Pancreatic cancer is one of the most aggressive
cancer types with a high mortality rate and low 5-year survival
rate despite all the advances in treatment (1,2). Patients
with resectable tumours generally have more prolonged survival than
patients with unresectable tumours, but only 10–20% of the patients
are diagnosed at a stage amenable to resection for lack of atypical
symptoms and early diagnostic biomarkers (3,4). Even
after curative resection, cancer recurs in the majority of patients
and about 90% of the cases develop distant metastasis. Distinct
somatic mutations in KRAS, CDKN2A, TP53 and SMAD4
genes are most prevalent in pancreatic cancer building up the
fundamental premise for the genetic alterations underlying
pancreatic tumorigenesis (5).
Autophagy is a highly regulated cellular pathway
involved in degradation and recycling of cellular elements.
Autophagy enables the cell to dissipate the redundant intracellular
components of the cell via lysosomal degradation and also recycle
the primary elements in order to combat any stress condition and
maintain cell viability (6). Salient
cargos for autophagy are damaged DNA, non-functional organelles,
protein aggregates, reactive oxygen species (ROS) and other
biochemical contents which otherwise can affect normal cellular
machinery (6). Thus, autophagy has a
crucial role to play in normal cellular physiology and thus its
aberration is highly correlated to diseases such as cancer.
Notably, it has been shown to play both oncogenic and
tumour-suppressive roles in modulating cancers of different organs
in different cellular conditions. Pancreatic ductal adenocarcinoma
(PDAC) is no exception and available evidence suggests an important
role of autophagy mainly towards the development of high-grade
pancreatic intraepithelial neoplasia (PanIN) and promotion of PDAC
(6). Therefore, considering the
importance of autophagy in pancreatic cancer, another aspect to be
focused in greater detail is the regulation of the phenomenon. This
particular area has also observed advancement and multiple studies
have coordinated between them to draw an informative picture of
cross talk between autophagy pathway genes and cancer-related genes
showing their regulation in pancreatic cancer. Key players in this
regulatory axis are the noncoding RNAs (ncRNAs), albeit the
noncoding module of autophagy regulators is almost a grey zone in
the context of pancreatic tumorigenesis. miRNAs are the major
ncRNAs playing a dual role acting as both pro- and anti-autophagic
during cancer initiation and development (7). Similarly, oncogenic or
tumour-suppressing roles of lncRNA in cancer have been identified
(8). Findings of previous studies
have also revealed an emerging role of lncRNAs in the regulation of
autophagy, further contributing to cancer development and
progression (9,10). Various types of RNA including the
transcripts of pseudogenes, lncRNAs, and circRNAs, can act as
competing endogenous RNA (ceRNA) by competing with mRNA for the
binding of miRNA and affect the gene expression at the
post-transcriptional level. Any changes in these biological
processes as a result of noncoding RNA dysfunction leads to changes
in the cellular homeostasis, which further affects pancreatic
tumorigenesis. In the present review, the mechanism of autophagy
was examined, evaluating the involvement of individual genes in
PDAC and elaborating existing information on the contribution of
miRNA, lncRNA and circular RNAs to the regulatory network
controlling the effect of autophagy on PDAC. The aim was to address
an important part of basic mechanistic aspects of pancreatic cancer
and help the scientific community to have an idea on how
autophagy-related genes and pathways could be targeted for
therapeutic purposes against pancreatic cancer.
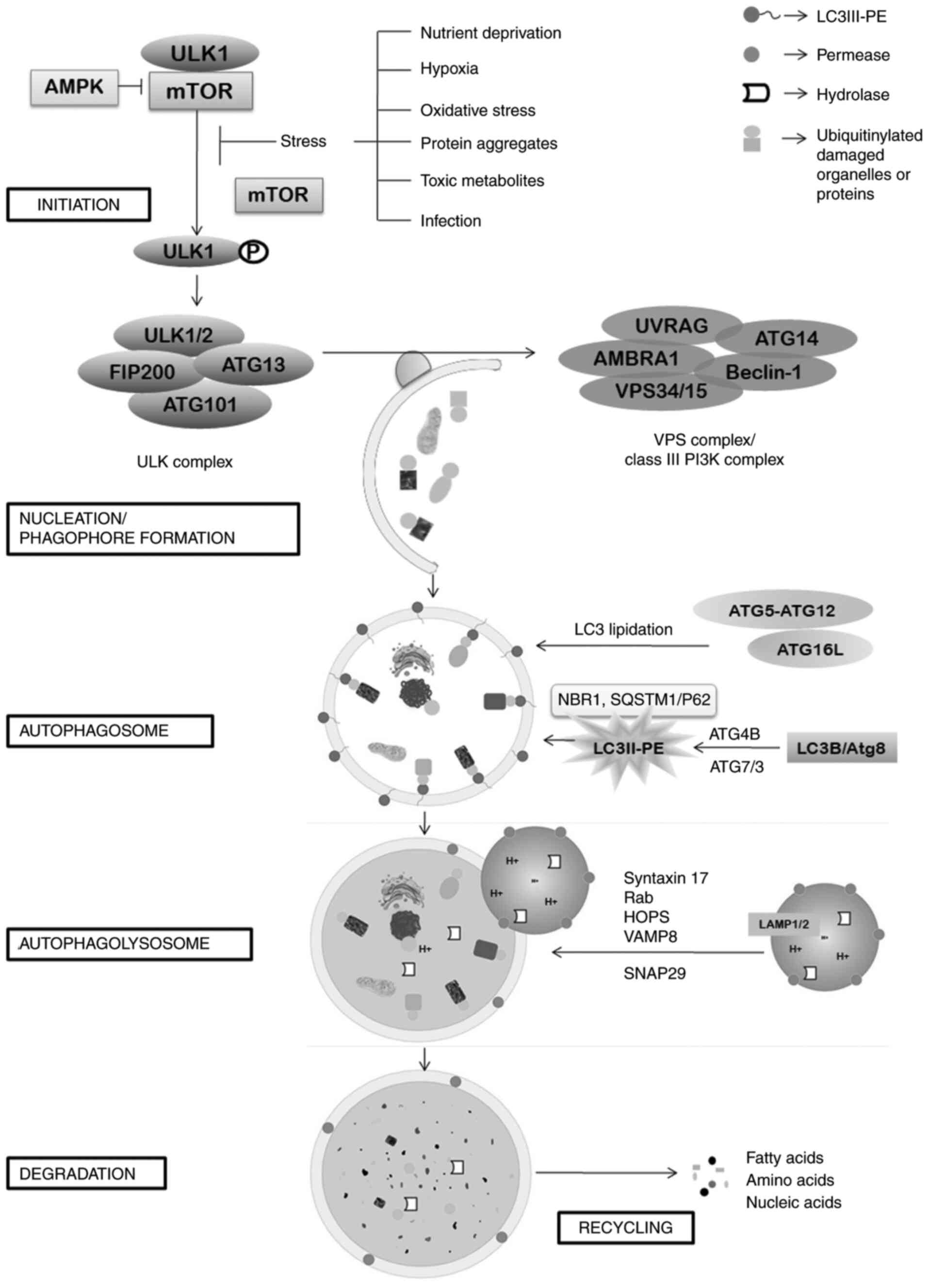
Autophagy, the self-eating mechanism of the body is
an evolutionarily conserved adaptive process in response to various
cellular stress conditions including nutrient deprivation, hypoxia,
oxidative stress, protein aggregates, toxic metabolites and
infection (11). This cytoprotective
machinery intends to degrade and recycle the damaged intracellular
constituents by means of lysosomal degradation. Following
induction, the process of autophagy begins with the formation of
ULK1 or ATG1 complex at the phagophore assembly site (PAS) on
endoplasmic reticulum. As a consequence of the stress conditions,
major cell growth regulator serine/threonine kinase mTOR (mTORC1
subtype only) is inhibited, which in turn results in
autophosphorylation and dissociation of ULK1 from mTOR, thereby
activating ULK1. This event is followed by several phosphorylation
cascades and eventually leads to the formation of ULK1 complex
consisting of ULK1, ULK2, ATG13, FIP200 (also known as RB1CC1), and
ATG101, which in turn activates a class III PI3K complex of VPS15,
VPS34 (PIK3C3), ATG14, Beclin-1 (Atg6), UVRAG (p63), and activating
molecule in BECN1-regulated autophagy protein 1 (AMBRA1)
(11). This nucleation of proteins on
the PAS site of an isolation membrane form a cup-shaped structure
and is termed phagophore. This contributing intracellular membrane
can be endoplasmic reticulum (ER)-exit sites (ERES), Golgi complex,
mitochondria, contact membrane of ER with Golgi body and
mitochondria, plasma membrane or recycling endosomes (12–18).
Whether preference between the membrane sources is based on any
specific criteria remains obscure; however, it is assumed to vary
depending on the cell type, stimulation for autophagy induction,
type of cargo to be carried and other substantial conditions. The
isolation membrane then elongates gradually to engulf the cargo or
the damaged cellular material and then fuses to form a
double-membrane bound autophagic vesicle, known as autophagosome.
The elongation and maturation steps are driven mainly by two
ubiquitin-like conjugation systems, the Atg12 and Atg8/LC3
(lipidation) conjugation systems. The Atg5-Atg12 conjugation system
forms a multimeric complex of Atg5-Atg7-Atg10-Atg12-Atg16L1 which
is critical to LC3 lipidation exhibiting as an E3-like ligase
(19). The second conjugating system
Atg8/LC3B-PE consists of Atg4B-atg7-Atg3 and the activated Atg8 is
then conjugated to a phospholipid, phosphatidylethanolamine (PE)
and thus the membrane-bound LC3B-IPE conjugate (LC3II) is formed on
both the autophagic membranes which is a prime feature for
autophagic vesicle formation. Microtubule-associated protein light
chain 3 (LC3/LC3B) and GABAA receptor-associated protein (GABARAP)
are the two conventional mammalian homologs of Atg8 (20). In addition to elongation and fusion of
the phagophore, Atg8/LCB-II act as a receptor for selective uptake
and degradation of poly-ubiquitinated protein aggregates. LC3B-II
interacts with ubiquitin-binding receptors p62/SQSTM1 and NBR1 and
other LC3-interacting regions (ILR domain) on the surface of the
cargo that promotes turnover of sequestered proteins (21). Intracellular membrane trafficking
proteins, Rab-GTPases, membrane-tethering fusion proteins such as
HOPS and SNARE complex of VAMP8, Syntaxin 17 and SNAP29, are in
charge of the motility and fusion of the autophagosome to lysosome,
forming autolysosome (22). After the
formation of autolysosome, LC3-II on the outer surface of
autophagosome is degraded by ATG4B to recycle it for further
autophagosome formation. Ultimately the cytosomal cargo is degraded
by the lysosomal proteases including cathepsins and other acid
hydrolases. Degraded products are then recycled through nutrient
transporters and used for cell growth. Fig. 1 shows the mechanism of autophagy.
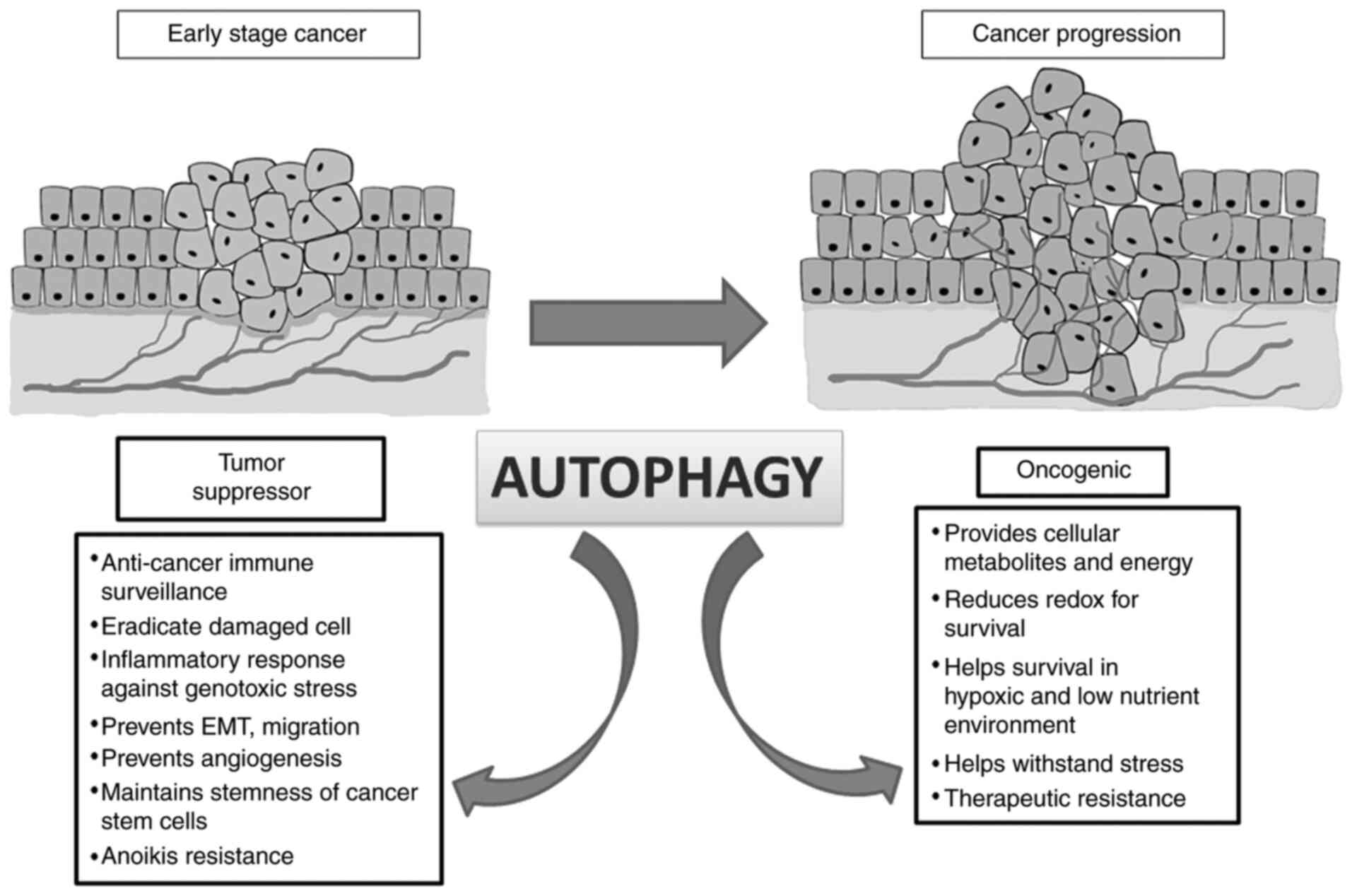
Autophagy has a dual role in cancer because
autophagic genes are both oncogenic and tumour suppressive
depending on type and stage of cancer. In the early stages of
tumorigenesis, autophagy acts in an onco-suppressive mode that is
vital for anticancer immunosurveillance by eradicating endogenous
ROS, oncogenic p62 protein aggregates, inflammatory response,
damaged cell and mount adequate measures against genotoxic stress
(23). Heterozygous disruption of
autophagy-execution gene Beclin-1 (BECN1) promotes
spontaneous malignancy. Monoallelic deletion of BECN1 gene
is evident in 75% of ovarian cancers, 50% of breast cancers and 40%
of prostate cancers (24). Thus, the
gene is demonstrated as a haplo-insufficient autophagic
tumour-suppressor gene (24).
Frameshift mutation in core autophagic proteins ATG2B, ATG5, ATG9B,
ATG12 and UVRAG is prevalent in gastric and colorectal cancer
patients with microsatellite instability (25). Loss-of-function mutations of these
genes restrain the genome-stabilizing effects of autophagy and make
the cells susceptible to tumorigenesis.
Autophagy also has a different role in cancer
progression by providing cellular metabolites for tumour growth and
energy requirement and perpetuating redox homeostasis for promoting
their survival. Substantial evidence demonstrates that autophagy
may cause resistance to cancer cells against therapeutic agents by
helping them to withstand the stress (26). Due to the high glucose demand of
cancer cells, glycolytic enzyme pyruvate kinase M1
(PKM1)-induced autophagy promotes malignancy in a
KRAS-G12D mouse model. Furthermore, embryonic fibroblast
cells from PKM1-ATG7 knockout mice have been shown to limit
tumour growth compared to the corresponding wild-type ATG7
cells (27). Autophagic paradox is
also apprehended in metastatic cascade. In early metastatic
episode, autophagy regulates EMT, tumor cell migration and
invasion. High LC3B expression has been correlated with metastasis
in hepatocellular carcinoma, and melanoma. An increased autophagy
gene signature expression is also associated with an aggressive and
invasive type of glioblastoma (28).
Emerging evidence also indicates the ability of autophagy to
maintain the stemness of cancer stem cells (CSCs). Increased
expression of stem cell marker CD44, mesenchymal marker vimentin,
other core stemness factors such as Forkhead box 3A
(FOXO3A), Sex determining region Y-box (SOX2), Nanog
Homeobox (NANOG), and STAT3 upon induction of
autophagy demonstrates the critical role of autophagy in
maintaining CSCs. Thus, autophagy maintains the pluripotency of the
stem cells and promotes their survival in a hypoxic and low
nutrient environment (29).
Accumulation of ROS in tumor endothelial cells (TECs) enhanced TEC
migration and upregulated angiogenic gene expression such as
VEGFR2. Autophagy is a prime mechanism of modulating redox
homeostasis of the endothelial cells and thus the process of tumor
angiogenesis is also influenced by this (30). Epigenetic modification of autophagy
regulators has been shown to impact the cancer progression.
Promoter hypermethylation of autophagy genes including ATG2B,
ATG4D, ATG9, ATG5, BECN1 and ULK2 appears to induce
tumour progression in several cancer types (31). Thus, the role of autophagy is a
double-edged sword in the framework of carcinogenesis (Fig. 2).
The link between autophagy and development of
pancreatic ductal adenocarcinoma is also an enigma to the
researchers. Findings suggest the autophagic response facilitates
the survival of pancreatic tumor cells. Primary pancreatic
malignant tumours and cell lines exhibit elevated LC3-II expression
and autophagosome count per cell, which suggests constitutive
activation of autophagy at the basal level (32). Additionally, genetic or pharmacologic
inhibition of autophagy led to increased ROS level, DNA damage and
metabolic deformity that ultimately resulted in robust tumour
regression prolonging their survival (6). Heterozygous disruption of ATG5
has been shown to promote tumour development and metastasis but
homozygous loss of ATG5 blocked tumorigenesis in oncogenic
KRAS expressing primary pancreatic cancer cell as well as
human PDAC samples (33). The finding
suggests a possible relationship between ATG5 dosage and its
function. Elevated precursor of nerve growth factor (proNGF)
expression provides anoikis resistance to pancreatic cancer cells
by promoting autophagic genes ATG1 and BECN1; giving
survival advantage to them (34).
However, the context-dependent aspect of autophagy has also been
portrayed in case of PDAC development. Status of p53 is considered
a cogent determinant of tumour-suppressive or tumour-promoting
outcome of autophagy. Humanized genetically modified mouse models
of PDAC lacking autophagy genes ATG5 or ATG7 impede
the progression of low-grade pre-malignant pancreatic lesions into
high grade, suggesting the protective role of autophagy in early
stages of tumorigenesis. By contrast, in mice having oncogenic
KRAS and lacking p53, deficient autophagy exerts a
pro-tumorigenic role (35).
Therapeutic resistance is also a notable feature of PDAC due to
autophagy. PDAC cells display culminated autophagic flux which is
pro-survival to the cancer cells. Constitutive activation and
nuclear transport of MiT/TFE family of transcription factors drives
the coherent gene network of autophagy and lysosomal catabolism in
PDAC cells (36). Sustained
anchorage-independent growth of PDAC cell largely depends on
concurrent mTORC1 inactivation and activated phosphatase for ULK1
(PP2A-B55α complex) (37). Ablation
of autophagy in PDAC cells affects the mitochondrial function that
dampens oxidative phosphorylation and ATP levels (38). ROS-induced autophagy promotes
cytosolic translocation of high-mobility group box 1 protein
(HMGB1) and its binding to beclin-1, which is a positive feedback
regulation of autophagy in PDAC cells, giving protection against
oxidative stress (39).
Tumour-stroma-associated pancreatic stellate cells (PSCs) produce
alanine through an autophagy-dependent mechanism that serves as an
alternative carbon source to support tumour growth in
nutrient-deprived condition (40).
YAP/TAZ signaling aids the process of autophagosome turnover and
also advocates the process of dedifferentiation into stem cell
population. Thus, the signaling pathway is found to be relevant in
linking autophagy and PDAC cancer stem cells (32). Considering the aforementioned studies,
it can be concluded that although pro- and anti-tumorigenic
function of autophagy in PDAC has been reported; mostly autophagy
has been seen to promote pancreatic carcinogenesis.
Noncoding RNAs (ncRNA) are functional molecules
lacking protein-coding regions and accounts for 98% of the
transcriptome (41). MicroRNAs
(miRNAs) and circular RNAs (circRNAs) come under the group of
highly conserved ncRNAs whereas long noncoding RNAs (lncRNAs) lack
general conservation across species (42). ncRNAs function as key regulators of
various biological and cellular processes including gene expression
at transcriptional and post-transcriptional level, DNA synthesis or
genome rearrangement and protection of genomes from foreign nucleic
acids (43). In the current review,
the important aspects of autophagy regulation by noncoding RNAs and
their impact on the mechanisms of pancreatic cancer are
discussed.
miRNAs are highly conserved, endogenous small
non-coding RNAs approximately 22 nt in length that bind 3′-UTR of
the target mRNA and regulate post-transcriptional gene expression
(44). Therefore, the underlying
mechanism of miRNA functioning is translational repression or
degradation of the target mRNA. Thus, miRNAs play a crucial role in
biological events including cell proliferation, differentiation,
metabolism and development, signal transduction, apoptotic cell
death, host-virus interaction, tumorigenesis and tumor progression
via miRNA-RNA-induced silencing complex (miRNA-RISC) (44). Along with mRNAs, miRNAs interact with
other noncoding RNAs such as lncRNAs and circRNAs to trigger their
decay and this forms a crosstalk and regulatory networks linking
the associated target genes (45).
miRNAs also participate in the regulation of autophagy genes at the
transcriptional and post-transcriptional level and modulate
different stages of autophagy (46).
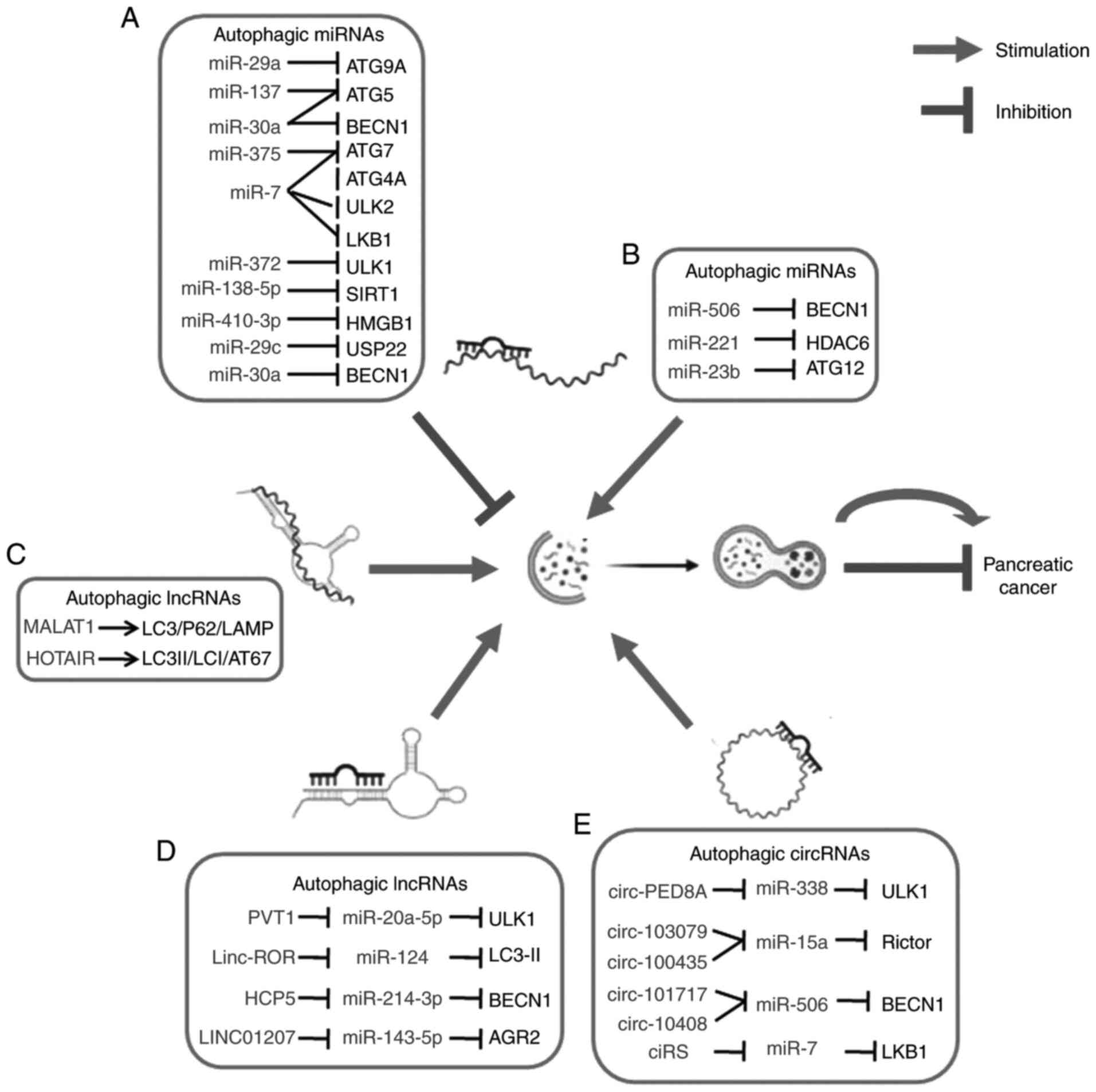
miR-30a is identified as one of the first autophagy-related ncRNAs
targeting BECN1 in a way that affects cellular processes in
various cancer cells (47). Several
studies have indicated that dysregulation of autophagy-related
miRNAs may be associated with the tumorigenesis of PC.
Several microRNAs can induce autophagy by targeting
anti-autophagic genes which subsequently affect the tumorigenesis
of pancreatic cancer. The tumour-suppressor role of miR-506 in PDAC
has been suggested in a study that triggered autophagic flux and
autophagy-related cell death through targeting the
STAT3-BCL2-BECN1 axis (48). miR-506 has been reported to
downregulate the expression of STAT3 which ultimately led to
inhibition of BCL2 and induction of BECN1. Another
microRNA, miR-221, also served as a tumour suppressor in pancreatic
cancer as it was significantly downregulated in highly invasive
pancreatic cancer cells and involved in autophagy-related
regulation in tumour cells of PDAC (49). Another interesting study demonstrated
that HDAC6 may serve as a target of miR-221 and miR-221 may
induce autophagy by suppressing HDAC6 expression and promoting
apoptosis in pancreatic cancer cells (50). Histone deacetylase-6 (HDAC6)
participates in the clearance of aggresomes by helping in the
retrograde transport of autophagosomes and lysosomes. Cells need
both HDAC6 and microtubule cytoskeleton for recruitment of the
Atg-group of proteins, damaged aggregates and lysosomes for
incorporation into aggresomes and use this transport mechanism to
enhance autophagic degradation of aggregated proteins (51). miR-23b can directly target an
important component of autophagy, ATG12, and promote
autophagy in pancreatic cancer cells (52). Table I
shows the list of miRNAs regulating autophagy in pancreatic
cancer.
MicroRNA-mediated translational repression of
autophagy genes impedes the process of autophagy which can further
modulate pancreatic malignancy. miR-29a can act as a potent
autophagy inhibitor in pancreatic cancer. miR-29a has been reported
to be significantly downregulated in pancreatic cancer cells and it
inhibits autophagy when overexpressed (53). Increased accumulation of
autophagosomes/autophagolysosomes and autophagy markers LC3B and
p62 and decreased autophagosome-lysosome fusion constitutes the
manifestation of the blockade of autophagy flux by miR-29a in
pancreatic cancer cells. miR-29a acts as a late-stage autophagy
inhibitor and restricts autophagosome-lysosome fusion by reducing
the expression of autophagy proteins, TFEB and ATG9A, essential for
lysosomal function and vesicular trafficking (53). Evidence has shown the prevention of
autophagy by miR-137 via targeting the 3′-UTR of ATG5 and
negatively regulating ATG5 expression in pancreatic cancer
cells (54). miR-7 targets several
autophagy-related genes, including LKB1, ULK2, ATG4A and
ATG7 and upregulates the LKB1-AMPK-mTOR signaling pathway to
reduce the supply of intracellular glucose to glycolysis in
pancreatic cancer. Thus, miR-7 can suppress pancreatic cancer
progression by inhibiting autophagy steps and vesicle elongation to
impair the activity of aerobic glycolysis (55). A tumor-suppressor role of miR-372 has
been reported in human pancreatic adenocarcinoma by regulating
autophagy, where miR-372 causes the downregulation of ULK1
expression in pancreatic cancer cell lines. Thus, the miR-372/ULK1
axis is involved in pancreatic cancer development by suppressing
cancer cell proliferation, migration, invasion and autophagy
(56). miR-138-5p can inhibit
autophagy and tumour cell growth in pancreatic cancer cells by
targeting serum starvation-induced autophagic flux. This particular
miRNA directly targets the 3′-untranslated region of
autophagy-related gene SIRT1 and suppresses its expression
level (57). Another miRNA miR-410-3p
targets 3′-UTR sequences of HMGB1, a primary regulator of
autophagy that binds to Beclin-1 and modulates Beclin1-PI3KC3
complex formation and is known to be involved in cancer development
via interfering with signaling pathways. In the gemcitabine-treated
PDAC cells, silencing of miR-410-3p promotes autophagic activation
and cell growth and suppresses cell apoptosis. miR-410-3p can also
attenuate chemoresistance to gemcitabine by inhibiting
HMGB1-induced autophagy in PDAC (58). In addition to these, miR-216a inhibits
beclin-1-mediated autophagy in pancreatic cancer and promotes
apoptosis of pancreatic cancer cells in response to radiation, thus
enhancing the radiosensitivity of pancreatic cancer cells (59). By contrast, miR-29c increases the
chemosensitivity of pancreatic cancer cells by inhibiting
USP22-mediated autophagy and cell survival by downregulating USP22
(60). There is evidence that
upregulated miR-375 suppresses autophagy and promotes apoptosis of
acinar cells by negatively regulating ATG7 in pancreatitis
(61). However, evidence also
suggests downregulation of miR-375 in several types of cancer,
including pancreatic cancer, having a role in cancer cell
proliferation. Thus, the tumor-suppressive role of miR-375 in
pancreatic cancer in the context of autophagy remains to be
investigated (62). Similarly,
miR-155 affects the PI3K/AKT/mTOR signaling pathway and impairs
pancreatic autophagy by targeting Rictor (RPTOR-independent
companion of MTOR complex 2) in pancreatitis (63). miR-9 has been reported to be
significantly downregulated in PDAC cells and overexpression of
miR-9 sensitized PDAC cells to doxorubicin via inhibition of
autophagy by directly targeting 3′-UTR of eIF5A2 transcript. eIF5A2
is known to be involved in the proliferation of some cancer cells
(64). The microRNA miR-30a directly
targets the autophagy genes ATG5 and BECN1 and
negatively regulates their expression to suppress autophagy.
However, miR-30a expression is suppressed in pancreatic cancer
cells by a transcriptional modulator protein YY1 (65,66).
Overall, a number of miRNAs function by suppression of autophagy.
However, in most of the cases expression of these miRNAs is
decreased in pancreatic cancer cells so that they obtain the
survival advantage.
lncRNAs are transcripts longer than 200 nucleotides
which do not encode proteins. There are over 15,000 lncRNAs present
across different species (67). The
lncRNA category includes antisense, intronic, intergenic molecules
as well as pseudogenes and retrotransposons. Gene regulatory mode
of function of lncRNAs is implemented by several mechanisms such as
epigenetic modification, aiding the assembly of transcriptional
modulators, sponging miRNAs and post-transcriptional modification
by interfering RNA-binding proteins to the target genes (68). There are reports that lncRNAs also
participate in the regulation of autophagy.
lncRNA acts as a sponge for miRNAs and regulates
miRNA at the transcriptional level as bioinformatics analysis has
identified miRNA recognition elements (MREs) on lncRNA sequences
(71). It is demonstrated that PVT1
can act as a ceRNA to sponge miR-20a-5p to upregulate ULK1 at the
post-trancriptional level and promote cytoprotective autophagy and
cell growth of PDAC cells with increased levels of LC3b-II. Thus,
overexpression of oncogenic PVT1 in PDAC is often associated with
poor prognosis and provides survival advantage to the
chemoresistant pancreatic cancer cells (72). Another oncogenic lncRNA, lincRNA-ROR
(linc-ROR) has been identified to be upregulated in pancreatic
cancer which acts as a ceRNA by sponging miR-124 and inducing
autophagy. Linc-ROR has been reported to be negatively correlated
with miR-124 expression in PDAC tissues and miR-124 directly
targets PTBP1, a splicing factor that switches the isoform
expression of PKM to PKM1 following a higher expression of LC3-II
(73). The lncRNA HLA complex P5
(HCP5) can also act as a ceRNA by sponging miR-214-3p to
target hepatoma-derived growth factor (HDGF) which leads to the
regulation of GEM-resistant pancreatic cancer cell proliferation,
invasion, migration, apoptosis, and autophagy. Previous findings
showed that the expression of HCP5 is upregulated in
pancreatic cancer tissues and negatively modulates miR-214-3p
expression. In addition, sh-HCP5 induces Beclin1, LC3-I/-II and a
decreased p62 expression whereas the opposite occurred in the case
of miR-214-3p inhibitor (74).
Similarly, overexpression of HOTAIR increased the ratio of
LC3-II/-I and the expression of ATG7, thereby enhancing the
formation of autophagosome which further promotes autophagy in
pancreatic cancer (70).
The lncRNAs are also involved in the crosstalk
between autophagy and apoptosis in pancreatic cancer. For example,
long intergenic non-protein coding RNA 1207 (LINC01207) has
been reported to be involved in autophagy and apoptosis via the
LINC01207/miR-143-5p/AGR2 axis in pancreatic cancer
cells (75). Previous findings have
shown upregulated expression of LINC01207 and AGR2,
while miR-143-5p was downregulated. AGR2 acts as a target
gene of miR-143-5p and binding of LINC01207 to miR-143-5p
upregulates AGR2 expression. Elevated AGR2 expression
also inhibits apoptosis in pancreatic cells by increasing the Bcl-2
expression. Thus, results of that study suggest that silencing of
LINC01207 can promote autophagy and apoptosis by sponging
miR-143-5p through an increase in LC3-II and Beclin-1 protein
expression while reducing the p62, AGR2 and ratio of Bcl-2/Bax
expression in pancreatic cancer (75). The aforementioned findings clearly
show the importance of lncRNAs in modulating autophagy in
pancreatic cancer.
Circular RNAs are an important member of the
noncoding RNA family. Circular RNAs (circRNAs) are a group of
abundant, conservative and highly stable novel type of endogenous
non-coding RNAs that are produced by back-splicing event and form a
three-dimensional covalently closed loop structure by linking 3′-
and 5′-ends (76). circRNAs can be
divided into three types such as exonic circRNAs, intronic circRNAs
and exon-intronic circRNAs (77).
Previous findings suggest that circRNAs have many biological
functions including miRNA sponges, protein sponges, enhancer of
protein function, protein scaffolding, protein recruiter and
template for translation, and can regulate several biological
processes related to tumour development, proliferation, apoptosis,
and invasion often through competitive binding (78,79).
Recent findings suggest potential ceRNA networks of circRNA and
miRNA are involved in the autophagy of PDAC which has been
predicted using bioinformatics analysis (80). High expression level of exosomal
circ-PED8A was reported to be associated with poor survival rate,
lymphatic invasion and TNM stage in pancreatic ductal
adenocarcinoma (81). circ-PDE8A
inhibits autophagy by acting as a ceRNA for miR-338 to promote
invasive metastasis through the MACC/MET/ERK or AKT pathways in
PDAC. circ-PDE8A also induces the invasive growth of PDAC cells by
upregulating MET and sponging miR-338 to regulate MACC1 (77,81). MACC1
has been shown to induce autophagy via the AMPK-ULK1 signaling
pathway (82). Similarly,
hsa_circ_103076 and hsa_circ_100435 were upregulated and associated
with miR-15a in PC. miR-15a can inhibit pancreatic cancer cell
proliferation and also induces autophagy by directly targeting
Rictor, a component of mTORC2 (83).
Therefore, hsa_circ_103076 and hsa_circ_100435 can induce autophagy
via functioning as miR-15a sponge (84). It has also been found that
hsa_circ_101717 and hsa_circ_10408 are upregulated in pancreatic
cancer tissues and both of them can exert a tumour suppression
function by sponging miR-506 which triggers autophagy-related cell
death via the STAT3-BCL2-BECN1 axis in PDAC (48). ciRS-7 has been reported to be one of
the few oncogenic circular RNAs which can inhibit tumour suppressor
miR-7 (85). ciRS-7 has been found to
be upregulated in PDAC. In addition, ciRS-7 could inhibit miR-7
activity which affects the proliferation and invasion of PDAC. As
mentioned earlier miR-7 can also suppress pancreatic cancer
progression via inhibiting the LKB1-AMPK-mTOR autophagy axis. We
can assume ciRS-7 may be partly associated in the regulation of
autophagy via miR-7 in PDAC (86)
(Table II). The field of circular
RNA is rapidly developing and the roles of circular RNAs in the
regulation of cancer are currently under investigation. Thus, the
correlation between circRNAs and their sponge effect on
autophagy-related miRNAs can provide new insight into the treatment
and prognosis of pancreatic cancer. The regulation of autophagy in
pancreatic cancer by ncRNAs is shown in Fig. 3.
Several miRNAs have the ability to modulate
autophagy-related proteins and thus regulate different stages of
autophagy in cancer. Some miRNAs which participate in autophagy
regulation are known to act as marker for tumor diagnosis (77). Several studies have demonstrated that
miRNA can regulate radiosensitivity in cancer cells by modulating
autophagy. For example, miR-214 increases radiosensitivity by
inhibiting ATG12-mediated autophagy (87), while miR-183-5p enhances
radio-resistance by targeting ATG5 in colorectal cancer
(88). ATG5 is also known to
be targeted by miR-137 to inhibit autophagy and chemo-sensitize PC
cells to doxorubicin (Dox) (54).
BECN1-mediated autophagy inhibition by miR-216a has been
reported to increase the radiosensitivity of pancreatic cancer
cells, where radiation therapy is a significant approach for
patients with unresectable malignancy (59). It has been demonstrated that the
miR-9/eIF5A2 axis regulates autophagy in PDAC to increase the
anti-cancer effect of doxorubicin in tumor cells (89). Furthermore, miR-29a can function as a
novel therapeutic agent as it sensitizes chemo-resistant cancer
cells to gemcitabine and decreases the invasive potential of
pancreatic cancer cells (53), while
miR-410-3p can attenuate chemoresistance to gemcitabine by
inhibiting HMGB1-induced autophagy in PDAC (58).
Several lncRNAs are also known to influence
radiosensitivity in cancer cells. lncRNA HOTAIR was found to
be highly expressed in pancreatic tumour tissues after radiotherapy
and knockdown of HOTAIR can increase radiosensitivity of pancreatic
cancer cells by regulating autophagy (70). Blockade of autophagy in pancreatic
cancer cells can sensitize it to gemcitabine and reduce the
activity of pancreatic cancer stem cells (90). lncRNA SNHG14 can enhance
gemcitabine resistance in PC by inhibiting cell apoptosis via the
SNHG14/miR-101/autophagy axis. SNHG14 has been
reported to sponge miR-101 where SNHG14 is upregulated while
miR-101 was downregulated in the PDAC tissues. Overexpression of
SNHG14 can increase autophagy-related proteins RAB5A and
ATG4D, thus enhancing PDAC cell progression (91). Understanding of these interactions
between noncoding RNAs and autophagy genes in cancer cells may be
helpful to design a potential therapeutic approach for pancreatic
cancer patients. There is, however, no reported study on the
specific aspects of circular RNAs and their therapeutic
implications involving autophagy pathways in pancreatic cancer;
mainly due to the fact that not much work has been performed using
such pathways.
Additionally, neoadjuvant-based systemic
chemotherapy has also been an important mode of treatment for solid
tumours and also for pancreatic cancer, promoting patient survival
(92,93). Evidence in breast cancer and
osteosarcoma shows that neoadjuvants suppress autophagy and
increase drug sensitivity of the malignant cells. Consequently,
well-known autophagy inhibitory drugs are being used as
neoadjuvants to increase the cytotoxic effect of anti-cancer drugs
or radiotherapy (94,95). We have seen thus far that noncoding
RNAs emerged as key regulators of autophagy and it is imperative
that there be cross-talk between neo-adjuvant chemotherapy, ncRNAs
and autophagy in cancer (96–98). However, to the best of our knowledge,
there has not been a single report for similar interaction studies
in pancreatic cancer. Therefore, the field holds true promise to
have these interactions explored in order to have meaningful
explanation of prognosis and treatment of pancreatic cancer. There
could be two possibilities: i) noncoding RNAs can be a predictive
biomarker of neoadjuvant therapy response. For example, lncRNAs,
microRNAs or circRNAs that promote autophagy can be a marker for
resistance to neoadjuvant therapy or ii) noncoding RNAs that are
known as important regulators of autophagy could serve as novel
therapeutic targets for systematic treatment with neoadjuvant
therapy molecules.
Autophagy is considered an important cellular
process having a significant role in the development of various
diseases, including cancer. In the current review we examined the
contribution of autophagy genes and key autophagic pathways in the
development and progression of pancreatic cancer. Moreover, we have
discussed in detail the regulatory role of ncRNAs in the process.
However, the field is expanding rapidly, especially, with the
identification of newer and newer lncRNAs and circRNAs, and the
demand to understand the mechanistic aspects is on the increase as
well. Thus, a significant part of our future effort should help
delineating the role of newly discovered lncRNAs and circRNAs, in
the factors they are interacting with, whether they are sponging
miRNAs or RBPs or whether their mechanism of action is through
modulation of transcription of autophagy genes or through their
post-transcriptional regulation. Another important aspect is
linking the basic mechanistic studies to the clinically relevant
ones where the diagnostic or therapeutic significance of these
molecules should be tested with much attention. Lastly, future
studies should utilize the recent advancement in technologies
addressing the global changes in gene expression pattern upon
alteration of key autophagy genes or pathways and then correlate
them with pancreatic cancer pathogenesis. Similarly, studies aiming
to determine autophagy-related coding and noncoding RNAs are
altered in different stages of pancreatic cancer or between
precursor lesions and malignancy could also open up new avenues
contributing to both enhancement of basic knowledge and
translation.
Not applicable.
The study was supported by intramural funding from
National Institute of Biomedical Genomics. MM, BS, and BC received
fellowship from University Grants Commission, Council for
Scientific and Industrial Research and Department of Biotechnology,
Government of India, respectively. Fellowship to SP was provided by
the Department of Biotechnology, Government of West Bengal.
Biorender.com was used to create the figure.
Not applicable.
MM and SP read the papers, interpreted the results,
wrote the review and drafted the manuscript with help from BS and
BC. SG conceptualized the study and developed the structure and
overall objectives. All the authors read, edited and approved the
final manuscript, revised it critically for important intellectual
content.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
Not applicable.
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Colombet M, Soerjomataram I,
Dyba T, Randi G, Bettio M, Gavin A, Visser O and Bray F: Cancer
incidence and mortality patterns in Europe: Estimates for 40
countries and 25 major cancers in 2018. Eur J Cancer. 103:356–387.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yang J, Ren B, Yang G, Wang H, Chen G, You
L, Zhang T and Zhao Y: The enhancement of glycolysis regulates
pancreatic cancer metastasis. Cell Mol Life Sci. 77:305–321. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Poruk KE, Firpo MA, Adler DG and Mulvihill
SJ: Screening for pancreatic cancer: Why, how, and who? Ann Surg.
257:17–26. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Devenport SN and Shah YM: Functions and
implications of autophagy in colon cancer. Cells. 8:13492019.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yang S, Wang X, Contino G, Liesa M, Sahin
E, Ying H, Bause A, Li Y, Stommel JM, Dell'antonio G, et al:
Pancreatic cancers require autophagy for tumor growth. Genes Dev.
25:717–729. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen L, Zhou Y, Sun Q, Zhou J, Pan H and
Sui X: Regulation of autophagy by MiRNAs and their emerging roles
in tumorigenesis and cancer treatment. Int Rev Cell Mol Biol.
334:1–26. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dey BK, Mueller AC and Dutta A: Long
non-coding RNAs as emerging regulators of differentiation,
development, and disease. Transcription. 5:e9440142014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sun T: Long noncoding RNAs act as
regulators of autophagy in cancer. Pharmacol Res. 129:151–155.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Choudhry H, Harris AL and McIntyre A: The
tumour hypoxia induced non-coding transcriptome. Mol Aspects Med.
47-48:35–53. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Levy JMM, Towers CG and Thorburn A:
Targeting autophagy in cancer. Nat Rev Cancer. 17:528–542. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Herrera-Cruz MS and Simmen T: Of yeast,
mice and men: MAMs come in two flavors. Biology Direct. 12:32017.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Puri C, Renna M, Bento CF, Moreau K and
Rubinsztein DC: Diverse autophagosome membrane sources coalesce in
recycling endosomes. Cell. 154:1285–1299. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ge L, Melville D, Zhang M and Schekman R:
The ER-Golgi intermediate compartment is a key membrane source for
the LC3 lipidation step of autophagosome biogenesis. Elife.
2:e009472013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Graef M, Friedman JR, Graham C, Babu M and
Nunnari J: ER exit sites are physical and functional core
autophagosome biogenesis components. Mol Biol Cell. 24:2918–2931.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ravikumar B, Moreau K, Jahreiss L, Puri C
and Rubinsztein DC: Plasma membrane contributes to the formation of
pre-autophagosomal structures. Nat Cell Biol. 12:747–757. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hailey DW, Rambold AS, Satpute-Krishnan P,
Mitra K, Sougrat R, Kim PK and Lippincott-Schwartz J: Mitochondria
supply membranes for autophagosome biogenesis during starvation.
Cell. 141:656–667. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Geng J, Nair U, Yasumura-Yorimitsu K and
Klionsky DJ: Post-Golgi Sec proteins are required for autophagy in
Saccharomyces cerevisiae. Mol Biol Cell. 21:2257–2269. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Otomo C, Metlagel Z, Takaesu G and Otomo
T: Structure of the human ATG12~ATG5 conjugate required for LC3
lipidation in autophagy. Nat Struct Mol Biol. 20:59–66. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tanida I: Autophagosome formation and
molecular mechanism of autophagy. Antioxid Redox Signal.
14:2201–2214. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lamark T, Svenning S and Johansen T:
Regulation of selective autophagy: The p62/SQSTM1 paradigm. Essays
Biochem. 61:609–624. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ganley IG: Autophagosome maturation and
lysosomal fusion. Essays Biochem. 55:65–78. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li X, He S and Ma B: Autophagy and
autophagy-related proteins in cancer. Mol Cancer. 19:122020.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh
H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, et al:
Promotion of tumorigenesis by heterozygous disruption of the beclin
1 autophagy gene. J Clin Invest. 112:1809–1820. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kang MR, Kim MS, Oh JE, Kim YR, Song SY,
Kim SS, Ahn CH, Yoo NJ and Lee SH: Frameshift mutations of
autophagy-related genes ATG2B, ATG5, ATG9B and ATG12 in gastric and
colorectal cancers with microsatellite instability. J Pathol.
217:702–706. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li YJ, Lei YH, Yao N, Wang CR, Hu N, Ye
WC, Zhang DM and Chen ZS: Autophagy and multidrug resistance in
cancer. Chin J Cancer. 36:522017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Morita M, Sato T, Nomura M, Sakamoto Y,
Inoue Y, Tanaka R, Ito S, Kurosawa K, Yamaguchi K, Sugiura Y, et
al: PKM1 confers metabolic advantages and promotes cell-autonomous
tumor cell growth. Cancer Cell. 33:355–367.e7. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mowers EE, Sharifi MN and Macleod KF:
Autophagy in cancer metastasis. Oncogene. 36:1619–1630. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
El Hout M, Cosialls E, Mehrpour M and
Hamai A: Crosstalk between autophagy and metabolic regulation of
cancer stem cells. Mol Cancer. 19:272020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kardideh B, Samimi Z, Norooznezhad F,
Kiani S and Mansouri K: Autophagy, cancer and angiogenesis: Where
is the link? Cell Biosci. 9:652019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bhol CS, Panigrahi DP, Praharaj PP,
Mahapatra KK, Patra S, Mishra SR, Behera BP and Bhutia SK:
Epigenetic modifications of autophagy in cancer and cancer
therapeutics. Semin Cancer Biol. 66:22–33. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
New M and Tooze S: The role of autophagy
in pancreatic cancer-recent advances. Biology (Basel).
9:72019.PubMed/NCBI
|
|
33
|
Gorgulu K, Diakopoulos KN, Ai J, Schoeps
B, Kabacaoglu D, Karpathaki AF, Ciecielski KJ, Kaya-Aksoy E, Ruess
DA, Berninger A, et al: Levels of the autophagy-related 5 protein
affect progression and metastasis of pancreatic tumors in mice.
Gastroenterology. 156:203–217.e20. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xu J, Song J, Yang X, Guo J, Wang T and
Zhuo W: ProNGF siRNA inhibits cell proliferation and invasion of
pancreatic cancer cells and promotes anoikis. Biomed Pharmacother.
111:1066–1073. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Rosenfeldt MT, O'Prey J, Morton JP, Nixon
C, MacKay G, Mrowinska A, Au A, Rai TS, Zheng L, Ridgway R, et al:
p53 status determines the role of autophagy in pancreatic tumour
development. Nature. 504:296–300. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Perera RM, Stoykova S, Nicolay BN, Ross
KN, Fitamant J, Boukhali M, Lengrand J, Deshpande V, Selig MK,
Ferrone CR, et al: Transcriptional control of autophagy-lysosome
function drives pancreatic cancer metabolism. Nature. 524:361–365.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wong PM, Feng Y, Wang J, Shi R and Jiang
X: Regulation of autophagy by coordinated action of mTORC1 and
protein phosphatase 2A. Nat Commun. 6:80482015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Biancur DE and Kimmelman AC: The
plasticity of pancreatic cancer metabolism in tumor progression and
therapeutic resistance. Biochim Biophys Acta Rev Cancer.
1870:67–75. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tang D, Kang R, Livesey KM, Zeh HJ III and
Lotze MT: High mobility group box 1 (HMGB1) activates an autophagic
response to oxidative stress. Antioxid Redox Signal. 15:2185–2195.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sousa CM, Biancur DE, Wang X, Halbrook CJ,
Sherman MH, Zhang L, Kremer D, Hwang RF, Witkiewicz AK, Ying H, et
al: Pancreatic stellate cells support tumour metabolism through
autophagic alanine secretion. Nature. 536:479–483. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gupta SK and Thum T: Non-coding RNAs as
orchestrators of autophagic processes. J Mol Cell Cardiol.
95:26–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Bejerano G, Pheasant M, Makunin I, Stephen
S, Kent WJ, Mattick JS and Haussler D: Ultraconserved elements in
the human genome. Science. 304:1321–1335. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Cech TR and Steitz JA: The noncoding RNA
revolution-trashing old rules to forge new ones. Cell. 157:77–94.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Huang Y, Shen XJ, Zou Q, Wang SP, Tang SM
and Zhang GZ: Biological functions of microRNAs: A review. J
Physiol Biochem. 67:129–139. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yamamura S, Imai-Sumida M, Tanaka Y and
Dahiya R: Interaction and cross-talk between non-coding RNAs. Cell
Mol Life Sci. 75:467–484. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhang J, Wang P, Wan L, Xu S and Pang D:
The emergence of noncoding RNAs as Heracles in autophagy.
Autophagy. 13:1004–1024. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhu H, Wu H, Liu X, Li B, Chen Y, Ren X,
Liu CG and Yang JM: Regulation of autophagy by a beclin 1-targeted
microRNA, miR-30a, in cancer cells. Autophagy. 5:816–823. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Sun L, Hu L, Cogdell D, Lu L, Gao C, Tian
W, Zhang Z, Kang Y, Fleming JB and Zhang W: MIR506 induces
autophagy-related cell death in pancreatic cancer cells by
targeting the STAT3 pathway. Autophagy. 13:703–714. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Tan X, Zhou L, Wang H, Yang Y, Sun Y, Wang
Z, Zhang X, Gao F and Li H: Differential expression profiles of
microRNAs in highly and weakly invasive/metastatic pancreatic
cancer cells. Oncol Lett. 16:6026–6038. 2018.PubMed/NCBI
|
|
50
|
Yang Y, Sun Y, Wang H, Li H, Zhang M, Zhou
L, Meng X, Wu Y, Liu P, Liu X, et al: MicroRNA-221 induces
autophagy through suppressing HDAC6 expression and promoting
apoptosis in pancreatic cancer. Oncol Lett. 16:7295–7301.
2018.PubMed/NCBI
|
|
51
|
Iwata A, Riley BE, Johnston JA and Kopito
RR: HDAC6 and microtubules are required for autophagic degradation
of aggregated huntingtin. J Biol Chem. 280:40282–4092. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Donadelli M and Palmieri M: Roles for
microRNA 23b in regulating autophagy and development of pancreatic
adenocarcinoma. Gastroenterology. 145:936–938. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Kwon JJ, Willy JA, Quirin KA, Wek RC, Korc
M, Yin XM and Kota J: Novel role of miR-29a in pancreatic cancer
autophagy and its therapeutic potential. Oncotarget. 7:71635–71650.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Wang ZC, Huang FZ, Xu HB, Sun JC and Wang
CF: MicroRNA-137 inhibits autophagy and chemosensitizes pancreatic
cancer cells by targeting ATG5. Int J Biochem Cell Biol. 111:63–71.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Gu DN, Jiang MJ, Mei Z, Dai JJ, Dai CY,
Fang C, Huang Q and Tian L: microRNA-7 impairs autophagy-derived
pools of glucose to suppress pancreatic cancer progression. Cancer
Lett. 400:69–78. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Chen H, Zhang Z, Lu Y, Song K, Liu X, Xia
F and Sun W: Downregulation of ULK1 by microRNA-372 inhibits the
survival of human pancreatic adenocarcinoma cells. Cancer Sci.
108:1811–1819. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Tian S, Guo X, Yu C, Sun C and Jiang J:
miR-138-5p suppresses autophagy in pancreatic cancer by targeting
SIRT1. Oncotarget. 8:11071–11082. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Xiong J, Wang D, Wei A, Ke N, Wang Y, Tang
J, He S, Hu W and Liu X: MicroRNA-410-3p attenuates gemcitabine
resistance in pancreatic ductal adenocarcinoma by inhibiting
HMGB1-mediated autophagy. Oncotarget. 8:107500–107512. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Zhang X, Shi H, Lin S, Ba M and Cui S:
MicroRNA-216a enhances the radiosensitivity of pancreatic cancer
cells by inhibiting beclin-1-mediated autophagy. Oncol Rep.
34:1557–1564. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Huang L, Hu C, Cao H, Wu X, Wang R, Lu H,
Li H and Chen H: MicroRNA-29c increases the chemosensitivity of
pancreatic cancer cells by inhibiting USP22 mediated autophagy.
Cell Physiol Biochem. 47:747–758. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Zhao SP, Yu C, Xiang KM, Yang MS, Liu ZL
and Yang BC: miR-375 inhibits autophagy and further promotes
inflammation and apoptosis of acinar cells by targeting ATG7.
Pancreas. 49:543–551. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Yan JW, Lin JS and He XX: The emerging
role of miR-375 in cancer. Int J Cancer. 135:1011–1018. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Zhang X, Chu J, Sun H, Zhao D, Ma B, Xue
D, Zhang W and Li Z: MiR-155 aggravates impaired autophagy of
pancreatic acinar cells through targeting Rictor. Acta Biochim
Biophys Sin (Shanghai). 52:192–199. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Cao TT, Lin SH, Fu L, Tang Z, Che CM,
Zhang LY, Ming XY, Liu TF, Tang XM, Tan BB, et al: Eukaryotic
translation initiation factor 5A2 promotes metabolic reprogramming
in hepatocellular carcinoma cells. Carcinogenesis. 38:94–104. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Wang T, Chen G, Ma X, Yang Y, Chen Y, Peng
Y, Bai Z, Zhang Z, Pei H and Guo W: MiR-30a regulates cancer cell
response to chemotherapy through SNAI1/IRS1/AKT pathway. Cell Death
Dis. 10:1532019. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Yang C, Zhang JJ, Peng YP, Zhu Y, Yin LD,
Wei JS, Gao WT, Jiang KR and Miao Y: A Yin-Yang 1/miR-30a
regulatory circuit modulates autophagy in pancreatic cancer cells.
J Transl Med. 15:2112017. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Derrien T, Johnson R, Bussotti G, Tanzer
A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG,
et al: The GENCODE v7 catalog of human long noncoding RNAs:
Analysis of their gene structure, evolution, and expression. Genome
Res. 22:1775–1789. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Bermudez M, Aguilar-Medina M,
Lizarraga-Verdugo E, Avendano-Felix M, Silva-Benitez E,
Lopez-Camarillo C and Ramos-Payán R: LncRNAs as regulators of
autophagy and drug resistance in colorectal cancer. Front Oncol.
9:10082019. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Li L, Chen H, Gao Y, Wang YW, Zhang GQ,
Pan SH, Ji L, Kong R, Wang G, Jia YH, et al: Long noncoding RNA
MALAT1 promotes aggressive pancreatic cancer proliferation and
metastasis via the stimulation of autophagy. Mol Cancer Ther.
15:2232–2243. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Wu C, Yang L, Qi X, Wang T, Li M and Xu K:
Inhibition of long non-coding RNA HOTAIR enhances radiosensitivity
via regulating autophagy in pancreatic cancer. Cancer Manag Res.
10:5261–5271. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Paraskevopoulou MD, Vlachos IS, Karagkouni
D, Georgakilas G, Kanellos I, Vergoulis T, Zagganas K, Tsanakas P,
Floros E, Dalamagas T and Hatzigeorgiou AG: DIANA-LncBase v2:
Indexing microRNA targets on non-coding transcripts. Nucleic Acids
Res. 44:D231–D238. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Huang F, Chen W, Peng J, Li Y, Zhuang Y,
Zhu Z, Shao C, Yang W, Yao H and Zhang S: LncRNA PVT1 triggers
Cyto-protective autophagy and promotes pancreatic ductal
adenocarcinoma development via the miR-20a-5p/ULK1 Axis. Mol
Cancer. 17:982018. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Li C, Zhao Z, Zhou Z and Liu R: Linc-ROR
confers gemcitabine resistance to pancreatic cancer cells via
inducing autophagy and modulating the miR-124/PTBP1/PKM2 axis.
Cancer Chemother Pharmacol. 78:1199–1207. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Liu Y, Wang J, Dong L, Xia L, Zhu H, Li Z
and Yu X: Long noncoding RNA HCP5 regulates pancreatic cancer
gemcitabine (GEM) resistance by sponging Hsa-miR-214-3p To target
HDGF. Onco Targets Ther. 12:8207–8216. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Liu C, Wang JO, Zhou WY, Chang XY, Zhang
MM, Zhang Y and Yang XH: Long non-coding RNA LINC01207 silencing
suppresses AGR2 expression to facilitate autophagy and apoptosis of
pancreatic cancer cells by sponging miR-143-5p. Mol Cell
Endocrinol. 493:1104242019. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Shao Y and Chen Y: Roles of circular RNAs
in neurologic disease. Front Mol Neurosci. 9:252016. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Jiang PC and Bu SR: Clinical value of
circular RNAs and autophagy-related miRNAs in the diagnosis and
treatment of pancreatic cancer. Hepatobiliary Pancreat Dis Int.
18:511–516. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Kristensen LS, Andersen MS, Stagsted LVW,
Ebbesen KK, Hansen TB and Kjems J: The biogenesis, biology and
characterization of circular RNAs. Nat Rev Genet. 20:675–691. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Zhong Y, Du Y, Yang X, Mo Y, Fan C, Xiong
F, Ren D, Ye X, Li C, Wang Y, et al: Circular RNAs function as
ceRNAs to regulate and control human cancer progression. Mol
Cancer. 17:792018. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Wei DM, Jiang MT, Lin P, Yang H, Dang YW,
Yu Q, Liao DY, Luo DZ and Chen G: Potential ceRNA networks involved
in autophagy suppression of pancreatic cancer caused by chloroquine
diphosphate: A study based on differentiallyexpressed circRNAs,
lncRNAs, miRNAs and mRNAs. Int J Oncol. 54:600–626. 2019.PubMed/NCBI
|
|
81
|
Li Z, Yanfang W, Li J, Jiang P, Peng T,
Chen K, Zhao X, Zhang Y, Zhen P, Zhu J and Li X: Tumor-released
exosomal circular RNA PDE8A promotes invasive growth via the
miR-338/MACC1/MET pathway in pancreatic cancer. Cancer Lett.
432:237–250. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Wu J, Zhang D, Li J, Deng X, Liang G, Long
Y, He X, Dai T and Ren D: MACC1 induces autophagy to regulate
proliferation, apoptosis, migration and invasion of squamous cell
carcinoma. Oncol Rep. 38:2369–2377. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Huang N, Wu J, Qiu W, Lyu Q, He J, Xie W,
Xu N and Zhang Y: MiR-15a and miR-16 induce autophagy and enhance
chemosensitivity of Camptothecin. Cancer Biol Ther. 16:941–948.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Guo S, Xu X, Ouyang Y, Wang Y, Yang J, Yin
L, Ge J and Wang H: Microarray expression profile analysis of
circular RNAs in pancreatic cancer. Mol Med Rep. 17:7661–7971.
2018.PubMed/NCBI
|
|
85
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Liu L, Liu FB, Huang M, Xie K, Xie QS, Liu
CH, Shen MJ and Huang Q: Circular RNA ciRS-7 promotes the
proliferation and metastasis of pancreatic cancer by regulating
miR-7-mediated EGFR/STAT3 signaling pathway. Hepatobiliary Pancreat
Dis Int. 18:580–586. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Hu JL, He GY, Lan XL, Zeng ZC, Guan J,
Ding Y, Qian XL, Liao WT, Ding YQ and Liang L: Inhibition of
ATG12-mediated autophagy by miR-214 enhances radiosensitivity in
colorectal cancer. Oncogenesis. 7:162018. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Zheng S, Zhong YF, Tan DM, Xu Y, Chen HX
and Wang D: miR-183-5p enhances the radioresistance of colorectal
cancer by directly targeting ATG5. J Biosci. 44:922019. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Wu Y, Tang Y, Xie S, Zheng X, Zhang S, Mao
J, Wang B, Hou Y, Hu L, Chai K and Chen W: Chimeric peptide
supramolecular nanoparticles for plectin-1 targeted miRNA-9
delivery in pancreatic cancer. Theranostics. 10:1151–1165. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Yang MC, Wang HC, Hou YC, Tung HL, Chiu TJ
and Shan YS: Blockade of autophagy reduces pancreatic cancer stem
cell activity and potentiates the tumoricidal effect of
gemcitabine. Mol Cancer. 14:1792015. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Zhang X, Zhao P, Wang C and Xin B: SNHG14
enhances gemcitabine resistance by sponging miR-101 to stimulate
cell autophagy in pancreatic cancer. Biochem Biophys Res Commun.
510:508–514. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Gaskill CE, Maxwell J, Ikoma N, Kim MP,
Tzeng CW, Lee JE and Katz MHG: History of preoperative therapy for
pancreatic cancer and the MD Anderson experience. J Surg Oncol.
123:1414–1422. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Brown ZJ and Cloyd JM: Trends in the
utilization of neoadjuvant therapy for pancreatic ductal
adenocarcinoma. J Surg Oncol. 123:1432–1440. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Cheng SW, Chen PC, Ger TR, Chiu HW and Lin
YF: GBP5 serves as a potential marker to predict a favorable
response in triple-negative breast cancer patients receiving a
taxane-based chemotherapy. J Pers Med. 11:1972021. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Saini H, Sharma H, Mukherjee S, Chowdhury
S and Chowdhury R: Verteporfin disrupts multiple steps of autophagy
and regulates p53 to sensitize osteosarcoma cells. Cancer Cell Int.
21:522021. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
YiRen H, YingCong Y, Sunwu Y, Keqin L,
Xiaochun T, Senrui C, Ende C, XiZhou L and Yanfan C: Long noncoding
RNA MALAT1 regulates autophagy associated chemoresistance via
miR-23b-3p sequestration in gastric cancer. Mol Cancer. 16:1742017.
View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Xiong H, Ni Z, He J, Jiang S, Li X, Gong
W, Zheng L, Chen S, Li B and Zhang N: LncRNA HULC triggers
autophagy via stabilizing Sirt1 and attenuates the chemosensitivity
of HCC cells. Oncogene. 36:3528–3540. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Tan S, Shi H, Ba M, Lin S, Tang H, Zeng X
and Zhang X: miR-409-3p sensitizes colon cancer cells to
oxaliplatin by inhibiting Beclin-1-mediated autophagy. Int J Mol
Med. 37:1030–1038. 2016. View Article : Google Scholar : PubMed/NCBI
|