Introduction
Thyroid carcinoma (THCA) is the most common type of
endocrine system malignancy and accounts for ~1% of all
malignancies (1). The incidence of
THCA has continued to increase worldwide over the past few decades
(2,3),
especially in women in China and the United States (4,5). THCA
originates from thyroid follicular or parafollicular cells and is
usually classified into differentiated THCA (follicular THCA and
papillary THCA), medullary THCA and undifferentiated THCA (6). The majority of THCAs exhibit relatively
low levels of malignancy, except for undifferentiated THCA (7, 8).
Most patients with differentiated THCA have a favorable prognosis
and rarely exhibit signs of distant metastasis. Only 1–3% of THCAs
are classified as undifferentiated THCAs (7,8); however,
the mortality rate of undifferentiated THCA is 14–50% and the
average survival time of patients with undifferentiated THCA is 3–5
months (8). The current prognosis of
patients with undifferentiated THCA is poor due to the rapid onset
and aggressiveness of the tumor, and the occurrence of widespread
pelvic metastasis (9). Thus, further
studies investigating the molecular mechanisms of invasion and
metastasis in undifferentiated THCA are urgently required.
Only 1–2% of the human genome transcript encodes
proteins, whereas >75% is transcribed into non-coding RNA
(ncRNA) (10). There are numerous
types of ncRNA, including long ncRNAs (lncRNAs), which are >200
nucleotides in length, and microRNAs (miRNAs/miRs) (11). Both lncRNAs and miRNAs have been
reported to be involved in the invasion and metastasis of tumor
cells. For instance, the lncRNA colorectal neoplasia differentially
expressed was discovered to promote glioma cell growth and invasion
through the mTOR signaling pathway (12). The lncRNA, H19, a host gene of
miR-675, was revealed to generate mature miR-675-3p and miR-675-5p,
which were subsequently demonstrated to be involved in tumor
metastasis (13). On the other hand,
miRNAs, which are 20–25 nucleotides in length, regulate the
expression of specific target genes that have roles in numerous
biological processes, such as tumor progression and survival
(14–17). Previous studies have reported that
miRNAs regulated proliferation and invasion in THCA. For instance,
Dong et al revealed that the knockdown of miR-363-3p
expression in papillary THCA inhibited tumor progression by
targeting NIN1 (RPN12) binding protein 1 homolog (18); and miR-219 was revealed to inhibit
cell viability and metastasis in papillary THCA by targeting EYA
transcriptional coactivator and phosphatase 2 (19). miRNAs have also been revealed to play
crucial roles in numerous tumor processes. Previous evidence
suggested that miR-34c-5p expression was closely associated with a
poor prognosis and unfavorable clinicopathological parameters
(20,21). Shen et al indicated that the
lncRNA KCNQ1 opposite strand/antisense transcript 1 sponged
miR-34c-5p to promote osteosarcoma growth via aldolase,
fructose-bisphosphate A-enhanced aerobic glycolysis (22). Numerous other studies have revealed
that miR-34c-5p was closely related to the development of numerous
types of human malignant cancer (23–25).
The functions of lncRNAs and miRNAs do not occur in
isolation. In fact, it has been well established that a regulatory
network composed of lncRNAs and miRNAs has an enhanced effect on
the regulation of cellular processes (26). miRNAs often inhibit protein
translation through binding to the 3′-untranslated region (UTR) of
target mRNAs, and lncRNAs can sponge miRNAs and repress this
process (27). It has been reported
that lncRNA CCDC144NL antisense RNA 1 sponged miR-143-3p and
regulated mitogen-activated protein kinase kinase kinase 7 by
acting as a competing endogenous RNA (ceRNA) in gastric cancer
(28).
In general, treatment strategies for cancer are
divided into three major categories: Surgical resection,
chemotherapy and radiotherapy (29).
Surgical resection is the primary treatment option recommended for
non-metastatic tumors. Metastatic cancers are treated with combined
therapies; however, these therapeutic regimens are often
insufficient to effectively treat metastatic tumors currently
(30). Therefore, there is an urgent
requirement to develop effective and novel approaches for the
treatment of cancer. Muhammad et al previously reported that
bitter melon extract inhibited breast cancer growth in a
preclinical model by inducing autophagic cell death (31). Cholesterol depletion by
methyl-β-cyclodextrin was demonstrated to increase
tamoxifen-induced cell death by enhancing its uptake in melanoma
(32). In addition,
methyl-β-cyclodextrin was revealed to enhance the sensitivity of
skin cancer cells to doxorubicin by regulating wild-type (Wt) p53
(33). Furthermore, numerous drugs
have been designed to treat cancer by targeting tumor-related
targets. In a previous study, cellular retinoic acid (RA) binding
protein 2 (CRABP2) was discovered to be an essential protein for
tumor growth. The abnormal expression of CRABP2 was revealed to be
associated with malignant human cancer types. In addition, the
knockdown of CRABP2 expression levels was revealed to suppress the
migration of cancer cells (34–36).
Previous studies on cancer cells have also revealed that the
delivery of RA by CRABP2 facilitated RAR-related orphan receptor A
transcriptional activity by enhancing differentiation and
apoptosis, and reducing proliferation (37,38).
The present study used datasets from The Cancer
Genome Atlas (TCGA) database to reveal that the relative expression
levels of LINC01816 were upregulated in patients with THCA compared
with healthy patients. The lncRNA long intergenic non-protein
coding RNA 1816 (LINC01816) was also revealed to promote
epithelial-mesenchymal transition (EMT), invasion and metastasis in
THCA tissues. Bioinformatics analysis subsequently identified
binding sites between miR-34c-5p and LINC01816 or the 3′-UTR of
CRABP2, which indicated the presence of a competing endogenous RNA
(ceRNA) regulatory mechanism. These findings suggested that ncRNAs
have the potential to become a novel research hotspot in the
context of cancer therapy.
Materials and methods
Patient samples
Matched paracarcinoma and cancer tissues were
obtained from 10 patients (Table S1)
undergoing surgery for THCA at The Affiliated Hospital of Zunyi
Medical University (Zunyi, China). The present study was approved
by the Zunyi Medical University Ethics Committee and informed
patient consent was obtained prior to participation.
Cell lines and culture
Human THCA cell lines, OCUT-2C, C643, 8305C and
CAL-62, as well as the normal thyroid cell line, HTori-3 were
purchased from the Institute of Cell Biology, Chinese Academy of
Sciences (Shanghai, China). OCUT-2C, C643, 8305C and CAL-62 were
cultured in DMEM supplemented with 1% antibiotics (100 U/ml
penicillin and 100 µg/ml streptomycin sulfates) and 10% FBS (all
from Gibco; Thermo Fisher Scientific, Inc.). HTori-3 was maintained
in DMEM/F12 supplemented with 1% glutamine and 10% FBS. Cells were
maintained in an incubator at 37°C in a humidified atmosphere with
5% CO2. In all experiments, cells were allowed to
acclimatize for 24 h prior to treatment. Cell lines were tested and
confirmed to be mycoplasma-free at the beginning and end of the
research.
Cell transfection
Cells were transfected upon reaching 60–70%
confluence with pCDNA3.1-LINC01816 or pCDNA3.1-CRABP2 plasmids,
miR-34c-5p mimics, miR-34c-5p inhibitor and their corresponding
controls using Lipofectamine® 3000 reagent (Invitrogen;
Thermo Fisher Scientific, Inc.), according to the manufacturer's
protocol. The miR-34c-5p mimics sequence was
5′-AGGCAGUGUAGUUAGCUGAUUGC-3′ and the sequence of the mimics
control was 5′-UUACUCGACACGUGUCAAGUUU-3′. The miR-34c-5p inhibitor
sequence was 5′-GCAAUCAGCUAACUACACUGCCU-3′ and the sequence of
inhibitor control was 5′-CAGUACUUUUGUGUAGUACAA-3′ The C643 or
OCUT-2C cells (2×105 cells/dish) were seeded into a 6-cm
cell culture dish and cultured to 60–70% confluence. The cells were
transfected with 1 µg plasmids or 30 nM miRNA mimics/inhibitor in
the 6-cm cell culture dish. The transfected cells were incubated at
37°C for 48 h. The transfected cells were harvested 48 h post
transfection and used for subsequent experiments.
Scratch wound healing assay
A wound healing assay was performed using C643 and
OCUT-2C cells to determine the levels of cell migration. Briefly,
cells (2×106 cells/well) were seeded into 24-well plates
and cultured to 100% confluence. Then, a linear scratch was made in
the cell monolayer using a sterile 200-µl pipette tip. The scratch
area was imaged at 0 and 24 h using an optical microscope (Olympus
Corporation). The scratch width was captured at a magnification of
×100. The area of each scratch wound was determined using ImageJ
software (v1.48; National Institutes of Health). The cell migratory
rate was calculated as: Migration rate (%)=(initial distance-24 h
scratch distance) ×100.
Cell invasion assay
The cell invasion assay was performed using a
Matrigel invasion chamber (Corning, Inc.) in a 24-well Transwell
plate (8 µm pore size). Briefly, the cell density was adjusted to
5×105 cells/ml. Then, 500 µl of serum-free medium
containing 3×104 cells was added into the upper chamber
of the Transwell plate. A volume of 750 µl medium supplemented with
10% FBS was added into the lower chamber. Following incubation at
37°C for 24 h, the invasive cells in the lower chamber were fixed
with 4% paraformaldehyde at room temperature for 20 min and
subsequently stained with 0.5% crystal violet at 37°C for 30 min; a
cotton swab was used to gently remove the cells on the upper
surface of the upper chamber. Stained cells were visualized using a
light microscope (magnification, ×100) in five randomly selected
fields of view from each group.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from tissues or cultured
cells using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) for mRNA or a miRNeasy Micro kit (for miRNA;
Qiagen GmbH) for miRNA, according to the manufacturers' protocols.
Then, 1 µg RNA was reverse transcribed into cDNA using a
Prime-Script RT-qPCR kit (Takara Bio, Inc.) for mRNA or a miScript
II RT kit (Qiagen GmbH) for miRNA, according to the manufacturer's
protocol. The primer sequences used were as follows: LINC01816
forward, 5′-CAGCTGTCTTTGTCTGGGGCGGCGG-3′ and reverse,
5′-GCCCCAGACAAAGACAGC-3′; miR-34c-5p forward,
5′-GAGGCAGTGTAGTTAGCTGA-3′ and reverse,
5′-TCCAGTTTTTTTTTTTTTTTGCAATC-3′; CRABP2 forward,
5′-ATGCCCAACTTCTCTGGCAACTGGA-3′ and reverse,
5′-CAGCATCACATTCACCC-3′; U6 forward, 5′-GTGCAGGGTCCGAGGT-3′ and
reverse, 5′-CTCGCTTCGGCAGCACA-3′; GAPDH forward,
5′-GGAGCGAGATCCCTCCAAAAT-3′ and reverse,
5′-GGCTGTTGTCATACTTCTCATGG-3′. RT-qPCR was subsequently performed
using a SYBR-Green PCR kit (Takara Bio, Inc.) on a LineGene 9600
Plus Real-Time PCR Detection system (Hangzhou Bioer Co., Ltd.).
Thermal cycler conditions were as follows: 1 cycle at 98°C for 3
min, 40 cycles at 98°C for 1 min, 60°C for 30 sec, and 72°C for 30
sec, 1 cycle of 72°C for 5 min. The relative expression levels of
mRNAs and miRNAs were calculated using the 2−ΔΔCq method
(39). mRNA expression levels were
normalized to GAPDH expression levels and miRNA expression levels
to U6 expression levels.
Dual-luciferase reporter assay
Cells were plated into 6-cm culture dishes at a
density of 5×105 cells/ml. The Wt or mutant (Mut) 3′-UTR
sequences of CRABP2 and LINC01816 were cloned into a psiCHECK2
luciferase reporter plasmid (Promega Corporation) containing
Renilla and firefly luciferase. The luciferase reporter
vectors and miR-34c-5p were co-transfected into C643 or OCUT-2C
cells using Lipofectamine 3000, according to the manufacturer's
protocol. Following 24 h of transfection, cells were lysed with 1X
lysis buffer (Beyotime Institute of Biotechnology) and the relative
luciferase activity was measured using a Dual-Luciferase Reporter
assay system (Beyotime Institute of Biotechnology). Firefly
luciferase activity was normalized to Renilla luciferase
activity.
RNA immunoprecipitation (RIP)
assay
RIP was performed using an RNA Binding Protein
Immunoprecipitation kit (EMD Millipore), according to the
manufacturer's protocol. Briefly, C643 and OCUT-2C cells were
digested and resuspended in PBS (1×107 cells in 2 ml
PBS). The RIP assay was performed using an anti-AGO2 antibody
(1:500; product code ab32381; Abcam) overnight at 4°C and an
anti-IgG antibody (1:500; product no. 3900S; Cell Signaling
Technology, Inc.) as the negative control. RT-qPCR was subsequently
performed to analyze the CRABP2 mRNA expression levels.
Immunofluorescence staining
Cells were seeded into 6-cm dishes at a density of
2×105 cells/dish. The cells were fixed with 4%
paraformaldehyde for 20 min at room temperature, permeabilized in
0.5% Triton X-100 for 1 h and blocked with 3% BSA at room
temperature for 2 h (Nanjing KeyGen Biotech Co., Ltd.).
Subsequently, the cells were incubated with the following primary
antibodies for 6 h at 4°C in humid conditions: Anti-E-cadherin
(1:100; product no. 14472S; Cell Signaling Technology, Inc.),
anti-vimentin (1:100; product no. 5741S; Cell Signaling Technology,
Inc.) and anti-CRABP2 (1:250; product code ab211927; Abcam).
Following the primary antibody incubation, the cells were incubated
with Alexa Fluor-594 anti-rabbit (1:500; cat. no. A-11012; Thermo
Fisher Scientific, Inc.) or anti-mouse secondary antibodies (1:500;
cat. no. A-11032; Thermo Fisher Scientific, Inc.) at room
temperature for 1 h. The cell nuclei were subsequently stained with
5 µg/ml DAPI (Beyotime Institute for Biotechnology) for 5 min.
Stained cells were visualized using an immunofluorescence
microscope at a magnification of ×400 (Olympus Corporation).
Western blotting
Total protein was extracted from cells using RIPA
lysis buffer (product no. P0013C; Beyotime Institute of
Biotechnology) supplemented with protease and phosphatase
inhibitors. Total protein concentration was quantified using a BCA
protein assay kit (Beyotime Institute of Biotechnology) and 20 µg
protein per lane was separated via 10% SDS-PAGE. The separated
proteins were subsequently transferred onto PVDF membranes (EMD
Millipore) and blocked with 5% BSA at 37°C for 1 h. The membranes
were then incubated overnight at 4°C with the following primary
antibodies: Anti-CRABP2 (1:1,000; product code ab211927; Abcam),
anti-E-cadherin (1:1,000; product no. 14472S; Cell Signaling
Technology, Inc.), anti-N-cadherin (1:1,000; product no. 13116S;
Cell Signaling Technology, Inc.), anti-vimentin (1:1,000; product
no. 5741S; Cell Signaling Technology, Inc.), anti-NME1 (1:1,000;
cat. no. 11086-2-AP; ProteinTech Group, Inc.), anti-AMF (1:1,000;
product code ab66340; Abcam), anti-cyclin D1 (1:1,000; product no.
55506S), anti-cyclin E1 (1:1,000; product no. 20808S), p27
(1:1,000; product no. 3686S), p21 (1:1,000; product no. 2947S),
cleaved caspase-3 (1:1,000; product no. 9664S), cleaved PARP
(1:1,000; product no. 5625S), Bax (1:1,000; product no. 5023S) and
Bcl-2 (1:1,000; product no. 15071S; all from Cell Signaling
Technology, Inc.) and anti-GAPDH (1:10,000; cat. no. 60004-1-AP;
ProteinTech Group, Inc.). Following the primary antibody
incubation, the PVDF membranes were washed 3 times with
TBS-Tween-20 (0.05%; 5 ml Tween-20 in 1,000 ml TBS) and then
incubated for an additional 2 h with a horseradish
peroxidase-conjugated secondary antibody (1:5,000; cat. no. BA1056;
Wuhan Boster Biological Technology, Ltd.) at room temperature for 2
h. Protein bands were visualized using an ECL Western Blotting
Detection kit (Beyotime Institute of Biotechnology). Densitometric
analysis was performed using ImageJ software (v1.48; National
Institutes of Health). Protein expression levels were normalized to
GAPDH expression levels.
Cell cycle analysis
A Cell Cycle kit (Beyotime, China) was used for cell
cycle analysis. Cells (1×106 cells/ml) were washed three
time by cold PBS and then fixed in 70% ethanol at −20°C for 24 h.
Subsequently, cells were washed with cold PBS and stained with
appropriate propidium iodide (PI) staining buffer containing 10%
RNase A at 37°C for 30 min in the dark. Cell cycle analysis was
performed using FACSCantoII cytometer (BD Biosciences). Analyses
were performed on Modifit LT 4.1 software (BD Biosciences).
Bioinformatics analysis
The expression levels of LINC01816 in different
types of cancer were obtained from the online GEPIA database
(40) (http://gepia.cancer-pku.cn/). The target predictions
between LINC01816/miR-34c-5p were analyzed using the online
starBase v2.0 database (41)
(http://starbase.sysu.edu.cn). The
corresponding target genes for miR-34c-5p were filtered out using
the TargetScan database (42)
(https://www.targetscan.org) and the KEGG
pathway analysis was performed using the Database for Annotation,
Visualization and Integrated Discovery (DAVID) database (43) (https://david-d.ncifcrf.gov/).
Statistical analysis
Statistical analysis was performed using GraphPad
Prism 8.0 software (GraphPad Software, Inc.) and SPSS 22.0 software
(IBM Corp.). All data are presented as the mean ± SD of ≥3
experimental repeats. Statistical differences between groups were
analyzed using unpaired Student's t-tests. P<0.05 was considered
to indicate a statistically significant difference.
Results
LINC01816 expression levels are
upregulated in undifferentiated THCA tissues and cell lines
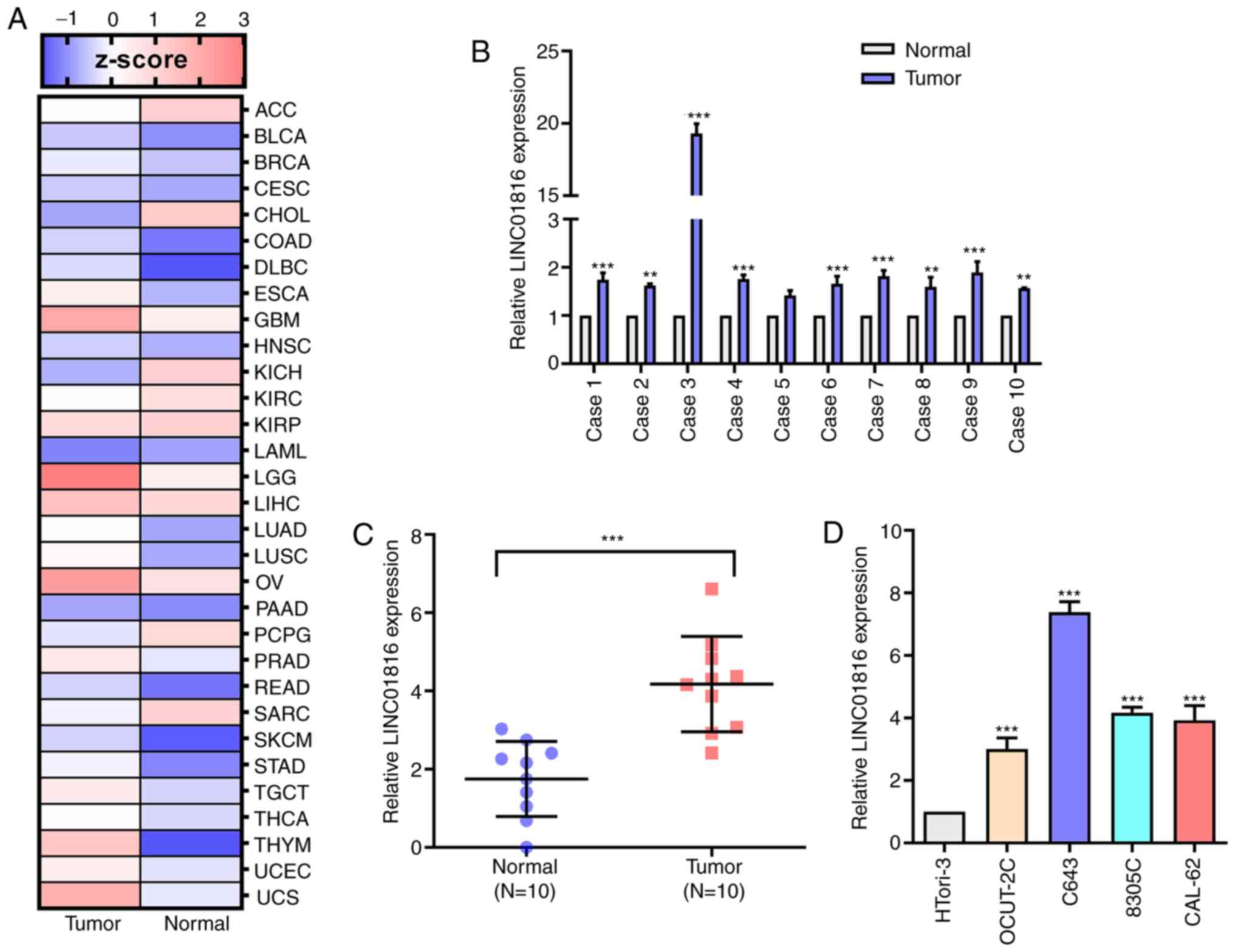
Analysis of THCA RNA sequencing datasets from TCGA
revealed that LINC01816 expression levels were upregulated in THCA
tissues compared with normal tissues (Fig. 1A). Thus, the expression levels of
LINC01816 in THCA and matched adjacent paracarcinoma thyroid
tissues obtained from 10 patients were analyzed. RT-qPCR analysis
of the 10 paired thyroid tissues revealed that the expression
levels of LINC01816 were upregulated in cancerous tissues compared
with noncancerous tissues (Fig. 1B and
C). Moreover, the RT-qPCR results indicated that the expression
levels of LINC01816 were significantly upregulated in the
undifferentiated THCA cell lines, C643, 8305C and CAL-62, compared
with the normal thyroid cell line, OCUT-2C (Fig. 1D).
LINC01816 promotes the EMT, migration
and invasion of undifferentiated THCA cells
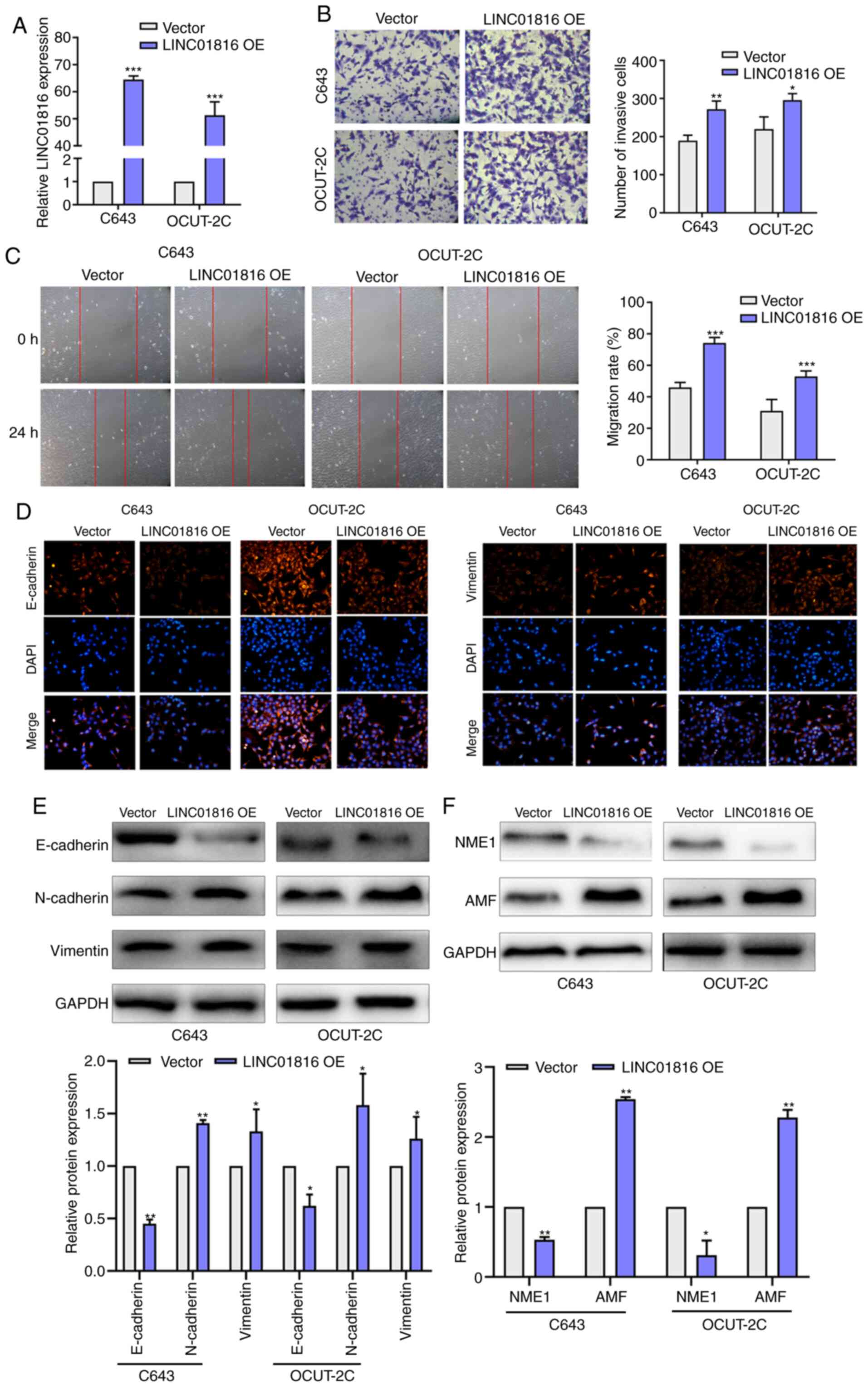
To investigate the role of LINC01816 in
undifferentiated THCA, pcDNA3.1-LINC01816 plasmids were transfected
into C643 and OCUT-2C cells to promote the overexpression of
LINC01816 (Fig. 2A). Transwell assays
were used to detect the cell invasive ability. As revealed in
Fig. 2B, the number of invasive cells
was increased in the LINC01816 overexpression group compared with
the vector group. The results of the wound healing assay also
demonstrated that the migratory rate was increased in cells
overexpressing LINC01816 compared with the vector group (Fig. 2C). Immunofluorescence staining
revealed that E-cadherin was downregulated while vimentin was
upregulated after LINC01816 overexpression in C643 and OCUT-2C
cells (Fig. 2D). In addition, the
protein expression levels of E-cadherin were downregulated in the
LINC01816 overexpression group, while the expression levels of
N-cadherin and vimentin were upregulated in the LINC01816
overexpression group (Fig. 2E).
Furthermore, the expression levels of NME1 were also downregulated
following the overexpression of LINC01818, while the expression
levels of AMF were upregulated, in C643 and OCUT-2C cells (Fig. 2F).
To determine the effects of LINC01818 on the
invasion and migration, cell Transwell and scratch wound healing
assay were performed in Htori-3 cells. LINC01816 promoted cell
invasion and migration in normal thyroid cells (Fig. S1). To determine the effects of
LINC01818 on the EMT process, LINC01818 expression was knocked down
in C643 cells. Following the knockdown of LINC01818, the expression
levels of E-cadherin were upregulated, while the expression levels
of N-cadherin and vimentin were downregulated (Fig. S2A). Similarly, the results of the
Transwell invasion and wound healing assays indicated that the
knockdown of LINC01816 expression suppressed cell invasion and
migration in C643 cells (Fig. S2B and
C). In addition, the cell cycle distribution was detected using
flow cytometry. The overexpression of LINC01816 resulted in
G2/M phase arrest (Fig.
S3A). Western blotting was performed to analyze the expression
levels of cell cycle-related proteins, including cyclin D1, cyclin
E1, p27 and p21. The expression levels of cyclin D1 and cyclin E
were downregulated following the overexpression of LINC01816, while
the expression levels of p27 and p21 were upregulated (Fig. S3B). Thus, it was suggested that the
cell cycle arrest in the G2/M phase may promote EMT. The
expression levels of the apoptosis-related proteins, cleaved
caspase-3 (c-caspase-3), cleaved poly (ADP-ribose) polymerase
(c-PARP), Bax and Bcl-2, were also analyzed using western blotting.
These results revealed that the changes in LINC01816 expression had
little effect on the levels of cell apoptosis in vitro
(Fig. S3C). These findings
aforementioned indicated that the upregulated expression levels of
LINC01816 may induce EMT, leading to an increase in cell
invasion.
miR-34c-5p is regulated by LINC01816
and inhibits EMT and cell migration
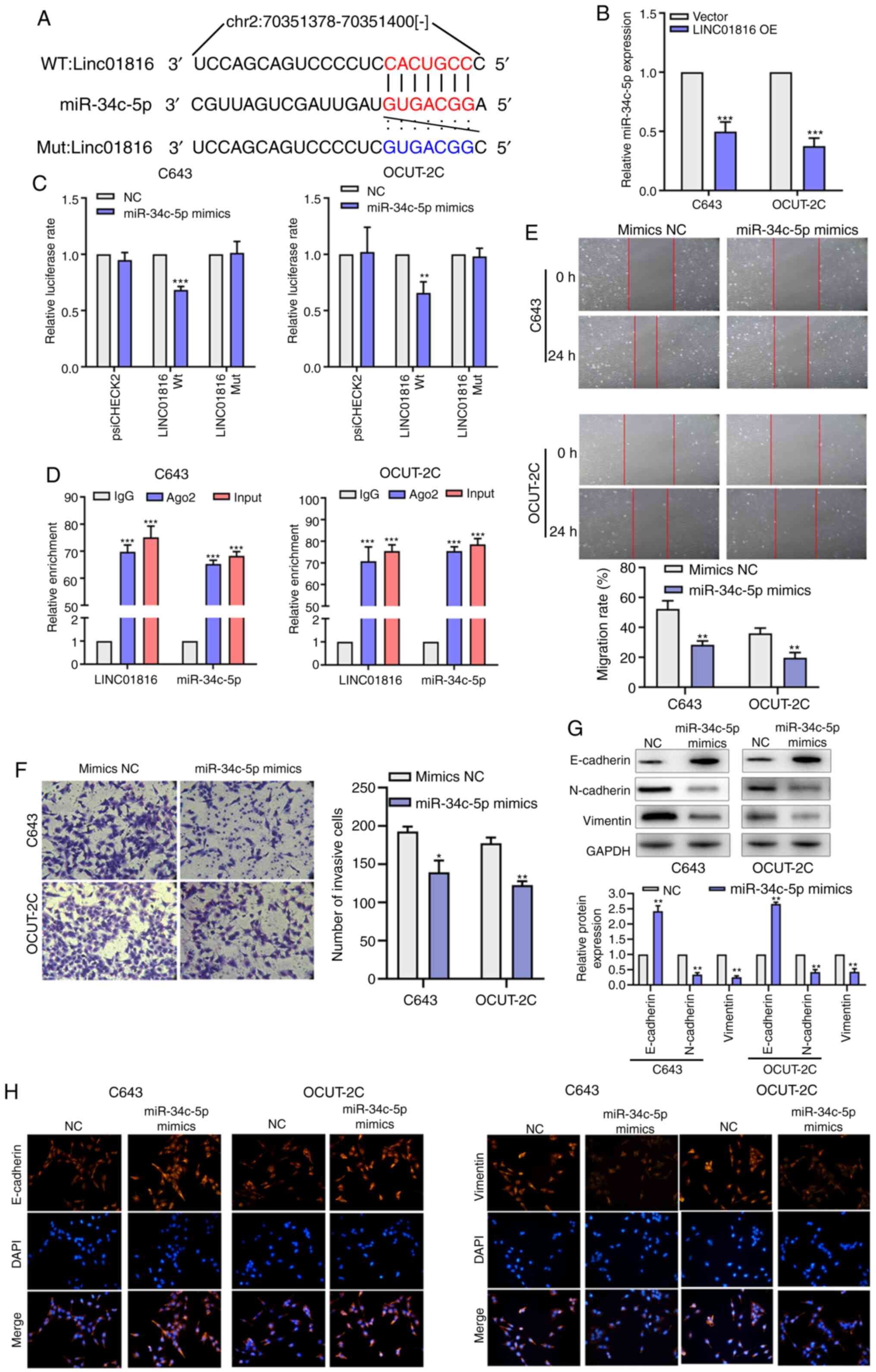
The bioinformatics online tool, starBase, was used
to predict that LINC01816 may act as a sponge by binding to
miR-34c-5p at the binding site located within chromosome 2:
70351393-70351399[-] (Fig. 3A).
Following the overexpression of LINC01816, the expression levels of
miR-34c-5p were demonstrated to be downregulated in C643 and
OCUT-2C cells (Fig. 3B). Moreover,
the results of the dual-luciferase reporter assay revealed that the
overexpression of miR-34c-5p reduced the relative luciferase
activity of LINC01816-Wt vectors, while no effect was observed on
the relative luciferase activity of LINC01816-Mut vectors (Fig. 3C). The RIP assay results also revealed
that LINC01816 and miR-34c-5p were markedly enriched in anti-Ago2
lysates, which suggested that LINC01816 may sponge miR-34c-5p
(Fig. 3D). As revealed in Fig. 3E and F, the results of the wound
healing assay and Transwell invasion assay revealed that the
migratory rate and number of invasive cells were decreased in cells
overexpressing miR-34c-5p compared with the mimics NC group. To
determine whether miR-34c-5p inhibited EMT in THCA, the protein
expression levels of E-cadherin, N-cadherin and vimentin were
analyzed by western blotting following the inhibition of miR-34c-5p
in C643 cells. The results revealed that the expression levels of
E-cadherin were downregulated, while the expression levels of
N-cadherin and vimentin were upregulated following the inhibition
of miR-34c-5p (Fig. S2A). Transwell
and wound healing assays were also performed and the results
demonstrated that the inhibition of miR-34c-5p expression promoted
cell invasion and migration in C643 cells (Fig. S2B and C). Conversely, the expression
levels of E-cadherin were significantly upregulated, while vimentin
and N-cadherin expression levels were downregulated in the
miR-34c-5p mimics group (Fig. 3G).
Immunofluorescence staining revealed that E-cadherin was
upregulated while vimentin was downregulated after miR-34c-5p
overexpression in C643 and OCUT-2C cells (Fig. 3H). These data indicated that
miR-34c-5p may attenuate the migratory and invasive abilities, and
the EMT of THCA cells, via sponging LINC01816.
CRABP2 is a target gene of miR-34c-5p
and is associated with EMT, migration and invasion
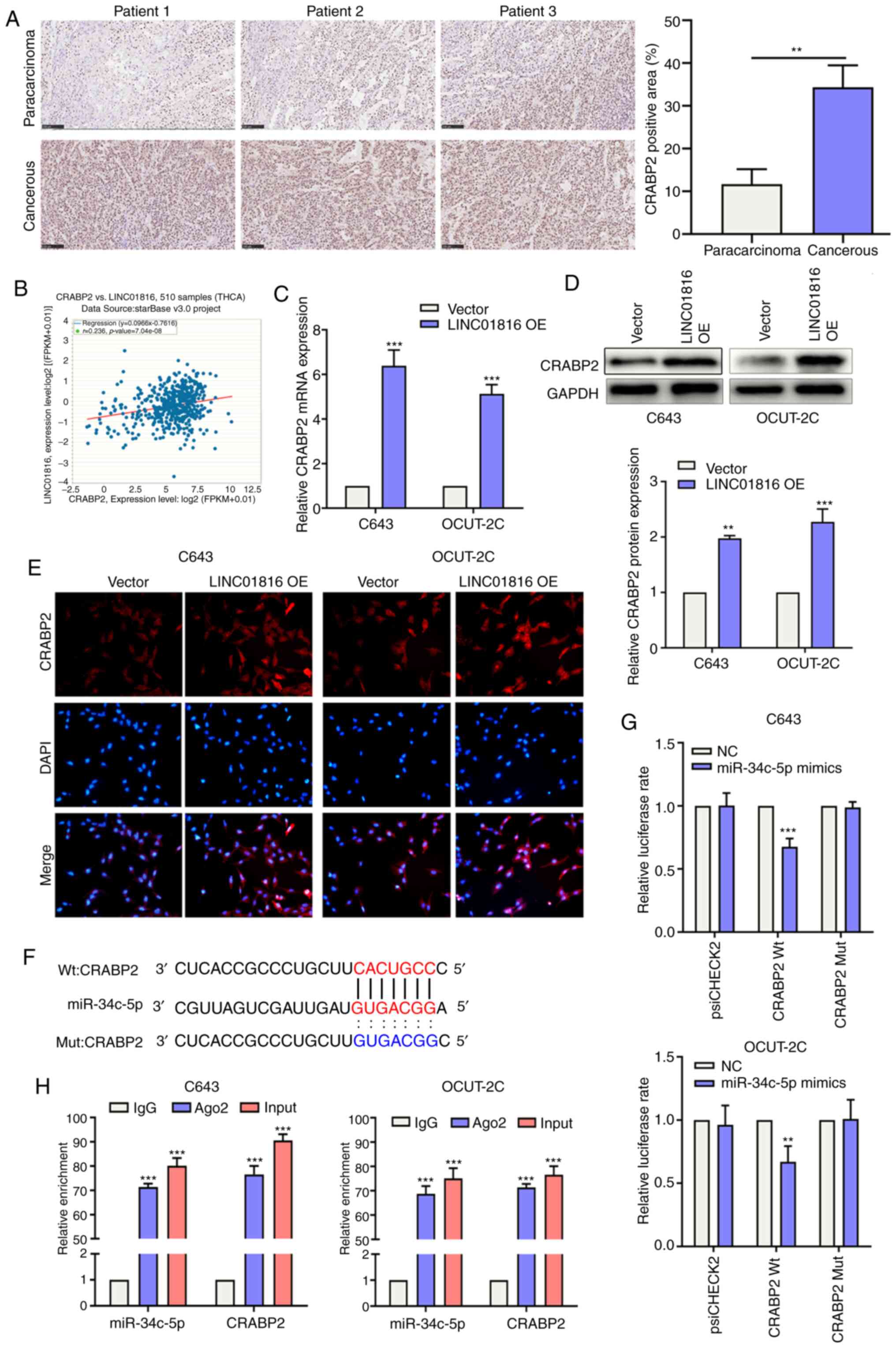
Immunohistochemistry was used to analyze the
expression levels of the CRABP2 protein in THCA and paracarcinoma
tissues of patients. As revealed in Fig.
4A, CRABP2 protein expression levels were significantly
upregulated in THCA tissues compared with paracarcinoma tissues.
starBase was used to determine the presence of a positive
association between CRABP2 and LINC01816 expression levels in THCA
(Fig. 4B). Following the
overexpression of LINC01816 in C643 and OCUT-2C cells, CRABP2
expression levels were revealed to be upregulated (Fig. 4C and D). The results of the
immunofluorescence analysis also validated these findings, as the
expression levels of CRABP2 were consistent with the results of the
aforementioned western blotting (Fig.
4E). The binding site between CRABP2 and miR-34c-5p is
presented in Fig. 4F. Subsequently,
luciferase reporter plasmids containing 3′-UTR-CRABP2-Wt/Mut were
constructed. The results of the dual-luciferase reporter assay
demonstrated that the overexpression of miR-34c-5p suppressed the
relative luciferase activity of the 3′-UTR-CRABP2-Wt vector, but
not of the 3′-UTR-CRABP2-Mut vector, in C643 and OCUT-2C cells
(Fig. 4G). The RIP assay results
demonstrated that miR-34c-5p overexpression significantly increased
the enrichment of CRABP2 in RIP-Ago2 compared with RIP-IgG in both
C643 and OCUT-2C cells (Fig. 4H). In
addition, other potential targets of miR-34c-5p were analyzed using
the DAVID database. The related signaling pathways of miR-34c-5p
target genes are presented in Fig.
S4. miR-34c-5p was identified to be mainly involved in the
following signaling pathways: ‘Thyroid hormone signaling pathway’
(44), ‘pathway in cancer’, ‘MAPK
signaling pathway’ (45) and ‘ErbB
signaling pathway’ (46), amongst
others. These pathways are known to be closely associated with
tumor processes. These results indicated that CRABP2 may be a
target gene of miR-34c-5p.
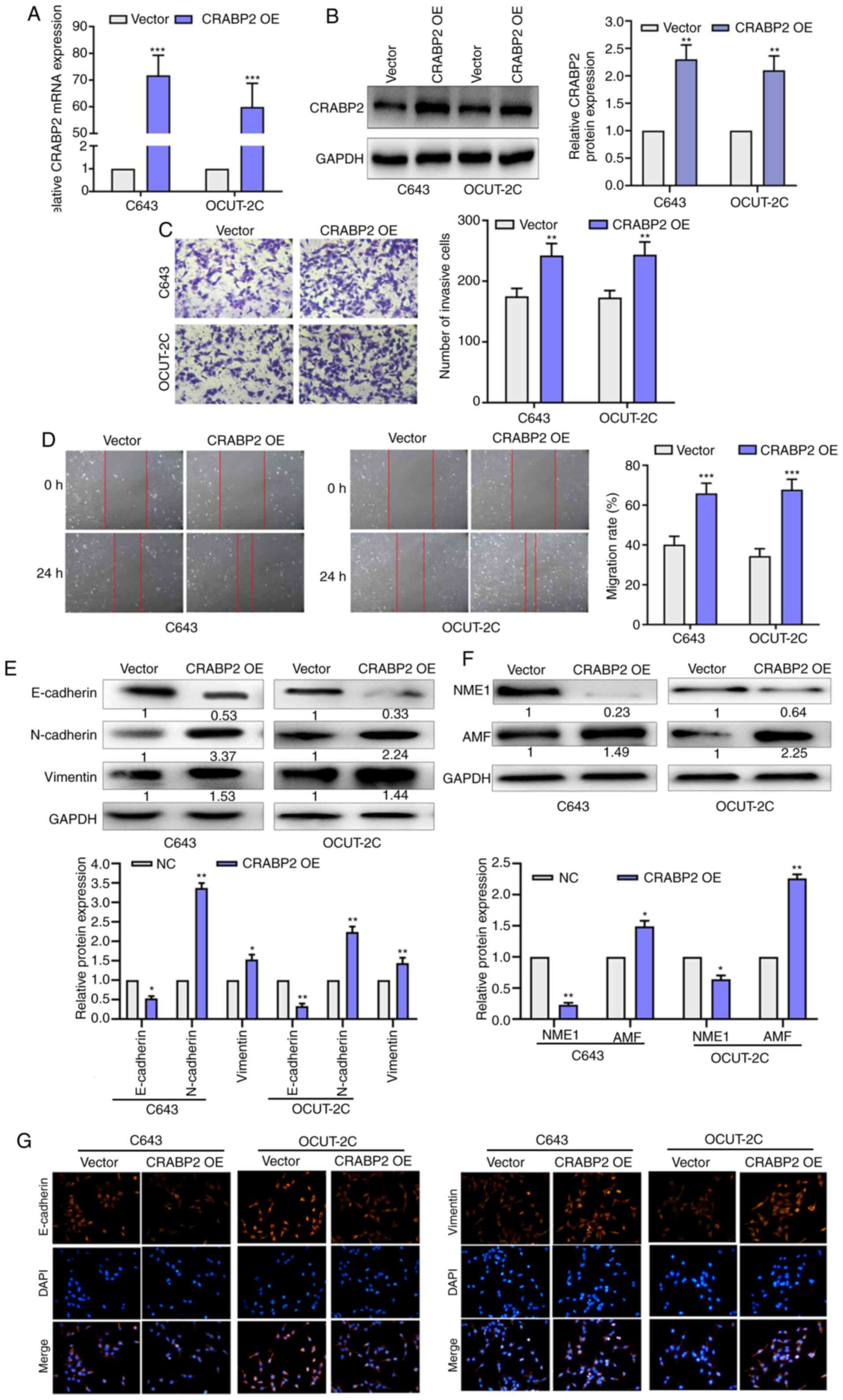
The transfection efficiency of the CRABP2
overexpression plasmid was determined using RT-qPCR and western
blotting. The mRNA and protein expression levels of CRABP2 were
increased after CRABP2 overexpression plasmid transfection in C643
and OCUT-2C cells (Fig. 5A and B). As
revealed in Fig. 5C, the number of
invasive cells was increased in the CRABP2 overexpression group
compared with the vector group. Wound healing assays were used to
determine the role of CRABP2 in EMT progression. As presented in
Fig. 5D, the migratory rate was
increased in the cells overexpressing CRABP2 compared with the
vector group. The protein expression levels of E-cadherin were
downregulated in the CRABP2 overexpression group, while the
expression levels of N-cadherin and vimentin were upregulated in
the CRABP2 overexpression group. The expression levels of NME1 were
also downregulated following the overexpression of CRABP2, while
AMF expression levels were upregulated in C643 and OCUT-2C cells
(Fig. 5E and F). Immunofluorescence
staining was used to further confirm these findings (Fig. 5G). The expression of E-cadherin was
decreased while the expression of vimentin was increased with
CRABP2 overexpression. These results indicated that the
overexpression of LINC01816 induced the EMT process and led to an
increase in cell migration. CRABP2 may be a target gene of
miR-34c-5p and may be associated with EMT, migration and invasion
in THCA.
Discussion
THCA has been classified as one of the fastest
growing solid malignant tumors in China in the past 20 years
(47). Although THCA has a low
mortality rate, patients with advanced disease cannot be cured and
thus, depend on drugs to maintain their quality of life (48). In the present study, analysis using
TCGA database revealed that LINC01816 expression levels were
significantly upregulated in THCA. In addition, LINC01816 knockdown
could significantly suppress the migratory and invasive abilities
of THCA. To clarify the mechanism of action of LINC01816 in the
regulation of THCA progression, bioinformatics analysis was first
used, and the results were subsequently verified using in
vitro cell line experiments. The results demonstrated that
miR-34c-5p shared a complementary binding site with LINC01816 and
its expression levels were regulated by LINC01816 in vitro.
LncRNAs are located in the cytoplasm, where they can sponge miRNAs
to regulate gene expression (49).
Previous studies have reported that miR-34c-5p was involved in
regulating tumor progression. For example, miR-34c-5p inhibited the
stemness of ovarian cancer and the development of drug resistance
by downregulating the amphiregulin/EGFR/ERK signaling pathway
(50). Wang et al determined
that miR-34c-5p targeted flotillin 2 to inhibit the proliferation,
migration and invasion of osteosarcoma cells, downregulate the
protein expression levels of cyclin D1, matrix metalloproteinase
(MMP)-2 and MMP-9, and upregulate the protein expression levels of
p21 (51). The signaling pathways
associated with miR-34c-5p target genes are known to be closely
associated with tumor processes (23–25). The
results of the present study also revealed that E-cadherin
expression levels were significantly downregulated following the
inhibition of miR-34c-5p. In addition, the transfection of THCA
cells with miR-34c-5p mimics reduced the migratory ability. These
data indicated that miR-34c-5p may serve as a tumor suppressor in
the regulation of tumor progression. In addition, the
overexpression of LINC01816 could reverse the inhibitory effect of
miR-34c-5p overexpression on THCA progression. Thus, these findings
indicated that LINC01816 may be involved in the regulation of THCA
as an oncogenic factor. Notably, the knockdown of miR-34c-5p
enhanced the effects of LINC01816.
CRABP2 is a cellular lipid-binding protein that is
involved in the transport of retinoic acid (52). The aberrant regulation of CRABP2 is a
common phenomenon in numerous different cancer types. CRABP2
expression levels were reported to be downregulated in prostate
cancer, esophageal squamous cell carcinoma, head and neck squamous
cell carcinoma and astrocytic glioma, amongst others. In contrast,
the upregulation of CRABP2 expression levels has been reported in
retinoblastoma, ovarian cancer, breast cancer, lung cancer and
uterine leiomyoma (53–56). However, to the best of our knowledge,
little is known about the role of CRABP2 in THCA. The results of
the present study revealed that the genetic knockdown of CRABP2
suppressed cell migration, invasion and EMT in C643 and OCUT-2C
cells. In addition, the expression levels of NME1 were revealed to
be downregulated following the overexpression of CRABP2. Further
experiments verified that CRABP2 expression levels were positively
associated with LINC01816 expression levels, and its expression was
regulated by LINC01816 and miR-34c-5p.
Collectively, the findings of the present study
provided significant evidence to suggest that LINC01816 expression
levels may be significantly upregulated in cancer tissues compared
with matched adjacent tissues. In addition, according to TCGA
database analysis, LINC01816 expression levels were determined to
be modulated by the miR-34c-5p/CRABP2 ceRNA regulatory network.
However, there are also limitations to the present study. For
example, the results were not verified using an in vivo
animal model or an independent cohort of patients. Therefore, these
issues will be addressed in future studies.
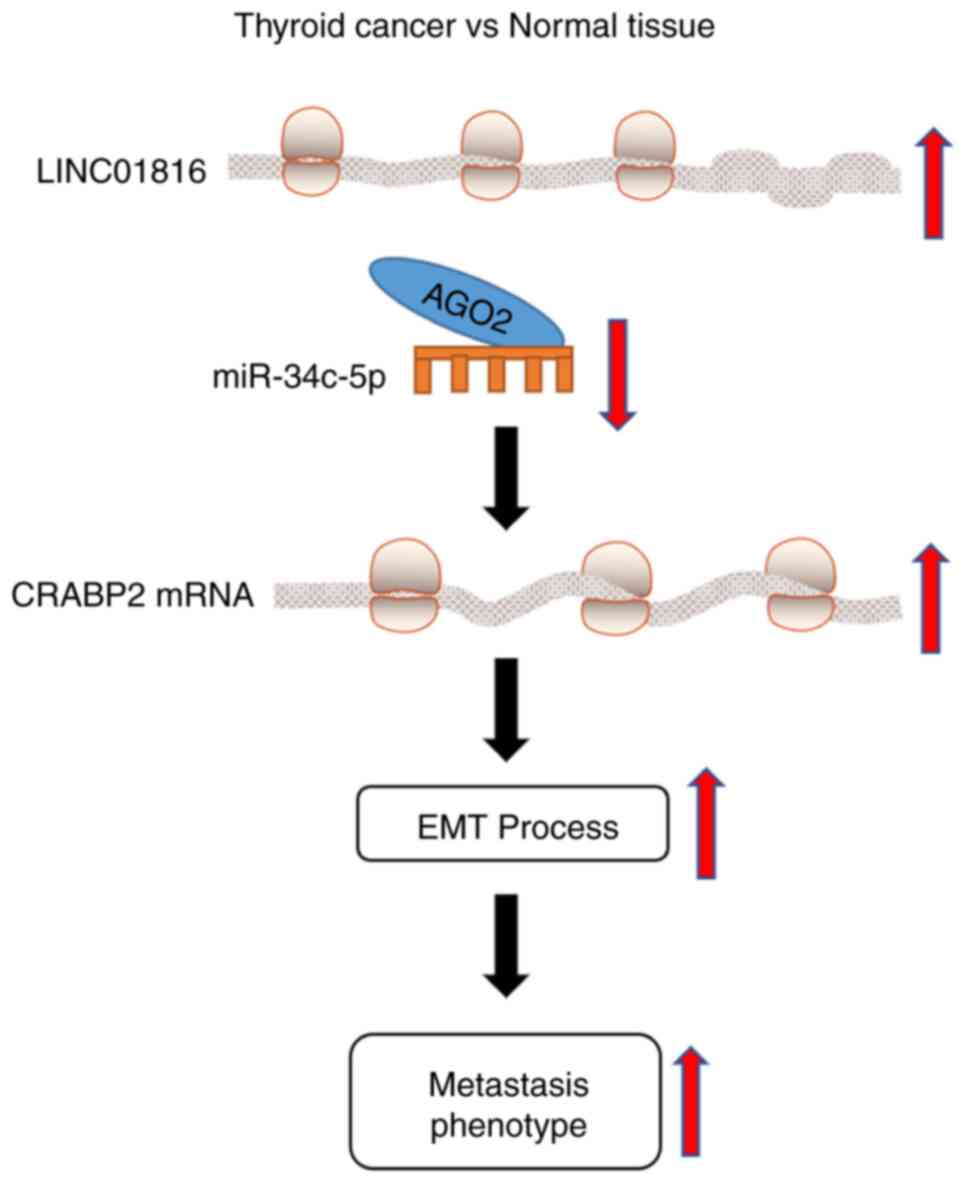
In conclusion, the findings of the present study
proposed a novel molecular mechanism for LINC01816 in regulating
the process of EMT in THCA. The results revealed that LINC01816
expression levels were upregulated in THCA, which may subsequently
serve a role in the migratory and EMT processes of THCA cells by
targeting the miR-34c-5p/CRABP2 axis (Fig. 6). Therefore, the
LINC01816/miR-34c-5p/CRABP2 axis may be of clinical significance in
THCA.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Author's contributions
HZ contributed to the design of the study, wrote the
manuscript and analyzed the data. XZ and YL revised the manuscript
and contributed to the design of the study. SL, WW and LZ acquired,
analyzed and interpreted the data. JZ made substantial
contributions to the conception and design of the present study and
revised the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
This present study was approved by the Ethics
Committee of the Affiliated Hospital of Zunyi Medical University
(Zunyi, China) and written informed consent was obtained from all
patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ciavardelli D, Bellomo M, Consalvo A,
Crescimanno C and Vella V: Metabolic alterations of thyroid cancer
as potential therapeutic targets. Biomed Res Int. 2017:25450312017.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Vaccarella S, Franceschi S, Bray F, Wild
CP, Plummer M and Dal Maso L: Worldwide thyroid-cancer epidemic?
The increasing impact of overdiagnosis. N Engl J Med. 375:614–617.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sanabria A, Kowalski LP, Shah JP, Nixon
IJ, Angelos P, Williams MD, Rinaldo A and Ferlito A: Growing
incidence of thyroid carcinoma in recent years: Factors underlying
overdiagnosis. Head Neck. 40:855–866. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Haramati S, Chapnik E, Sztainberg Y, Eilam
R, Zwang R, Gershoni N, McGlinn E, Heiser PW, Wills AM, Wirguin I,
et al: miRNA malfunction causes spinal motor neuron disease. Proc
Natl Acad Sci USA. 107:13111–13116. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Thomson DW and Dinger ME: Endogenous
microRNA sponges: Evidence and controversy. Nat Rev Genet.
17:272–283. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lacouture ME, Ciccolini K, Kloos RT and
Agulnik M: Overview and management of dermatologic events
associated with targeted therapies for medullary thyroid cancer.
Thyroid. 24:1329–1340. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kim H, Kim YN, Kim HI, Park SY, Choe JH,
Kim JH, Kim JS, Chung JH, Kim TH and Kim SW: Preoperative serum
thyroglobulin predicts initial distant metastasis in patients with
differentiated thyroid cancer. Sci Rep. 7:169552017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang M, Qiu S and Qin J: Baicalein induced
apoptosis and autophagy of undifferentiated thyroid cancer cells by
the ERK/PI3K/Akt pathway. Am J Transl Res. 11:3341–3352.
2019.PubMed/NCBI
|
|
9
|
Jung CW, Han KH, Seol H, et al: Expression
of cancer stem cell markers and epithelial-mesenchymal
transition-related factors in anaplastic thyroid carcinoma.
International journal of clinical and experimental pathology.
8:560–568. 2015.PubMed/NCBI
|
|
10
|
Sandberg K, Samson WK and Ji H: Decoding
noncoding RNA: da Vinci redux? Circ Res. 113:240–241. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Diamantopoulos MA, Tsiakanikas P and
Scorilas A: Non-coding RNAs: The riddle of the transcriptome and
their perspectives in cancer. Ann Transl Med. 6:2412018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dai M, Li S and Qin X: Colorectal
neoplasia differentially expressed: A long noncoding RNA with an
imperative role in cancer. Onco Targets Ther. 11:3755–3763. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ogoyama M, Ohkuchi A, Takahashi H, Zhao D,
Matsubara S and Takizawa T: LncRNA H19-derived miR-675-5p
accelerates the invasion of extravillous trophoblast cells by
inhibiting GATA2 and subsequently activating matrix
metalloproteinases. Int J Mol Sci. 22:12372021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Iorio MV and Croce CM: MicroRNA
dysregulation in cancer: Diagnostics, monitoring and therapeutics.
A comprehensive review. EMBO Mol Med. 4:143–159. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Muhammad N, Bhattacharya S, Steele R and
Ray RB: Anti-miR-203 suppresses ER-positive breast cancer growth
and stemness by targeting SOCS3. Oncotarget. 7:58595–58605. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yanaihara N and Harris CC: MicroRNA
involvement in human cancers. Clin Chem. 59:1811–1812. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rosignolo F, Memeo L, Monzani F, Colarossi
C, Pecce V, Verrienti A, Durante C, Grani G, Lamartina L, Forte S,
et al: MicroRNA-based molecular classification of papillary thyroid
carcinoma. Int J Oncol. 50:1767–1777. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dong S, Xue S, Sun Y, Han Z, Sun L, Xu J
and Liu J: MicroRNA-363-3p downregulation in papillary thyroid
cancer inhibits tumor progression by targeting NOB1. J Investig
Med. 69:66–74. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu HM, Dong AB, Hua H, Sun YH, Wang JR,
Yu QQ, Zhang JH and Sun WH: MicroRNA-219 inhibits cell viability
and metastasis in papillary thyroid carcinoma by targeting EYA2.
Eur Rev Med Pharmacol Sci. 24:9556–9564. 2020.PubMed/NCBI
|
|
20
|
Li N, Mao D, Cao Y, Li H, Ren F and Li K:
Downregulation of SIRT6 by miR-34c-5p is associated with poor
prognosis and promotes colon cancer proliferation through
inhibiting apoptosis via the JAK2/STAT3 signaling pathway. Int J
Oncol. 52:1515–1527. 2018.PubMed/NCBI
|
|
21
|
Liang M, Yu S, Tang S, Bai L, Cheng J, Gu
Y, Li S, Zheng X, Duan L, Wang L, et al: A panel of plasma exosomal
mirnas as potential biomarkers for differential diagnosis of
thyroid nodules. Front Genet. 11:4492020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shen Y, Xu J, Pan X, Zhang Y, Weng Y, Zhou
D and He S: LncRNA KCNQ1OT1 sponges miR-34c-5p to promote
osteosarcoma growth via ALDOA enhanced aerobic glycolysis. Cell
Death Dis. 11:2782020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yanokura M, Banno K and Aoki D:
MicroRNA-34b expression enhances chemosensitivity of endometrial
cancer cells to paclitaxel. Int J Oncol. 57:1145–1156.
2020.PubMed/NCBI
|
|
24
|
Lin BZ, Wan SY, Lin MY, Chang CH, Chen TW,
Yang MH and Lee YJ: Involvement of differentially expressed
microRNAs in the PEGylated liposome encapsulated
188rhenium-mediated suppression of orthotopic
hypopharyngeal tumor. Molecules. 25:36092020. View Article : Google Scholar
|
|
25
|
Wang H, Bian S and Yang CS: Green tea
polyphenol EGCG suppresses lung cancer cell growth through
upregulating miR-210 expression caused by stabilizing HIF-1α.
Carcinogenesis. 32:1881–1889. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kazimierczyk M, Kasprowicz MK, Kasprzyk ME
and Wrzesinski J: Human long noncoding RNA interactome: detection,
characterization and function. Int J Mol Sci. 21:10272020.
View Article : Google Scholar
|
|
27
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fan H, Ge Y, Ma X, Li Z, Shi L, Lin L,
Xiao J, Chen W, Ni P, Yang L and Xu Z: Long non-coding RNA
CCDC144NL-AS1 sponges miR-143-3p and regulates MAP3K7 by acting as
a competing endogenous RNA in gastric cancer. Cell Death Dis.
11:5212020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yu H, Zhong X, Gao P, Shi J, Wu Z, Guo Z,
Wang Z and Song Y: The potential effect of metformin on cancer: An
umbrella review. Front Endocrinol (Lausanne). 10:6172019.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bayat Mokhtari R, Homayouni TS, Baluch N,
Morgatskaya E, Kumar S, Das B and Yeger H: Combination therapy in
combating cancer. Oncotarget. 8:38022–38043. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Muhammad N, Steele R, Isbell TS, Philips N
and Ray RB: Bitter melon extract inhibits breast cancer growth in
preclinical model by inducing autophagic cell death. Oncotarget.
8:66226–66236. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mohammad N, Malvi P, Meena AS, Singh SV,
Chaube B, Vannuruswamy G, Kulkarni MJ and Bhat MK: Cholesterol
depletion by methyl-β-cyclodextrin augments tamoxifen induced cell
death by enhancing its uptake in melanoma. Mol Cancer. 13:2042014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mohammad N, Singh SV, Malvi P, Chaube B,
Athavale D, Vanuopadath M, Nair SS, Nair B and Bhat MK: Strategy to
enhance efficacy of doxorubicin in solid tumor cells by
methyl-β-cyclodextrin: Involvement of p53 and Fas receptor ligand
complex. Sci Rep. 5:118532015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gupta A, Williams BR, Hanash SM and Rawwas
J: Cellular retinoic acid-binding protein II is a direct
transcriptional target of MycN in neuroblastoma. Cancer Res.
66:8100–8108. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gupta A, Kessler P, Rawwas J and Williams
BR: Regulation of CRABP-II expression by MycN in Wilms tumor. Exp
Cell Res. 314:3663–3668. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Favorskaya I, Kainov Y, Chemeris G,
Komelkov A, Zborovskaya I and Tchevkina E: Expression and clinical
significance of CRABP1 and CRABP2 in non-small cell lung cancer.
Tumour Biol:. 35:10295–10300. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Donato LJ and Noy N: Suppression of
mammary carcinoma growth by retinoic acid: Proapoptotic genes are
targets for retinoic acid receptor and cellular retinoic
acid-binding protein II signaling. Cancer Res. 65:8193–8199. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Budhu AS and Noy N: Direct channeling of
retinoic acid between cellular retinoic acid-binding protein II and
retinoic acid receptor sensitizes mammary carcinoma cells to
retinoic acid-induced growth arrest. Mol Cell Biol. 22:2632–2641.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42((Database Issue)): D92–D97. 2013.PubMed/NCBI
|
|
42
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:e050052015. View Article : Google Scholar
|
|
43
|
Jiao X, Sherman BT, Huang da W, Stephens
R, Baseler MW, Lane HC and Lempicki RA: DAVID-WS: A stateful web
service to facilitate gene/protein list analysis. Bioinformatics.
28:1805–1806. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Jonklaas J, Nsouli-Maktabi H and Soldin
SJ: Endogenous thyrotropin and triiodothyronine concentrations in
individuals with thyroid cancer. Thyroid. 18:943–952. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Jiang W, Cai F, Xu H, Lu Y, Chen J, Liu J,
Cao N, Zhang X, Chen X, Huang Q, et al: Extracellular signal
regulated kinase 5 promotes cell migration, invasion and lung
metastasis in a FAK-dependent manner. Protein Cell. 11:825–845.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Mahtouk K, Jourdan M, De Vos J, Hertogh C,
Fiol G, Jourdan E, Rossi JF and Klein B: An inhibitor of the EGF
receptor family blocks myeloma cell growth factor activity of
HB-EGF and potentiates dexamethasone or anti-IL-6 antibody-induced
apoptosis. Blood. 103:1829–1837. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Brito JP, Morris JC and Montori VM:
Thyroid cancer: Zealous imaging has increased detection and
treatment of low risk tumours. BMJ. 347:f47062013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lirov R, Worden FP and Cohen MS: The
treatment of advanced thyroid cancer in the age of novel targeted
therapies. Drugs. 77:733–745. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Rashid F, Shah A and Shan G: Long
non-coding RNAs in the cytoplasm. Genomics Proteomics
Bioinformatics. 14:73–80. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Tung SL, Huang WC, Hsu FC, Yang ZP, Jang
TH, Chang JW, Chuang CM, Lai CR and Wang LH: miRNA-34c-5p inhibits
amphiregulin-induced ovarian cancer stemness and drug resistance
via downregulation of the AREG-EGFR-ERK pathway. Oncogenesis.
6:e3262017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wang Y, Wang X, Tang J, Su X and Miao Y:
The study of mechanism of miR-34c-5p targeting FLOT2 to regulate
proliferation, migration and invasion of osteosarcoma cells. Artif
Cells Nanomed Biotechnol. 47:3559–3568. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Yuan J, Tang Z, Yang S and Li K: CRABP2
promotes myoblast differentiation and is modulated by the
transcription factors MyoD and Sp1 in C2C12 cells. PLoS One.
8:e554792013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Okuducu AF, Janzen V, Ko Y, Hahne JC, Lu
H, Ma ZL, Albers P, Sahin A, Wellmann A, Scheinert P and Wernert N:
Cellular retinoic acid-binding protein 2 is down-regulated in
prostate cancer. Int J Oncol. 27:1273–1282. 2005.PubMed/NCBI
|
|
54
|
Wu JI, Lin YP, Tseng CW, Chen HJ and Wang
LH: Crabp2 promotes metastasis of lung cancer cells via HuR and
integrin β1/FAK/ERK signaling. Sci Rep. 9:8452019. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Li YT, Tian XT, Wu ML, Zheng X, Kong QY,
Cheng XX, Zhu GW, Liu J and Li H: Resveratrol suppresses the growth
and enhances retinoic acid sensitivity of anaplastic thyroid cancer
cells. Int J Mol Sci. 19:10302018. View Article : Google Scholar
|
|
56
|
Liu RZ, Li S, Garcia E, Glubrecht DD, Poon
HY, Easaw JC and Godbout R: Association between cytoplasmic CRABP2,
altered retinoic acid signaling, and poor prognosis in
glioblastoma. Glia. 64:963–976. 2016.PubMed/NCBI
|