Introduction
Cutaneous squamous cell carcinoma (cSCC) is the
second most common type of non-melanoma skin cancer (1). cSCC has been revealed to develop from a
pre-malignant condition, such as actinic keratosis (AK) or Bowen's
disease (BD) (2,3). Several risk factors have been closely
associated with the incidence of cSCC, such as ultraviolet (UV)
radiation, smoking, and chemical and air pollutants (4). In particular, UV radiation induces DNA
damage in cells and genetic mutations in genes such as p53, which
was discovered to result in abnormalities in cell migration,
invasion and metastasis (5).
Mutations in p53 have been regarded as a pathological mechanism of
UV radiation-induced AK and BD, and it has been hypothesized that
the early alterations due to UV damage may promote the
tumorigenesis and progression of cSCC (6). Although multiple risk factors are
associated with a high susceptibility to cSCC development, the
exact molecular mechanisms underlying human cSCC progression remain
unknown. In total, approximately 5% of patients with cSCC are
diagnosed with lymph node metastasis or metastasis in other organs
(7). The conventional treatments of
cSCC currently include surgical intervention, radiotherapy and
chemotherapy; however, the prognosis of metastatic cSCC remains
poor with a 1 year survival rate of 44–56% (8).
Long non-coding RNAs (lncRNAs) are non-protein
coding transcripts of >200 nucleotides in length (9). Increasing evidence has reported that
lncRNAs serve a crucial role in the tumorigenesis and progression
of numerous types of cancer, and have exhibited both oncogenic or
tumor-suppressive functions (10,11). H19,
located on chromosome 11p15.5, is a long non-coding RNA (lncRNA)
which was initially termed as an oncofetal transcript (12). The aberrant expression of H19 was
revealed to participate in the pathogenesis of several types of
human cancer, including bladder, breast, thyroid and hepatocellular
carcinoma (13–15). In addition, a previous study suggested
that H19 may be regarded as a biomarker for numerous types of
cancer and a potential therapeutic target (16). Several recent studies have
demonstrated that H19 served as an oncogene. For example, in
cholangiocarcinoma, H19 exerted its oncogenic role via sponging
let-7a/let-7b to regulate cell migration and invasion (17). In colorectal cancer, the
overexpression of H19 and microRNA (miRNA/miR)-138 promoted the
migration and invasion of colorectal cancer cells and upregulated
the expression levels of high mobility group AT-hook 1 (HMGA1)
(18). In addition, H19 altered the
expression levels of cascades of epithelial-mesenchymal transition
(EMT)-related makers to promote the proliferation, migration and
invasion of melanoma cancer cells, which was suggested to influence
the prognosis (19). Furthermore, H19
was reported to function as a precursor of miR-675 which was
reported in several cancers types, including breast, non-small cell
lung cancer and colon cancer (20).
H19 can generate miR-675 in a classic manner depending on Drosha
and Dicer splicing, and is located in exosomes, such as
endosome-derived extracellular vesicles containing proteins, lipids
and RNAs (21,22). However, the pathogenic mechanisms of
H19 in cSCC remain poorly understood. The present study
hypothesized that H19 may promote cSCC tumorigenesis and metastasis
and regulate cSCC cell migration and invasion.
In the present study, the expression levels of H19
in clinical sample tissues from patients with cSCC were analyzed,
and the potential functional role of H19 in cSCC was also
investigated. The expression levels of H19 and miR-675 were
explored in SCC cell lines. In addition, the potential molecular
mechanisms underlying the effects of H19 and miR-675 on the EMT
process in cSCC were investigated. To the best of our knowledge,
the present study provided the first evidence of the interaction
between H19, miR-675, p53 and the EMT process in cSCC, and
highlighted the potential oncogenic role of the H19/miR-675 axis in
cSCC.
Materials and methods
Patient studies
A total of 60 paired cSCC adjacent normal tissues
were collected from patients with cSCC at the Department of
Dermatology, Xinhua Hospital (Shanghai, China) between January,
2018 and August, 2020. The medical records of the patients were
retrospectively reviewed for inclusion and exclusion criteria. The
inclusion criteria were as follows: i) Patients who suffered from
cSCC; ii) first onset without any prior treatment; and iii)
patients who had a complete and standardized postoperative
pathology report and follow-up information. The exclusion criteria
were as follows: i) Patients who had undergone systemic or local
therapy prior to surgical resection, preoperative radiotherapy or
chemotherapy or immunotherapy and incomplete medical records; ii)
patients who suffered from other skin diseases concurrently; and
iii) patients who lacked a complete and standardized postoperative
pathology report and follow-up information. The patients had an
average age of 68±16.20 (range, 36 to 97 years); 29 patients were
male (age range, 36 to 89 years) and 31 patients were female (age
range, 42 to 97 years). The present study protocol was approved by
The Human Ethics Committee at our institute. The acquisition and
storage of tissues was approved by the Ethics Committee of Xinhua
Hospital, School of Medicine, Shanghai Jiao Tong University
(Shanghai, China). Written informed consent was obtained from all
the patients prior to participation, and the study was conducted
according to the principles of the Declaration of Helsinki. All
collected samples were snap frozen in liquid nitrogen and then
stored at −80°C until required for further experimentation.
Cell lines and culture
cSCC cell lines, SCL1 (cat. no. CC-Y1669) and A431
(cat. no. CC-Y1033), and the keratinocyte cell line, HaCaT (cat.
no. CC-Y1177), were obtained from Ek-Bioscience; all cells were
mycoplasma-free. A431 and HaCaT cells were cultured in DMEM (Gibco;
Thermo Fisher Scientific, Inc.), while SCL1 cells were cultured in
RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.). All cells
were supplemented with 10% FBS (Gibco; Thermo Fisher Scientific,
Inc.) and 1% penicillin-streptomycin (Invitrogen; Thermo Fisher
Scientific, Inc.). All cell lines were incubated in a humidified
incubator containing 5% CO2 at 37°C.
Cell transfection
Specific small interfering RNAs (siRNAs) targeting
H19 (20 nM H19-siRNA-1 and 20 nM H19-siRNA-2), 20 nM p53-siRNA, 20
nM negative control (NC) siRNA, 3 µg pcDNA3.1-control plasmids, 3
µg pcDNA3.1-H19 and 3 µg pcDNA3.1-p53 overexpression plasmids were
all designed and synthesized by Hanbio Biotechnology Co., Ltd.
miR-675 mimic (50 nM) and miR-675 inhibitor oligonucleotides (100
nM), as well as mimic-NC (50 nM) and inhibitor-NC (100 nM), were
synthesized by Guangzhou RiboBio Co., Ltd. Cells were plated in
6-well plates (1×105 cells/well) and incubated at 37°C
and 5% CO2 until the cells reached 60–70% confluence.
Then cells were transfected with equal concentrations of oligo
fragments diluted in Opti-MEM/reduced serum medium (Gibco; Thermo
Fisher Scientific, Inc.) using Lipofectamine® 3000
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to
the manufacturer's instructions. Once the two solutions were mixed
and incubated for 5 min at room temperature, the mixture was plated
into a six-well plate. After incubation for 3 h at 37°C, 1 ml of
DMEM containing 10% FBS and no antibiotics was added to each well.
Subsequently, media were replaced with normal growth media 24 h
after transfection. The sequences are listed in Table I.
 | Table I.Sequences designed for the study. |
Table I.
Sequences designed for the study.
| Name | Sequences
(5′-3′) |
|---|
| H19-siRNA-1 |
CUGGACUCAUCAUCAAUAA |
| H19-siRNA-2 |
GAACCCACAACAUGAAAGA |
| p53-siRNA |
GACUCCAGUGGUAAUCUAC |
| NC-siRNA |
UUCUCCGAACGUGUCACGU |
| H19 |
CAGCCCAACATCAAAGACA |
| p53 |
GAGGTTGGCTCTGACTGTACC |
| pcDNA3.1-NC |
CTAGAGAACCCACTGCTTAC |
| miR-675-5p
mimics |
UGGUGCGGAGAGGGCCCACAGUG |
| NC for the
miR-675-5p mimics |
UUUGUACUACACAAAAGUACUG |
| miR-675-5p
inhibitors |
CACUGUGGGCCCUCUCCGCACCA |
| NC for the
miR-675-5p inhibitors |
CAGUACUUUUGUGUAGUACAAA |
RNA extraction and quantitative
real-time polymerase chain reaction (RT-qPCR) analysis
Total RNA was extracted from the tissues and cells
for the generation of single-stranded cDNA using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). The first
complementary deoxyribonucleic acid (cDNA) strand of H19 was
synthesized using an EZ-press RNA Purification kit (EZBioscience).
The reverse transcription of miR-675 was performed using the miRNA
1st Strand cDNA Synthesis kit (Vazyme Biotech Co., Ltd.) according
to the manufacturer's protocol. Reverse transcription conditions
were as follows: 42°C for 15 min and 95°C for 1 min. Quantitative
PCR was subsequently performed using the 2X SYBR Green qPCR Master
mix (EZBioscience) on a QuantStudio™ 3 Real-Time PCR system
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The
thermocycling conditions were as follows: Hot-start DNA polymerase
activation (95°C, 5 min); 40 cycles (95°C, 15 sec; and 60°C, 30
sec); and last melt curve analysis (95°C, 15 sec; 60°C, 1 min;
95°C, 30 sec; and 60°C, 15 sec). The following primer pairs were
used for the qPCR: H19 forward, 5′-CTCCCTCTTCTTCTTTTTCATC-3′ and
reverse, 5′-CGCACACTCGTACTGAGACT-3′; miR-675-5p forward,
5′-TTGGTGGTGCGGAGAG-3′ and reverse, 5′-AGTGCGTGTCGTGGAGTC-3′;
β-actin forward, 5′-AAGGTGACAGCAGTCGGTT-3′ and reverse,
5′-TGTGTGGACTTGGGAGAGG-3′; and U6 forward,
5′-CTTCGGCAGCACATATACTA-3′ and reverse, 5′-AACTGGTGTCGTGGAGTC-3′.
Relative expression levels were calculated using the
2−∆∆Cq method (23).
β-actin was used as the reference gene for lncRNA and mRNA, and U6
was the internal reference gene for miRNA. Experiments were
repeated ≥3 times with five replicate wells per sample.
Western blotting
Proteins were extracted from cells with RIPA lysis
buffer (Beyotime Institute of Biotechnology). Total protein was
quantified using a BCA protein assay (Beyotime Institute of
Biotechnology) with multi-volume spectrophotometer system (Epoch;
BioTek Instruments, Inc.) and proteins (40 µg) were separated via
10% SDS-PAGE. The separated proteins were subsequently transferred
onto PVDF membranes (EMD Millipore) and blocked with 10% non-fat
milk for 1 h at room temperature. The membranes were then incubated
with the following primary antibodies overnight at 4°C: Anti-p53
(1:1,000; product no. 9282T; Cell Signaling Technology, Inc.),
anti-Bax (1:1,000; product no. 2772T; Cell Signaling Technology,
Inc.), anti-Bcl-2 (1:1,000; product no. 4223T; Cell Signaling
Technology, Inc.), anti-E-cadherin (1:1,000; product code ab231303;
Abcam), anti-vimentin (1:1,000; product code ab92547; Abcam),
anti-N-cadherin (1:1,000; ab76011; Abcam) and anti-GAPDH (1:1,000;
product no. AF1186; Beyotime Institute of Biotechnology). The
following day, the membranes were washed with TBST three times (10
min each) and incubated with secondary antibodies (anti-rabbit)
(1:1,000; product no. A0208; Beyotime Institute of Biotechnology)
at room temperature for 1.5 h. Protein bands were visualized using
an ECL kit (Beyotime Institute of Biotechnology). ImageJ software
1.8.0 (National Institutes of Health) was used to quantify the
integrated density of the bands.
Cell Counting Kit-8 (CCK-8) assay
Cell proliferation was analyzed using a CCK-8 assay
(Dojindo Molecular Technologies, Inc.) according to the
manufacturer's protocol. Briefly, transfected cells were seeded
into 96-well plates (5×103 cells/well) and incubated for
24, 48, 72, 96 or 120 h. Following the incubation, CCK-8 reagent
(10 µl/well) was added to each well at 37°C. The optical density
(OD) values were detected at a wavelength of 450 nm using a
microplate reader (Bio-Rad Laboratories, Inc.) after 2 h.
Experiments were repeated ≥3 times with five replicate wells per
sample.
Wound healing assay
Cells were seeded into six-well plates at a density
of 2.0×105 cells/well to determine the migratory
ability. Upon the cells reaching 80–90% confluence, a 100-µl
pipette tip was used to produce a single scratch in the cell
monolayer of each well; PBS was used to remove the non-adherent
cells following scratching. The cells were subsequently cultured in
fresh culture medium in an incubator at 37°C. An inverted
microscope was used to observe the migratory distance of the cells
into the scratch area at 0, 12 and 24 h. The assay was repeated
three times.
Transwell migration and invasion
assays
Transwell migration and invasion assays were
performed using a Transwell chamber (Corning, Inc.). Briefly,
transfected cells at a density of 5.0×104 suspended in
serum-free medium were seeded into the upper chamber of the
Transwell plate. The lower chambers were filled with 500 µl
RPMI-1640 medium supplemented with 12% fetal bovine (FBS) serum.
Following incubation for 24 h at 37°C, the cells remaining in the
upper chamber were removed, while the cells in the lower chamber
were fixed with 4% paraformaldehyde for 15 min and stained with
0.1% crystal violet at room temperature for 30 min. An inverted
light microscope (Olympus Inverted Microscope) was used to count
cell migration/invasion in randomly selected fields of view to
semi-quantify the mean migration and invasion distances. Invasion
assays were conducted in accordance with the aforementioned
procedures except that the lower chambers were precoated with the
diluted Matrigel. After incubation for 24 h for migration and 20 h
for invasion at 37°C, the cells on the upper surface were wiped off
and cells on the lower surface were stained with 0.1% crystal
violet at room temperature for 30 min. The average number of
migrated and invasive cells was counted and photographed under a
light microscope (magnification, ×200).
Terminal deoxynucleotidyl transferase
dUTP nick end labeling assay
Apoptosis-related DNA fragmentation was analyzed by
One Step TUNEL Apoptosis Assay kit (Beyotime Institute of
Biotechnology) according to the manufacturer's protocol. Briefly,
cells (1×106 cells/well) were cultured on the
coverslips, washed with PBS and fixed with ice-cold 4%
paraformaldehyde for 30 min at room temperature. Subsequently, the
terminal deoxynucleotidyl transferase (a template-independent
polymerase) was used to incorporate nucleotides at the DNA cleavage
site. Following the incubation, the nuclei were stained with DAPI
at room temperature for 1 h and fluorescence images were obtained
in three different fields of view of each coverslip using a
fluorescence microscope (magnification, ×200).
Dual-luciferase reporter assay
Wild-type (WT) H19 and p53 3′-untranslated region
(UTR) sequences containing miR-675 binding sites were separately
cloned into the pGL4-basic vectors (Promega Corporation) to
generate pGL4-H19-WT and pGL4-p53-WT vectors, respectively. In
addition, the miR-675 binding site was separately mutated into H19
and p53 fragments, which were then cloned into pGL4 vectors to
obtain pGL4-H19-mutant (MUT) and pGL4-p53-MUT vectors,
respectively. In brief, HaCaT cells were seeded in 96-well plates
and reached 70% confluence overnight. Then HaCaT cells were
transfected with the Renilla luciferase reporter plasmids
using Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). Following 24 h of transfection, the relative
luciferase activities were measured using a Dual-Luciferase
Reporter assay system (Promega Corporation), according to the
manufacturer's protocol. Renilla luciferase activity was
normalized to firefly luciferase expression. All experiments were
performed in triplicate and independently repeated three times.
Statistical analysis
Online publicly available algorithms (microRNA.org) were used to predict the targets of
miR-675 (24). GraphPad Prism 7
(GraphPad Software, Inc.) was used to analyze the data; measurement
data were expressed as the mean ± standard deviation (x ± s).
Differences between groups were compared using Student's unpaired
t-test or ANOVA followed by Sidak's post hoc test. P<0.05 was
considered to indicate a statistically significant difference.
Results
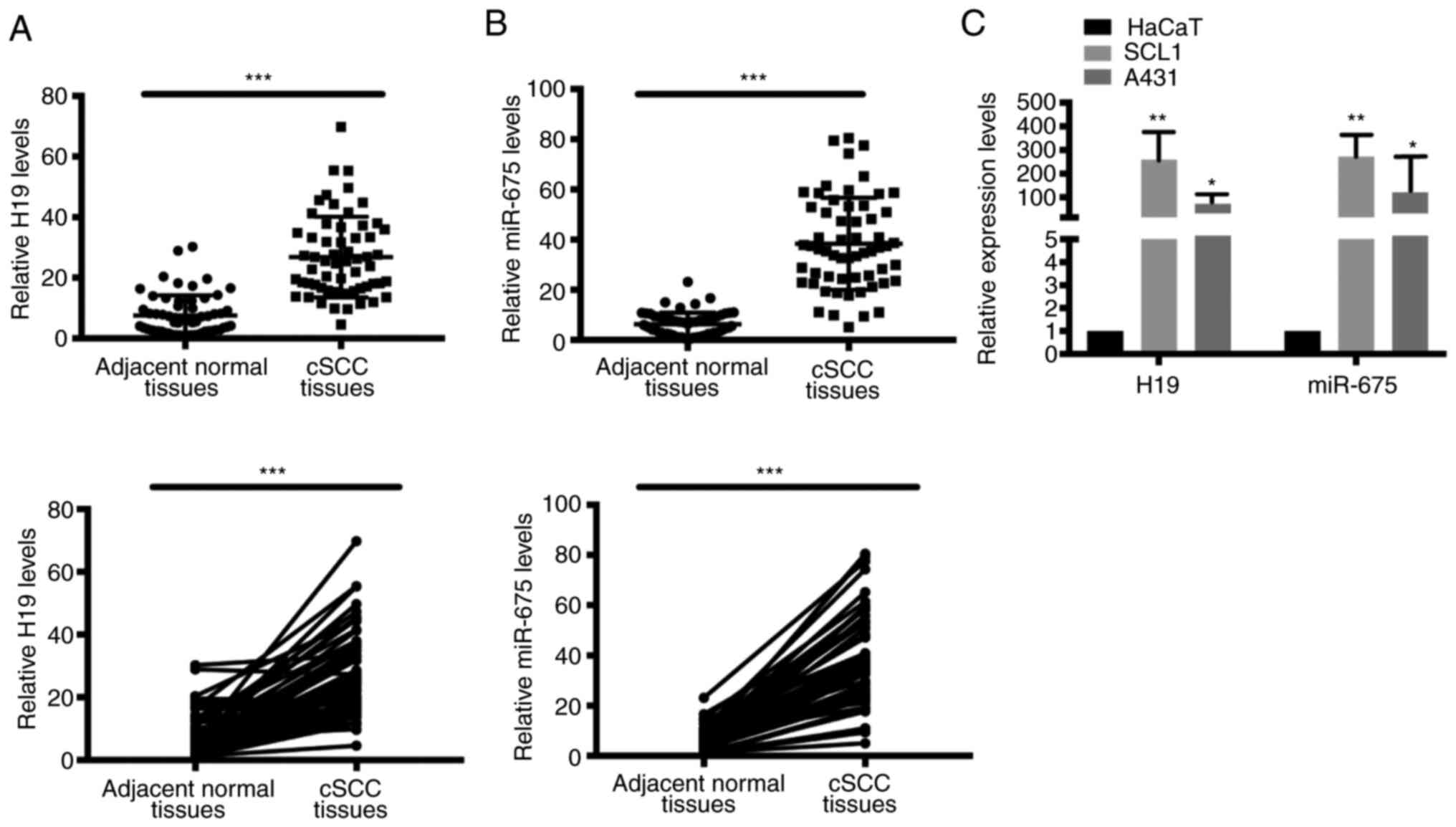
H19 and miR-675 expression levels are
upregulated in cSCC tissues and cell lines
To determine the potential function of H19 and
miR-675 in cSCC, the mRNA expression levels of H19 and miR-675 in
cSCC tissues and cell lines were analyzed using RT-qPCR. A total of
60 patient samples were used in the present research. Both H19 and
miR-675 expression levels were significantly upregulated in tumor
tissues from cSCC compared with adjacent normal tissues (Fig. 1A and B). Similarly, the expression
levels of H19 and miR-675 were also upregulated in the SCC cell
lines, SCL1 and A431 (Fig. 1C).
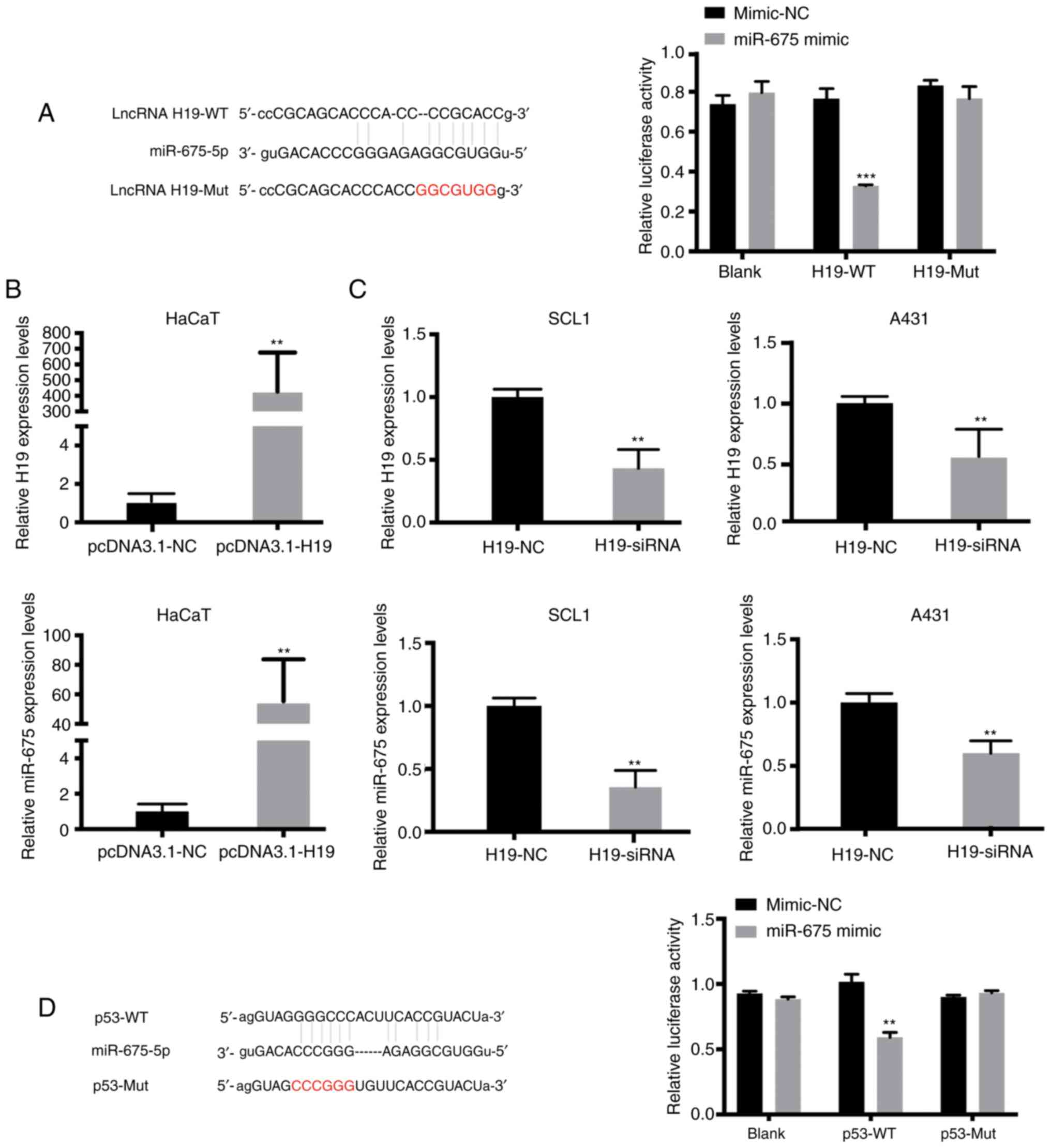
miR-675 targets both H19 and p53 in
cSCC
Using online publicly available algorithms
(microRNA.org), miR-675 targets were predicted.
The results revealed the putative binding site of miR-675 in the
3′UTR of H19 and p53. To determine whether H19 may interact with
miR-675 to affect the molecular mechanism of cSCC cell migration
and invasion, dual-luciferase reporter gene assays were performed.
The results revealed that the relative luciferase activity was
significantly reduced in HaCaT cells co-transfected with the H19-WT
vector and miR-675 mimic compared with the cells in the H19-WT +
mimic-NC and H19-Mut + miR-675 mimic groups (Fig. 2A). As anticipated, the overexpression
of H19 increased the expression levels of miR-675 in HaCaT cells
(Fig. 2B). Since H19 expression was
upregulated in SCL1 and A431 cells, these two cell lines were
transfected with H19-siRNA. RT-qPCR analysis was used to verify the
successful siRNA-mediated knockdown of H19 expression, and it was
subsequently demonstrated that the inhibition of H19 significantly
downregulated the expression levels of miR-675 in both cSCC cell
lines (Fig. 2C). Moreover, microRNA.org was used to predict that miR-675 could
bind to the 3′-UTR of p53. Dual-luciferase reporter assays were
subsequently performed to verify the association between miR-675
and p53. The relative luciferase activity was significantly
decreased in HaCaT cells in the p53-WT + miR-675 mimic group
compared with the p53-Mut + miR-675 mimic and p53-WT + mimic-NC
groups (P<0.05; Fig. 2D). These
findings suggested that miR-675 may target both H19 and p53 in
HaCaT and SCC cells.
H19/miR-675 axis promotes the
proliferation, migration and invasion, but inhibits apoptosis in
HaCaT cells
Both H19 and miR-675 were hypothesized to regulate
cell proliferation, migration and invasion in cSCC. Therefore,
CCK-8, wound healing and Transwell assays were conducted to
determine the involvement of the H19/miR-675 axis in these
biological functions. The overexpression of H19 significantly
increased the proliferation of HaCaT cells. Conversely, following
the confirmation of the successful siRNA-mediated knockdown of H19,
the proliferation of HaCaT cells transfected with H19-siRNA was
revealed to be significantly decreased (Fig. 3A). To determine whether miR-675 served
a role in H19-induced cSCC cell migration, HaCaT cells were
co-transfected with H19 siRNA and miR-675 mimic, and the migration
was analyzed using wound healing and Transwell assays. The miR-675
expression levels were demonstrated to be upregulated following the
transfection with miR-675 mimics. Following the co-transfection of
miR-675 mimic and H19-siRNA, the migration was decreased in HaCaT
cells (Fig. 3B). Moreover, H19-siRNA
could reverse the miR-675 mimic-induced increase in migration and
invasion of HaCaT cells, as determined using Transwell assays
(Fig. 3C). The levels of apoptosis
were also detected using a TUNEL assay following the transfection
of HaCaT cells with the miR-675 mimic and H19-siRNA. As revealed in
Fig. 3D, H19-siRNA could partially
reverse the miR-675 mimic-induced reduction in apoptosis compared
with the mimic-NC group (Fig.
3D).
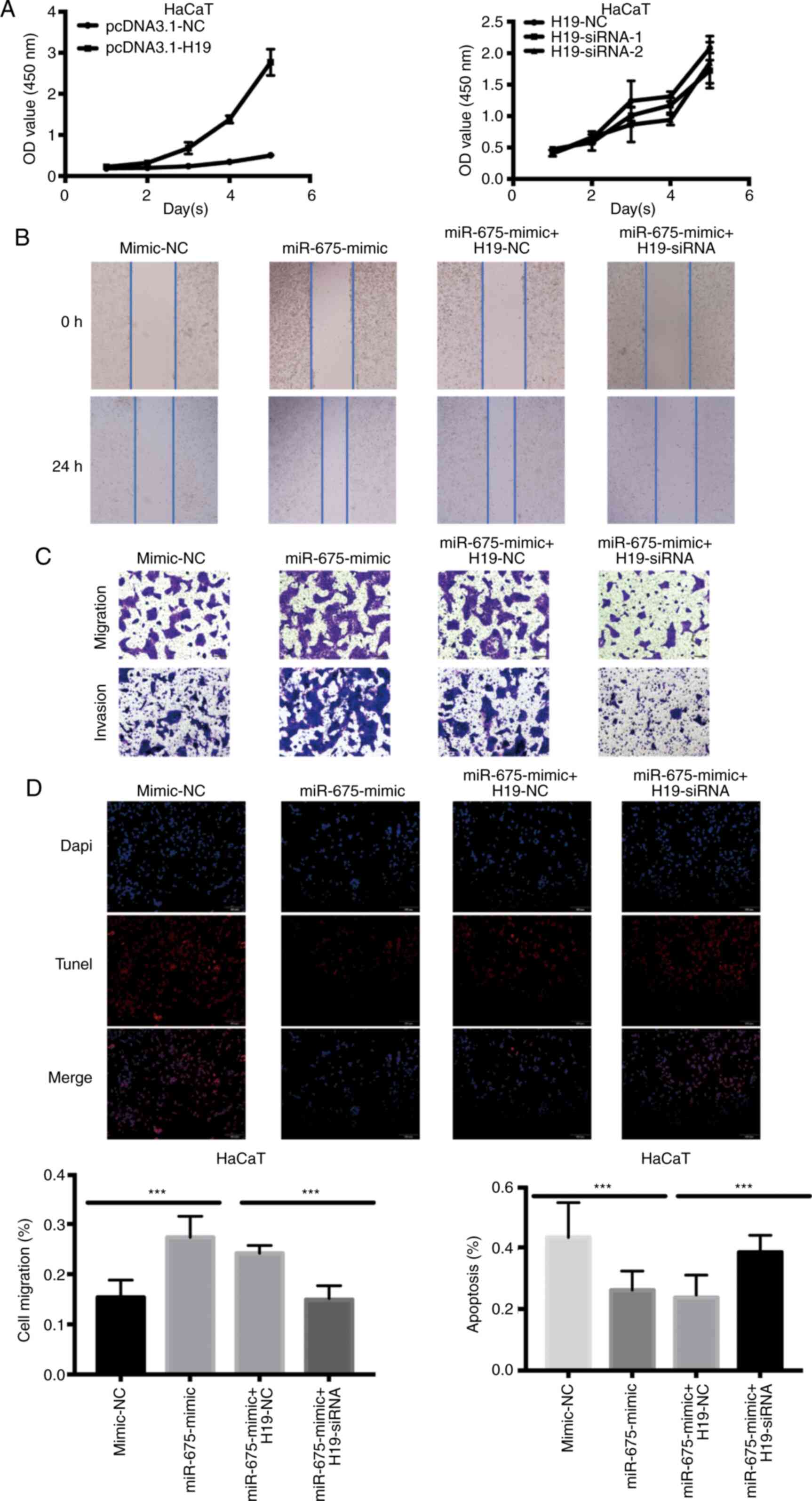 | Figure 3.Overexpression of H19 and miR-675
promotes HaCaT cell proliferation, migration and invasion, and
inhibits apoptosis. (A) Proliferation of HaCaT cells transfected
with pcDNA3.1-H19 and H19-siRNA was analyzed using a Cell Counting
Kit-8 assay. (B) A wound healing assay was used to determine the
cell migration of HaCaT cells transfected with mimic-NC, miR-675
mimic, miR-675 mimic + H19-NC and miR-675 mimic + H19-siRNA.
Magnification, ×200. (C) Migration and invasion of HaCaT cells
transfected with mimic-NC, miR-675 mimic, miR-675 mimic + H19-NC
and miR-675 mimic + H19-siRNA were analyzed using Transwell assays.
Magnification, ×200. (D) TUNEL assays were used to determine
apoptosis in HaCaT cells transfected with mimic-NC, miR-675 mimic,
miR-675 mimic + H19-NC and miR-675 mimic + H19-siRNA.
Magnification, ×200. ***P<0.001. miR, microRNA; siRNA, small
interfering RNA; NC, negative control. |
H19 and miR-675 knockdown suppresses
the proliferation, migration and invasion, and promotes apoptosis
in SCC cells
To further validate that H19 bound to and
sequestered miR-675 to regulate cell migration and proliferation,
CCK-8, wound healing and Transwell assays were performed following
the overexpression of H19 and knockdown of miR-675 in SCC cells.
Following H19-siRNA transfection, the results of the CCK-8 assay
revealed that the cell proliferation ability was significantly
decreased (Fig. 4A). In addition, the
overexpression of H19 counteracted the miR-675 inhibitor-induced
inhibition of cell migration, as observed in the wound healing
(Fig. 4B) and Transwell (Fig. 4C) assays, which clarified that H19
could rescue the inhibitive effect of miR-675 inhibitor on cSCC
cell migration and invasion. The results of the TUNEL assay
demonstrated that pcDNA3.1-H19 reversed the increase in cell
apoptosis induced by the miR-675 inhibitor in cSCC cells (Fig. 4D). Thus, the co-transfection with
miR-675 inhibitor and pcDNA3.1-H19 could markedly reverse the
effect of the miR-675 inhibitor on the proliferation, migration and
invasion of cSCC cells. Furthermore, the transfection with
pcDNA3.1-H19 could weaken the effects of the miR-675 inhibitor on
cSCC cell apoptosis.
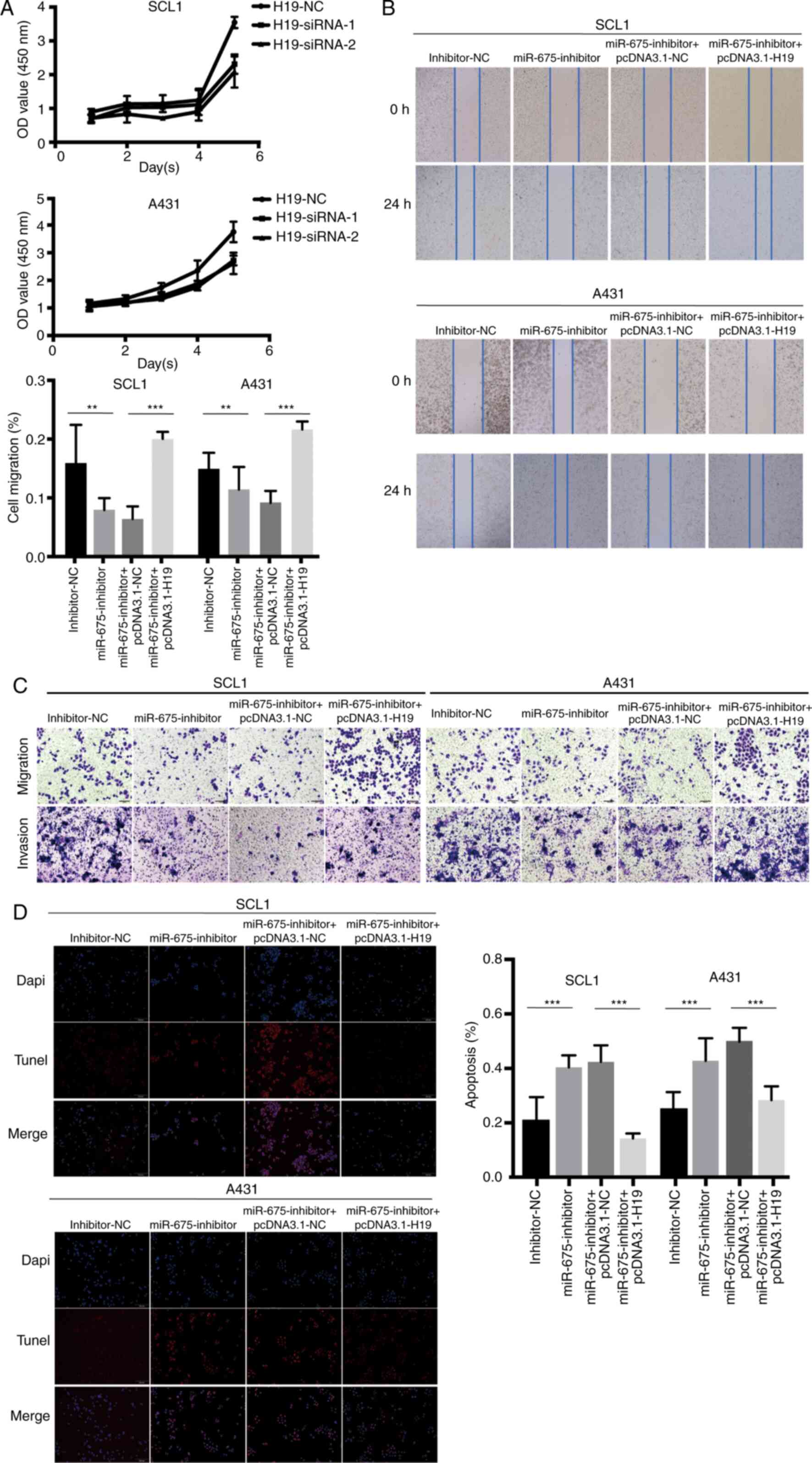 | Figure 4.H19 and miR-675 knockdown inhibits
cSCC cell proliferation, migration and invasion, and promotes
apoptosis. (A) Proliferation of SCL1 and A431 cells transfected
with H19-siRNAs was detected using a Cell Counting Kit-8 assay. (B)
A wound healing assay was used to analyze the migration of cSCC
cells transfected with inhibitor-NC, miR-675 inhibitor,
miR-675-inhibitor + pcDNA3.1-NC and miR-675 inhibitor +
pcDNA3.1-H19. Magnification, ×200. (C) Migration and invasion of
cSCC cells transfected with inhibitor-NC, miR-675 inhibitor,
miR-675-inhibitor + pcDNA3.1-NC and miR-675 inhibitor +
pcDNA3.1-H19 were analyzed using Transwell assays. Magnification,
×200. (D) TUNEL assays were used to determine apoptosis in cSCC
cells transfected with inhibitor-NC, miR-675 inhibitor,
miR-675-inhibitor + pcDNA3.1-NC and miR-675 inhibitor +
pcDNA3.1-H19. Magnification, ×200. **P<0.01 and ***P<0.001.
miR, microRNA; siRNA, small interfering RNA; NC, negative control;
cSCC, cutaneous squamous cell carcinoma. |
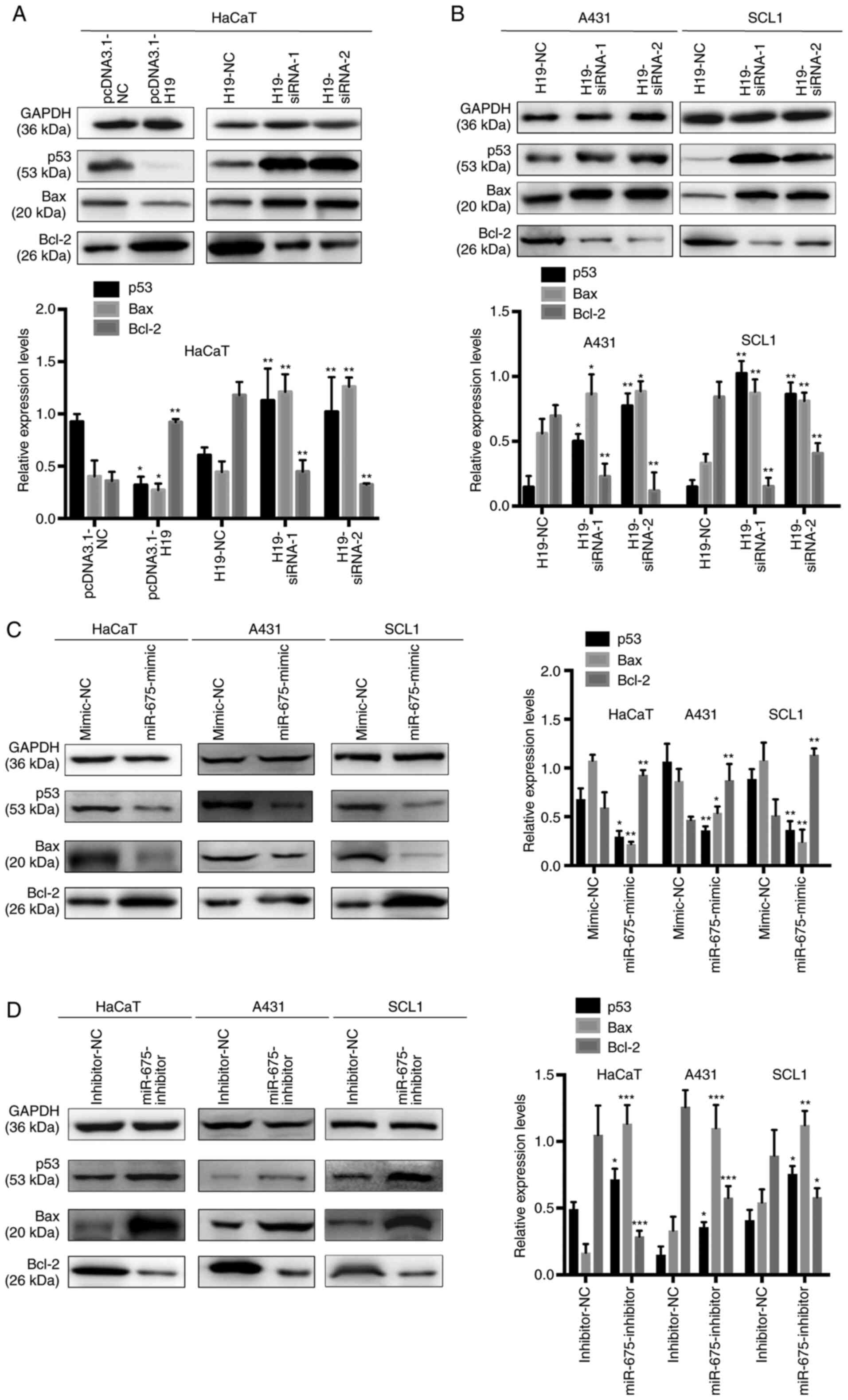
p53 is a direct target gene of the
H19/miR-675 axis in cSCC
To determine the efficiency of the overexpression or
knockdown of H19 on the expression levels of p53 in HaCaT, SCL1 and
A431 cells, gain-of-the-function experiments were performed in
HaCaT cells overexpressing H19 and loss-of-function experiments
were performed by transfecting H19-siRNA into HaCaT, SCL1 and A431
cells. A previous study demonstrated that WT p53 directly
upregulated the expression levels and subsequently activated Bax,
which suggested the presence of a complex interaction between Bax,
Bcl-2 and p53 during cell proliferation and apoptosis (25). In HaCaT cells, p53 and Bax expression
levels were significantly downregulated, while Bcl-2 expression
levels were upregulated following the overexpression of H19.
Conversely, the transfection with H19-siRNA significantly
upregulated the expression levels of p53 and Bax, while decreasing
the expression levels of Bcl-2 in HaCaT, SCL1 and A431 cells
(Fig. 5A and B). These results
indicated that the alterations in H19 expression levels may be
associated with the expression levels of miR-675, which may
subsequently regulate p53 expression in cSCC.
To further determine the underlying regulatory
mechanism, miR-675 was overexpressed using a miR-675 mimic and
knocked down with a miR-675 inhibitor in HaCaT, SCL1 and A431 cells
(Fig. 5C and D), and the expression
levels of p53, Bax and Bcl-2 were analyzed using western blotting.
The expression levels of p53 and Bax were downregulated following
the transfection with the miR-675 mimic and upregulated following
the transfection with the miR-675 inhibitor. In addition, while
Bcl-2 expression levels were upregulated following the transfection
with the miR-675 mimic, the expression levels were downregulated
following the transfection with the miR-675 inhibitor. These
results suggested that p53 may be a target of miR-675 and may be
negatively regulated by the H19/miR-675 axis.
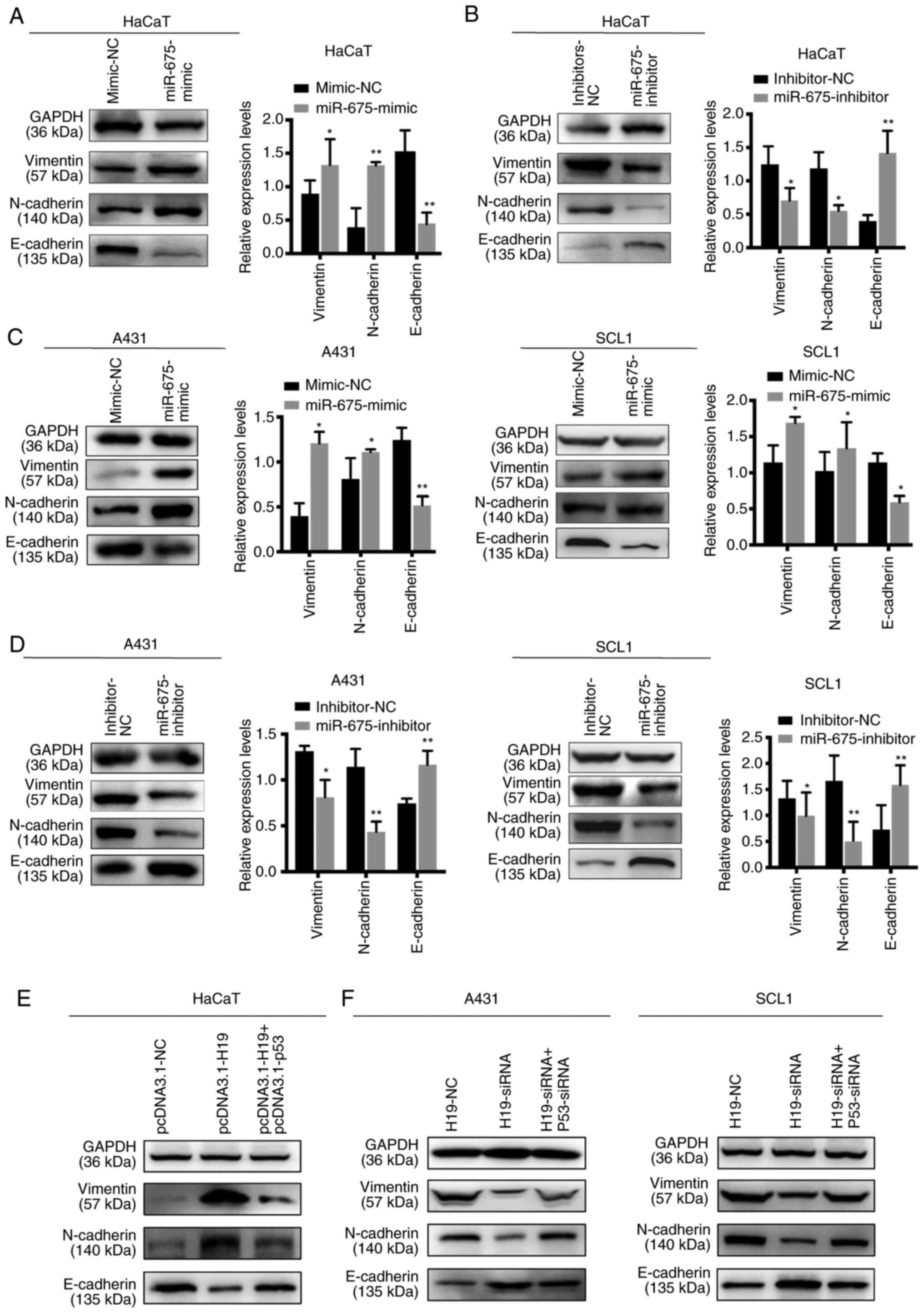
Effect of H19 and miR-675 on the EMT
of cSCC cell lines
EMT is an important process for tumor invasion and
metastasis. Thus, western blotting was used to determine the
effects of H19 and miR-675 on the expression levels of the
EMT-related markers, E-cadherin, vimentin and N-cadherin. The
overexpression of miR-675 in HaCaT cells upregulated the expression
levels of N-cadherin and vimentin, and downregulated the expression
levels of E-cadherin. Conversely, the knockdown of miR-675 markedly
downregulated N-cadherin and vimentin expression levels, while the
expression levels of E-cadherin were upregulated (Fig. 6A and B). Similarly, in SCL1 and A431
cells transfected with the miR-675 mimic, the protein expression
levels of N-cadherin and vimentin were upregulated, while those of
E-cadherin were downregulated (Fig. 6C
and D). The opposite trends were observed following the
transfection of cells with the miR-675 inhibitor. Furthermore, p53
could reverse the effects of H19 on the EMT processes. HaCaT cells
co-transfected with pcDNA3.1-H19 and pcDNA3.1-p53 suppressed the
progression of EMT (Fig. 6E).
Furthermore, cSCC cells co-transfected with H19-siRNA and p53-siRNA
could promote the EMT process inhibited by H19-siRNA (Fig. 6F). Collectively, these findings
suggested that H19 and miR-675 may promote the EMT process in
cSCC.
Discussion
To the best of our knowledge, the present study was
the first to report that the expression levels of H19 were
upregulated in cSCC tissues and cells compared with normal adjacent
tissues and keratinocyte cells, respectively. In addition, the
present study determined the functional association between H19 and
p53; and hypothesized that miR-675, a transcript of H19, may serve
an intermediary role in affecting the expression levels of p53 and
EMT-related markers. The results of the cell functional assays
demonstrated that both H19 and miR-675 expression levels were
significantly upregulated in cSCC cell lines. Moreover, the
knockdown of H19 and miR-675 expression inhibited cSCC cell
proliferation, migration and invasion, and partially downregulated
p53 expression levels. Furthermore, western blotting revealed that
the effect of silencing of H19 expression on the EMT process could
be reversed by p53 in cSCC cell lines. Altogether, these data
suggested that the upregulation of H19 may promote cSCC
tumorigenesis and progression.
Cutaneous squamous cell carcinoma originates from
keratinocytes and is the second most common type of non-melanoma
skin tumor (26). An increasing
number of studies have reported an association between lncRNAs and
cSCC (27). Accumulating evidence has
revealed that H19, as a cancer-related lncRNA, served an important
role in physiological activities and pathological mechanisms
(16,28,29). For
example, H19 was reported to act as an oncogene in bladder,
gastric, breast and thyroid cancer, in addition to glioma (15,30–33). In
colorectal cancer, H19 promoted the cell proliferation, migration
and invasion of cancer cells through sponging miR-138 and
subsequently upregulating the expression levels of HMGA1 (18). In cholangiocarcinoma, upregulated
expression levels of H19 were associated with IκB phosphorylation
and the transport of the p50/p65 heterodimer to the nucleus, which
led to the upregulation of the expression levels of genes involved
in the NF-κB signaling pathway (34).
In addition, H19 expression levels were also revealed to be
downregulated in hepatocellular cancer (35). However, the underlying mechanisms of
H19 dysregulation in cSCC remain poorly understood. The findings of
the present study verified the effects of lncRNA H19 on promoting
the tumorigenesis of cSCC cells, in which the upregulation of H19
expression promoted cSCC cell proliferation, migration and invasion
and prevented apoptosis.
A previous study suggested that H19 may act as a
precursor of miR-675 at the post-transcriptional level (20). In non-small cell lung cancer cells,
the knockdown of H19 markedly downregulated miR-675 expression
levels. Furthermore, the H19/miR-675 axis was discovered to be
upregulated post-hypoxia exposure (COPD) and thus, was suggested as
a potential novel therapeutic tool for the management of non-small
cell lung cancer (36).
The roles of lncRNAs in cSCC have been previously
investigated to improve the current understanding of mutations in
cSCC and lay the foundations for targeted therapy in cSCC (37). A recent review summarized that the
expression levels of lncRNAs, such as p38 inhibited cSCC-associated
lincRNA, long intergenic non-protein coding RNA (LINC) 00319,
testis associated oncogenic lncRNA, AK144841, metastasis associated
lung adenocarcinoma transcript 1, LINC01048 and HOX transcript
antisense RNA were upregulated, while the expression levels of
TINCR ubiquitin domain containing, LINC00520 and growth arrest
specific 5 were downregulated in cSCC (27). However, to the best of our knowledge,
the function of H19 has not been reported in cSCC. Furthermore,
previous studies have also revealed that several miRNAs, such as
miR-21, miR-205, miR-365, miR-31, miR-135b, miR-142, and miR-186,
exerted oncogenic functions while other miRNAs, including miR-20a,
miR-203, miR-181a, miR-125b, miR-34a, miR-148a, miR-214, miR-124,
miR-204, and miR-199a served as tumor suppressors (38). However, the underlying mechanisms of
miR-675 in cSCC remain largely unknown. The results of the present
study indicated that the overexpression of H19 may upregulate
miR-675 expression levels, and that the knockdown of H19 inhibited
cell growth, migration and cell invasion by regulating miR-675
expression in cSCC cell lines.
EMT is a crucial process during the progression of
various types of cancer and was discovered to serve an important
role in the primary epithelial cancer and metastasis into the
adjacent organs and blood vessels (39). E-cadherin is localized on the surfaces
of epithelial cells in regions of cell-cell contact known as
adherens junctions, and has been reported to be closely associated
with tumor invasiveness and cancer metastasis (40,41).
Vimentin was revealed to regulate cell migration through recycling
of endocytosed cell adhesion receptors (42). In addition, N-cadherin acts as a
mesenchymal marker (43). Thus, the
present study aimed to determine the association between H19 and
EMT in cSCC by investigating the expression levels of these
proteins. The western blotting results revealed that the expression
levels of these important EMT markers, E-cadherin, N-cadherin and
vimentin were significantly affected by the H19/miR-675 axis in
cSCC cell lines. Furthermore, the findings suggested that the
overexpression of p53 rescued the effects induced by H19 in cSCC.
Therefore, the results of the present study indicated that the
H19/miR-675 axis may promote metastasis by affecting the EMT
process. To the best of our knowledge, the present study was the
first to provide evidence of the function of the H19/miR-675/p53
axis in cSCC.
In conclusion, the findings of the present study
suggested that H19 may act as an oncogenic lncRNA in cSCC
proliferation, migration and invasion via modulating miR-675, and
therefore may be associated with a poor prognosis in cSCC. These
data suggested that H19 may represent a potential target for cSCC
treatment and offered novel insights for further diagnostic and
therapeutic medical research to investigate the role of H19 in
cSCC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The analyzed data sets generated during the study
are available from the corresponding author on reasonable
request.
Authors' contributions
WZ and KZ conducted the research and performed the
experiments. WZ, KZ, XZ and CW performed the data collection and
analyses. WZ and KZ designed the experiments and supervised the
procedures and wrote the manuscript. XZ and DD performed the
surgical excision of skin lesions of patients, collected clinical
tissues samples and summarized the information of patients. ZY
proposed the research direction, revising the manuscript for
important intellectual content. All authors read and approved the
manuscript and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Xinhua Hospital, School of Medicine, Shanghai Jiao
Tong University (Shanghai, China). All patients provided written
informed consent prior to participation.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Motaparthi K, Kapil JP and Velazquez EF:
Cutaneous squamous cell carcinoma: Review of the eighth edition of
the American Joint Committee on Cancer staging guidelines,
prognostic factors, and histopathologic variants. Adv Anat Pathol.
24:171–194. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ackerman AB and Mones JM: Solar (actinic)
keratosis is squamous cell carcinoma. Br J Dermatol. 155:9–22.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Koike Y, Yozaki M, Kuwatsuka Y and Utani
A: Epithelial-mesenchymal transition in Bowen's disease when
arising de novo and acquiring invasive capacity. J Dermatol.
45:748–750. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ishitsuka Y, Kawachi Y, Taguchi S,
Maruyama H, Nakamura Y, Fujisawa Y, Furuta J, Nakamura Y, Ishii Y
and Otsuka F: Pituitary tumor-transforming gene 1 as a
proliferation marker lacking prognostic value in cutaneous squamous
cell carcinoma. Exp Dermatol. 22:318–322. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Graindorge S, Cognat V, Johann To Berens
P, Mutterer J and Molinier J: Photodamage repair pathways
contribute to the accurate maintenance of the DNA methylome
landscape upon UV exposure. PLoS Genet. 15:e10084762019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fernandez Figueras MT: From actinic
keratosis to squamous cell carcinoma: Pathophysiology revisited. J
Eur Acad Dermatol Venereol. 31 (Suppl 2):S5–S7. 2017. View Article : Google Scholar
|
|
7
|
Lobl M, Grinnell M, Phillips A, Abels J
and Wysong A: The correlation between immunohistochemistry findings
and metastasis in squamous cell carcinoma: A review. Dermatol Surg.
Nov 3–2020.(Epub ahead of print). doi:
10.1097/DSS.0000000000002850. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tian K, Liu W, Zhang J, Fan X, Liu J, Zhao
N, Yao C and Miao G: MicroRNA-125b exerts antitumor functions in
cutaneous squamous cell carcinoma by targeting the STAT3 pathway.
Cell Mol Biol Lett. 25:122020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cerk S, Schwarzenbacher D, Adiprasito JB,
Stotz M, Hutterer GC, Gerger A, Ling H, Calin GA and Pichler M:
Current status of long non-coding RNAs in human breast cancer. Int
J Mol Sci. 17:14852016. View Article : Google Scholar
|
|
10
|
Hao S, Yao L, Huang J, He H, Yang F, Di Y,
Jin C and Fu D: Genome-wide analysis identified a number of
dysregulated long noncoding RNA (lncRNA) in human pancreatic ductal
adenocarcinoma. Technol Cancer Res Treat. 17:15330346177484292018.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ponzio G, Rezzonico R, Bourget I, Allan R,
Nottet N, Popa A, Magnone V, Rios G, Mari B and Barbry P: A new
long noncoding RNA (lncRNA) is induced in cutaneous squamous cell
carcinoma and down-regulates several anticancer and cell
differentiation genes in mouse. J Biol Chem. 292:12483–12495. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pachnis V, Belayew A and Tilghman SM:
Locus unlinked to alpha-fetoprotein under the control of the murine
raf and Rif genes. Proc Natl Acad Sci USA. 81:5523–5527. 1984.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhou W, Ye XL, Xu J, Cao MG, Fang ZY, Li
LY, Guan GH, Liu Q, Qian YH and Xie D: The lncRNA H19 mediates
breast cancer cell plasticity during EMT and MET plasticity by
differentially sponging miR-200b/c and let-7b. Sci Signal.
10:eaak95572017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wu J, Qin Y, Li B, He WZ and Sun ZL:
Hypomethylated and hypermethylated profiles of H19DMR are
associated with the aberrant imprinting of IGF2 and H19 in human
hepatocellular carcinoma. Genomics. 91:443–450. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li X, Li Q, Jin X, Guo H and Li Y: Long
non-coding RNA H19 knockdown inhibits the cell viability and
promotes apoptosis of thyroid cancer cells through regulating the
PI3K/AKT pathway. Exp Ther Med. 18:1863–1869. 2019.PubMed/NCBI
|
|
16
|
Ghafouri-Fard S, Esmaeili M and Taheri M:
H19 lncRNA: Roles in tumorigenesis. Biomed Pharmacother.
123:1097742020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang WT, Ye H, Wei PP, Han BW, He B, Chen
ZH and Chen YQ: LncRNAs H19 and HULC, activated by oxidative
stress, promote cell migration and invasion in cholangiocarcinoma
through a ceRNA manner. J Hematol Oncol. 9:1172016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yang Q, Wang X, Tang C, Chen X and He J:
H19 promotes the migration and invasion of colon cancer by sponging
miR-138 to upregulate the expression of HMGA1. Int J Oncol.
50:1801–1809. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shi G, Li H, Gao F and Tan Q: lncRNA H19
predicts poor prognosis in patients with melanoma and regulates
cell growth, invasion, migration and epithelial-mesenchymal
transition in melanoma cells. Onco Targets Ther. 11:3583–3595.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cai X and Cullen BR: The imprinted H19
noncoding RNA is a primary microRNA precursor. RNA. 13:313–316.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Simons M and Raposo G: Exosomes-vesicular
carriers for intercellular communication. Curr Opin Cell Biol.
21:575–581. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kim NH, Lee CH and Lee AY: H19 RNA
downregulation stimulated melanogenesis in melasma. Pigment Cell
Melanoma Res. 23:84–92. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Betel D, Wilson M, Gabow A, Marks DS and
Sander C: The microRNA.org resource: Targets and expression.
Nucleic Acids Res. 36((Database Issue)): D149–D153. 2008.PubMed/NCBI
|
|
25
|
Miyashita T and Reed JC: Tumor suppressor
p53 is a direct transcriptional activator of the human bax gene.
Cell. 80:293–299. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sand M, Bechara FG, Gambichler T, Sand D,
Bromba M, Hahn SA, Stockfleth E and Hessam S: Circular RNA
expression in cutaneous squamous cell carcinoma. J Dermatol Sci.
83:210–218. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang Y, Sun B, Wen X, Hao D, Du D, He G
and Jiang X: The roles of lncRNA in cutaneous squamous cell
carcinoma. Front Oncol. 10:1582020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li J, Su T, Zou C, Luo W, Shi G, Chen L,
Fang C and Li C: Long non-coding RNA H19 regulates porcine
satellite cell differentiation through miR-140-5p/SOX4 and DBN1.
Front Cell Dev Biol. 8:5187242020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lecerf C, Peperstraete E, Le Bourhis X and
Adriaenssens E: Propagation and maintenance of cancer stem cells: A
major influence of the long non-coding RNA H19. Cells. 9:26132020.
View Article : Google Scholar
|
|
30
|
Hua Q, Lv X, Gu X, Chen Y, Chu H, Du M,
Gong W, Wang M and Zhang Z: Genetic variants in lncRNA H19 are
associated with the risk of bladder cancer in a Chinese population.
Mutagenesis. 31:531–538. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu G, Xiang T, Wu QF and Wang WX: Long
noncoding RNA H19-derived miR-675 enhances proliferation and
invasion via RUNX1 in gastric cancer cells. Oncol Res. 23:99–107.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Peperstraete E, Lecerf C, Collette J,
Vennin C, Raby L, Völkel P, Angrand PO, Winter M, Bertucci F,
Finetti P, et al: Enhancement of breast cancer cell aggressiveness
by lncRNA H19 and its Mir-675 derivative: Insight into shared and
different actions. Cancers (Basel). 12:17302020. View Article : Google Scholar
|
|
33
|
Hu Q, Yin J, Zeng A, Jin X, Zhang Z, Yan W
and You Y: H19 functions as a competing endogenous RNA to regulate
EMT by sponging miR-130a-3p in glioma. Cell Physiol Biochem.
50:233–245. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yang S, Fang F, Yu X, Yang C, Zhang X,
Wang L, Zhu L, Shao K and Zhu T: Knockdown of H19 inhibits the
pathogenesis of acne vulgaris by targeting the miR-196a/TLR2/NF-κB
axis. Inflammation. 43:1936–1947. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Iempridee T: Long non-coding RNA H19
enhances cell proliferation and anchorage-independent growth of
cervical cancer cell lines. Exp Biol Med (Maywood). 242:184–193.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zheng ZH, Wu DM, Fan SH, Zhang ZF, Chen GQ
and Lu J: Upregulation of miR-675-5p induced by lncRNA H19 was
associated with tumor progression and development by targeting
tumor suppressor p53 in non-small cell lung cancer. J Cell Biochem.
120:18724–18735. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
García-Sancha N, Corchado-Cobos R,
Pérez-Losada J and Cañueto J: MicroRNA dysregulation in cutaneous
squamous cell carcinoma. Int J Mol Sci. 20:21812019. View Article : Google Scholar
|
|
39
|
Singh A and Settleman J: EMT, cancer stem
cells and drug resistance: An emerging axis of evil in the war on
cancer. Oncogene. 29:4741–4751. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lee JM, Dedhar S, Kalluri R and Thompson
EW: The epithelial-mesenchymal transition: New insights in
signaling, development, and disease. J Cell Biol. 172:973–981.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gumbiner BM: Regulation of
cadherin-mediated adhesion in morphogenesis. Nat Rev Mol Cell Biol.
6:622–634. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ivaska J, Vuoriluoto K, Huovinen T, Izawa
I, Inagaki M and Parker PJ: PKCepsilon-mediated phosphorylation of
vimentin controls integrin recycling and motility. EMBO J.
24:3834–3845. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wang L and Yu P: miR-300 promotes
proliferation and EMT-mediated colorectal cancer migration and
invasion by targeting p53. Oncol Rep. 36:3225–3232. 2016.
View Article : Google Scholar : PubMed/NCBI
|