Introduction
Liver cancer is a common type of neoplasia and has
the second highest rate of cancer-associated mortality in the world
(1). More than 90% of primary liver
cancer cases are hepatocellular carcinoma (HCC). In China, the
5-year survival of HCC is only 12.1% (2). Etiologically, hepatitis B and C virus
infection-associated fibrosis and cirrhosis are the most common
risk factors for the development of liver cancer (3). Although therapeutic approaches, such as
surgical resection, chemotherapy and immunotherapy, have advanced,
liver cancer remains a major threat (4) because many patients with liver cancer
are diagnosed at advanced stage and prone to development of
multi-drug resistance. Hence, novel therapeutic targets for
intervention and more reliable biomarkers for early diagnosis of
liver cancer are urgently needed (4).
MicroRNAs (miRNAs or miRs), ~23 nucleotides in
length, are key members of the non-coding RNA family (5). miRNAs control expression of their target
mRNAs principally by binding to the 3′-untranslated region (3′-UTR)
to inhibit the translation of mRNA and decrease its half-life.
miR-922 is a tumor-promoting gene that promotes the development and
progression of tumors (6,7). miR-922 expression levels are decreased
in breast cancer (8). By contrast,
miR-922 expression is significantly upregulated in hepatocellular
carcinoma (HCC) tissue and promotes proliferation of HCC cells by
targeting cylindromatosis (CYLD) to enhance c-Myc and cyclin D1
expression levels and inhibit retinoblastoma (Rb) phosphorylation
(8). However, the role of miR-922 in
regulating the malignant behavior of liver cancer cells is still
unclear.
cAMP response element binding protein 1
(CREB1) is a member of the leucine zipper transcription
factor family and enhances HCC progression by promoting
angiogenesis and resistance to apoptosis (9). Upregulated CREB1 expression
levels and phosphorylation have been detected in HCC tissue
(10). Furthermore, CREB1 also
mediates hepatitis B virus X protein-cortactin interactions to
promote malignant behavior of HCC cells (11). AT-rich interactive domain 2
(ARID2) is a tumor suppressor and member of the
switch/sucrose non-fermentable chromatin remodeling complex
(12). ARID2 knockout disrupts
DNA damage responses by inhibiting recruitment of xeroderma
pigmentosum complementation group G (12). To the best of our knowledge, however,
there is no information on whether miR-922 targets and regulates
ARID2 expression and its effect on malignant behavior of
liver cancer cells.
The present study validated miR-922 expression
levels in 10 pairs of liver cancer and adjacent tissue, liver
cancer cells and non-tumor hepatocytes. The aim was to determine
how CREB1 regulates miR-922 expression levels in liver
cancer cells. The impact of ARID2, a potential target of
miR-922, on the malignant behaviors of liver cancer cells was also
investigated both in vitro and in vivo.
Materials and methods
Clinical samples
The present study was approved by the Ethics
Committee of the Third Xiangya Hospital of Central South University
and all patients provided written informed consent. Liver cancer
and matched adjacent (distance, >3 cm) non-tumor hepatic tissue
(10 pairs) were collected from patients with liver cancer during
surgical treatment between September 2018 and September 2019 in The
Third Xiangya Hospital of Central South University, China (Table I). All samples were collected with
signed informed consent. Patients were diagnosed based on the
practice guidelines of the American Association for the Study of
Liver Diseases (13). Liver specimens
were evaluated by pathologists and clinical stage was determined
according to the Barcelona Clinic Liver Cancer classification
(14). Exclusion criteria were as
follows: i) patients ≤18 or ≥70 years of age or without full civil
capacity to provide informed consent; ii) history of preoperative
anticancer radiotherapy or chemotherapy, biological, immune or
traditional Chinese medicine therapy; iii) incomplete postoperative
follow-up data and iv) history of another organ malignancy or
systemic immune disease.
 | Table I.Expression levels of mir-922 and
ARID2 in patients with hepatocellular carcinoma. |
Table I.
Expression levels of mir-922 and
ARID2 in patients with hepatocellular carcinoma.
|
|
|
| miR-922 | ARID2 |
|---|
|
|
|
|
|
|
|---|
| Characteristic |
| n | Mean ± SD | P-value | Mean ± SD | P-value |
|---|
| Sex | Male | 7 | 9.366±3.475 | 0.842 | 0.301±0.163 | 0.315 |
|
| Female | 3 | 8.986±2.209 |
| 0.194±0.048 |
|
| Age, years | ≤50 | 4 | 7.806±2.489 | 0.238 | 0.374±0.173 | 0.053 |
|
| >50 | 6 | 10.216±3.162 |
| 0.199±0.070 |
|
| AFP, ng/ml | <20 | 2 | 6.803±0.139 | 0.027 | 0.479±0.107 | 0.010 |
|
| ≥20 | 8 | 9.864±3.105 |
| 0.216±0.097 |
|
| HBsAg | Positive | 10 | 9.252±3.028 |
| 9.252±3.028 |
|
|
| Negative | 0 |
|
|
|
|
| Cirrhosis | Positive | 8 | 9.924±3.024 | 0.020 | 0.299±0.146 | 0.195 |
|
| Negative | 2 | 6.562±0.633 |
| 0.146±0.021 |
|
| Tumor size, cm | ≤5 | 7 | 8.312±2.263 | 0.141 | 0.314±0.149 | 0.130 |
|
| >5 | 3 | 11.446±3.947 |
| 0.161±0.046 |
|
| Tumor number | 1 | 8 | 8.686±3.155 | 0.039 | 0.298±0.147 | 0.210 |
|
| ≥2 | 2 | 11.517±0.167 |
| 0.149±0.163 |
|
| Tumor stage | I/II | 6 | 7.687±1.956 | 0.035 | 0.340±0.146 | 0.047 |
|
| III/IV | 4 | 11.599±2.980 |
| 0.161±0.381 |
|
| Distant
metastasis | No | 8 | 8.303±2.444 | 0.037 | 0.321±0.131 | 0.116 |
|
| Yes | 2 | 13.050±2.150 |
| 0.150±0.016 |
|
Cell culture and transfection
Human liver cancer HepG2, MHCC97H, MHCC97L and
non-tumor hepatic THLE-2 cells were purchased from the Cell Bank of
China (Shanghai, China). THLE-2 cells were cultured in RPMI-1640;
other cells were cultured in DMEM supplemented with 10% fetal
bovine serum (all Thermo Fisher Scientific, Inc.) at 37°C in 5%
CO2. The authenticity of all cell lines was tested by
STR (Shanghai Yihe Applied Biotechnology Co., Ltd. and Guangzhou
Cellcook Biotech Co., Ltd.).
HepG2 and MHCC97L cells were transfected with
control plasmid (pcDNA3.1-NC) or plasmid for the expression of
ARID2 [pcDNA3.1-ARID2 (OE), GeneCopoeia, Inc.] using
Lipofectamine® 3000 (Thermo Fisher Scientific, Inc.) and
treated with 1 mg/ml G418 for 3 weeks to generate control
HepG2/MHCC97L-overexpression (OE)-negative control (NC) or ARID
stable expressing HepG2/MHCC97L-ARID2-OE cells.
HepG2/MHCC97L, HepG2/MHCC97L-NC and HepG2/MHCC97L-ARID2-OE
cells were transfected with control scramble miRNA, miR-922 mimics
or miR-922 inhibitor (Guangzhou RiboBio Co., Ltd.) for 48 h at
37°C. In addition, HepG2/MHCC97L cells were transduced with control
lentivirus or lentivirus for expression of ARID-specific short
hairpin (sh)RNA [pGPU6/GFP-ARID2 (sh)] in the presence of 4 µg/ml
puromycin for 4 days to generate control HepG2/MHCC97LshRNA-NC and
ARID stably silencing HepG2/MHCC97L-ARID2sh cells.
Subsequent experiments were performed 48 h after transfection.
Reverse transcription-quantitative (RT-q)PCR. Total
RNA was extracted from individual groups of cells (THLE-2, HepG2,
MHCC97H and MHCC97L) using TRIzol® reagent and reverse
transcribed into cDNA using a PrimeScript RT reagent kit
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. The relative levels of miRNA and mRNA
transcripts to control U6 and GAPDH were quantified by RT-qPCR in
duplicate using SYBR Premix Ex Taq (Takara Biotechnology Co., Ltd.)
and specific primers (Table II).
Thermocycling conditions were as follows: Initial denaturation at
94°C for 10 min, followed by 40 cycles of 94°C for 30 sec, 60°C for
30 sec and 72°C for 10 sec and final extension at 72°C for 8 min.
The data were analyzed by 2−ΔΔCq method (15).
 | Table II.Primer sequences. |
Table II.
Primer sequences.
| Gene | Sequence
(5′→3′) |
|---|
| GAPDH | Forward |
caatgaccccttcattgacc |
|
| Reverse |
gacaagcttcccgttctcag |
| β catenin | Forward |
atgactcgagctcagagggt |
|
| Reverse |
attgcacgtgtggcaagttc |
| microRNA-922 | Forward |
gcagcagagaataggactacgtc |
|
| Reverse |
tggtgtcgtggagtcg |
| CREB1
promoter region |
|
|
| 0-200
bp from TSS | Forward |
agtaagagggccagggttca |
|
| Reverse |
atgtcccaggtttcctccct |
| 200-400
bp from TSS | Forward |
ggactgaaaatgatccgctgc |
|
| Reverse |
ctgcagctgtctcttacccc |
| 400-600
bp from TSS | Forward |
gccggtagagaaggaaaggt |
|
| Reverse |
gttgtccttcctggtactcca |
|
800-1,000 bp from TSS | Forward |
cggagtccagaatcgaaccc |
|
| Reverse |
ctcggcctcctaaagtggtg |
|
1,000-1,200 bp from TSS | Forward |
tactagggaggctgaggcag |
|
| Reverse |
gagtctagcccttgtcgcc |
|
1,200-1,400 bp from TSS | Forward |
gagagggctgcagagttcac |
|
| Reverse |
tctacagcagggattcgcac |
|
1,400-1,600 bp from TSS | Forward |
catatttccaggggtccggg |
|
| Reverse |
cccgatactgtggcaccttg |
|
1,600-1,800 bp from TSS | Forward |
cagcagaccttcttccccag |
|
| Reverse |
cctggctctgctcttgacc |
|
1,800-2,000 bp from TSS | Forward |
tttccaccaagtcgccgac |
|
| Reverse |
cggctttcgcatcggtaaag |
Cell proliferation assay
The HepG2 and MHCC97L cell proliferation was
determined by Cell Counting Kit-8 (CCK-8) assay (Dojindo Molecular
Technologies, Inc.) according to the manufacturer's protocol. The
absorbance at 450 nm was measured using a microplate reader
following incubation for 1–2 h.
Wound healing assay
After serum-starved HepG2 and MHCC97L cells in each
group reached 100% confluence, they were wounded with a plastic
tip. The cells were cultured for 24 h at 37°C. Wound width was
measured at 0 and 24 h under a light microscope (magnification,
×100).
Transwell invasion assay
The HepG2 and MHCC97L cells (1×105
cells/well) were starved overnight, then cultured in the upper
chamber pre-coated (37°C for 30 min) with Matrigel (BD Biosciences)
of 24-well Transwell plates (pore size, 8 µm; Corning, Inc.). The
bottom chamber was filled with complete medium supplemented with
10% FBS (Thermo Fisher Scientific, Inc.). The cells were cultured
for 48 h at 37°C, fixed with 10% glutaraldehyde at 4°C for 30 min
and stained with 1% crystal violet for 20 min at room temperature,
followed by photoimaging under a light microscope (Olympus
Corporation) at 100× magnification.
Colony formation assay
Each group of cells (300 cells/well) was cultured in
6-well plates for 2 weeks at 37°C. The formed cell colonies were
fixed with 4% formaldehyde for 15 min at room temperature and
stained with 1% crystal violet for 20 min at room temperature,
followed by counting in a blinded manner.
Chromatin immunoprecipitation
(ChIP)
Potential binding of CREB1 to the promoter
region of miR-299 was determined by ChIP assay as previously
described (16). Briefly, MHCC97L and
HepG2 cells were transfected with scramble control or
CREB1-specific shRNA (Table
II) for 48 h and the efficacy of CREB1 silencing was
verified by western blotting as aforementioned. The control and
CREB1-silenced cells were cross-linked with 1% formaldehyde
for 10 min at room temperature and sonicated on ice to generate
~500-bp DNA fragments. Following centrifugation (14,000 × g; 10
min; 4°C), the obtained soluble chromatin samples were reacted with
anti-CREB 1 (1:10; cat. no. ab31387; Abcam), anti-H3K27me3 (1:10;
cat. no. ab6002; Abcam), anti-H3K27AC (1:10; cat. no. ab4729;
Abcam) or control IgG (1:10; cat. no. ab171870; Abcam) overnight at
4°C. The immunocomplex was precipitated by Protein A Agarose/Salmon
Sperm DNA (50% Slurry) beads and eluted. DNA fragments were
analyzed by qPCR, as aforementioned, using specific primers
(Table II).
Luciferase reporter gene assay
Hsa-miR-922 mimics and hsa-miR-922 inhibitor were
designed and synthesized by Shanghai GenePharma Co., Ltd. as
follows: Mimics forward, 5′-GCAGCAGAGAAUAGGACUACGUC-3′ and reverse,
5′-CGUAGUCCUAUUCUCUGCUGCUU-3′; inhibitor
5′-GACGUAGUCCUAUUCUCUGCUGC-3′ and NC forward,
5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse,
5′-ACGUGACACGUUCGGAGAATT-3′. Reporter plasmid pmirGLO
Dual-Luciferase miRNA Target Expression Vector was obtained from
Promega Corporation. Transfection was performed using
Lipofectamine® 2000 (cat. no. 11668030; Thermo Fisher
Scientific, Inc.) and Dual-Glo® Luciferase Assay system
(cat. no. E2920; Promega Corporation) was used for luciferase
assay. DNA fragments for the 3′UTR of fidgetin, microtubule
severing factor (FIGN) and ARID2 were cloned and the
specific motifs for miR-922 binding were mutated using QuikChange
Site-Directed Mutagenesis kit (Stratagene; Agilent Technologies,
Inc.), according to the manufacturer's instructions. The wild-type
(WT) and mutant (MT) 3′UTR of FIGN and ARID2 were
cloned into the 3′-end of firefly luciferase of the Dual-luciferase
Target Vector (Promega Corporation). Subsequently, MHCC97L cells
were co-transfected with miR-922 mimic, together with the plasmid
for WT or MT FIGN and ARID2 reporter and plasmid for
Renilla luciferase expression for 48 h. After 48 h, the
medium in the 96-well plate was discarded before washing twice with
PBS. A total of 100 µl diluted 1X Dual-Glo Luciferase Assay Reagent
was added to each well before shaking at 25°C for 15 min.
Luminescence activity was detected and normalized to that of
Renilla luciferase.
In addition, the miR-922 promoter region (2,000 bp
upstream of transcription initiation site) was cloned into the
Dual-luciferase Target Vector to control firefly luciferase
expression. Then, 293T cells were co-transfected with plasmids for
the firefly luciferase reporter, CREB1 and Renilla
luciferase expression. The regulatory effect of CREB1 on the
miR-922 promoter-controlled luciferase expression was determined by
Dual-Luciferase Reporter Assay system. The control cells received
plasmids for firefly luciferase reporter and Renilla
luciferase expression, as well as an empty plasmid without enhanced
CREB1 expression.
In situ hybridization
Digoxigenin (Dig)-labelled probe
(5′-GCAGCAGAGAAUAGGACUACGUC-3′) for miR-922 was designed and
synthesized by BersinBi. The distribution of miR-922 transcripts in
liver cancer tissue was determined by in situ hybridization
using an In Situ Hybridization kit (Wuhan Boster Biological
Technology Ltd.), according to the manufacturer's protocol.
Briefly, liver cancer and adjacent non-cancer tissue sections
(thickness, 4 µm) were dewaxed, rehydrated and treated with 3%
H2O2 for 10 min at room temperature to
inactivate endogenous enzymes. The sections were digested with
pepsin in 3% citric acid at 37°C for 30 min and fixed with 4%
paraformaldehyde for 5 min at room temperature. After being washed,
sections were treated with pre-hybridization solution at 38–42°C
for 2–4 h and hybridized in the presence or absence (negative
control) of the probe at 38–42°C overnight. The sections were
washed with 2X saline-sodium citrate and reacted with biotinylated
mouse anti-Dig (1:2,000; cat. no. D15041; Bellancom) at 37°C for 60
min. After being washed, the sections were incubated with
streptavidin biotin-peroxidase complex for 60 min at room
temperature and reacted with biotinylated peroxidase for 20 min at
37°C, then visualized with 3,3′-diaminobenzidine (DAB) at room
temperature for 10 min and counterstained with hematoxylin. The
sections were examined under a light microscope (magnification,
×400).
Western blotting
The HepG2 and MHCC97L cells were harvested and lyzed
in RIPA lysis buffer (cat. no. P0013B; Beyotime Institute of
Biotechnology), followed by centrifugation (9,000 × g; 4°C; 10
min). After determining the protein concentrations using a BCA kit
(Abcam), cell lysates (50 µg/lane) were separated by SDS-PAGE on
12% gels and transferred onto polyvinylidene fluoride membranes
(Amersham Pharmacia Biotech; Cytiva). The immunoblot membranes were
blocked with 5% fat-free dry milk in TBST (0.1% Tween-20) buffer at
37°C for 2 h and incubated with primary antibodies overnight at 4°C
(ARID2, 1:1,000, cat. no. #82342, Cell Signaling Technology,
Inc.; FIGN, 1:1,000, cat. no. ab122238, Abcam). After being washed,
the membranes were reacted at room temperature for 1 h with
peroxidase-conjugated secondary antibodies (1:10,000; cat. no.
ab6721; Abcam). The signals were visualized using enhanced
chemiluminescence reagents (GE Healthcare) and quantified by
densitometric analysis using ImageJ software (version 1.48;
National Institutes of Health).
RNA immunoprecipitation (RIP)
A Magna RIP RNA-Binding Protein Immunoprecipitation
kit (cat. no. 17-701; Merck Sharp & Dohme) was used for RIP
assay according to the manufacturer's instructions. Briefly,
miR-922-expressing cells were fixed with 2% formaldehyde for 5 min
at room temperature, lysed and sonicated, followed by
centrifugation at 12,000 × g and 4°C for 1 min. The supernatants
were collected and incubated with ARID2 antibody (1:1,000,
cat. no. #82342; Cell Signaling Technology, Inc.) and FIGN
antibody (1:1,000, cat. no. ab122238, Abcam); overnight at 4°C. The
immunocomplex was precipitated using protein A/G Dynabeads; after
being washed, the immunocomplex was digested with proteinase K.
Finally, RNA was extracted with TRIzol and the relative levels of
miR-922 in the immunocomplex were determined by RT-qPCR, as
aforementioned, using specific primers (forward,
5′-ATGGCGTTTTCCCTCTCC-3′ and reverse,
5′-TGACGTAGTCCTATTCTCTGC-3′).
Cell apoptosis analysis
The number of apoptotic HepG2 and MHCC97L cells was
determined by flow cytometry using an Annexin V-FITC Apoptosis
Detection kit (Beyotime Institute of Biotechnology), according to
the manufacturer's instructions. Briefly, cells were stained with
Annexin V-FITC and PI in the dark. After being washed, cells were
analyzed by flow cytometry using an Attune Nxt flow cytometer (BD
Biosciences) and FlowJo™ software (version 10.7; BD
Biosceinces).
Bioinformatics analysis
Genecards (genecards.org/) and Jaspar (jaspar.genereg.net/) were used to predict
transcription factor binding to the miR-922 promoter region. In
order to determine the potential role of CREB1 expression in
liver cancer, CREB1 expression levels were searched in liver
cancer (n=371) and non-tumor tissue samples (n=50) of The Cancer
Genome Atlas (TCGA) database (portal.gdc.cancer.gov/). The association between
CREB1 protein expression levels and the overall survival
(OS) of 360 patients with liver cancer was analyzed in The Human
Protein Atlas (proteinatlas.org/).
Tumor xenograft
Animal experiments were approved by the Ethical
Committee for Animal Research of The Third Xiangya Hospital of
Central South University. A total of 25 male Balb/c nude mice (age,
8 weeks; weight, 18–20 g) were obtained from Charles River
Laboratories, Inc. and housed in a specific pathogen-free room
(temperature, 26°C; humidity, 50%; 10/14-h light/dark cycle) with
free access to autoclaved food and water. Individual mice were
injected subcutaneously with 1×107 MHCC97H,
MHCC97H-ARID2-NC, MHCC97H-ARID2-sh, MHCC97H-NC-OE or
MHCC97H-ARID2-OE cells (n=5/group). After 4 weeks, mice were
anesthetized via intraperitoneal injection of 2% pentobarbital
sodium (30 mg/kg body weight). The mice were checked for deep
anesthesia, including moderate breathing, cardiovascular depression
and complete muscle relaxation, and euthanized by cervical
dislocation. Tumors were dissected and images were captured. Tumor
volume and weight were measured.
ELISA
The levels of serum VEGF and TNF-α in mice were
measured by ELISA using VEGF (cat. no. SEA143Mu; Uscnks, Inc.) and
TNF-α ELISA kits (cat. no. RAF129R, Biovendor, Inc.). The samples
were tested in triplicate and the minimum detectable concentration
for VEGF and TNF-α was 10 pg/ml.
Immunohistochemistry
The expression levels of Bax, Bcl-2, PCNA, Cyclin
D1, MMP3, MMP9 and ARID2 in tumor tissue from mice
were analyzed by immunohistochemistry. Briefly, tissues were fixed
with 4% formaldehyde for 24 h at room temperature and
paraffin-embedded at 54°C for 4 h. The tissue sections (4 µm) were
deparaffinized, rehydrated and subjected to antigen retrieval in
sodium citrate buffer in a microwave. After being washed, the
sections were incubated overnight at 4°C with primary antibodies
against Bax (1:200; cat. no. ab32503; Abcam), Bcl-2 (1:500; cat.
no. ab32124; Abcam), PCNA (1:200; cat. no. ab92729; Abcam), Cyclin
D1 (1:400; cat. no. ab16663; Abcam), MMP3 (1:200; cat. no.
ab227755; Abcam), MMP9 (1:200; cat. no. ab119906; Abcam) and
ARID2 (1:400; cat. no. #82342; Cell Signaling Technology,
Inc.). The sections were reacted with horseradish
peroxidase-conjugated secondary antibodies for 20 min at room
temperature and the immunocomplex was viewed using a DAB Detection
IHC kit (cat. no. ab64264; Abcam) according to the manufacturer's
instructions and counterstained with hematoxylin for 10 sec at room
temperature. Images were captured under a light microscope
(magnification, ×400) and analyzed using Image Pro-Plus (version
6.0; Media Cybernetics, Inc.).
Statistical analysis
Data are expressed as mean ± SD (n=3). All
experiments were repeated three times. Statistical analysis was
performed using SPSS 22.0 (IBM Corp). Paired student's t-test and
one-way ANOVA followed by post hoc Tukey's test were used for
comparisons between two or multiple groups, respectively. The
survival data were estimated by the Kaplan-Meier method and
analyzed by log-rank test. P<0.05 was considered to indicate a
statistically significant difference.
Results
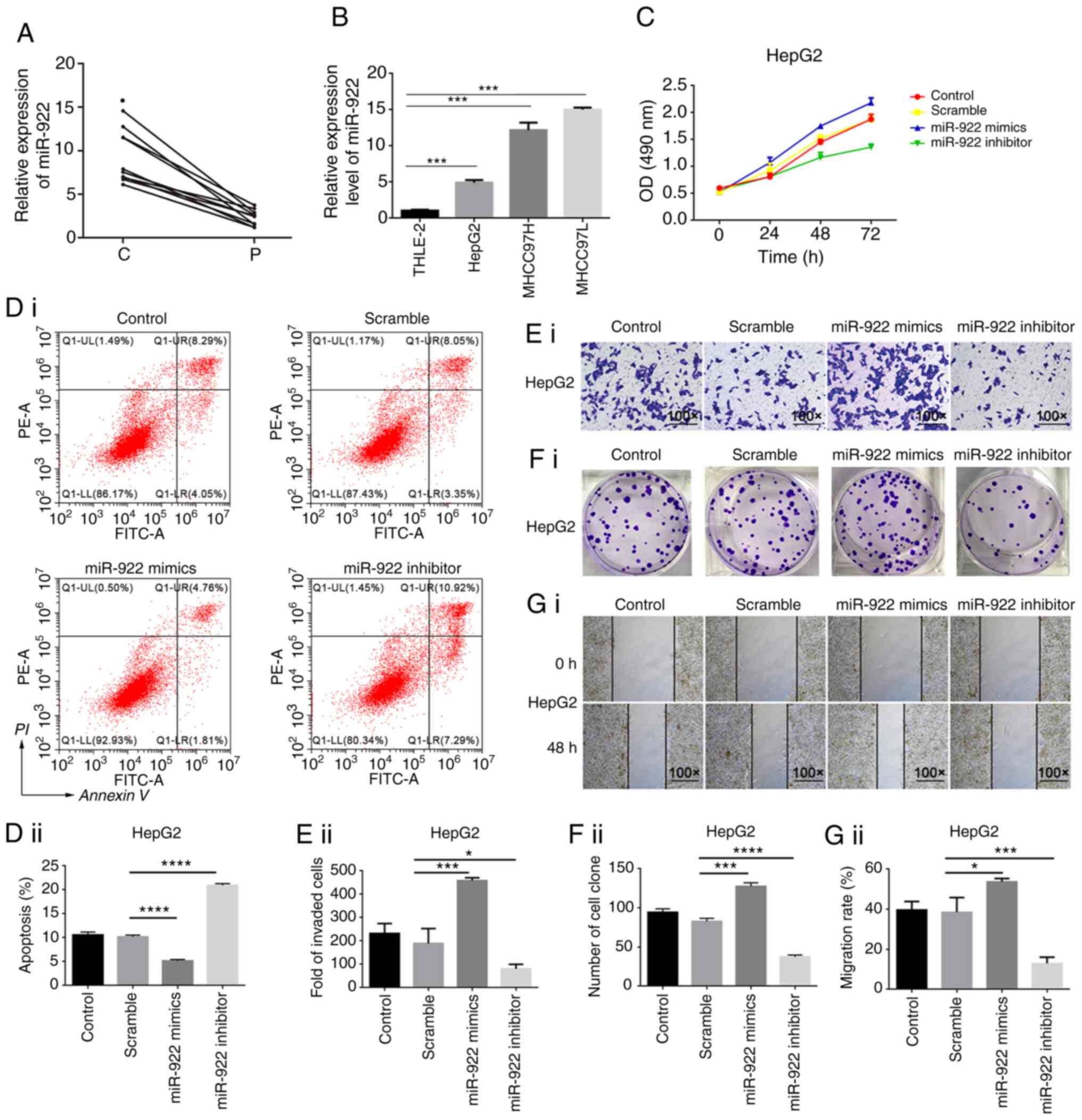
Elevated miR-922 transcripts in liver
cancer tissue
In order to reveal the function of miRNAs in liver
cancer progression, differentially expressed miRNAs in liver cancer
tissue were screened; miR-922 expression levels were significantly
elevated in liver cancer tissue. Expression levels of miR-922 were
assessed in 10 pairs of liver cancer and matched adjacent tissue
(Table I). miR-922 expression levels
in liver cancer tissue were notably higher than in matched adjacent
tissue (Fig. 1A, P<0.05).
Similarly, miR-922 expression increased in HepG2, MHCC97H and
MHCC97L cells, relative to non-tumor hepatic THLE-2 cells (Fig. 1B). Hence, upregulated miR-922
expression occurred in liver cancer tissue and may participate in
the pathogenesis of liver cancer.
miR-922 enhances malignant behavior of
liver cancer cells
In order to determine the effect of altered miR-922
expression on malignant behavior of liver cancer cells, the
regulatory role of miR-922 in the proliferation, clonogenicity,
wound healing, invasion and apoptosis of HepG2 and MHCC97L cells
was assessed in vitro. In comparison with control
HepG2/MHCC97L and HepG2-NC/MHCC97L-NC, miR-922 over-expression
significantly increased proliferation, clonogenicity, would healing
and invasion of HepG2 and MHCC97L cells, but decreased the number
of apoptotic HepG2/MHCC97L cells (Figs.
1C-G and S1). By contrast,
transfection with miR-922 inhibitor exhibited opposite effects on
malignant behavior in HepG2 and MHCC97L cells. miR-922 mimics or
inhibitor was introduced into HepG2 and MHCC97L to overexpress or
inhibit the expression of miR-922, respectively. The efficacy of
overexpression or inhibition of miR-922 was confirmed by RT-PCR
method (Fig. S5).
CREB1 promotes miR-922
transcription
Bioinformatics analysis was performed to predict the
potential binding of transcription factors to the promoter region
of miR-922 using the GeneCard database; results predicted binding
of CREB1 to the promoter region of miR-922. Accordingly, it
was speculated that CREB1 may enhance expression levels of
miR-922 in liver cancer cells. CREB1 is a key transcription
factor that promotes the development and progression of tumors
(17). In order to determine whether
CREB1 could regulate miR-922 transcription, the potential
binding of CREB1 to the miR-922 promoter region was assessed
by ChIP assay. Anti-CREB1 antibody precipitated chromatins
containing the miR-922 promoter region, indicating that
CREB1 bound to the miR-922 promoter region (Fig. 2A). Similarly, luciferase reporter
assay indicated that co-transfection with the plasmid for
CREB1 expression significantly increased miR-922 promoter
activity in 293T cells (Fig. 2B).
Furthermore, CREB1 silencing significantly decreased
enrichment of H3K27Ac but elevated that of H3K27me3 in 293T cells
(Fig. 2C and D). In situ
hybridization indicated that the expression levels of miR-922 in
liver cancer tissue were higher than in adjacent non-tumor tissue
(Fig. 2E). Similarly, miR-922
expression levels in liver cancer tissue were significantly higher
than in non-tumor liver tissue in TCGA database (Fig. 2G). Consistently, immunohistochemistry
revealed significantly higher levels of CREB1 in liver
cancer tissue compared with non-tumor tissue (Fig. 2F). Higher levels of CREB1
expression were significantly associated with a shorter period of
OS in patients with liver cancer in The Human Protein Atlas
database (P<0.001; Fig. 2H).
Together, these data indicated that higher levels of CREB1
enhanced miR-922 expression levels, promoting progression and poor
prognosis of liver cancer.
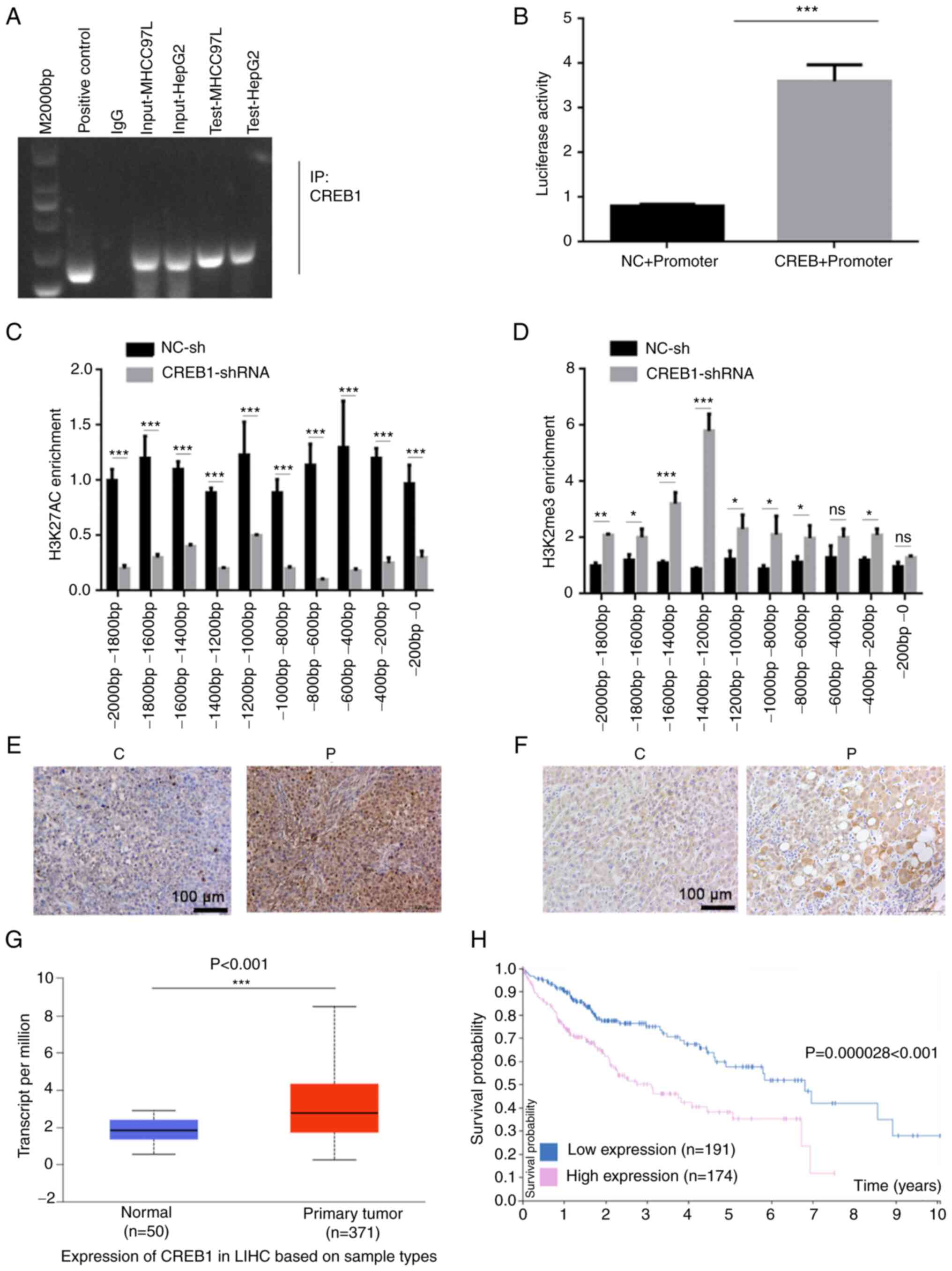 | Figure 2.CREB1 stimulates miR-922
transcription in liver cancer. (A) ChIP-PCR indicated that
CREB1 bound to the miR-922 promoter region. (B) Luciferase
assay demonstrated that induction of CREB1 expression
enhanced miR-922 promoter-controlled luciferase expression in 293T
cells. ChIP-PCR analyzed enrichment of (C) H3K27AC histone
acetyltransferase and (D) H3K27me3 histone methyltransferase on
different fragments of the CREB1 promoter region. (E) In
situ hybridization demonstrated increased expression levels of
miR-922 in liver cancer tissue compared with adjacent non-tumor
tissue. (F) Immunohistochemistry showed increased CREB1
expression levels in liver cancer tissue compared with adjacent
non-tumor tissue. (G) miR-922 expression levels are increased in
LIHC tissue in The Cancer Genome Atlas database. (H) Higher levels
of CREB1 expression were associated with a shorter overall
survival of patients with liver cancer in The Human Protein Atlas
database. *P<0.05, **P<0.01, ***P<0.001. CREB1,
cAMP response element binding protein 1; miR, microRNA; ChIP,
chromatin immunoprecipitation; NC, negative control; sh, short
hairpin; C, cancer tissue; P, para-carcinoma tissue; ns, not
significant; LIHC, liver hepatocellular carcinoma. |
miR-922 targets ARID2 in liver cancer
cells
Potential targeted genes of miR-922 were studied via
bioinformatics analysis. Among the predicted targeted genes of
miR-922, miR-922 may target FIGN and ARID2 (data not
shown). Given that FIGN and ARID2 regulate the
development of malignant tumors (12,18), their
expression levels were analyzed in 10 pairs of liver cancer tissue
by RT-qPCR. FIGN mRNA transcripts notably increased, whereas
ARID2 mRNA transcripts were decreased in liver cancer tissue
compared with non-tumor tissue (Fig.
3A, P<0.05). A similar pattern of FIGN and ARID2
protein expression was observed in these tissues by western
blotting (Fig. 3B). Moreover,
upregulated FIGN and downregulated ARID2 expression levels
were detected in HepG2, MHCC97H and MHCC97L cells compared with in
non-tumor hepatic THLE-2 cells (Fig.
3C). In order to clarify the targeting association, WT or MT
3′UTR of FIGN and ARID2 was cloned into the
luciferase reporter plasmid to generate WT or MT FIGN and
ARID2 luciferase reporter plasmid, respectively. Following
co-transfection, dual luciferase reporter assay indicated that
co-transfection with miR-922 mimics significantly decreased
ARID2-regulated, but not MT-ARID2-regulated,
luciferase activity in MHCC97L cells (P<0.01); WT-FIGN or
MT-FIGN-regulated luciferase activity was not affected in MHCC97L
cells (Fig. 3D). These data suggest
that miR-922 may target ARID2 to decrease its expression in
liver cancer. RIP assays using anti-AGO2 detected miR-922 in HepG2
and MHCC97L cells (Fig. 3E),
indicating that miR-922 existed in the miR-922/AGO2 complex. These
data suggest that miR-922 may target ARID2 and decrease its
expression to modulate malignant behavior of liver cancer
cells.
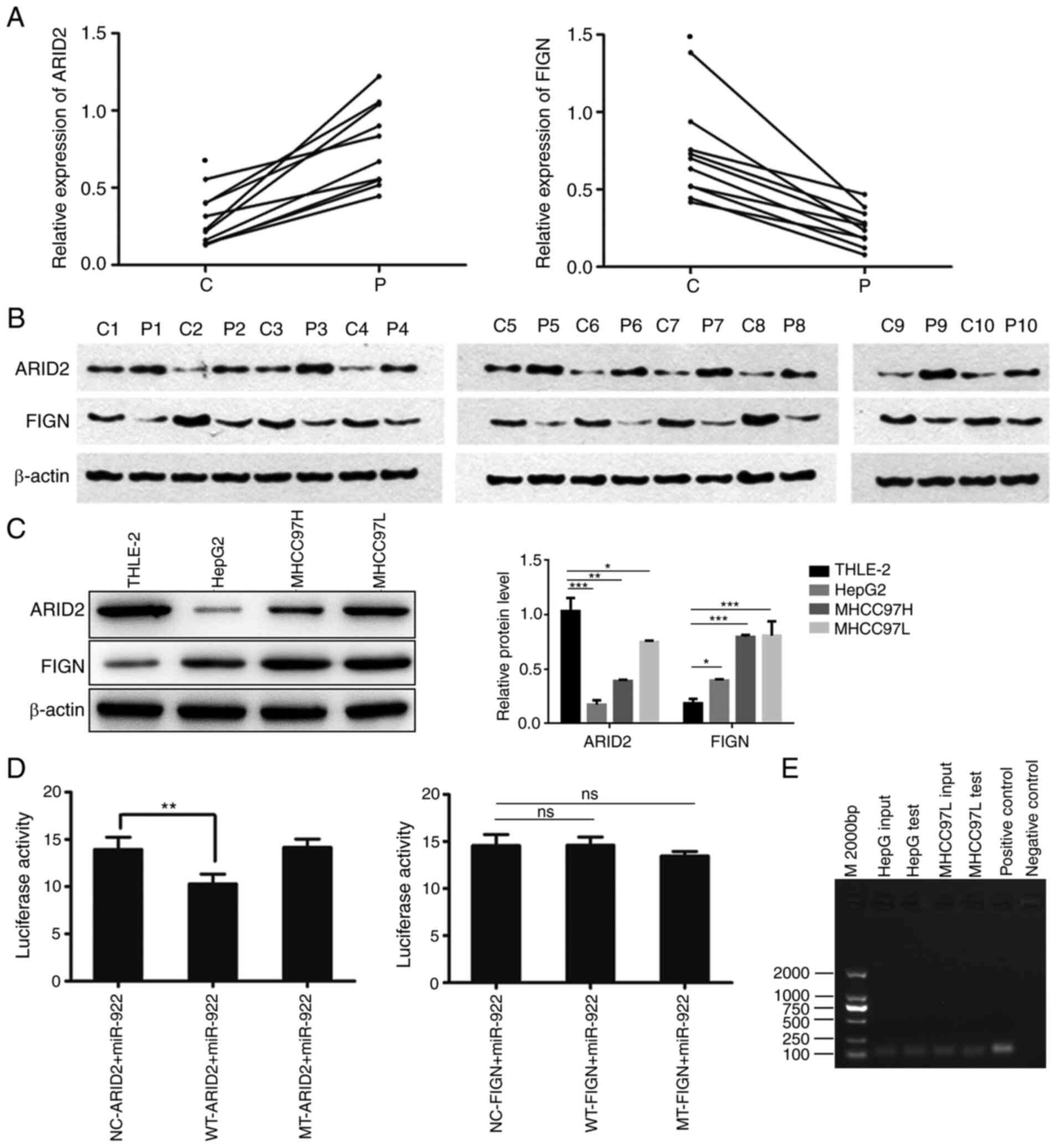 | Figure 3.ARID2 is a potential target of
miR-922 in liver cancer. Relative levels of FIGN and ARID2
expression levels in 10 pairs of liver cancer and adjacent
non-tumor tissue were determined by reverse
transcription-quantitative PCR and western blot analysis. WT or MT
3'UTR of FIGN and ARID2 were cloned into luciferase
reporter vector to generate WT or MT FIGN and ARID2
luciferase reporter plasmids, respectively. Luciferase reporter
gene assay was performed in MHCC97L cells following transfection
with miR-922 mimics and plasmid. The presence of miR-922 in
miRNA/AGO2 complex was determined by RNA immunoprecipitation using
anti-AGO2 antibody. (A) FIGN and ARID2 mRNA
transcripts in liver cancer tissue. FIGN and ARID2 protein
expression levels in (B) liver cancer tissue and (C) HepG2, MHCC97H
and MHCC97L and non-tumor hepatocyte THLE-2 cells. (D) Luciferase
activity. (E) Representative images of agarose gel electrophoresis
of PCR products. Data are presented as the mean ± SD (n=3).
*P<0.05, **P<0.01, ***P<0.001. C, cancer tissue; P,
para-carcinoma tissue; ARID2, AT-rich interactive domain 2;
miR, microRNA; FIGN, fidgetin, microtubule severing factor; WT,
wild-type; MT, mutant; NC, negative control; ns, not
significant. |
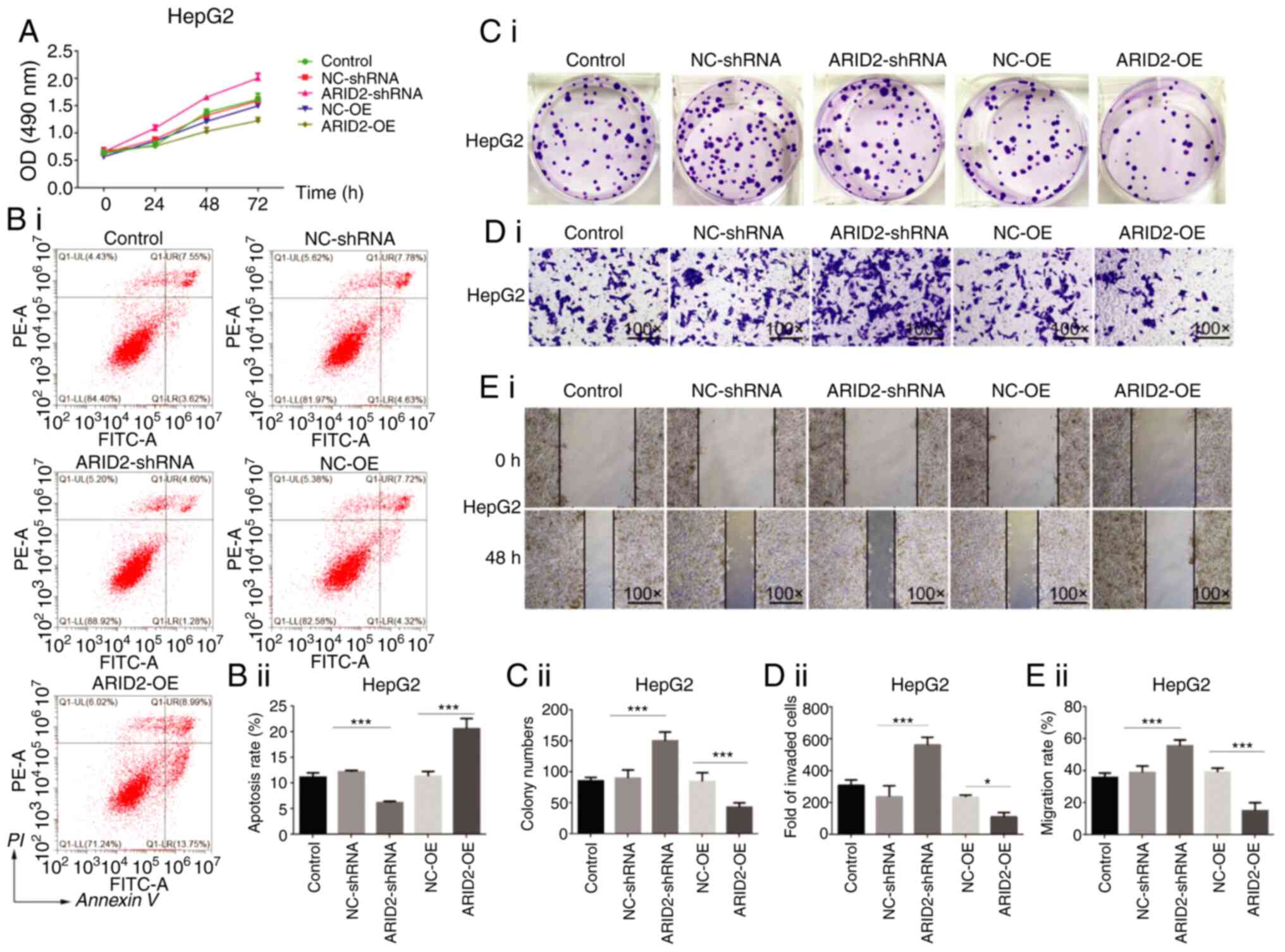
Altered ARID2 expression modulates
malignant behavior of liver cancer cells
ARID2 is a tumor suppressor and serves as a
genetic modulator during the progression of several types of
cancers, including liver cancer (12). The regulatory role of ARID2 in
the malignant behavior of HepG2 and MHCC97L cells was investigated
by CCK-8, colony formation, wound healing, apoptosis and Transwell
invasion assays in vitro. The ARID2 cDNA sequence was
cloned into pcDNA3.1 vector for gene over-expression and three
ARID2 specific shRNA were cloned into pGUP6 vector for
stable gene-silence. The protein level of ARID2 in HEK293
was determined after transduction with either ARID2-pcDNA3.1
or sh-ARID2 plasmid. As shown in figure, the ARID2
expression was significantly increased in ARID2-pcDNA3.1
group and was dramatically reduced in sh-ARID2 group
compared to the control group. Among three ARID2 specific
shRNA, sh-ARID2−2 achieved most robust knockdown efficiency,
which then was used in following experiment (Fig. S6). In comparison with the control
NC-OE and NC-sh cells, ARID2 over-expression significantly
decreased proliferation, clonogenicity, wound healing and invasion,
but increased the number of apoptotic HepG2 and MHCC97L cells
(Figs. 4 and S2). By contrast, ARID2 silencing
exhibited opposite effects on the malignant behavior of HepG2 and
MHCC97L cells.
Similarly, ARID2 over-expression in MHCC97L
cells significantly decreased tumor volume and weight in
vivo, whereas ARID2 silencing in MHCC97L cells did not
significantly alter tumor volume and weight in mice (Fig. 5A-C). Furthermore, ARID2
over-expression significantly decreased miR-922 expression levels,
whereas ARID2 silencing increased its expression in
xenografts (Fig. 5D), suggesting that
ARID2 may regulate miR-922 expression. In addition,
ARID2 over-expression significantly decreased serum levels
of VEGF and TNF-α, whereas ARID2 silencing elevated the
serum levels of VEGF and TNF-α in tumor-bearing mice (Fig. 5E and F). Immunohistochemistry revealed
that ARID2 over-expression increased ARID2 and Bax
expression levels, but decreased those of cyclin D1, PCNA, BcL-2,
MMP3 and MMP9 in liver cancer xenografts (Figs. 5G and S3). By contrast, ARID2 silencing
exhibited opposite effects on expression levels in liver cancer
xenografts (Figs. 5G and S3). Collectively, these data demonstrated
that ARID2 mitigated malignant behavior of liver cancer
cells.
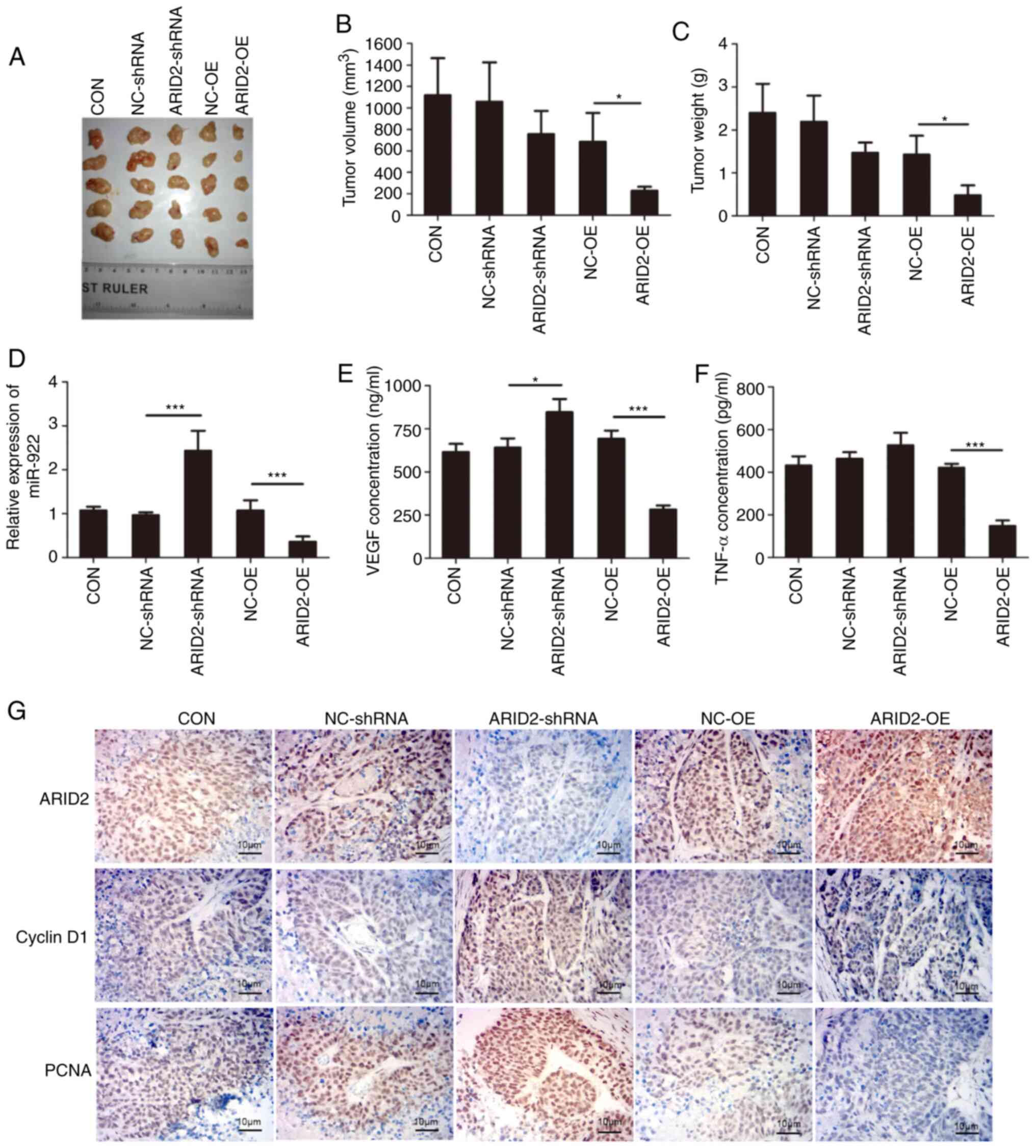 | Figure 5.Altered ARID2 expression
modulates the growth of xenograft liver cancer tumor in mice. Male
Balb/c nude mice were injected subcutaneously with 1×107
cells; 28 days later, tumor tissue was dissected and volume and
weight were measured. The relative levels of miR-922 expression in
tumor tissue were determined by reverse transcription-quantitative
PCR and the levels of serum VEGF and TNF-α in mice were measured by
ELISA. ARID2, Bcl-2, Bax, PCNA, Cyclin D1, MMP3 and
MMP9 expression levels in tumor tissue were characterized by
immunohistochemistry. (A) Tumor-bearing mice. Tumor (B) volume and
(C) weight. (D) Relative levels of miR-922 transcripts in tumor
tissue. ELISA analysis of serum (E) VEGF and (F) TNF-α levels in
mice. (G) Immunohistochemistry analysis of ARID2, Bcl-2,
Bax, PCNA, cyclin D1, MMP3 and MMP9 expression levels
in tumor tissue. Data are presented as the mean ± SD (n=3).
*P<0.05, ***P<0.001. ARID2, ARID2, AT-rich interactive
domain 2; PCNA, proliferating cell nuclear antigen; CON, control;
NC, negative control; sh, short hairpin; OE, over-expression. |
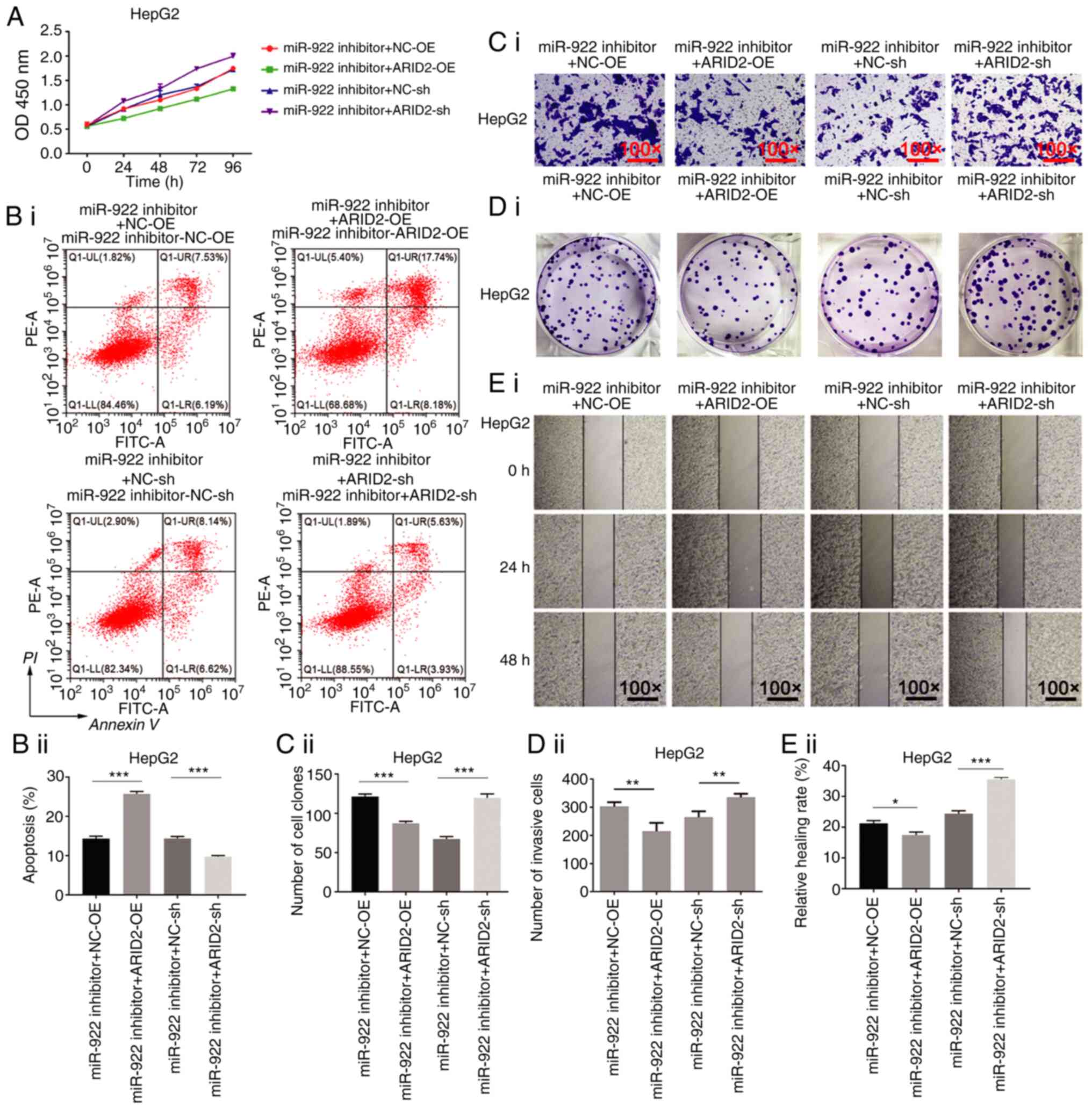
Altered ARID2 expression modulates
miR-922 inhibitor-decreased malignant behavior of liver cancer
cells
Finally, it was determined whether altered
ARID2 expression modulates miR-922 inhibitor-decreased
malignant behavior of liver cancer cells. ARID2
over-expression enhanced miR-922 inhibitor-decreased proliferation,
clonogenicity, invasion and wound healing of HepG2 cells and
increased the number of apoptotic HepG2 cells (Fig. 6). By contrast, ARID2 silencing
mitigated miR-922 inhibitor-decreased malignant behavior of HepG2
cells (Fig. 6). Similar effects of
altered ARID2 expression on miR-922 inhibitor-decreased
malignant behavior were observed in MHCC97L cells (Fig. S4). These data indicated that
miR-922-regulated ARID2 expression was key for control of
malignant behavior of liver cancer cells.
Discussion
A previous study verified that miRNAs regulate
hepatocarcinogenesis (19).
miR-221/222, miR-34a and miR-224 serve as oncogenes to promote
tumor cell growth (20,21), while miR-122 and miR-375 suppress
tumor progression by inhibiting liver cancer cell invasion and
metastasis (17,22). Previous studies have shown that
miR-922 expression is upregulated in liver cancer (23) and miR-922 promotes the proliferation
of liver cancer cells by targeting CYLD to enhance c-Myc and
cyclin D1 expression levels and inhibit Rb phosphorylation
(7). However, the molecular
mechanisms underlying the oncogenic role of miR-922 are still
unclear.
The present study found significantly elevated
levels of miR-922 transcripts in liver cancer tissue compared with
non-tumor adjacent tissue, consistent with a previous study
(7). In addition, miR-922 expression
levels were upregulated in liver cancer cell lines compared with
non-tumor hepatocytes. Induction of miR-922 over-expression
enhanced malignant behavior, rapid proliferation, strong
clonogenicity, lower numbers of apoptotic cells, potent would
healing and invasion ability in liver cancer cells. All these data
indicate that miR-922 may serve as an oncogenic factor to promote
malignant behavior of liver cancer cells.
Multiple factors regulate the transcription of
miRNAs, including genetic abnormality, epigenetic regulation and
transcription factors (24).
CREB1, a leucine zipper-type transcription factor, regulates
numerous types of malignancy (25). A
previous study demonstrated that CREB1 transcriptionally
regulates a number of miRNAs (17).
The present data indicated that CREB1 bound to the miR-922
promoter region and stimulated its transcription. Inhibition of
CREB1 decreased H3K27 acetylation upstream of the miR-922
promoter region but enhanced repressive H3K27 trimethylation. These
results suggested that CREB1 may serve as a transcription
factor to induce miR-922 expression in liver cancer cells. Future
studies should investigate the potential role of other
transcription factors in regulating miR-922 expression in liver
cancer. In addition, CREB1 was upregulated in liver cancer
tissue and positively associated with miR-922 expression levels.
Increased CREB1 expression levels were associated inversely
with OS of patients with liver cancer. Hence, high levels of
CREB1 and miR-922 may be valuable for the prognosis of liver
cancer.
In order to identify the targeted genes of miR-922
and the underlying mechanisms, potential targeted genes of miR-922
were predicted; miR-922 targeted ARID2. ARID2 is a
tumor suppressor and subunit of SWitch/sucrose non-fermentable
complex B (26). Given that
ARID2 mutations are associated with the development of HCC
(27–29), targeting ARID2 by miR-922 may
promote malignant behavior of HCC. ARID2 over-expression
eliminated malignant behavior of liver cancer cells whereas
ARID2 silencing had opposite effects, consistent with a
previous report (30). In addition,
ARID2 over-expression significantly mitigated or abrogated
miR-922-promoted malignant behavior of liver cancer cells.
ARID2 over-expression also enhanced Bax expression levels,
but decreased those of Bcl-2, PCNA, cyclin D1, MMP3 and
MMP9 in liver cancer tumors, which explained why
ARID2 over-expression inhibited growth of implanted liver
cancer xenografts in mice. Previous studies have shown that
ARID2 is targeted by miR-376c-3p, miR-208-3p and miR-155 in
HCC (30–32). Accordingly, the present findings
extended previous observations and indicate that the
miR-922/ARID2 axis is key for regulating malignant behavior
of liver cancer cells (33) and that
miR-922 may collaborate with other miRNAs to attenuate ARID2
expression levels, which promotes development and progression of
liver cancer. Further investigation is required to determine
whether miR-922 directly interacts with ARID2 mRNA in liver
cancer cells.
Together, the present data indicated significantly
upregulated miR-922 expression levels in liver cancer tissue and
cells; its elevated expression was associated with CREB1
expression levels in liver cancer tissue. Both in vitro and
in vivo experiments demonstrated that miR-922 enhanced
malignancy of liver cancer by promoting tumor growth and cell
proliferation, wound healing and invasion. Mechanistically, these
findings may provide novel insights into the
CREB1/miR-922/ARID2 interaction network in liver
cancer progression. Therefore, miR-922 may be a valuable diagnostic
and prognostic biomarker for liver cancer; targeting the
CREB1/miR-922/ARID2 axis may represent a new
therapeutic strategy for intervention of liver cancer. The present
study had limitations, including limited sample size of patients
with HCC with chronic hepatitis B, but not with other risk factors,
such as hepatitis B core and alcoholic liver disease. Therefore,
further studies with a larger population of patients with liver
cancer with diverse risk factors are warranted to validate the role
of the CREB1/ARID2/miR-922 axis in the progression of
liver cancer.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from
Hunan Provincial Natural Science Foundation (grant no. 2019JJ40495)
and Guangxi Provincial Natural Science Foundation (grant no.
2018GXNSFDA281003).
Availability of data and materials
The datasets used and analyzed during the current
study are available from the corresponding author upon reasonable
request.
Authors' contributions
XL and ZL designed the study. XL, HZ, PZ, HYZ, LC
and JG performed the experiments. XL and YY collected the data. HZ,
PZ, YY, LC and JG analyzed the data. XL, HZ, PZ, YY, HYZ, LC and JG
interpreted the data. XL and ZL drafted the manuscript. HZ and PZ
revised the manuscript. All authors read and approved the final
manuscript. All authors confirm the authenticity of all the raw
data.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the Third Xiangya Hospital of Central South University
(approval no. 2020-S362). All participants provided written
informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
ARID2
|
AT-rich interactive domain 2
|
|
CREB1
|
cAMP response element binding protein
1
|
|
HCC
|
hepatocellular carcinoma
|
|
miRNA
|
microRNA
|
References
|
1
|
Singh AK, Kumar R and Pandey AK:
Hepatocellular carcinoma: Causes, mechanism of progression and
biomarkers. Curr Chem Genom Transl Med. 12:9–26. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhu X, Tang Z and Sun HC: Targeting
angiogenesis for liver cancer: Past, present, and future. Genes
Dis. 7:328–335. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sagnelli E, Potenza N, Onorato L, Sagnelli
C, Coppola N and Russo A: Micro-RNAs in hepatitis B virus-related
chronic liver diseases and hepatocellular carcinoma. World J
Hepatol. 10:558–570. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sengupta S and Parikh ND: Biomarker
development for hepatocellular carcinoma early detection: Current
and future perspectives. Hepat Oncol. 4:111–122. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rupaimoole R, Calin GA, Lopez-Berestein G
and Sood AK: miRNA deregulation in cancer cells and the tumor
microenvironment. Cancer Discov. 6:235–246. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shayimu P, Wang JB, Liu L, Tuerdi R, Yu CG
and Yusufu A: miR-922 regulates apoptosis, migration, and invasion
by targeting SOCS1 in gastric cancer. Kaohsiung J Med Sci.
36:178–185. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu J, Su Z, Zeng Y, Zhang H, Yang S and
Liu G: miR-922 regulates CYLD expression and promotes the cell
proliferation of human hepatocellular carcinoma. Oncol Rep.
37:1445–1450. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sevinc ED, Cecener G, Ak S, Tunca B, Egeli
U, Gokgoz S, Tolunay S and Tasdelen I: Expression and clinical
significance of miRNAs that may be associated with the FHIT gene in
breast cancer. Gene. 590:278–284. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Abramovitch R, Tavor E, Jacob-Hirsch J,
Zeira E, Amariglio N, Pappo O, Rechavi G, Galun E and Honigman A: A
pivotal role of cyclic AMP-responsive element binding protein in
tumor progression. Cancer Res. 64:1338–1346. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kovach SJ, Price JA, Shaw CM, Theodorakis
NG and McKillop IH: Role of cyclic-AMP responsive element binding
(CREB) proteins in cell proliferation in a rat model of
hepatocellular carcinoma. J Cell Physiol. 206:411–419. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li Y, Fu Y, Hu X, Sun L, Tang D, Li N,
Peng F and Fan XG: The HBx-CTTN interaction promotes cell
proliferation and migration of hepatocellular carcinoma via CREB1.
Cell Death Dis. 10:4052019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Oba A, Shimada S, Akiyama Y, Nishikawaji
T, Mogushi K, Ito H, Matsumura S, Aihara A, Mitsunori Y, Ban D, et
al: ARID2 modulates DNA damage response in human hepatocellular
carcinoma cells. J Hepatol. 66:942–951. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Marrero JA, Kulik LM, Sirlin CB, Zhu AX,
Finn RS, Abecassis MM, Roberts LR and Heimbach JK: Diagnosis,
staging, and management of hepatocellular carcinoma: 2018 practice
guidance by the American association for the study of liver
diseases. Hepatology. 68:723–750. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tellapuri S, Sutphin PD, Beg MS, Singal AG
and Kalva SP: Staging systems of hepatocellular carcinoma: A
review. Indian J Gastroenterol. 37:481–491. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhu P, Wang Y, Wu J, Huang G, Liu B, Ye B,
Du Y, Gao G, Tian Y, He L and Fan Z: LncBRM initiates YAP1
signalling activation to drive self-renewal of liver cancer stem
cells. Nat Commun. 7:136082016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang YW, Chen X, Ma R and Gao P:
Understanding the CREB1-miRNA feedback loop in human malignancies.
Tumour Biol. 37:8487–8502. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rad R, Rad L, Wang W, Strong A, Ponstingl
H, Bronner IF, Mayho M, Steiger K, Weber J, Hieber M, et al: A
conditional piggyBac transposition system for genetic screening in
mice identifies oncogenic networks in pancreatic cancer. Nat Genet.
47:47–56. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xu J, Li J, Zheng TH, Bai L and Liu ZJ:
MicroRNAs in the occurrence and development of primary
hepatocellular carcinoma. Adv Clin Exp Med. 25:971–975. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pineau P, Volinia S, McJunkin K, Marchio
A, Battiston C, Terris B, Mazzaferro V, Lowe SW, Croce CM and
Dejean A: miR-221 overexpression contributes to liver
tumorigenesis. Proc Natl Acad Sci USA. 107:264–269. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Varnholt H: The role of microRNAs in
primary liver cancer. Ann Hepatol. 7:104–113. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
He XX, Chang Y, Meng FY, Wang MY, Xie QH,
Tang F, Li PY, Song YH and Lin JS: MicroRNA-375 targets AEG-1 in
hepatocellular carcinoma and suppresses liver cancer cell growth in
vitro and in vivo. Oncogene. 31:3357–3369. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu Y, Chen SH, Jin X and Li YM: Analysis
of differentially expressed genes and microRNAs in alcoholic liver
disease. Int J Mol Med. 31:547–554. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lee YS and Dutta A: MicroRNAs in cancer.
Annu Rev Pathol. 4:199–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hu PC, Li K, Tian YH, Pan WT, Wang Y, Xu
XL, He YQ, Gao Y, Wei L and Zhang JW: CREB1/Lin28/miR-638/VASP
interactive network drives the development of breast cancer. Int J
Biol Sci. 15:2733–2749. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hodges C, Kirkland JG and Crabtree GR: The
many roles of BAF (mSWI/SNF) and PBAF complexes in cancer. Cold
Spring Harb Perspect Med. 6:a0269302016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Linhares AC, Stupka JA, Ciapponi A,
Bardach AE, Glujovsky D, Aruj PK, Mazzoni A, Rodriguez JA, Rearte
A, Lanzieri TM, et al: Burden and typing of rotavirus group A in
Latin America and the Caribbean: Systematic review and
meta-analysis. Rev Med Virol. 21:89–109. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhao H, Wang J, Han Y, Huang Z, Ying J, Bi
X, Zhao J, Fang Y, Zhou H, Zhou J, et al: ARID2: A new tumor
suppressor gene in hepatocellular carcinoma. Oncotarget. 2:886–891.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
You J, Yang H, Lai Y, Simon L, Au J and
Burkart AL: AT-rich interactive domain 2, p110α, p53, and β-catenin
protein expression in hepatocellular carcinoma and
clinicopathologic implications. Hum Pathol. 46:583–592. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang L, Wang W, Li X, He S, Yao J, Wang
X, Zhang D and Sun X: MicroRNA-155 promotes tumor growth of human
hepatocellular carcinoma by targeting ARID2. Int J Oncol.
48:2425–2434. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang Y, Chang W, Chang W, Chang X, Zhai S,
Pan G and Dang S: MicroRNA-376c-3p facilitates human hepatocellular
carcinoma progression via repressing AT-rich interaction domain 2.
J Cancer. 9:4187–4196. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yu P, Wu D, You Y, Sun J, Lu L, Tan J and
Bie P: miR-208-3p promotes hepatocellular carcinoma cell
proliferation and invasion through regulating ARID2 expression. Exp
Cell Res. 336:232–241. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu Q, Yang P, Tu K, Zhang H, Zheng X, Yao
Y and Liu Q: TPX2 knockdown suppressed hepatocellular carcinoma
cell invasion via inactivating AKT signaling and inhibiting MMP2
and MMP9 expression. Chin J Cancer Res. 26:410–417. 2014.PubMed/NCBI
|