Glioma, the most prevalent primary malignant cancer
of the central nervous system of adults (1), is characterized by difficulty of early
diagnosis, a high recurrence rate and a poor prognosis, especially
for advanced and high-grade types (2–4). The World
Health Organization has classified gliomas into four grades based
on their histopathology and clinical prognosis: Grades I and II are
routinely viewed as low-grade gliomas, while grades III and IV are
deemed as high-grade gliomas (5).
Glioblastoma multiforme (GBM), the most malignant and aggressive
type, has a median survival time of 12–14 months after initial
diagnosis, longer than that of only pancreatic and lung cancer
(6). As research has progressed in
recent years, medical technology has constantly improved. However,
due to the rapid proliferation, high invasive potential and
radio/chemotherapeutic resistance of GBM, current treatments,
including surgical resection, radiotherapy and chemotherapy, do not
have optimal effects, and patients with GBM still have a poor
prognosis (7). Therefore, it is
necessary to clarify in detail the pathogenetic mechanisms of
glioma to achieve improved therapeutic effects and longer survival
times in patients after initial diagnosis.
Long non-coding RNAs (lncRNAs) are transcripts that
contain >200 nucleotides (nt), but lack protein-coding capacity
(8). The structure of lncRNAs is
typically similar to that of mRNAs, which have 5′-m7G caps and
3′-poly(A) tails (9), but they are
more tissue-specific than mRNAs, indicating that lncRNAs may have
specific biological roles and functional mechanisms (10). Recently, an increasing number of
studies has reported that lncRNAs participate in a variety of
cellular physiological processes, including stemness,
tumorigenesis, proliferation, invasion, angiogenesis and drug
resistance, by regulating gene expression at the epigenetic,
transcriptional and post-transcriptional levels (11,12). It
has been demonstrated that most lncRNAs can recruit regulatory
complexes through RNA-protein interactions to affect the expression
levels of nearby genes, while some lncRNAs can also function as
local regulators (13).
Overexpression, deficiency or mutation of lncRNA genes has been
reported to be associated with numerous human diseases, such as
cancer, cardiovascular diseases, metabolic diseases and
inflammation (14–17). Similarly, in glioma, progressive
evidence has illustrated that abnormal expression levels of lncRNAs
are closely associated with the occurrence and development of
glioma and other malignant phenotypes (12).
Technological advancements, especially the
completion of the human genome sequencing, have allowed the
discovery of an increasing number of lncRNAs with different targets
and functions (18); however, the
specific mechanisms and functions of lncRNAs remain unclear. The
present review summarizes the functions and mechanisms of lncRNAs
at the molecular level in glioma and provides some prospects for
their use in the therapy and diagnosis of glioma.
Previous human genome studies have reported that
lncRNAs are transcripts produced by RNA polymerase II (RNAPol II)
that contain >200 nt, but lack an open reading frame (ORF) for
translation into proteins (19,20).
Accumulating studies have found that lncRNAs are important players
at almost every level of gene function and regulation (11,14,21). Based
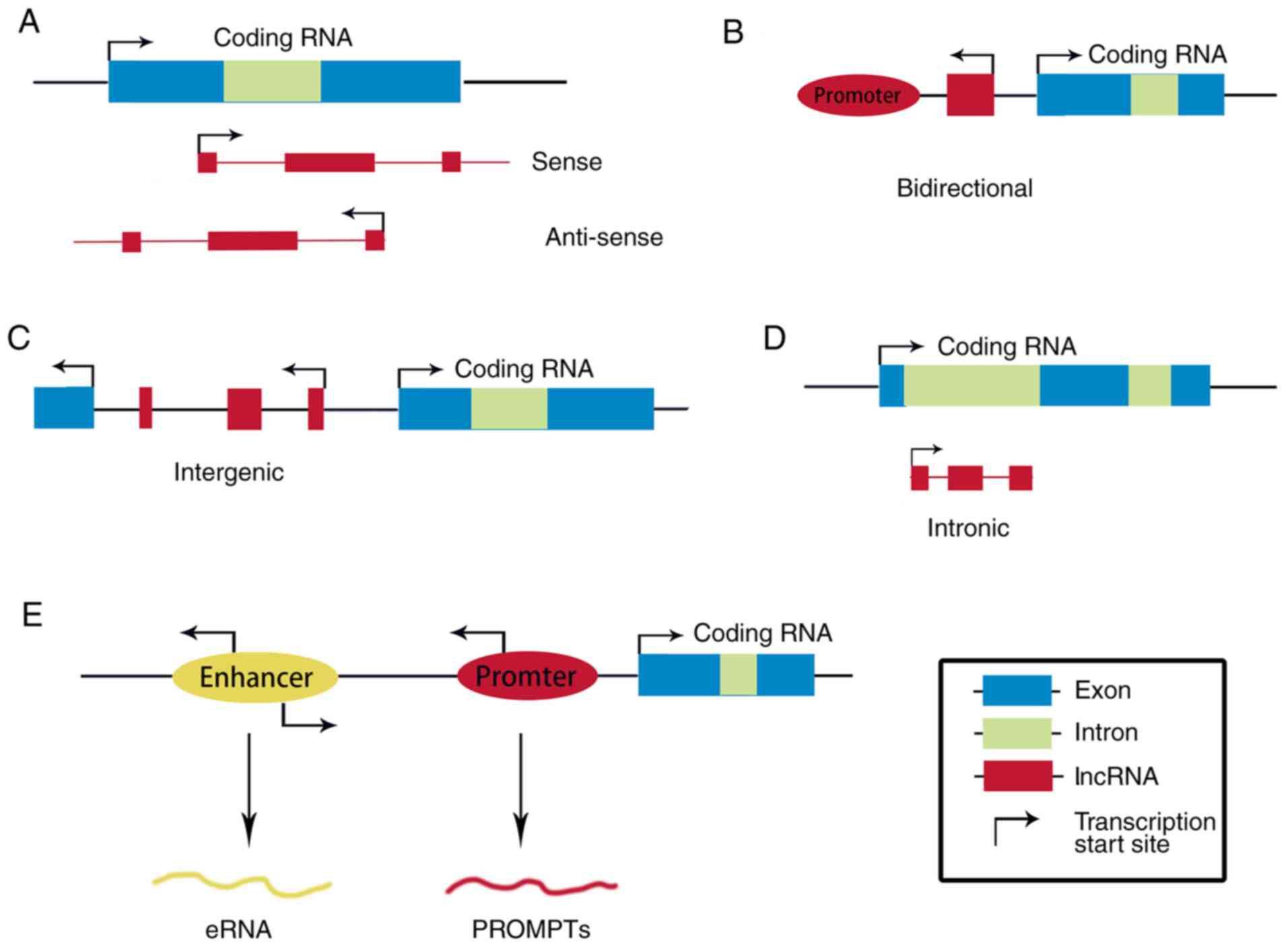
on their genomic location relative to neighboring protein-coding
genes and their molecular characteristics, lncRNAs can be
classified into five categories: Sense, antisense, bidirectional
(22), intronic (23) and intergenic [long intergenic ncRNAs
(lincRNAs)] (8) (Fig. 1). Through the classification of
unspliced and spliced lncRNAs from mouse and human embryonic stem
cells (24), most lncRNAs are either
localized to enhancer regions (~20%), called enhancer RNAs (eRNAs)
(25,26), or associated with upstream antisense
RNAs, which are derived from loci near transcription start sites
(TSSs) of coding RNAs (60–70%) (24,27). The
remaining lncRNAs are derived from transcripts that overlap with
coding sequences (~5%) or from more distal, unannotated regions
(~5%) (27). The latter lncRNAs are
usually called lincRNAs (27,28). Moreover, eRNAs and promoter upstream
transcripts, which are transcribed from enhancers and promoters,
respectively, are functionally similar to regulatory DNA molecules
(23). For example, eRNAs can promote
the interactions of enhancers and promoters to activate target
genes (29). However, although a
number of lncRNAs are associated with annotated genomic regions,
some intervening lncRNAs come from separate transcriptional
elements that do not overlap with coding sequences or enhancers;
these loci have their own promoters and can function through
chromatin modifications as protein-coding genes (30). The lncRNAs metastasis-associated lung
adenocarcinoma transcript 1 (MALAT1) and nuclear enriched abundant
transcript 1 (NEAT1), which are well-known structural intervening
lncRNAs, belong to this category (14). Considering the close association
between the structures and locations of lncRNAs and their stability
and functional mechanisms (14), the
identification of lncRNA secondary structures and classification is
anticipated to serve a key role in the research and clinical
application of lncRNAs (19).
Aberrant expression levels of lncRNAs can mediate
cell biological processes, such as proliferation, stemness, drug
resistance and angiogenesis, and accelerate the progression of
glioma malignancy (31). Therefore,
an increasing number of studies have focused on the analysis of
lncRNA gene expression profiles in GBM to identify the detailed
mechanisms. By comparing the expression levels of mRNAs and lncRNAs
between GBM and normal brain tissues, Han et al (32) found that 654 lncRNAs were upregulated
and 654 lncRNAs were downregulated in GBM. Moreover, 104 matched
lncRNA-mRNA pairs were identified, and 90 lncRNAs and 81
lncRNA-mRNA pairs were found to be differentially expressed
(32). Chen et al (33) used the significant analysis of
microarray (SAM) method in a training dataset to analyze the
differential expression of lncRNAs between GBM and normal brain
tissues, identifying 299 lncRNAs with differential expression, of
which 133 were upregulated and 166 were downregulated in GBM
compared with in normal brain tissues (33). The SAM method was then used to analyze
the differential expression of lncRNAs between low-grade and
high-grade gliomas in the training dataset, and 47 lncRNAs were
found to be differentially expressed between low-grade and
high-grade gliomas (33). By
comparing the expression levels of mRNAs and lncRNAs between normal
brain tissues and GBM, Li et al (34) found that 398 lncRNAs were
differentially expressed and 1,995 mRNAs were dysregulated in GBM.
Among these differentially expressed lncRNAs, 98 participated in 32
gene functions and 30 molecular pathways associated with
tumorigenesis, development and metastasis of glioma (34,35).
Aberrant expression levels of lncRNAs in glioma have
been identified to serve a crucial role in the tumorigenesis,
proliferation and invasion of glioma cells (31). Moreover, abnormal lncRNA expression
profiles in clinical glioma specimens are closely associated with
histological differentiation and malignancy grade, which have
crucial clinical significance in early glioma diagnosis of
subclassifications and in patient prognosis (36).
In glioma, lncRNAs can regulate gene expression at
the epigenetic level before transcription by recruiting chromatin
modifiers such as enhancer of zeste homolog 2 (EZH2)/polycomb
repressive complex 2 (PRC2) (37) and
WD repeat domain 5/trithorax group proteins (38) to a specific genomic location as
scaffolds to regulate the trimethylation or acetylation of histone
H3 (31). Therefore, lncRNAs can
participate in the regulation of glioma phenotypes, such as
tumorigenic behaviors, proliferation, invasion and drug resistance
(37,39).
The lncRNA NEAT1 is distributed mainly in the cell
nucleus and has two transcripts, NEAT1_1 (3.7 kb) and NEAT1_2 (23
kb) (40). The lncRNA NEAT1 has been
demonstrated to promote the occurrence of numerous types of cancer,
such as colorectal cancer, breast cancer, liver cancer and glioma
(41–44). A study on the detailed mechanism of
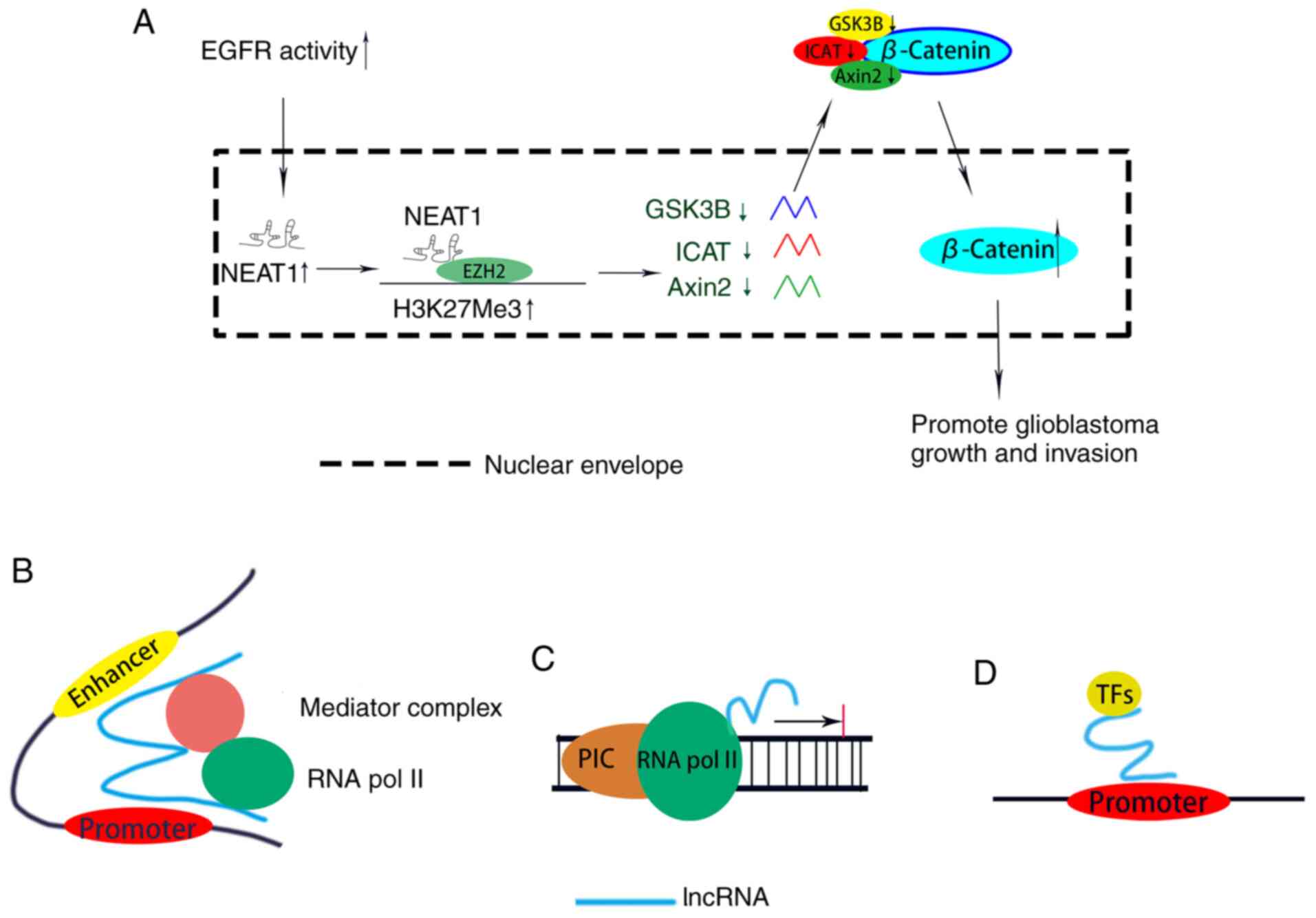
lncRNAs have suggested that the lncRNA NEAT1 can be activated by
the upstream EGFR signaling pathway; in addition, it can act as a
scaffold to recruit and interact with the chromosome modification
enzyme EZH2 (37). Moreover, the
interaction between the lncRNA NEAT1 and EZH2 can promote histone
H3 trimethylation in the promoter regions of Axin2, inhibitor of
β-catenin and T-cell factor and glycogen synthase kinase 3β, which
are negative regulatory factors of the WNT/β-catenin signaling
pathway (45,46), to silence these downstream target
genes, thereby activating the WNT/β-catenin signaling pathway to
promote glioma tumorigenesis and proliferation (37) (Fig. 2A).
Similarly, in neuroblastoma, the lncRNA neuroblastoma-associated
transcript-1 (NBAT-1) functions as a scaffold to recruit and
interact with EZH2 to downregulate the expression levels of
NBAT-1/EZH2 target genes, such as SRY-box transcription factor 9
(SOX9), oncostatin M receptor and versican, to decrease the risk of
neuroblastoma (47). The lncRNA
temozolomide-associated lncRNA in GBM recurrence (TALC), with a
total length of 418 nt and containing two exons, is highly
expressed in temozolomide (TMZ)-resistant glioma cells (39). The lncRNA TALC, induced by
AKT-mediated TMZ resistance in GBM, can control the acetylation of
histone H3 on lysine 27 (H3K27) in the promoter regions of O6
methylguanine-DNA methyltransferase (MGMT) to trap microRNA
(miRNA/miR)-20b-3p, activate c-MET and increase MGMT expression
(39). The lncRNA ZFAT antisense RNA
1 (ZFAT-AS1), derived from an imprinted gene located on the long
arm of the human genome, can bind to the relevant EZH2 subunit of
the PRC2 complex to catalyze histone H3K27 methylation to inhibit
transcription of the downstream gene caudal type homeobox 2 (CDX2),
in turn promoting glioma cell proliferation, migration and invasion
(48).
Transcription is an important cellular physiological
process that transfers DNA genetic material to the cytoplasm as RNA
(49). Based on their patterns of
interaction with proteins, lncRNAs regulate transcriptional
processes via three mechanisms.
The transcription of most genes involves the
interaction of a proximal promoter with more distant enhancer
elements (50). Enhancers are usually
located far from the transcriptional initiation site and interact
with tissue-specific transcription factors that perform their
function to modulate the differential expression of genes (51). Kim et al (52) found that some ncRNAs can be
bidirectionally transcribed from activated enhancers, and the
expression levels of these eRNAs are associated with the activity
of the enhancer. Follow-up studies have shown that eRNAs may exert
enhancer-like effects, such as remodeling chromatin, promoting
chromatin accessibility (53) and
bridging a distal enhancer with a proximal promoter (54). A class of lncRNAs similar to eRNAs is
composed of activating ncRNAs (ncRNA-as), which have a
transcriptional activation function; these lncRNAs are transcribed
from independent loci, not from enhancers, and compose a class of
functional molecules that can regulate the enhancing effect
(25,55). Depletion of lncRNAs may lead to
elevated expression levels of adjacent protein-coding genes at
numerous loci in the human genome (25,55). This
promotion of gene expression is mediated by RNA, and studies have
shown that ncRNA-as can moderate this RNA-dependent transcriptional
responsiveness in cis (25,55). These enhancer-like effects of lncRNAs
may be functional mechanisms broadly used to modulate gene
expression. Moreover, both eRNA and ncRNA-as can link the enhancer
and promoter element of the coding gene as a scaffold for a protein
complex, thereby regulating the transcription process (56) (Fig.
2B).
Transcription factors, which are a class of DNA
binding proteins, can specifically bind to specific sequences in
the TSS of protein-coding genes to modulate the transcription
process (61). Accumulating evidence
has demonstrated that lncRNAs can interact with transcription
factors at the promoter regions of coding genes to regulate
transcription (62). For instance,
the lncRNA PANDA, transcribed from the CDKN1A promoter, interacts
with the nuclear transcription factor Y subunit α or PRCs (PRC1 and
PRC2) to either accelerate or suppress senescence (63) (Fig.
2D).
In glioma, lncRNAs generally bind to transcription
factors at the promoter region of target genes to regulate
transcription. For instance, the lncRNA paxillin interacting
protein 1-antisense RNA 1 (PAXIP1-AS1), a critical mediator of cell
death, has been found to recruit the transcription factor ETS
proto-oncogene 1 (ETS1) to the promoter region of kinesin family
member 14 (KIF14) to upregulate its expression (64). Thus, the lncRNA PAXIP1-AS1 promotes
glioma cell migration, invasion and angiogenesis via the
PAXIP1-AS1/ETS1/KIF14 axis (64).
Furthermore, the lncRNA growth arrest-specific transcription 5
(GAS5), a member of the 5′ terminal oligopyrimidine class of genes
(65), can inhibit tumorigenesis by
recruiting the transcription factor TFAP2A to its promoter region
under physiological conditions (66).
However, an indel genetic polymorphism of the lncRNA GAS5 increases
glioma susceptibility by blocking the binding of the transcription
factor TFAP2A (66).
In addition to participating in transcriptional and
epigenetic regulation, lncRNAs often modulate gene expression by
post-transcriptional regulation (11). A number of published results have
shown that lncRNAs can act as competing endogenous RNAs (ceRNAs) to
decrease the expression levels of specific mRNAs (67), mediate alternative splicing (68) and affect mRNA stability (69) and intercellular communication
(70). This section focusses on these
functions to summarize the post-transcriptional mechanisms of
lncRNAs in glioma.
ceRNAs are a class of transcripts that can affect
the expression levels of mRNAs at the post-transcriptional level by
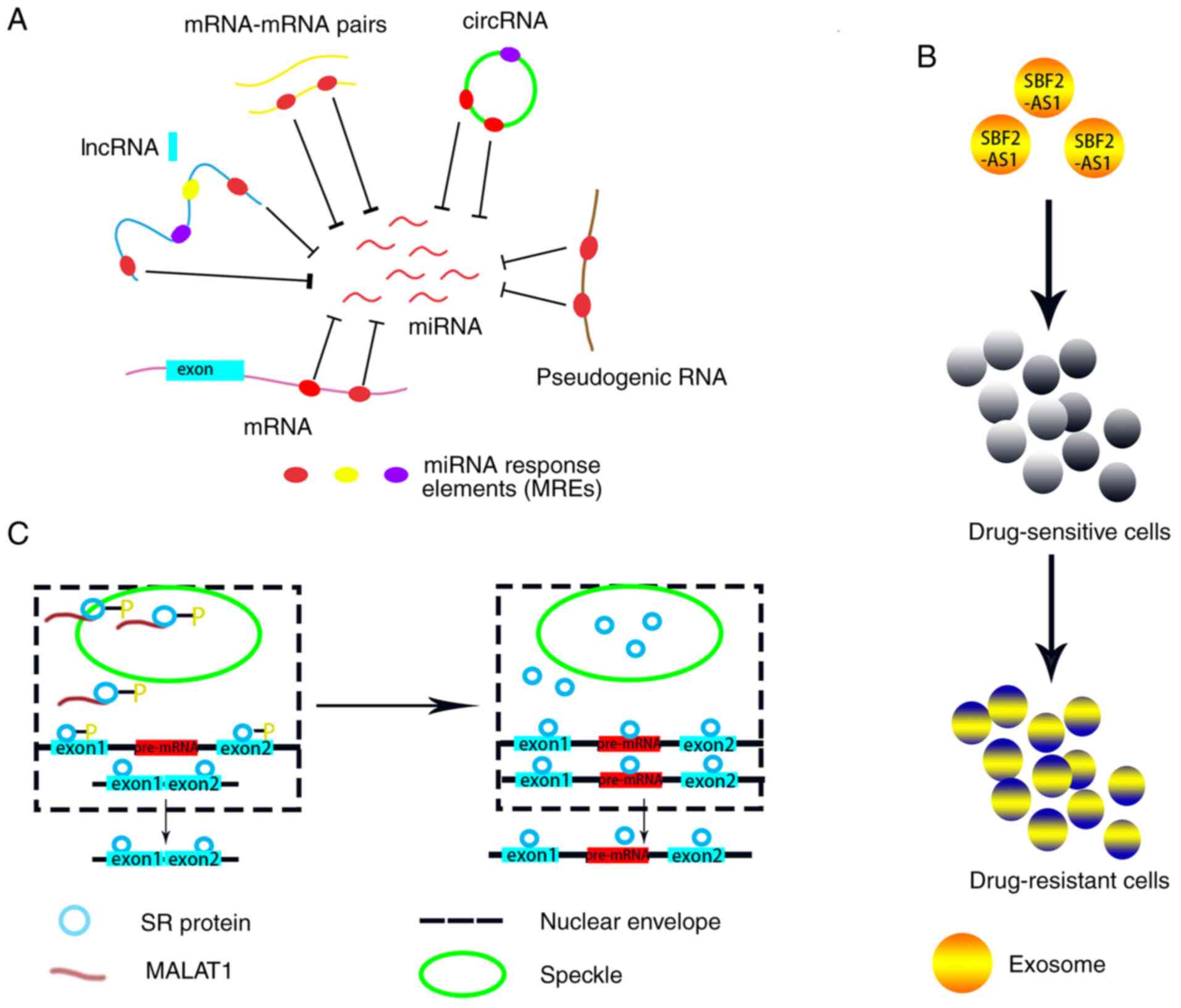
competing for binding to miRNAs (67). In 2011, a review proposed the ceRNA
hypothesis, which states that mRNAs contain miRNA response elements
(MREs) to which miRNAs can specifically bind, and that ncRNA
species, including lncRNAs, circular RNAs (circRNAs), pseudogene
RNAs and mRNA-mRNA pairs, also contain MREs and can potentially
compete for a limited pool of miRNAs to regulate gene expression
(71) (Fig.
3A). Subsequently, accumulating evidence has shown that lncRNAs
may function as ceRNAs to sponge miRNAs and prevent them from
interacting with their downstream target genes, thereby silencing
these genes to affect the expression levels of the corresponding
proteins (18,72–75).
The lncRNA CRNDE, encoded by the colorectal
neoplasia differentially expressed gene, has been shown to be the
most highly upregulated lncRNA among 129 differentially expressed
lncRNAs in glioma (76). Li et
al (75) demonstrated that the
lncRNA CRNDE can act as a ceRNA to interact with miR-136-5p, thus
competitively inhibiting miR-136-5p-mediated inhibition of Wnt-2
and Bcl-2. This event leads to an increase in the
post-transcriptional expression levels of Wnt-2 and Bcl-2, and in
the activation of the PI3K/AKT/mTOR signaling pathway (75). Moreover, Zheng et al (77,78) found
that the lncRNA CRNDR can facilitate the proliferation, invasion
and migration of glioma cells, and decrease their apoptosis through
competitive inhibition of miR-384 and miR-186. Similarly, the
lncRNA X-inactive specific transcript (XIST), transcribed from the
X inactivation centre (79), can also
act as a ceRNA of miR-126 to regulate the IRS1/PI3K/Akt signaling
pathway in order to promote the viability, migration, invasion,
apoptosis resistance and glucose metabolism of GBM cells (80). The lncRNAs HOTAIR and MEG3 can promote
or suppress, respectively, the proliferation, invasion and
migration of glioma cells by acting as ceRNAs of miR-141 and
miR-19a (74).
In summary, lncRNAs act as ceRNAs through a
ceRNA/miRNA/mRNA regulatory axis at the post-trans-criptional
level. The concept of a ceRNA/miRNA/mRNA regulatory pathway offers
a novel concept for studying the underlying molecular mechanisms of
glioma, as well as improves the understanding of lncRNAs and
identifies a specific and sensitive profile of interactions between
lncRNAs and mRNAs, which may contribute to the development of
methods for both the earlier diagnosis and targeted therapy of
glioma.
Alternative splicing of pre-mRNAs serves a crucial
role in the regulation and diversity of gene functions, and is used
by higher eukaryotes to increase the complexity of the
transcriptome and proteome (81).
Alternative splicing is mediated mainly by trans-acting protein
factors, including serine/arginine-rich (SR)-associated proteins,
heterogeneous nuclear ribonucleoproteins, the SR family of nuclear
phosphoproteins (SR proteins) and small nuclear ribonucleoproteins
(82). Among these factors, SR
proteins, a class of proteins that can specifically bind to RNA,
typically serve a key role in alternative splicing (83). Some results have shown that lncRNAs in
glioma can modulate alternative splicing by controlling these
trans-acting protein factors (68).
The lncRNA MALAT1 is one of the most abundant
lncRNAs in normal human physiological tissues (84). Initially, lncRNA MALAT1 was known as a
prognostic molecular marker of advanced lung cancer (85); however, previous studies have found an
association between MALAT1 and other types of cancer, such as
pancreatic cancer, prostate cancer, breast cancer, glioma and
leukemia (86–90). Tripathi et al (91) reported that the lncRNA MALAT1 can bind
to SR proteins and act as a molecular sponge to modulate the
phosphorylation levels of SR proteins. By regulating the
phosphorylation status of SR proteins, the lncRNA MALAT1 can
indirectly mediate the intranuclear transfer of SR proteins between
nuclear speckles and transcription sites to control their
distribution to nuclear speckles, thereby regulating alternative
splicing (91) (Fig. 3C). Furthermore, by modulating the
activation levels of SR proteins, MALAT1 regulates alternative
splicing, as well as controlling other post-transcriptional gene
regulatory mechanisms associated with SR proteins, such as
translation, nonsense-mediated decay and RNA export (92,93).
Regulation of mRNA stability and protein
modification are important processes in post-transcriptional
regulation. From the perspective of modulating mRNA stability,
lncRNAs can either enhance mRNA stability by forming protective
lncRNA-mRNA duplexes (94) or
accelerate mRNA degradation by recruiting RNA-binding proteins,
such as polypyrimidine tract binding protein 1 (PTBP1), to target
pre-mRNAs in order to promote mRNA degradation (95). For instance, the lncRNA PTB-AS can
directly bind to the PTBP1 3′-untranslated region via
staphylococcal nuclease domain-containing 1 to stabilize PTBP1
mRNA, which significantly promotes the proliferation and migration
of glioma cells (96). The lncRNA
FMR1 autosomal homolog 1 can maintain the stability of miR-17-92a-1
cluster host gene mRNA to upregulate the downstream protein TAL
bHLH transcription factor 1 in order to regulate the biological
behavior of glioma cells (97).
Moreover, lncRNAs can directly interact with key
proteins of signaling pathways, thus influencing their expression
levels and regulating their functions (31). For example, in addition to acting as a
ceRNA, the lncRNA CRNDE can also bind to the P70S6K protein, a
direct downstream effector of the mTOR signaling pathway, and
enhance its phosphorylation level, suggesting that CRNDE may
modulate the mTOR signaling pathway by modifying this downstream
protein (98).
Previous studies have shown that lncRNAs can serve
important roles in intercellular communication. Barile and Vassalli
(99) found that lncRNAs can be
incorporated into exosomes and secreted into recipient cells
passing through blood vessels, thereby controlling target signaling
pathways and regulating cell phenotypes. Exosomes are the most
clearly defined vesicles known to date; these vesicles have
diameters ranging between 40 and 150 nm, and can be secreted by
numerous different types of cells (100). Their promising diagnostic and
therapeutic potential and value have received increasing attention,
particularly in cancer, such as glioma, breast cancer, prostate
cancer and pancreatic cancer (101–106).
Accumulated evidence has demonstrated that exosomes can decelerate
lncRNA degradation in the circulation and that exosomal lncRNAs can
be utilized for early diagnosis of cancer (107). Moreover, a number of studies have
confirmed that exosomal lncRNAs can function as intercellular
carriers to transmit cellular messenger molecules, including
lncRNAs (108–110).
For instance, a recent study identified a lncRNA,
lncSBF2-AS1 (ENSG00000246273), that can be activated by the
transcription factor ZEB1 and promote the TMZ resistance of glioma
cells (110). Zhang et al
(110) revealed that lncSBF2-AS1
released from TMZ-resistant glioma cells via exosomes can endow
neighboring TMZ-sensitive cells with a TMZ-resistant phenotype; by
contrast, deficiency of exosomal lncSBF2-AS1 can partially reverse
the drug resistance phenotype of the parental cells, suggesting
that exosomal lncSBF2-AS1 can induce TMZ resistance via
intercellular communication (Fig.
3B). Similarly, the lncRNA HOTAIR can be secreted into adjacent
cells via serum exosomes and modulate TMZ resistance through the
miR-519a-3p/ribonucleotide reductase catalytic subunit M1 axis
(111).
In addition to the aforementioned functional
mechanisms, lncRNAs have been proven to have other functions in
normal and cancer tissues, such as encoding functional peptides. As
mass spectrometry, deep RNA sequencing and bioinformatics
techniques have improved, accumulating evidence suggests that
lncRNAs that were previously considered non-coding may have the
ability to encode small biologically active peptides (112). lncRNAs may have small ORFs (sORFs)
that can be translated into small peptides containing <100 amino
acids (aa) (113). Some studies have
identified that these functional peptides encoded by lncRNA sORFs
can regulate biological processes and influence tumorigenesis,
proliferation, invasion and metastasis.
Based on these functional mechanisms of lncRNAs, we
speculate that lncRNAs may function as novel biomarkers and
therapeutic targets in glioma to improve patient prognosis. For
example, lncRNAs have highly specific expression patterns in
different cells and tissues; thus, they may be used to distinguish
different subtypes of glioma and evaluate patient prognosis
(10). A previous study revealed that
using small interfering RNAs to target tumor-associated lncRNAs
achieves therapeutic effects (118).
Moreover, given that lncRNAs always interact with other molecules
to regulate gene expression, the binding sites of these
interactions may become new therapeutic targets using methods such
as a peptide nuclein acid-based strategy, which can block the
interaction between lncRNA HOTAIR and EZH2, subsequently decreasing
HOTAIR-EZH2 complex activity (119).
Based on the differential expression of lncRNAs between normal and
glioma tissues, we speculate that lncRNAs may act as potential
biomarkers to diagnose glioma in early stages. Considering that
exosomes are extremely stable and are readily accessible in nearly
all types of human biofluids, and that lncRNAs can be secreted into
the circulation through packaging into exosomes (120), exosomal lncRNAs may be one of the
most promising biomarkers. Since exosomes can cross the blood-brain
barrier, the strategy of exploiting exosomes to deliver
glioma-suppressive lncRNAs to target sites may be a promising
therapeutic option for glioma (121).
Currently, accumulating evidence has proven that
lncRNAs are closely associated with the malignant progression of
cancer and serve important roles in the onset and progression of
glioma. Therefore, the present review described the classification
of lncRNAs, the functional mechanisms of lncRNAs in glioma at the
epigenetic, transcriptional and post-transcriptional levels, and
the ability of lncRNAs to encode functional peptides. However,
numerous questions about lncRNAs remain unanswered. Considering the
significance of lncRNAs in numerous physiological processes and
their close association with the occurrence of diseases, research
on lncRNAs is expected to grow exponentially in the future. We
predict that further exploration will focus on the following
aspects: Detecting the secondary structures of lncRNA interaction
sites; investigating lncRNA binding patterns to seek new RNA-based
targets; establishing complete ceRNA/miRNA/mRNA regulatory networks
and translating these findings from theories to clinical
applications; improving the identification and isolation of
tumor-specific exosomal lncRNAs, further revealing the detailed
mechanism underlying the intercellular transfer of exosomal
lncRNAs; increasing the intracellular uptake efficiency and the
relative stability of lncRNA-based drugs; accurately delivering
lncRNAs to target sites to enhance the therapeutic effects of
lncRNA-associated drugs; further studying the encoding function of
lncRNAs; and evaluating the clinical potential of functional
polypeptides. The understanding of lncRNAs remains incomplete, but
the clinical potential of lncRNAs is worth exploring. The present
review summarized new insights into the functional mechanisms of
lncRNAs from different aspects and may be useful for future
research in similar areas.
Not applicable.
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81572490 and
81172405), the Tianjin Science and Technology Committee (grant no.
18JCZDJC98600) and the Science and Technology fund of Tianjin
Binhai New Area Health and Family Planning Commission (grant no.
2018BWKZ003).
Not applicable.
XC was a major contributor in writing the
manuscript. GG provided the major ideas and outlines, and gave the
final approval of the version to be published. YL contributed to
conception and design, and acquisition of data. SW contributed to
acquisition of data and revision of the text. YZ contributed to
acquisition of data and revision of the figures. YL and QH
confirmed the authenticity of the data. QH was the corresponding
author and primarily responsible for revision of the manuscript.
All authors read and approved the final manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Louis DN, Perry A, Reifenberger G, von
Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD,
Kleihues P and Ellison DW: The 2016 world health organization
classification of tumors of the central nervous system: A summary.
Acta Neuropathol. 131:803–820. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Reni M, Mazza E, Zanon S, Gatta G and
Vecht CJ: Central nervous system gliomas. Crit Rev Oncol Hematol.
113:213–234. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen R, Smith-Cohn M, Cohen AL and Colman
H: Glioma subclassifications and their clinical significance.
Neurotherapeutics. 14:284–297. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Alexander BM and Cloughesy TF: Adult
glioblastoma. J Clin Oncol. 35:2402–2409. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Louis DN, Ohgaki H, Wiestler OD, Cavenee
WK, Burger PC, Jouvet A, Scheithauer BW and Kleihues P: The 2007
WHO classification of tumours of the central nervous system. Acta
Neuropathol. 114:97–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Taylor OG, Brzozowski JS and Skelding KA:
Glioblastoma multiforme: An overview of emerging therapeutic
targets. Front Oncol. 9:9632019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cheng J, Meng J, Zhu L and Peng Y:
Exosomal noncoding RNAs in Glioma: Biological functions and
potential clinical applications. Mol Cancer. 19:662020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ulitsky I and Bartel DP: lincRNAs:
Genomics, evolution, and mechanisms. Cell. 154:26–46. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lagarde J, Uszczynska-Ratajczak B,
Santoyo-Lopez J, Gonzalez JM, Tapanari E, Mudge JM, Steward CA,
Wilming L, Tanzer A, Howald C, et al: Extension of human lncRNA
transcripts by RACE coupled with long-read high-throughput
sequencing (RACE-Seq). Nat Commun. 7:123392016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Deveson IW, Hardwick SA, Mercer TR and
Mattick JS: The dimensions, dynamics, and relevance of the
mammalian noncoding transcriptome. Trends Genet. 33:464–478. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bonasio R and Shiekhattar R: Regulation of
transcription by long noncoding RNAs. Annu Rev Genet. 48:433–455.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yan Y, Xu Z, Li Z, Sun L and Gong Z: An
insight into the increasing role of lncRNAs in the pathogenesis of
gliomas. Front Mol Neurosci. 10:532017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Engreitz JM, Haines JE, Perez EM, Munson
G, Chen J, Kane M, McDonel PE, Guttman M and Lander ES: Local
regulation of gene expression by lncRNA promoters, transcription
and splicing. Nature. 539:452–455. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Quinn JJ and Chang HY: Unique features of
long non-coding RNA biogenesis and function. Nat Rev Genet.
17:47–62. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huang CK, Kafert-Kasting S and Thum T:
Preclinical and clinical development of noncoding RNA therapeutics
for cardiovascular disease. Circ Res. 126:663–678. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kato M and Natarajan R: Epigenetics and
epigenomics in diabetic kidney disease and metabolic memory. Nat
Rev Nephrol. 15:327–345. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Atianand MK, Caffrey DR and Fitzgerald KA:
Immunobiology of long noncoding RNAs. Annu Rev Immunol. 35:177–198.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cheng Z, Li Z, Ma K, Li X, Tian N, Duan J,
Xiao X and Wang Y: Long non-coding RNA XIST promotes glioma
tumorigenicity and angiogenesis by acting as a molecular sponge of
miR-429. J Cancer. 8:4106–4116. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Qian X, Zhao J, Yeung PY, Zhang QC and
Kwok CK: Revealing lncRNA structures and interactions by
sequencing-based approaches. Trends Biochem Sci. 44:33–52. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mercer TR and Mattick JS: Structure and
function of long noncoding RNAs in epigenetic regulation. Nat
Struct Mol Biol. 20:300–307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Schmitt AM and Chang HY: Long noncoding
RNAs in cancer pathways. Cancer Cell. 29:452–463. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Rinn JL and Chang HY: Genome regulation by
long noncoding RNAs. Annu Rev Biochem. 81:145–166. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
St Laurent G, Wahlestedt C and Kapranov P:
The Landscape of long noncoding RNA classification. Trends Genet.
31:239–251. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sigova AA, Mullen AC, Molinie B, Gupta S,
Orlando DA, Guenther MG, Almada AE, Lin C, Sharp PA, Giallourakis
CC and Young RA: Divergent transcription of long noncoding RNA/mRNA
gene pairs in embryonic stem cells. Proc Natl Acad Sci USA.
110:2876–2881. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ørom UA, Derrien T, Beringer M, Gumireddy
K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q,
et al: Long noncoding RNAs with enhancer-like function in human
cells. Cell. 143:46–58. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
de Santa F, Barozzi I, Mietton F,
Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei CL and
Natoli G: A large fraction of extragenic RNA pol II transcription
sites overlap enhancers. PLoS Biol. 8:e10003842010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Guttman M, Amit I, Garber M, French C, Lin
MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al:
Chromatin signature reveals over a thousand highly conserved large
non-coding RNAs in mammals. Nature. 458:223–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ulitsky I, Shkumatava A, Jan CH, Sive H
and Bartel DP: Conserved function of lincRNAs in vertebrate
embryonic development despite rapid sequence evolution. Cell.
147:1537–1550. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Fitz J, Neumann T, Steininger M, Wiedemann
EM, Garcia AC, Athanasiadis A, Schoeberl UE and Pavri R:
Spt5-mediated enhancer transcription directly couples enhancer
activation with physical promoter interaction. Nat Genet.
52:505–515. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cabili MN, Trapnell C, Goff L, Koziol M,
Tazon-Vega B, Regev A and Rinn JL: Integrative annotation of human
large intergenic noncoding RNAs reveals global properties and
specific subclasses. Genes Dev. 25:1915–1927. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Peng Z, Liu C and Wu M: New insights into
long noncoding RNAs and their roles in glioma. Mol Cancer.
17:612018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Han L, Zhang K, Shi Z, Zhang J, Zhu J, Zhu
S, Zhang A, Jia Z, Wang G, Yu S, et al: lncRNA profile of
glioblastoma reveals the potential role of lncRNAs in contributing
to glioblastoma pathogenesis. Int J Oncol. 40:2004–2012.
2012.PubMed/NCBI
|
|
33
|
Chen G, Cao Y, Zhang L, Ma H, Shen C and
Zhao J: Analysis of long non-coding RNA expression profiles
identifies novel lncRNA biomarkers in the tumorigenesis and
malignant progression of gliomas. Oncotarget. 8:67744–67753. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li Q, Jia H, Li H, Dong C, Wang Y and Zou
Z: lncRNA and mRNA expression profiles of glioblastoma multiforme
(GBM) reveal the potential roles of lncRNAs in GBM pathogenesis.
Tumour Biol. 37:14537–14552. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xi J, Sun Q, Ma L and Kang J: Long
non-coding RNAs in glioma progression. Cancer Lett. 419:203–209.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Malissovas N, Ninou E, Michail A and
Politis PK: Targeting long non-coding RNAs in nervous system
cancers: New insights in prognosis, diagnosis and therapy. Curr Med
Chem. 26:5649–5663. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen Q, Cai J, Wang Q, Wang Y, Liu M, Yang
J, Zhou J, Kang C, Li M and Jiang C: Long noncoding RNA NEAT1,
regulated by the EGFR pathway, contributes to glioblastoma
progression through the WNT/β-catenin pathway by scaffolding EZH2.
Clin Cancer Res. 24:684–695. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Quagliata L, Matter MS, Piscuoglio S,
Arabi L, Ruiz C, Procino A, Kovac M, Moretti F, Makowska Z,
Boldanova T, et al: Long noncoding RNA HOTTIP/HOXA13 expression is
associated with disease progression and predicts outcome in
hepatocellular carcinoma patients. Hepatology. 59:911–923. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wu P, Cai J, Chen Q, Han B, Meng X, Li Y,
Li Z, Wang R, Lin L, Duan C, et al: lnc-TALC promotes
O6-methylguanine-DNA methyltransferase expression via
regulating the c-Met pathway by competitively binding with
miR-20b-3p. Nat Commun. 10:20452019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ghafouri-Fard S and Taheri M: Nuclear
enriched abundant transcript 1 (NEAT1): A long non-coding RNA with
diverse functions in tumorigenesis. Biomed Pharmacother. 111:51–59.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhao W, Li W, Jin X, Niu T, Cao Y, Zhou P
and Zheng M: Silencing long non-coding RNA NEAT1 enhances the
suppression of cell growth, invasion, and apoptosis of bladder
cancer cells under cisplatin chemotherapy. Int J Clin Exp Pathol.
12:549–558. 2019.PubMed/NCBI
|
|
42
|
Wu Y, Yang L, Zhao J, Li C, Nie J, Liu F,
Zhuo C, Zheng Y, Li B, Wang Z and Xu Y: Nuclear-enriched abundant
transcript 1 as a diagnostic and prognostic biomarker in colorectal
cancer. Mol Cancer. 14:1912015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Fujimoto A, Furuta M, Totoki Y, Tsunoda T,
Kato M, Shiraishi Y, Tanaka H, Taniguchi H, Kawakami Y, Ueno M, et
al: Whole-genome mutational landscape and characterization of
noncoding and structural mutations in liver cancer. Nat Genet.
48:500–509. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Jiang X, Zhou Y, Sun AJ and Xue JL: NEAT1
contributes to breast cancer progression through modulating miR-448
and ZEB1. J Cell Physiol. 233:8558–8566. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Jho EH, Zhang T, Domon C, Joo CK, Freund
JN and Costantini F: Wnt/beta-catenin/Tcf signaling induces the
transcription of Axin2, a negative regulator of the signaling
pathway. Mol Cell Biol. 22:1172–1183. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Tago K, Nakamura T, Nishita M, Hyodo J,
Nagai S, Murata Y, Adachi S, Ohwada S, Morishita Y, Shibuya H and
Akiyama T: Inhibition of Wnt signaling by ICAT, a novel
beta-catenin-interacting protein. Genes Dev. 14:1741–1749.
2000.PubMed/NCBI
|
|
47
|
Pandey GK, Mitra S, Subhash S, Hertwig F,
Kanduri M, Mishra K, Fransson S, Ganeshram A, Mondal T, Bandaru S,
et al: The risk-associated long noncoding RNA NBAT-1 controls
neuroblastoma progression by regulating cell proliferation and
neuronal differentiation. Cancer Cell. 26:722–737. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhang F, Ruan X, Ma J, Liu X, Zheng J, Liu
Y, Liu L, Shen S, Shao L, Wang D, et al: DGCR8/ZFAT-AS1 promotes
CDX2 transcription in a PRC2 complex-dependent manner to facilitate
the malignant biological behavior of glioma cells. Mol Ther.
28:613–630. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
de Klerk E and t Hoen PA: Alternative mRNA
transcription, processing, and translation: Insights from RNA
sequencing. Trends Genet. 31:128–139. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ong CT and Corces VG: Enhancer function:
New insights into the regulation of tissue-specific gene
expression. Nat Rev Genet. 12:283–293. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Field A and Adelman K: Evaluating enhancer
function and transcription. Annu Rev Biochem. 89:213–234. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Kim TK, Hemberg M, Gray JM, Costa AM, Bear
DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et
al: Widespread transcription at neuronal activity-regulated
enhancers. Nature. 465:182–187. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Mousavi K, Zare H, Dell'orso S, Grontved
L, Gutierrez-Cruz G, Derfoul A, Hager GL and Sartorelli V: eRNAs
promote transcription by establishing chromatin accessibility at
defined genomic loci. Mol Cell. 51:606–617. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Li W, Notani D, Ma Q, Tanasa B, Nunez E,
Chen AY, Merkurjev D, Zhang J, Ohgi K, Song X, et al: Functional
roles of enhancer RNAs for oestrogen-dependent transcriptional
activation. Nature. 498:516–520. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Ørom UA, Derrien T, Guigo R and
Shiekhattar R: Long noncoding RNAs as enhancers of gene expression.
Cold Spring Harb Symp Quant Biol. 75:325–331. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Dykes IM and Emanueli C: Transcriptional
and post-transcriptional gene regulation by long non-coding RNA.
Genomics Proteomics Bioinformatics. 15:177–186. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Long Y, Wang X, Youmans DT and Cech TR:
How do lncRNAs regulate transcription? Sci Adv. 3:eaao21102017.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zovoilis A, Cifuentes-Rojas C, Chu HP,
Hernandez AJ and Lee JT: Destabilization of B2 RNA by EZH2
activates the stress response. Cell. 167:1788–1802.e13. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Hernandez AJ, Zovoilis A, Cifuentes-Rojas
C, Han L, Bujisic B and Lee JT: B2 and ALU retrotransposons are
self-cleaving ribozymes whose activity is enhanced by EZH2. Proc
Natl Acad Sci USA. 117:415–425. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Espinoza CA, Allen TA, Hieb AR, Kugel JF
and Goodrich JA: B2 RNA binds directly to RNA polymerase II to
repress transcript synthesis. Nat Struct Mol Biol. 11:822–829.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Lambert SA, Jolma A, Campitelli LF, Das
PK, Yin Y, Albu M, Chen X, Taipale J, Hughes TR and Weirauch MT:
The human transcription factors. Cell. 172:650–665. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Hudson WH and Ortlund EA: The structure,
function and evolution of proteins that bind DNA and RNA. Nat Rev
Mol Cell Biol. 15:749–760. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Hung T, Wang Y, Lin MF, Koegel AK, Kotake
Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P, et al:
Extensive and coordinated transcription of noncoding RNAs within
cell-cycle promoters. Nat Genet. 43:621–629. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Xu H, Zhao G, Zhang Y, Jiang H, Wang W,
Zhao D, Yu H and Qi L: Long non-coding RNA PAXIP1-AS1 facilitates
cell invasion and angiogenesis of glioma by recruiting
transcription factor ETS1 to upregulate KIF14 expression. J Exp
Clin Cancer Res. 38:4862019. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Schneider C, King RM and Philipson L:
Genes specifically expressed at growth arrest of mammalian cells.
Cell. 54:787–793. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Yuan J, Zhang N, Zheng Y, Chen YD, Liu J
and Yang M: lncRNA GAS5 indel genetic polymorphism contributes to
glioma risk through interfering binding of transcriptional factor
TFAP2A. DNA Cell Biol. 37:750–757. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Qi X, Zhang DH, Wu N, Xiao JH, Wang X and
Ma W: ceRNA in cancer: Possible functions and clinical
implications. J Med Genet. 52:710–718. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Amirkhah R, Naderi-Meshkin H, Shah JS,
Dunne PD and Schmitz U: The intricate interplay between epigenetic
events, alternative splicing and noncoding RNA deregulation in
colorectal cancer. Cells. 8:9292019. View Article : Google Scholar
|
|
69
|
Zhang XZ, Liu H and Chen SR: Mechanisms of
long non-coding RNAs in cancers and their dynamic regulations.
Cancers (Basel). 12:12452020. View Article : Google Scholar
|
|
70
|
Li Y, Yin Z, Fan J, Zhang S and Yang W:
The roles of exosomal miRNAs and lncRNAs in lung diseases. Signal
Transduct Target Ther. 4:472019. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Cesana M, Cacchiarelli D, Legnini I,
Santini T, Sthandier O, Chinappi M, Tramontano A and Bozzoni I: A
long noncoding RNA controls muscle differentiation by functioning
as a competing endogenous RNA. Cell. 147:358–369. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Sa L, Li Y, Zhao L, Liu Y, Wang P, Liu L,
Li Z, Ma J, Cai H and Xue Y: The Role of HOTAIR/miR-148b-3p/USF1 on
regulating the permeability of BTB. Front Mol Neurosci. 10:1942017.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Qin N, Tong GF, Sun LW and Xu XL: Long
noncoding RNA MEG3 suppresses glioma cell proliferation, migration,
and invasion by acting as a competing endogenous RNA of miR-19a.
Oncol Res. 25:1471–1478. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Li DX, Fei XR, Dong YF, Cheng CD, Yang Y,
Deng XF, Huang HL, Niu WX, Zhou CX, Xia CY and Niu CS: The long
non-coding RNA CRNDE acts as a ceRNA and promotes glioma malignancy
by preventing miR-136-5p-mediated downregulation of Bcl-2 and Wnt2.
Oncotarget. 8:88163–88178. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Zhang X, Sun S, Pu JK, Tsang AC, Lee D,
Man VO, Lui WM, Wong ST and Leung GK: Long non-coding RNA
expression profiles predict clinical phenotypes in glioma.
Neurobiol Dis. 48:1–8. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Zheng J, Li XD, Wang P, Liu XB, Xue YX, Hu
Y, Li Z, Li ZQ, Wang ZH and Liu YH: CRNDE affects the malignant
biological characteristics of human glioma stem cells by negatively
regulating miR-186. Oncotarget. 6:25339–25355. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Zheng J, Liu X, Wang P, Xue Y, Ma J, Qu C
and Liu Y: CRNDE promotes malignant progression of glioma by
attenuating miR-384/PIWIL4/STAT3 axis. Mol Ther. 24:1199–1215.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Brockdorff N, Bowness JS and Wei G:
Progress toward understanding chromosome silencing by Xist RNA.
Genes Dev. 34:733–744. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Cheng Z, Luo C and Guo Z:
lncRNA-XIST/microRNA-126 sponge mediates cell proliferation and
glucose metabolism through the IRS1/PI3K/Akt pathway in glioma. J
Cell Biochem. 121:2170–2183. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Ule J and Blencowe BJ: Alternative
splicing regulatory networks: Functions, mechanisms, and evolution.
Mol Cell. 76:329–345. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Shepard PJ and Hertel KJ: The SR protein
family. Genome Biol. 10:2422009. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Blencowe BJ: Alternative splicing: New
insights from global analyses. Cell. 126:37–47. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Sun Y and Ma L: New insights into long
non-coding RNA MALAT1 in cancer and metastasis. Cancers (Basel).
11:2162019. View Article : Google Scholar
|
|
85
|
Ji P, Diederichs S, Wang W, Böing S,
Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et
al: MALAT-1, a novel noncoding RNA, and thymosin beta4 predict
metastasis and survival in early-stage non-small cell lung cancer.
Oncogene. 22:8031–8041. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Arun G and Spector DL: MALAT1 long
non-coding RNA and breast cancer. RNA Biol. 16:860–863. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Chang J, Xu W, Du X and Hou J: MALAT1
silencing suppresses prostate cancer progression by upregulating
miR-1 and downregulating KRAS. Onco Targets Ther. 11:3461–3473.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Le L, Chen H, Gao Y, Wang YW, Zhang GQ,
Pan SH, Ji L, Kong R, Wang G, Jia YH, et al: Long noncoding RNA
MALAT1 promotes aggressive pancreatic cancer proliferation and
metastasis via the stimulation of autophagy. Mol Cancer Ther.
15:2232–2243. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Liao K, Lin Y, Gao W, Xiao Z, Medina R,
Dmitriev P, Cui J, Zhuang Z, Zhao X, Qiu Y, et al: Blocking lncRNA
MALAT1/miR-199a/ZHX1 axis inhibits glioblastoma proliferation and
progression. Mol Ther Nucleic Acids. 18:388–399. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Wen F, Cao YX, Luo ZY, Liao P and Lu ZW:
lncRNA MALAT1 promotes cell proliferation and imatinib resistance
by sponging miR-328 in chronic myelogenous leukemia. Biochem
Biophys Res Commun. 507:1–8. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Tripathi V, Ellis JD, Shen Z, Song DY, Pan
Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al: The
nuclear-retained noncoding RNA MALAT1 regulates alternative
splicing by modulating SR splicing factor phosphorylation. Mol
Cell. 39:925–938. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Long JC and Caceres JF: The SR protein
family of splicing factors: Master regulators of gene expression.
Biochem J. 417:15–27. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Stamm S: Regulation of alternative
splicing by reversible protein phosphorylation. J Biol Chem.
283:1223–1227. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Huang B, Song JH, Cheng Y, Abraham JM,
Ibrahim S, Sun Z, Ke X and Meltzer SJ: Long non-coding antisense
RNA KRT7-AS is activated in gastric cancers and supports cancer
cell progression by increasing KRT7 expression. Oncogene.
35:4927–4936. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Zhang L, Yang Z, Trottier J, Barbier O and
Wang L: Long noncoding RNA MEG3 induces cholestatic liver injury by
interaction with PTBP1 to facilitate shp mRNA decay. Hepatology.
65:604–615. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Zhu L, Wei Q, Qi Y, Ruan X, Wu F, Li L,
Zhou J, Liu W, Jiang T, Zhang J, et al: PTB-AS, a novel natural
antisense transcript, promotes glioma progression by improving
PTBP1 mRNA stability with SND1. Mol Ther. 27:1621–1637. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Cao S, Zheng J, Liu X, Liu Y, Ruan X, Ma
J, Liu L, Wang D, Yang C, Cai H, et al: FXR1 promotes the malignant
biological behavior of glioma cells via stabilizing MIR17HG. J Exp
Clin Cancer Res. 38:372019. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Wang Y, Wang Y, Li J, Zhang Y, Yin H and
Han B: CRNDE, a long-noncoding RNA, promotes glioma cell growth and
invasion through mTOR signaling. Cancer Lett. 367:122–128. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Barile L and Vassalli G: Exosomes: Therapy
delivery tools and biomarkers of diseases. Pharmacol Ther.
174:63–78. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Jiang L, Gu Y, Du Y and Liu J: Exosomes:
Diagnostic biomarkers and therapeutic delivery vehicles for cancer.
Mol Pharm. 16:3333–3349. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
D'Asti E, Chennakrishnaiah S, Lee TH and
Rak J: Extracellular vesicles in brain tumor progression. Cell Mol
Neurobiol. 36:383–407. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Katsuda T, Kosaka N and Ochiya T: The
roles of extracellular vesicles in cancer biology: Toward the
development of novel cancer biomarkers. Proteomics. 14:412–425.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Vader P, Breakefield XO and Wood MJ:
Extracellular vesicles: Emerging targets for cancer therapy. Trends
Mol Med. 20:385–393. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Jabbari N, Akbariazar E, Feqhhi M,
Rahbarghazi R and Rezaie J: Breast cancer-derived exosomes: Tumor
progression and therapeutic agents. J Cell Physiol. 235:6345–6356.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Lorenc T, Klimczyk K, Michalczewska I,
Słomka M, Kubiak-Tomaszewska G and Olejarz W: Exosomes in prostate
cancer diagnosis, prognosis and therapy. Int J Mol Sci.
21:21182020. View Article : Google Scholar
|
|
106
|
Sun W, Ren Y, Lu Z and Zhao X: The
potential roles of exosomes in pancreatic cancer initiation and
metastasis. Mol Cancer. 19:1352020. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Zhu L, Li J, Gong Y, Wu Q, Tan S, Sun D,
Xu X, Zuo Y, Zhao Y, Wei YQ, et al: Exosomal tRNA-derived small RNA
as a promising biomarker for cancer diagnosis. Mol Cancer.
18:742019. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Shah S, Wittmann S, Kilchert C and
Vasiljeva L: lncRNA recruits RNAi and the exosome to dynamically
regulate pho1 expression in response to phosphate levels in fission
yeast. Genes Dev. 28:231–244. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Han M, Gu Y, Lu P, Li J, Cao H, Li X, Qian
X, Yu C, Yang Y, Yang X, et al: Exosome-mediated lncRNA AFAP1-AS1
promotes trastuzumab resistance through binding with AUF1 and
activating ERBB2 translation. Mol Cancer. 19:262020. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Zhang Z, Yin J, Lu C, Wei Y, Zeng A and
You Y: Exosomal transfer of long non-coding RNA SBF2-AS1 enhances
chemoresistance to temozolomide in glioblastoma. J Exp Clin Cancer
Res. 38:1662019. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Yuan Z, Yang Z, Li W, Wu A, Su Z and Jiang
B: Exosome-mediated transfer of long noncoding RNA HOTAIR regulates
temozolomide resistance by miR-519a-3p/RRM1 axis in glioblastoma.
Cancer Biother Radiopharm. Jul 24–2020.(Epub ahead of print). doi:
10.1089/cbr.2019.3499. View Article : Google Scholar
|
|
112
|
Li LJ, Leng RX, Fan YG, Pan HF and Ye DQ:
Translation of noncoding RNAs: Focus on lncRNAs, pri-miRNAs, and
circRNAs. Exp Cell Res. 361:1–8. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Wu P, Mo Y, Peng M, Tang T, Zhong Y, Deng
X, Xiong F, Guo C, Wu X, Li Y, et al: Emerging role of
tumor-related functional peptides encoded by lncRNA and circRNA.
Mol Cancer. 19:222020. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Wang Y, Wu S, Zhu X, Zhang L, Deng J, Li
F, Guo B, Zhang S, Wu R, Zhang Z, et al: lncRNA-encoded polypeptide
ASRPS inhibits triple-negative breast cancer angiogenesis. J Exp
Med. 217:jem.20190950. 2020. View Article : Google Scholar
|
|
115
|
Begum S, Yiu A, Stebbing J and Castellano
L: Novel tumour suppressive protein encoded by circular RNA,
circ-SHPRH, in glioblastomas. Oncogene. 37:4055–4057. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Zhang M, Huang N, Yang X, Luo J, Yan S,
Xiao F, Chen W, Gao X, Zhao K, Zhou H, et al: A novel protein
encoded by the circular form of the SHPRH gene suppresses glioma
tumorigenesis. Oncogene. 37:1805–1814. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Zhang M, Zhao K, Xu X, Yang Y, Yan S, Wei
P, Liu H, Xu J, Xiao F, Zhou H, et al: A peptide encoded by
circular form of LINC-PINT suppresses oncogenic transcriptional
elongation in glioblastoma. Nat Commun. 9:44752018. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Matsui M and Corey DR: Non-coding RNAs as
drug targets. Nat Rev Drug Discov. 16:167–179. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Özeş AR, Wang Y, Zong X, Fang F, Pilrose J
and Nephew KP: Therapeutic targeting using tumor specific peptides
inhibits long non-coding RNA HOTAIR activity in ovarian and breast
cancer. Sci Rep. 7:8942017. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Bullock MD, Silva AM, Kanlikilicer-Unaldi
P, Filant J, Rashed MH, Sood AK, Lopez-Berestein G and Calin GA:
Exosomal non-coding RNAs: Diagnostic, prognostic and therapeutic
applications in cancer. Noncoding RNA. 1:53–68. 2015.PubMed/NCBI
|
|
121
|
H Rashed M, Bayraktar E, K Helal G,
Abd-Ellah MF, Amero P, Chavez-Reyes A and Rodriguez-Aguayo C:
Exosomes: From garbage bins to promising therapeutic targets. Int J
Mol Sci. 18:5382017. View Article : Google Scholar
|