Introduction
Colon cancer is one of the most common malignant
tumors (1–3). Every year, there are >945,000 new
cases of colon cancer worldwide, and ~492,000 deaths (1–4). Although
there have been numerous improvements in tumor therapy, colon
cancer remains one of the leading causes of cancer-associated
deaths in the world (1,2). In patients with colon cancer, metastasis
is the main cause of mortality (5).
It is a huge challenge for both clinicians and researchers to find
novel therapeutic approaches for colon cancer (6).
Early growth response gene 1 (Egr-1) is a
transcription factor that serves important roles in tumor
processes, including cell proliferation, migration, invasion,
differentiation and apoptosis (7–10). Egr-1
expression is dysregulated in several types of tumor, such as
leukemia (11), pancreatic tumor
(12), gastric tumor (9), thyroid tumor (13), liver cancer (14) and glioma (15), and is a positive prognostic factor for
the survival of patients with several types of cancer, such as
gastric cancer (16); therefore,
Egr-1 is suggested to be a new biomarker and a promising
therapeutic target (17). However,
the function of Egr-1 in the progression of colon cancer is not
fully understood.
Egr-1 is a ubiquitous transcription factor and
regulates various target genes by recognizing and binding GC-rich
regions in the promoter region of target genes (18–20). Egr-1
can exert both positive and negative transcription roles in
regulating different target genes (21). The dual characters of Egr-1 in
regulating target genes may be due to the co-factors recruited by
Egr-1. Egr-1 interacts with transcriptional coactivators, such as
cyclic AMP response element-binding protein/p300, to trigger the
transcription of several genes (22,23), while
by binding with histone deacetylases, Egr-1 also serves an
inhibitory role in regulating target genes (24).
Cyclin-dependent kinase-like 1 (CDKL1), located on
chromosome 14q21.3, is a member of the CDKL kinase family (25,26). CDKL1
expression is upregulated in breast cancer (27), gastric cancer (28), melanoma (29) and colon cancer (30). Moreover, knockdown of CDKL1 in these
types of cancer induces apoptosis and/or inhibits cell
proliferation, migration and invasion (27–30).
Therefore, CDKL1 may serve as a promising therapeutic target;
however, the mechanism by which CDKL1 expression is regulated is
unknown. To the best of our knowledge, there is no report on the
role of CDKL1 in colon cancer progression. Therefore, the present
study aimed to investigate whether Egr-1 regulated CDKL1 expression
and the role of Egr-1-regulated CDKL1 in colon cancer
progression.
The present study aimed to illustrate the function
and the underlying mechanism of Egr-1-mediated CDKL1 regulation in
colon cancer cell proliferation, migration and invasion, and to
verify whether Egr-1/CDKL1 may be promising prognostic biomarkers
and therapeutic targets for colon cancer.
Materials and methods
Survival analysis
The association of Egr-1 expression and the survival
of 279 patients with colon adenocarcinoma was analyzed using The
Cancer Genome Atlas (TCGA) in the UALCAN website (http://ualcan.path.uab.edu/), which is a
comprehensive, user-friendly and interactive web resource for
analyzing cancer OMICS data. ‘TCGA Gene Analysis’ was chosen and
‘Egr-1’ was scanned in TCGA dataset of colon adenocarcinoma.
Samples were categorized into two groups: High expression (with TPM
values above upper quartile) and Low/Medium expression (with TPM
values below upper quartile) (31).
The survival curve of the effect of Egr-1 expression on the
survival of patients with colon adenocarcinoma was then presented.
The Kaplan-Meier method was used for survival analysis.
Cell lines
Since colon adenocarcinoma and colon carcinoma are
the two main types of colon cancer, the present study used two cell
lines, namely SW480 (a colon adenocarcinoma cell line) and HCT-116
(a colon carcinoma cell line). SW480 and HCT-116 cells were
purchased from the American Type Culture Collection (ATCC) and were
cultured in DMEM containing 10% FBS (both Gibco; Thermo Fisher
Scientific, Inc.) at 37°C in an incubator with 5%
CO2.
Egr-1-knockdown in colon cancer
cells
The lentivirus (Lv)-shEgr1 was constructed by
Shanghai GeneChem Co., Ltd., using a 3rd generation packing system.
A total of 2 µg of the packing vector containing short hairpin RNA
with the sequence 5′-GGCATACCAAGATCCACTT-3′ targeting Egr-1 and 4
µg envelope plasmids mix were transfected into 293 cells (ATCC)
using Lipofectamine® 2000 reagent (Invitrogen; Thermo
Fisher Scientific, Inc.). After the cells were cultured at 37°C for
48 h, the lentivirus particles were collected and concentrated
using ultracentrifugation at 100,000 × g at 4°C for 30 min.
Egr-1-knockdown was performed by infecting the Lv-shEgr1 at MOI of
10 into SW480 and HCT-116 cells. Briefly, cells were cultured in a
6-well plate, until the confluence of the cells reached 80%.
Lv-shEgr1 was added into the medium. After 24 h of transduction,
the medium containing Lv-shEgr1 was replaced with a medium without
lentivirus and cultured for 48 h at 37°C. Cells infected with
Lv-shEgr1 were selected with 10 µg/µl puromycin.
Overexpression of Egr-1 or CDKL1 in
colon cancer cells
Lv-oeEgr1 was constructed by Shanghai GeneChem Co.,
Ltd., using a 3rd generation packing system. A total of 2 µg of the
packing vector containing the Egr-1 coding sequence and 4 µg
envelope plasmids mix were transfected into 293 cells using
Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). After the cells were cultured at 37°C for 48 h,
the lentivirus particles were collected and concentrated using
ultracentrifugation at 100,000 × g at 4°C for 30 min.
Overexpression of Egr-1 was performed by infecting the lentivirus
Lv-oeEgr1 at MOI of 10 into SW480 and HCT-116 cells. Briefly, cells
were cultured in a 6-well plate, until the confluence of the cells
reached 80%, Lv-oeEgr1 was added into the medium. After 24 h of
transduction, the medium containing Lv-oeEgr1 was replaced with a
medium without lentivirus, and cultured for 48 h culture at 37°C.
Cells infected with Lv-oeEgr1 were selected with 10 µg/µl
puromycin.
Overexpression of CDKL1 was performed with the same
procedure as overexpression of Egr-1, except that the lentivirus
was Lv-CDKL1 containing the coding region of CDKL1.
Western blot assay
Proteins were extracted from SW480 and HCT-116 cells
using a RIPA lysis buffer kit (EMD Millipore). The protein
concentration was measured using a Pierce Rapid Gold BCA kit
(Pierce; Thermo Fisher Scientific, Inc.). A total of 20 µg
protein/lane was subjected to 10% SDS-PAGE to separate the proteins
according to their molecular weight, and the proteins were then
transferred to a nitrocellulose membrane (EMD Millipore). After
blocking with 5% skimmed milk at 20°C for 30 min, the membrane was
incubated with primary antibody for 16 h at 4°C, followed by
incubation with peroxidase-conjugated secondary antibody (1:10,000;
cat. no. A0208; Beyotime Institute of Biotechnology) at 20°C for 1
h. The membrane was finally incubated in ECL reagents (EMD
Millipore) and visualized using a chemiluminescence imager (Bio-Rad
Laboratories, Inc.). The Egr-1 primary antibody was purchased from
Cell Signaling Technology, Inc. (cat. no. 4154), while the CDKL1
(cat. no. ab136129) and β-actin (cat. no. ab8227) primary
antibodies were purchased from Abcam. All the primary antibodies
were diluted at 1:1,000 in 5% skimmed milk.
Reverse transcription-quantitative
(RT-q)PCR
RNA was extracted from cells using TRzol reagent
(cat. no. RA101-01; Beijing Biomed Gene Technology Co., Ltd.)
according to the manufacturer's protocol. An
ultraviolet-spectrophotometer (Thermo Fisher Scientific, Inc.) was
used to measure the mRNA concentration. mRNA (2 µg) was reverse
transcribed into cDNA using a First Strand cDNA Synthesis kit (cat.
no. K1622; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. qPCR was performed in a PCR instrument
(Applied Biosystems; Thermo Fisher Scientific, Inc.) using a SYBR
Green Master Mix Reagent (Thermo Fisher Scientific, Inc.). β-actin
was used as the reference gene. The thermocycling conditions were
as follows: 30 sec denaturation at 95°C, followed by 40 cycles of
denaturation at 95°C for 15 sec and annealing and extension at 60°C
for 1 min. The primers used were as follows: Egr-1 sense,
5′-GAACAACCCTACGAGCACCTG−3′ and antisense,
5′-GCCACAAAGTGTTGCCACTG-3′; CDKL1 sense,
5′-CGAATGCTCAAGCAACTCAAGC-3′ and antisense,
5′-GCCAAGTTATGCTCTTCACGAG-3′; and β-actin sense,
5′-GTGGACATCCGCAAAGAC−3′ and antisense, 5′-AAAGGGTGTAACGCAACTA-3′.
The data were analyzed using the 2−ΔΔCq relative
expression method (32).
Construction of CDKL1
promoter-luciferase reporter
The human CDKL1 promoter sequence was obtained from
GenBank (www.ncbi.nlm.nih.gov/gene/). CDKL1 promoter (−1,500 bp
to +100 bp, the translational starting site as +1) was amplified
from the genomic DNA of SW480 cells using a high-fidelity DNA
polymerase (New England BioLabs, Inc.) using the following
conditions: 30 sec denaturation at 95°C, followed by 35 cycles of
denaturation at 95°C for 15 sec and annealing and extension at 65°C
for 1.5 min. The primers used were as follows: Sense,
5′-CCAAGCTTGTAGAGGAAAGTGCTGACTTT-3′ and antisense,
5′-CCTCGAGGTGTCCCTGTTTCTACATTTGA-3. After PCR amplification, the
product was cut with HindIII and XhoI restriction
enzymes (New England BioLabs, Inc.) and inserted into the
pGL3-Basic plasmid (Promega Corporation) between the HindIII
and XhoI sites. The promoter-luciferase reporter was named
pGL3-CDKL1.
Luciferase assay
SW480 or HCT-116 cells transfected with shCon,
shEgr-1, oeCon or oeEgr-1 were seeded into a 12-well plate at
20,000 cells/well. After 24 h of culture, the pGL3-CDKL1 plasmid (1
µg) was transfected into cells using Lipofectamine® 2000
reagent (Invitrogen; Thermo Fisher Scientific, Inc.). After another
24 h of culture following transfection, cells were lysed and mixed
with the luciferin substrate of a Luciferase Assay System kit (cat.
no. E1500; Promega Corporation); luciferase activity was determined
with a luminometer (Thermo Fisher Scientific, Inc.).
Chromatin immunoprecipitation (ChIP)
assay
The ChIP assay was performed using a ChIP assay kit
(Beyotime Institute of Biotechnology) following the manufacturer's
protocol. Briefly, SW480 or HCT-116 cells were cross-linked by
being incubated at 25°C for 10 min in a thermostatic incubator
(Thermo Fisher Scientific, Inc.), and then the cross-link was
stopped by glycin. The genomic DNA was sheared into fragments of
200–1,000 bp by 300 W ultra-sonication at 4°C for 5 min. A total of
500 µl cell lysate per IP reaction was incubated with 10 µg
anti-Egr-1 antibody (cat. no. 4154; Cell Signaling Technology,
Inc.) or anti-Flag antibody (cat. no. 14793; Cell Signaling
Technology, Inc.) for 16 h at −4°C, followed by incubation with
Protein A/G MagBeads (Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocol. After the cross-linking was unfastened
with 0.2 µM NaCl at 65°C for 6 h, the DNA was subjected to PCR with
a Taq DNA Polymerase (Beyotime Institute of Biotechnology) using
the following conditions: 30 sec denaturation at 95°C, followed by
35 cycles of denaturation at 95°C for 15 sec, annealing at 55°C for
1 min and extension at 72°C for 1 min. The primers for amplifying
the fragments of CDKL1 promoter were: Sense,
5′-GCAAATCTAGCAGGTCTGC-3′ and antisense,
5′-GTTCTAATTTCGGGTCTCA-3′.
Transwell assay
For migration, 50,000 cells were seeded into the
upper chambers containing serum-free DMEM. The lower reservoir
contained DMEM with 15% FBS. After 10 h of culture at 37°C, the
cells in the upper chambers were removed, and the cells that
crossed the membrane were stained with crystal violet reagent
(Beyotime Institute of Biotechnology) at 25°C for 5 min. The
stained cells were observed under a light microscope (Olympus
Corporation) at ×200 magnification and counted in six randomly
selected visual fields.
The invasion assays were performed using the same
procedures as the migration assays, except that the upper surface
of the membrane was precoated with 20 µg Matrigel (Sigma-Aldrich;
Merck KGaA) at 37°C for 2 h and the number of seeded cells was
100,000 cells/chamber.
Cell Counting Kit-8 (CCK-8) cell
proliferation assay
Cells were seeded into 96-well plates at 5,000
cells/well. After 0, 24 and 48 h of culture at 37°C, 10 µl CCK-8
reagent (Dojindo Molecular Technologies, Inc.) was added into each
well, and the plate was incubated at 37°C for 2 h. The absorbance
was measured at 450 nm using a microplate reader (Thermo Fisher
Scientific, Inc.).
Statistical analysis
Data were analyzed with SPSS software version 19.0
(IBM Corp.). The log-rank test was used to analyze the association
between Egr-1 expression and patient survival. Data of qPCR,
Transwell, CCK-8 and luciferase assays were repeated three times
and presented as the mean ± SD, and differences were analyzed by
one-way ANOVA followed by Tukey's post-hoc test. P<0.05 was
considered to indicate a statistically significant difference.
Results
Association of Egr-1 expression and
the survival of patients with colon adenocarcinoma
UALCAN is a comprehensive, user-friendly and
interactive web resource for analyzing cancer OMICS data (31). Based on UALCAN, the present study
analyzed the association of Egr-1 expression with the survival of
patients with colon adenocarcinoma, which is one of the main types
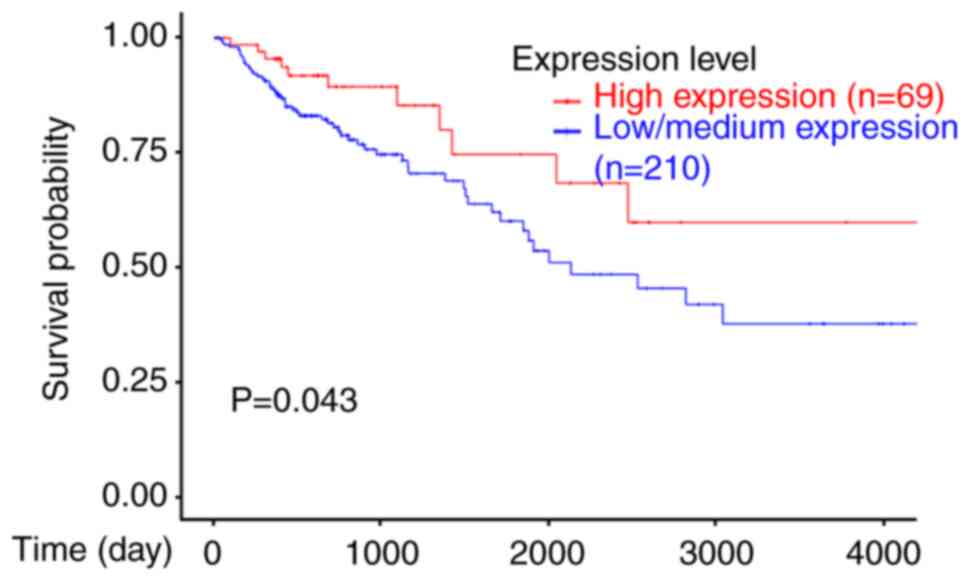
of colon cancer. As shown in Fig. 1,
patients with higher Egr-1 expression exhibited an improved
survival rate than patients with lower Egr-1 expression. These data
indicated that Egr-1 may function as a prognostic biomarker for
improved survival of patients with colon adenocarcinoma. Since
there is no data about colon carcinoma, another main type of colon
cancer, the association of Egr-1 expression with the survival of
patients with colon carcinoma was not analyzed.
Knockdown of Egr-1 induces colon
cancer cell proliferation, migration and invasion
Egr-1-knockdown was achieved by infecting SW480 and
HCT-116 cells with shEgr-1 lentivirus or shCon lentivirus used as
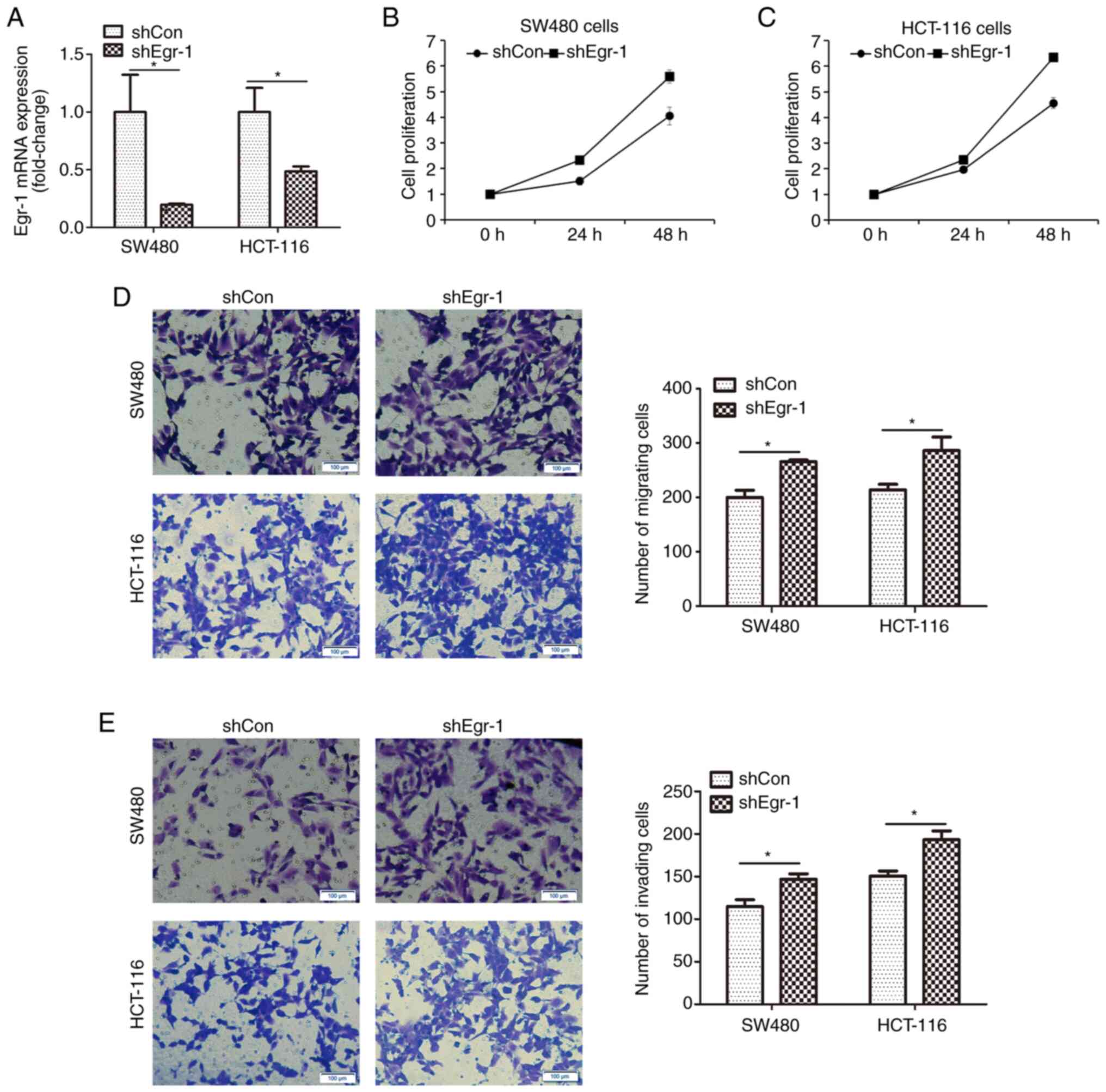
the negative control. Fig. 2A
indicates that after infection, Egr-1 mRNA expression was
significantly decreased in both cell lines, indicating that Egr-1
was successfully knocked down. Cells with or without
Egr-1-knockdown were subjected to CCK-8 assay. As shown in Fig. 2B and C, knockdown of Egr-1 increased
the proliferation of SW480 and HCT-116 cells. Furthermore, cell
migration and invasion were measured by Transwell assay. As shown
in Fig. 2D, the migratory capacity of
both SW480 and HCT-116 cells was significantly improved by
Egr-1-knockdown; similarly, the invasive capacity of both cell
lines was significantly enhanced (Fig.
2E). These results indicated the inhibitory role of Egr-1 in
colon cancer progression.
Egr-1 negatively regulates CDKL1
expression
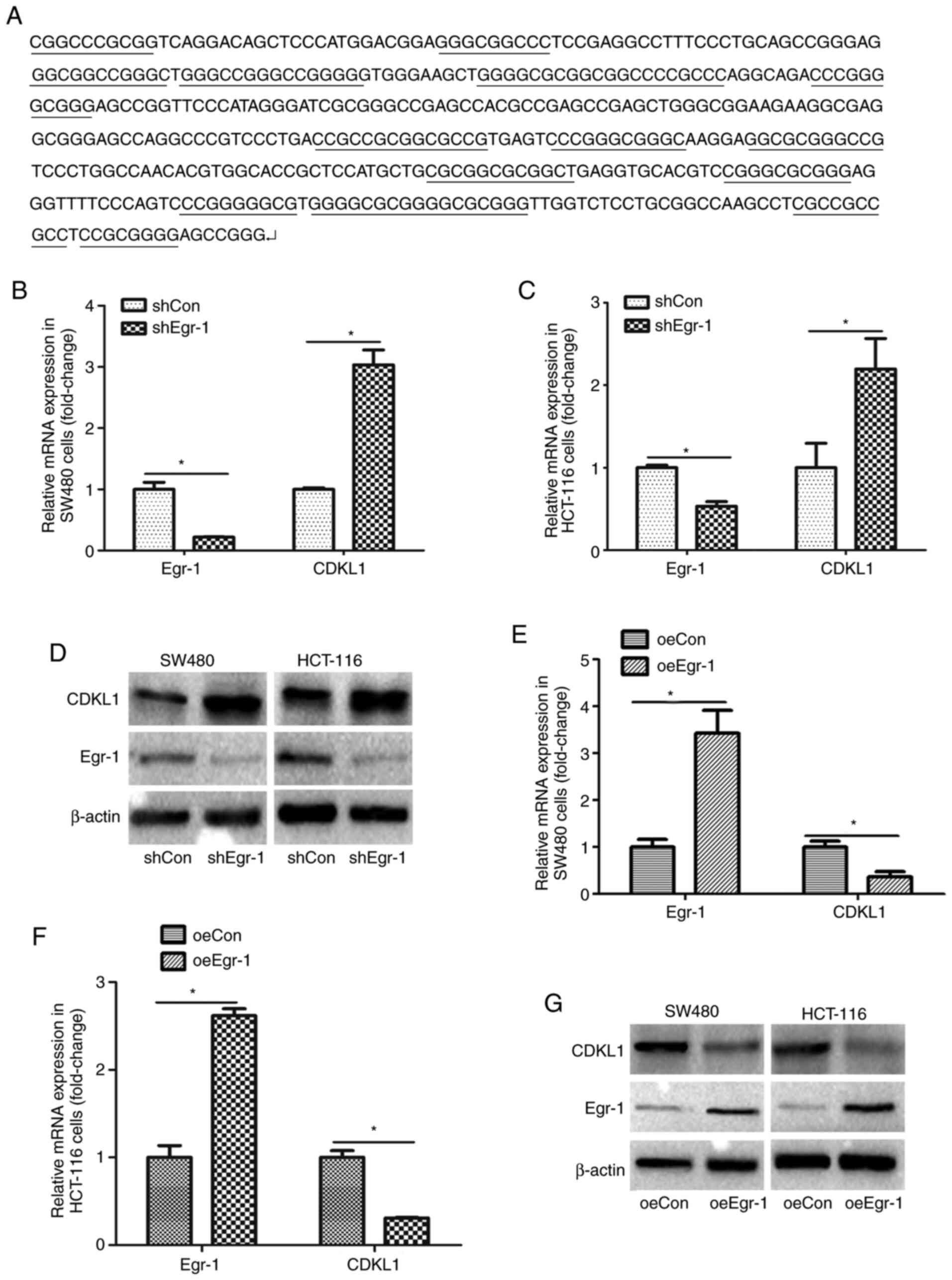
The CDKL1 promoter sequence was obtained from
GenBank (www.ncbi.nlm.nih.gov/gene/). In some regions of the
CDKL1 promoter, the content of G and C reached >80%, and there
were several segments containing continuous GC, indicating that
CDKL1 possessed a GC-rich promoter (Fig.
3A). Therefore, it was hypothesized that CDKL1 may be a target
gene of Egr-1. CDKL1 expression was first analyzed following
Egr-1-knockdown. The mRNA of SW480 and HCT-116 cells was extracted,
and CDKL1 expression was measured using RT-qPCR. As shown in
Fig. 3B and C, CDKL1 mRNA expression
was significantly higher in cells with Egr-1-knockdown.
Additionally, western blot analysis indicated that CDKL1 protein
expression was upregulated following Egr-1-knockdown (Fig. 3D). Furthermore, CDKL1 expression was
detected under Egr-1 overexpression in SW480 and HCT-116 cells. As
shown in Fig. 3E-G, both CDKL1 mRNA
and protein expression was decreased when Egr-1 was overexpressed.
These results indicated that CDKL1 was negatively regulated by
Egr-1.
Egr-1 regulates colon cancer cell
proliferation, migration and invasion through CDKL1
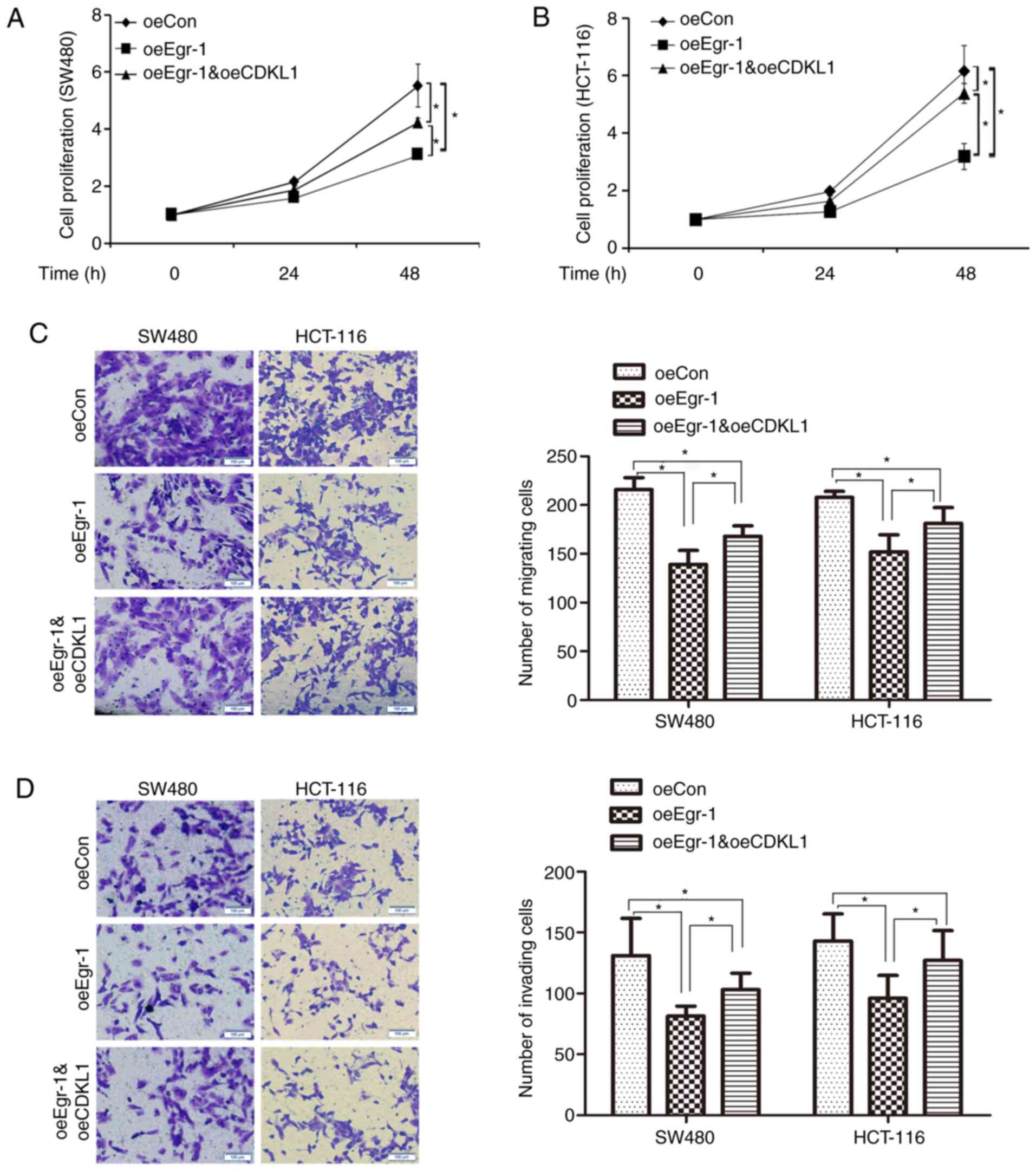
As CDKL1 was identified as a target gene of Egr-1,
whether Egr-1 regulated colon cancer cell proliferation, migration
and invasion through CDKL1 was further analyzed. CDKL1 was
overexpressed in colon cancer cells with Egr-1 overexpression, and
cell proliferation, migration and invasion were measured. As shown
in Fig. 4A and B, Egr-1
overexpression significantly inhibited the proliferation of SW480
and HCT-116 cells, while the inhibitory role of Egr-1 was reversed
by CDKL1 overexpression. As shown in Fig.
4C and D, Egr-1 overexpression inhibited the migration and
invasion of SW480 and HCT-116 cells, while the inhibitory role of
Egr-1 was reversed by CDKL1 overexpression. Therefore, Egr-1 may
inhibit colon cancer progression at least partially through
downregulating CDKL1.
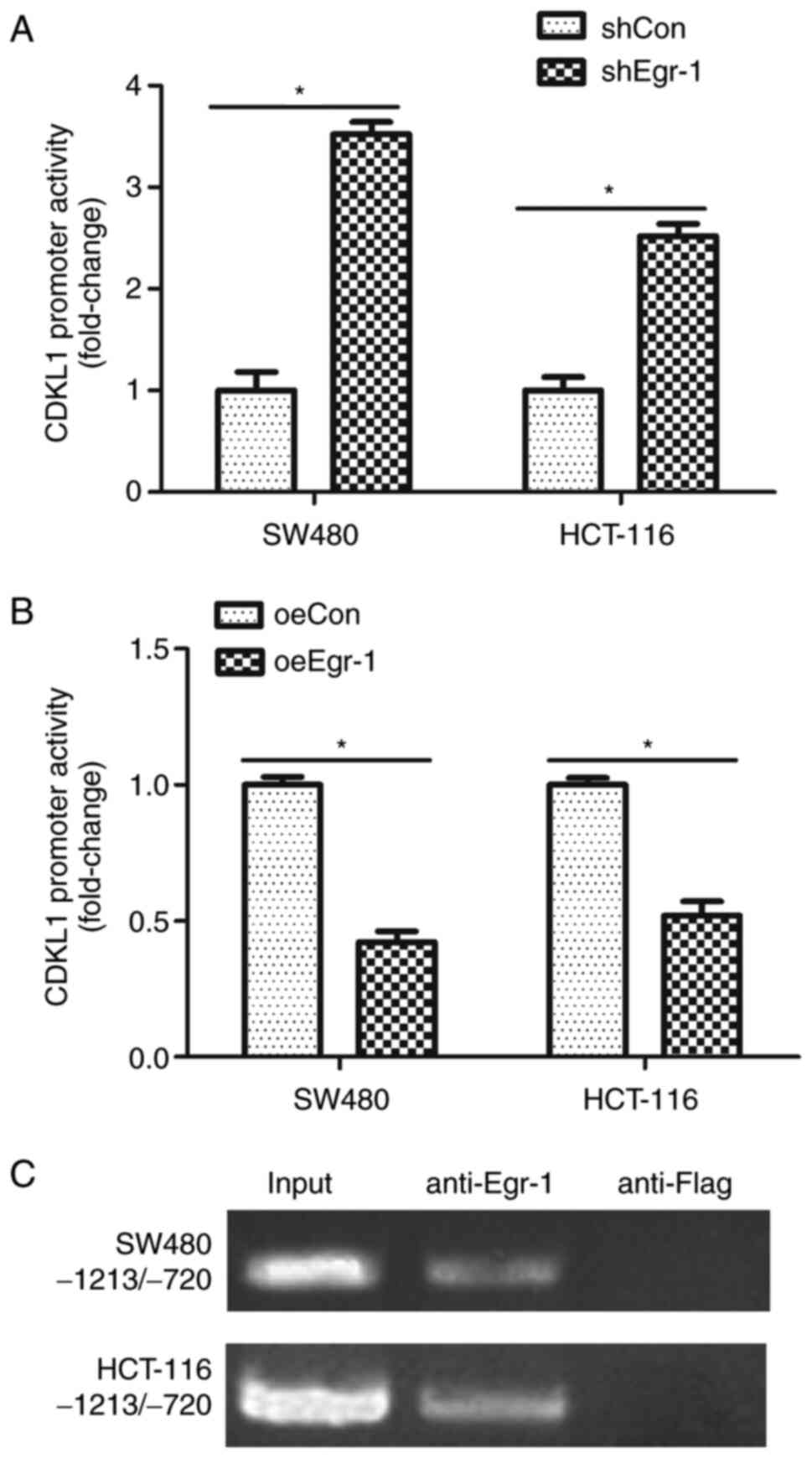
Egr-1 negatively regulates CDKL1
expression through modulating the promoter activity
As Egr-1 overexpression downregulated CDKL1
expression and correspondingly inhibited colon cancer cell
proliferation, migration and invasion, it was necessary to reveal
the mechanism by which Egr-1 regulated CDKL1. A CDKL1
promoter-reporter, pGL3-CDKL1, was constructed in the present
study. The luciferase assay showed that knockdown of Egr-1
significantly increased CDKL1 promoter activity in both SW480 and
HCT-116 cells (Fig. 5A); however,
overexpression of Egr-1 significantly inhibited CDKL1 promoter
activity in both SW480 and HCT-116 cells (Fig. 5B). A ChIP assay was also performed,
revealing that Egr-1 could bind to the CDKL1 promoter region in
both SW480 and HCT-116 cells (Fig.
5C). These results indicated that Egr-1 regulated CDKL1 at the
transcriptional level.
Discussion
The present study demonstrated that Egr-1 regulated
colon cancer cell proliferation, migration and invasion through
CDKL1. Additionally, the current results indicated that Egr-1
modulated CDKL1 expression at the transcriptional level.
Egr-1 has been previously reported to be
overexpressed in colon cancer (33).
Egr-1 is involved in tumor progression via regulating cell
proliferation, apoptosis, cell migration and invasion (8–10).
Considering the important roles of Egr-1 in tumor progression, it
has been suggested as a promising target for tumor therapy. The
present study demonstrated that high Egr-1 expression was
associated with improved patient survival and inhibited colon
cancer cell proliferation, migration and invasion. However, there
are several controversial reports about the function of Egr-1 in
tumor progression. A study by Sun et al (9) has reported that Egr-1 promotes the
proliferation and invasion of gastric cancer cells. Additionally,
Scharnhorst et al (34)
reported the tumor-promoting role of Egr-1 in Wilms' tumor. In
colon cancer, it is reported that Egr-1 is associated with
lymphovascular invasion, lymph node and distant metastasis,
indicating a positive role in tumor progression (35). However, in another study, Egr-1
exerted a tumor-suppressive role and participated in tolfenamic
acid-induced apoptosis in colon cancer cells (33). The conclusions of the present study
are in agreement with the latter study and suggested the
tumor-suppressive role of Egr-1 in colon cancer. The dual functions
of Egr-1 in the regulation of tumor progression may due to the
individual difference of cell types or patients.
Egr-1 participates in tumor progression by
regulating several targets. For example, Egr-1 suppresses breast
cancer cell proliferation via downregulating Cyclin Ds (36). However, the mechanisms by which Egr-1
regulates colon cancer cell progression remain unclear. In the
current study, it was revealed that the inhibitory roles of Egr-1
in cell proliferation, migration and invasion were partially
abolished by CDKL1, indicating that Egr-1 participates in colon
cancer progression via regulating CDKL1.
In the present study, Egr-1-knockdown increased
CDKL1 mRNA and protein expression, while overexpression of Egr-1
decreased CDKL1 mRNA and protein expression, indicating that Egr-1
may be an inhibitor of CDKL1 expression. CDKL1 was identified as a
novel Egr-1 target gene. Additionally, the present study revealed
that Egr-1 inhibited CDKL1 expression by blocking CDKL1
transcription. Egr-1 binds to GC-rich regions at the promoter of
target genes (18–20). There are several GC-rich regions at
the CDKL1 promoter, and the current study has demonstrated that
Egr-1 could bind to the promoter of CDKL1; however, a limitation of
the present study is that the accurate binding site was not
determined. To the best of our knowledge, the current study is the
first to suggest that Egr-1 may regulate CDKL1 transcription. In
addition to Egr-1, there are other transcription factors that can
bind to GC-rich region of promoters, such as specific protein 1
(Sp1), GC binding factor 2 and Kruppel-like factors (37–39);
therefore, these transcription factors may also regulate CDKL1 at
the transcriptional level. The binding sites of some transcription
factors may overlap in a promoter, such as the binding sites of Sp1
and Egr-1 overlapping in the promoter of nasopharyngeal
carcinoma-associated gene 6, inducing competitive binding of the
related transcription factors (40).
Whether the binding sites of the aforementioned transcription
factors are overlapping should be further investigated in future
studies.
In conclusion, the present study indicated that
Egr-1 bound to the promoter of CDKL1 and inhibited its expression,
and correspondingly inhibited colon cancer cell proliferation,
migration and invasion. The current study suggested that
Egr-1/CDKL1 may be used as promising prognostic biomarkers and
therapeutic targets for colon cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Natural
Science Foundation of Taizhou People's Hospital (grant no.
ZL201803).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the Figshare repository, https://figshare.com/s/1720cea2192d08d21b5f.
Authors' contributions
SS performed the cell biological experiments, such
as CCK-8, Transwell and cell infection assays. MJ performed the
western blot and RT-qPCR assays. JL constructed the pGL3-CDKL1
plasmid. XL performed the luciferase activity assay. HL analyzed
the association of Egr-1 expression with the survival of patients
with colon adenocarcinoma. DW performed the ChIP assay and
confirmed the authenticity of all the raw data. CX performed
statistical analysis and confirmed the authenticity of all the raw
data. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD, Goding Sauer A,
Fedewa SA, Butterly LF, Anderson JC, Cercek A, Smith RA and Jemal
A: Colorectal cancer statistics, 2020. CA Cancer J Clin.
70:145–164. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2020. CA Cancer J Clin. 70:7–30. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Brody H: Colorectal cancer. Nature.
521:S12015. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Weitz J, Koch M, Debus J, Hohler T, Galle
PR and Buchler MW: Colorectal cancer. Lancet. 365:153–165. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Vatandoust S, Price TJ and Karapetis CS:
Colorectal cancer: Metastases to a single organ. World J
Gastroenterol. 21:11767–11776. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wrobel P and Ahmed S: Current status of
immunotherapy in metastatic colorectal cancer. Int J Colorectal
Dis. 34:13–25. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Park YS, Kim HS, Kim JH, Choi SH, Kim DS,
Ryoo ZY, Kim JY and Lee S: NAB2-STAT6 fusion protein mediates cell
proliferation and oncogenic progression via EGR-1 regulation.
Biochem Biophys Res Commun. 526:287–292. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yi L, Lyn YJ, Peng C, Zhu RL, Bai SS, Liu
L, Wang PX, Zhou H and Dong Y: Sinomenine inhibits fibroblast-like
synoviocyte proliferation by regulating α7nAChR expression via
ERK/Egr-1 pathway. Int Immunopharmacol. 56:65–70. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sun T, Tian H, Feng YG, Zhu YQ and Zhang
WQ: Egr-1 promotes cell proliferation and invasion by increasing
β-catenin expression in gastric cancer. Dig Dis Sci. 58:423–430.
2013.PubMed/NCBI
|
|
10
|
Lee BS, Kang S, Kim KA, Song YJ, Cheong
KH, Cha HY and Kim CH: Met degradation by SAIT301, a Met monoclonal
antibody, reduces the invasion and migration of nasopharyngeal
cancer cells via inhibition of EGR-1 expression. Cell Death Dis.
5:e11592014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu P, Li J, Lu H and Xu B: Thalidomide
inhibits leukemia cell invasion and migration by upregulation of
early growth response gene 1. Leuke Lymphoma. 50:109–113. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Skorokhod A, Bachmann J, Giese NA,
Martignoni ME and Krakowski-Roosen H: Real-imaging cDNA-AFLP
transcript profiling of pancreatic cancer patients: Egr-1 as a
potential key regulator of muscle cachexia. BMC Cancer. 12:2652012.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kong Y, Yin J, Fu Y, Chen Y, Zhou Y and
Geng X: Suppression of Elk1 inhibits thyroid cancer progression by
mediating PTEN expression. Oncol Rep. 40:1769–1776. 2018.PubMed/NCBI
|
|
14
|
Kim SO, Kwon JI, Jeong YK, Kim GY, Kim ND
and Choi YH: Induction of Egr-1 is associated with anti-metastatic
and anti-invasive ability of beta-lapachone in human
hepatocarcinoma cells. Biosci Biotechnol Biochem. 71:2169–2176.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Calogero A, Porcellini A, Lombari V,
Fabbiano C, Arcella A, Miscusi M, Ponti D and Ragona G: Sensitivity
to cisplatin in primary cell lines derived from human glioma
correlates with levels of EGR-1 expression. Cancer Cell Int.
11:52011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Myung E, Park YL, Kim N, Chung CY, Park
HB, Park HC, Myung DS, Kim JS, Cho SB, Lee WS and Joo YE:
Expression of early growth response-1 in human gastric cancer and
its relationship with tumor cell behaviors and prognosis. Pathol
Res Pract. 209:692–699. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li TT, Liu MR and Pei DS: Friend or foe,
the role of EGR-1 in cancer. Med Oncol. 37:72019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sukhatme VP, Cao XM, Chang LC, Tsai-Morris
CH, Stamenkovich D, Ferreira PC, Cohen DR, Edwards SA, Shows TB,
Curran T, et al: A zinc finger-encoding gene coregulated with c-fos
during growth and differentiation, and after cellular
depolarization. Cell. 53:37–43. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Thiel G, Muller I and Rossler OG:
Expression, signaling and function of Egr transcription factors in
pancreatic β-cells and insulin-responsive tissues. Mol Cell
Endocrinol. 388:10–19. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kubosaki A, Tomaru Y, Tagami M, Arner E,
Miura H, Suzuki T, Suzuki M, Suzuki H and Hayashizaki Y:
Genome-wide investigation of in vivo EGR-1 binding sites in
monocytic differentiation. Genome Biol. 10:R412009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Thiel G and Cibelli G: Regulation of life
and death by the zinc finger transcription factor Egr-1. J Cell
Physiol. 193:287–292. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Silverman ES, Du J, Williams AJ,
Wadgaonkar R, Drazen JM and Collins T:
cAMP-response-element-binding-protein-binding protein (CBP) and
p300 are transcriptional co-activators of early growth response
factor-1 (Egr-1). Biochem J. 336:183–189. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Vo N and Goodman RH: CREB-binding protein
and p300 in transcriptional regulation. J Biol Chem.
276:13505–13508. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cartron PF, Blanquart C, Hervouet E,
Gregoire M and Vallette FM: HDAC1-mSin3a-NCOR1, Dnmt3b-HDAC1-Egr1
and Dnmt1-PCNA-UHRF1-G9a regulate the NY-ESO1 gene expression. Mol
Oncol. 7:452–463. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Meyerson M, Enders GH, Wu CL, Su LK, Gorka
C, Nelson C, Harlow E and Tsai LH: A family of human cdc2-related
protein kinases. EMBO J. 11:2909–2917. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yen SH, Kenessey A, Lee SC and Dickson DW:
The distribution and biochemical properties of a Cdc2-related
kinase, KKIALRE, in normal and Alzheimer brains. J Neurochem.
65:2577–2584. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tang L, Gao Y, Yan F and Tang J:
Evaluation of cyclin-dependent kinase-like 1 expression in breast
cancer tissues and its regulation in cancer cell growth. Cancer
Biother Radiopharm. 27:392–398. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sun W, Yao L, Jiang B, Shao H, Zhao Y and
Wang Q: A role for Cdkl1 in the development of gastric cancer. Acta
Oncol. 51:790–796. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Song Z, Lin J, Sun Z, Ni J and Sha Y:
RNAi-mediated downregulation of CDKL1 inhibits growth and
colony-formation ability, promotes apoptosis of human melanoma
cells. J Dermatol Sci. 79:57–63. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Qin C, Ren L, Ji M, Lv S, Wei Y, Zhu D,
Lin Q, Xu P, Chang W and Xu J: CDKL1 promotes tumor proliferation
and invasion in colorectal cancer. Onco Targets Ther. 10:1613–1624.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lee SH, Bahn JH, Choi CK, Whitlock NC,
English AE, Safe S and Baek SJ: ESE-1/EGR-1 pathway plays a role in
tolfenamic acid-induced apoptosis in colorectal cancer cells. Mol
Cancer Ther. 7:3739–3750. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Scharnhorst V, Menke AL, Attema J,
Haneveld JK, Riteco N, van Steenbrugge GJ, van der Eb AJ and
Jochemsen AG: EGR-1 enhances tumor growth and modulates the effect
of the Wilms' tumor 1 gene products on tumorigenicity. Oncogene.
19:791–800. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Myung DS, Park YL, Kim N, Chung CY, Park
HC, Kim JS, Cho SB, Lee WS, Lee JH and Joo YE: Expression of early
growth response-1 in colorectal cancer and its relation to tumor
cell proliferation and apoptosis. Oncol Rep. 31:788–794. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wei LL, Wu XJ, Gong CC and Pei DS: Egr-1
suppresses breast cancer cells proliferation by arresting cell
cycle progression via down-regulating CyclinDs. Int J Clin Exp
Pathol. 10:10212–10222. 2017.PubMed/NCBI
|
|
37
|
Ma Y, Ren Y and Guan J: Knockdown of GC
binding factor 2 by RNA interference inhibits invasion and
migration of vascular smooth muscle cells. Mol Med Rep.
20:1781–1789. 2019.PubMed/NCBI
|
|
38
|
Kou XX, Hao T, Meng Z, Zhou YH and Gan YH:
Acetylated Sp1 inhibits PTEN expression through binding to PTEN
core promoter and recruitment of HDAC1 and promotes cancer cell
migration and invasion. Carcinogenesis. 34:58–67. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Buttar NS, DeMars CJ, Lomberk G, Rizvi S,
Bonilla-Velez J, Achra S, Rashtak S, Wang KK, Fernandez-Zapico ME
and Urrutia R: Distinct role of Kruppel-like factor 11 in the
regulation of prostaglandin E2 biosynthesis. J Biol Chem.
285:11433–11444. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liu M, Wang X, Peng Y, Shen S and Li G:
Egr-1 regulates the transcription of NGX6 gene through a Sp1/Egr-1
overlapping site in the promoter. BMC Mol Biol. 15:142014.
View Article : Google Scholar : PubMed/NCBI
|