Previous
studies have revealed that lncRNAs regulate their target genes by
binding with RBPs, including EZH2 (17,18). It
has been reported that CRNDE lncRNA suppresses its target gene
dual-specificity phosphatase 5 by regulating EZH2 expression in CRC
(17). Similarly, Linc00460 inhibits
cell proliferation and induces apoptosis by repressing its target
gene Kruppel-like factor 2 in CRC (18).
The present study aimed to assess the role and
mechanism of LOC284454 in HCC cell migration and proliferation. The
assays were performed to determine whether LOC284454 may serve as a
promising biomarker and novel therapeutic target for patients with
HCC.
Materials and methods
HCC and noncancerous tissue
specimens
The present study complied with the principles of
the Declaration of Helsinki and was approved by the Ethics
Committee of Xi'an Jiaotong University (Xi'an, China). A total of
90 paired human cancer and noncancerous tissue specimens (≥2 cm
away from the tumor margin) were collected from patients who were
histopathologically and clinically diagnosed with HCC and who
underwent surgical excision between January 2013 and January 2018
at the Second Affiliated Hospital of Xi'an Jiaotong University. The
samples were retained in liquid nitrogen after operation. The
clinicopathological characteristics of the patients (median age, 55
years; age range, 30–69 years) are listed in Table I. Tumor stages were determined
according to the 2002 American Joint Committee on
Cancer/International Union against Cancer Tumor/Lymph Node
Metastasis/Distal Metastasis (TNM) classification system (19,20). The
longest follow-up duration of the patients was 90.1 months. The
overall survival (OS) rate of all patients was 25.6% at 5 years.
Receiver operating characteristic curve analysis determined that
the cut-off value of LOC284454 was 3.27. The cohort of patients
with HCC with LOC284454 expression >3.27 were designated as the
LOC284454-high group, and the cohort of patients with LOC284454
expression <3.27 was designated as the LOC284454-low group. A
total of 15 human non-tumor liver samples were collected at the
Second Affiliated Hospital of Xi'an Jiaotong University in October
2020. None of the individuals had HCC or other malignancies
Table SI shows the clinical
characteristics of these individuals (median age, 36 years; age
range, 27–57 years).
 | Table I.Association between LOC284454
expression and clinicopathological features in patients with
hepatocellular carcinoma. |
Table I.
Association between LOC284454
expression and clinicopathological features in patients with
hepatocellular carcinoma.
|
|
| LOC284454, n |
|
|---|
|
|
|
|
|
|---|
| Variable | Total number of
patients, n (%) | Low expression | High
expression | P-value |
|---|
| Age, years |
|
|
| 0.479 |
|
<50 | 37 (41.1) | 15 | 22 |
|
|
≥50 | 53 (58.9) | 20 | 33 |
|
| Sex |
|
|
|
|
|
Female | 13 (14.4) | 4 | 9 | 0.373 |
|
Male | 77 (85.6) | 31 | 46 |
|
| Tumor size, cm |
|
|
| 0.033a |
|
<5 | 35 (38.9) | 9 | 26 |
|
| ≥5 | 55 (61.1) | 26 | 29 |
|
| Tumor
multiplicity |
|
|
| 0.040a |
|
Single | 64 (71.1) | 29 | 35 |
|
|
Multiple | 26 (28.9) | 6 | 20 |
|
| Microscopic
vascular invasion |
|
|
|
<0.01b |
| No | 51 (56.7) | 33 | 18 |
|
|
Yes | 39 (43.3) | 2 | 37 |
|
| Stage |
|
|
|
<0.01b |
|
I–II | 51 (56.7) | 33 | 18 |
|
|
III–IV | 39 (43.3) | 2 | 37 |
|
Cell lines and cell culture
The FuHeng Cell Center provided the normal
hepatocyte cells (THLE-2 cells), the human liver carcinoma cells
(HepG2, Hep3B and MHCC-97L cells) and 293T cells. The cells were
grown in DMEM (Invitrogen; Thermo Fisher Scientific, Inc.) with 10%
FBS (Gibco; Thermo Fisher Scientific, Inc.) in an incubator at 37°C
with 5% CO2, and STR profiling was used for
authentication.
LOC284454 and EZH2 overexpression lentiviral
vectors, and short hairpin (sh)RNA-encoding lentiviral vectors
directed against LOC284454, E-cadherin and EZH2 and their
respective non-targeting negative control (NC) vectors were
designed and provided by Shanghai GenePharma Co., Ltd., with the
following sequences: LOC284454 target vector,
5′-GCAGCUUUUCAGAAAUCU-3′ and NC vector, 5′-TCGTACTCGTCTTCGAT-3′;
E-cadherin target vector, 5′-GGUGGAGGAAGGACCAUUA-3′ and NC vector,
5′-AUAGCCCUGUAAUGCUUU-3′; EZH2 target vector,
5′-GGACCAUUAUAUUCCUUA-3′ and NC vector, 5′-AGUGAAACCUUGGCUAGCU-3′.
Lentiviral transfection was conducted according to the
manufacturer's protocol. When MHCC-97L or Hep3B cells seeded in
6-well plates reached 70% confluence, a total of 0.5 µg lentiviral
construct and 10 µg polybrene (Sigma-Aldrich; Merck KgaA) were
added to the medium. An MOI value of 10 was used for ov-LOC284454
and ov-EZH2, and an MOI of 8 for the others lentiviral vectors.
After 12 h, the medium was replaced with fresh medium containing 1
µg/ml puromycin. Subsequently, the cells were collected after 72 h
for further experiments.
Reverse transcription-quantitative PCR
(RT-qPCR)
TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) was used to extract RNA from specimens or
cells. The RNA was then reverse transcribed into cDNA using the
PrimeScript RT Reagent kit (Takara Bio, Inc.) according to the
manufacturer's protocol, and Oligo dT was used as a complementary
universal primer. Subsequently, qPCR experiments were performed
using the SYBR Premix kit (Takara Bio, Inc.). The relative mRNA
expression against GAPDH expression was assessed using
2−ΔΔCq method (21). The
optimal results were obtained when the amplification curve
exhibited a typical S-type amplification and the curve was smooth.
The reaction conditions were as follows: One cycle of preheating
for 10 min at 92°C, followed by 40 cycles at 90°C for 15 sec, 55°C
for 50 sec and 70°C for 30 sec, and finally one cycle at 97°C for
90 sec, 60°C for 3 min and 94°C for 10 sec. The primer sequences
are shown in Table SII.
MTT assays
Parental and transfected MHCC-97L or Hep3B cells
were seeded into 96-well plates (2,000 cells/well). A total of 15
µl MTT was added to each well at the indicated time points (0, 24,
48 or 72 h). The cells treated with MTT were then incubated for
another 4 h at room temperature, and the solution in each well was
replaced with fresh medium containing 150 µl DMSO. The results were
measured at an absorbance of 492 nm using a microplate reader
(BioTek Instruments, Inc.; Agilent Technologies, Inc.).
Western blotting
The concentration of the protein samples extracted
from MHCC-97L or Hep3B cells using RIPA lysis buffer (Thermo Fisher
Scientific, Inc.) was measured using a BCA kit (Beyotime Institute
of Biotechnology). The protein samples (25 µg/lane) were resolved
via 4–20% SDS-PAGE (GenScript), electronically transferred to PVDF
membranes, blocked in 5% skimmed milk at 37°C for 1 h and incubated
overnight at 4°C with the appropriate primary antibodies, including
anti-E-cadherin (1:500; cat. no. 14472), anti-N-cadherin (1:300;
cat. no. 13116), anti-vimentin (1:300; cat. no. 574), anti-EZH2
(1:500; cat. no. 5246) and anti-GAPDH (1:3,000; cat. no. 5174). The
primary antibodies were all purchased from Cell Signaling
Technology, Inc. Protein expression levels were normalized to those
of the internal control (GAPDH). Subsequently, the membranes were
incubated with goat anti-rabbit secondary antibodies (cat. no.
bs-40295G-IRDye8) for E-cadherin, N-cadherin, vimentin and EZH2,
and goat anti-mouse secondary antibodies (cat. no.
bs-40296G-IRDye8) for GAPDH (both 1:10,000; BIOSS) at 37°C for 2 h.
The intensity of the protein signal was observed using an ECL
Detection System (Thermo Fisher Scientific, Inc.). ImageJ version
1.6.0 software (National Institutes of Health) was used to quantify
the intensity of the protein bands.
Transwell assays
Transwell chambers (8-µm; EMD Millipore) were used
for examining cell migration and invasion. To measure cell
invasion, the membranes were precoated with 200 mg/ml
Matrigel® (BD Biosciences) for ~30 min at room
temperature. MHCC-97L or Hep3B cells (1×104) suspended
in 200 µl DMEM without FBS were seeded into the upper chambers of
Transwell inserts placed in 24-well plates. As the attractant for
cell invasion and migration, 600 µl DMEM with 10% FBS was added to
the bottom chambers. After incubation for 48 h at 37°C, the cells
that invaded the membrane were stained with 0.1% crystal violet
solution (Amresco, LLC) for 30 min at room temperature. The invaded
cells in at least three random fields of view at ×200 magnification
were counted using a fluorescence microscope (Olympus
Corporation).
RNA immunoprecipitation (RIP)
assays
The EZMagna RIP kit (EMD Millipore) was used to
conduct RIP assays according to the manufacturer's instructions. A
total of 3×107 MHCC-97L or Hep3B cells transfected with
ov-LOC284454 or ov-EZH2 for 48 h were collected and incubated
overnight at 4°C with RIP buffer in the EZMagna RIP kit containing
antibody against Argonaute2 (Anti-Ago2; 1:1,000; cat. no. ab32381;
Abcam) or anti-IgG (1:1,000; cat. no. ab109489; Abcam) and 5 μg of
magnetic beads coated with anti-Argonaute2 (Ago2). After incubation
with protease K buffer at 55°C for 30 min, the immunoprecipitated
RNA was extracted using the RNeasy MinElute Cleanup kit (EMD
Millipore) and reverse transcribed using Prime-Script RT Master Mix
(Takara Bio, Inc.). Finally, the purified RNAs were measured by
qPCR assays, as aforementioned. lncDUXAP10 was used as the negative
control since it has been shown not to bind with EHZ2 (22).
Chromatin immunoprecipitation
(ChIP)
ChIP assays were performed using a Pierce Agarose
ChIP kit (cat. no. 26156; Pierce; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. MHCC-97L or Hep3B cells
were collected in a tube for centrifugation at 1,000 × g for 2 min
at 4°C, were then treated with 1% formaldehyde and collected with
lysis buffer (cat. no. P0013G; Beyotime Institute of
Biotechnology). Ultrasonication (20 KHz; 10 times at room
temperature; 10 sec each time with 3 sec intervals) was used to
break DNA fragments. The remaining lysates were immunoprecipitated
with normal rabbit IgG antibody (1:500; cat. no. sc-2025; Santa
Cruz Biotechnology, Inc.) and then immunoprecipitated with Protein
G Agarose Beads and incubated overnight at 4°C with gentle shaking.
Subsequently, DNA crosslinking was performed with buffer made up of
50 mmol/l NaCl, 10 mmol/l KCl and 4.2 mmol/l
Na2HPO4, and 20 mg/ml Proteinase K, and the
DNA was purified with a PCR Purification kit (cat. no. 28104;
Qiagen GmbH). Finally, RT-qPCR assays were performed as
aforementioned to amplify the precipitated DNAs.
RNA pull-down assay
After the lysates were collected from MHCC-97L and
Hep3B cells transfected with a biotin-labeled EZH2-WT or EZH2-MUT
probe, which were designed and provided by Shanghai GenePharma Co.,
Ltd, a pull-down assay was performed using the Pierce Magnetic
RNA-Protein Pull-Down kit (Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. Briefly, 200 µl cell
lysates were combined with streptavidin-coated A/G plus magnetic
beads (Invitrogen; Thermo Fisher Scientific, Inc.) for 2 h at room
temperature. Subsequently, the mixture was centrifuged at 1,000 × g
for 1 min at 4°C and washed with ice-cold washing buffer (20 mM
Tris-base pH 7.4, 350 mM KCl and 0.02% NP-40), low-salt buffer and
then high-salt buffer. After the beads bound with the magnetic
frame, total RNA was extracted using TRIzol and measured via
RT-qPCR, as aforementioned.
Fluorescence in situ hybridization
(FISH) assay
The Ribo™ FISH kit (Guangzhou RiboBio Co., Ltd.) was
used for the RNA FISH assay according to the manufacturer's
protocol. MHCC-97L and Hep3B cells were fixed with 4%
paraformaldehyde for 10 min at 25°C, and then permeabilized with
0.5% PBS for 10 min at 4°C. Next, the cells were blocked with
prehybridization buffer for 30 min at 37°C and then the cells were
washed with saline sodium citrate buffer solution and the nuclei
were stained with DAPI at 37°C for 1 min (Beyotime Institute of
Biotechnology). The air-dried cells were incubated further with 40
nM of the FISH probe designed and synthesized by Guangzhou RiboBio
Co., Ltd., in hybridization buffer at 95°C for 1 min. Fluorescence
was observed using an LSM 880 confocal laser scanning microscope
(Zeiss GmbH; magnification, ×400). The relative fluorescence
intensity was quantified using ImageJ software version 1.51
(National Institutes of Health).
Luciferase reporter assay
293T cells (5×103/well) were seeded in a
24-well plate and then co-transfected with Renilla
luciferase vector (phRL-TK; Promega Corporation), wild-type (wt)
LOC284454 or mutant (mut) LOC284454 vector, wt or mut EZH2 vector,
and pGL3-E-cadherin-3′-untranlated region reporter vector (Shanghai
GenePharma Co., Ltd.). After transfection using
Lipofectamine® 2000 (Invitrogen; Thermp Fisher
Scientific, Inc.) for 24 h, luciferase activity was measured using
the Dual-Luciferase Reporter Assay System (Promega Corporation).
The relative luciferase activity was normalized to Renilla
luciferase activity.
Statistical analysis
SPSS 18.0 software (SPSS, Inc.) was used to perform
statistical analyses. The assays were performed three times, and
the results are shown as the mean ± SD. χ2 analyses were
performed to analyze the association between clinicopathological
features and LOC284454 expression. Kaplan-Meier survival analysis
with the log-rank test was conducted to estimate the association
between LOC284454 expression and the prognosis of patients. One-way
ANOVA with Dunnett's multiple comparisons test was used for
comparisons among multiple groups and Student's t-test was used to
compare the values between two groups. The comparisons between
paired tumor and normal tissues were analyzed with a paired t-test,
while comparisons between cells with an unpaired t-test. Spearman's
correlation analysis was used to assess the correlation between
LOC284454 and E-cadherin expression in HCC. *P<0.05 was
considered to indicate a statistically significant difference.
Results
LOC284454 expression is aberrantly
elevated in HCC and has prognostic significance in patients with
HCC
First, LOC284454 expression was investigated in 90
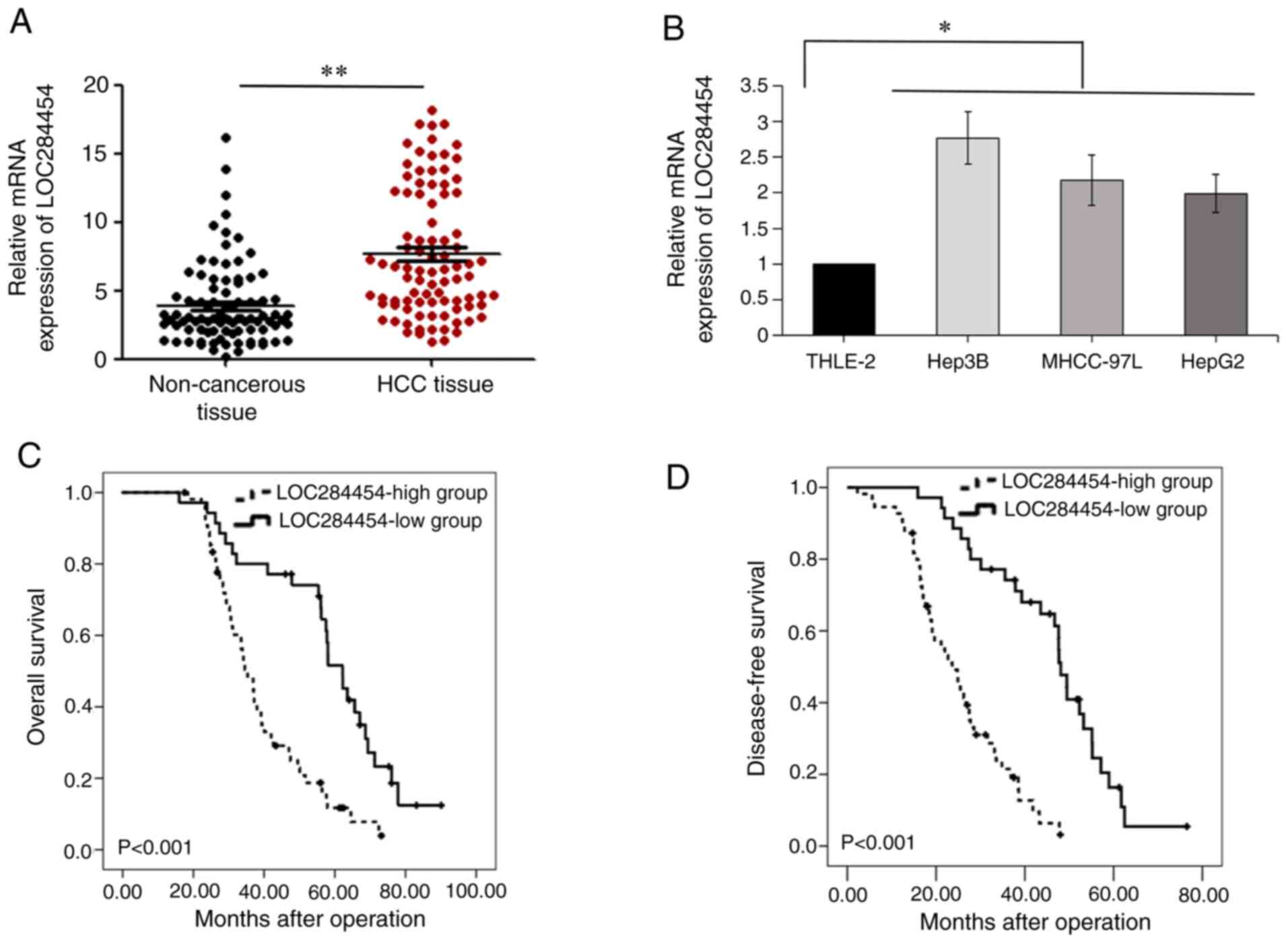
paired HCC and non-cancerous tissues using RT-qPCR, revealing that
LOC284454 expression was significantly elevated in HCC samples
compared with in non-cancerous samples (P<0.05; Fig. 1A). Furthermore, RT-qPCR analysis
suggested that LOC284454 expression was higher in liver cancer
cells, including HepG2, MHCC-97L and Hep3B cells, compared with
that in normal hepatocytes (THLE-2 cells) (P<0.05; Fig. 1B).
To evaluate whether LOC284454 expression had
clinical importance in HCC, the association between LOC284454
expression and clinicopathological features of patients with HCC
was analyzed. As shown in Table I,
the results demonstrated that LOC284454 expression was associated
with microscopic vascular invasion (P<0.01), tumor multiplicity
(P=0.04), advanced TNM stage (P<0.01) and larger tumor size
(P=0.033) in patients with HCC. Kaplan-Meier analysis revealed that
compared with lower LOC284454 expression, increased LOC284454
expression indicated a shorter OS (P<0.001; Fig. 1C) and shorter disease-free survival
(P<0.001; Fig. 1D) of patients
with HCC. A total of 15 human non-tumor liver samples were also
collected and RT-qPCR was performed. The results suggested that the
expression levels of LOC284454 in human non-tumor liver samples and
non-cancerous tissues from patients with HCC were not significantly
different (Fig. S1).
LOC284454 promotes HCC cell invasion
and migration
LOC284454 expression in MHCC-97L and Hep3B cells was
significantly higher than that in THLE-2 cells. The shRNA-encoding
lentivirus targeting LOC284454 (sh-LOC284454) or its NC lentivirus
(sh-Ctrl) were transfected into MHCC-97L cells and Hep3B cells.
RT-qPCR revealed that LOC284454 expression was significantly
decreased in cells transfected with sh-LOC284454 compared with
those transfected with sh-Ctrl (P<0.01; Fig. 2A). Additionally, FISH assays were
performed in HCC cells, revealing that LOC284454 was mainly located
in the nucleus and that LOC284454 expression was decreased in
MHCC-97L cells and Hep3B cells transfected with sh-LOC284454
(Fig. S2). The MTT assay suggested
that MHCC-97L and Hep3B cell proliferation in the sh-LOC284454
group was significantly decreased than that in the sh-Ctrl group
(P<0.01; Fig. 2B). In addition,
the change in HCC cell migration and invasion induced by the
knockdown of LOC284454 was evaluated using Transwell assays. The
results confirmed that MHCC-97L and Hep3B cell migration and
invasion were significantly decreased by knockdown of LOC284454
(P<0.01; Fig. 2C). These results
suggested that LOC284454 enhanced the malignant biological behavior
of HCC cells, especially invasion and migration.
EMT is well known as an essential mechanism and
primary process of tumor metastasis (23,24). Thus,
it was hypothesized that LOC284454 promoted HCC cell migration by
inducing EMT. RT-qPCR (Fig. 2D) and
western blot analysis (Fig. 2E) were
performed to assess the change in the expression levels of EMT
markers in MHCC-97L and Hep3B cells. The results indicated that
E-cadherin expression was significantly increased by
LOC284454-knockdown, while N-cadherin and vimentin expression was
not affected (P<0.05; Fig. 2D and
E). The current data demonstrated that LOC284454 facilitates
migration and invasion by promoting EMT in HCC cells.
LOC284454 inhibits E-cadherin
To identify the mechanism of LOC284454 in HCC, the
present study attempted to identify the downstream effector of
LOC284454. Downregulation of E-cadherin is essential for EMT, and
the current results suggested that E-cadherin expression was
significantly affected upon LOC284454-knockdown. RT-qPCR was
performed to evaluate the correlation between LOC284454 expression
and N-cadherin, vimentin or E-cadherin expression in HCC tissues.
Spearman's correlation analysis (Fig.
3A) revealed a significant negative correlation between
LOC284454 and E-cadherin expression (P<0.001; r=−0.846);
however, there was a positive correlation between LOC284454 and
N-cadherin (P=0.001; r=0.352) or vimentin (P<0.001; r=0.373)
expression. These results suggested that LOC284454 may enhance EMT
of HCC cells by suppressing E-cadherin expression.
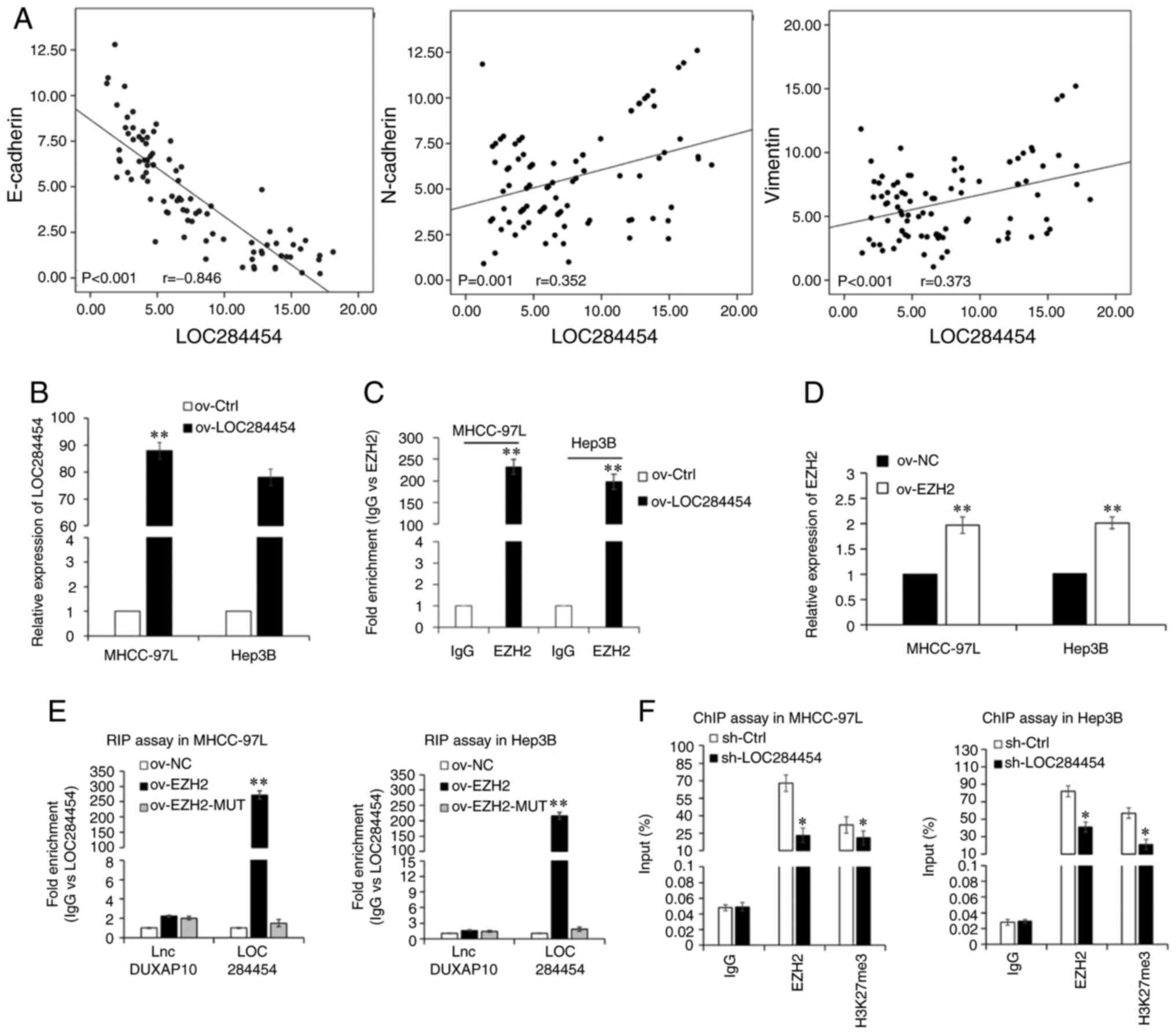 | Figure 3.LOC284454 binds with EZH2. (A)
Spearman's correlation analysis of the association between the
expression levels of LOC284454 and E-cadherin, N-cadherin or
vimentin in HCC tissues. (B) RT-qPCR assays were used to analyze
LOC284454 expression. (C) RIP assay revealed the enrichment of EZH2
mRNA. (D) RT-qPCR of EZH2 expression in MHCC-97L and Hep3B cells
transfected with ov-EZH2. (E) RIP assay revealed the enrichment of
LOC284454. (F) ChIP assay revealed whether EZH2 could bind to the
E-cadherin promoter. *P<0.05; **P<0.01 vs. Ctrl/NC. RIP, RNA
immunoprecipitation; lnc, long non-coding; ov, overexpression;
Ctrl, control; NC, negative control; EZH2, enhancer of zeste
homolog 2; ChIP, chromatin immunoprecipitation; RT-qPCR, reverse
transcription-quantitative PCR; sh, short hairpin. |
LOC284454 binds to EZH2
Numerous studies have elucidated that lncRNAs
modulate their target genes by binding with RBPs and polycomb
repressive complex2 (PRC2) (25–27). RBP
EZH2 is the core subunit of PRC2, is involved in the regulation of
lncRNAs on their downstream effectors and participates in the
downregulation of E-cadherin in cancer metastasis (28,29). Thus,
RIP experiments were performed to assess the interaction between
LOC284454 and EZH2. First, the LOC284454 overexpression lentiviral
vector (ov-LOC284454) or its NC lentiviral vector (ov-Ctrl) were
transfected into HCC cells. RT-qPCR revealed that LOC284454
expression was significantly increased in HCC cells transfected
with ov-LOC284454 compared with in those transfected with ov-Ctrl
(P<0.01; Fig. 3B). The RIP assay
results corroborated the aforementioned hypothesis that LOC284454
could bind to EZH2 in HCC cells (P<0.01; Fig. 3C). Considering that Ago2 is known as a
core component of the RNA-induced silencing complex (30), further RIP assays were performed using
Ago2 antibodies. Ago2 protein-RNA complexes were precipitated from
MHCC-97L and Hep3B cells, and the results revealed that endogenous
LOC284454 was preferentially enriched in Ago2 RIPs compared with
control IgG antibody RIPs (P<0.01; Fig. S3A). Moreover, Ago2 RIP samples were
significantly enriched for endogenous EZH2, suggesting that
LOC284454 and EZH2 may be in the same Ago2 complex in MHCC-97L and
Hep3B cells (P<0.01; Fig. S3A).
Furthermore, the overexpression EZH2 lentiviral vector (ov-EZH2) or
its NC lentiviral vector (ov-NC) were transfected into MHCC-97L and
Hep3B cells. RT-qPCR revealed that EZH2 expression was
significantly increased in HCC cells expressing ov-EZH2 compared
with in those transfected with ov-NC (P<0.01; Fig. 3D). EZH2- mutational lentiviral vectors
(EZH2-MUT) were also developed, and RIP assays were performed in
which lncDUXAP10 was used as the negative control since it has been
shown not to bind with EHZ2 (22).
The RIP assays suggested that LOC284454 was more enriched in
MHCC-97L and Hep3B cells transfected with the ov-EZH2 vector than
in those transfected with the EZH2-MUT or ov-NC vector, further
suggesting a direct interaction between EZH2 and LOC284454 in HCC
cells (P<0.01; Fig. 3E). In
addition, biotin-labeled pull-down followed by RT-qPCR analysis
revealed that a wt biotin-labeled EZH2 probe (Bio-EZH2-probe-WT)
pulled down LOC284454 from the RNA-protein complex, while
Bio-EZH2-probe-MUT did not pull down LOC284454 (P<0.01; Fig. S3B). Consistently, the results of the
dual-luciferase reporter assays revealed that LOC284454 and EZH2
interacted with E-cadherin (P<0.01; Fig. S3C). To reveal whether LOC284454 could
modulate the enrichment of EZH2 in the E-cadherin mRNA promoter
region, ChIP assays were performed. The ChIP assay results
suggested that LOC284454-knockdown significantly decreased the
enrichment of EZH2 in the promoter region of E-cadherin, suggesting
that LOC284454 and EZH2 may be enriched in the E-cadherin promoter
region (P<0.01; Fig. 3F).
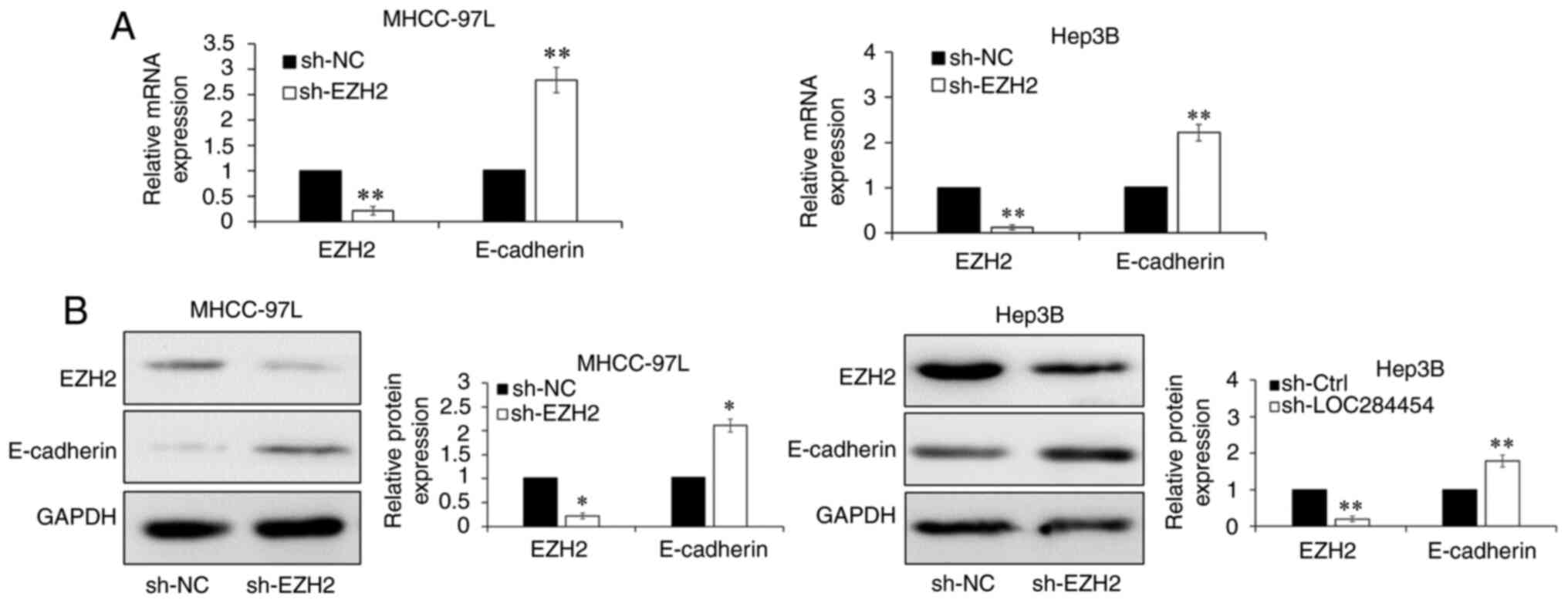
The shRNA-encoding lentiviral vector targeting EZH2
(sh-EZH2) or its NC lentiviral vector (sh-NC) were transfected into
MHCC-97L and Hep3B cells. The RT-qPCR results indicated that EZH2
expression was significantly decreased in HCC cells transfected
with sh-EZH2 compared with in those transfected with sh-0NC
(P<0.01; Fig. 4A). RT-qPCR
(P<0.01; Fig. 4A) and western blot
analysis (P<0.05; Fig. 4B)
suggested that E-cadherin expression was significantly upregulated
upon EZH2-knockdown. To investigate whether EZH2 was essential for
LOC284454-dependent E-cadherin expression downregulation, the
sh-EZH2 lentiviral vector was transfected into MHCC-97L and Hep3B
cells overexpressing LOC284454 (ov-LOC284454+sh-EZH2). The results
of RT-qPCR and western blot analysis revealed that LOC284454 and
EZH2 expression was increased upon the transfection of
ov-LOC284454, and decreased after co-transfection with sh-EZH2
vectors into HCC cells, while E-cadherin expression was not
downregulated by overexpression of LOC284454 when EZH2 was knocked
down in MHCC-97L and Hep3B cells (Fig.
S4). These results demonstrated that LOC284454 suppressed
E-cadherin expression by binding with EZH2.
E-cadherin is essential for the
oncogenic function of LOC284454
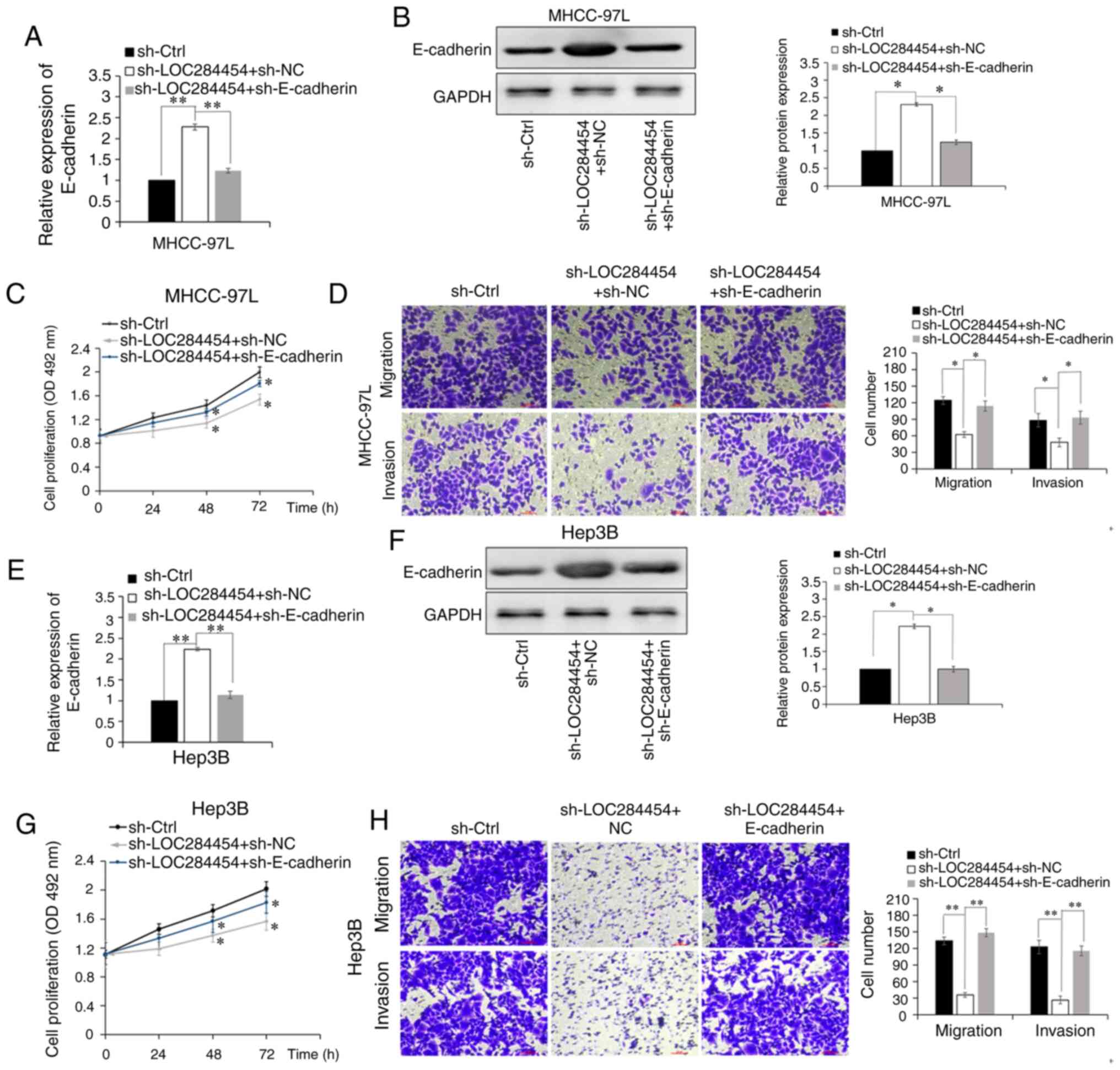
To highlight the significance of E-cadherin in the
function of LOC284454, rescue assays were performed. RT-qPCR
results confirmed that transfection with sh-E-cadherin
significantly decreased E-cadherin expression compared with the
negative control group (sh-Ctrl) in MHCC-97L cells and Hep3B cells
(P<0.01; Fig. S5). RT-qPCR
(P<0.01; Fig. 5A) and western blot
analysis (P<0.05; Fig. 5B)
revealed that compared with cells transfected with
sh-LOC284454+sh-NC, E-cadherin expression was significantly
downregulated in LOC284454-knockdown MHCC-97L cells transfected
with sh-E-cadherin (sh-LOC284454 + sh-E-cadherin). The MTT assay
revealed that cell proliferation was attenuated after E-cadherin
expression was downregulated by transfection with sh-E-cadherin in
LOC284454-knockdown MHCC-97L cells (P<0.01; Fig. 5C). The Transwell assay revealed that
E-cadherin-knockdown reversed the inhibitory effect of
LOC284454-knockdown on the invasion and migration of MHCC-97L cells
(P<0.01; Fig. 5D). In addition,
inhibition of E-cadherin in LOC284454-knockdown Hep3B cells
exhibited similar results (Fig.
5E-H). These results strongly suggested that E-cadherin may be
essential for the oncogenic functions of LOC284454 in HCC.
Discussion
The present study revealed that LOC284454 expression
was aberrantly elevated in HCC tissues and cells, and increased
LOC284454 expression was associated with a poor prognosis in
patients with HCC. Furthermore, the present results revealed that
LOC284454 acted as an oncogenic factor, promoting HCC cell invasion
and migration by decreasing E-cadherin expression, which is
essential for EMT and metastasis of HCC cells. Mechanistically,
LOC284454 could bind to EZH2 mRNA and subsequently inhibit
E-cadherin by enriching in its promoter region. Furthermore, rescue
assays demonstrated that E-cadherin was essential for the oncogenic
functions of LOC284454 in HCC cell invasion and migration. The
current results suggested that the LOC284454/EZH2/E-cadherin axis
may be a promising therapeutic target for patients with HCC.
Previous studies reported that lncRNAs had a significant impact in
various types of human cancer, including renal cell carcinoma, HCC
and nasopharyngeal carcinoma (4–6).
Therefore, the discovery of new hepatocarcinogenesis-associated
lncRNAs and further understanding of the role of lncRNAs during the
occurrence and development of HCC may help to reveal the pathogenic
mechanisms of HCC. Recently, the importance of LOC284454 in human
tumors has gained more attention. However, the expression pattern
and the importance of LOC284454 in HCC has not been previously
reported. The present experiments in HCC samples and cells revealed
that LOC284454 expression was aberrantly increased in HCC compared
with in normal samples and cells. Further clinical investigation
demonstrated that elevated LOC284454 expression predicted more
aggressive clinical features and a poor prognosis in patients with
HCC. This was in accordance with previous studies that confirmed
that numerous lncRNAs are abnormally expressed and are used as
independent molecular markers in HCC. For example, the lncRNA SNHG1
was reported to be differentially expressed in HCC and served as an
independent molecular marker of HCC [hazard ratio (HR) =2.977]
(31). Elevated expression levels of
HOXD-AS1 lncRNA was also found in HCC and was found to be
associated with a poor prognosis; multivariate regression analysis
revealed that HOXD-AS1 expression was an independent and
significant factor for OS (HR=1.552) (32). Upregulated lncCSMD1-1 expression was
reported to be associated with an advanced stage of HCC and with
worse outcomes in patients with HCC (HR=3.80) (33). In the present study, LOC284454 was
knocked down in MHCC-97L and Hep3B cells and then MTT and Transwell
assays were performed to test the function of LOC284454 in HCC
cells. LOC284454 was discovered to facilitate HCC cell migration,
invasion and proliferation. To the best of our knowledge, the
present study is the first to reveal that LOC284454 may be of
immense importance as a prognostic biomarker in HCC.
There is an increasing amount of evidence that
lncRNAs work as pro-tumorigenic or tumor-suppressing factors upon
the modulation of their target genes by forming complexes with RBPs
(34–36). The present study demonstrated that
LOC284454 promoted hepatocarcinogenesis by repressing E-cadherin
expression, verifying the association between the pro-tumorigenic
effects of LOC284454 and its regulatory effect on E-cadherin.
Similar findings in other studies have revealed that numerous
lncRNAs serve as oncogenic factors in multiple types of human
cancer, such as bladder, gastric and non-small cell lung cancer, by
impairing E-cadherin expression at the transcriptional level,
including H19 lncRNA, RP11-789C1.1 lncRNA and FEZF1-AS1 lncRNA
(37–39). In the present study, RIP assays were
performed and confirmed that LOC284454 could bind with EZH2. The
RT-qPCR results suggested that LOC284454 and EZH2 inhibited
E-cadherin expression. Subsequently, ChIP assays confirmed that
LOC284454 could bind with EZH2 and then suppress E-cadherin
expression at the transcriptional level. Several other independent
studies have also unveiled a direct association and regulation
between EZH2 and E-cadherin. Liu et al (40) revealed that EZH2 facilitated
metastasis and decreased E-cadherin expression in renal cell
carcinoma. Additionally, EZH2 was confirmed to form a complex with
histone deacetylase 1/2 and Snail, and then to bind with the
E-cadherin promoter with Snail in NPC (41). Linc-UBC1 was reported to promote the
metastasis of esophageal squamous cell carcinoma by enhancing EZH2
and then suppressing E-cadherin expression (42). To the best of our knowledge, the
current data revealed for the first time that LOC284454 promoted
HCC cell migration and invasion by binding with EZH2 and then
repressing the downstream target gene E-cadherin.
E-cadherin serves an important tumor-suppressing
role in HCC by regulating cell differentiation, polarity, invasion,
migration and stem cell-like properties (43,44). The
silencing or inhibition of E-cadherin frequently occurs in the
initial phases of cancer cell invasion and migration, and has a
significant impact on HCC development and metastasis (45). Furthermore, downregulation of
E-cadherin expression in human cancer has been identified as a
critical fundamental mechanism and major characteristic of EMT
(46). Qiu et al (47) reported that E-cadherin expression is
decreased or silenced in breast cancer samples and cells, and that
P2Y2 receptor-facilitated breast cancer metastasis depends on
regulation of E-cadherin. Furthermore, E-cadherin is essential for
the microenvironment in tumors by regulating tumor radioresistance
and EMT (48). Additionally,
E-cadherin was confirmed to participate in the oncogenic function
of N-myc downstream-regulated gene-1 and transforming growth
factor-β in pancreatic cancer (49).
In the present study, a rescue assay was performed and found that
E-cadherin was essential for the oncogenic effects of LOC284454 on
the invasion and migration of HCC cells. It is possible that
E-cadherin is not the only downstream regulator of LOC284454.
Further investigations of the downstream regulation mechanisms of
LOC284454 are required in the future. However, to the best of our
knowledge, the present study was the first to reveal a direct
interaction between EZH2 and LOC284454 in HCC by RIP, suggesting
that LOC284454 may modulate the enrichment of EZH2 in the
E-cadherin mRNA promoter region, which may markedly inhibit
E-cadherin expression. Thus, the findings of the present study may
contribute to the understanding of the pathogenesis of HCC.
In summary, the present study identified the role of
LOC284454 in HCC and proposed that the LOC284454/EZH2/E-cadherin
signaling pathway may be a novel therapeutic target for patients
with HCC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was funded by the National Natural
Science Foundation of China (grant no. 82003122) and the Natural
Science Foundation of Shaanxi Province (grant no. 2019JQ-128).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
LH collected the clinical samples, performed most of
the experiments and designed the study. WZ performed the
experiments and the statistical analysis. FW assisted with the
design of the study and drafting of the manuscript. The
authenticity of the raw data has been assessed by WZ and FW. All
authors have read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Xi'an Jiaotong University (Xi'an, China), and written
informed consent was provided by participants before the
examination phase. The study complied with the principles of the
Declaration of Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
lncRNAs
|
long non-coding RNAs
|
|
HCC
|
hepatocellular carcinoma
|
|
CRC
|
colorectal cancer
|
|
NPC
|
nasopharyngeal carcinoma
|
|
EMT
|
epithelial-mesenchymal transition
|
|
RBP
|
RNA-binding protein
|
|
EZH2
|
enhancer of zeste homolog 2
|
|
OS
|
overall survival
|
|
PRC2
|
polycomb repressive complex 2
|
References
|
1
|
Chaudhari VA, Khobragade K, Bhandare M and
Shrikhande SV: Management of fibrolamellar hepatocellular
carcinoma. Chin Clin Oncol. 7:512018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Thompson AI, Conroy KP and Henderson NC:
Hepatic stellate cells: Central modulators of hepatic
carcinogenesis. BMC Gastroenterol. 15:632015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Petruzziello A: Epidemiology of hepatitis
B virus (HBV) and hepatitis C virus (HCV) related hepatocellular
carcinoma. Open Virol J. 12:26–32. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jiang X and Liu W: Long noncoding RNA
highly upregulated in liver cancer activates p53-p21 pathway and
promotes nasopharyngeal carcinoma cell growth. DNA Cell Biol.
36:596–602. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liao K, Qian Z, Zhang S, Chen B, Li Z,
Huang R, Cheng L, Wang T, Yang R, Lan J, et al: The LGMN pseudogene
promotes tumor progression by acting as a miR-495-3p sponge in
glioblastoma. Cancer Lett. 490:111–123. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
García-Venzor A, Mandujano-Tinoco EA,
Ruiz-Silvestre A, Sánchez JM, Lizarraga F, Zampedri C,
Melendez-Zajgla J and Maldonado V: lncMat2B regulated by severe
hypoxia induces cisplatin resistance by increasing DNA damage
repair and tumor-initiating population in breast cancer cells.
Carcinogenesis. 41:1485–1497. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang Y, Huang W, Yuan Y, Li J, Wu J, Yu
J, He Y, Wei Z and Zhang C: Long non-coding RNA H19 promotes
colorectal cancer metastasis via binding to hnRNPA2B1. J Exp Clin
Cancer Res. 39:1412020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Das M, Renganathan A, Dighe SN, Bhaduri U,
Shettar A, Mukherjee G, Kondaiah P and Satyanarayana Rao MR:
DDX5/p68 associated lncRNA LOC284454 is differentially expressed in
human cancers and modulates gene expression. RNA Biol. 15:214–230.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fan C, Wang J, Tang Y, Zhang S, Xiong F,
Guo C, Zhou Y, Li Z, Li X, Li Y, et al: Upregulation of long
non-coding RNA LOC284454 may serve as a new serum diagnostic
biomarker for head and neck cancers. BMC Cancer. 20:9172020.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fan C, Tang Y, Wang J, Wang Y, Xiong F,
Zhang S, Li X, Xiang B, Wu X, Guo C, et al: Long non-coding RNA
LOC284454 promotes migration and invasion of nasopharyngeal
carcinoma via modulating the Rho/Rac signaling pathway.
Carcinogenesis. 40:380–391. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nieto MA, Huang RY, Jackson RA and Thiery
JP: EMT: 2016. Cell. 166:21–45. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Creighton CJ, Gibbons DL and Kurie JM: The
role of epithelial-mesenchymal transition programming in invasion
and metastasis: A clinical perspective. Cancer Manag Res.
5:187–195. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Han LL, Jia L, Wu F and Huang C: Sirtuin6
(SIRT6) promotes the EMT of hepatocellular carcinoma by stimulating
autophagic degradation of E-cadherin. Mol Cancer Res. 17:2267–2280.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhou W, Gong L, Wu Q, Xing C, Wei B, Chen
T, Zhou Y, Yin S, Jiang B, Xie H, et al: PHF8 upregulation
contributes to autophagic degradation of E-cadherin,
epithelial-mesenchymal transition and metastasis in hepatocellular
carcinoma. J Exp Clin Cancer Res. 37:2152018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lee CH, Yu JR, Granat J, Saldaña-Meyer R,
Andrade J, LeRoy G, Jin Y, Lund P, Stafford JM, Garcia BA, et al:
Automethylation of PRC2 promotes H3K27 methylation and is impaired
in H3K27M pediatric glioma. Genes Dev. 33:1428–1440. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ding J, Li J, Wang H, Tian Y, Xie M, He X,
Ji H, Ma Z, Hui B, Wang K and Ji G: Long noncoding RNA CRNDE
promotes colorectal cancer cell proliferation via epigenetically
silencing DUSP5/CDKN1A expression. Cell Death Dis. 8:e29972017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lian Y, Yan C, Xu H, Yang J, Yu Y, Zhou J,
Shi Y, Ren J, Ji G and Wang K: A novel lncRNA, LINC00460, affects
cell proliferation and apoptosis by regulating KLF2 and CUL4A
expression in colorectal cancer. Mol Ther Nucleic Acids.
12:684–697. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kumar MB, Lu JJ, Loh KS, Chong LM, Soo R,
Goh BC, Tan KS and Shakespeare TP: Tailoring distant metastatic
imaging for patients with clinically localized undifferentiated
nasopharyngeal carcinoma. Int J Radiat Oncol Biol Phys. 58:688–693.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cantù G, Bimbi G, Miceli R, Mariani L,
Colombo S, Riccio S, Squadrelli M, Battisti A, Pompilio M and Rossi
M: Lymph node metastases in malignant tumors of the paranasal
sinuses: Prognostic value and treatment. Arch Otolaryngol Head Neck
Surg. 134:170–177. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang Q, Wang WW, Xu TH and Xu ZF: Highly
expressed long non-coding RNA DUXAP10 promotes proliferation of
ovarian cancer. Eur Rev Med Pharmacol Sci. 22:314–321.
2018.PubMed/NCBI
|
|
23
|
Oka H, Shiozaki H, Kobayashi K, Inoue M,
Tahara H, Kobayashi T, Takatsuka Y, Matsuyoshi N, Hirano S,
Takeichi M, et al: Expression of E-cadherin cell adhesion molecules
in human breast cancer tissues and its relationship to metastasis.
Cancer Res. 53:1696–1701. 1993.PubMed/NCBI
|
|
24
|
Schipper JH, Frixen UH, Behrens J, Unger
A, Jahnke K and Birchmeier W: E-cadherin expression in squamous
cell carcinomas of head and neck: Inverse correlation with tumor
dedifferentiation and lymph node metastasis. Cancer Res.
51:6328–6337. 1991.PubMed/NCBI
|
|
25
|
Li X, Zhang F, Ma J, Ruan X, Liu X, Zheng
J, Liu Y, Cao S, Shen S, Shao L, et al: NCBP3/SNHG6 inhibits GBX2
transcription in a histone modification manner to facilitate the
malignant biological behaviour of glioma cells. RNA Biol. 26:1–17.
2020. View Article : Google Scholar
|
|
26
|
Zhang F, Ruan X, Ma J, Liu X, Zheng J, Liu
Y, Liu L, Shen S, Shao L, Wang D, et al: DGCR8/ZFAT-AS1 promotes
CDX2 transcription in a PRC2 complex-dependent manner to facilitate
the malignant biological behavior of glioma cells. Mol Ther.
28:613–630. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang X, Goodrich KJ, Conlon EG, Gao J,
Erbse AH, Manley JL and Cech TR: C9orf72 and triplet repeat
disorder RNAs: G-quadruplex formation, binding to PRC2 and
implications for disease mechanisms. RNA. 25:935–947. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
He W, Yu Y, Huang W, Feng G and Li J: The
pseudogene DUXAP8 promotes colorectal cancer cell proliferation,
invasion, and migration by inducing epithelial-mesenchymal
transition through interacting with EZH2 and H3K27me3. Onco Targets
Ther. 13:11059–11070. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen WW, Qi JW, Hang Y, Wu JX, Zhou XX,
Chen JZ, Wang J and Wang HH: Simvastatin is beneficial to lung
cancer progression by inducing METTL3-induced m6A modification on
EZH2 mRNA. Eur Rev Med Pharmacol Sci. 24:4263–4270. 2020.PubMed/NCBI
|
|
30
|
Chen Y, Yang F, Fang E, Xiao W, Mei H, Li
H, Li D, Song H, Wang J, Hong M, et al: Circular RNA circAGO2
drives cancer progression through facilitating HuR-repressed
functions of AGO2-miRNA complexes. Cell Death Differ. 26:1346–1364.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gu X, Li H, Sha L and Zhao W: A prognostic
model composed of four long noncoding RNAs predicts the overall
survival of Asian patients with hepatocellular carcinoma. Cancer
Med. 9:5719–5730. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang H, Huo X, Yang XR, He J, Cheng L,
Wang N, Deng X, Jin H, Wang N, Wang C, et al: STAT3-mediated
upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer
metastasis by regulating SOX4. Mol Cancer. 16:1362017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu J, Xu R, Mai SJ, Ma YS, Zhang MY, Cao
PS, Weng NQ, Wang RQ, Cao D, Wei W, et al: lncRNA CSMD1-1 promotes
the progression of Hepatocellular Carcinoma by activating MYC
signaling. Theranostics. 10:7527–7544. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang J, Choi JM, Holehouse AS, Lee HO,
Zhang X, Jahnel M, Maharana S, Lemaitre R, Pozniakovsky A, Drechsel
D, et al: A molecular grammar governing the driving forces for
phase separation of Prion-like RNA binding proteins. Cell.
174:688–699.e16. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Dominguez D, Freese P, Alexis MS, Su A,
Hochman M, Palden T, Bazile C, Lambert NJ, Van Nostrand EL, Pratt
GA, et al: Sequence, structure, and context preferences of human
RNA binding proteins. Mol Cell. 70:854–867.e9. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Holmqvist E and Vogel J: RNA-binding
proteins in bacteria. Nat Rev Microbiol. 16:601–615. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhu Z, Xu L, Wan Y, Zhou J, Fu D, Chao H,
Bao K and Zeng T: Inhibition of E-cadherin expression by lnc-RNA
H19 to facilitate bladder cancer metastasis. Cancer Biomark.
22:275–281. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chen Z, Wu J, Huang W, Peng J, Ye J, Yang
L, Yuan Y, Chen C, Zhang C, Cai S, et al: Long non-coding RNA
RP11-789C1.1 suppresses epithelial to mesenchymal transition in
gastric cancer through the RP11-789C1.1/miR-5003/E-cadherin axis.
Cell Physiol Biochem. 47:2432–2444. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
He R, Zhang FH and Shen N: lncRNA
FEZF1-AS1 enhances epithelial-mesenchymal transition (EMT) through
suppressing E-cadherin and regulating WNT pathway in non-small cell
lung cancer (NSCLC). Biomed Pharmacother. 95:331–338. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liu L, Xu Z, Zhong L, Wang H, Jiang S,
Long Q, Xu J and Guo J: Enhancer of zeste homolog 2 (EZH2) promotes
tumour cell migration and invasion via epigenetic repression of
E-cadherin in renal cell carcinoma. BJU Int. 117:351–362. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Tong ZT, Cai MY, Wang XG, Kong LL, Mai SJ,
Liu YH, Zhang HB, Liao YJ, Zheng F, Zhu W, et al: EZH2 supports
nasopharyngeal carcinoma cell aggressiveness by forming a
co-repressor complex with HDAC1/HDAC2 and Snail to inhibit
E-cadherin. Oncogene. 31:583–594. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Niu G, Zhuang H, Li B and Cao G: Long
noncoding RNA linc-UBC1 promotes tumor invasion and metastasis by
regulating EZH2 and repressing E-cadherin in esophageal squamous
cell carcinoma. J BUON. 23:157–162. 2018.PubMed/NCBI
|
|
43
|
Petrova YI, Schecterson L and Gumbiner BM:
Roles for E-cadherin cell surface regulation in cancer. Mol Biol
Cell. 27:3233–3244. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wong SHM, Fang CM, Chuah LH, Leong CO and
Ngai SC: E-cadherin: Its dysregulation in carcinogenesis and
clinical implications. Crit Rev Oncol Hematol. 121:11–22. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Onder TT, Gupta PB, Mani SA, Yang J,
Lander ES and Weinberg RA: Loss of E-cadherin promotes metastasis
via multiple downstream transcriptional pathways. Cancer Res.
68:3645–3654. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhai B, Yan HX, Liu SQ, Chen L, Wu MC and
Wang HY: Reduced expression of E-cadherin/catenin complex in
hepatocellular carcinomas. World J Gastroenterol. 14:5665–5673.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Qiu Y, Liu Y, Li WH, Zhang HQ, Tian XX and
Fang WG: P2Y2 receptor promotes the migration and invasion of
breast cancer cells via EMT-related genes Snail and E-cadherin.
Oncol Rep. 39:138–150. 2018.PubMed/NCBI
|
|
48
|
Theys J, Jutten B, Habets R, Paesmans K,
Groot AJ, Lambin P, Wouters BG, Lammering G and Vooijs M:
E-Cadherin loss associated with EMT promotes radioresistance in
human tumor cells. Radiother Oncol. 99:392–397. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Menezes SV, Fouani L, Huang MLH, Geleta B,
Maleki S, Richardson A, Richardson DR and Kovacevic Z: The
metastasis suppressor, NDRG1, attenuates oncogenic TGF-β and NF-κB
signaling to enhance membrane E-cadherin expression in pancreatic
cancer cells. Carcinogenesis. 40:805–818. 2019. View Article : Google Scholar : PubMed/NCBI
|