Introduction
CRC is one of the most common malignancies of the
gastrointestinal tract, ranking third among common causes of
cancer-related mortality worldwide (1). Over the past decade, substantial
improvements have been made in the diagnosis and management of CRC,
achieving an annual decline of ~3% in CRC deaths (2). Unfortunately, CRC still causes ~600,000
deaths annually (3). Metastatic
lesions, particularly liver metastases, are the main cause of
CRC-related mortality (4). Similar to
most solid tumors, CRC has a complex pathogenesis involving
inactivation of tumor suppressor genes and activation of oncogenes
(5). Although increasing evidence has
partly uncovered the regulatory mechanism underlying CRC
tumorigenesis, the exact mechanism underlying cancer metastasis
remains elusive. Therefore, in-depth study of the molecules
implicated in the metastasis and recurrence of CRC is crucial for
improving the prognosis and treatment of the patients.
MicroRNAs (miRNAs), a class of small non-coding RNAs
with a length of 19–24 nucleotides, mainly exert their biological
effects by binding to the 3′-untranslated region (3′-UTR) of target
mRNAs to cause mRNA degradation or translational repression at the
post-transcriptional level (6,7). miRNAs
have been identified as tumor suppressors or oncogenes in multiple
cancers, including CRC (8–10). Mounting evidence has also revealed
that dysregulation of miRNAs is associated with tumor growth, cell
proliferation and migration (11,12).
Recent studies have demonstrated that miR-129 is expressed at low
levels in several cancers, including renal cell carcinoma (13), bladder cancer (14) and gastric cancer (15), in which miR-129 has been confirmed to
act as a tumor suppressor. In addition, miR-129 has been reported
to promote apoptosis and chemosensitivity to 5-fluorouracil in CRC
(16). Those studies indicated that
miR-129 is implicated in CRC progression, but its role and
potential regulatory mechanisms in CRC metastasis has yet to be
fully elucidated.
Sex-determining region Y-related high-mobility
group-box 4 (SOX4), a member of the SOX transcription factor
family, is involved in embryonic development and cell
differentiation (17,18). SOX4 overexpression in CRC was
previously demonstrated to be correlated with poor prognosis and
recurrence (19), whereas knockdown
of SOX4 was reported to suppress CRC cell proliferation and
invasion (20). SOX4 is a potential
target for multiple miRNAs in CRC, including miR-320 (21), miR-363-3p (22) and miR-539 (23). In addition, miR-129-5p was shown to
suppress cell proliferation and invasion via regulating the
SOX4/Wnt/β-catenin signaling pathway in chondrosarcoma (24). Kang et al (25) reported that miR-129-2 inhibits cell
proliferation and migration by targeting SOX4 in esophageal
carcinoma. However, whether miR-129 can inhibit the malignant
phenotypes of CRC by regulating SOX4 has not been reported to
date.
Therefore, the aim of the present study was to
elucidate the interaction between miR-129 and SOX4 in CRC. The
expression of miR-129 and SOX4 was assessed in CRC tissues and cell
lines, and it was investigated whether the effects of miR-129 on
CRC cell proliferation, migration, invasion and
epithelial-to-mesenchymal transition (EMT) were mediated through
targeting SOX4 and activation of the nuclear factor (NF)-κB
signaling pathway.
Materials and methods
Tissue samples
A cohort of 60 CRC tissue samples and matched
tumor-adjacent tissue samples were acquired from patients
undergoing a surgical procedure between May 2017 and May 2018,
according to the institutional guidelines. The clinical
characteristics of the 60 patients with CRC are summarized in
Table I. For all patients, original
diagnosis and tumor grading were conducted in a blinded manner by
two experienced pathologists according to the American Joint
Committee on Cancer (AJCC) pathologic tumor-node metastasis (TNM)
classification system (26). The
protocol of the present study was approved by the Ethics Committee
of the First Affiliated Hospital of Gannan Medical University.
Informed consent was obtained from all the patients. The tissue
samples were collected during surgery and immediately stored in
liquid nitrogen.
 | Table I.Clinical characteristics of 60
patients with colorectal cancer. |
Table I.
Clinical characteristics of 60
patients with colorectal cancer.
| Variables | No. |
|---|
| Age, years |
|
|
<60 | 39 |
|
≥60 | 21 |
| Sex |
|
|
Male | 33 |
|
Female | 27 |
| Tumor stage |
|
|
I–II | 29 |
|
III–IV | 31 |
| Lymph node
metastasis |
|
|
Yes | 35 |
| No | 25 |
Cell culture
The human CRC cell lines SW620 (CL-0225) and SW480
(CL-0223) were obtained from Procell Life Science & Technology
Co., Ltd., and the normal colorectal cell line NCM460 (BNF-3068)
was purchased from the Cell Bank of Type Culture Collection of the
Chinese Academy of Sciences (Shanghai, China). All cells were
cultured in DMEM (HyClone, Cytiva) supplemented with 10% FBS
(Gibco; Thermo Fisher Scientific, Inc.), and 1%
penicillin-streptomycin (Beijing Solarbio Science & Technology
Co., Ltd.) in a humidified incubator with 5% CO2 at
37°C.
Transfection
miR-129 mimic, inhibitor and negative control (NC)
were purchased from Guangzhou RiboBio Co., Ltd. At 24 h before
transfection, SW620 and SW480 cells were seeded in 6-well plates at
a density of 2×105 cells/well. The cells in each well
were transfected with a solution of 50 pmol/ml miR-129 mimic or
inhibitor using Lipofectamine™ 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. Fresh
medium was replaced following transfection for 6 h. Transfection
efficiency was determined using reverse transcription-quantitative
PCR (RT-qPCR) assay following transfection for 48 h. The sequences
of the oligonucleotides were as follows: miR-129 mimic:
5′-CUUUUUGCGGUCUGGGCUUGC-3′; miR-129 inhibitor:
5′-GCAAGCCCAGACCGCAAAAAG-3′; and miR-NC:
5′-AGUGCUAGUAGCUUUGCAUGU-3′.
RT-qPCR analysis
Total RNA was extracted from CRC cell lines, CRC
tissues and matched tumor-adjacent tissues using TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) for miRNA or
mRNA analysis. To quantify mRNA expression, cDNA was synthesized
using a PrimeScript™ RT reagent kit (Takara Biotechnology Co.,
Ltd.) according to the manufacturer's protocol at 37°C for 15 min
and 85°C for 5 sec. SYBR® Premix Ex Taq II (Takara
Biotechnology Co., Ltd.) was used to perform RT-qPCR in an Applied
Biosystems 7500 Real-Time PCR system (Applied Biosystems; Thermo
Fisher Scientific, Inc.). The thermocycling conditions were as
follows: 3 min at 95°C; 40 cycles at 95°C for 5 sec and 60°C for 30
sec, followed by 72°C for 30 sec. To quantify miRNA expression,
reverse transcription and qPCR were performed using Bulge-Loop™
miRNA RT-qPCR Primer and Bulge-Loop™ miRNA RT-qPCR Starter kit
(Guangzhou RiboBio Co., Ltd). The thermocycling conditions of qPCR
were as follows: 95°C for 10 min, followed by 40 cycles of 95°C for
2 sec, 60°C for 20 sec and 70°C for 10 sec. The 2−∆∆Cq
method was used for comparative quantitation (27). GAPDH and U6 small nuclear RNA were
used as internal normalization controls.
Western blot analysis
Total protein was extracted from CRC cells using
radioimmunoprecipitation assay lysis buffer (Wuhan Boster
Biological Technology, Ltd.) containing protease inhibitors (Roche
Diagnostics) according to the manufacturer's protocol. Total
protein concentrations were measured using a bicinchoninic acid
protein assay kit (Wuhan Boster Biological Technology, Ltd.). Equal
amounts (20 µg) of protein were separated with 10% SDS-PAGE and
then transferred onto PVDF membranes (EMD Millipore). The membranes
were blocked by 5% skimmed milk in TBST for 1 h at room
temperature, followed by incubation at 4°C overnight with primary
antibodies against SOX4 (ab80261; 1:500), P21 (ab227443; 1:1,000),
P27 (ab215434; 1:1,000), cyclin D1 (ab226977; 1:1,000), matrix
metallopeptidase (MMP)-2 (ab37150; 1:1,000), MMP-9 (ab38898;
1:1,000), E-cadherin (ab15148; 1:500), N-cadherin (ab18203; 1:500),
vimentin (ab137321;1:1,000), ataxia telangiectasia-mutated (ATM;
ab17995; 1:5,000), P50 (ab131546; 1:1,000), NF-κB inhibitor α
(IkBα; ab97783; 1:2,000), p-IkBα (ab24784; 1:2,000), NF-κBp65
(ab16502; 1:1,000), p-NF-κBp65 (ab86299; 1:2,000) and GAPDH
(ab9485; 1:2,000), all purchased from Abcam. Subsequently, the
membranes were washed with Tris-buffered saline with Tween-20 and
incubated with HRP-conjugated goat anti-rabbit immunoglobulin G
secondary antibody (BA1054; Wuhan Boster Biological Technology,
Ltd.) at a dilution of 1:5,000 for 1 h at room temperature.
Enhanced chemiluminescence reagent (EMD Millipore) was used to
detect the signals on the membranes using Bio-Rad ChemiDocTMMP
system (Bio-Rad Laboratories, Inc.). Image-ProPlus software v6.0
(Media Cybernetics, Inc.) was used to quantify bands, and GAPDH was
used as a loading control.
Immunofluorescence assay
NCM460, SW620 and SW480 cells were plated in 12-well
plates and incubated overnight for adherence. Briefly, the cells
were fixed in 100% methanol for 5 min at room temperature and then
permeabilized with 0.1% Triton X-100 and incubated in 5% BSA
(Thermo Fisher Scientific, Inc.) to block non-specific
protein-protein interactions. The cells were then incubated with
primary rabbit anti-SOX4 antibody (ab80261; 1:500; Abcam) overnight
at 4°C. The cells were washed twice with PBS/0.1% Tween-20 and
incubated with a secondary antibody (green) of
DyLight®488 goat anti-rabbit IgG-H&L (ab96899;
1:250) for 1 h at room temperature. Cell nuclei were counterstained
with 4′,6-diamidino-2-phenylindole (DAPI) staining solution. All
images were captured using an Olympus FV-1000 confocal microscope
(Olympus Corporation) at a magnification of ×200, and adjusted for
brightness using ImageJ software v6.0 (National Institutes of
Health).
Cell proliferation and colony
formation assay
To evaluate cell viability, NCM460, SW620 or SW480
cells (6×103/well) were seeded in 96-well culture plates
and incubated in an atmosphere containing 5% CO2 at
37°C. After the cells had reached 70% confluence, they were
transfected with miR-129 mimic or inhibitor and maintained for 24,
48 and 72 h. Subsequently, 10 µl MTT diluted in fresh medium (90
µl) was added to each well and incubated for an additional 4 h at
37°C. Subsequently, the culture medium was removed and 110 µl
formazan was added into each well to dissolve the formed crystals
with agitation for 10 min. The absorbance of each well was measured
at 490 nm using a microplate reader (Thermo Fisher Scientific,
Inc.). All experiments were performed in triplicate.
To determine colony formation ability, NCM460, SW620
or SW480 cells (1,000 cells/dish) were seeded in 3.5-cm cell
culture dishes and incubated in complete DMEM at 37°C for 2 weeks.
Subsequently, the colonies were fixed with 20% methanol for 10 min,
followed by staining with 0.1% crystal violet solution (Beyotime
Institute of Biotechnology) for 5 min at room temperature. The
number of formed colonies (>50 cells) was counted under an
inverted microscope (Nikon Eclipse TE2000; Nikon Corporation) at a
magnification of ×40. Plating efficiency (PE) was calculated as
follows: PE=(number of colonies formed/number of plated cells)
×100%. Three independent experiments were performed.
Wound healing assay
To assess the tumor cell migration ability, a wound
healing assay was performed. Briefly, cells were evenly plated in
6-well culture plates and allowed to reach 70% confluence. An
artificial wound across the cell monolayer was made using a 200-µl
sterile pipette tip. The cells were then washed with PBS thrice and
incubated in serum-free DMEM. To visualize migrating cells and
wound healing, images were captured at 0 and 48 h using an inverted
microscope (Nikon Eclipse TE2000; Nikon Corporation). at a
magnification of ×200.
Transwell migration and invasion
assays
Cell migration assay was performed using a 24-well
Transwell chamber (Corning, Inc.) with 8.0-µm pore membranes.
Briefly, NCM460, SW620 or SW480 cells (1×105) were
suspended in a serum-free DMEM and then plated in the upper chamber
(200 µl). The lower chamber was filled with 600 µl DMEM
supplemented with 10% FBS. After being incubated for 48 h at 37°C
under 5% CO2, non-migrated cells were wiped with a
cotton swab, and the migrated cells on the lower surface were fixed
with 70% ethanol for 20 min at room temperature and stained with
0.1% crystal violet solution for 15 min at room temperature.
Finally, the migrated cells were photographed under an inverted
microscope (Nikon Eclipse TE2000; Nikon Corporation) in five
randomly selected fields at a magnification of ×200 and counted
using ImageJ software v6.0 (National Institutes of Health). The
invasion assay was performed in a similar manner, except that the
Transwell chambers were coated with Matrigel (100 µg/ml at 37°C; BD
Biosciences).
Cell cycle assay
SW620 or SW480 cells were harvested at 48 h
post-transfection by centrifugation at 800 × g at room temperature
for 5 min. The collected cells were suspended in 1 ml PBS
(1×106/ml) and then centrifuged at 800 × g at room
temperature for 2 min to remove the supernatant. Subsequently, the
cells were fixed with 70% ethanol (500 µl) at 4°C overnight. Prior
to staining, the cells were washed with PBS and then suspended in
100 µl RNase A solution (Beijing Solarbio Science & Technology
Co., Ltd.) and bathed in 37°C for 30 min, then stained with
propidium iodide (PI; 400 µl; Beijing Solarbio Science &
Technology Co., Ltd.) for 5 min at 4°C in the dark. Alterations in
DNA content were analyzed using a FACSCalibur™ flow cytometer (BD
Biosciences) within 1 h and the data were analyzed using FlowJo
10.07 software (Tree Star, Inc.).
Luciferase reporter assay
Bioinformatics software, including TargetScan
(http://www.targetscan.org/), mirDB
(http://mirdb.org/) and DIANA TOOLS (http://diana.imis.athena-innovation.gr),
was used to predict the putative target genes of miR-129 in SW620
or SW480 cells. The SOX4 3′-UTR encompassing the wild-type (WT) or
mutant (Mut) fragments of the miR-129 binding site was amplified by
PCR and then cloned into a pmirGlO Dual-luciferase miRNA Target
Expression Vector (Promega Corporation) to form the reporter
vector, namely SOX4 WT and SOX4 Mut, respectively.
For luciferase reporter assays, SW620 or SW480 cells
were plated in 24-well plates (5×104 cells/well) and
allowed to reach 50% confluence. Then, the cells were
co-transfected with 2 µg/ml of SOX4 WT or SOX4 Mut, together with
100 ng/ml of Renilla luciferase pRL-TK plasmid (Promega
Corporation) and 50 pmol/ml of miR-129 mimic or corresponding NC
using Lipofectamine™ 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). The Dual-Luciferase Reporter Assay kit (Promega
Corporation) was used to calculate the luciferase activity at 48 h
post-transfection. Firefly luciferase activity was normalized to
Renilla luciferase activity for each tested well.
Microarray analysis
Microarray analysis was conducted to detect the
expression profiles of miRNAs in CRC using CRC tissue samples and
matched tumor-adjacent tissue samples. miRNAs isolated from
clinical specimens were labeled by Cy3 and hybridized with Agilent
Human miRNA Microarrays (Agilent Technologies, Inc.) following the
manufacturer's instructions. Microarrays were scanned by an Agilent
SureScan Microarray Scanner and then analyzed with GeneSpringGX
software 11.0 (Agilent Technologies, Inc.). The differentially
expressed miRNAs were screened with fold change >2 and
P<0.05.
Statistical analysis
Statistical analyses were performed using SPSS 20.0
software (IBM Corp.). Values are presented as mean ± standard
deviation (SD) from three separate experiments. Differences among
multiple groups were compared by one-way ANOVA followed by Tukey's
post hoc test, and differences between tumor specimens and matched
tumor-adjacent tissue samples were compared by paired Student's
t-test. *P<0.05 and **P<0.01 were considered to indicate
statistically significant differences.
Results
miR-129 is downregulated and SOX4 is
increased in CRC tissues and cell lines
To identify miRNAs potentially implicated in CRC,
microarray expression profiles were analyzed. Three human CRC
microarray datasets were selected and analyzed for miRNAs
consistently aberrantly expressed between CRC and normal tissues.
miR-129 was found to be significantly decreased in CRC. RT-qPCR,
western blot or immunofluorescence assays were performed to
evaluate the changes in the expression levels of miR-129 and SOX4
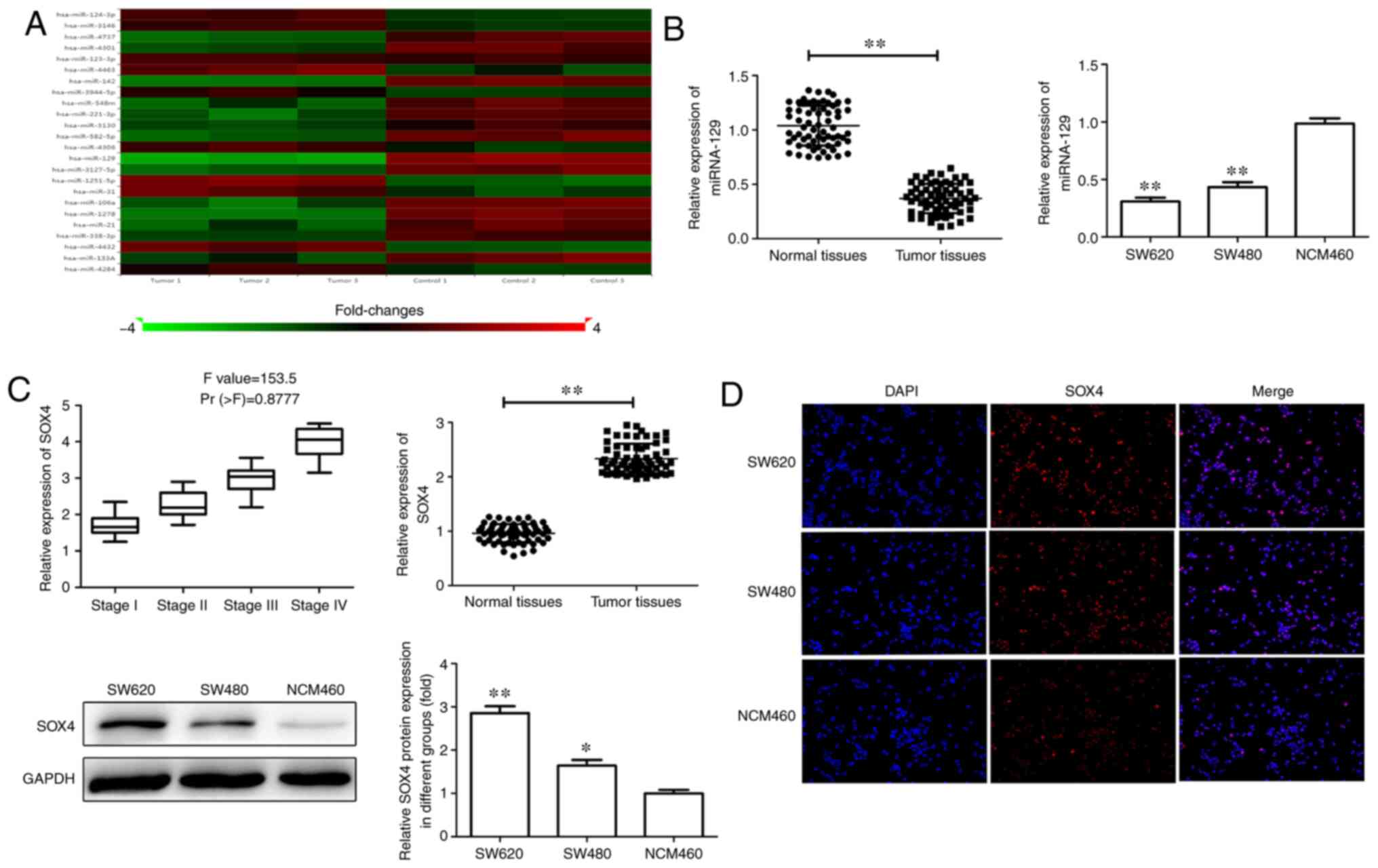
in CRC tissues and cell lines. As shown in Fig. 1A, miR-129 expression was significantly
decreased in CRC tissues compared with that in adjacent tissues.
Similarly, SW620 and SW480 cells also exhibited significantly lower
expression of miR-129 compared with that in NCM460 cells (Fig. 1B). Furthermore, SOX4 expression was
measured in CRC tissues and cell lines and was found to be
significantly upregulated at both the mRNA and protein levels in
CRC tissues and cell lines. Moreover, elevated SOX4 expression was
positively correlated with advanced TNM stage (Fig. 1B-D). These data demonstrated miR-129
expression was decreased whereas SOX4 expression was increased in
CRC, suggesting that miR-129 and SOX4 may play a key role in CRC
tumorigenesis and progression.
Proliferation, migration and invasion
of CRC cells
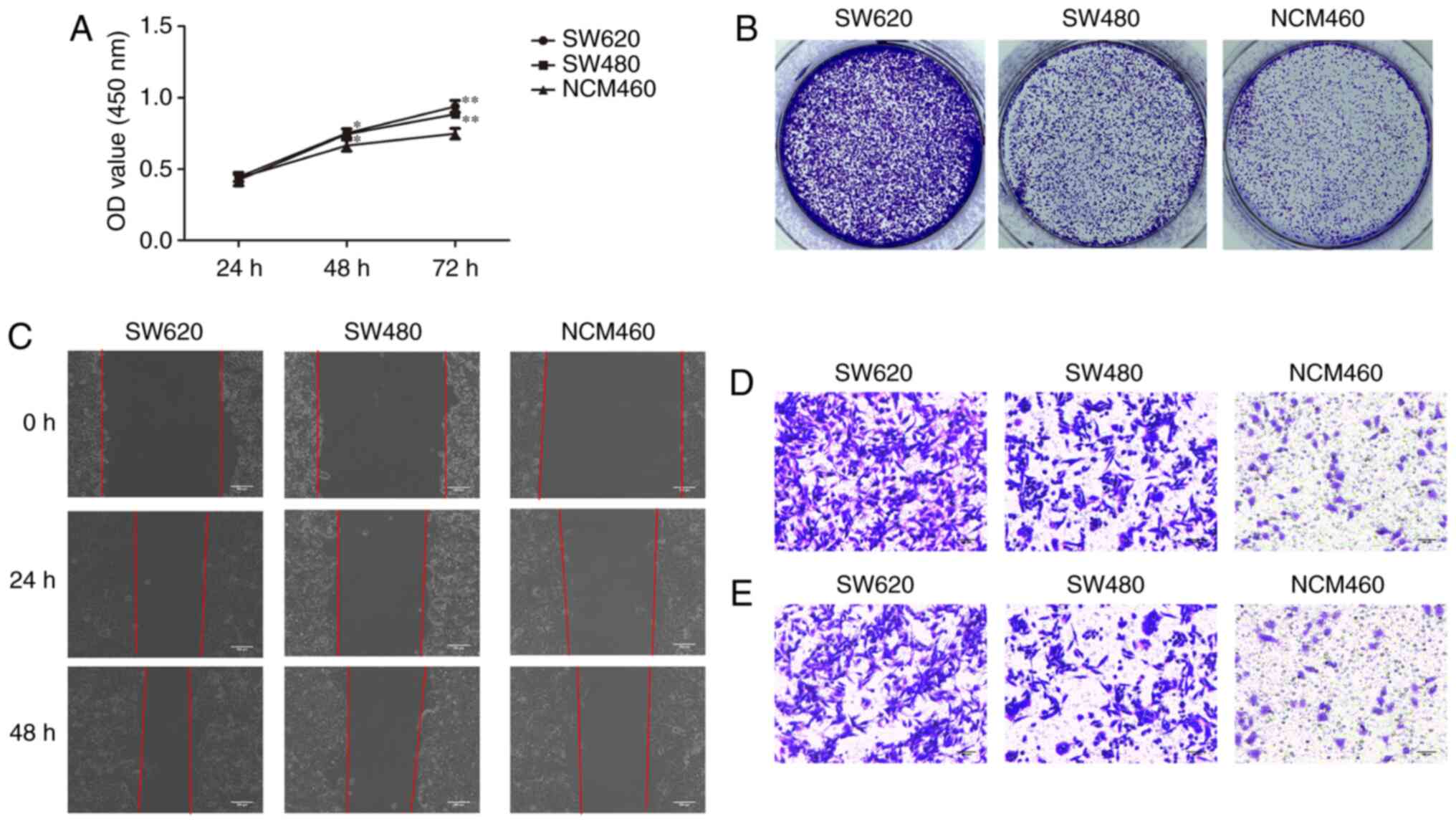
To investigate the malignant behavior of CRC cells,
the proliferation, migration and invasion of CRC cell lines and a
normal colorectal cell line were examined. As shown in Fig. 2, the proliferation, migration and
invasion capacity of SW620 and SW480 cells was significantly higher
compared with that of NCM460 cells. In addition, compared with
SW480 cells, SW620 cells exhibited a significantly higher
proliferation, migration and invasion capacity, indicating that
SW620 cells display a more malignant behavior.
Cell proliferation is suppressed by
miR-129 overexpression and promoted by miR-129 inhibition
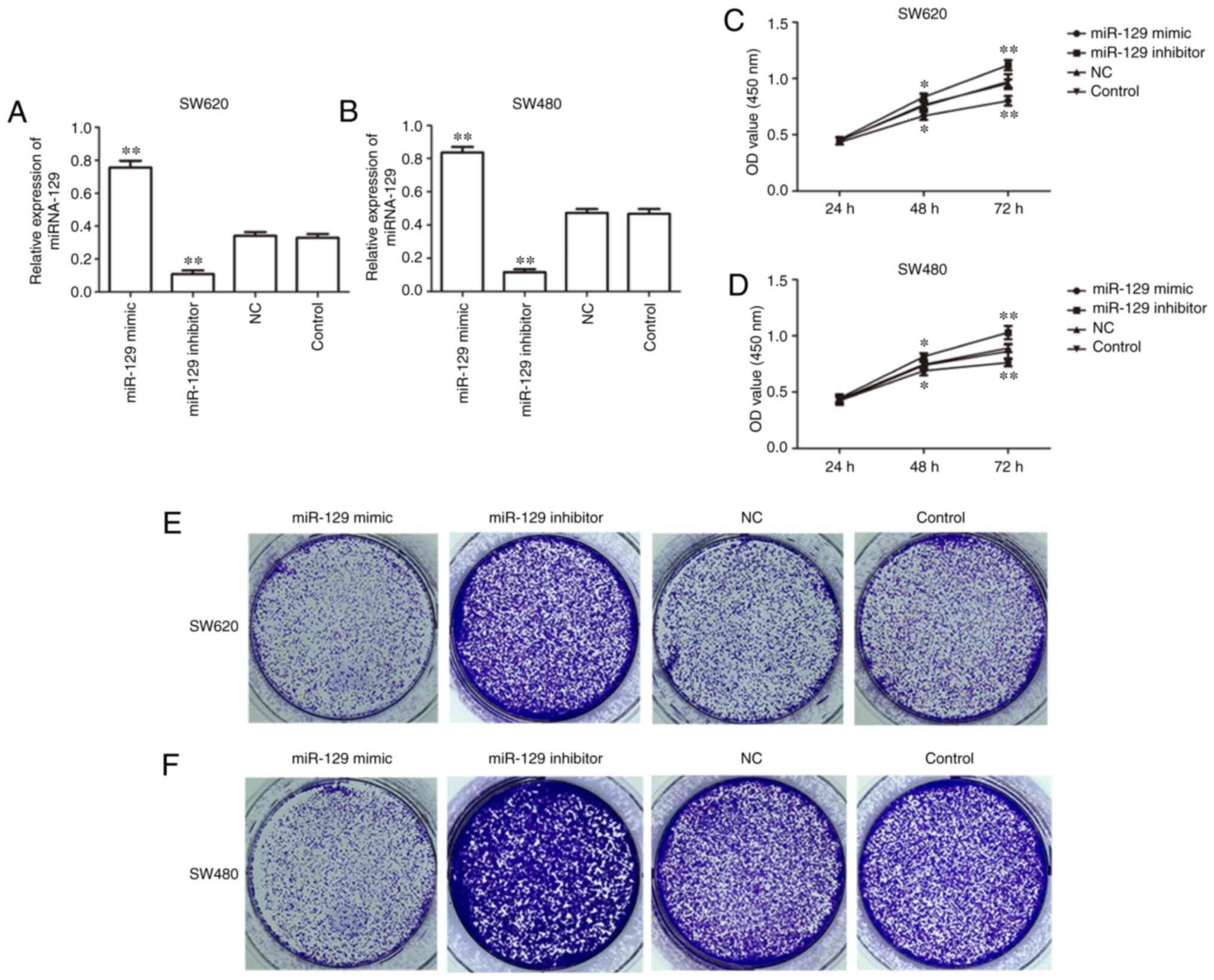
To investigate the effect of miR-129 on human CRC
cells, gain-of-function and loss-of-function experiments were
performed to verify the biological role of miR-129. miR-129
expression was shown to be markedly upregulated following
transfection with miR-129 mimic and downregulated following
transfection with miR-129 inhibitor in SW620 and SW480 cells
(Fig. 3A and B). RT-qPCR analysis was
performed to evaluate the transfection efficiency and the result
indicated a high transfection efficiency in SW620 and SW480 cells.
Furthermore, MTT assay revealed that miR-129 overexpression
inhibited whereas miR-129 inhibition promoted the proliferation of
SW620 and SW480 cells (Fig. 3C and
D). In addition, colony formation assay demonstrated that
miR-129 overexpression decreased the number of colonies formed,
while miR-129 inhibition increased the colony number in SW620 and
SW480 cells (Fig. 3E and F).
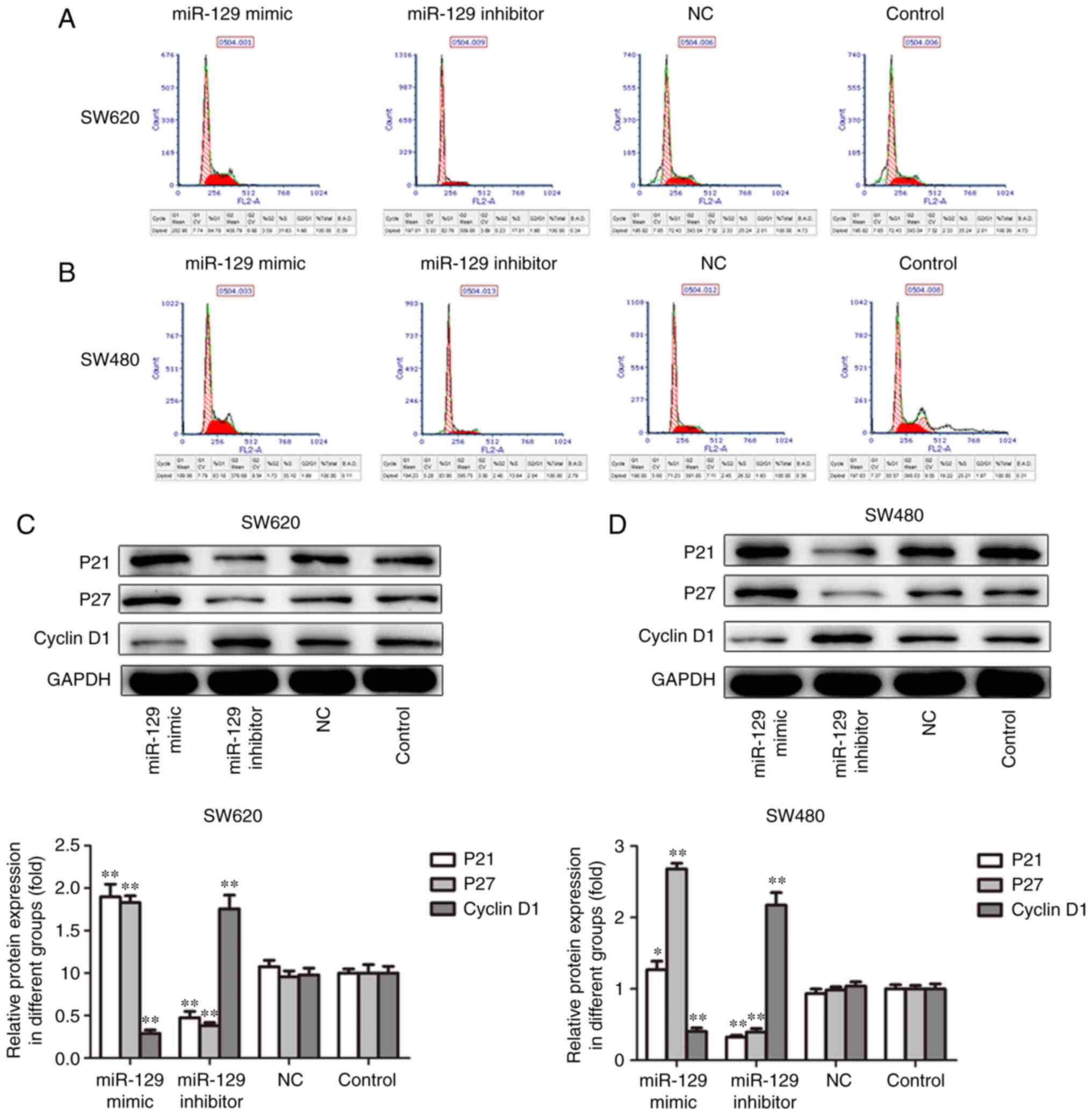
In order to further confirm the aforementioned
results, the present study investigated the effects of miR-129 on
cell cycle and associated proteins. As shown in Fig. 4A and B, miR-129 overexpression
increased the percentage of cells in the S phase, whereas miR-129
inhibition increased the percentages of SW620 and SW480 cells in
the S phase. In addition, miR-129 overexpression decreased the
expression of cyclin D1 and increased the expression of P21 and P27
proteins, while the inhibition of miR-129 exerted the opposite
effects on the expression of cell cycle-related proteins in SW620
and SW480 cells. Taken together, these data suggest that miR-129
inhibits the CRC cell proliferation ability in vitro.
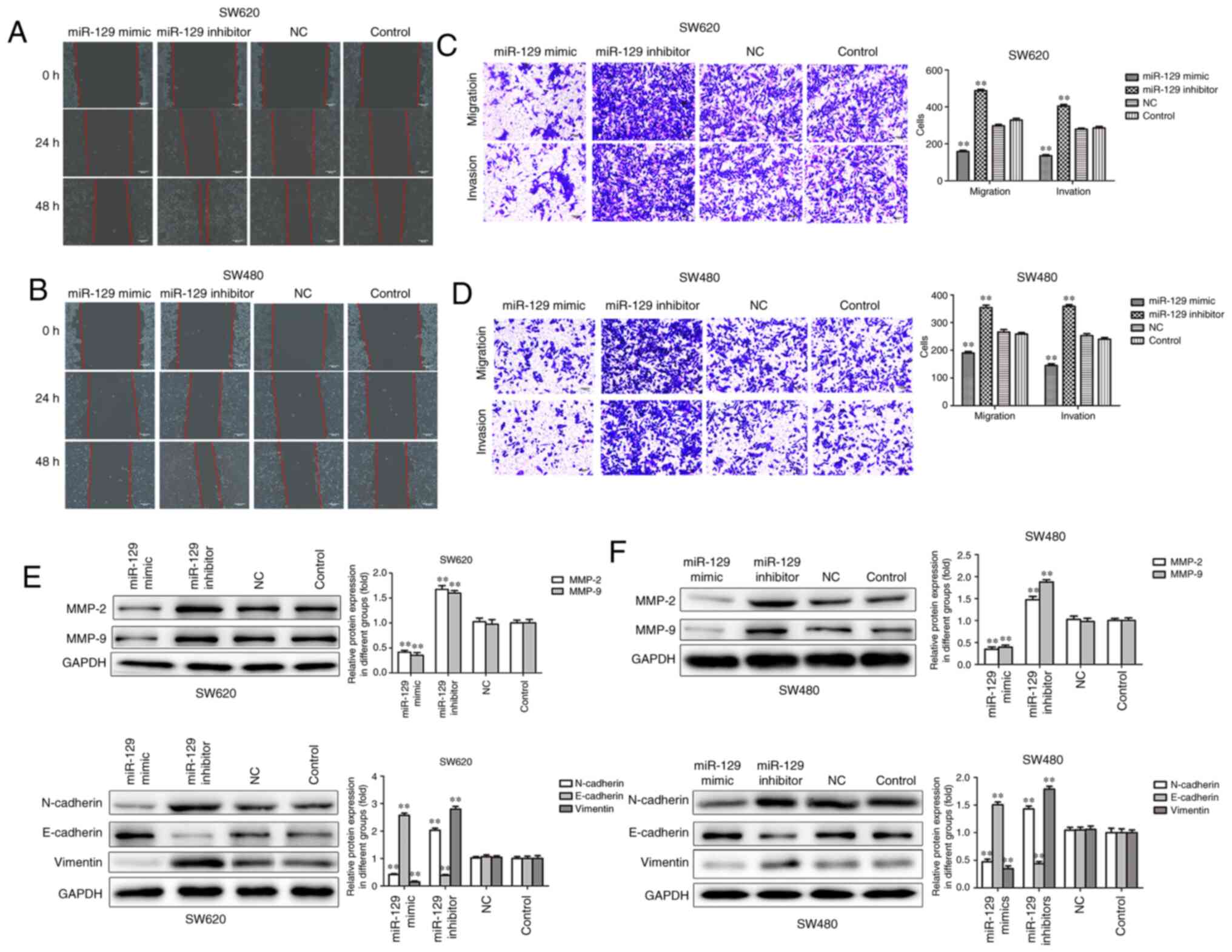
Overexpression of miR-129 suppresses
migration, invasion and EMT of CRC cells
Sequentially, wound healing and Transwell assays
were performed to elucidate the role of miR-129 in cell migration
and invasion. The wound healing and Transwell assays revealed that
the migration and invasion of SW620 and SW480 CRC cells were
inhibited by miR-129 mimic and increased by miR-129 inhibitor
(Fig. 5A-D). In addition, MMP-2 and
MMP-9 protein levels were significantly decreased by miR-129 mimic
and increased by miR-129 inhibitor (Fig.
5E and F).
EMT is considered to be an essential part of the
process of tumor cell metastatic dissemination (28). Thus, it was herein investigated
whether miR-129 is involved in EMT. As shown in Fig. 5E and F, miR-129 overexpression was
able to significantly increase the expression of the epithelial
marker E-cadherin and decrease the expression of the mesenchymal
markers N-cadherin and vimentin, while the inhibition of miR-129
exerted the opposite effects on the expression of EMT-related
proteins in SW620 and SW480 cells. Taken together, these results
indicate that miR-129 may act as a tumor suppressor through
inhibiting CRC cell migration, invasion and EMT.
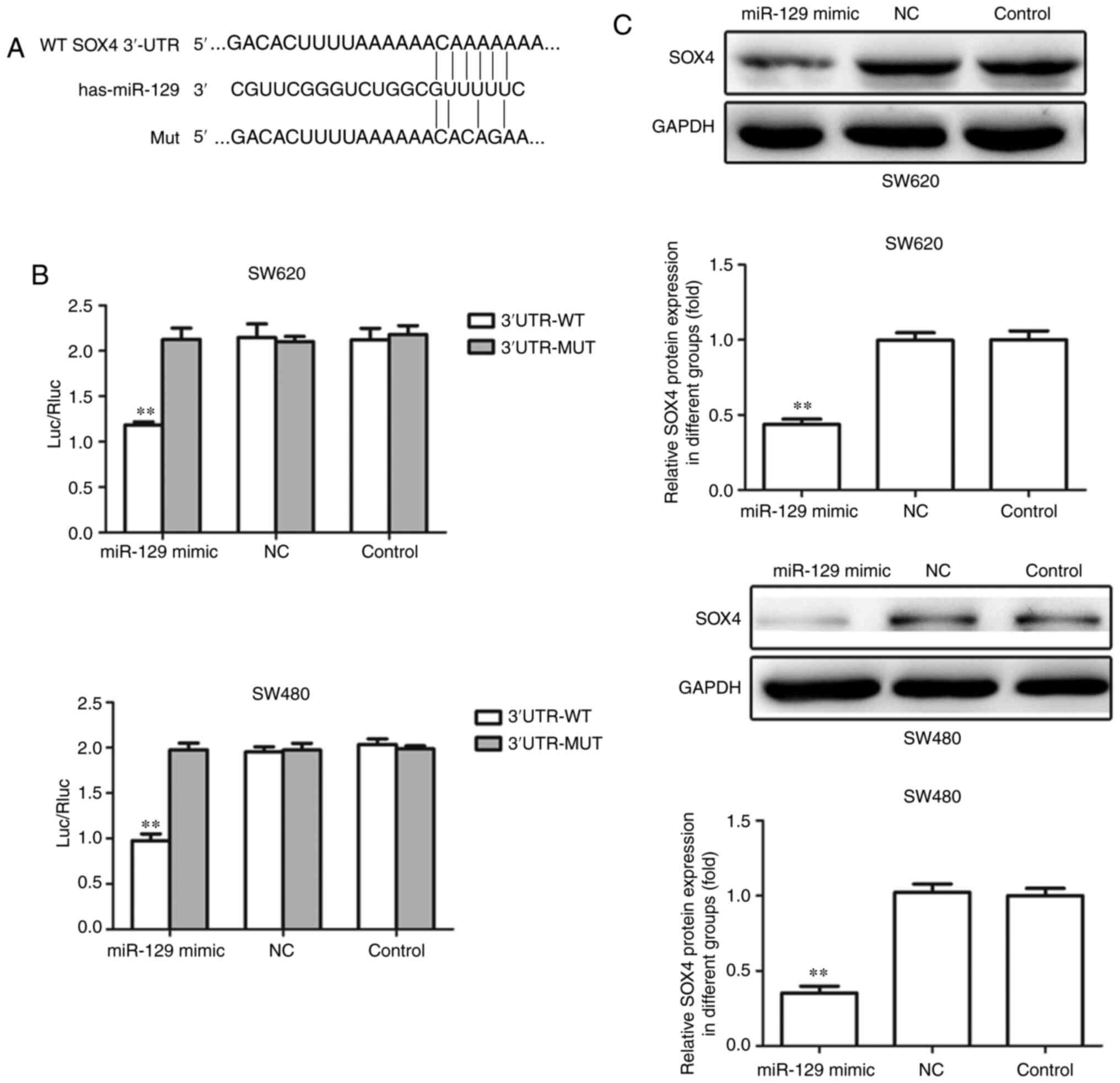
SOX4 is a direct target of miR-129 in
CRC cell lines
To further investigate the molecular mechanism
underlying the regulatory role of miR-129 in the tumorigenesis and
progression of CRC, bioinformatics databases were employed. As a
result, the 3′-UTR of SOX4 was found to contain the complementary
binding sequences for miR-129 (Fig.
6A). To investigate whether miR-129 directly binds to the
3′-UTR of SOX4 mRNA, a luciferase reporter vector containing this
region with miR-129 binding sites was constructed. As shown in
Fig. 6B, luciferase reporter assays
indicated that transfection of miR-129 mimic indeed suppressed the
activity of SOX4 3′-UTR region, whereas the luciferase activity of
the mutant type of SOX4 3′-UTR exhibited no obvious change.
Furthermore, the protein expression level of SOX4 in SW620 and
SW480 cells decreased significantly upon transfection of miR-129
mimic (Fig. 6C). These data suggest
that miR-129 targets SOX4 and suppresses its expression in CRC cell
lines.
miR-129 overexpression may inhibit CRC
cell proliferation, invasion and EMT by activating the NF-κB
signaling pathway
RT-qPCR and western blot assays were conducted to
further investigate the potential signaling pathway associated with
the regulatory effect of miR-129 on CRC cell proliferation,
invasion and EMT. The results demonstrated that miR-129
overexpression resulted in an upregulation of ATM and P50 mRNA
expression, while the inhibition of miR-129 resulted in a
downregulation of ATM and P50 mRNA expression, while the expression
of IkBα and NF-κB mRNA exhibited no obvious change in either group
in SW620 and SW480 cells (Fig. 7A and
B). Western blotting revealed that miR-129 mimic increased the
expression of the ATM, P50, p-IkBα and p-NF-κBp65 proteins, while
miR-129 inhibitor significantly decreased the expression of these
proteins. Although the expression of IkBα and NF-κBp65 exhibited no
obvious change in either group, the p-IkBα/IkBα and
p-NF-κBp65/NF-κBp65 ratios were significantly increased following
miR-129 overexpression, while miR-129 inhibition exerted the
opposite effects in SW620 and SW480 cells (Fig. 7C and D).
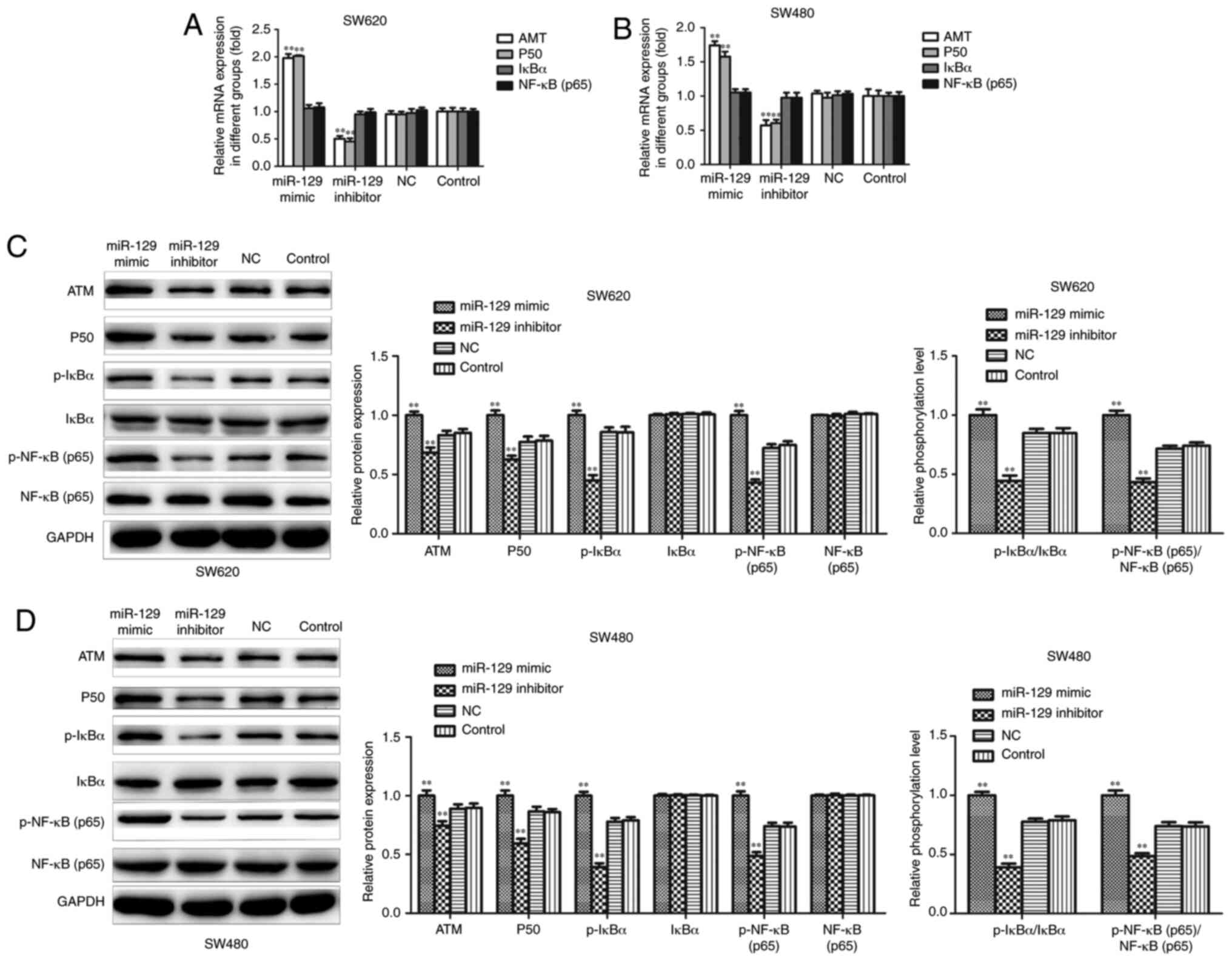 | Figure 7.Effects of miR-129 on NF-κB signaling
pathway in CRC cell lines. SW620 and SW480 cells were transfected
with miR-129 mimic or inhibitor for 48 h. (A and B) Expressions of
ATM, P50, IkBα and NF-κB mRNA were determined using reverse
transcription-quantitative PCR assay. (C and D) Expressions of ATM,
P50, p-IkBα, IkBα, p-NF-κB (p65) and NF-κB (p65) proteins were
detected by western blotting assay. The experiment was repeated
three times, results were shown as means ± standard deviation.
**P<0.01 vs. NC group. CRC, colorectal cancer; NF-κB, nuclear
factor-κB; ATM, ataxia telangiectasia, mutated; IkBα, NF-κB
inhibitor α; NC, negative control. |
Discussion
CRC remains one of the most common malignant tumors
of the gastrointestinal tract, despite significant advances in the
diagnosis and treatment (29). Local
recurrence and distant metastasis are the main causes of poor
survival in patients with advanced CRC (30). Therefore, it is urgent to elucidate
the mechanisms underlying the occurrence and metastasis of CRC in
order to design effective treatments. Several miRNAs were recently
identified as important regulators of tumorigenesis, angiogenesis,
invasion and metastasis (31). In
addition, miRNAs have been considered as potential therapeutic
targets and biomarkers for a number of human cancers, including CRC
(32,33). The focus of the present study was the
abnormal expression of miR-129 and the regulatory function of
miR-129 in cell proliferation, invasion and EMT via the SOX4/NF-κB
pathway.
Previous studies have demonstrated that miR-129 is
abnormally regulated in various malignant tumors, including
hepatocellular carcinoma (34),
glioma (35), non-small cell lung
cancer (36), and gastric cancer
(15), and participates in the
pathogenic processes. Recently, miR-129 was reported to be
increased in CRC tissues and blood samples, and the high expression
level of miR-129 contributes to cell proliferation and migration
via targeting estrogen receptor beta (37). Inversely, Karaayvaz et al
(16) reported that miR-129 was
specifically decreased in CRC and functions as a tumor suppressor
to promote cell apoptosis and chemosensitivity. Thus, further
investigation is required to identify the expression and mechanisms
of action of miR-129 in CRC. In the present study, it was observed
that miR-129 was downregulated in CRC clinical tissues and cell
lines, and these results were consistent with those of other
large-sample studies (38). Moreover,
gain-of-function experiments demonstrated that miR-129
overexpression suppressed cell proliferation and induced cell cycle
arrest at the S phase. Increased metastatic ability is a hallmark
of human cancers (31). The present
study revealed that the overexpression of miR-129 suppressed cell
migration and invasion, while miR-129 knockdown increased the
metastatic ability of CRC cells. EMT is closely associated with the
occurrence of tumor metastasis, which is accompanied by the loss of
membrane E-cadherin expression and increased levels of mesenchymal
markers, such as vimentin (39). The
present study demonstrated that the overexpression of miR-129
reduced N-cadherin and vimentin but increased E-cadherin expression
in CRC cells, while miR-129 knockdown exerted the opposite effects.
Taken together, these data demonstrated that miR-129 inhibited the
progression of CRC by suppressing the proliferation, migration,
invasion and EMT of CRC cells.
It is well known that miRNAs regulate cell
biological behavior via directly regulating target genes. A number
of target genes of miR-129 have been identified (33,34,39,40).
SOX4, a critical oncogenic protein, has been shown to be involved
in the progression and metastasis of various malignant tumors
(41,42). Previous studies have indicated that
the overexpression of SOX4 in CRC is regulated via miRNA-mediated
post-transcriptional mechanisms that prevent cell proliferation,
migration and invasion (43,44). However, the association between
miR-129 and SOX4 in CRC remains unclear. In the present study, it
was demonstrated that SOX4 is a direct target of miR-129 in CRC
cells, which was supported by data from the luciferase and western
blot assays. These data indicated that miR-129 may regulate cell
migration, invasion and EMT through targeting SOX4 in CRC.
The NF-κB pathway, a critical regulator of cell
apoptosis, mediates important biological processes, including
tumorigenesis (45). Lan et al
(46) reported that miR-15a/16
enhanced radiation sensitivity by targeting the TLR1/NF-κB
signaling pathway in non-small cell lung cancer. Ren et al
(47) demonstrated that miR-210-3p
promoted prostate cancer cell EMT and bone metastasis by activating
the NF-κB signaling pathway. The results of the present study
revealed that miR-129 overexpression promoted the expression of
NF-κB pathway-related proteins, while the inhibition of miR-129
exerted the opposite effect. These results indicated that miR-129
inhibited the malignant phenotype of CRC, possibly by activating
the NF-κB signaling pathway. Thus, it may be inferred that SOX4 is
involved in the activation of the NF-κB pathway through miR-129,
which will be investigated further in future studies.
In summary, the present study demonstrated that
miR-129 expression was downregulated while SOX2 expression was
upregulated in CRC. The findings further revealed that miR-129 may
function as a tumor suppressor by inhibiting cell proliferation,
migration, invasion and EMT in CRC cells via targeting SOX4, and
the potential mechanism may involve activation of the NF-κB
signaling pathway. These data may provide novel insight into the
molecular mechanism underlying CRC progression, and the
upregulation of miR-129 may be a promising strategy for the
treatment of CRC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
LW designed the experiments. ZC and TZ were the
major contributors to the writing of the manuscript. JZ, YT and BL
performed the experiments. All authors have read and approved the
final version of the manuscript.
Ethics approval and consent to
participate
The protocol of the present study was approved by
the Ethics Committee of the First Affiliated Hospital of Gannan
Medical University. Informed consent was obtained from all the
patients.
Patient consent for publication
Not applicable.
Competing interests
All the authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CRC
|
colorectal cancer
|
|
SOX4
|
sex-determining region Y-related
high-mobility group-box 4
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
|
EMT
|
epithelial-to-mesenchymal
transition
|
|
NF-κB
|
nuclear factor-κB
|
|
3′-UTR
|
3′-untranslated region
|
|
miRNAs
|
microRNAs
|
|
NC
|
negative control
|
|
PE
|
plating efficiency
|
|
Mut
|
mutant
|
|
WT
|
wild-type
|
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fan C, Lin B, Huang Z, Cui D, Zhu M, Ma Z,
Zhang Y, Liu F and Liu Y: MicroRNA-873 inhibits colorectal cancer
metastasis by targeting ELK1 and STRN4. Oncotarget. 10:4192–4204.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Brenner H, Kloor M and Pox CP: Colorectal
cancer. Lancet. 383:1490–1502. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siegel RL, Miller KD, Fedewa SA, Ahnen DJ,
Meester RGS, Barzi A and Jemal A: Colorectal cancer statistics,
2017. CA Cancer J Clin. 67:177–193. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liang Z, Li X, Liu S, Li C, Wang X and
Xing J: miR-141-3p inhibits cell proliferation, migration and
invasion by targeting TRAF5 in colorectal cancer. Biochem Biophys
Res Commun. 514:699–705. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Du T and Zamore PD: Beginning to
understand microRNA function. Cell Res. 17:661–663. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ambros V and Chen X: The regulation of
genes and genomes by small RNAs. Development. 134:1635–1641. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Voorhoeve PM: MicroRNAs: Oncogenes, tumor
suppressors or master regulators of cancer heterogeneity? Biochim
Biophys Acta. 1805:72–86. 2010.PubMed/NCBI
|
|
9
|
Cai C, Ashktorab H, Pang X, Zhao Y, Sha W,
Liu Y and Gu X: MicroRNA-211 expression promotes colorectal cancer
cell growth in vitro and in vivo by targeting tumor suppressor
CHD5. PLoS One. 7:e297502012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang L, Dong Y, Zhu N, Tsoi H, Zhao Z, Wu
CW, Wang K, Zheng S, Ng SS, Chan FK, et al: MicroRNA-139-5p exerts
tumor suppressor function by targeting NOTCH1 in colorectal cancer.
Mol Cancer. 13:1242014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bertoli G, Cava C, Diceglie C, Martelli C,
Rizzo G, Piccotti F, Ottobrini L and Castiglioni I: MicroRNA-567
dysregulation contributes to carcinogenesis of breast cancer,
targeting tumor cell proliferation, and migration. Breast Cancer
Res Treat. 161:605–616. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yu H, Duan P, Zhu H and Rao D: miR-613
inhibits bladder cancer proliferation and migration through
targeting SphK1. Am J Transl Res. 9:1213–1221. 2017.PubMed/NCBI
|
|
13
|
Chen X, Ruan A, Wang X, Han W, Wang R, Lou
N, Ruan H, Qiu B, Yang H and Zhang X: miR-129-3p, as a diagnostic
and prognostic biomarker for renal cell carcinoma, attenuates cell
migration and invasion via downregulating multiple
metastasis-related genes. J Cancer Res Clin Oncol. 140:1295–1304.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dyrskjøt L, Ostenfeld MS, Bramsen JB,
Silahtaroglu AN, Lamy P, Ramanathan R, Fristrup N, Jensen JL,
Andersen CL, Zieger K, et al: Genomic profiling of microRNAs in
bladder cancer: miR-129 is associated with poor outcome and
promotes cell death in vitro. Cancer Res. 69:4851–4860. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yu X, Song H, Xia T, Han S, Xiao B, Luo L,
Xi Y and Guo J: Growth inhibitory effects of three miR-129 family
members on gastric cancer. Gene. 532:87–93. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Karaayvaz M, Zhai H and Ju J: miR-129
promotes apoptosis and enhances chemosensitivity to 5-fluorouracil
in colorectal cancer. Cell Death Dis. 4:e6592013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Poncy A, Antoniou A, Cordi S, Pierreux CE,
Jacquemin P and Lemaigre FP: Transcription factors SOX4 and SOX9
cooperatively control development of bile ducts. Dev Biol.
404:136–148. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jang SM, Kim JW, Kim CH, An JH, Johnson A,
Song PI, Rhee S and Choi KH: KAT5-mediated SOX4 acetylation
orchestrates chromatin remodeling during myoblast differentiation.
Cell Death Dis. 6:e18572015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Andersen CL, Christensen LL, Thorsen K,
Schepeler T, Sørensen FB, Verspaget HW, Simon R, Kruhøffer M,
Aaltonen LA, Laurberg S and Ørntoft TF: Dysregulation of the
transcription factors SOX4, CBFB and SMARCC1 correlates with
outcome of colorectal cancer. Br J Cancer. 100:511–523. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang B, Li Y, Tan F and Xiao Z: Increased
expression of SOX4 is associated with colorectal cancer
progression. Tumour Biol. 37:9131–9137. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Vishnubalaji R, Hamam R, Yue S, Al-Obeed
O, Kassem M, Liu FF, Aldahmash A and Alajez NM: MicroRNA-320
suppresses colorectal cancer by targeting SOX4, FOXM1, and FOXQ1.
Oncotarget. 7:35789–35802. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hu F, Min J, Cao X, Liu L, Ge Z, Hu J and
Li X: miR-363-3p inhibits the epithelial-to-mesenchymal transition
and suppresses metastasis in colorectal cancer by targeting Sox4.
Biochem Biophys Res Commun. 474:35–42. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhao J, Xu J and Zhang R: MicroRNA-539
inhibits colorectal cancer progression by directly targeting SOX4.
Oncol Lett. 16:2693–2700. 2018.PubMed/NCBI
|
|
24
|
Zhang P, Li J, Song Y and Wang X:
miR-129-5p inhibits proliferation and invasion of chondrosarcoma
cells by regulating SOX4/Wnt/β-catenin signaling pathway. Cell
Physiol Biochem. 42:242–253. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kang M, Li Y, Liu W, Wang R, Tang A, Hao
H, Liu Z and Ou H: miR-129-2 suppresses proliferation and migration
of esophageal carcinoma cells through downregulation of SOX4
expression. Int J Mol Med. 32:51–58. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wong CE, Yu JS, Quigley DA, To MD, Jen KY,
Huang PY, Del Rosario R and Balmain A: Inflammation and Hras
signaling control epithelial-mesenchymal transition during skin
tumor progression. Genes Dev. 27:670–682. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liang Z, Li X, Liu S, Li C, Wang X and
Xing J: miR-141-3p inhibits cell proliferation, migration and
invasion by targeting TRAF5 in colorectal cancer. Biochem Biophys
Res Commun. 514:699–705. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kalluri R and Weinberg RA: The basics of
epithelial-mesenchymal transition. Clin Invest. 119:1420–1428.
2009. View Article : Google Scholar
|
|
30
|
Yang W, Lee DY and Ben-David Y: The roles
of microRNAs in tumorigenesis and angiogenesis. Int J Physiol
Pathophysiol Pharmacol. 3:140–155. 2011.PubMed/NCBI
|
|
31
|
Jiang C, Wang H, Zhou L, Jiang T, Xu Y and
Xia L: MicroRNA-212 inhibits the metastasis of nasopharyngeal
carcinoma by targeting SOX4. Oncol Rep. 38:82–88. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ma Y, Zhang P, Wang F, Zhang H, Yang J,
Peng J, Liu W and Qin H: miR-150 as a potential biomarker
associated with prognosis and therapeutic outcome in colorectal
cancer. Gut. 61:1447–1453. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhai J, Qu S, Li X, Zhong J, Chen X, Qu Z
and Wu D: miR-129 suppresses tumor cell growth and invasion by
targeting PAK5 in hepatocellular carcinoma. Biochem Biophys Res
Commun. 464:161–167. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yang Y, Huang JQ, Zhang X and Shen LF:
miR-129-2 functions as a tumor suppressor in glioma cells by
targeting HMGB1 and is down-regulated by DNA methylation. Mol Cell
Biochem. 404:229–239. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li J, Wang H, Ke H and Ni S: miR-129
regulates MMP9 to control metastasis of non-small cell lung cancer.
Tumour Biol. 36:5785–5790. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ya G, Wang H, Ma Y, Hu A, Ma Y, Hu J and
Yu Y: Serum miR-129 functions as a biomarker for colorectal cancer
by targeting estrogen receptor (ER) β. Pharmazie. 72:107–112.
2017.PubMed/NCBI
|
|
37
|
Lin J, Shi Z, Yu Z and He Z: LncRNA
HIF1A-AS2 positively affects the progression and EMT formation of
colorectal cancer through regulating miR-129-5p and DNMT3A. Biomed
Pharmacother. 98:433–439. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Thomson S, Petti F, Sujka-Kwok I, Mercado
P, Bean J, Monaghan M, Seymour SL, Argast GM, Epstein DM and Haley
JD: A systems view of epithelial-mesenchymal transition signaling
states. Clin Exp Metastasis. 28:137–155. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liu Q, Jiang J, Fu Y, Liu T, Yu Y and
Zhang X: miR-129-5p functions as a tumor suppressor in gastric
cancer progression through targeting ADAM9. Biomed Pharmacother.
105:420–427. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Setijono SR, Park M, Kim G, Kim Y, Cho KW
and Song SJ: miR-218 and miR-129 regulate breast cancer progression
by targeting Lamins. Biochem Biophys Res Commun. 496:826–833. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Jafarnejad SM, Ardekani GS, Ghaffari M,
Martinka M and Li G: Sox4 mediated dicer expression is critical for
suppression of melanoma cell invasion. Oncogene. 32:2131–2139.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ramezani-Rad P, Geng H, Hurtz C, Chan LN,
Chen Z, Jumaa H, Melnick A, Paietta E, Carroll WL, Willman CL, et
al: SOX4 enables oncogenic survival signals in acute lymphoblastic
leukemia. Blood. 121:148–155. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yeh YM, Chuang CM, Chao KC and Wang LH:
MicroRNA-138 suppresses ovarian cancer cell invasion and metastasis
by targeting SOX4 and HIF-1α. Int J Cancer. 133:867–878. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Li Y, Chen P, Zu L, Liu B, Wang M and Zhou
Q: MicroRNA-338-3p suppresses metastasis of lung cancer cells by
targeting the EMT regulator Sox4. Am J Cancer Res. 6:127–140.
2016.PubMed/NCBI
|
|
45
|
Slattery ML, Mullany LE, Sakoda L,
Samowitz WS, Wolff RK, Stevens JR and Herrick JS: The NF-κB
signalling pathway in colorectal cancer: Associations between
dysregulated gene and miRNA expression. J Cancer Res Clin Oncol.
144:269–283. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Lan F, Yue X, Ren G, Li H, Ping L, Wang Y
and Xia T: miR-15a/16 enhances radiation sensitivity of non-small
cell lung cancer cells by targeting the TLR1/NF-κB signaling
pathway. Int J Radiat Oncol Biol Phys. 91:73–81. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ren D, Yang Q, Dai Y, Guo W, Du H, Song L
and Peng X: Oncogenic miR-210-3p promotes prostate cancer cell EMT
and bone metastasis via NF-κB signaling pathway. Mol Cancer.
16:1172017. View Article : Google Scholar : PubMed/NCBI
|