Introduction
Breast cancer is one of the most commonly diagnosed
cancer types and the second leading cause of cancer-related
mortality in women world-wide (1,2). Although
early detection and systemic therapy significantly improve the
outcome of patients with breast cancer, the survival rate of
patients with metastatic breast cancer remains relatively low, with
a 5-year survival of <25% (3,4).
Therefore, it is important to understand the underlying molecular
mechanism of breast carcinogenesis and progression, as well as to
identify novel biomarkers and therapeutic target molecules for
early diagnosis and individualized therapy.
MicroRNAs (miRNAs) are endogenous small non-coding
RNAs, ~22 nucleotides in length, that are involved in gene
silencing through translational repression or mRNA degradation by
binding to the 3′-untranslated regions (3′-UTRs) of target genes
(5). Previous studies have reported
that miRNAs participate in multiple biological functions and are
involved in various physiological processes, including cell
proliferation, differentiation, metabolism, senescence and
apoptosis (6,7). Aberrant expression of miRNAs has been
shown to be involved in the development and progression of multiple
human cancer types (8). In humans,
miRNA (miR)-454 is located on chromosome 17q22, and it has two main
mature forms: miR-454-5p and miR-454-3p. miR-454 has been
identified as either an oncogene or a tumor suppressor in various
types of cancer, including pancreatic cancer (9,10), colon
cancer (11), melanoma (12), chondrosarcoma (13), glioma (14–16),
hepatocellular carcinoma (17,18), lung
cancer (19–21), laryngeal cancer (22), ovarian cancer (23), prostate cancer (24), gastric cancer (25–28),
cervical cancer (29), bladder cancer
(30), oral squamous cell carcinoma
(31), osteosarcoma (32), renal carcinoma (33) and breast cancer (34–36).
Although several studies have reported that miR-454-3p could
function as an oncogene in breast cancer, the role of miR-454-5p
remains unknown.
Epithelial-mesenchymal transition (EMT) is a
biological process through which epithelial cells lose cell
polarity and cell-cell adhesion; cells also acquire a fibroblastic
morphotype with invasive and migratory properties during tissue
fibrosis, embryonic development and cancer progression (37,38). EMT
contributes to cancer progression, metastasis and therapeutic
resistance in all types of human cancer, which may correlate with
unfavorable outcomes of patients with cancer (39). Therefore, targeting components of EMT
signaling is considered as a promising strategy in cancer therapy.
Previous studies have shown that miRNAs participate in EMT
regulation during breast cancer progression (40,41).
The present study aimed to investigate the role of
miR-454-5p in breast cancer progression both in vitro and
in vivo, and explored the potential mechanism of miR-454-5p
on the regulation of EMT in breast cancer.
Materials and methods
Cell lines and culturing
The human breast epithelial cell line MCF10A and
breast cancer cell lines MCF7, T47D, BT549, MDA-MB-231 and SKBR3
were obtained from The Cell Bank of Type Culture Collection of The
Chinese Academy of Sciences. MCF10A cells were cultured in DMEM/F12
(Hyclone; Cytiva) supplemented with 5% horse serum (Thermo Fisher
Scientific, Inc.), 10 µg/ml insulin (Sigma-Aldrich; Merck KGaA), 20
ng/ml epidermal growth factor (R&D Systems, Inc.), 0.1 µg/ml
cholera toxin (Sigma-Aldrich; Merck KGaA) and 0.5 µg/ml
hydrocortisone (Sigma-Aldrich; Merck KGaA). BT549, MDA-MB-231 and
SKBR3 cells were maintained in RPMI-1640 (Hyclone; Cytiva)
supplemented with 10% FBS (Thermo Fisher Scientific, Inc.). MCF7
and T47D cells were cultured in DMEM (Hyclone; Cytiva) with 10%
FBS. All cells were supplemented with 100 mg/ml streptomycin
(Hyclone; Cytiva) and 100 IU/ml penicillin (Hyclone; Cytiva) and
cultured in an atmosphere containing 5% CO2 at 37°C.
Transfections
miR-454-5p mimics (5′-GAAGUAAAGGGGCAAGAUAGGGC-3′)
and negative control (5′-UUCUCCGAACGUGUCACGUUU-3′) oligonucleotides
were purchased from Guangzhou RiboBio Co., Ltd. Transient
transfection was conducted using Lipofectamine® 3000
(Thermo Fisher Scientific, Inc.) according to the manufacturer's
instructions. Cell were seeded in a 6-well plate at a density of
2×105 cells/well and then transfected with 200 pmol
oligonucleotides at 37°C for 24 h. Experiments were performed 48 h
after transfection.
The miR-454-5p antisense strand
(5′-GCCCUAUCUUGCCCCUUUACUUC-3′) or inhibitor control
(5′-UCUACUCUUUCUAGGAGGUUGUGA-3′; anti-Control) was cloned into the
pEZX-AM04 lentiviral vector (GeneCopoeia, Inc.) and short hairpin
RNA (sh)FoxJ2 (5′-GCAAGCCACGATACAGCTATT-3′) or shControl
(5′-CCTAAGGTTAAGTCGCCCTCG-3′) was cloned into the pLKO.1 lentiviral
vector (GeneCopoeia, Inc.). 293T cells were transfected with 10 µg
lentiviral vectors and 10 µg packaging vectors when the density of
cells reached 80–90%. After transfection for 48 h at 37°C,
supernatants containing virus particles were harvested and
purified. The lentiviral particles were used to infect targeted
cells at 60% confluency with a multiplicity of infection of 30 and
stable cells were selected with 800 mg/ml puromycin. The FoxJ2
knockdown efficiencies were confirmed by reverse
transcription-quantitative PCR (RT-qPCR) and western blotting
(Fig. S1).
RT-qPCR
Total RNA was extracted from breast cancer cells
using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. The
isolated RNA was reverse transcribed into cDNA using a PrimeScript™
RT Master Mix kit (Takara Biotechnology Co., Ltd.) according to the
manufacturer's protocol. qPCR analysis was performed using SYBR
Green Mix (Takara Biotechnology Co., Ltd.) on a CFX96 Touch™
Real-time PCR System (Bio-Rad Laboratories, Inc.); the
thermocycling conditions were as follows: Initial denaturation for
8 min at 95°C; followed by 40 cycles of two-step PCR at 95°C for 20
sec and 60°C for 1 min. The expression of mRNA or miRNA was
normalized to GAPDH or U6, respectively. Data were analyzed using
the 2−ΔΔCq method (42).
The qPCR primers are listed in Table
I.
 | Table I.Primer sequences used in quantitative
PCR. |
Table I.
Primer sequences used in quantitative
PCR.
| Name | Primer sequence
(5′→3′) |
|---|
| FoxJ2 | F:
ATGGCTTCTGACCTAGAGAGTAG |
|
| R:
CTGCCTCGTCTTTGCTCAGG |
| E-cadherin | F:
CGAGAGCTACACGTTCACGG |
|
| R:
GGGTGTCGAGGGAAAAATAGG |
| GAPDH | F:
ATGACCCCTTCATTGACCTCA |
|
| R:
GAGATGATGACCCTTTTGGCT |
| E-cadherin R1 | F:
CTGTACAGAGCATTTATGGCTCAA |
|
| R:
TGTCTCCCTATGCTGTTGTGG |
| E-cadherin R2 | F:
GAGTCTCTTGAACCCGGCA |
|
| R:
CCACTGAGCTAGCAGCCTAAT |
| E-cadherin R3 | F:
CACTCCAGCTTGGGTGAAAGA |
|
| R:
GGCTCACTAAGACCTGGGAT |
Western blotting
Total protein was isolated from cells using RIPA
lysis buffer containing PMSF (both from Cell Signaling Technology,
Inc.). Protein concentrations were determined using a BCA Protein
Assay kit (Thermo Fisher Scientific, Inc.). All samples (40 µg per
lane) were separated by SDS-PAGE on 10% gels and then transferred
to polyvinylidene fluoride membranes (EMD Millipore). After
blocking with 5% skimmed milk for 1 h at room temperature, the
membrane was incubated with primary antibodies overnight at 4°C,
followed by washing with PBS and incubation with horseradish
peroxidase-conjugated goat anti-rabbit (cat. no. 7074) or horse
anti-mouse (cat. no. 7076) (both at a dilution of 1:2,000; Cell
Signaling Technology, Inc.) secondary antibodies for 1 h at room
temperature. The bands were visualized with ECL reagent (EMD
Millipore). Antibodies against N-cadherin (cat. no. sc-393933),
E-cadherin (cat. no. sc-71008), Vimentin (cat. no. sc-80975), FoxJ2
(cat. no. sc-514265) (all from Santa Cruz Biotechnology, Inc.) and
GAPDH (cat. no. 5174; Cell Signaling Technology, Inc.) were used at
a dilution of 1:1,000.
Luciferase reporter assay
For FoxJ2 3′-UTR activity analysis, the 3′-UTR of
FoxJ2 mRNA and the corresponding sequence with mutations (FoxJ2-1:
5′-GCTGGCAAAGAAATGGATAGGGA-3′ to
5′-GCTGGCAAAGAAATGGATAAA A-3′; FoxJ2-2:
5′-GAAGTAAAGGGGCAAGATAGGGC-3′ to
5′-GAAGTAAAGGGGCAAGATAAAAC-3′) were cloned into a
psiCHEK2 luciferase reporter plasmid (Promega Corporation).
MDA-MB-231 cells were seeded into a 24-well plate at
1×104 cells/well and co-transfected with psiCHEK2-FoxJ2
(500 ng) and miR-454-5p mimics (50 pmol) or negative control using
Lipofectamine 3000 for 48 h at 37°C. For E-cadherin promoter
activity analysis, the E-cadherin promoter was cloned into a
pGL3-basic luciferase reporter plasmid (Promega Corporation) and
was transfected into MDA-MB-231 cells stably expressing
anti-miR-454-5p or shFoxJ2, as well as into the control cells using
Lipofectamine 3000 for 48 h at 37°C. Luciferase activity was
analyzed after transfection for 48 h and the cell lysates were
measured for luciferase activity using Dual-Luciferase Reporter
Assay System (Promega Corporation) according to the manufacturer's
instructions. The firefly luciferase activity was normalized to
Renilla luciferase activity.
Chromatin immunoprecipitation (ChIP)
assay
Stably transfected MDA-MB-231 cells were
cross-linked with 1% formaldehyde for 15 min at room temperature.
After quenching with 125 mM glycine, the cells were collected,
washed and sonicated in RIPA buffer. The cross-linked lysate was
sonicated (power, 20 W; duration, 30 sec/cycle for 40 cycles;
temperature, 5–6°C) to obtain 500–1,500 bp fragments, which were
immunoprecipitated with IgG or anti-FoxJ2 antibody (cat. no.
sc-514265; Santa Cruz Biotechnology, Inc.) at 4°C for 3 h. This was
followed by incubation with 50 µl protein A/G beads overnight at
4°C. qPCR was used to identify and quantify the precipitated DNA.
Primers for ChIP assays are presented in Table I.
Colony formation assay
Breast cancer cells were seeded into six-well plates
at 500 cells/well. Transient transfection with miR-454-5p mimics
was conducted using Lipofectamine® 3000, aforementioned,
or MDA-MB-231 cells stably expressing anti-miR-454-5p or shFoxJ2
were used, as well as their control cells. Cells were cultured for
20 days at 37°C, and colonies were washed with PBS, fixed with 4%
paraformaldehyde for 15 min at room temperature, and stained with
hematoxylin. The colonies with >50 cells were counted under a
light microscope at ×100 magnification.
Cell morphology
MDA-MB-231 cells stably expressing anti-miR-454-5p
or shFoxJ2, as well as their control cells, were seeded in 6-well
plates at a density of 5×104 cells/well and cultured at
37°C for 24 h. Cell morphology was examined under a light
microscope at ×100 magnification.
Cell viability assay
The Cell Counting Kit-8 (CCK-8; Dojindo Molecular
Technologies, Inc.) was used to determine cell viability, according
to the manufacturer's protocols. Cells were seeded into a 96-well
plate at 5×103 cells/well. After transfection for 24 h,
CCK-8 reagent (10 µl) was added to the medium and incubated at 37°C
for 2 h. The absorbance of each sample was measured using a
microplate reader (Thermo Fisher Scientific, Inc.) at 450 nm.
Immunofluorescence
Cells were plated at a density of 2×104
on glass coverslips, washed with ice-cold PBS and fixed in 4%
formaldehyde solution for 15 min at room temperature. The
coverslips were blocked with 2% BSA (Cell Signaling Technology,
Inc.) in PBS for 30 min at room temperature and incubated with a
primary antibody against E-cadherin (1:500; cat. no. sc-71008;
Santa Cruz Biotechnology, Inc.) overnight at 4°C. Subsequently,
cells were incubated with Alexa Fluor® 488-conjugated
anti-mouse secondary antibody (1:300; cat. no. 4408; Cell Signaling
Technology, Inc.) for 1 h and then stained with DAPI for 10 min,
both at room temperature. The coverslips were washed with PBS and
observed under a fluorescence microscope at ×400 magnification.
Cell cycle analysis
Cells were seeded into a 6-well plate at a density
of 5×104 cells/well. After culture at 37°C for 24 h,
cells were collected and fixed with 70% ethanol for 24 h at −20°C,
followed by incubation with 10 µg/ml PI and 50 µg/ml RNase (BD
Biosciences) on ice for 15 min. Cells were then assessed by flow
cytometry using a BD FACSCalibur flow cytometer (BD
Biosciences).
Transwell assays
Cell invasion and migration were assessed using BD
Transwell chambers pre-coated with or without Matrigel,
respectively (BD Biosciences). Cells were seeded (2×104
cells/well) into the upper Transwell chamber containing RPMI-1640
medium without serum; RPMI-1640 medium with 10% FBS was added to
the bottom chamber. After incubation for 16 h at 37°C, the invaded
or migrated cells on the lower surface were fixed in 4%
formaldehyde solution for 15 min at room temperature, and
subsequently stained with crystal violet for 15 min at room
temperature. Images of the invaded or migrated cells were captured
under a light microscope at ×100 magnification.
Bioinformatics analysis
TargetScan Human release 7.2. (http://www.targetscan.org) was used for prediction of
miR-454-5p targets. The top three genes containing two potential
miR-454-5p target sites were selected for further study.
LASAGNA-Search 2.0 (https://www.bitnos.com/info/lasagna-search) was used
to predict FoxJ2 binding sites on the E-cadherin promoter
region.
Validation of gene expression and
outcome by TCGA database
The expression levels of miR-454-5p were analyzed in
breast cancer tissues (n=749) and normal breast tissues (n=76)
obtained from The Cancer Genome Atlas (TCGA) database by using
UALCAN (http://ualcan.path.uab.edu/index.html). The
relationship between miR-454-5p and FoxJ2 was determined by ENCORI
(http://starbase.sysu.edu.cn). The
prognostic value of miR-454-5p expression was examined by using the
online database, KM-Plotter (www.kmplot.com/mirpower). Patients were divided into
two groups by ‘auto select best cutoff’ feature and assessed using
a Kaplan-Meier survival plot.
Xenografts
Female BALB/c-nude mice (n=12; age, 5 weeks; weight,
22 g) were purchased from Vital River Lab Animal Technology
Company. The mice were maintained in a specific pathogen-free
environment with a 10/14-h light/dark cycle in 40–60% humidity at
27°C and had ad libitum access to food and water. The
experimental procedures were approved by the Animal Experimentation
Ethics Committee of Qingdao Central Hospital (Qingdao, China). A
total of 1×106 stably transfected MDA-MB-231 cells were
injected subcutaneously into the right mammary fat pad. Tumor
volume was calculated using the following equation: (Length ×
width2)/2. The mice were sacrificed on day 35 after
tumor implantation, and the tumors were excised and weighed. The
tumor burden did not exceed the recommended dimensions (tumor
diametermax ≤16.5 mm; volumemax ≤1,200
mm3). The animals were anesthetized (60 mg/kg ketamine
and 5.0 mg/kg xylazine) and then sacrificed by cervical
dislocation.
Statistical analysis
Statistical analyses were performed using SPSS 20.0
(IBM Corp.). Data are presented as the mean ± SD from at least
three independent experiments. The unpaired Student's t-test was
used to compare differences between two groups, and one-way ANOVA
followed by Tukey's multiple comparison post hoc test was used when
comparing >2 groups. P<0.05 was considered to indicate a
statistically significant difference.
Results
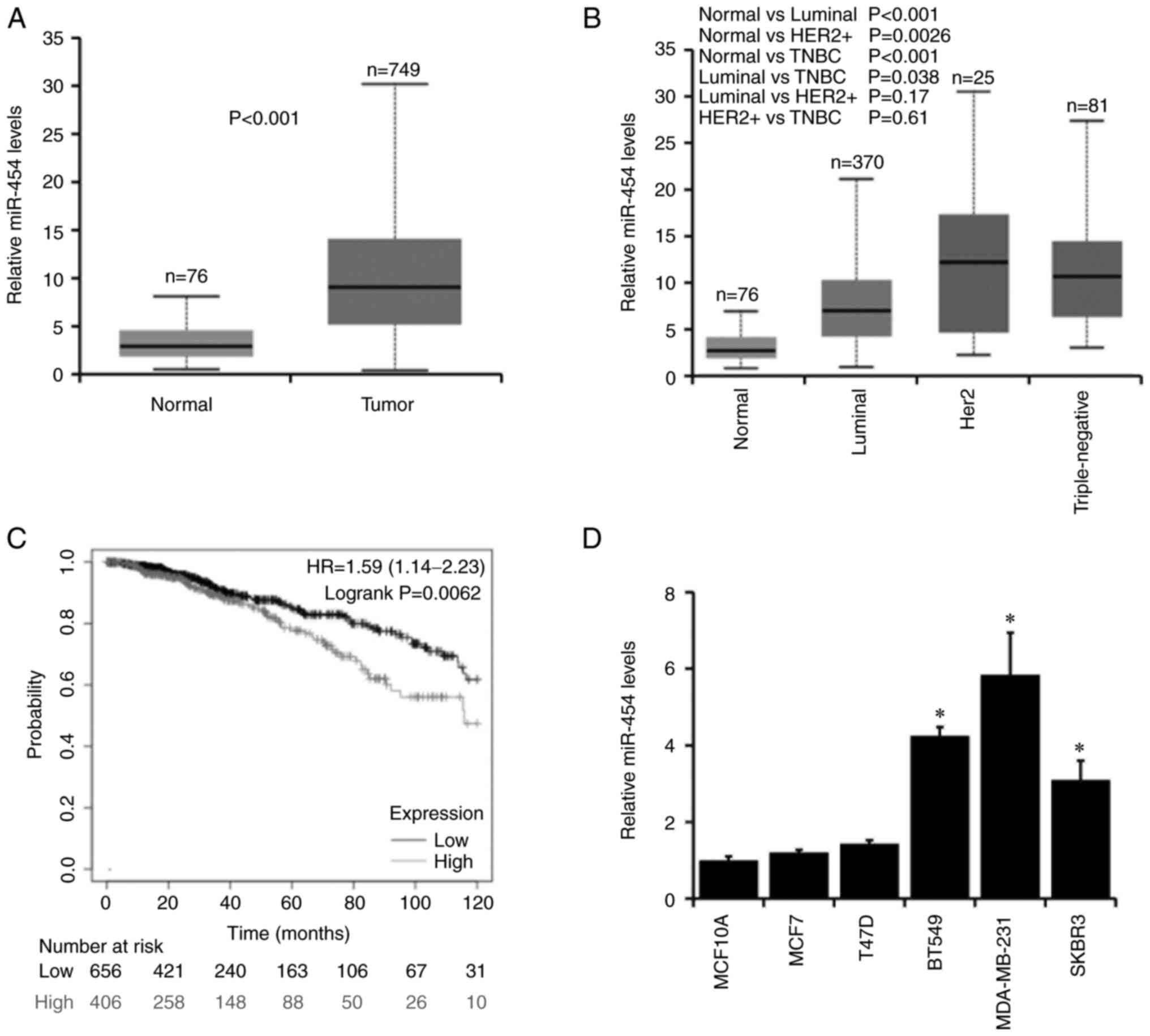
miR-454-5p is upregulated in breast
cancer and is associated with poor prognosis
To determine the role of miR-454-5p in breast
cancer, the expression levels of miR-454-5p were analyzed in breast
cancer tissues (n=749) and normal breast tissues (n=76) obtained
from The Cancer Genome Atlas (TCGA) database by using UALCAN.
Increased miR-454-5p expression levels were observed in breast
cancer compared with normal tissues (Fig.
1A), and miR-454-5p expression was significantly upregulated in
triple-negative and Her2+ breast cancer types (Fig. 1B). Furthermore, high miR-454-5p
expression was associated with a poor prognosis in patients with
breast cancer as determined using the KM-Plotter database (Fig. 1C). Subsequently, the expression levels
of miR-454-5p were examined in five breast cancer cell lines (MCF7,
T47D, BT549, MDA-MB-231 and SKBR3) and a normal breast epithelial
cell line MCF10A. The expression levels of miR-454-5p were
significantly higher in BT549, MDA-MB-231 and SKBR3 breast cancer
cell lines compared with MCF10A cells (Fig. 1D). Taken together, these results
indicated that miR-454-5p may serve an important role in breast
cancer progression.
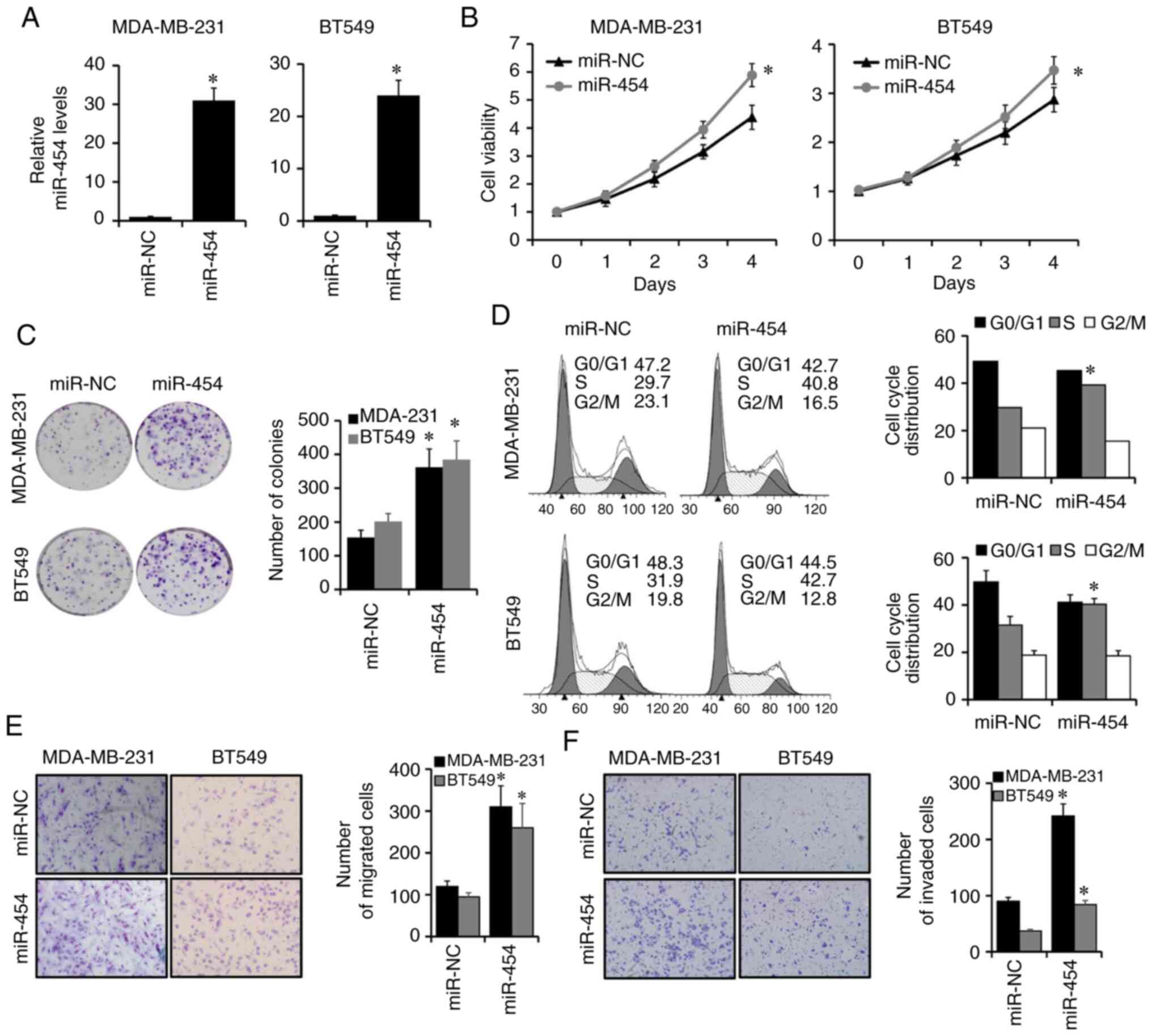
Overexpression of miR-454-5p promotes
breast cancer cell proliferation, migration and invasion
As miR-454-5p was upregulated in BT549, MDA-MB-231
and SKBR3 compared to MCF10A cells, aforementioned, which suggested
that miR-454-5p may serve an important role in these cell lines,
MDA-MB-231 and BT549 were choose to further study. To evaluate the
potential role of miR-454-5p in breast cancer progression,
miR-454-5p mimics or negative controls were transfected into
MDA-MB-231 and BT549 cells. The expression of miR-454-5p was
effectively elevated in MDA-MB-231 and BT549 cells after
transfection with miR-454-5p mimics compared with
miR-NC-transfected cells, as determined by RT-qPCR (Fig. 2A). CCK-8 and colony formation assays
were conducted to evaluate the effects of miR-454-5p on breast
cancer proliferation. It was found that the overexpression of
miR-454-5p significantly enhanced cell viability and the number of
colonies in MDA-MB-231 and BT549 cells compared with the miR-NC
group (Fig. 2B and C, respectively).
Furthermore, the proportion of cells at the S phase was
significantly increased in miR-454-5p-overexpressing MDA-MB-231 and
BT549 cells compared with that of control cells (Fig. 2D). Transwell analyses were performed
to assess the effects of miR-454-5p on breast cancer cell migration
and invasion (Fig. 2E and F,
respectively). The number of migrated and invaded cells was
significantly increased in miR-454-5p-overexpressing MDA-MB-231 and
BT549 cells compared with those in control cells. These results
suggested that miR-454-5p may function as an oncogene in breast
cancer progression.
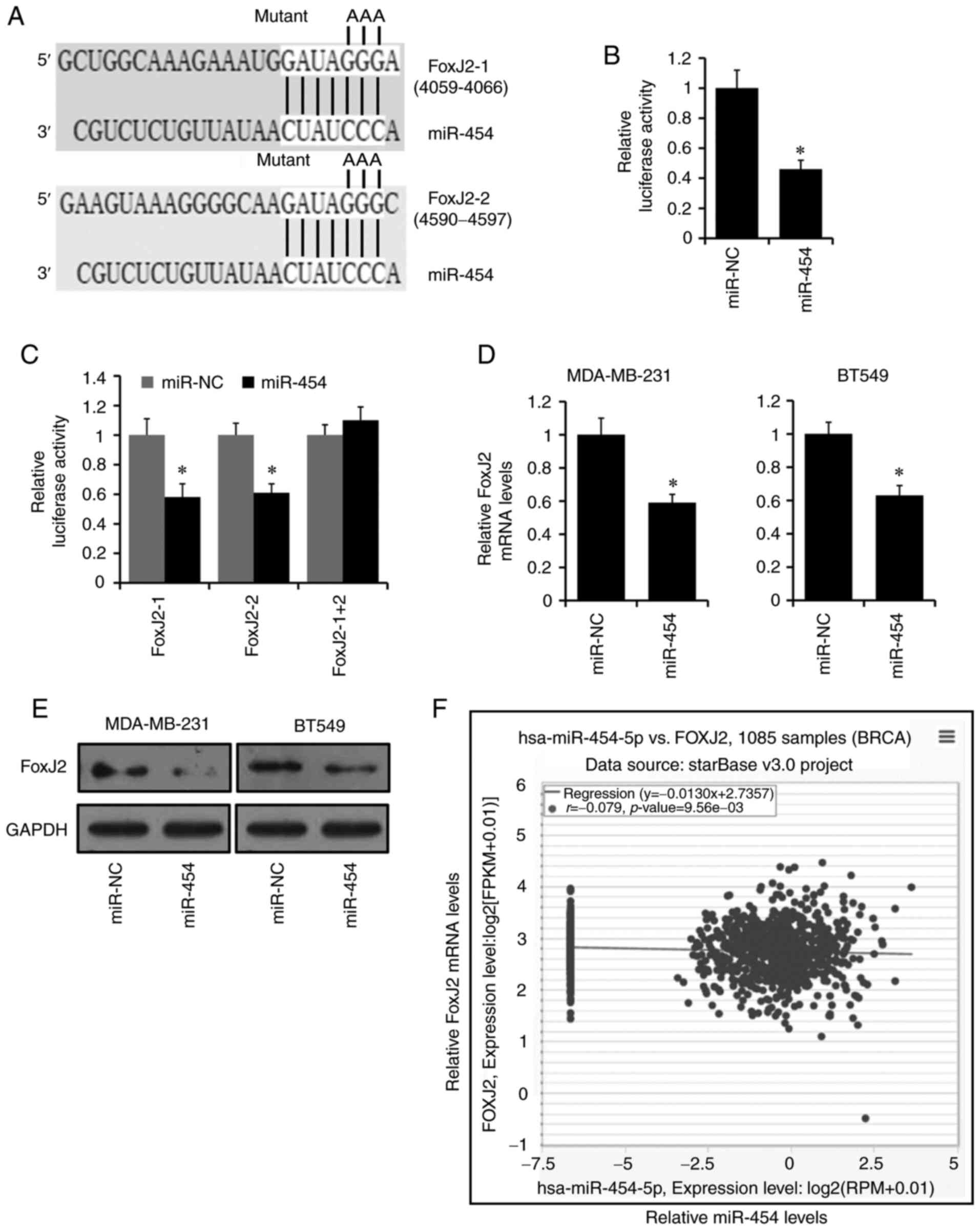
FoxJ2 is a target of miR-454-5p
TargetScan was used to predict target genes of
miR-454-5p, and the top three genes containing two potential target
sites in their 3′-UTRs were ATXN7L3B, RIPPLY3 and FoxJ2 (Fig. 3A). Of the three genes, only FoxJ2 has
been reported to be involved in cancer progression (43–46);
therefore, it was selected for further study. The luciferase
reporter assay was used to determine whether miR-454-5p could
directly bind to the 3′-UTR of FoxJ2. The two putative miR-454-5p
target sites in the 3′-UTR of FoxJ2 were cloned into the psiCHEK2
reporter plasmid and subsequently transfected into MDA-MB-231 cells
along with miR-454-5p mimics or negative control. The luciferase
activity of the FoxJ2 3′-UTR reporter construct was significantly
decreased in the miR-454-5p-transfected MDA-MB-231 cells compared
with luciferase activity in the miR-NC cells (Fig. 3B). This effect was abolished in the
mutated FoxJ2 3′-UTR group in which both target sites for
miR-454-5p were inactivated (FoxJ-1 + 2) by site-directed
mutagenesis (Fig. 3C). The expression
levels of FoxJ2 were notably decreased in MDA-MB-231 and BT549
cells transfected with miR-454-5p mimics compared with the
expression levels in control cells, as demonstrated by RT-qPCR
(Fig. 3D) and western blotting
(Fig. 3E).
To further investigate the relationship between
miR-454-5p and FoxJ2, the expression levels of miR-454-5p and FoxJ2
from TCGA database was analyzed by ENCORI. As presented in Fig. 3F, the expression of miR-454-5p
exhibited a negative co-expression trend with FoxJ2. Thus, these
results support the bioinformatics prediction that FoxJ2 was a
direct target of miR-454-5p.
Silencing of miR-454-5p inhibits
breast cancer proliferation, migration and invasion by upregulating
FoxJ2 expression
To further confirm the regulation of the
miR-454-5p/FoxJ2 axis in breast cancer progression, stably
transfected MDA-MB-231 cell lines overexpressing the
anti-miR-454-5p inhibitor with or without shFoxJ2 co-transfection,
as well as anti-Control cells, were generated. The expression of
miR-454-5p was significantly downregulated in miR-454-5p-silenced
MDA-MB-231 cells compared with that of control cells, as determined
via RT-qPCR (Fig. 4A). The expression
of FoxJ2 was upregulated in miR-454-5p-silenced MDA-MB-231 cells,
whereas it was notably downregulated in miR-454-5p and
FoxJ2-silenced MDA-MB-231 cells compared with that of control
cells, as determined using western blotting (Fig. 4B). It was also found that knockdown of
FoxJ2 significantly reversed the inhibitory effects of miR-454-5p
silencing on the proliferation, migration and invasion of
MDA-MB-231 cells as determined via CCK-8 (Fig. 4C), colony formation (Fig. 4D), cell cycle (Fig. 4E) and Transwell assays (Fig. 4F), respectively.
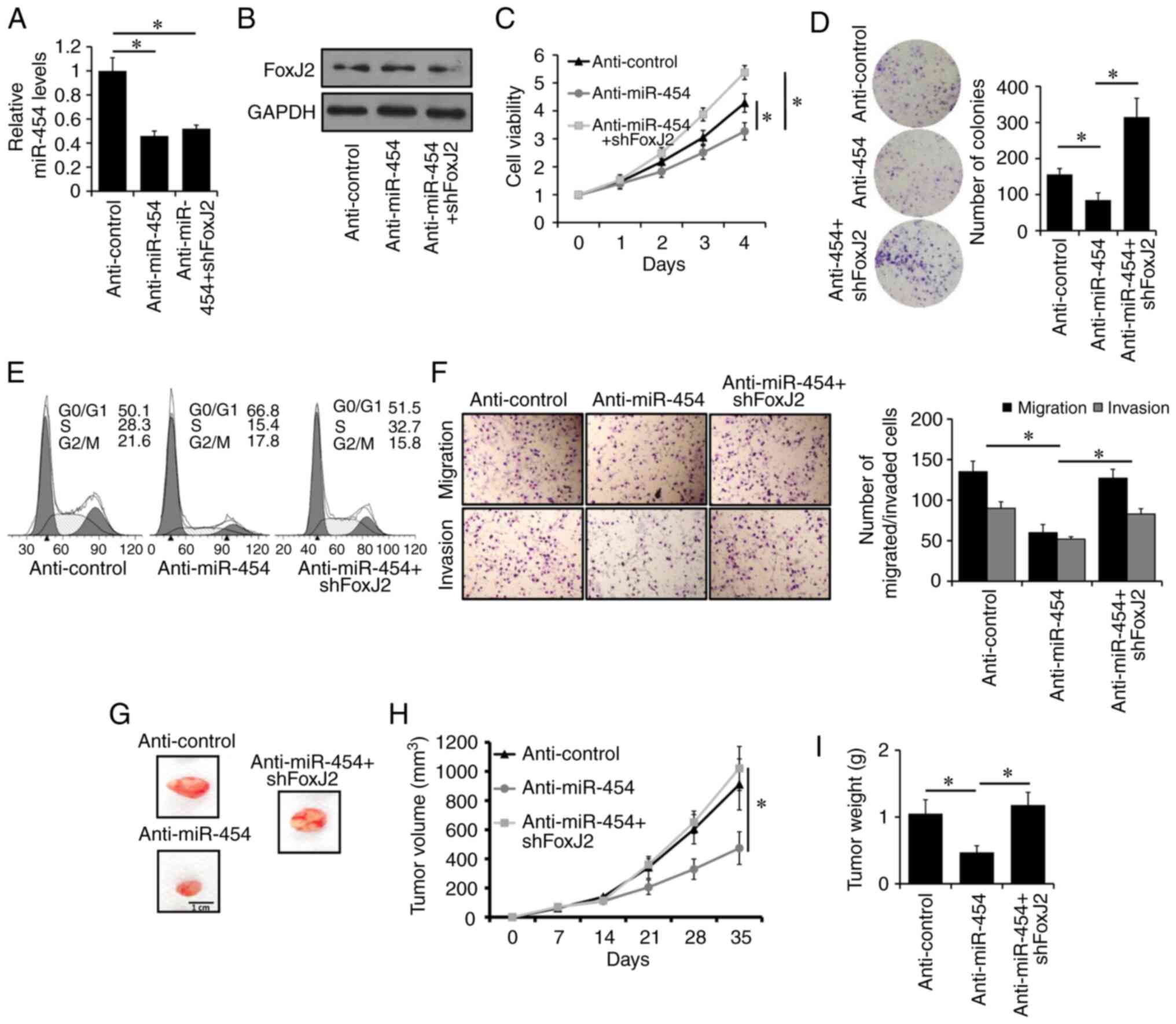 | Figure 4.Silencing of miR-454-5p inhibits
breast cancer progression through the upregulation of FoxJ2.
Expression levels of (A) miR-454-5p and (B) FoxJ2 protein in
MDA-MB-231 cells stably expressing anti-miR-454-5p with or without
shFoxJ2 transfection, or anti-Control cells, were examined by
reverse transcription-quantitative PCR or western blotting,
respectively. (C) Cell viability and (D) colony formation in
MDA-MB-231 cells stably expressing anti-miR-454-5p with or without
shFoxJ2 transfection, or anti-Control cells, was assessed using a
Cell Counting Kit-8 assay or colony formation assay, respectively.
(E) Cell cycle analysis of MDA-MB-231 cells stably expressing
anti-miR-454-5p with or without shFoxJ2 transfection, or
anti-Control cells. (F) Migration and invasion of MDA-MB-231 cells
stably expressing anti-miR-454-5p with or without shFoxJ2
transfection, or anti-Control cells, was assessed by Transwell or
Matrigel analysis, respectively. (G) Representative images of the
tumors formed by MDA-MB-231 cells stably expressing anti-miR-454-5p
with or without shFoxJ2 transfection, or anti-Control cells, at the
time of harvest. Tumor (H) volume and (I) weight of xenografted
mice injected with MDA-MB-231 cells stably expressing
anti-miR-454-5p with or without shFoxJ2 transfection, or
anti-Control cells, at the indicated times. *P<0.05. FoxJ2,
forkhead box J2; miR, microRNA; sh, short hairpin RNA. |
To investigate the biological effect of the
miR-454-5p/FoxJ2 axis on breast cancer progression in vivo,
MDA-MB-231-control, MDA-MB-231-anti-454 and MDA-MB-231-anti-454 +
shFoxJ2 cells were injected into the mammary fat pads of female
BALB/c-nude mice. As presented in Fig.
4G-I, tumor volumes and weights were significantly decreased in
mice injected with MDA-MB-231-anti-miR-454 cells compared with
those in mice injected with MDA-MB-231-control cells. Moreover, it
was identified that knockdown of FoxJ2 could abolish the inhibitory
effects of miR-454-5p silencing on tumor growth. Collectively,
these results indicated that silencing of miR-454-5p may inhibit
breast cancer progression through the upregulation of FoxJ2
expression.
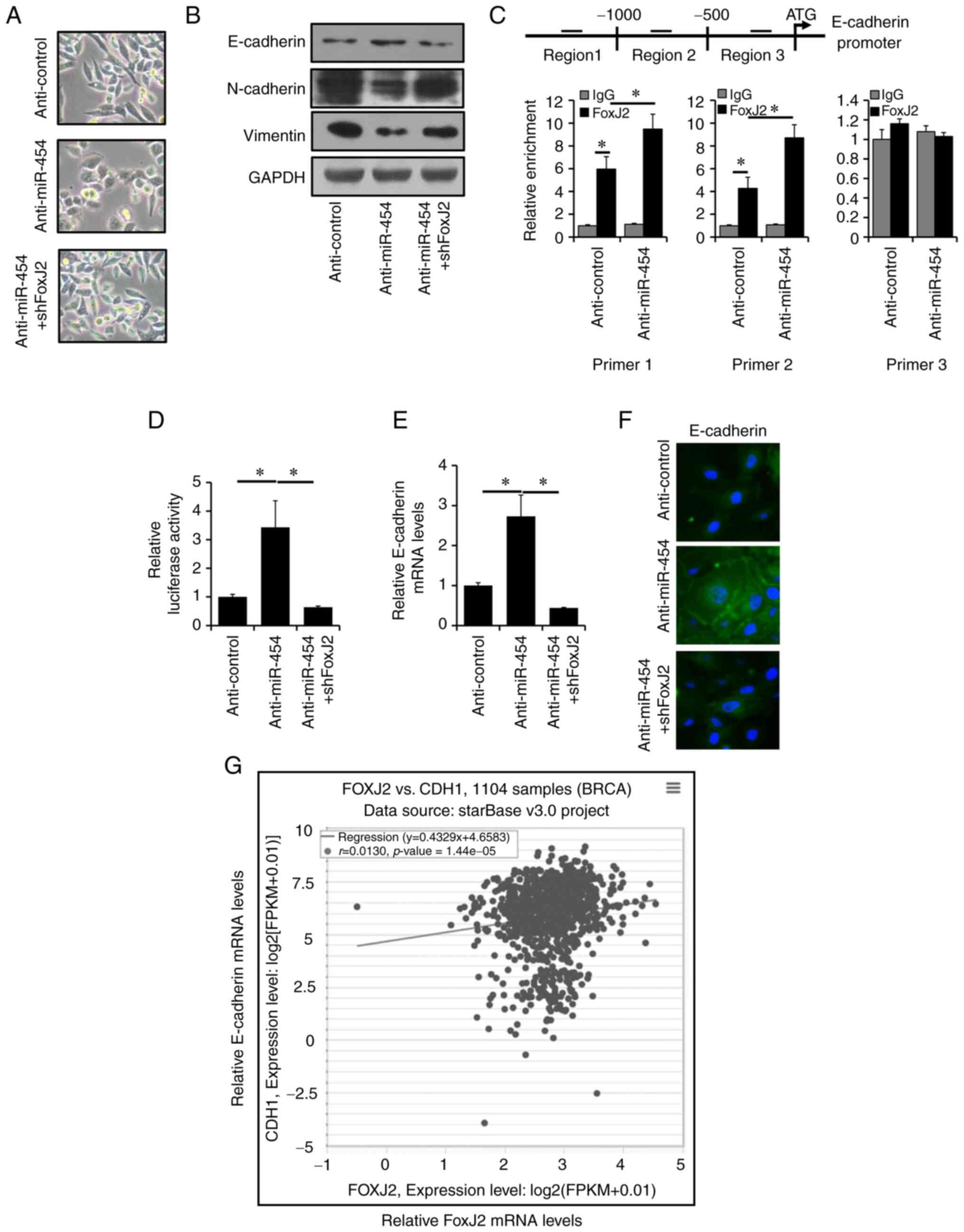
Silencing of miR-454-5p reverses EMT
through the transcriptional upregulation of E-cadherin
Accumulating evidence has suggested that EMT could
promote progression in breast cancer (41). Therefore, it was investigated whether
miR-454-5p regulated EMT to affect breast cancer progression. It
was identified that miR-454-5p-silenced MDA-MB-231 cells had a
cobblestone-like morphology, whereas anti-miR-454-5p + shFoxJ2
MDA-MB-231 cells and anti-Control cells maintained their
spindle-like fibroblast morphology (Fig.
5A). In addition, it was demonstrated that the expression of
epithelial marker E-cadherin was increased in miR-454-5p-silenced
MDA-MB-231 cells, whereas the expression levels of mesenchymal
markers Vimentin and N-cadherin were decreased in
miR-454-5p-silenced MDA-MB-231 cells compared with the expression
levels in control cells, as determined by western blotting
(Fig. 5B). Moreover, knockdown of
FoxJ2 could reverse this effect caused by miR-454-5p silencing
(Fig. 5B). In total, three putative
FoxJ2 binding sites on the E-cadherin promoter region were
identified using LASAGNA-Search (47). ChIP analysis results demonstrated that
FoxJ2 bound to region 1 and region 2 from the E-cadherin promoter
region (Fig. 5C). Luciferase assay
results indicated that knockdown of FoxJ2 significantly abolished
miR-454-5p depletion-induced E-cadherin promoter activity (Fig. 5D). E-cadherin mRNA expression levels
were increased in miR-454-5p-silenced MDA-MB-231 cells, whereas
this effect was abolished by FoxJ2 knockdown as shown by RT-qPCR
(Fig. 5E) and immunofluorescence
(Fig. 5F). E-cadherin mRNA was
determined to be positive co-expression trend with FoxJ2 mRNA by
ENCORI (Fig. 5G). Together, these
results indicated that miR-454-5p may regulated EMT through the
transcriptional upregulation of E-cadherin by FoxJ2.
Discussion
The results from present study demonstrated that
miR-454-5p was upregulated in breast cancer compared with normal
breast epithelial tissue and cell lines. Furthermore, the results
suggested that miR-454-5p may promote breast cancer progression
both in vitro and in vivo. FoxJ2 was identified as a
direct target of miR-454-5p, and FoxJ2 may transactivate the
expression of E-cadherin. Moreover, silencing of miR-454-5p
reversed EMT through the transactivation of E-cadherin by FoxJ2.
The present results suggested that miR-454-5p may be a potential
target for breast cancer therapy.
Accumulating evidence has revealed that the
dysregulation of miRNAs is involved in carcinogenesis and cancer
progression in all types of human cancer, including breast cancer
(41,48). Abnormal expression of miR-454 has been
observed in a variety of human cancer types, suggesting that
miR-454 may serve an important role in cancer development and
progression (9–36). miR-454-3p has been demonstrated to
function as an oncogene, and high expression of miR-454-3p may be
associated with unfavorable outcome in triple-negative breast
cancer (36,49). Moreover, miR-454-3p has been shown to
promote breast cancer metastasis through the suppression of
Wnt/β-catenin signaling antagonists (35). Consistent with these previous reports,
the present study demonstrated that miR-454-5p was upregulated in
breast cancer and that miR-454-5p may promote breast cancer
progression both in vitro and in vivo, suggesting
that miR-454-5p may function as an oncogene in breast cancer.
Fox protein family members share a highly conserved
common forkhead DNA-binding domain and are widespread from yeast to
humans (50). These transcription
factors are crucial players in multiple cellular processes,
including differentiation, proliferation, metabolism, migration and
apoptosis, and are often dysregulated in cancer development and
progression (50). FoxJ2 belongs to
the human Fox gene family and serves an important role in
embryogenesis and carcinogenesis (51). FoxJ2 has also been reported to
function as a tumor suppressor in multiple types of cancer,
including colorectal cancer (52),
hepatocellular carcinoma (53),
non-small lung cancer (43),
extrahepatic cholangiocarcinoma (44), glioma (45) and breast cancer (46). In line with the aforementioned
results, the present findings demonstrated that FoxJ2 was a target
of miR-454-5p and that miR-454-5p may promote breast cancer
progression through the downregulation of FoxJ2 expression.
EMT, a main driver of tumor metastasis, is defined
as a process by which epithelial cells lose their cell polarity and
cell-cell adhesions, resulting in changes to cell morphology and
enhanced cell migratory and invasive abilities (37). Vimentin, N-cadherin and E-cadherin are
generally considered as EMT markers. During the EMT process, the
expression of E-cadherin (an epithelial marker) is decreased,
whereas the expression levels of Vimentin and N-cadherin
(mesenchymal markers) are increased. Several studies have reported
that FoxJ2 can inhibit cancer migration, invasion and the EMT
phenotype (43–44,52,53).
Consistent with these findings, the present study identified a
decrease in Vimentin and N-cadherin expression levels, and an
increase in E-cadherin expression in miR-454-5p-silenced cells,
whereas knockdown of FoxJ2 reversed these effects, suggesting that
miR-454-5p may induce an EMT phenotype in breast cancer.
Furthermore, it was revealed that FoxJ2 could bind to the
E-cadherin promoter region and transactivated E-cadherin
expression. Thus, the present results suggested that miR-454-5p
promoted breast cancer progression through the induction of an EMT
phenotype by downregulating the FoxJ2/E-cadherin axis.
In conclusion, the present study demonstrated that
miR-454-5p was upregulated in breast cancer. It was suggested that
miR-454-5p may promote breast cancer progression through the
induction of EMT by targeting the FoxJ2/E-cadherin axis. Therefore,
miR-454-5p may serve as a novel therapeutic target and prognostic
predictor for patients with breast cancer.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
Not applicable.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
CPW and YZY contributed to writing the manuscript
collection and analysis of data. HZ, LJX, YW, and QTW collected and
interpreted data. QM contributed to study conception and design as
well as revising and approving the final version of the manuscript.
CPW and QM confirm the authenticity of all the raw data. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The experimental procedures were approved by the
Animal Experimentation Ethics Committee of Qingdao Central Hospital
(Qingdao, China; approval no. KY202014301).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2020. CA Cancer J Clin. 70:7–30. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
DeSantis CE, Ma J, Gaudet MM, Newman LA,
Miller KD, Goding Sauer A, Jemal A and Siegel RL: Breast cancer
statistics, 2019. CA Cancer J Clin. 69:438–451. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bishop AJ, Ensor J, Moulder SL, Shaitelman
SF, Edson MA, Whitman GJ, Bishnoi S, Hoffman KE, Stauder MC, Valero
V, et al: Prognosis for patients with metastatic breast cancer who
achieve a no-evidence-of-disease status after systemic or local
therapy. Cancer. 121:4324–4332. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liang Y, Zhang H, Song X and Yang Q:
Metastatic heterogeneity of breast cancer: Molecular mechanism and
potential therapeutic targets. Semin Cancer Biol. 60:14–27. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Schmiedel JM, Klemm SL, Zheng Y, Sahay A,
Blüthgen N, Marks DS and van Oudenaarden A: Gene expression.
MicroRNA control of protein expression noise. Science. 348:128–132.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Loh HY, Norman BP, Lai KS, Rahman N,
Alitheen NBM and Osman MA: The regulatory role of MicroRNAs in
breast cancer. Int J Mol Sci. 20:49402019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fan Y, Shi C, Li T and Kuang T:
microRNA-454 shows anti-angiogenic and anti-metastatic activity in
pancreatic ductal adenocarcinoma by targeting LRP6. Am J Cancer
Res. 7:139–147. 2017.PubMed/NCBI
|
|
10
|
Fan Y, Xu LL, Shi CY, Wei W, Wang DS and
Cai DF: MicroRNA-454 regulates stromal cell derived factor-1 in the
control of the growth of pancreatic ductal adenocarcinoma. Sci Rep.
6:227932016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li W, Feng Y, Ma Z and Lu L: Expression of
miR-454-3p and its effect on proliferation, invasion and metastasis
of colon cancer. Nan Fang Yi Ke Da Xue Xue Bao. 38:1421–1426.
2018.(In Chinese). PubMed/NCBI
|
|
12
|
Sun L, Wang Q, Gao X, Shi D, Mi S and Han
Q: MicroRNA-454 functions as an oncogene by regulating PTEN in
uveal melanoma. FEBS Lett. 589:2791–2796. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bao X, Ren T, Huang Y, Sun K, Wang S, Liu
K, Zheng B and Guo W: Knockdown of long non-coding RNA HOTAIR
increases miR-454-3p by targeting Stat3 and Atg12 to inhibit
chondrosarcoma growth. Cell Death Dis. 8:e26052017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zuo J, Yu H, Xie P, Liu W, Wang K and Ni
H: miR-454-3p exerts tumor-suppressive functions by down-regulation
of NFATc2 in glioblastoma. Gene. 710:233–239. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shao N, Xue L, Wang R, Luo K, Zhi F and
Lan Q: miR-454-3p is an exosomal biomarker and functions as a tumor
suppressor in glioma. Mol Cancer Ther. 18:459–469. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fang B, Zhu J, Wang Y, Geng F and Li G:
MiR-454 inhibited cell proliferation of human glioblastoma cells by
suppressing PDK1 expression. Biomed Pharmacother. 75:148–152. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li Y, Jiao Y, Fu Z, Luo Z, Su J and Li Y:
High miR-454-3p expression predicts poor prognosis in
hepatocellular carcinoma. Cancer Manag Res. 11:2795–2802. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhou L, Qu YM, Zhao XM and Yue ZD:
Involvement of miR-454 overexpression in the poor prognosis of
hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 20:825–829.
2016.PubMed/NCBI
|
|
19
|
Jin C, Lin T and Shan L: Downregulation of
Calbindin 1 by miR-454-3p suppresses cell proliferation in nonsmall
cell lung cancer in vitro. Cancer Biother Radiopharm. 34:119–127.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu S, Ge X, Su L, Zhang A and Mou X:
MicroRNA-454 inhibits nonsmall cell lung cancer cells growth and
metastasis via targeting signal transducer and activator of
transcription-3. Mol Med Rep. 17:3979–3986. 2018.PubMed/NCBI
|
|
21
|
Zhu DY, Li XN, Qi Y, Liu DL, Yang Y, Zhao
J, Zhang CY, Wu K and Zhao S: MiR-454 promotes the progression of
human non-small cell lung cancer and directly targets PTEN. Biomed
Pharmacother. 81:79–85. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cui X, Xiao D, Cui Y and Wang X:
Exosomes-derived long non-coding RNA HOTAIR reduces laryngeal
cancer radiosensitivity by regulating microRNA-454-3p/E2F2 axis.
Onco Targets Ther. 12:10827–10839. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang DY, Li N and Cui YL: Long non-coding
RNA CCAT1 sponges miR-454 to promote chemoresistance of ovarian
cancer cells to cisplatin by regulation of surviving. Cancer Res
Treat. 52:798–814. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fu Q, Gao Y, Yang F, Mao T, Sun Z, Wang H,
Song B and Li X: Suppression of microRNA-454 impedes the
proliferation and invasion of prostate cancer cells by promoting
N-myc downstream-regulated gene 2 and inhibiting WNT/β-catenin
signaling. Biomed Pharmacother. 97:120–127. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang C, Liu J, Pan X, Peng C, Xiong B,
Feng M and Yang X: miR-454 promotes survival and induces
oxaliplatin resistance in gastric carcinoma cells by targeting
CYLD. Exp Ther Med. 19:3604–3610. 2020.PubMed/NCBI
|
|
26
|
Jiang D, Li H, Xiang H, Gao M, Yin C, Wang
H, Sun Y and Xiong M: Long chain non-coding RNA (lncRNA) HOTAIR
knockdown increases miR-454-3p to suppress gastric cancer growth by
targeting STAT3/Cyclin D1. Med Sci Monit. 25:1537–1548. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xu G, Zhu H, Zhang M and Xu J: Histone
deacetylase 3 is associated with gastric cancer cell growth via the
miR-454-mediated targeting of CHD5. Int J Mol Med. 41:155–163.
2018.PubMed/NCBI
|
|
28
|
Song Z, Li W, Wang L, Jia N and Chen B:
MicroRNA-454 inhibits tumor cell proliferation, migration and
invasion by downregulating zinc finger Eboxbinding homeobox 1 in
gastric cancer. Mol Med Rep. 16:9067–9073. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Song Y, Guo Q, Gao S and Hua K: miR-454-3p
promotes proliferation and induces apoptosis in human cervical
cancer cells by targeting TRIM3. Biochem Biophys Res Commun.
516:872–879. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang S, Zhang G, Zheng W, Xue Q, Wei D,
Zheng Y and Yuan J: MiR-454-3p and miR-374b-5p suppress migration
and invasion of bladder cancer cells through targetting ZEB2.
Biosci Rep. 38:BSR201814362018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Guo JY, Wang YK, Lv B and Jin H: miR-454
performs tumor-promoting effects in oral squamous cell carcinoma
via reducing NR3C2. J Oral Pathol Med. 49:286–293. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Niu G, Li B, Sun J and Sun L: miR-454 is
down-regulated in osteosarcomas and suppresses cell proliferation
and invasion by directly targeting c-Met. Cell Prolif. 48:348–355.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wu X, Ding N, Hu W, He J, Xu S, Pei H, Hua
J, Zhou G and Wang J: Down-regulation of BTG1 by miR-454-3p
enhances cellular radiosensitivity in renal carcinoma cells. Radiat
Oncol. 9:1792014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li X, Hou L, Yin L and Zhao S: LncRNA XIST
interacts with miR-454 to inhibit cells proliferation, epithelial
mesenchymal transition and induces apoptosis in triple-negative
breast cancer. J Biosci. 45:452020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ren L, Chen H, Song J, Chen X, Lin C,
Zhang X, Hou N, Pan J, Zhou Z, Wang L, et al: MiR-454-3p-mediated
Wnt/β-catenin signaling antagonists suppression promotes breast
cancer metastasis. Theranostics. 9:449–465. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li Q, Liu J, Meng X, Pang R and Li J:
MicroRNA-454 may function as an oncogene via targeting AKT in
triple negative breast cancer. J Biol Res (Thessalon). 24:102017.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kalluri R and Weinberg RA: The basics of
epithelial-mesenchymal transition. J Clin Invest. 119:1420–1428.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Rastaldi MP: Epithelial-mesenchymal
transition and its implications for the development of renal
tubulointerstitial fibrosis. J Nephrol. 19:407–412. 2006.PubMed/NCBI
|
|
39
|
Dongre A and Weinberg RA: New insights
into the mechanisms of epithelial-mesenchymal transition and
implications for cancer. Nat Rev Mol Cell Biol. 20:69–84. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ding L, Gu H, Xiong X, Ao H, Cao J, Lin W,
Yu M, Lin J and Cui Q: MicroRNAs involved in carcinogenesis,
prognosis, therapeutic resistance and applications in human
triple-negative breast cancer. Cells. 8:14922019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Petri BJ and Klinge CM: Regulation of
breast cancer metastasis signaling by miRNAs. Cancer Metastasis
Rev. 39:837–886. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yang Q, Cao X, Tao G, Zhou F, Zhao P, Shen
Y and Chen X: Effects of FOXJ2 on TGF-β1-induced
epithelial-mesenchymal transition through Notch signaling pathway
in non-small lung cancer. Cell Biol Int. 41:79–83. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Qiang Y, Wang F, Yan S, Zhang H, Zhu L and
Chen Z, Tu F, Wang D, Wang G, Wang W and Chen Z: Abnormal
expression of Forkhead Box J2 (FOXJ2) suppresses migration and
invasion in extrahepatic cholangiocarcinoma and is associated with
prognosis. Int J Oncol. 46:2449–2458. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Qiu X, Ji B, Yang L, Huang Q, Shi W, Ding
Z, He X, Ban N, Fan S, Zhang J and Tian Y: The role of FoxJ2 in the
migration of human glioma cells. Pathol Res Pract. 211:389–397.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wang Y, Yang S, Ni Q, He S, Zhao Y, Yuan
Q, Li C, Chen H, Zhang L, Zou L, et al: Overexpression of forkhead
box J2 can decrease the migration of breast cancer cells. J Cell
Biochem. 113:2729–2737. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Lee C and Huang CH: LASAGNA-Search 2.0:
Integrated transcription factor binding site search and
visualization in a browser. Bioinformatics. 30:1923–1925. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Crudele F, Bianchi N, Reali E, Galasso M,
Agnoletto C and Volinia S: The network of non-coding RNAs and their
molecular targets in breast cancer. Mol Cancer. 19:612020.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Cao ZG, Li JJ, Yao L, Huang YN, Liu YR, Hu
X, Song CG and Shao ZM: High expression of microRNA-454 is
associated with poor prognosis in triple-negative breast cancer.
Oncotarget. 7:64900–64909. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Jin Y, Liang Z and Lou H: The emerging
roles of fox family transcription factors in chromosome
replication, organization, and genome stability. Cells. 9:2582020.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Kehn K, Berro R, Alhaj A, Bottazzi ME, Yeh
WI, Klase Z, Van Duyne R, Fu S and Kashanchi F: Functional
consequences of cyclin D1/BRCA1 interaction in breast cancer cells.
Oncogene. 26:5060–5069. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Qiang Y, Feng L, Wang G, Liu J, Zhang J,
Xiang L, Su C, Zhang S, Xie X and Chen E: miR-20a/Foxj2 axis
mediates growth and metastasis of colorectal cancer cells as
identified by integrated analysis. Med Sci Monit. 26:e9235592020.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhang H, Tang QF, Sun MY, Zhang CY, Zhu
JY, Shen YL, Zhao B, Shao ZY, Zhang LJ and Zhang H: ARHGAP9
suppresses the migration and invasion of hepatocellular carcinoma
cells through up-regulating FOXJ2/E-cadherin. Cell Death Dis.
9:9162018. View Article : Google Scholar : PubMed/NCBI
|