Introduction
Breast cancer (BC) is the leading cause of
cancer-related deaths among women worldwide (1). The incidence and mortality rates of BC
also rank first among female malignant tumours, posing a serious
threat to the health and lives of women (2). Although great progress has been made in
the treatment of BC in recent years, the overall survival rate of
BC patients has not improved significantly due to recurrence and
metastasis, and the incidence of BC in China has been on the rise
in the past decade (3). Therefore,
increasing attention has been given to exploring relevant potential
molecular markers to further improve the understanding of the
pattern of BC proliferation and to develop future treatment
measures to control BC progression.
As an important member of the matrix
metalloproteinase (MMP) family, MMP11 regulates a series of
physiological processes and signalling events, manipulates some
biologically active molecules on the cell surface, and alters the
biological behaviour of cells, playing an important role in the
tumour microenvironment (4,5). Moreover, accumulated studies have
revealed that MMPs are closely related to tumorigenesis and that
the most functionally significant MMP is MMP11, which is
overexpressed in tumours and is involved in the proliferation and
malignant development of tumour cells (6,7). A
previous study revealed that high expression of MMP11 in BC was
associated with a poor prognosis in patients (8). MMP11 was revealed to promote tumour
malignancy by inhibiting apoptosis and by promoting migration and
invasion, but the underlying mechanism remains unclear (9,10).
In the present study, the expression level of MMP11
was first analysed in breast cancer tissues and adjacent tissues
using clinical samples, and the relationship between MMP11 and
clinical prognosis was explored. Furthermore, MTT and colony
formation assays were used to confirm the role of MMP11 on breast
cancer cell proliferation. Functional enrichment analysis and
immunoprecipitation were used to confirm the mechanisms of MMP11 in
the regulation of breast cancer cell proliferation. Finally,
xenograft assays were further performed to demonstrate the role of
MMP11 in breast cancer proliferation and growth in vivo.
Materials and methods
Patients and clinical samples
In total, 17 pairs of BC and paracancer (PC) tissues
were collected from patients (aged 55–65 years old) at Hubei Cancer
Hospital (Wuhan, China) between December 2018 and December 2019. BC
patients did not receive any chemotherapy or endocrine therapy
prior to tumour removal. Immediately after surgery, all tissues
were frozen in liquid nitrogen and maintained at −80°C until
further analysis. In addition, three of the 17 pairs of BC tissues
and their adjacent tissues were randomly selected for mRNA
transcriptome sequencing (Illumina; PE-401-3001; Shanghai GeneChem
Co., Ltd.). All clinical samples were collected with written
informed consent from the patients, and this protocol was approved
by the Ethics Committee of the Hubei Cancer Hospital.
Cell culture
Human breast epithelial cells (MCF10A) and human BC
cell lines (MCF7, MDA231, MDA468 and MX1) were purchased from BeiNa
Bio. MCF10A cells were maintained in RPMI-1640 medium supplemented
with 10% FBS (both from Gibco; Thermo Fisher Scientific, Inc.) and
1% penicillin and streptomycin (ScienCell Research Laboratories,
Inc.) in a humidified incubator with 5% CO2 at 37°C.
MCF7, MDA231, MDA468 and MX1 were maintained in Dulbecco's modified
Eagle's medium (DMEM) (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% FBS and 1% penicillin and streptomycin in a
humidified incubator with 5% CO2 at 37°C. All cell lines
were confirmed to be free of mycoplasma contamination.
Cycloheximide (CHX), MG132 and chloroquine, which were used for
cell-based assays, were purchased from MCE. SRI-011381 (TGF-β
agonist) was purchased from Selleck Chemicals.
Transfection and infection
The MMP11 overexpression (OE) plasmid and
corresponding negative control (NC) plasmid were synthesized by
Shanghai GeneChem Co., Ltd. Smad family member 2 small interfering
(si)RNA (si-Smad2) and the corresponding NC siRNA were purchased
from Guangzhou RiboBio Co., Ltd. The sequences of siRNAs are as
follows: Smad2 sense, 5′-CCAGGUGGUGAAGAAUCUATT-3′, and antisense,
5′-UAGAUUCUUCACCACCUGGTT-3′; NC siRNA sense,
5′-AAAAATTCAAGACUUGGAGCU-3′ and antisense,
5′-UCUUGTTUUUUUAGCUCCAAG-3′. siRNAs at a final concentration of 50
nM and plasmids at a dose of 1.6 µg per well (12-well
plates) were transfected into cells at 37°C for 48 h using
Lipofectamine 3000 reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. Subsequent assays
were carried out after 72 h of transfection. The MMP11 lentivirus
(Lv-sh) and empty control vector (Lv-NC) were constructed by
Shanghai GeneChem Co., Ltd. The aforementioned recombinant
lentiviruses were stably transfected into BC cell lines (MCF7 and
MDA231) with an MOI of 2, and then the transfected cells were used
for subsequent experiments after 3 days of puromycin selection with
a final concentration of 2 µg/ml.
3-(4,5)-Dimethylthiahiazo(-z-y1)-3,5-di-phenyltetrazolium
bromide (MTT) assay for cell proliferation
To assess cell viability, the cells were plated in
96-well plates at a density of 5×103 cells/well, and
then subjected to the MTT assay (Sigma-Aldrich; Merck KGaA). Then,
20 µl of MTT solution (5 mg/ml) was added to each well.
After 4 h of incubation at 37°C, the culture medium in the well was
removed, 150 µl dimethyl sulfoxide (DMSO) was added to each
well, and the optical density was measured at 490 nm. The
experiment was replicated three times independently. Each condition
was plated in triplicate.
Colony formation assay
Seventy-two hours after transfection of siRNA or
MMP11 lentivirus, 500 cells were inoculated into 6-well plates and
grown in complete medium for two weeks. Colonies were fixed with 4%
paraformaldehyde at room temperature for 15 min and stained with
0.1% crystal violet at room temperature for 30 min in 100% ethanol
and then counted and photographed (D850; Nikon Corporation). All
colony formation assays were conducted in triplicate.
Western blot analysis and
immunohistochemistry
Protein lysates of tumour tissues or cultured cells
were prepared using RIPA buffer (Sigma-Aldrich; Merck KGaA)
containing 1% phenylmethanesulfonyl fluoride (PMSF) and phosphatase
inhibitors. Western blot analysis was performed as previously
described (11). The antibodies used
were as follows: MMP11 (product code ab119284; 1:1,000 dilution;
Abcam), Smad2 (cat. no. 12570-1-AP; 1:1,000 dilution), Smad3 (cat.
no. 25494-1-AP; 1:1,000 dilution), and β-actin (cat. no.
20536-1-AP; 1:1,000 dilution) primary antibodies and goat
anti-rabbit IgG secondary antibody (cat. no. SA00001-2; 1:5,000
dilution; all from ProteinTech Group, Inc.). For
immunohistochemistry (IHC), the aforementioned antibodies MMP11 and
Smad2 (1:1,000 dilution) were used. The specific
immunohistochemistry protocol was performed as previously described
(12).
RNA isolation, reverse
transcription-quantitative (RT-qPCR)
Total RNA from tissues or cultured cell lines was
extracted using TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's
instructions. RT-qPCR was carried out using the Prime-Script RT kit
(TaKaRa Biotechnology Co., Ltd.). Gene expression levels were
established by RT-qPCR using SYBR Premix Ex Taq (TaKaRa
Biotechnology Co., Ltd.) and normalized to the endogenous level of
β-actin as a control. The thermocycling conditions were as follows:
Initial denaturation at 95°C for 30 sec, and then the PCR
reactions: 95°C for 5 sec, 60°C for 35 sec, and 60 cycles. The
primers used were as follows: MMP11 forward,
5′-CCATGTAATATCTAGATAAGGTCGGA-3′ and reverse,
5′-GAGTCAAGGTCGGGTGCGTGGGAAG-3′; β-actin forward
5′-CCAAGGCCAACCGCGAGAAGATGAC-3′ and reverse
5′-AGGGTACATGGTGGTGCCGCCAGAC-3′. All these experiments were
performed in triplicate on a Bio-Rad CFX96 instrument (Bio-Rad
Laboratories, Inc.), and the relative expression values were
calculated using the 2−ΔΔCq method (12).
Immunoprecipitation
MCF7 cells were transfected as previously indicated
and then lysed in lysis buffer. The samples were centrifuged at
4000 × g at 4°C for 10 min, and the supernatants were retained.
Then, the supernatants were incubated with anti-MMP11 (1 µg/ml)
or IgG (1 µg/ml; product no. 6990; Cell Signaling
Technology, Inc.) at 4°C for 4 h and protein A/G-Sepharose beads
(10 µl) for 2 h at 4°C. The immunocomplexes were washed
three times, boiled in sample buffer, and subjected to western
blotting with anti-Smad2 antibody.
Xenograft assay
MCF7 cells (3×106/mouse, 5/group)
transfected with Lv-MMP11 or Lv-NC were injected subcutaneously
into the right flank region of male BALB/c nude mice (6 weeks old;
weight, ~15 g; 10 in total). All nude mice were kept in a Specific
Pathogen Free (SPF) environment with controllable light (12-h
light/dark cycle), temperature and humidity, with food and water
available ad libitum. The mice were monitored weekly and the
tumour volume was assessed; the long diameter of the tumour did not
exceed 2 cm. The formula for the tumour volume was
V=LxW2xπ/6 (V, volume; L, long diameter of the
tumour; W, short diameter of the the tumour). Approximately 4 weeks
after injection, according to institutional ethical guidelines, the
mice were anesthetized with isoflurane and sacrificed by humanely
cervical dislocation, and the weights of the tumours were recorded.
The mouse experiments and the handling of animals were performed
according to the Institutional and Animal Care and Use Committee of
Hubei Cancer Hospital and the NIH Guide for the Care and Use of
Laboratory Animals.
Bioinformatics analysis
The R ‘limma’ Bioconductor package was used to
screen the differentially expressed genes (DEGs) between KIRC and
adjacent tissues based on the following criteria: Fold change (FC),
|log2(FC)|>1; and false discovery rate (FDR) <0.05. Adjusted
P<0.05 was used to define a gene as a DEG. An online database
(https://www.UniProt.org) was used to predict
MMP11-related molecules. The Database for Annotation, Visualization
and Integrated Discovery (DAVID) v6.8 (david-d. ncifcrf.gov/) was
used to analyze functional enrichment among DEGs. In addition, only
those Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways with
P≤0.05 and ≥10 enriched genes were considered significant. Online
database analysis (http://kmplot.com) was used to
further investigate the relationship between MMP11 and the
prognosis of BC patients.
Statistical analysis
All statistical analyses were performed using SPSS
22.0 (IBM Corp.), and figures were produced using GraphPad Prism
6.0 (GraphPad Software, Inc). For parametric variables, paired and
unpaired Student's t-test and one-factor ANOVA followed by Tukey's
post hoc test were utilized to analyse the results of the animal
experiments. Survival was assessed using the Kaplan-Meier method
and analysed using the log-rank test. Univariate analyses were
carried out using the Cox proportional hazards model. All
experimental data are expressed as the mean ± SD of at least
five independent experiments. In addition, the BC data set of The
Cancer Genome Atlas (TCGA) (http://cancergenome.nih.gov/) of the University of
North Carolina TCGA Genome Characterization Center was used for
clinical correlation analysis (13).
The differences were considered statistically significant when
P<0.05.
Results
Overexpression of MMP11 in BC
tissues
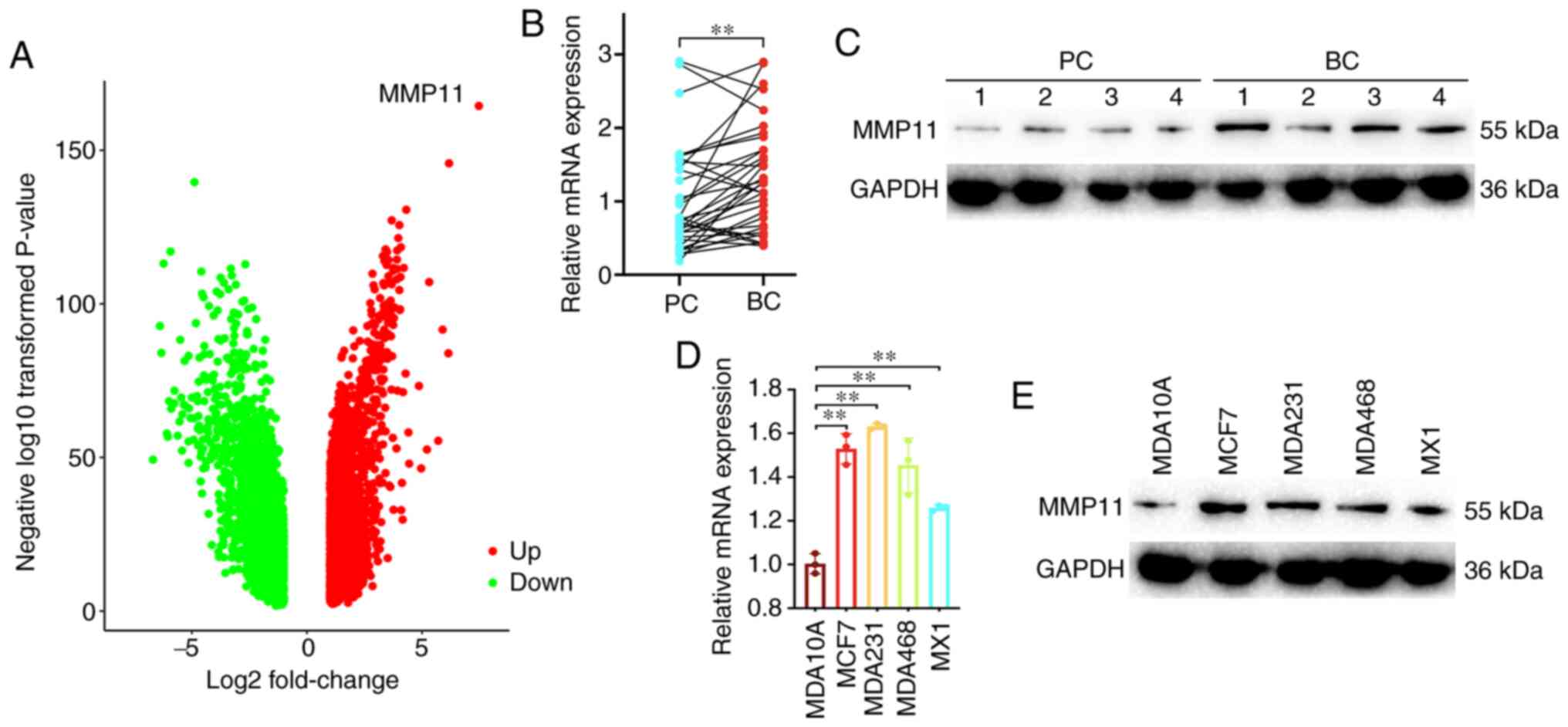
To explore gene expression patterns, transcriptome
sequencing of clinical BC samples and their adjacent tissues was
performed. The sequencing results revealed that MMP11 was the most
upregulated gene (Fig. 1A). To
confirm the gene overexpression of MMP11, 34 paired of BC and
adjacent PC tissues were used. Based on the RT-qPCR analysis, MMP11
mRNA was overexpressed in tumour tissues (Fig. 1B). Western blotting was also performed
in paired BC and PC tissues, and comparison of the expression
levels revealed that MMP11 was highly expressed in tumour tissues
(Fig. 1C). In addition, the
expression level of MMP11 was significantly higher in the four BC
(MCF7, MDA231, MDA468, MX1) cell lines than in the normal human
breast epithelial cell line (MDA10A) (Fig. 1D and E).
MMP11 expression is significantly
associated with BC progression and poor prognosis
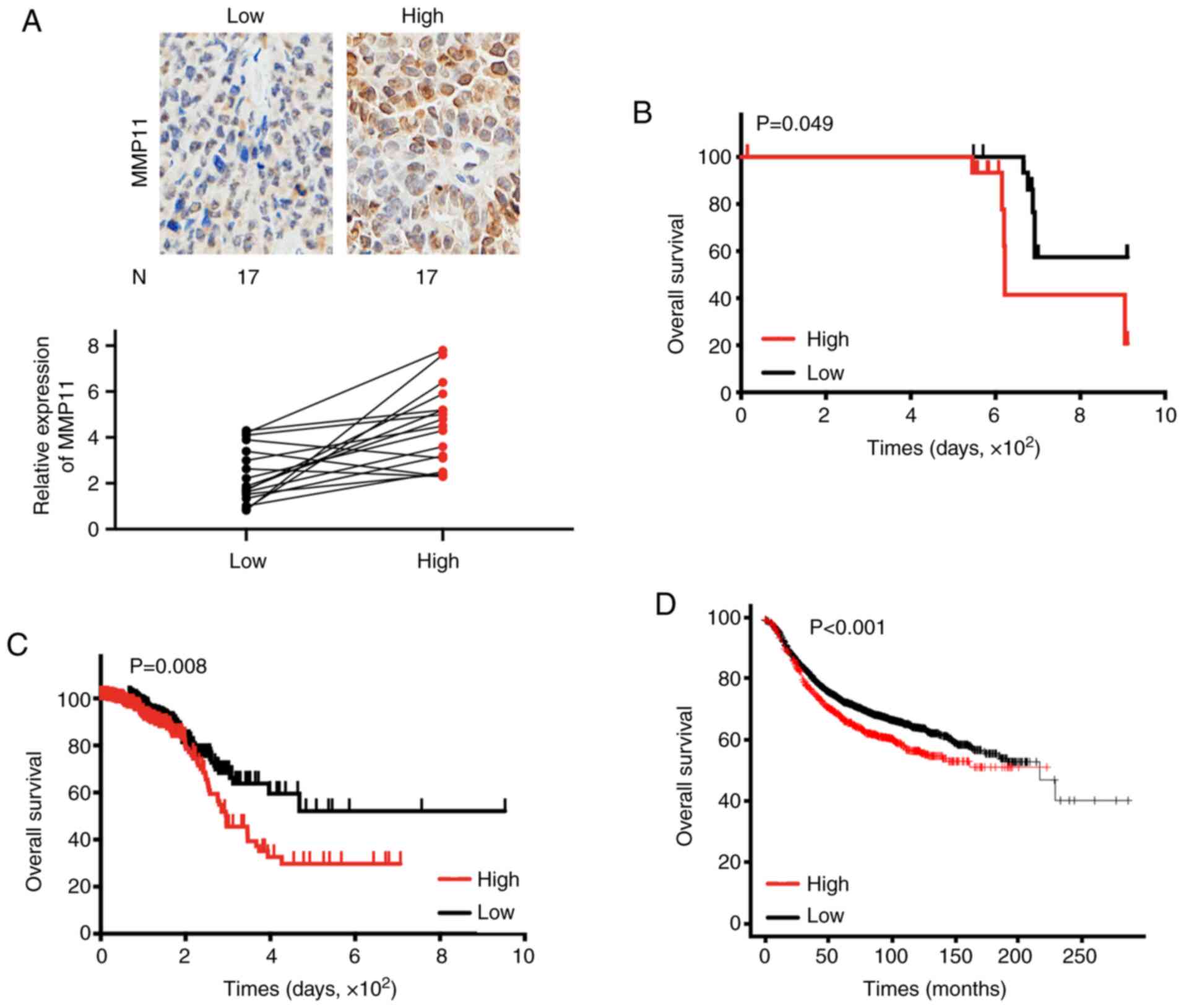
First, 34 BC specimens were assessed by RT-qPCR and
IHC, and it was revealed that the mRNA and protein expression
levels of MMP11 were basically the same (Fig. 2A). Then, patient clinicopathological
characteristics were analysed to determine the association between
MMP11 expression and clinical factors. The results revealed that
only tumour stage was significantly associated with MMP11
expression (P=0.037) (Table I).
Additionally, Kaplan-Meier survival analysis confirmed that high
MMP11 expression was associated with poor OS in BC patients
(Fig. 2B). To further expand the
clinical samples and analyse the clinical significance of MMP11,
the BC dataset from TCGA was downloaded and analysed. As revealed
in Table II, a systematic analysis
revealed that high MMP11 expression was significantly associated
with progesterone receptor status (P=0.045), HER2 receptor status
(P=0.027), tumour stage (P=0.003), and pathologic stage (P=0.004)
in BC. Moreover, Kaplan-Meier survival analysis confirmed that the
OS rate of patients with low MMP11 expression was significantly
higher than that of patients with high MMP11 expression (P=0.008)
(Fig. 2C). Online database analysis
(http://kmplot.com) also confirmed that high expression
of MMP11 was closely related to poor OS in BC (P<0.001)
(Fig. 2D) (14).
 | Table I.Clinical significance of MMP11 in
BC. |
Table I.
Clinical significance of MMP11 in
BC.
|
| MMP11 | High expression | Low expression | P-value |
|---|
| Age (years) | >60 | 7 | 8 | 0.73 |
|
| <60 | 10 | 9 |
|
| Neoplasm
subdivision | Left | 9 | 10 | 0.73 |
|
| Right | 8 | 7 |
|
| Oestrogen
receptor | Positive | 12 | 14 | 0.419 |
|
| Negative | 5 | 3 |
|
| Progesterone
receptor | Positive | 10 | 11 | 0.724 |
|
| Negative | 7 | 6 |
|
| HER2 receptor | Positive | 9 | 5 | 0.163 |
|
| Negative | 8 | 12 |
|
| Menopause
status | Pre | 3 | 7 | 0.132 |
|
| Post | 14 | 10 |
|
| Pathologic
stage | 0–1 | 7 | 13 | 0.037 |
|
| 2–3 | 10 | 4 |
|
| T | 0–1 | 9 | 14 | 0.067 |
|
| 2–3 | 8 | 3 |
|
| N | 0–1 | 12 | 12 | 1 |
|
| 2–3 | 5 | 5 |
|
| M | 0 | 15 | 17 | 0.145 |
|
| 1 | 2 | 0 |
|
 | Table II.Clinical significance of MMP11 in BC
based on TCGA data. |
Table II.
Clinical significance of MMP11 in BC
based on TCGA data.
|
| MMP11 | High
expression | Low expression | P-value |
|---|
| Age (years) | >60 | 186 | 201 | 0.492 |
|
| <60 | 170 | 203 |
|
| Sex | Female | 387 | 379 | 0.764 |
|
| Male | 5 | 4 |
|
| Neoplasm
subdivision | Left | 208 | 206 | 0.839 |
|
| Right | 184 | 177 |
|
| Oestrogen
receptor | Positive | 299 | 290 | 0.756 |
|
| Negative | 85 | 87 |
|
| Progesterone
receptor | Positive | 267 | 235 | 0.045 |
|
| Negative | 117 | 140 |
|
| HER2 receptor | Positive | 72 | 54 | 0.027 |
|
| Negative | 196 | 231 |
|
| Menopause
status | Pre | 98 | 94 | 0.480 |
|
| Post | 231 | 250 |
|
| Pathologic
stage | 0–1 | 248 | 279 | 0.004 |
|
| 2–3 | 133 | 95 |
|
| T | 0–1 | 279 | 315 | 0.003 |
|
| 2–3 | 112 | 77 |
|
| N | 0–1 | 319 | 301 | 0.320 |
|
| 2–3 | 66 | 75 |
|
| M | 0 | 339 | 336 | 0.655 |
|
| 1 | 10 | 8 |
|
Overexpression of MMP11 promotes the
proliferation of BC cells
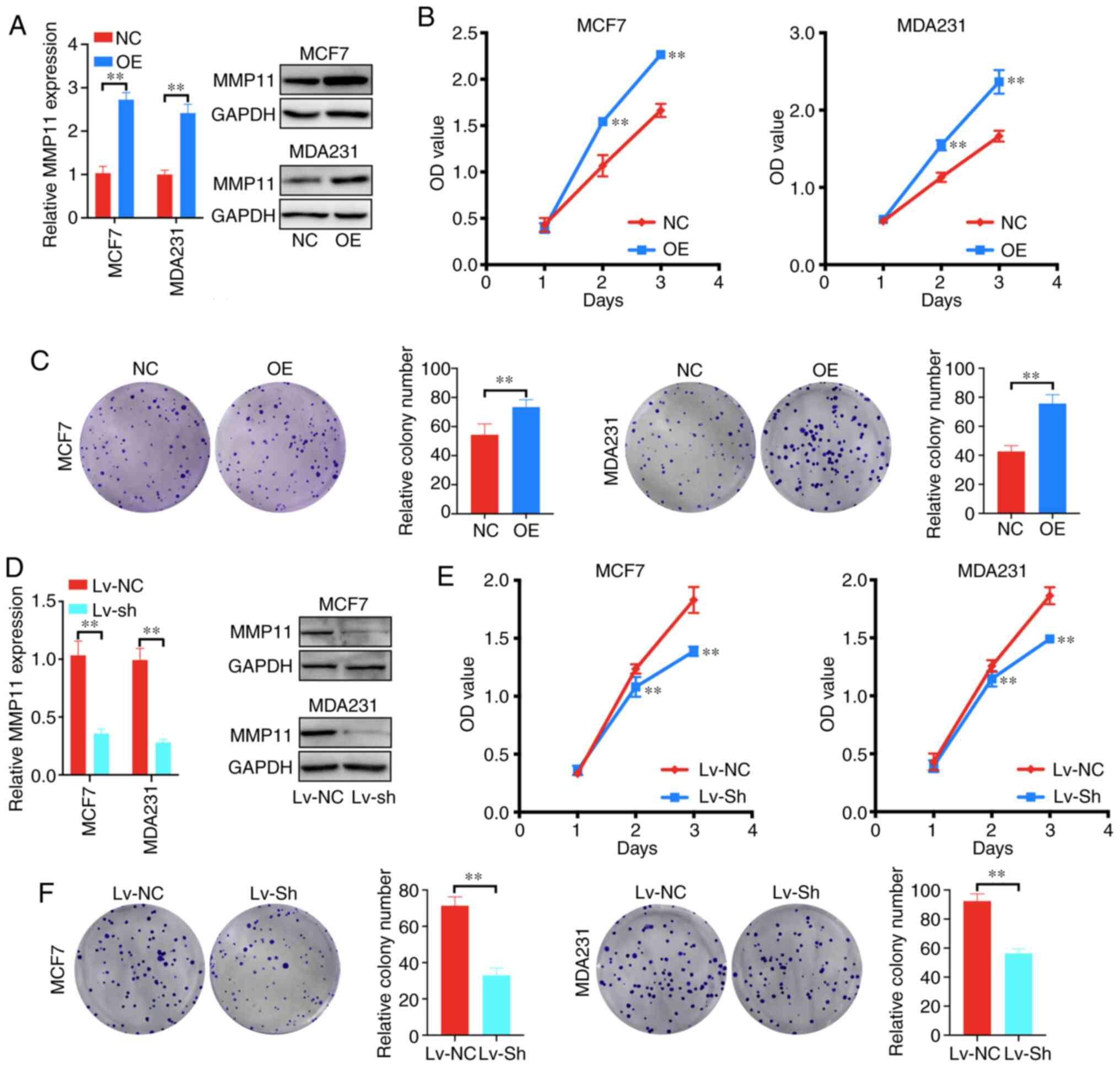
To explore whether MMP11 affects the biological
behaviour of BC, cell proliferation and colony formation assays
were applied. First, it was determined that OE plasmid transfection
significantly increased the mRNA and protein expression of MMP11 in
MCF7 and MDA231 cells compared with that in the control group
(Fig. 3A). Further results revealed
that MMP11 overexpression significantly increased the proliferation
and colony formation of MCF7 and MDA231 cells (Fig. 3B and C). In addition, to further
demonstrate the role of MMP11 in BC, MMP11 Lv-shRNA was used to
determine whether MMP11 knockdown could inhibit BC cell
development. Lv-shRNA significantly reduced the expression of MMP11
mRNA and protein (Fig. 3D). Moreover,
knockdown of MMP11 markedly inhibited the proliferation and colony
formation of MCF7 and MDA231 cells (Fig.
3E and F). Therefore, these results indicated that
overexpression of MMP11 promoted the proliferation of BC cells.
MMP11 regulates the TGF-β signalling
pathway
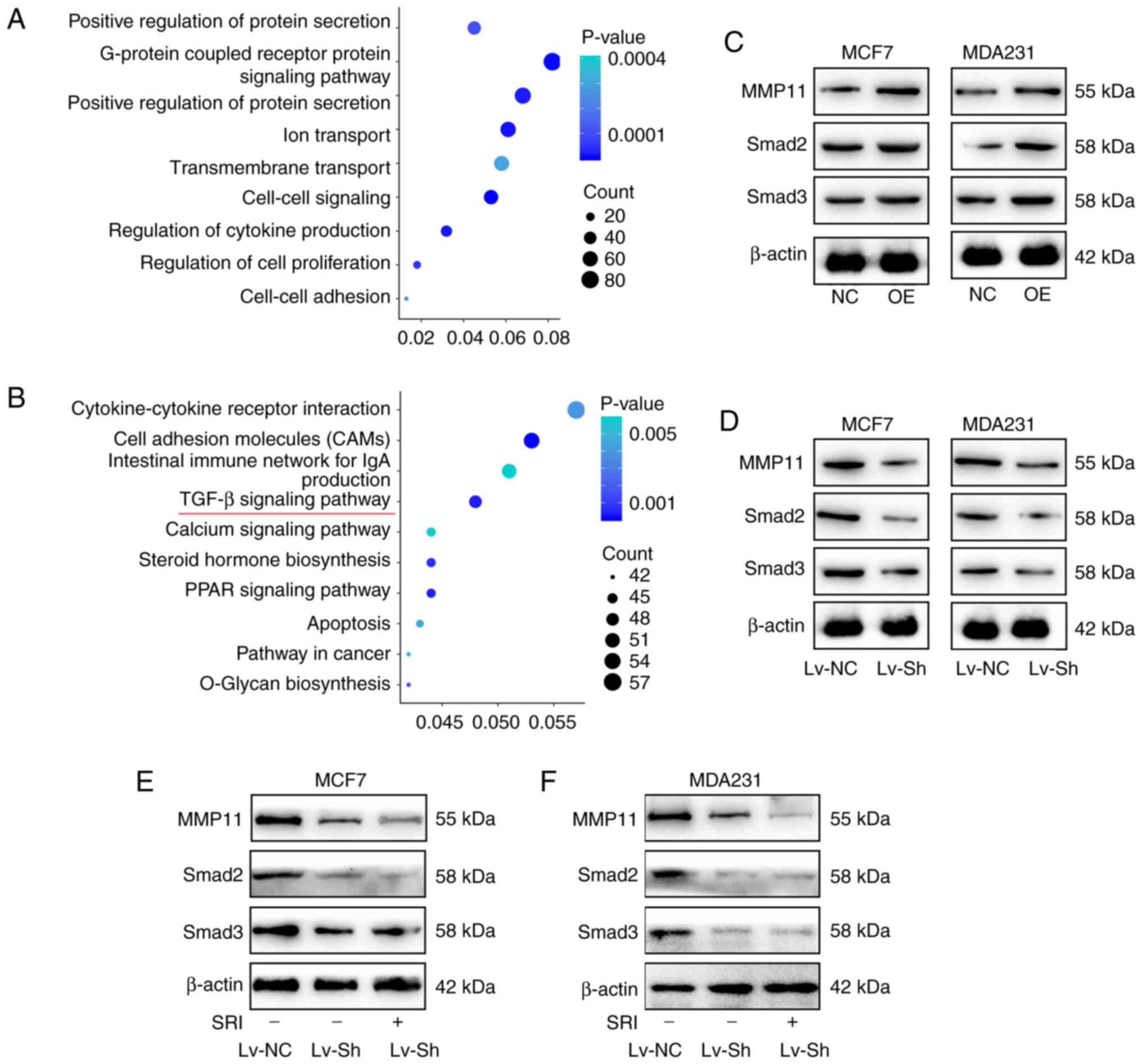
To further investigate the biological importance of
MMP11 in BC, we used an online database (https://www.UniProt.org) to predict MMP11-related
molecules and performed Gene Ontology (GO) and Kyoto Encyclopedia
of Genes and Genomes (KEGG) analyses (https://david-d.ncifcrf.gov) (15,16). The
biological processes of MMP11-related molecules were mainly
enriched in ‘cell-cell signalling’, ‘regulation of cell
proliferation’, and ‘cell-cell adhesion’ (Fig. 4A). In addition, certain signalling
pathways associated with breast carcinogenesis were identified to
be related to MMP11 in the KEGG pathway analysis; these pathways
included the ‘TGF-β signalling pathway’, ‘PPAR signalling pathway’,
‘apoptosis’ pathway and ‘pathways in cancer’ (Fig. 4B). Then, the TGF-β signalling pathway,
which was the most closely related to cell growth and proliferation
in our analysis, was investigated. Smad2 and Smad3 are two key
molecules in the TGF-β signalling pathway (17). The present results revealed that MMP11
overexpression increased the expression of Smad2 and Smad3
(Fig. 4C), while knockdown of MMP11
inhibited Smad2 and Smad3 expression (Fig. 4D), and this inhibition could not be
increased by TGF-β inducers (Fig. 4E and
F), which revealed that MMP11 promoted the expression of Smad2
and Smad3 independent on TGF-β.
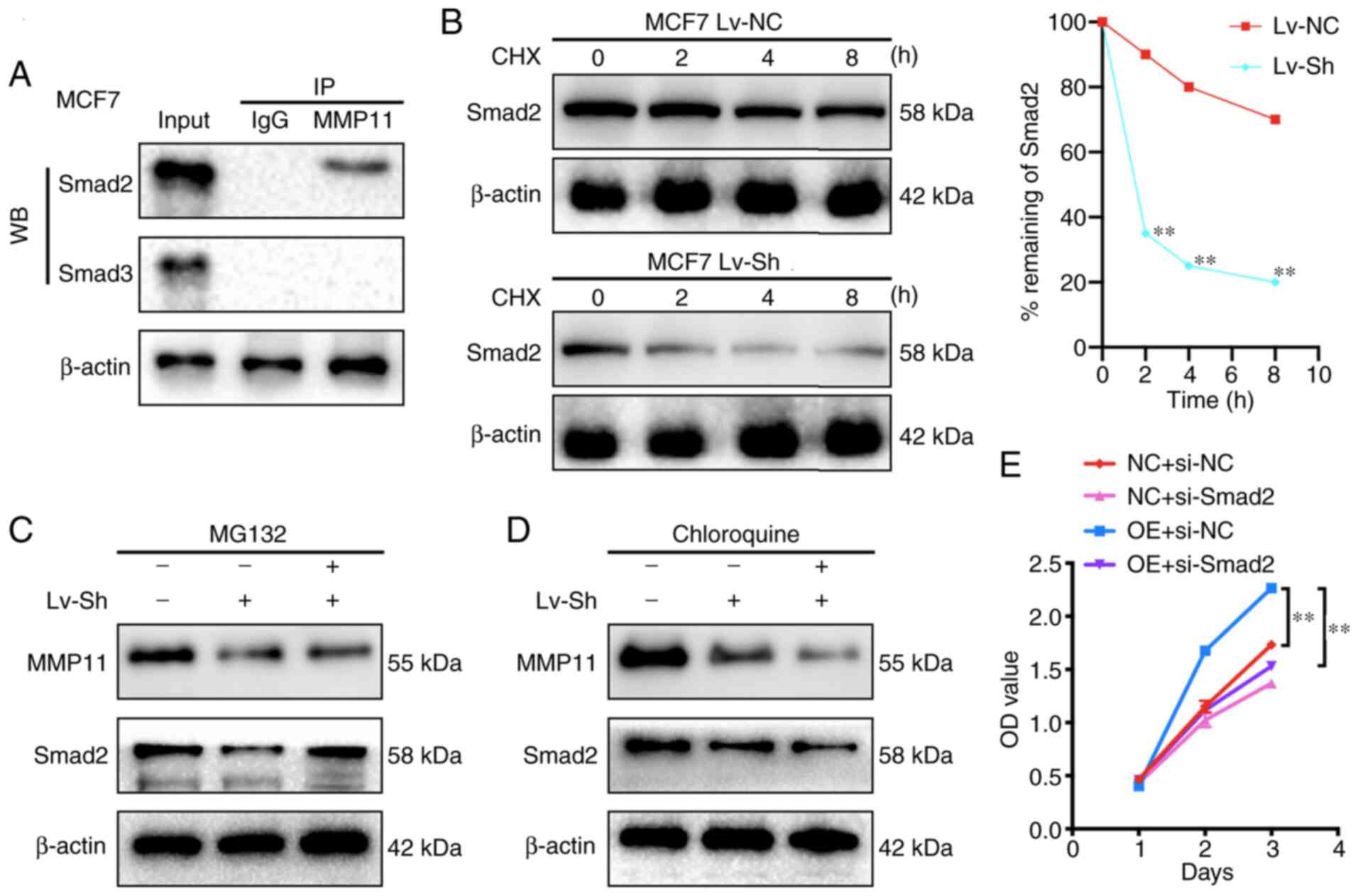
MMP11 binds to Smad2 and inhibits its
degradation
To verify the interaction of Smad2 and Smad3 with
MMP11, first, changes in the mRNA expression of Smad2 and Smad3
were detected after MMP11 overexpression by RT-qPCR and it was
revealed that there was no significant difference in the mRNA
expression of Smad2 and Smad3 between the MMP11 OE group and the NC
group (data not shown). It was further attempted to use anti-MMP11
antibodies to immunoprecipitate Smad2 or Smad3 from MCF7 cells,
which exhibit endogenous expression of MMP11, Smad2 and Smad3.
Notably, it was revealed that Smad2 was detected in the MMP11
immune complex by western blot analysis (Fig. 5A). Therefore, it was speculated that
MMP11 could affect the protein degradation of Smad2. To further
verify our conjecture, a CHX chase experiment was performed. The
results revealed that transfection of MCF7 cells with MMP11 Lv-sh
led to a marked reduction in the half-life of Smad2 compared with
that of Lv-NC transfection (Fig. 5B),
indicating that the decrease in MMP11 expression promoted the
degradation of Smad2. Furthermore, MCF7 cells were transfected with
MMP11 Lv-sh and then treated with the proteasome inhibitor MG132 or
autophagy inhibitor chloroquine. The present results revealed that
MG132 effectively abrogated the effect of MMP11 Lv-sh on Smad2
protein degradation, whereas chloroquine had no such effect
(Fig. 5C and D), which confirmed that
MMP11 inhibited Smad2 degradation through the ubiquitin proteasome
pathway. Finally, the MMP11 OE plasmid and Smad2-siRNA were
co-transfected into MCF7 cells, and the MMP11
overexpression-mediated induction of cell proliferation was
significantly inhibited by si-Smad2 (Fig.
5E), which verified that Smad2 was a critical downstream factor
that mediated the ability of MMP11 to promote BC cell
proliferation.
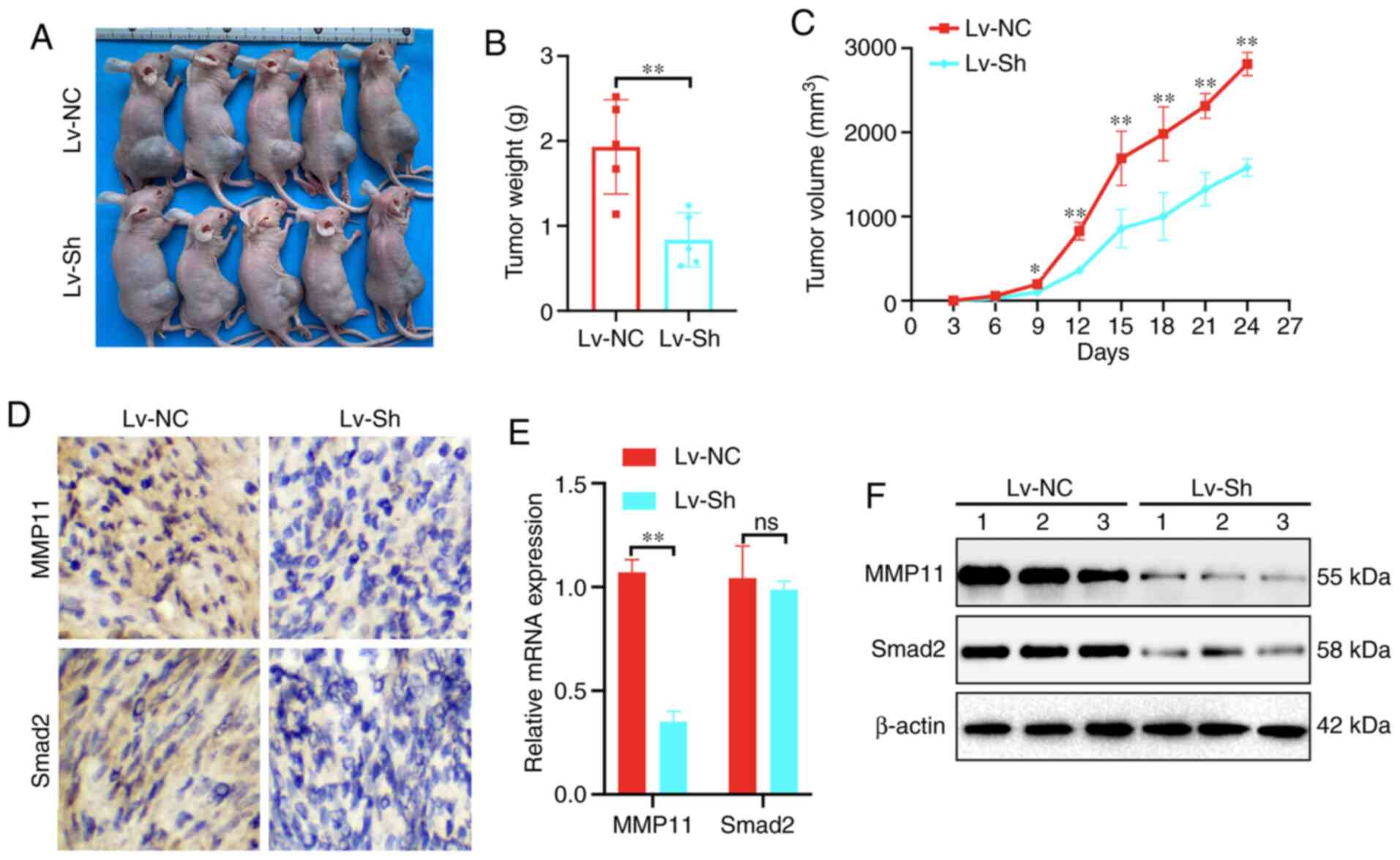
Decreased MMP11 inhibits BC growth in
vivo
To further clarify the biological role of MMP11
in vivo, MCF7 cells stably transfected with Lv-NC or Lv-sh
of MMP11 were implanted as xenografts in BALB/c nude mice (Fig. 6A). Compared with those in the Lv-NC
group, the tumour weight and growth of the Lv-sh group were
significantly inhibited (Fig. 6B and
C). According to IHC, RT-qPCR and western blot assay results,
the expression of MMP11 and Smad2 in implanted tumours of the Lv-sh
group was significantly lower than that in the Lv-NC group
(Fig. 6D-F). These results further
indicated that MMP11 played an essential role in promoting BC
through Smad2.
Discussion
BC treatment has come a long way in recent decades.
However, BC is a challenging disease, and the OS rate has not
significantly improved (18). If
reliable potential molecular markers are available at the time of
initial diagnosis, the prognosis and treatment outcomes of BC
patients may be considerably improved. Therefore, there is a need
to expand the current understanding of the molecular mechanisms of
BC progression and to identify new biomarkers. There is ample
evidence that MMP11 is a regulator of gene expression and complex
pathways in cancer (19–21). In the present study, the relationship
between MMP11 with clinicopathological parameters and total
survival of BC was analysed. In addition, GO and KEGG pathway
enrichment analyses were performed to assess gene function.
Specifically, it was determined that high expression of MMP11 in BC
was significantly associated with tumour stages and a poor
prognosis. In addition, using bioinformatics analysis, it was
demonstrated that MMP11 was associated with multiple signalling
pathways related to tumour malignancy.
The present study first explored the biological
roles of MMP11 in BC. A previous study have revealed that MMP11 is
significantly associated with the regulation of cell proliferation
in BC (22). The present study
revealed that MMP11 was overexpressed at the mRNA and protein
levels in BC tissues compared with PC tissues and that the
expression levels of MMP11 in four PC cell lines was significantly
higher than those in a normal human cell line. In addition, MMP11
overexpression was also revealed to significantly enhance the
proliferative ability of MCF7 and MDA231 cells. To further
demonstrate the roles of MMP11 in vivo, xenograft tumour
formation experiments were conducted. The experimental results were
consistent with those of the in vitro assays, that is, MMP11
knockdown significantly inhibited BC growth in vivo. These
results further demonstrated the cancer-promoting role of MMP11 in
BC.
To further explore the underlying mechanism,
bioinformatics analysis was conducted and it was revealed that cell
adhesion molecules played an important role in the process of MMP11
promoting BC, however a previous study reported that they had a
stronger relationship with tumour metastasis (23). Our initial purpose was to study tumour
proliferation, thus, we first focused on the TGF-β signalling
pathway. TGF-β superfamily signalling plays a critical role in the
regulation of cell growth, differentiation, and development in a
wide range of biological systems. In general, TGF-β/activin
signalling is initiated by ligand-induced oligomerization of
serine/threonine receptor kinases and phosphorylation of the
cytoplasmic signalling molecules Smad2 and Smad3. Moreover, in
certain contexts, TGF-β signalling could also affect
Smad-independent pathways, including the MEK/Erk, SAPK/JNK, and p38
MAPK pathways (24–26). In the present study, mechanistically,
it was revealed that MMP11 could significantly regulate the protein
expression levels of Smad2 and Smad3. We first tried to explain its
mechanism at the transcriptional level, however it was revealed
that the mRNA levels of Smad2 and Smad3 were not significantly
affected by changes in MMP11 expression. Based on the literature
(5), it was also revealed that MMP11
does not function as a transcription factor. Therefore, it was
hypothesized that MMP11 mainly affects the degradation of Smad2 and
Smad3 protein. CHX and MG132 inhibition assays revealed that MMP11
could stabilize the Smad2 protein structure by inhibiting its
ubiquitination-mediated degradation. Unfortunately, the direct
connection between MMP11 and Smad3 has not been explored clearly,
and this is the goal of our next study. In addition, notably, our
analysis revealed that the aberrant expression of MMP11 was also
related to cell adhesion molecules and apoptosis, which indicated
that MMP11 played an important role in the development of BC.
In conclusion, the present results revealed that
MMP-11 itself promoted BC cell proliferation, thereby promoting BC
development. Mechanistically, the cancer-promoting roles of MMP11
in BC are related to the increased stability of the Smad2 protein
and the activation of the TGF-β signalling pathway. Conversely,
MMP11 inhibition in BC cells greatly impaired their ability to
proliferate and grow. Moreover, it was concluded that the knockdown
of MMP11 could reverse its pro-tumorigenic effects in vivo.
Therefore, the MMP11 gene is proposed as a target for BC
therapy.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and analysed during the current
study were made available by the TCGA Genome Data Analysis Center
of the Broad Institute in 2016. All other datasets used during the
present study are available from the corresponding author upon
reasonable request.
Authors' contributions
GW conceived and designed the study. YZ participated
in the design of the study and performed bioinformatics analysis.
XL, PZ, and GP provided their advice during the process of the
research. YZ and XL performed the data analysis and wrote the
manuscript. YZ, PZ, and GP performed the cell line validation
assay. All authors reviewed and edited the manuscript and read and
approved the final manuscript.
Ethics approval and consent to
participate
All clinical samples were collected with written
informed consent from the patients, and this protocol was approved
by the Ethics Committee of the Hubei Cancer Hospital (Wuhan,
China). The mouse experiments and the handling of animals were
performed according to the Institutional and Animal Care and Use
Committee of Hubei Cancer Hospital and the NIH Guide for the Care
and Use of Laboratory Animals.
Patient consent for publication
Not applicable.
Patient competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Colombet M, Soerjomataram I,
Dyba T, Randi G, Bettio M, Gavin A, Visser O and Bray F: Cancer
incidence and mortality patterns in Europe: Estimates for 40
countries and 25 major cancers in 2018. Eur J Cancer. 103:356–387.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Torre LA, Siegel RL, Ward EM and Jemal A:
Global cancer incidence and mortality rates and trends-an update.
Cancer Epidemiol Biomarkers Prev. 25:16–27. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liu N, Johnson KJ and Ma CX: Male breast
cancer: An updated surveillance, epidemiology, and end results data
analysis. Clin Breast Cancer. 18:e997–e1002. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pittayapruek P, Meephansan J, Prapapan O,
Komine M and Ohtsuki M: Role of matrix metalloproteinases in
photoaging and photocarcinogenesis. Int J Mol Sci. 17:8682016.
View Article : Google Scholar
|
|
5
|
Motrescu ER and Rio MC: Cancer cells,
adipocytes and matrix metalloproteinase 11: A vicious tumor
progression cycle. Biol Chem. 389:1037–1041. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Scheau C, Badarau IA, Costache R, Caruntu
C, Mihai GL, Didilescu AC, Constantin C and Neagu M: The role of
matrix metalloproteinases in the epithelial-mesenchymal transition
of hepatocellular carcinoma. Anal Cell Pathol (Amst).
2019:94239072019.PubMed/NCBI
|
|
7
|
Zhang X, Huang S, Guo J, Zhou L, You L,
Zhang T and Zhao Y: Insights into the distinct roles of MMP-11 in
tumor biology and future therapeutics (Review). Int J Oncol.
48:1783–1793. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Han J, Choi YL, Kim H, Choi JY, Lee SK,
Lee JE, Choi JS, Park S, Choi JS, Kim YD, et al: MMP11 and CD2 as
novel prognostic factors in hormone receptor-negative,
HER2-positive breast cancer. Breast Cancer Res Treat. 164:41–56.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Eiro N, Cid S, Fernández B, Fraile M,
Cernea A, Sánchez R, Andicoechea A, DeAndrés Galiana EJ, González
LO, Fernández-Muñiz Z, et al: MMP11 expression in intratumoral
inflammatory cells in breast cancer. Histopathology. 75:916–930.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Roscilli G, Cappelletti M, De Vitis C,
Ciliberto G, Napoli A, Ruco L, Mancini R and Aurisicchio L:
Circulating MMP11 and specific antibody immune response in breast
and prostate cancer patients. J Transl Med. 12:542014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang MW, Liu J, Liu Q, Xu QH, Li TF, Jin S
and Xia TS: LncRNA SNHG7 promotes the proliferation and inhibits
apoptosis of gastric cancer cells by repressing the P15 and P16
expression. Eur Rev Med Pharmacol Sci. 21:4613–4622.
2017.PubMed/NCBI
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Goldman MJ, Craft B, Hastie M, Repečka K,
McDade F, Kamath A, Banerjee A, Luo Y, Rogers D, Brooks AN, et al:
Visualizing and interpreting cancer genomics data via the Xena
platform. Nat Biotechnol. 38:675–678. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Györffy B, Lanczky A, Eklund AC, Denkert
C, Budczies J, Li Q and Szallasi Z: An online survival analysis
tool to rapidly assess the effect of 22,277 genes on breast cancer
prognosis using microarray data of 1809 patients. Breast Cancer Res
Treat. 123:725–731. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen L, Yang T, Lu DW, Zhao H, Feng YL,
Chen H, Chen DQ, Vaziri ND and Zhao YY: Central role of
dysregulation of TGF-β/Smad in CKD progression and potential
targets of its treatment. Biomed Pharmacother. 101:670–681. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tao Z, Shi A, Lu C, Song T, Zhang Z and
Zhao J: Breast cancer: Epidemiology and etiology. Cell Biochem
Biophys. 72:333–338. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li Z, Ding S, Zhong Q, Li G, Zhang Y and
Huang XC: Significance of MMP11 and P14(ARF) expressions in
clinical outcomes of patients with laryngeal cancer. Int J Clin Exp
Med. 8:15581–15590. 2015.PubMed/NCBI
|
|
20
|
Kou YB, Zhang SY, Zhao BL, Ding R, Liu H
and Li S: Knockdown of MMP11 inhibits proliferation and invasion of
gastric cancer cells. Int J Immunopathol Pharmacol. 26:361–370.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Han HB, Gu J, Zuo HJ, Chen ZG, Zhao W, Li
M, Ji DB, Lu YY and Zhang ZQ: Let-7c functions as a metastasis
suppressor by targeting MMP11 and PBX3 in colorectal cancer. J
Pathol. 226:544–555. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
González de Vega R, Clases D,
Fernández-Sánchez ML, Eiró N, González LO, Vizoso FJ, Doble PA and
Sanz-Medel A: MMP-11 as a biomarker for metastatic breast cancer by
immunohistochemical-assisted imaging mass spectrometry. Anal
Bioanal Chem. 411:639–646. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Janiszewska M, Primi MC and Izard T: Cell
adhesion in cancer: Beyond the migration of single cells. J Biol
Chem. 295:2495–2505. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Horbelt D, Denkis A and Knaus P: A
portrait of transforming growth factor β superfamily signalling:
Background matters. Int J Biochem Cell Biol. 44:469–474. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ikushima H and Miyazono K: TGFbeta
signalling: A complex web in cancer progression. Nat Rev Cancer.
10:415–424. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Velapasamy S, Dawson CW, Young LS,
Paterson IC and Yap LF: The dynamic roles of TGF-β signalling in
EBV-associated cancers. Cancers (Basel). 10:2472018. View Article : Google Scholar
|