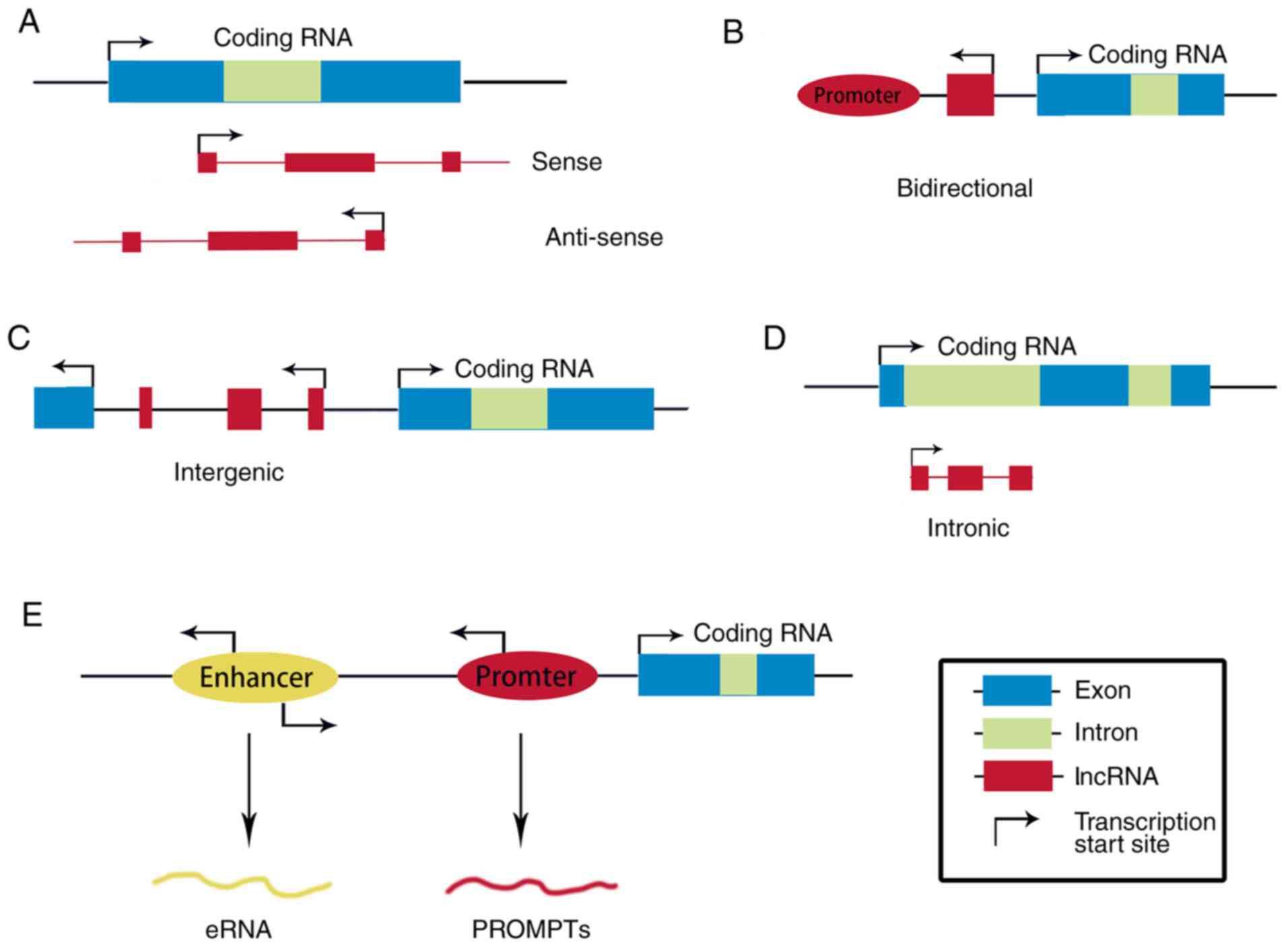
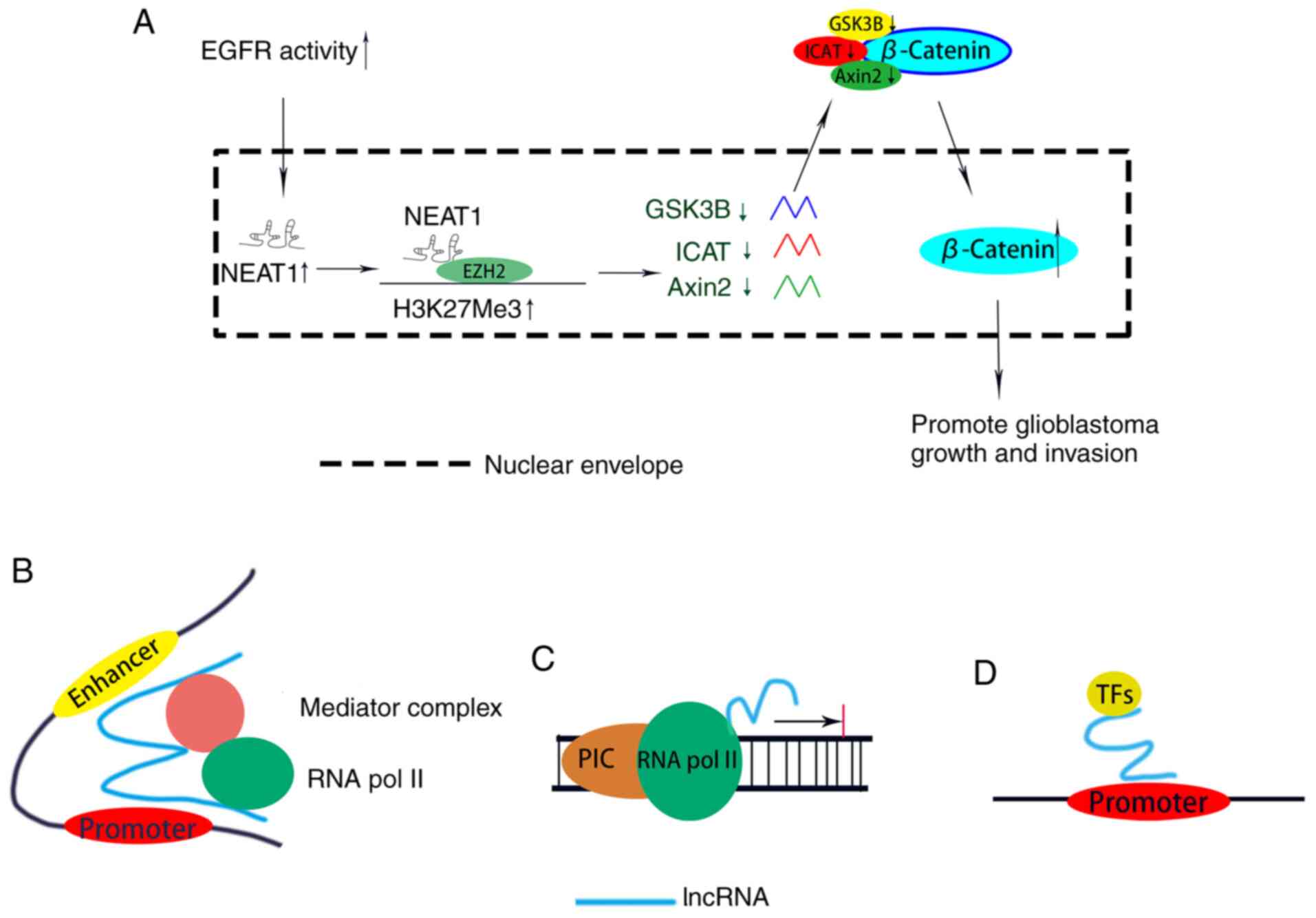
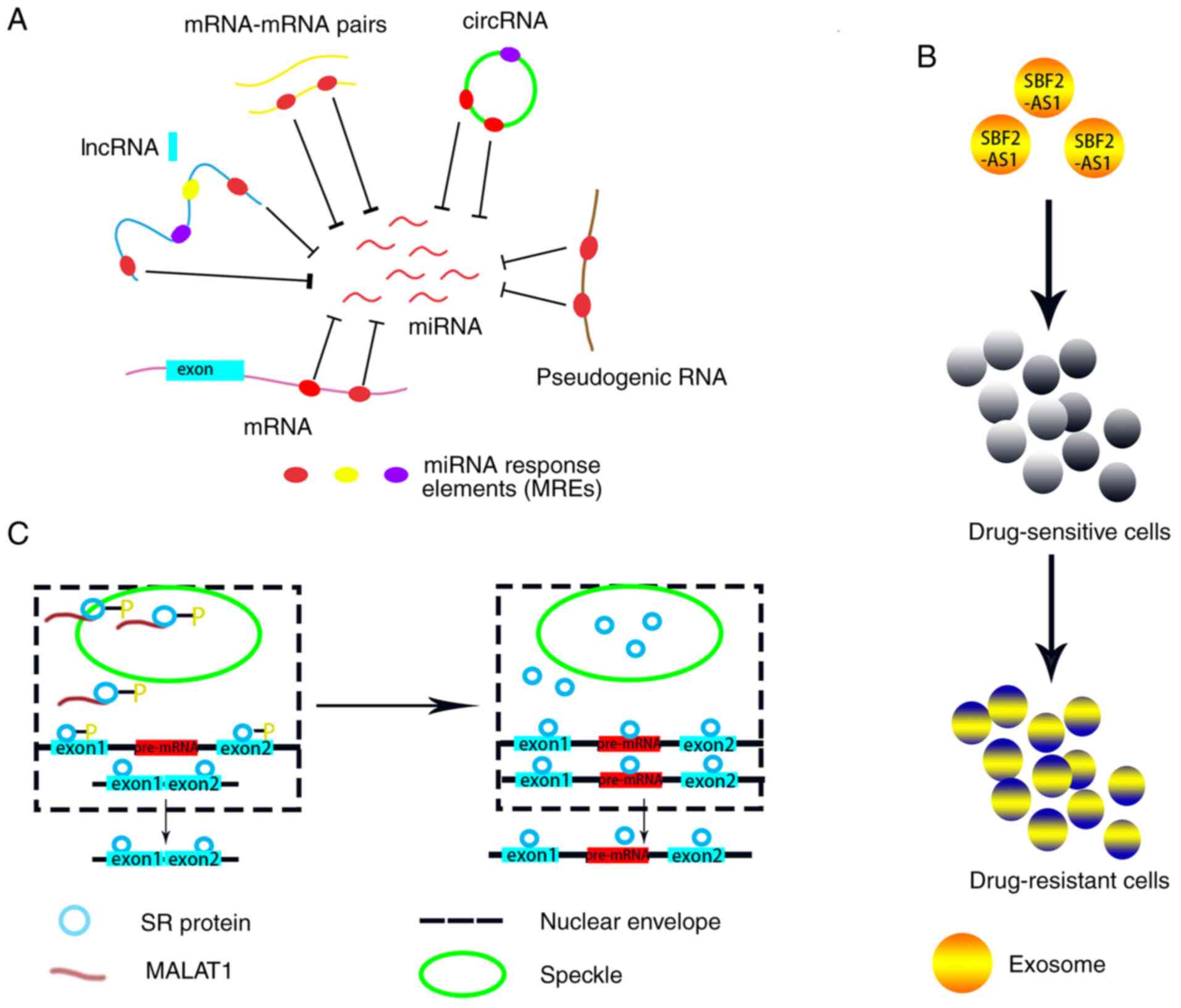
|
1
|
Louis DN, Perry A, Reifenberger G, von
Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD,
Kleihues P and Ellison DW: The 2016 world health organization
classification of tumors of the central nervous system: A summary.
Acta Neuropathol. 131:803–820. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Reni M, Mazza E, Zanon S, Gatta G and
Vecht CJ: Central nervous system gliomas. Crit Rev Oncol Hematol.
113:213–234. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen R, Smith-Cohn M, Cohen AL and Colman
H: Glioma subclassifications and their clinical significance.
Neurotherapeutics. 14:284–297. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Alexander BM and Cloughesy TF: Adult
glioblastoma. J Clin Oncol. 35:2402–2409. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Louis DN, Ohgaki H, Wiestler OD, Cavenee
WK, Burger PC, Jouvet A, Scheithauer BW and Kleihues P: The 2007
WHO classification of tumours of the central nervous system. Acta
Neuropathol. 114:97–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Taylor OG, Brzozowski JS and Skelding KA:
Glioblastoma multiforme: An overview of emerging therapeutic
targets. Front Oncol. 9:9632019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cheng J, Meng J, Zhu L and Peng Y:
Exosomal noncoding RNAs in Glioma: Biological functions and
potential clinical applications. Mol Cancer. 19:662020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ulitsky I and Bartel DP: lincRNAs:
Genomics, evolution, and mechanisms. Cell. 154:26–46. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lagarde J, Uszczynska-Ratajczak B,
Santoyo-Lopez J, Gonzalez JM, Tapanari E, Mudge JM, Steward CA,
Wilming L, Tanzer A, Howald C, et al: Extension of human lncRNA
transcripts by RACE coupled with long-read high-throughput
sequencing (RACE-Seq). Nat Commun. 7:123392016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Deveson IW, Hardwick SA, Mercer TR and
Mattick JS: The dimensions, dynamics, and relevance of the
mammalian noncoding transcriptome. Trends Genet. 33:464–478. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bonasio R and Shiekhattar R: Regulation of
transcription by long noncoding RNAs. Annu Rev Genet. 48:433–455.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yan Y, Xu Z, Li Z, Sun L and Gong Z: An
insight into the increasing role of lncRNAs in the pathogenesis of
gliomas. Front Mol Neurosci. 10:532017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Engreitz JM, Haines JE, Perez EM, Munson
G, Chen J, Kane M, McDonel PE, Guttman M and Lander ES: Local
regulation of gene expression by lncRNA promoters, transcription
and splicing. Nature. 539:452–455. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Quinn JJ and Chang HY: Unique features of
long non-coding RNA biogenesis and function. Nat Rev Genet.
17:47–62. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huang CK, Kafert-Kasting S and Thum T:
Preclinical and clinical development of noncoding RNA therapeutics
for cardiovascular disease. Circ Res. 126:663–678. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kato M and Natarajan R: Epigenetics and
epigenomics in diabetic kidney disease and metabolic memory. Nat
Rev Nephrol. 15:327–345. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Atianand MK, Caffrey DR and Fitzgerald KA:
Immunobiology of long noncoding RNAs. Annu Rev Immunol. 35:177–198.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cheng Z, Li Z, Ma K, Li X, Tian N, Duan J,
Xiao X and Wang Y: Long non-coding RNA XIST promotes glioma
tumorigenicity and angiogenesis by acting as a molecular sponge of
miR-429. J Cancer. 8:4106–4116. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Qian X, Zhao J, Yeung PY, Zhang QC and
Kwok CK: Revealing lncRNA structures and interactions by
sequencing-based approaches. Trends Biochem Sci. 44:33–52. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mercer TR and Mattick JS: Structure and
function of long noncoding RNAs in epigenetic regulation. Nat
Struct Mol Biol. 20:300–307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Schmitt AM and Chang HY: Long noncoding
RNAs in cancer pathways. Cancer Cell. 29:452–463. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Rinn JL and Chang HY: Genome regulation by
long noncoding RNAs. Annu Rev Biochem. 81:145–166. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
St Laurent G, Wahlestedt C and Kapranov P:
The Landscape of long noncoding RNA classification. Trends Genet.
31:239–251. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sigova AA, Mullen AC, Molinie B, Gupta S,
Orlando DA, Guenther MG, Almada AE, Lin C, Sharp PA, Giallourakis
CC and Young RA: Divergent transcription of long noncoding RNA/mRNA
gene pairs in embryonic stem cells. Proc Natl Acad Sci USA.
110:2876–2881. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ørom UA, Derrien T, Beringer M, Gumireddy
K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q,
et al: Long noncoding RNAs with enhancer-like function in human
cells. Cell. 143:46–58. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
de Santa F, Barozzi I, Mietton F,
Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei CL and
Natoli G: A large fraction of extragenic RNA pol II transcription
sites overlap enhancers. PLoS Biol. 8:e10003842010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Guttman M, Amit I, Garber M, French C, Lin
MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al:
Chromatin signature reveals over a thousand highly conserved large
non-coding RNAs in mammals. Nature. 458:223–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ulitsky I, Shkumatava A, Jan CH, Sive H
and Bartel DP: Conserved function of lincRNAs in vertebrate
embryonic development despite rapid sequence evolution. Cell.
147:1537–1550. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Fitz J, Neumann T, Steininger M, Wiedemann
EM, Garcia AC, Athanasiadis A, Schoeberl UE and Pavri R:
Spt5-mediated enhancer transcription directly couples enhancer
activation with physical promoter interaction. Nat Genet.
52:505–515. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cabili MN, Trapnell C, Goff L, Koziol M,
Tazon-Vega B, Regev A and Rinn JL: Integrative annotation of human
large intergenic noncoding RNAs reveals global properties and
specific subclasses. Genes Dev. 25:1915–1927. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Peng Z, Liu C and Wu M: New insights into
long noncoding RNAs and their roles in glioma. Mol Cancer.
17:612018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Han L, Zhang K, Shi Z, Zhang J, Zhu J, Zhu
S, Zhang A, Jia Z, Wang G, Yu S, et al: lncRNA profile of
glioblastoma reveals the potential role of lncRNAs in contributing
to glioblastoma pathogenesis. Int J Oncol. 40:2004–2012.
2012.PubMed/NCBI
|
|
33
|
Chen G, Cao Y, Zhang L, Ma H, Shen C and
Zhao J: Analysis of long non-coding RNA expression profiles
identifies novel lncRNA biomarkers in the tumorigenesis and
malignant progression of gliomas. Oncotarget. 8:67744–67753. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li Q, Jia H, Li H, Dong C, Wang Y and Zou
Z: lncRNA and mRNA expression profiles of glioblastoma multiforme
(GBM) reveal the potential roles of lncRNAs in GBM pathogenesis.
Tumour Biol. 37:14537–14552. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xi J, Sun Q, Ma L and Kang J: Long
non-coding RNAs in glioma progression. Cancer Lett. 419:203–209.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Malissovas N, Ninou E, Michail A and
Politis PK: Targeting long non-coding RNAs in nervous system
cancers: New insights in prognosis, diagnosis and therapy. Curr Med
Chem. 26:5649–5663. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen Q, Cai J, Wang Q, Wang Y, Liu M, Yang
J, Zhou J, Kang C, Li M and Jiang C: Long noncoding RNA NEAT1,
regulated by the EGFR pathway, contributes to glioblastoma
progression through the WNT/β-catenin pathway by scaffolding EZH2.
Clin Cancer Res. 24:684–695. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Quagliata L, Matter MS, Piscuoglio S,
Arabi L, Ruiz C, Procino A, Kovac M, Moretti F, Makowska Z,
Boldanova T, et al: Long noncoding RNA HOTTIP/HOXA13 expression is
associated with disease progression and predicts outcome in
hepatocellular carcinoma patients. Hepatology. 59:911–923. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wu P, Cai J, Chen Q, Han B, Meng X, Li Y,
Li Z, Wang R, Lin L, Duan C, et al: lnc-TALC promotes
O6-methylguanine-DNA methyltransferase expression via
regulating the c-Met pathway by competitively binding with
miR-20b-3p. Nat Commun. 10:20452019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ghafouri-Fard S and Taheri M: Nuclear
enriched abundant transcript 1 (NEAT1): A long non-coding RNA with
diverse functions in tumorigenesis. Biomed Pharmacother. 111:51–59.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhao W, Li W, Jin X, Niu T, Cao Y, Zhou P
and Zheng M: Silencing long non-coding RNA NEAT1 enhances the
suppression of cell growth, invasion, and apoptosis of bladder
cancer cells under cisplatin chemotherapy. Int J Clin Exp Pathol.
12:549–558. 2019.PubMed/NCBI
|
|
42
|
Wu Y, Yang L, Zhao J, Li C, Nie J, Liu F,
Zhuo C, Zheng Y, Li B, Wang Z and Xu Y: Nuclear-enriched abundant
transcript 1 as a diagnostic and prognostic biomarker in colorectal
cancer. Mol Cancer. 14:1912015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Fujimoto A, Furuta M, Totoki Y, Tsunoda T,
Kato M, Shiraishi Y, Tanaka H, Taniguchi H, Kawakami Y, Ueno M, et
al: Whole-genome mutational landscape and characterization of
noncoding and structural mutations in liver cancer. Nat Genet.
48:500–509. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Jiang X, Zhou Y, Sun AJ and Xue JL: NEAT1
contributes to breast cancer progression through modulating miR-448
and ZEB1. J Cell Physiol. 233:8558–8566. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Jho EH, Zhang T, Domon C, Joo CK, Freund
JN and Costantini F: Wnt/beta-catenin/Tcf signaling induces the
transcription of Axin2, a negative regulator of the signaling
pathway. Mol Cell Biol. 22:1172–1183. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Tago K, Nakamura T, Nishita M, Hyodo J,
Nagai S, Murata Y, Adachi S, Ohwada S, Morishita Y, Shibuya H and
Akiyama T: Inhibition of Wnt signaling by ICAT, a novel
beta-catenin-interacting protein. Genes Dev. 14:1741–1749.
2000.PubMed/NCBI
|
|
47
|
Pandey GK, Mitra S, Subhash S, Hertwig F,
Kanduri M, Mishra K, Fransson S, Ganeshram A, Mondal T, Bandaru S,
et al: The risk-associated long noncoding RNA NBAT-1 controls
neuroblastoma progression by regulating cell proliferation and
neuronal differentiation. Cancer Cell. 26:722–737. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhang F, Ruan X, Ma J, Liu X, Zheng J, Liu
Y, Liu L, Shen S, Shao L, Wang D, et al: DGCR8/ZFAT-AS1 promotes
CDX2 transcription in a PRC2 complex-dependent manner to facilitate
the malignant biological behavior of glioma cells. Mol Ther.
28:613–630. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
de Klerk E and t Hoen PA: Alternative mRNA
transcription, processing, and translation: Insights from RNA
sequencing. Trends Genet. 31:128–139. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ong CT and Corces VG: Enhancer function:
New insights into the regulation of tissue-specific gene
expression. Nat Rev Genet. 12:283–293. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Field A and Adelman K: Evaluating enhancer
function and transcription. Annu Rev Biochem. 89:213–234. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Kim TK, Hemberg M, Gray JM, Costa AM, Bear
DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et
al: Widespread transcription at neuronal activity-regulated
enhancers. Nature. 465:182–187. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Mousavi K, Zare H, Dell'orso S, Grontved
L, Gutierrez-Cruz G, Derfoul A, Hager GL and Sartorelli V: eRNAs
promote transcription by establishing chromatin accessibility at
defined genomic loci. Mol Cell. 51:606–617. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Li W, Notani D, Ma Q, Tanasa B, Nunez E,
Chen AY, Merkurjev D, Zhang J, Ohgi K, Song X, et al: Functional
roles of enhancer RNAs for oestrogen-dependent transcriptional
activation. Nature. 498:516–520. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Ørom UA, Derrien T, Guigo R and
Shiekhattar R: Long noncoding RNAs as enhancers of gene expression.
Cold Spring Harb Symp Quant Biol. 75:325–331. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Dykes IM and Emanueli C: Transcriptional
and post-transcriptional gene regulation by long non-coding RNA.
Genomics Proteomics Bioinformatics. 15:177–186. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Long Y, Wang X, Youmans DT and Cech TR:
How do lncRNAs regulate transcription? Sci Adv. 3:eaao21102017.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zovoilis A, Cifuentes-Rojas C, Chu HP,
Hernandez AJ and Lee JT: Destabilization of B2 RNA by EZH2
activates the stress response. Cell. 167:1788–1802.e13. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Hernandez AJ, Zovoilis A, Cifuentes-Rojas
C, Han L, Bujisic B and Lee JT: B2 and ALU retrotransposons are
self-cleaving ribozymes whose activity is enhanced by EZH2. Proc
Natl Acad Sci USA. 117:415–425. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Espinoza CA, Allen TA, Hieb AR, Kugel JF
and Goodrich JA: B2 RNA binds directly to RNA polymerase II to
repress transcript synthesis. Nat Struct Mol Biol. 11:822–829.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Lambert SA, Jolma A, Campitelli LF, Das
PK, Yin Y, Albu M, Chen X, Taipale J, Hughes TR and Weirauch MT:
The human transcription factors. Cell. 172:650–665. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Hudson WH and Ortlund EA: The structure,
function and evolution of proteins that bind DNA and RNA. Nat Rev
Mol Cell Biol. 15:749–760. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Hung T, Wang Y, Lin MF, Koegel AK, Kotake
Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P, et al:
Extensive and coordinated transcription of noncoding RNAs within
cell-cycle promoters. Nat Genet. 43:621–629. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Xu H, Zhao G, Zhang Y, Jiang H, Wang W,
Zhao D, Yu H and Qi L: Long non-coding RNA PAXIP1-AS1 facilitates
cell invasion and angiogenesis of glioma by recruiting
transcription factor ETS1 to upregulate KIF14 expression. J Exp
Clin Cancer Res. 38:4862019. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Schneider C, King RM and Philipson L:
Genes specifically expressed at growth arrest of mammalian cells.
Cell. 54:787–793. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Yuan J, Zhang N, Zheng Y, Chen YD, Liu J
and Yang M: lncRNA GAS5 indel genetic polymorphism contributes to
glioma risk through interfering binding of transcriptional factor
TFAP2A. DNA Cell Biol. 37:750–757. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Qi X, Zhang DH, Wu N, Xiao JH, Wang X and
Ma W: ceRNA in cancer: Possible functions and clinical
implications. J Med Genet. 52:710–718. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Amirkhah R, Naderi-Meshkin H, Shah JS,
Dunne PD and Schmitz U: The intricate interplay between epigenetic
events, alternative splicing and noncoding RNA deregulation in
colorectal cancer. Cells. 8:9292019. View Article : Google Scholar
|
|
69
|
Zhang XZ, Liu H and Chen SR: Mechanisms of
long non-coding RNAs in cancers and their dynamic regulations.
Cancers (Basel). 12:12452020. View Article : Google Scholar
|
|
70
|
Li Y, Yin Z, Fan J, Zhang S and Yang W:
The roles of exosomal miRNAs and lncRNAs in lung diseases. Signal
Transduct Target Ther. 4:472019. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Cesana M, Cacchiarelli D, Legnini I,
Santini T, Sthandier O, Chinappi M, Tramontano A and Bozzoni I: A
long noncoding RNA controls muscle differentiation by functioning
as a competing endogenous RNA. Cell. 147:358–369. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Sa L, Li Y, Zhao L, Liu Y, Wang P, Liu L,
Li Z, Ma J, Cai H and Xue Y: The Role of HOTAIR/miR-148b-3p/USF1 on
regulating the permeability of BTB. Front Mol Neurosci. 10:1942017.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Qin N, Tong GF, Sun LW and Xu XL: Long
noncoding RNA MEG3 suppresses glioma cell proliferation, migration,
and invasion by acting as a competing endogenous RNA of miR-19a.
Oncol Res. 25:1471–1478. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Li DX, Fei XR, Dong YF, Cheng CD, Yang Y,
Deng XF, Huang HL, Niu WX, Zhou CX, Xia CY and Niu CS: The long
non-coding RNA CRNDE acts as a ceRNA and promotes glioma malignancy
by preventing miR-136-5p-mediated downregulation of Bcl-2 and Wnt2.
Oncotarget. 8:88163–88178. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Zhang X, Sun S, Pu JK, Tsang AC, Lee D,
Man VO, Lui WM, Wong ST and Leung GK: Long non-coding RNA
expression profiles predict clinical phenotypes in glioma.
Neurobiol Dis. 48:1–8. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Zheng J, Li XD, Wang P, Liu XB, Xue YX, Hu
Y, Li Z, Li ZQ, Wang ZH and Liu YH: CRNDE affects the malignant
biological characteristics of human glioma stem cells by negatively
regulating miR-186. Oncotarget. 6:25339–25355. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Zheng J, Liu X, Wang P, Xue Y, Ma J, Qu C
and Liu Y: CRNDE promotes malignant progression of glioma by
attenuating miR-384/PIWIL4/STAT3 axis. Mol Ther. 24:1199–1215.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Brockdorff N, Bowness JS and Wei G:
Progress toward understanding chromosome silencing by Xist RNA.
Genes Dev. 34:733–744. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Cheng Z, Luo C and Guo Z:
lncRNA-XIST/microRNA-126 sponge mediates cell proliferation and
glucose metabolism through the IRS1/PI3K/Akt pathway in glioma. J
Cell Biochem. 121:2170–2183. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Ule J and Blencowe BJ: Alternative
splicing regulatory networks: Functions, mechanisms, and evolution.
Mol Cell. 76:329–345. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Shepard PJ and Hertel KJ: The SR protein
family. Genome Biol. 10:2422009. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Blencowe BJ: Alternative splicing: New
insights from global analyses. Cell. 126:37–47. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Sun Y and Ma L: New insights into long
non-coding RNA MALAT1 in cancer and metastasis. Cancers (Basel).
11:2162019. View Article : Google Scholar
|
|
85
|
Ji P, Diederichs S, Wang W, Böing S,
Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et
al: MALAT-1, a novel noncoding RNA, and thymosin beta4 predict
metastasis and survival in early-stage non-small cell lung cancer.
Oncogene. 22:8031–8041. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Arun G and Spector DL: MALAT1 long
non-coding RNA and breast cancer. RNA Biol. 16:860–863. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Chang J, Xu W, Du X and Hou J: MALAT1
silencing suppresses prostate cancer progression by upregulating
miR-1 and downregulating KRAS. Onco Targets Ther. 11:3461–3473.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Le L, Chen H, Gao Y, Wang YW, Zhang GQ,
Pan SH, Ji L, Kong R, Wang G, Jia YH, et al: Long noncoding RNA
MALAT1 promotes aggressive pancreatic cancer proliferation and
metastasis via the stimulation of autophagy. Mol Cancer Ther.
15:2232–2243. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Liao K, Lin Y, Gao W, Xiao Z, Medina R,
Dmitriev P, Cui J, Zhuang Z, Zhao X, Qiu Y, et al: Blocking lncRNA
MALAT1/miR-199a/ZHX1 axis inhibits glioblastoma proliferation and
progression. Mol Ther Nucleic Acids. 18:388–399. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Wen F, Cao YX, Luo ZY, Liao P and Lu ZW:
lncRNA MALAT1 promotes cell proliferation and imatinib resistance
by sponging miR-328 in chronic myelogenous leukemia. Biochem
Biophys Res Commun. 507:1–8. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Tripathi V, Ellis JD, Shen Z, Song DY, Pan
Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al: The
nuclear-retained noncoding RNA MALAT1 regulates alternative
splicing by modulating SR splicing factor phosphorylation. Mol
Cell. 39:925–938. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Long JC and Caceres JF: The SR protein
family of splicing factors: Master regulators of gene expression.
Biochem J. 417:15–27. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Stamm S: Regulation of alternative
splicing by reversible protein phosphorylation. J Biol Chem.
283:1223–1227. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Huang B, Song JH, Cheng Y, Abraham JM,
Ibrahim S, Sun Z, Ke X and Meltzer SJ: Long non-coding antisense
RNA KRT7-AS is activated in gastric cancers and supports cancer
cell progression by increasing KRT7 expression. Oncogene.
35:4927–4936. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Zhang L, Yang Z, Trottier J, Barbier O and
Wang L: Long noncoding RNA MEG3 induces cholestatic liver injury by
interaction with PTBP1 to facilitate shp mRNA decay. Hepatology.
65:604–615. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Zhu L, Wei Q, Qi Y, Ruan X, Wu F, Li L,
Zhou J, Liu W, Jiang T, Zhang J, et al: PTB-AS, a novel natural
antisense transcript, promotes glioma progression by improving
PTBP1 mRNA stability with SND1. Mol Ther. 27:1621–1637. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Cao S, Zheng J, Liu X, Liu Y, Ruan X, Ma
J, Liu L, Wang D, Yang C, Cai H, et al: FXR1 promotes the malignant
biological behavior of glioma cells via stabilizing MIR17HG. J Exp
Clin Cancer Res. 38:372019. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Wang Y, Wang Y, Li J, Zhang Y, Yin H and
Han B: CRNDE, a long-noncoding RNA, promotes glioma cell growth and
invasion through mTOR signaling. Cancer Lett. 367:122–128. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Barile L and Vassalli G: Exosomes: Therapy
delivery tools and biomarkers of diseases. Pharmacol Ther.
174:63–78. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Jiang L, Gu Y, Du Y and Liu J: Exosomes:
Diagnostic biomarkers and therapeutic delivery vehicles for cancer.
Mol Pharm. 16:3333–3349. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
D'Asti E, Chennakrishnaiah S, Lee TH and
Rak J: Extracellular vesicles in brain tumor progression. Cell Mol
Neurobiol. 36:383–407. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Katsuda T, Kosaka N and Ochiya T: The
roles of extracellular vesicles in cancer biology: Toward the
development of novel cancer biomarkers. Proteomics. 14:412–425.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Vader P, Breakefield XO and Wood MJ:
Extracellular vesicles: Emerging targets for cancer therapy. Trends
Mol Med. 20:385–393. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Jabbari N, Akbariazar E, Feqhhi M,
Rahbarghazi R and Rezaie J: Breast cancer-derived exosomes: Tumor
progression and therapeutic agents. J Cell Physiol. 235:6345–6356.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Lorenc T, Klimczyk K, Michalczewska I,
Słomka M, Kubiak-Tomaszewska G and Olejarz W: Exosomes in prostate
cancer diagnosis, prognosis and therapy. Int J Mol Sci.
21:21182020. View Article : Google Scholar
|
|
106
|
Sun W, Ren Y, Lu Z and Zhao X: The
potential roles of exosomes in pancreatic cancer initiation and
metastasis. Mol Cancer. 19:1352020. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Zhu L, Li J, Gong Y, Wu Q, Tan S, Sun D,
Xu X, Zuo Y, Zhao Y, Wei YQ, et al: Exosomal tRNA-derived small RNA
as a promising biomarker for cancer diagnosis. Mol Cancer.
18:742019. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Shah S, Wittmann S, Kilchert C and
Vasiljeva L: lncRNA recruits RNAi and the exosome to dynamically
regulate pho1 expression in response to phosphate levels in fission
yeast. Genes Dev. 28:231–244. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Han M, Gu Y, Lu P, Li J, Cao H, Li X, Qian
X, Yu C, Yang Y, Yang X, et al: Exosome-mediated lncRNA AFAP1-AS1
promotes trastuzumab resistance through binding with AUF1 and
activating ERBB2 translation. Mol Cancer. 19:262020. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Zhang Z, Yin J, Lu C, Wei Y, Zeng A and
You Y: Exosomal transfer of long non-coding RNA SBF2-AS1 enhances
chemoresistance to temozolomide in glioblastoma. J Exp Clin Cancer
Res. 38:1662019. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Yuan Z, Yang Z, Li W, Wu A, Su Z and Jiang
B: Exosome-mediated transfer of long noncoding RNA HOTAIR regulates
temozolomide resistance by miR-519a-3p/RRM1 axis in glioblastoma.
Cancer Biother Radiopharm. Jul 24–2020.(Epub ahead of print). doi:
10.1089/cbr.2019.3499. View Article : Google Scholar
|
|
112
|
Li LJ, Leng RX, Fan YG, Pan HF and Ye DQ:
Translation of noncoding RNAs: Focus on lncRNAs, pri-miRNAs, and
circRNAs. Exp Cell Res. 361:1–8. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Wu P, Mo Y, Peng M, Tang T, Zhong Y, Deng
X, Xiong F, Guo C, Wu X, Li Y, et al: Emerging role of
tumor-related functional peptides encoded by lncRNA and circRNA.
Mol Cancer. 19:222020. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Wang Y, Wu S, Zhu X, Zhang L, Deng J, Li
F, Guo B, Zhang S, Wu R, Zhang Z, et al: lncRNA-encoded polypeptide
ASRPS inhibits triple-negative breast cancer angiogenesis. J Exp
Med. 217:jem.20190950. 2020. View Article : Google Scholar
|
|
115
|
Begum S, Yiu A, Stebbing J and Castellano
L: Novel tumour suppressive protein encoded by circular RNA,
circ-SHPRH, in glioblastomas. Oncogene. 37:4055–4057. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Zhang M, Huang N, Yang X, Luo J, Yan S,
Xiao F, Chen W, Gao X, Zhao K, Zhou H, et al: A novel protein
encoded by the circular form of the SHPRH gene suppresses glioma
tumorigenesis. Oncogene. 37:1805–1814. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Zhang M, Zhao K, Xu X, Yang Y, Yan S, Wei
P, Liu H, Xu J, Xiao F, Zhou H, et al: A peptide encoded by
circular form of LINC-PINT suppresses oncogenic transcriptional
elongation in glioblastoma. Nat Commun. 9:44752018. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Matsui M and Corey DR: Non-coding RNAs as
drug targets. Nat Rev Drug Discov. 16:167–179. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Özeş AR, Wang Y, Zong X, Fang F, Pilrose J
and Nephew KP: Therapeutic targeting using tumor specific peptides
inhibits long non-coding RNA HOTAIR activity in ovarian and breast
cancer. Sci Rep. 7:8942017. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Bullock MD, Silva AM, Kanlikilicer-Unaldi
P, Filant J, Rashed MH, Sood AK, Lopez-Berestein G and Calin GA:
Exosomal non-coding RNAs: Diagnostic, prognostic and therapeutic
applications in cancer. Noncoding RNA. 1:53–68. 2015.PubMed/NCBI
|
|
121
|
H Rashed M, Bayraktar E, K Helal G,
Abd-Ellah MF, Amero P, Chavez-Reyes A and Rodriguez-Aguayo C:
Exosomes: From garbage bins to promising therapeutic targets. Int J
Mol Sci. 18:5382017. View Article : Google Scholar
|