Introduction
Lung cancer is the most common malignancy worldwide
and poses a serious threat to human life and health (1). In addition, non-small-cell lung cancer
(NSCLC) accounts for ~85% of total lung cancer diagnoses (2). NSCLC is often asymptomatic at the early
stage of the disease and most patients are diagnosed when the
disease has progressed to the intermediate or late stage (2,3). Although
an increasing number of diagnostic and therapeutic strategies have
been developed, the overall survival rate remains unsatisfactory
(4). Some patients still die due to
distant metastasis and drug resistance (5,6). Thus, it
is key to investigate the potential molecular mechanisms of NSCLC
and to identify novel treatment approaches to improve outcomes in
patients with NSCLC.
RNA N6-methyladenosine (m6A) modification is the
most abundant conserved post-translational modification in
eukaryotic organisms (7). It
primarily regulates gene expression, mRNA stability, alternative
splicing, translation efficiency and primary microRNA (miRNA)
processing (8,9). The dynamic modification of m6A is
regulated by two types of catalytic proteins (m6A ‘writers’ and
‘erasers’) (10). The ‘writers’
primarily include methyltransferase-like (METTL)3, METTL14 and
Wilms tumor 1 associated protein (11,12).
‘Erasers’, such as alkylation repair homolog protein 5 and fat mass
and obesity-associated protein, serve as demethylases to reverse
methylation (13). A previous study
demonstrated that m6A modulators exhibit various biological
functions in different processes, such as embryonic growth, stem
cell differentiation, circadian rhythm and tumor progression
(14).
METTL3 is a core methyltransferase for m6A
modification (15). Certain studies
have reported that METTL3 serves an oncogenic role in gastric,
colorectal and bladder cancer, as well as hepatocellular carcinoma
(15–18). By contrast, a recent study showed that
METTL3 is a tumor suppressor in kidney cancer (19). Nevertheless, the biological function
of METTL3 and its underlying regulatory mechanisms in NSCLC have
not been sufficiently described.
The aim of the present study was to investigate
METTL3 expression in NSCLC cells and human NSCLC tissue and to
determine the role of METTL3 in NSCLC progression both in
vitro and in vivo. The results of the present study may
provide a novel therapeutic target for treatment of NSCLC.
Materials and methods
Patients and specimen collection
A total of 77 NSCLC tissue samples and paired
adjacent normal tissue samples (distance, ≥3 cm) were obtained from
patients undergoing surgery from May 2018 to January 2020 at the
First Affiliated Hospital of Xinxiang Medical University (Xinxiang,
China). Of these patients, 44 were male (mean age, 60.22 years) and
33 were female (mean age, 58.6 years). The histological type of the
tumors was assessed using the World Health Organization
Classification (20) by two
independent pathologists. Ethics approval was granted by the Ethics
Committee of the First Affiliated Hospital of Xinxiang Medical
University (approval no. KN201808002). Oral consent was obtained
from patients. All experiments were performed in accordance with
approved guidelines and regulations (21).
Cell culture
Human NSCLC cell lines (A549, PC9, H1299, H1975 and
HCC827) and normal pulmonary epithelial cells (BEAS-2B) were
obtained from the American Type Culture Collection. A549, PC9,
H1299, H1975 and HCC827 cells were cultured in RPMI-1640 medium
(Invitrogen; Thermo Fisher Scientific, Inc.). BEAS-2B cells were
cultured in DMEM (Invitrogen; Thermo Fisher Scientific, Inc.). All
media were supplemented with 10% FBS (Gibco; Thermo Fisher
Scientific, Inc.) and all cells were incubated at 37°C with 5%
CO2.
Establishment of stable cell
lines
Stable cell lines were constructed using lentivirus
system. The plasmids used to make stably overexpressed/knockdown
cell lines were based on the pcDNA3/Flag-METTL3 plasmid (Addgene;
cat. no. 53739) or short hairpin (sh)RNA-METTL3 (GenePharma Co.,
Ltd.). To generate lentivirus particles, H1299 or H1975 cells were
co-transfected with plasmids together with Trans-lentiviral
packaging plasmids using Trans-lentiviral Packaging kit
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. Lentivirus particles were harvested at
48 h post transfection and concentrated using Lenti-X concentrator
according to manufactory instruction (Clontech Laboratories, Inc.).
Stable cell lines was transduced with lentivirus particles in
serum-free medium (Invitrogen; Thermo Fisher Scientific, Inc.)
containing 8 µg/ml polybrene (Sigma-Aldrich; Merck KGaA) followed
by selection with 2 µg/ml puromycin for ≥10 days. The cells with
stable overexpression or knockdown of METTL3 were s identified by
RT-qPCR and immunoblot analysis. The target sequences of shRNAs
were as follows: shMETTL3#1, 5′-GCCAAGGAACAATCCATTGTT-3′;
shMETTL3#2, 5′-GCTGCACTTCAGACGAATTAT-3′; and shMETTL3#,
5′GCTTACTATCTAGCATCACAT-3′.
RNA isolation and reverse
transcription-quantitative (RT-q)PCR
Total RNA was isolated from cells using
TRIzol® Reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's instructions. Then, 1 µg
total RNA was reverse-transcribed to complementary DNA using
PrimeScript™ RT Master Mix (Takara Bio, Inc.). The reaction
conditions were 37°C for 15 min and 85°C for 15 sec. mRNA
expression was measured using a TB Green™ kit (Takara Bio, Inc.),
according to the manufacturer's instructions. Quantification of the
target and reference (GAPDH) genes was performed in triplicate
using a LightCycler® 480 II (Roche Diagnostics GmbH)
with an initial denaturing step at 95°C for 30 sec, followed by 40
cycles of 95°C for 5 sec and 60°C for 30 sec. The primers used were
as follows: GAPDH forward, 5′-CTGGGCTACACTGAGCACC-3′ and reverse,
5′-GGAACGCTTCACGAATTTG-3′; and METTL3 forward,
5′-AAGTGGTCGTTGAGGGCAATG-3′ and reverse, 5′-GCTTGGCGTGTGGTCTTT-3′.
All primers were synthesized by Shanghai Shenggong Biology
Engineering Technology Service, Ltd.. The analysis was conducted
with the 2−ΔΔCq quantification method (22).
Cell viability assay
Cells were seeded in 96-well plates at a density of
2,000 cells/well. Cells were incubated at 37°C in an incubator with
5% CO2 for 0, 24, 48 and 72 h. Cell Counting Kit-8
(CCK-8; Sigma-Aldrich; Merck KGaA) was used to measure cell
viability according to the manufacturer's instructions. The
absorbance was measured at a wavelength of 450 nm using a plate
reader (Bio-Rad Laboratories, Inc.).
Apoptosis
Apoptosis was measured using flow cytometry with a
FITC-Annexin V/PI detection kit (Wanleibio Co., Ltd.). A total of
1×106 cells per well was seeded in 6-well plates and
cells were then harvested after culturing for 48 h at 37°C.
Afterwards, the cells were stained in the dark with FITC-Annexin V
and PI at room temperature for 15 min. Subsequently, the
percentages of apoptotic cells were measured using FACS flow
cytometry (BD Biosciences) and analyzed using FlowJo 8.6.3 (Tree
Star, Inc.).
Cell migration assay
Cell migration assays were performed using a cell
migration assay kit (Corning, Inc.) and Transwell inserts. Briefly,
2×104 cells were seeded in the upper chamber of a Boyden
chamber (without serum in the RPMI-1640 medium, Invitrogen; Thermo
Fisher Scientific, Inc.). The lower portion of the chamber was
filled with 600 µl RPMI-1640 medium (Invitrogen; Thermo Fisher
Scientific, Inc.) supplemented with 20% FBS (Gibco; Thermo Fisher
Scientific, Inc.). Following incubation for 24 h at 37°C, migrated
cells in the lower chambers were fixed with 4% paraformaldehyde for
15 min at room temperature and stained with 0.05% crystal violet
(Sigma-Aldrich; Merck KGaA) for 10 min at room temperature. Cells
were counted in five randomly selected fields of view under a light
microscope (Leica GmbH DMI4000B) at 200× magnification.
m6A qPCR
m6A qPCR was performed as reported previously
(23) to assess the relative
abundance of the selected mRNA in m6A antibody immunoprecipitation
(IP) and input samples. Isolated m6A-RIP RNA was quantified by
qPCR. The primers used were as follows: Bcl-2 forward,
5′-ATCGCCCTGTGGATGACTGAGT-3′ and reverse:
5′-GCCAGGAGAAATCAAACAGAGGC-3′.
m6A sequencing (m6A-seq)
Total RNA was isolated and purified using
PolyATtract mRNA Isolation Systems kit (cat. no. Z5310; Promega
Corporation). m6A-seq was performed as previously described
(24). The NEBNext Ultra Directional
RNA Library Prep kit (cat. no. E7420L; Illumina, Inc.) was used for
library construction. Clustered libraries were sequenced with an
Illumina HiSeq 4000 (Illumina, Inc.) by Novogene Co., Ltd.
Western blotting
Total protein was extracted from cells using RIPA
buffer containing protease and phosphatase inhibitor cocktail (both
from Sigma-Aldrich; Merck KGaA). The protein concentration was
determined with a BCA Protein Assay kit (Beyotime Institute of
Biotechnology). Cellular proteins were separated by 10% SDS-PAGE
and then transferred to PVDF membranes. The membranes were blocked
with 5% non-fat milk and 0.1% Tween-20 in Tris-buffered saline at
room temperature for 2 h. Membranes were probed at 4°C overnight
with primary antibodies against METTL3 (1:2,000; cat. no. 86132S;
Cell Signaling Technology, Inc.), Bcl-2 (1:1,000; cat. no. ab32124;
Abcam) and GAPDH (1:5,000; cat. no. 5174S; Cell Signaling
Technology, Inc.). After washing in PBST (0.5% Tween-20). Three
times, the blots were incubated with horseradish peroxidase
(HRP)-conjugated secondary antibodies (1:1,000; cat. no. BA1070;
Wuhan Boster Biological Technology, Ltd.) at room temperature for 1
h. The signal was visualized using ECL western blotting detection
reagents (Tanon Science and Technology Co., Ltd). Protein
expression levels were quantified using ImageJ software (version
1.50; National Institutes of Health).
Experimental mouse techniques
A total of 15 female BALB/c nude mice (age, 6 weeks;
weight, 18–20 g) were purchased from the Model Animal Center at
Nanjing University (Nanjing, China) and raised under pathogen-free
conditions in the Xinxiang Medical University Animal Center
(Xinxiang, China). The mice were housed with filtered air, 12 h
light/dark cycle, constant temperature (25°C), relative humidity
(50±5%) and free access to food and water. In order to establish a
human NSCLC xenograft model, 5×106 H1299,
sh-METTL3-H1299 and METTL3 stably overexpressed H1299 cells
(2×106 per mouse) were subcutaneously injected into
mice. Tumor growth was observed daily. Tumor volume was calculated
as follows: 0.5× (length × width2). At 24 days
post-inoculation, the maximum diameter exhibited by a single
subcutaneous tumor was 15 mm and mice were anesthetized by
intraperitoneal administration of sodium pentobarbital (50 mg/kg),
then sacrificed by cervical dislocation. Tumors were harvested and
weighed for histological analysis. Lungs were extracted and fixed
for 24 h at room temperature using 10% (v/v) neutral-buffered
formalin and transferred to 70% ethanol until embedding in paraffin
then sectioned at 3 µm. For hematoxylin-eosin staining, sections
were incubated with hematoxylin for 2 min and eosin for 1 min. The
experimental procedures were approved by the Laboratory Animal Care
Committee at Xinxiang Medical College (approval no.
XXMU-2020-146367).
Immunohistochemistry (IHC) assay
Tissue sections were incubated with primary
antibodies against METTL3 (1:50; cat. no. 86132S; Cell Signaling
Technology, Inc.), Bcl-2 (1:50; cat. no. ab32124; Abcam) and Ki67
(1:100; cat. no. ab16667; Abcam) at 4°C overnight. Following
washing with PBST containing 0.05% Tween-20, the sections were
incubated with HRP-conjugated anti-rabbit secondary antibody
(1:5,000; cat. no. BM3894; Wuhan Boster Biological Technology,
Ltd.) at room temperature for 1 h. The sections were developed with
0.05% 3-diaminobenzidine tetrahydrochloride at room temperature for
10 sec, which was followed by counterstaining with 10% Mayer's
hematoxylin for 4 min at room temperature. IHC results were
analyzed by two experienced pathologists. A total of five random
fields under a light microscope (Leica GmbH DMI4000B) at 200×
magnification were chosen to calculate the percentage of positively
stained cells vs. the total number of tumor cells. The staining
proportion of the positive cells was divided into four groups as
follows: -, 0 positive cells observed; +, <30% vs. positive
tumor cells observed; ++, 30–60% vs. positive tumor cells observed;
and +++: >60% vs. positive tumor cells observed. The - and +
groups were classed as METTL3-low expression group, while the ++
and +++ groups were classed as METTL3-high expression group for
subsequent analysis. Low- and high-expression groups were combined
following scoring and analyzed by Fisher's exact t-test.
Bioinformatics analysis
The Gene Expression Profiling Interactive Analysis
(GEPIA) database (gepia.cancer-pku.cn/) was used to evaluate the
mRNA expression levels of METTL3 in NSCLC and normal tissue. Log
rank test was used for Kaplan-Meier analysis. The Kaplan-Meier
plotting tool (kmplot.com/analysis/) was used to evaluate the
relapse-free survival of patients with NSCLC expressing low or high
levels of METTL3. The association between METTL3 and Bcl-2 mRNA
expression levels in NSCLC was analyzed using GEPIA database
(gepia.cancer-pku.cn/) and Pearson's
correlation coefficient.
Statistical analysis
A total of three independent repeats of experiments
were performed. All data are shown as the mean ± SD. Data were
analyzed using SPSS software 22.0 (IBM Corp.). Associations between
two groups were evaluated using χ2 test and Pearson's
correlation analysis. Comparisons between two groups were performed
using Student's unpaired two-tailed or paired t-test. Multiple
groups were analyzed using one-way ANOVA followed by Bonferroni's
post hoc test for pairwise comparisons. P<0.05 was considered to
indicate a statistically significant difference.
Results
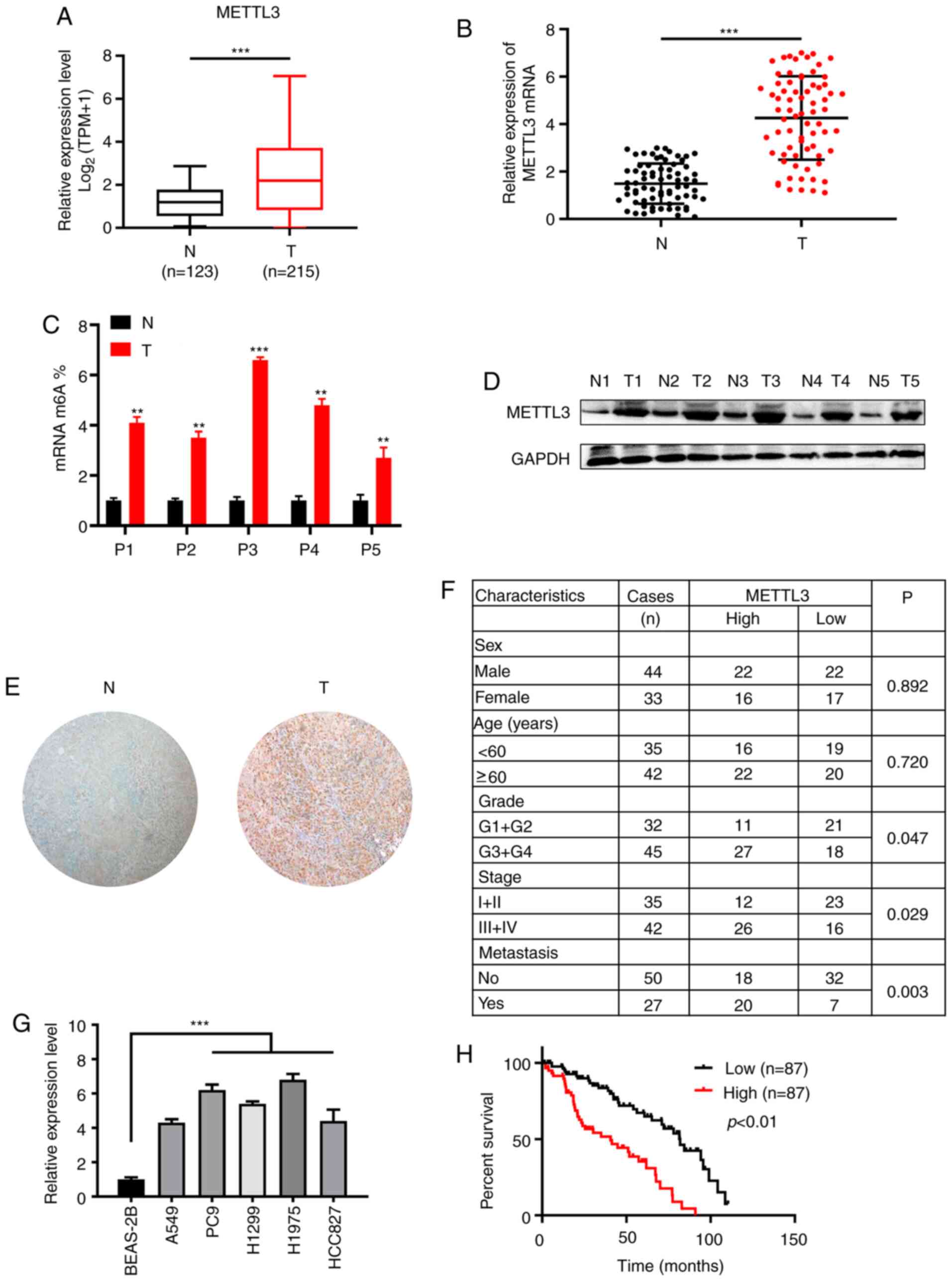
METTL3 expression is elevated in NSCLC
tissue and cells
First, it was determined that the expression of
METTL3 was upregulated in the NSCLC cohort using bioinformatics
tools (gepia.cancer-pku.cn/; Fig.
1A). In addition, RT-qPCR analysis of METTL3 mRNA expression in
samples from patients with NSCLC showed that the expression of
METTL3 was significantly higher in NSCLC tumor samples than in
normal paracancerous tissue (Fig.
1B). m6A quantitative analysis showed that the m6A content
level was significantly higher in tumor than in normal
paracancerous tissue (Fig. 1C).
Western blot and IHC analysis showed that the protein expression of
METTL3 was increased in NSCLC compared with normal paracancerous
tissue (Fig. 1D and E). Clinical and
pathological data are shown in Fig.
1F. A significant positive association between high METTL3
expression and tumor differentiation, tumor stage and metastasis
was observed. These results indicated that METTL3 served a
significant role in NSCLC development and progression.
Consistently, METTL3 expression was increased in the NSCLC cell
lines compared with the normal human lung epithelial cell line
BEAS-2B (Fig. 1G). Kaplan-Meier
survival analysis indicated that patients with tumors with high
METTL3 expression had a poor survival rate and outcome (Fig. 1H).
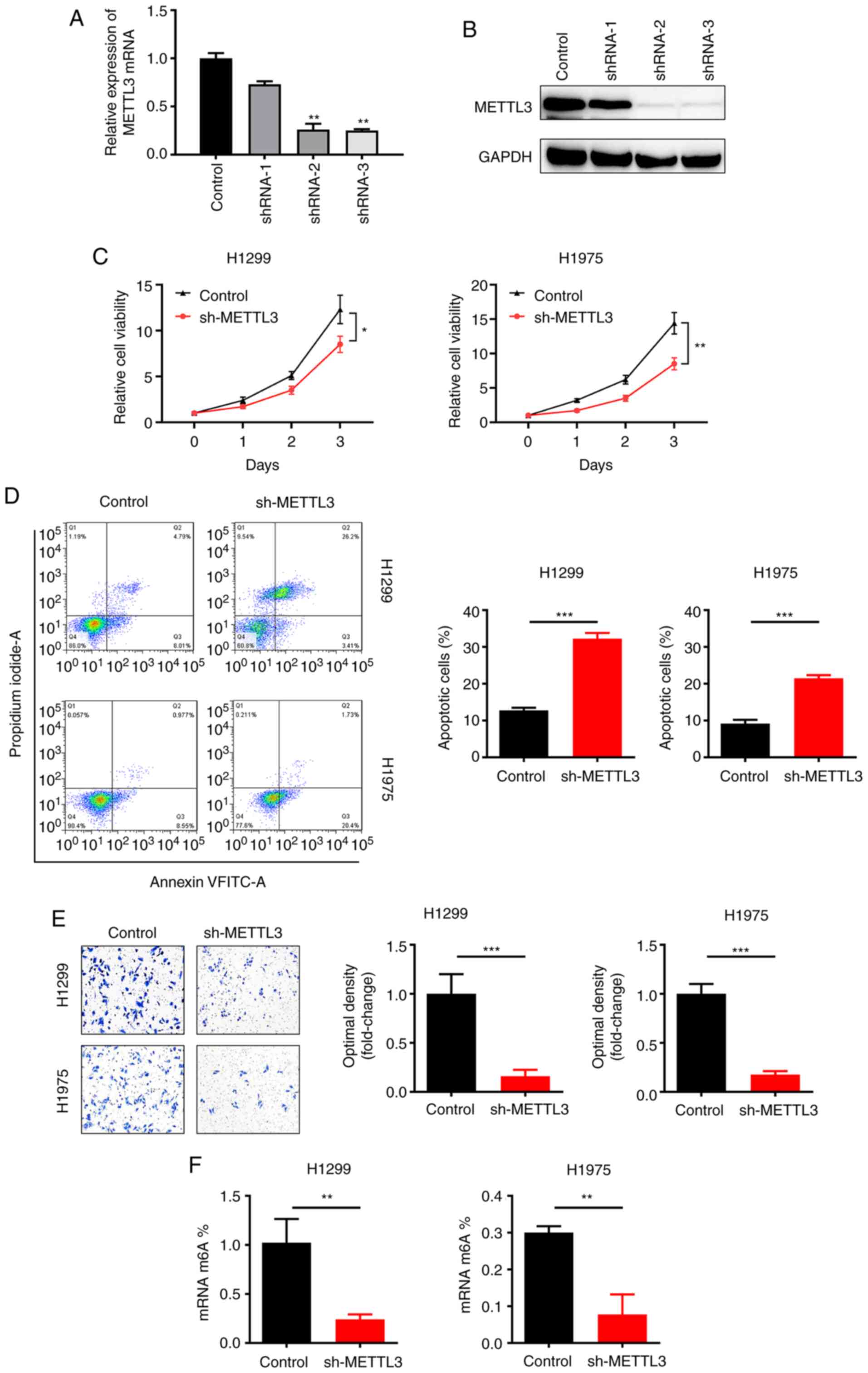
METTL3 knockdown inhibits NSCLC cell
viability and migration in vitro
In order to investigate the functional role of
METTL3 in NSCLC, METTL3 was knocked down using three independent
shRNAs (shRNA-1, shRNA-2 and shRNA-3) in an NSCLC cell line
(H1299). Efficient knockdown of METTL3 was confirmed by RT-qPCR
(Fig. 2A) and western blotting
(Fig. 2B). The CCK-8 assay results
showed significantly decreased viability of NSCLC cell lines
(H1299, H1975) after silencing METTL3 (Fig. 2C). Moreover, flow cytometry assay
revealed that METTL3 silencing accelerated apoptosis in NSCLC cell
lines (H1299, H1975; Fig. 2D).
Migration assay revealed that METTL3 silencing repressed migration
(Fig. 2E). m6A-qPCR validation showed
that the percentage of m6A content was significantly decreased when
METTL3 was knocked down in H1299 and H1975 cell lines (Fig. 2F). Collectively, these results
indicated that silencing METTL3 suppressed viability and migration
and promoted apoptosis in NSCLC cell lines.
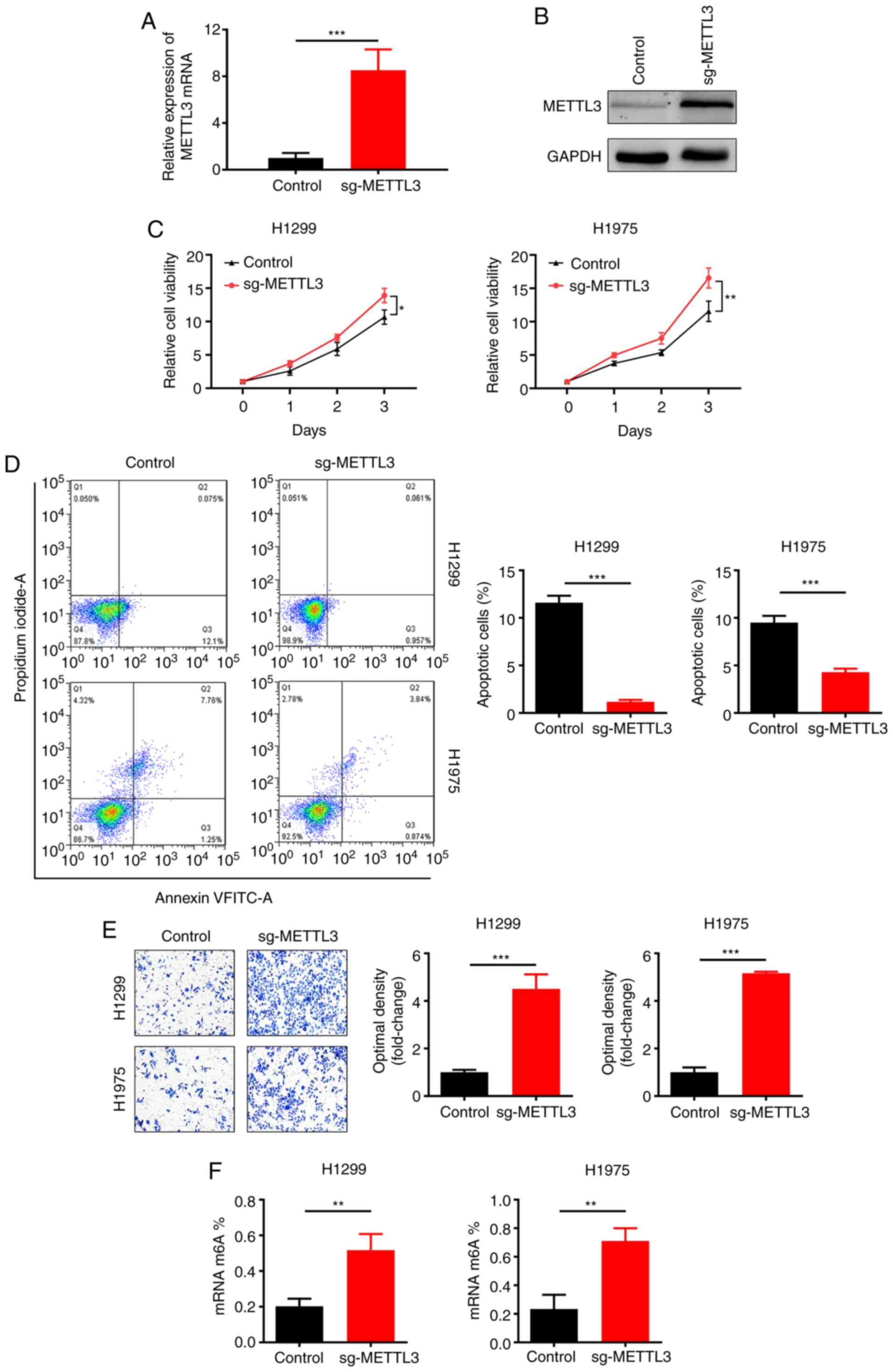
METTL3 overexpression promotes NSCLC
cell viability and migration in vitro
Next, stable METTL3-overexpressing NSCLC cell lines
(H1299 and H1975) were constructed by lentiviral infection.
Upregulation of METTL3 expression was confirmed by RT-qPCR
(Fig. 3A) and western blotting
(Fig. 3B). METTL3 overexpression
significantly promoted NSCLC cell line (H1299 and H1975) viability,
as determined by CCK-8 assay (Fig.
3C). Moreover, flow cytometry revealed that overexpression of
METTL3 inhibited the apoptosis of NSCLC cell lines (H1299 and
H1975; Fig. 3D). Furthermore, METTL3
overexpression significantly increased migration of NSCLC cell
lines in the Transwell assay (Fig.
3E). Quantitative analysis of m6A showed that METTL3
overexpression increased m6A content (Fig. 3F). Based on these results, it was
concluded that METTL3 overexpression promoted NSCLC cell viability
and migration.
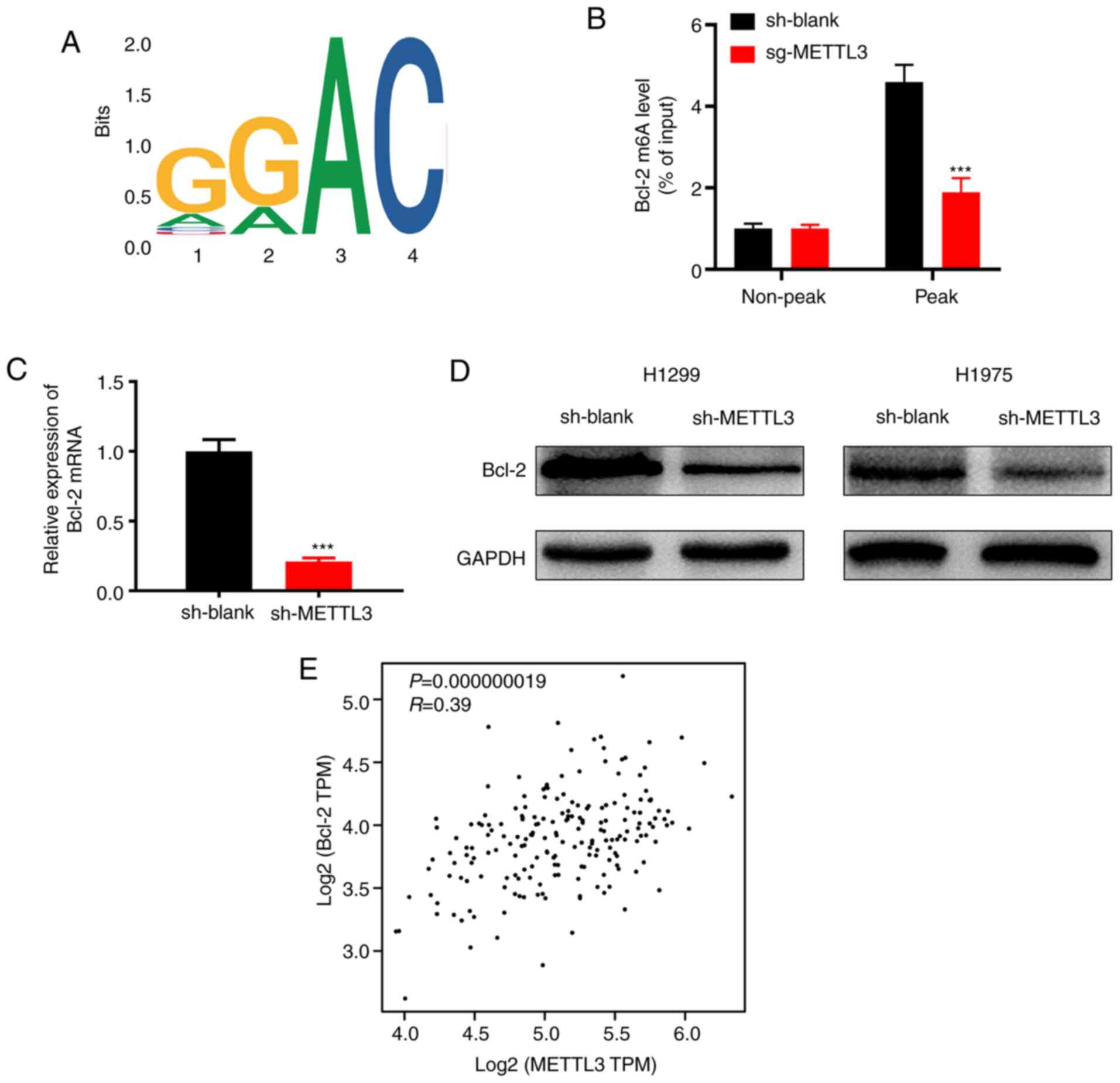
Bcl-2 is a downstream target of
METTL3
A previous study reported that Bcl-2 may be mediated
by METTL3 in breast cancer (21). In
order to identify the underlying molecular mechanisms by which
METTL3 promotes NSCLC progression, m6A-seq was performed. The
results showed that the GGAC motif was highly enriched within m6A
sites in the immunopurified RNA (Fig.
4A). In order to verify that METTL3 targets Bcl-2 mRNA for m6A
modification, ethylated RNA immunoprecipitation qPCR was performed
to validate the m6A-seq data. The results showed that METTL3
knockdown significantly decreased m6A levels of Bcl-2 mRNA
(Fig. 4B). RT-qPCR analysis showed
that Bcl-2 expression was significantly decreased at the mRNA level
following METTL3 knockdown in H1299 and H1975 cells (Fig. 4C). Bcl-2 protein expression was
significantly downregulated in METTL3-knockdown H1299 and H1975
cells (Fig. 4D). Then, the online
bioinformatics tool GEPIA database (gepia.cancer-pku.cn/) was used
for correlation analysis and our results showed that the METTL3
expression level also showed a positive correlation with Bcl-2 in
tumors from individuals with NSCLC (Fig.
4E). Thus, it was concluded that Bcl-2 was a target of
METTL3.
METTL3 facilitates NSCLC tumorigenesis
by enhancing expression of Bcl-2 in vivo
In order to confirm the potential contribution of
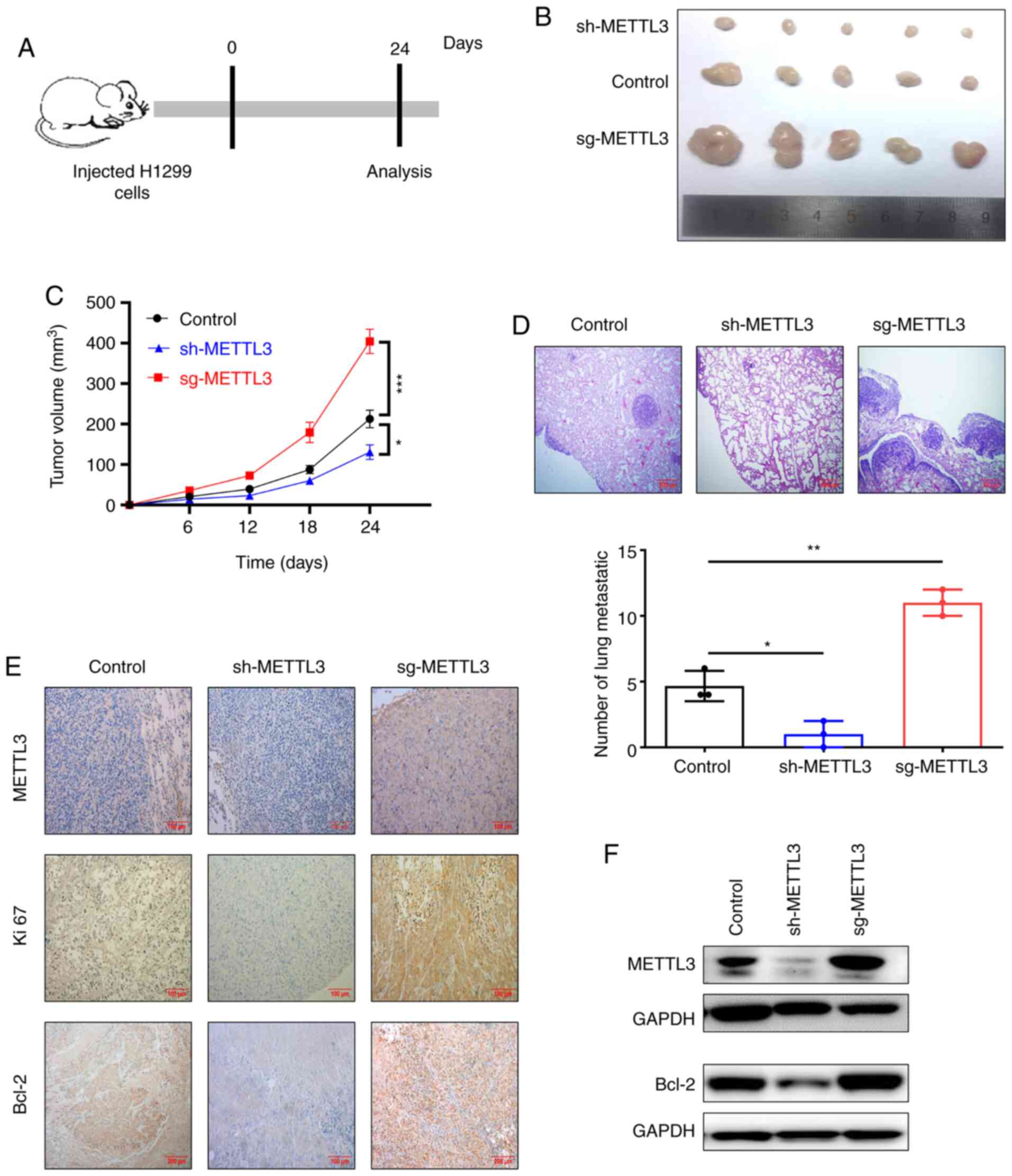
METTL3 to tumor progression in vivo, BALB/c nude mice were
subcutaneously injected with H1299 cells with METTL3 overexpression
or knockdown to develop an NSCLC xenograft model. After 24 days,
the tumors were removed, photographed and weighed (Fig. 5A and B). Tumors arising from cells
with METTL3 overexpression were significantly larger and heavier
than those arising from control cells (Fig. 5C). By contrast, knockdown of METTL3
significantly repressed tumor growth (volume) compared with that of
tumors arising from control cells (Fig.
5C). Hematoxylin-eosin staining indicated that knockdown of
METTL3 decreased the number of metastatic nodules, while
overexpression of METTL3 increased the number of lung metastatic
nodules compared with control (Fig.
5D). In order to confirm whether METTL3 promoted tumor growth
in vivo by regulating the expression of Bcl-2, IHC staining
and western blot analysis were performed. The results indicated
that METTL3 overexpression increased Bcl-2 expression and METTL3
knockdown inhibited Bcl-2 expression (Fig. 5E-F). Collectively, these results
suggested that METTL3-mediated m6A modification promoted NSCLC
progression by enhancing expression of Bcl-2 in vivo.
Discussion
Recently, the role of m6A modification in mRNA
translation has emerged as a hot spot in the field of epigenetics
(25). Emerging evidence indicates
that m6A modification serves important and diverse biological
functions in multiple cellular processes, such as RNA synthesis,
processing, translation and metabolism (26,27). m6A
modification regulates the tumorigenesis of cancers (28,29). For
example, in breast cancer tumorigenesis, m6A demethylase ALKBH5
mediates m6A modification in the 3′-UTR of NANOG mRNA (30). Therefore, more detailed work is needed
to define the role and underlying mechanisms of m6a modification in
certain types of cancer.
Certain studies have demonstrated that METTL3 serves
a regulatory role in a variety of cancer types, such as breast,
bladder and lung cancer, as well as hepatocellular carcinoma
(31–35). For example, METTL3 can act as an
oncogene in bladder tumorigenesis (36); however, METTL3 and METTL14 also serve
as tumor suppressors in hepatocellular carcinoma (37,38).
Certain studies have reported that METTL3 contributes to
transforming growth factor-β-induced epithelial-mesenchymal
transition of lung cancer cells (39). An additional study revealed that there
is a significant correlation between METTL3 expression and the
occurrence of chronic obstructive pulmonary disease (40). However, the role of METTL3-mediated
m6A modification in NSCLC initiation and progression is not fully
known. The present study demonstrated that METTL3 expression was
significantly upregulated in NSCLC. The role of METTL3 in promoting
NSCLC progression was demonstrated both in vitro and in
vivo. Bcl-2 was shown to be a direct target of METTL3-mediated
m6A modification in NSCLC. Finally, the present findings shed new
light on m6A methylation-mediated Bcl-2 overexpression during
progression of NSCLC and identified that METTL3 serves an oncogenic
role as an m6A ‘writer’ in NSCLC. Thus, METTL3 is a promising
therapeutic target for NSCLC treatment.
A recent study indicated that METTL3 regulates
expression of various mRNAs in bladder cancer, including MYC, NF-κB
and AF4/FMR2 family member 4 (41).
Hence, it was hypothesized that METTL3 may promote the
translational efficiency of Bcl-2 mRNA to regulate NSCLC
progression. Similarly, in breast cancer, METTL3-mediated m6A
modification of Bcl-2 promotes its expression (27). Furthermore, METTL3 and Bcl-2
expression levels were positively correlated. Certain studies have
revealed that METTL3 overexpression directly mediates m6A
methylation and inhibits apoptosis in various types of cancer
(42,43). For example, overexpression of Bcl-2
blocks adriamycin-induced apoptosis in bladder cancer cells
(44). Another report revealed that
high Bcl-2 expression is associated with recurrence and poor
survival in gastric cancer (45–46).
Taken together, the results of the present study
highlight the critical role of METTL3 in NSCLC progression. METTL3
regulated cellular growth, survival and migration in NSCLC. METTL3
promoted NSCLC progression by modulating the level of Bcl-2. This
finding may contribute to understanding of the oncogenic role of
METTL3, which may be an effective therapeutic target for NSCLC.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Henan
Provincial Medical Science and Technology Research Joint
Co-Construction Project (grant no. LHGJ20190485).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the Sequence Read Archive
repository, accession no. PRJNA721237.
Authors' contributions
The project was designed and conceived by YZ and CD.
SL, YZ and TZ analyzed the data. YZ wrote the manuscript. YZ and CD
guided the study and revised the manuscript. YZ, SL, TZ and CD
confirm the authenticity of all the raw data. All authors read and
approved the final version of the manuscript.
Ethics approval and consent to
participate
Ethics approval was granted by the Ethics Committee
of the First Affiliated Hospital of Xinxiang Medical University
(approval no. KN201808002). Oral consent was obtained from
patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Neal RD, Sun F, Emery JD and Callister ME:
Lung cancer. BMJ. 365:l17252019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Herbst RS, Morgensztern D and Boshoff C:
The biology and management of nonsnon-small cell lung cancer.
Nature. 553:446–454. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Reck M and Rabe KF: Precision diagnosis
and treatment for advanced non-small-cell lung cancer. N Engl J
Med. 377:849–861. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tandberg DJ, Tong BC, Ackerson BG and
Kelsey CR: Surgery versus stereotactic body radiation therapy for
stage I non-small cell lung cancer: A comprehensive review. Cancer.
124:667–678. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wu SG and Shih JY: Management of acquired
resistance to EGFR TKI-targeted therapy in advanced non-small cell
lung cancer. Mol Cancer. 17:382018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu WJ, Du Y, Wen R, Yang M and Xu J: Drug
resistance to targeted therapeutic strategies in non-small cell
lung cancer. Pharmacol Ther. 206:1074382020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dai D, Wang H, Zhu L, Jin H and Wang X:
N6-methyladenosine links RNA metabolism to cancer progression. Cell
Death Dis. 9:1242018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Maity A and Das B: N6-methyladenosine
modification in mRNA: Machinery, function and implications for
health and diseases. FEBS J. 283:1607–1630. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
He L, Li H, Wu A, Peng Y, Shu G and Yin G:
Functions of N6-methyladenosine and its role in cancer. Mol Cancer.
18:1762019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR
and Qian SB: Dynamic m(6)A mRNA methylation directs translational
control of heat shock response. Nature. 526:591–594. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang
L, Jia G, Yu M, Lu Z, Deng X, et al: A METTL3-METTL14 complex
mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem
Biol. 10:93–95. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ping XL, Sun BF, Wang L, Xiao W, Yang X,
Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, et al: Mammalian WTAP is
a regulatory subunit of the RNA N6-methyladenosine
methyltransferase. Cell Res. 24:177–189. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Roundtree IA, Evans ME, Pan T and He C:
Dynamic RNA modifications in gene expression regulation. Cell.
169:1187–1200. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sun T, Wu R and Ming L: The role of m6A
RNA methylation in cancer. Biomed Pharmacother. 112:1086132019.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lin S, Choe J, Du P, Triboulet R and
Gregory RI: The m(6)A Methyltransferase METTL3 promotes translation
in human cancer cells. Mol Cell. 62:335–345. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Peng W, Li J, Chen R, Gu Q, Yang P, Qian
W, Ji D, Wang Q, Zhang Z, Tang J and Sun Y: Upregulated METTL3
promotes metastasis of colorectal Cancer via miR-1246/SPRED2/MAPK
signaling pathway. J Exp Clin Cancer Res. 38:3932019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang Q, Chen C, Ding Q, Zhao Y, Wang Z,
Chen J, Jiang Z, Zhang Y, Xu G, Zhang J, et al: METTL3-mediated
m6A modification of HDGF mRNA promotes gastric cancer
progression and has prognostic significance. Gut. 69:1193–1205.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen M, Wei L, Law CT, Tsang FH, Shen J,
Cheng CL, Tsang LH, Ho DW, Chiu DK, Lee JM, et al: RNA
N6-methyladenosine methyltransferase-like 3 promotes liver cancer
progression through YTHDF2-dependent posttranscriptional silencing
of SOCS2. Hepatology. 67:2254–2270. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tao Z, Zhao Y and Chen X: Role of
methyltransferase-like enzyme 3 and methyltransferase-like enzyme
14 in urological cancers. PeerJ. 8:e95892020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Travis WD, Brambilla E, Burke AP, Marx A
and Nicholson AG: Introduction to the 2015 world health
organization classification of tumors of the lung, pleura, thymus
and heart. J Thorac Oncol. 10:1240–1242. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ni WJ and Leng XM: Down-regulated miR-495
can target programmed cell death 10 in ankylosing spondylitis. Mol
Med. 26:502020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang S, Zhao BS, Zhou A, Lin K, Zheng S,
Lu Z, Chen Y, Sulman EP, Xie K, Bögler O, et al: m6A
Demethylase ALKBH5 maintains tumorigenicity of glioblastoma
stem-like cells by sustaining FOXM1 expression and cell
proliferation program. Cancer Cell. 31:591–606.e6. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dominissini D, Moshitch-Moshkovitz S,
Salmon-Divon M, Amariglio N and Rechavi G: Transcriptome-wide
mapping of N(6)-methyladenosine by m(6)A-seq based on
immunocapturing and massively parallel sequencing. Nat Protoc.
8:176–189. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zaccara S, Ries RJ and Jaffrey SR:
Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell
Biol. 20:608–624. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Niu Y, Lin Z, Wan A, Chen H, Liang H, Sun
L, Wang Y, Li X, Xiong XF, Wei B, et al: RNA N6-methyladenosine
demethylase FTO promotes breast tumor progression through
inhibiting BNIP3. Mol Cancer. 18:462019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang H, Xu B and Shi J: N6-methyladenosine
METTL3 promotes the breast cancer progression via targeting Bcl-2.
Gene. 22:1440762020. View Article : Google Scholar
|
|
28
|
Zhang H, Shi X, Huang T, Zhao X, Chen W,
Gu N and Zhang R: Dynamic landscape and evolution of m6A
methylation in human. Nucleic Acids Res. 48:6251–6264. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liang Z, Kidwell RL, Deng H and Xie Q:
Epigenetic N6-methyladenosine modification of RNA and DNA regulates
cancer. Cancer Biol Med. 17:9–19. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang C, Samanta D, Lu H, Bullen JW, Zhang
H, Chen I, He X and Semenza GL: Hypoxia induces the breast cancer
stem cell phenotype by HIF-dependent and ALKBH5-mediated
m6A-demethylation of NANOG mRNA. Proc Natl Acad Sci USA.
113:E2047–E2056. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu ZX, Li LM, Sun HL and Liu SM: Link
Between m6A modification and cancers. Front Bioeng Biotechnol.
6:892018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zeng C, Huang W, Li Y and Weng H: Roles of
METTL3 in cancer: Mechanisms and therapeutic targeting. J Hematol
Oncol. 13:1172020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cai X, Wang X, Cao C, Gao Y, Zhang S, Yang
Z, Liu Y, Zhang X, Zhang W and Ye L: HBXIP-elevated
methyltransferase METTL3 promotes the progression of breast cancer
via inhibiting tumor suppressor let-7g. Cancer Lett. 415:11–19.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lin Y, Wei X, Jian Z and Zhang X: METTL3
expression is associated with glycolysis metabolism and sensitivity
to glycolytic stress in hepatocellular carcinoma. Cancer Med.
9:2859–2867. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang H, Deng Q, Lv Z, Ling Y, Hou X, Chen
Z, Dinglin X, Ma S, Li D, Wu Y, et al: N6-methyladenosine induced
miR-143-3p promotes the brain metastasis of lung cancer via
regulation of VASH1. Mol Cancer. 18:1812019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jin H, Ying X, Que B, Wang X, Chao Y,
Zhang H, Yuan Z, Qi D, Lin S, Min W, et al:
N6-methyladenosine modification of ITGA6 mRNA promotes
the development and progression of bladder cancer. EBioMedicine.
47:195–207. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ma JZ, Yang F, Zhou CC, Liu F, Yuan JH,
Wang F, Wang TT, Xu QG, Zhou WP and Sun SH: METTL14 suppresses the
metastatic potential of hepatocellular carcinoma by modulating
N6-methyladenosine-dependent primary MicroRNA
processing. Hepatology. 65:529–543. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liu X, Qin J, Gao T, Li C, Chen X, Zeng K,
Xu M, He B, Pan B, Xu X, et al: Analysis of METTL3 and METTL14 in
hepatocellular carcinoma. Aging (Albany NY). 12:21638–21659. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wanna-Udom S, Terashima M, Lyu H, Ishimura
A, Takino T, Sakari M, Tsukahara T and Suzuki T: The m6A
methyltransferase METTL3 contributes to Transforming Growth
Factor-beta-induced epithelial-mesenchymal transition of lung
cancer cells through the regulation of JUNB. Biochem Biophys Res
Commun. 524:150–155. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Huang X, Lv D, Yang X, Li M and Zhang H:
m6A RNA methylation regulators could contribute to the occurrence
of chronic obstructive pulmonary disease. J Cell Mol Med.
24:12706–12715. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Cheng M, Sheng L, Gao Q, Xiong Q, Zhang H,
Wu M, Liang Y, Zhu F, Zhang Y, Zhang X, et al: The m6A
methyltransferase METTL3 promotes bladder cancer progression via
AFF4/NF-κB/MYC signaling network. Oncogene. 38:3667–3680. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yang Z and Jiang X, Li D and Jiang X:
HBXIP promotes gastric cancer via METTL3-mediated MYC mRNA m6A
modification. Aging (Albany NY). 12:24967–24982. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Xiang S, Liang X, Yin S, Liu J and Xiang
Z: N6-methyladenosine methyltransferase METTL3 promotes colorectal
cancer cell proliferation through enhancing MYC expression. Am J
Transl Res. 12:1789–1806. 2020.PubMed/NCBI
|
|
44
|
Chui K and Zhang Z: Bcl-2 overexpression
inhibits generation of intracellular reactive oxygen species and
blocks adriamycin-induced apoptosis in bladder cancer cells. Asian
Pac J Cancer Prev. 14:895–901. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Xu G and Tang X: Troxerutin (TXN)
potentiated 5-Fluorouracil (5-Fu) treatment of human gastric cancer
through suppressing STAT3/NF-κB and Bcl-2 signaling pathways.
Biomed Pharmacother. 92:95–107. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kim R, Emi M, Tanabe K and Toge T:
Therapeutic potential of antisense Bcl-2 as a chemosensitizer for
cancer therapy. Cancer. 101:2491–502. 2004. View Article : Google Scholar : PubMed/NCBI
|