Introduction
Colorectal cancer (CRC) is one of the most common
causes of cancer-related mortality in the world. The mean 5-year
survival rate of CRC is estimated to be <10% if metastasis
occurs, and as high as 90% if the cancer is detected at an early
stage (1,2). Several methods are currently available
for detecting CRC, such as fecal immunochemical testing (FIT),
fecal occult blood testing, and the most important measure,
colonoscopy. However, the tolerance of colonoscopy remains low in
the general population, due to the troublesome bowel preparation
and the risk of complications. Although progress has been achieved
in the research of antitumor drugs, the current curative effect for
CRC is far from expected due to the late stage of disease at
diagnosis and the unselected patients. This highlights the need to
identify novel methods or effective biomarkers to diagnose CRC at
an early stage and determine the patient's response to
individualized treatment.
Definition of epigenetics
Accumulation of genetic and epigenetic changes
ultimately lead to the initiation and progression of cancer
(2,3).
Genetic mutations have long been considered a major cause of
cancer, but more recently, epigenetic changes have also been
suggested to be important factors in cancer development (4). The unified definition of epigenetics
remains ambiguous; however, more researchers support Holliday's
definition as it offers two aspects of epigenetics, that is, ‘the
study of the changes in gene expression, which occurs in organisms
with differentiated cells, and the mitotic inheritance of given
patterns of gene expression’ and ‘nuclear inheritance, which is not
based on differences in DNA sequence’. The streamlined definition
is ‘the study of changes in gene function that are mitotically
and/or meiotically heritable and that do not entail a change in the
DNA sequence’ (5).
The study of DNA forms the cornerstone of genetics,
while epigenetics is the study of changes in gene expression that
do not involve alterations in the underlying DNA sequence. Common
epigenetic changes in different cancer types include abnormal DNA
methylation, abnormal histone modifications and changes in the
expression levels of numerous non-coding RNAs (6). Abnormal DNA methylation was the earliest
identified modification and is the most widely studied epigenetic
change (7).
Tumor-derived cell-free DNA
(cfDNA)
For hypermethylated DNA to be a valid biomarker, it
should be available via minimally invasive procedures and from
tumor-remote media. Moreover, it must be effective in detecting the
disease at an early stage. Based on these characteristics, research
on cancer-specific hypermethylated genes has focused on
tumor-derived cfDNA.
Cancer cells release nucleic acids, proteins,
vesicles and other biological components into the blood and other
body fluids (8). Among these
potential biomarkers, tumor-derived cfDNA has been examined over
the past decade as an important tool in oncology precision
medicine. Abnormal DNA methylation is one of the characteristics of
numerous cancer type, and most importantly, it can be detected in
cfDNA in blood, urine, feces and other biological samples.
Moreover, cfDNA methylation is used for early detection of cancer,
minimal residual disease surveillance, prediction of treatment
response and prognosis and tracing of tissue origin.
DNA methylation markers have numerous advantages
(9). Firstly, DNA methylation occurs
early in tumorigenesis and can be tissue- and cancer-type specific.
Secondly, it is consistent across multiple genomic regions and can
be detected using numerous CpG dinucleotides. Finally, and most
importantly, methylation patterns are often associated with the
origin of specific cancer type, and thus, can be used to reveal the
tissue origin.
The landmark for cfDNA methylation analysis was the
development of a screening test for CRC, based on the methylated
septin 9 (mSEPT9), which was approved by the Food and Drug
Administration (FDA) in 2016 (10).
At the same time, accumulating evidence verified the value of cfDNA
methylation as a biomarker for the diagnosis and evaluation of
cancer.
Characteristics of CRC DNA methylation
Currently, the molecular pathogenesis of CRC mainly
includes the following processes: Chromosomal instability,
microsatellite instability (MSI), epigenetic instability [such as
CpG island methylator phenotype (CIMP)], altered tumor
microenvironments and an altered metabolic state (11).
The current level of the understanding of epigenomic
alterations in CRC is less than that of gene mutations, but
research in this area has advanced rapidly in recent years. Similar
to other malignant tumor cells, CRC cells also show two different
DNA methylation changes. One is global hypomethylation, and the
other is promoter methylation of specific genes (12). Accumulating evidence suggests the
existence of a group of CRC subsets with high levels and distinct
patterns of DNA methylation, known as CIMP (2). CIMP was defined as a molecular subclass
of CRC in 1999, which was a significant advancement in the
understanding of the molecular mechanisms of CRC formation.
Overall, hypomethylation is associated with increased genomic
instability, suggesting that the maintenance of methylation may be
chemoprophylaxis (13).
Hypermethylation of cfDNA from CRC and other types of cancer in the
blood or stool has been considered as a potential non-invasive
cancer biomarker, which may exhibit high sensitivity and
specificity in some cases (10,14). In
addition, with further research on the mechanism of tumor
methylation, the reversal of the DNA methylation abnormalities by
targeting the maintenance DNA methylation mechanism has become a
potential therapeutic method (4).
Methylation detection methods
DNA methylation can be detected using a variety of
methods, such as methylation-specific polymerase chain reaction
(15), DNA sequencing, MethyLight
(16), methylation-specific melting
curve analysis (17), pyrosequencing
(18), microarray analysis (19) and liquid chromatography (LC). Notably,
LC was the first platform used to quantify global DNA methylation,
which was represented by the total 5-methyldeoxycytidine content in
DNA samples (20). With advances in
technology, LC coupled with mass spectrometry provides an accurate
and highly sensitive method for overall DNA methylation
quantification (21).
Value of methylation markers
Several methylation markers have been found to be
associated with the initiation and progression of CRC (2,3,22). Some of these markers show potential
for early detection (Table I),
prognostic evaluation (Table II),
and even prediction of treatment response to CRC. The status of
methylation markers is associated with various conditions, such as
tumor size, grade or metastasis (Fig.
1). Therefore, methylation markers, such as
O6-methylguanine-DNA methyltransferase (MGMT) and long
interspersed nucleotide element-1 (LINE-1) (23,24), may
have a cross-over role in the prognostic assessment of CRC,
prediction of treatment response and even early diagnosis, in
different conditions. Thus, to achieve the best predictive effect,
the examination of multiple methylation markers, multitarget stool
DNA (MT-sDNA) and fecal occult blood testing should be
combined.
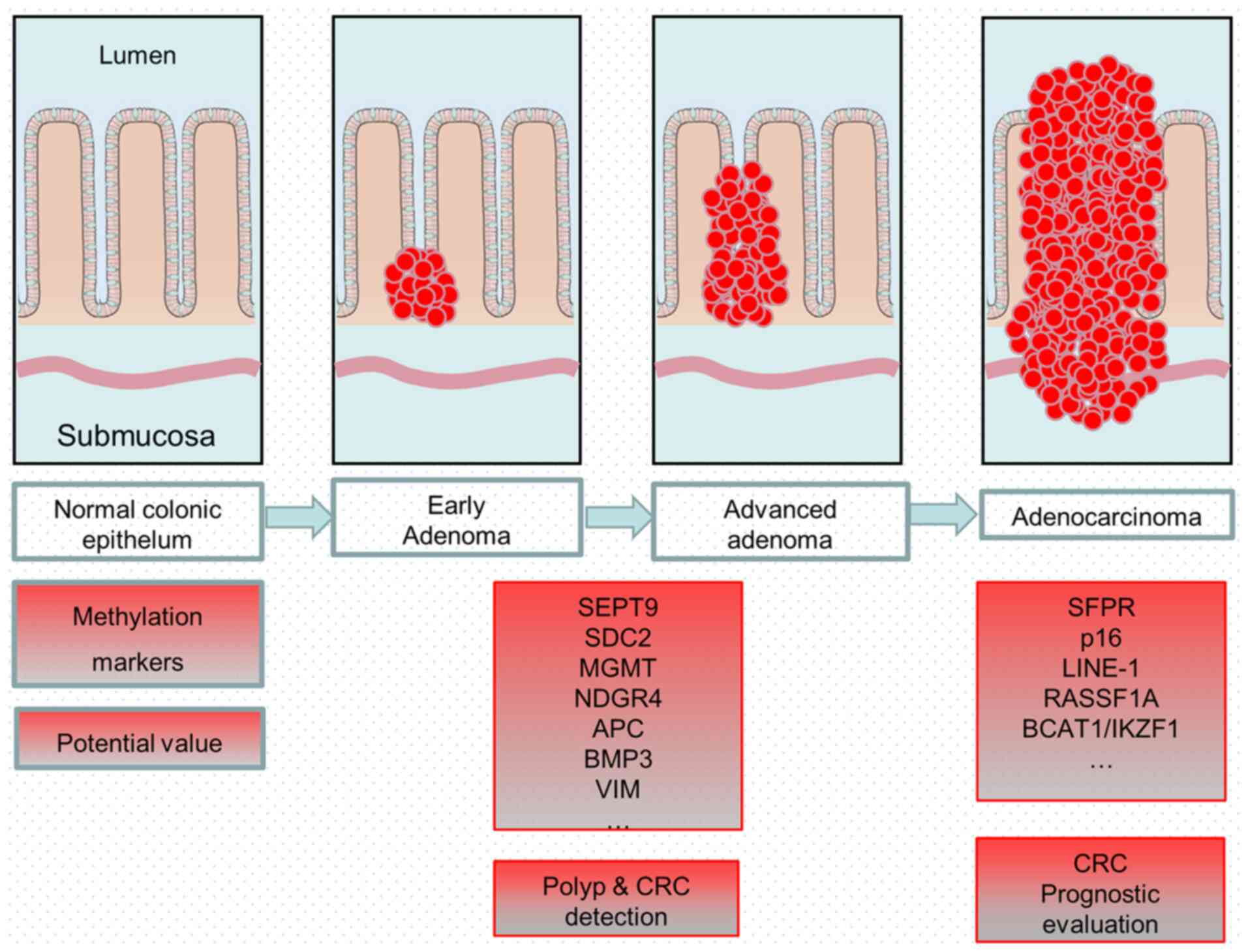 | Figure 1.A model of the progression from
adenoma to adenocarcinoma and various methylation markers that show
potential value in the early detection or prognostic evaluation of
CRC. CRC, colorectal cancer; SEPT9, septin 9; SDC2, syndecan 2;
MGMT, O6-methylguanine-DNA methyltransferase; NDGR4,
N-Myc downstream regulated gene 4; APC, adenomatous polyposis coli;
BMP3, bone morphogenetic protein 3; VIM, vimentin; SFPR, secreted
frizzled-related protein; LINE-1, long interspersed nucleotide
element-1; RASSF1A, RAS association domain family protein 1; BCAT1,
branched chain amino acid transaminase 1; IKZF1, IKAROS family zinc
finger 1. |
 | Table I.DNA-methylation markers used for the
screening and early detection of colorectal cancer. |
Table I.
DNA-methylation markers used for the
screening and early detection of colorectal cancer.
| Methylated DNA | Authors, year | Country | Sensitivity % | Specificity % | Detection
method | (Refs.) |
|---|
| SEPT9 | Church et
al, 2014 | USA | 68.2 | 78.8 | MSP | (30) |
|
| Song et al,
2016 | China | 75.1 | 95.1 | MSP | (31) |
| SDC2 | Oh et al,
2017 | Republic of
Korea | 90.9 | 90 | MSP | (35) |
|
MGMT/SEPT9 | Freitas et
al, 2018 | Portugal | 93.8 | 82 | MSP | (42) |
| NDRG4 | Bagheri et
al, 2020 | Iran | 86 | 92 | MSP | (46) |
|
NDRG4/SNAP91/FIT | Rademakers et
al, 2021 | The
Netherlands | 86 | 96 | MSP | (47) |
|
APC/FOXA1/RASSF1A | Nunes et al,
2018 | Portugal | 78.4 | 69.9 | MSP | (54) |
| BMP3 | Loh et al,
2008 | Australia | 56.66 | 93.3 | MSP | (56) |
|
| Rokni et al,
2018 | Iran | 40 | 94 | MSP | (58) |
| VIM | Mojtabanezhad
Shariatpanahi et al, 2018 | Iran | 52 | 88 |
| (48) |
|
| Lu et al,
2014 | China | 41.1 | 85 | MSP | (49) |
 | Table II.DNA-methylation markers used for
prognostic evaluation in colorectal cancer. |
Table II.
DNA-methylation markers used for
prognostic evaluation in colorectal cancer.
| Methylated DNA | Authors, year | Country | Prognostic
value | (Refs.) |
|---|
| SFRP | Bagci et al,
2016 | Turkey | Associated with
lymph node invasion and OS | (72) |
|
| Boughanem et
al, 2020 | Spain | Increases CRC cell
proliferation and tumor growth | (70) |
| p16 | Karam et al,
2018 | Egypt | Correlated with
Dukes' stage, lymph node metastasis and CEA levels | (74) |
|
| Kim et al,
2016 | Korea | Predictive of
clinical outcome in metastatic CRC patients | (77) |
| LINE-1 | Barchitta et
al, 2014 | Italy | Associated with
poor prognosis, survival and advanced stage | (82) |
|
| Jiang et al,
2021 | USA | Indicative of the
CRC tumorigenesis pathway | (81) |
|
| Boughanem et
al, 2020 | Spain | Associated with
survival rates | (84) |
|
BCAT1/IKZF1 | Pedersen et
al, 2015 | Australia | Positively
correlated with the degree of invasion | (89) |
| RASSF1A | Sun et al,
2017 | China | Affects the
sensitivity of patients with CRC to oxaliplatin-based
chemotherapy | (97) |
Screening and early detection markers
SEPT9
The SEPT9 gene is a tumor-suppressor gene,
which is closely associated with the development of tumors and
other types of human diseases. SEPT9 is involved in a
variety of important physiological processes, such as cytokinesis,
DNA repair, cell migration and apoptosis (25). Abnormal methylation reduces the
activity of SEPT9 gene transcription, leading to
dysregulated gene expression and abnormal physical function, which
may eventually lead to the development of cancer (26).
Toth et al (27) reported that the protein expression
level of SEPT9 in CRC was significantly lower compared with that in
normal epithelial cells, and that the mRNA expression level of
SEPT9 was decreased during the transformation from adenoma
to CRC. Moreover, Wasserkort et al (28) examined the methylation status of SEPT9
in different types of colon lesions and their adjacent tissue, and
suggested that hypermethylation of SEPT9 may be a late event
in the progression of adenomas to CRC. This may be the reason why
the sensitivity of SEPT9 gene methylation detection in
adenoma is lower compared with that of CRC. A recent meta-analysis
revealed that plasma mSEPT9 was of high diagnostic value for CRC
and was significantly correlated with CRC stage (29). In addition, survival analysis
indicated that there was a negative correlation between SEPT9
methylation levels and disease-free survival (DFS) after CRC
surgery (30). Song et al
(31) also reported that an SEPT9
assay displayed 75.1% sensitivity and 95.1% specificity for CRC
detection. Notably, the detection rate for CRC stage 0 was 57%, for
stage I it was 64%, and for stage II it was 88%, which was valuable
for population screening, alone or combined with other methods
(31).
In a previous study that included 1,544 CRC samples
(stages I–IV), a prospective evaluation of SEPT9 trial was
retrospectively conducted. The sensitivity and specificity of CRC
were reported at 68.2 and 78.8%, respectively, while the
sensitivity for advanced adenoma was 21.6%. The main performance
characteristics of mSEPT9 were demonstrated by this study (30), and mSEPT9 was eventually approved by
the FDA for CRC screening (10).
Syndecan 2 (SDC2) precursor
The SDC2 protein is a membrane protein that serves a
role in cell proliferation and migration, and helps maintain cell
integrity. The SDC2 gene is not expressed in epithelial
cells of normal colonic tissues, but is expressed in mesenchymal
cells (32).
Important evidence has been discovered using
different analyses of methylation conducted in comprehensive
experiments. For example, CpG sites of SDC2 abnormal
methylation have been observed in the tumor tissues of the majority
of patients with CRC, thereby showing the significant potential of
SDC2 methylation in the early detection of CRC. In addition,
the level of SDC2 DNA methylation in feces is closely
associated with the occurrence of CRC, but not with clinical stage
(33,34). Other studies have reported that
SDC2 methylation may be highly sensitive in the detection of
both advanced adenoma and CRC. Oh et al (35) examined the methylation of the
SDC2 gene in primary tumors, adenomatous polyps,
hyperplastic polyps and normal tissues, and identified that the
positive SDC2 methylation was 100, 90.6, 94.1 and 0%,
respectively. SDC2 methylation was also significantly
elevated depending on the severity of the lesion. The overall
sensitivity for CRC and small polyps was 90.0 and 33.3%,
respectively, and the specificity was 90.9% for SDC2
methylation in fecal DNA. Some previous studies indicated that the
diagnostic efficacy of SDC2 methylation had a sensitivity of
77.4–81.1% and a specificity of 88.2–98% in fecal DNA for CRC
(36,37). Thus, SDC2 methylation may be a
promising non-invasive biomarker for the early detection of CRC. It
has also been reported that the combination of multiple biomarkers
may be an effective strategy for improving the sensitivity and
specificity of early cancer diagnosis (38).
MGMT
MGMT, which encodes five exons and four
introns, is located at 10q26 on chromosome 10 and the MGMT protein
acts as a DNA repair enzyme (16).
Abnormal hypermethylation of the MGMT promoter is associated with a
lack of mRNA expression, accompanied by the loss of protein content
and enzyme activity (16,23). In total, ~40% of metastatic CRCs
exhibit silencing of the MGMT gene, which results in a
corresponding inhibition of protein synthesis (39). Previous studies have reported that
MGMT promoter methylation was the main cause of MGMT
gene expression disorder (40).
Shima et al (40) evaluated 855 cases of stage I–IV CRC
for MGMT using two methods, methylation-specific PCR and
immunohistochemistry. The results demonstrated that the methylation
rate was 38% and the loss of MGMT expression was 37%, and the
consistency of the two methods was 81%. Sartore-Bianchi et
al (41) screened the molecular
characteristics of 2,044 patients with mCRC and then found that the
MGMT promoter hypermethylation proportion was 48.7%. In
another study that included 70 patients with CRC, MGMT
hypermethylation was detected in serum-free circulating DNA, and
MGMT promoter hypermethylation was observed in 90% of CRC
cases, while no MGMT hypermethylation was found in the serum
of healthy subjects (23). Freitas
et al (42) reported that a
combination of methylated MGMT and SEPT9 present a
93.8% sensitivity, 82.0% specificity, respectively, for CRC
detection in tissue samples. Therefore, it was suggested that MGMT
may be used as a clinical biomarker for early diagnosis of CRC.
However, there are some differences in the positive rate of
MGMT methylation in the aforementioned studies and, thus,
future studies are required to verify the findings.
N-Myc downstream regulated gene 4
(NDGR4)
NDRG4, is involved in a variety of biological
activities, such as cell proliferation, differentiation,
development and stress. The protein sequences encoded by this
family have a 52–65% homology and are highly conserved in the
evolution of various species (43),
with α/β hydrolase folding. Previously, researchers have identified
NDRG4 promoter CpG island methylation as a potential and one of the
most accurate marker for CRC detection (44), and this has been verified by an
independent study (45).
Bagheri et al (46) revealed that the sensitivity and
specificity of the NDRG4 gene in the diagnosis of CRC were 86 and
92%, respectively, and the proportion of methylation was
proportional to CRC staging. Rademakers et al (47) combined types of biomarkers to
establish a panel for the highest diagnostic potential, resulting
in the combination of SNAP91/NDRG4/FIT as the best performing, with
a sensitivity and specificity of 86.0 and 96.0%, respectively.
Moreover, these authors demonstrated that the sensitivity and
specificity of the NDRG4 gene were sufficient to serve as a
novel, non-invasive marker for CRC screening. A meta-analysis also
showed that NDRG4 could be considered as an important marker for
the diagnosis of CRC (48).
In a previous study, the positive rates of
methylated NDRG4 in cancer tissues, precancerous tissues,
blood, urine and feces were shown to be 81, 8.3, 54.8, 72.6 and
76.2%, respectively. It was also confirmed that methylated
NDRG4 in the feces and urine had high sensitivity and
specificity. Additionally, this new method is expected to be a
promising and potential marker for the early diagnosis of CRC due
to the ease of collection of urine or feces samples (49,50).
Adenomatous polyposis coli (APC)
APC is a tumor-suppressor gene that was discovered
during the research of familial adenomatous polyposis. Furthermore,
hypermethylation of APC promoters has been frequently observed in
sporadic and familial CRC types over the past two decades (51).
A previous study of patients with CRC examined the
methylation status of APC and found that APC methylation
occurred 33% of the time in patients with CRC (52). And recent research also reported that
in CRC patient under 50 years of age, the rate of APC promoter
methylation was 40% (53). Nunes
et al reported the methylation levels of APC combined with
FOXA1 and RASSF1A to diagnose CRC in stages 0, I and II with 78.4%
sensitivity, 69.9% specificity, respectively, but the result needs
further validation because the CRC samples were only 37 (54). Moreover, methylated APC promoters were
found to be significantly associated with later stages and an older
age. The incidence of APC methylation in patients with mCRC was
53.7%. That study demonstrated that APC promoter hypermethylation
is a common epigenetic event in patients with both early and
metastatic CRC, and it serves an important role in the development
of CRC (52). A meta-analysis by
Liang et al (55) investigated
APC hypermethylation in the early diagnosis of CRC. This
study included 24 articles and 2,025 patients with CRC. The
analysis demonstrated that APC promoter hypermethylation was
an early cancerous event of CRC and, thus, may be a noteworthy
diagnostic indicator of early CRC.
Bone morphogenetic protein 3
(BMP3)
The first important evidence of BMP3 inactivation in
early polyp formation and colon tumor development was reported in
2008 by Loh et al (56). These
authors reported that the percentage of aberrant BMP3
hypermethylation in colorectal tumors was 55% (33/60). This result
suggested that BMP3 may be a potential marker for the early
detection of neoplastic lesions. In subsequent studies, researchers
have focused on the diagnostic value, such as the sensitivity and
specificity, of methylated BMP3 in CRC. For example,
Houshmand et al (57) examined
the methylation status of the BMP3 gene in CRC tissue
samples, and detected a sensitivity of 56.66% and specificity of
93.3%. In addition, Rokni et al (58) detected the methylation status of the
BMP3 gene in plasma DNA samples from 50 patients with
histologically diagnosed polyps or tumors and 50 healthy
individuals. Their results showed that the frequency of BMP3
methylated DNA was significantly higher in patients with polyps
compared with that in healthy controls, with a sensitivity of 40%
and specificity of 94%. In a study by Kisiel et al (59), the combination of abnormal methylated
DNA markers with BMP3 and NDRG4 had a sensitivity of
100% and a specificity of 89% for CRC and high-grade dysplasia.
These findings suggest that methylated BMP3 may be a useful
biomarker for CRC detection, but that it should be combined with
other biomarkers due to the lack of specificity. A typical example
of a combination method is the application of MT-sDNA test, which
consists of fecal hemoglobin, quantitative detection of methylation
BMP3 and NDRG4, β mutant KRAS and β-actin tests. The sensitivity
for curable stage CRC ranges from 93 to 100%, while its specificity
ranges from 87 to 93%. Due to this reliable performance, it was
approved by the US FDA (60).
Vimentin (VIM)
VIM encodes a cytoskeletal protein that is
considered to be involved in cancer invasion and metastasis.
VIM promoter methylation has been observed in CRC (61), such as in the fecal DNA of patients
with CRC (62). VIM promoter
methylation has been detected in stool at a sensitivity of 52% and
specificity of 88% (61). Lu et
al (49) reported that the
promoter methylation levels of VIM in patients with CRC were 41.1%
and the specificity was 85.0%. These results suggest that promoter
methylated VIM in fecal samples was sensitive, specific and
non-invasive for CRC screening and, therefore, it has been
commercialized as a non-invasive method for early CRC detection
(63).
In addition, previous studies have suggested that
the hypermethylation of the following genes, including ALX homeobox
4 (ALX4), neurogenin 1 (NeuroG1), helicase like
transcription factor (HLTF), hyperpigmentation progressive 1
(HPP1), WNT inhibitory factor 1 (WIF1), Ras
association domain family member 2a (RASSF2a), GATA binding
protein 4 (GATA4), β-1,4-glucuronytransferase 1
(B4GAT1), proline rich membrane anchor 1 (PRIMA1),
APC, ATM serine/threonine kinase, glutathione S-transferase π 1
(GSTP1) and tissue factor pathway inhibitor 2(TFPI2)
(63,64), have been analyzed in fecal, blood and
urine samples from patients with CRC and used for the early
detection of CRC. From the potential methylation markers,
methylated VIM, SEPT9, BMP3, SDC2, NDRG4 and a combination of
methylated branched chain amino acid transaminase 1 (BCAT1)/IKAROS
family zinc finger 1 (IKZF1) have been proven to be reliable and
accurate, and have been approved for clinical use (10,60,63,65).
As shown by a recent meta-analysis (66), the value of a single hypermethylated
DNA promoter region as a marker for CRC screening is limited, but
the combination of indicators demonstrates significant potential.
Therefore, the combination of different detection indicators must
be utilized to balance their respective diagnostic performance,
thus achieving the best diagnostic effect with regards to
specificity and sensitivity.
Prognostic biomarkers
At present, the most effective method for evaluating
the prognosis of patients with CRC is based on the histological
characteristics of the tumor, such as pathological type and stage.
However, the survival time of patients with CRC at the same stage
is heterogeneous; thus, a more accurate method is required to
determine the prognosis of these patients. In recent years,
numerous clinical studies have investigated the feasibility of
using specific methylated DNA markers to evaluate the prognosis of
patients with CRC.
Secreted frizzled-related protein
(SFRP)
Researchers have observed abnormal methylation of
SFRPs in various types of human cancer. The activation of the Wnt
pathway induced by the loss of SFRP gene expression is one
of the most important mechanisms for tumorigenesis and cancer
development (67). In humans, there
are five types of SFRPs (SFRP1, 2, 3, 4 and 5). Among the SFRP
family members, SFRP1 and SFRP2 have been the most extensively
researched in human cancer, and studies have shown that
SFRP1 and SFRP2 promoter methylation may contribute
to the risk of CRC (68).
The loss of SFRP expression induced by DNA
methylation is the primary mechanism causing the silencing of SFRP
and it is associated with tumor formation in CRC (69,70). Kumar
et al (71) evaluated the
promoter methylation status of the SFRP1 gene in 54 cases
with stage II–III CRC. It was found that SFRP1 gene
methylation was associated with lymph node invasion (P=0.05) and a
poorer mean overall survival (OS). Moreover, these authors
indicated that SFRP1 gene methylation was a prognostic marker in
CRC (71). It has also been shown
that the hypermethylation rate of the SFRP2 promoter in
patients with CRC was 66.7% (72). In
addition, a recent meta-analysis of the SFRP family revealed a
significant association between hypermethylation of SFRP1 and
cancer risk (67). A review by van
Loom et al (73) revealed that
numerous studies have reported promotor hypermethylation
downregulated SFRP2 gene expression in several types of
cancer. These authors discussed the role of SFRP2 in tumor
angiogenesis and noncanonical Wnt signaling, and suggested its
potential as an anti-angiogenic therapeutic target or an effective
prognostic marker in cancer.
p16
p16 is a tumor-suppressor gene frequently studied
due to its significant function in the cell cycle. There are three
main mechanisms of p16 inactivation: Gene mutation, homozygous
deletion and 5′-CpG island methylation, which is one of the major
causative factors of various human cancers (74).
Ye et al (75)
detected p16 methylation in CRC tissues and adjacent normal
tissues, and the results demonstrated that p16 methylation was
higher in CRC tissues compared with adjacent normal tissues.
Moreover, Lee et al (76)
found that hypermethylation of the p16 gene was
significantly associated with lymph node metastasis in CRC, and
reported that the hypermethylation frequency of the p16 gene
was 32.3%. However, p16 gene promoter hypermethylation frequency
was detected at 15.1% in CRC, and this rate was lower than that
previously determined (72). This
variation may be caused by the different sensitivity of detection
methods, which can be further verified in future studies. Karam
et al (74) reported a
sensitivity of 55.38%, specificity of 98.5% and diagnostic accuracy
of 77.7% in patients with CRC. Furthermore, p16 methylation was
significantly correlated with age, sex, Dukes'stage, lymph node
metastasis, carcinoembryonic antigen levels, a shortened time to
progression and overall survival (75,77).
Therefore, it may be associated with prognosis and may thus serve
as a prognostic biomarker.
Long interspersed nucleotide element-1
(LINE-1)
LINEs are retrotransposable elements identified in
numerous eukaryotic genomes. These are highly methylated in normal
somatic cells and, therefore, are mostly inhibited, thus preventing
their potential to cause genomic instability (78). Full-length LINE-1 can lead to adenoma
formation and cancer progression, and the activity of LINE-1 is
dependent on the epigenetic regulation of its promoter (79). Hypomethylation of the LINE-1 promoter
has been observed in gastrointestinal cancer as well as other types
of cancer (79,80).
Jiang et al (81) examined promoter methylation of
LINE-1 in adenomas and sessile serrated lesions (SSLS); the
results showed that high-grade dysplasia (HGD) and increasing size
of adenoma were associated with decreased LINE-1
methylation. Shademan et al (78) quantified promoter methylation and
LINE-1 transcripts in three stages of CRC, non-advanced adenoma,
advanced adenoma and adenocarcinoma. Their results demonstrated
that the methylation of the LINE-1 promoter in non-advanced
adenoma was significantly higher compared with that in advanced
adenoma and adenocarcinoma. The correlation analysis also revealed
a decrease in LINE-1 promoter methylation, genomic
polymorphism insertion and an increase in LINE-1 transcription in
advanced adenomas, suggesting that early and late polyps may
involve some main pathogenetic mechanisms that ultimately lead to
cancer.
Bachitta et al (82) conducted a meta-analysis in 2014, which
described the role of LINE-1 hypomethylation in human
cancer. It has also been shown that LINE-1 hypomethylation in CRC
was associated with poor prognosis, survival and advanced stage,
and that it could be a prognosis predictor of CRC (83,84). In
addition to its prognosis value, a recent study reported that
LINE-1 hypomethylation was effective in the early detection
of CRC (85). Hence, LINE-1
hypomethylation may be a promising marker for the prognostic and
early detection of CRC.
BCAT1/IKZF1
BCAT1 controls the metabolism of branched-chain
amino acids, which are essential nutrients for growth. Abnormal
methylation of the BCAT1 gene locus has been observed in CRC
and various pathologic conditions (86). IKZF1 encodes a DNA-binding
protein, which regulates the cell cycle. In CRC, IKZF1
promoter methylation is associated with the loss of regulation of
cell proliferation and differentiation (87).
Mitchell et al (88) demonstrated that the expression levels
of the two genes, BCAT1 and IKZF1, were low in
healthy human plasma (3.5 and 4.9%, respectively) but were
significantly increased in patients with CRC; therefore, these two
genes were selected as biomarkers for CRC detection. In a previous
study that included 2,127 samples, 85/129 cases of CRC had a
sensitivity of 66%. For stages I, II, III and IV, the positive
rates were 38, 69, 73 and 94%, respectively. It was also found that
the positive rate was positively correlated with the degree of
invasion (89). In a trial involving
1,381 volunteers, which examined FIT and BCAT1/IKZF1 DNA
methylation, the results demonstrated that the sensitivity of the
BCAT1/IKZF1 methylation to CRC was 62%, with a specificity of 92%,
which was higher compared with the commonly used positive threshold
of FIT. When combining FIT (cut-off 10 µg Hb/g) with the
BCAT1/IKZF1 blood test, the sensitivity for cancer was 89%, with an
improved specificity of 74% (90).
From research on recurrent CRC, postoperative patients who were
BCAT1/IKZF1-positive were found to have an increased risk of
residual disease and subsequent recurrence. It has been shown that
the BCAT1/IKZF1 test was more sensitive to recurrence compared with
the conventional CEA test, and in the case of positive test, the
recurrence rate was twice as high as that of CEA (91,92). The
BCAT1/IKZF1 test is a novel blood test that have been approved
under the commercial name ‘COLVERA’ for CRC (65), and could be used for monitoring
recurrence and as a non-invasive diagnostic method.
RAS association domain family protein
1 (RASSF1A)
RASSF1A is a tumor-suppressor gene that
serves a role in regulating cell proliferation and apoptosis, and
is involved in numerous types of cancer, including CRC. More
importantly, it can be detected early in liquid biopsies (93). RASSF1A is subject to epigenetic
regulation and suppression, the main regulatory mechanism being via
DNA methylation (94). It has been
reported that the development of adenomatous polyps and colorectal
laterally spreading tumors (LSTs) are precursor lesions of CRC, and
80–90% of CRCs develop via these factors (95). In a previous study, Ni et al
(96) detected the degree of
RASSF1A methylation in patients with LSTs. The results
indicated that RASSF1A methylation was lower in patients
with LSTs compared with CRC cases, but was higher than that in
polypoid adenomas. Thus, it was suggested that RASSF1A has the
potential to be used for the early diagnosis of CRC and may enable
timely intervention.
Promoter methylation of RASSF1A can affect
the sensitivity of patients with CRC to oxaliplatin-based
chemotherapy. In a study by Sun et al (97), it was identified that RASSF1A
methylation was independently correlated with the prognosis of
patients treated with oxaliplatin-based chemotherapy, while another
meta-analysis revealed that RASSF1A hypermethylation was a
risk factor and a potential prognostic biomarker in CRC (98). Moreover, it was indicated that the
methylation of RASSF1A may be a prognostic marker for stage
II and III CRC cases. In addition, with further research on the
mechanism of methylation, aberrant methylation may be a promising
and novel target to improve chemotherapy efficacy.
Predictive biomarkers for response to
treatment
Due to the existence of chemotherapy resistance,
patients show different responses to chemotherapy; thus, their
prognosis also varies. Therefore, it is important to identify an
effective prognostic evaluation index. In recent years, numerous
studies have been conducted to evaluate the biomarkers of the
5-fluorouracil (5-FU)-based chemotherapy response in patients with
CRC, but most have not completed the 1/2 discovery phase and will
not be discussed in this study (63).
However, some methylation markers that may be useful for predicting
the response to treatment of CRC have been suggested in previous
studies.
Human mutL homolog 1 (hMLH1)
hMLH1 is considered to be an important
member of the mismatch repair gene, and it encodes a variety of DNA
repair enzymes to cooperate in the recognition and repair of DNA
mismatches (99). Abnormal
hypermethylation of the hMLH1 promoter CpG island has been shown to
be closely associated with the occurrence of CRC, and its
epigenetic changes may affect DNA stability (100). In total, ~15% of CRC cases exhibit
high levels of MSI, reflecting the dysfunction of the
post-replication DNA mismatch repair system, which mainly occurs
via HMLH1 gene silencing mediated by CpG methylation (101). It has been reported that a MLH1
promoter hypermethylation assay is a cost-effective method for
identifying microsatellite-high (MSI-H) in patients with sporadic
CRC (102).
Fu et al (103) evaluated 115 patients with stage II
CRC who were assigned to four groups:
CIMP+/MLH1-unmethylated,
CIMP+/MLH1-methylated,
CIMP−/MLH1-unmethylated or
CIMP−/MLH1-methylated. The results demonstrated that the
CIMP+/MLH1-unmethylated group was associated with higher
aggressiveness and poorer prognosis. DFS and OS were predicted by
CIMP/MLH1 methylation status, with the shortest DFS and OS observed
in the unmethylated CIMP+/MLH1 group. This study revealed that the
CIMP combined with MLH1 methylation status was meaningful for tumor
subtype classification in patients with stage II CRC. In 2018, a
meta-analysis, which that included 47 studies with 4,296 cases and
2,827 controls, suggested that hMLH1 methylation was
associated with an increase in CRC risk (99).
Furthermore, Kuan et al (104) demonstrated that patients with CRC at
an advanced stage who exhibited hMLH1 methylation in tumor tissue
had a higher risk of CRC recurrence compared with patients with
local invasion who had an unmethylated status. It has also been
reported that the methylation of hMLH1 has significant
potential in predicting treatment response, such as that to
5-FU-based chemotherapy, and survival (105,106).
It was also observed in an in vitro study that hMLH1 might
play a key role in cancer stem cells response to 5-FU (107).
Wnt5A
Wnt signaling is a term used to describe a group of
signaling pathways that regulate processes essential to
physiological functions, including cell proliferation and
differentiation. It is also closely associated with CRC (108). Wnt5a/b expression is associated with
epithelial-to-mesenchymal transformation (EMT), and an increase in
its gene expression has been observed in numerous malignancies,
including CRC (109). EMT is a
process that enhances the migration of epithelial cells (110). In cancer, EMT increases the ability
of cancer cells to escape from the primary tumor, allowing them to
infiltrate into neighboring tissues.
In most patients with CRC, genes that regulate the
Wnt pathway display genomic and epigenomic abnormalities. The
decrease of Wnt pathway inhibitor protein can facilitate the growth
and metastasis of CRC, which is an important factor in determining
patient prognosis (111). Wnt5A is
one of the negative regulators of the Wnt pathway and is often
inhibited by promoter methylation in CRC (112). Hibi et al (113) reported that the Wnt5A gene
was detected in 35% of primary CRC cases and that it was methylated
from the early stages of CRC. Furthermore, Kim et al
(112) collected tissues from 194
patients with metastatic or recurrent CRC and detected the
methylation status of Wnt5A. The results indicated that the
methylation frequency of Wnt5A was 32.0%, which was similar
to those of a previous study (113).
Jiang et al (114) also found
that, in 5-FU-treated CRC cases, the Wnt5A methylation
status was significantly associated with longer progression-free
survival and improved drug response, suggesting that 5-FU was more
effective in CRC cases with Wnt5A hypermethylation.
Therefore, the Wnt5A methylation status may predict the 5-FU
treatment response in patients with CRC.
Methylation-based therapies in CRC
In recent years, the comprehensive treatment of CRC
has shown progress, but the patient response to treatment remains
limited. The benefits that patients derive from systemic treatment
are often compromised by resistance to conventional chemotherapy
agents, such as 5-FU, oxaliplatin and irinotecan (115). CRC occurs as a result of gradual
genetic and epigenetic changes and their long-term accumulation.
These epigenetic modifications are reversible. It has been
suggested that combined traditional treatments with epigenetic
agents may represent novel relevant therapeutic targets and may
help to reverse drug resistance (116,117).
DNA methylation is considered to be an epigenetic
change that is closely associated with the occurrence and
development of CRC (118). It is
catalyzed by DNA methyltransferases (DNMTs), which can be divided
into three groups according to their function: DNMT1, which is the
most abundant and important isoenzyme (119), DNMT2 and DNMT3 (DNMT3A and DNMT3B).
DNMTs regulate gene expression and the deregulation of DNA
methylation. On that basis, DNMT inhibitors (DNMTi) have been
widely researched in the prevention and treatment of CRC (115). In CRC cell lines and animal models,
inhibition of DNMT1 and DNMT3B reduced the overall genomic
methylation rates by 95%, leading to re-expression of
tumor-suppressor genes, and was associated with the induction of
apoptosis, decreased cell proliferation and reduced stem cell
function, and helped to overcome oxaliplatin resistance to enhance
the effectiveness of anti-CRC therapy (120,121).
In several clinical trials, DNMTi have been used as a novel
treatment for patients with CRC, particularly those with high
levels of DNA methylation (118,122).
The combined use of methylation-based therapy and standard
chemotherapeutics showed improved treatment efficacy, without
increasing toxicity (122). The
latest research has also shown that the detection of tumor DNA
methylation and the timely use of epigenetic therapies, including
DNMTi, may help expand the therapeutic armamentarium (4). However, additional complete results are
required and should be validated by further trials.
Current situation and difficulties
Although DNA methylation biomarkers show potential
in contributing to the early diagnosis of cancer, and while some
have already been approved for detecting CRC, such as the
methylated SEPT9 DNA plasma assay and MT-sDNA (123), with large numbers of trials having
demonstrated their effectiveness in CRC screening (10,14,60,124),
there remain multiple issues to be overcome before these can be
widely used in the clinical setting. For example, potential
candidate methylation markers should be validated using a large
number of high-quality studies and epigenomic analysis.
Furthermore, standardized methods are required, particularly in
quantifying methylation data in clinical trials. In addition,
clinical trials must aim to detect the true significance of the
methylation markers.
Chemotherapy and radiotherapy may also cause
epigenetic changes in patients with CRC, and the clinical
implications of this observation are unknown and should be further
evaluated (125). Although some of
the underlying mechanisms have not yet been fully determined,
researchers have begun to study the clinical use of epigenetic
drugs, such as radiosensitizer therapy or antitumor therapy.
However, the clinical use of epigenetic drugs may not ultimately
produce the desired results.
Conclusion
Research on DNA methylation is promising. The
application of DNA methylation biomarkers is expected to serve an
important role in non-invasive testing in order to improve the
acceptability of CRC screening in the general population. Moreover,
these DNA methylation changes may contribute to the assessment of
treatment response and adjustment of therapeutic strategies in CRC.
It has been shown that the identification of these biomarkers may
help stratify patients and, potentially, facilitate the development
of precision medicine. However, although methylation markers have
great potential, some limitations must be overcome and the true
significance of biomarkers should be further validated before their
wide use in the clinical setting.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Clinical
Research Fund project of Zhejiang Medical Association (grant no.
2019ZYC-A182) and the Science and Technology Plan Project of
Taizhou (grant no. 1902ky37).
Availability of data and materials
All information included in this review is
documented by relevant references.
Authors' contributions
TF designed the review and collected the literature
data; CK drafted the manuscript. TF and CK reviewed the literature
findings and results. TF and CK read and approved the manuscript
for publication.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Liu Y, Guo F, Zhu X, Guo W, Fu T and Wang
W: Death domain-associated protein promotes colon cancer metastasis
through direct interaction with ZEB1. J Cancer. 11:750–758. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang C, Liu Y, Guo W, Zhu X, Ahuja N and
Fu T: MAPT promoter CpG island hypermethylation is associated with
poor prognosis in patients with stage II colorectal cancer. Cancer
Manag Res Volume. 11:7337–7343. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jones PA, Ohtani H, Chakravarthy A and De
Carvalho DD: Epigenetic therapy in immune-oncology. Nat Rev Cancer.
19:151–161. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bates SE: Epigenetic therapies for cancer.
N Engl J Med. 383:650–663. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Deans C and Maggert KA: What do you mean,
‘epigenetic’? Genetics. 199:887–896. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Al AN, Tupper C and Jialal I: Genetics,
epigenetic mechanism. 2021.PubMed/NCBI
|
|
7
|
Okugawa Y, Grady WM and Goel A: Epigenetic
alterations in colorectal cancer: Emerging biomarkers.
Gastroenterology. 149:1204–1225.e12. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
van der Pol Y and Mouliere F: Toward the
early detection of cancer by decoding the epigenetic and
environmental fingerprints of cell-free DNA. Cancer Cell.
36:350–368. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Luo H, Wei W, Ye Z, Zheng J and Xu RH:
Liquid biopsy of methylation biomarkers in cell-free DNA. Trends
Mol Med. 27:482–500. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Peterse E, Meester R, de Jonge L, Omidvari
AH, Alarid-Escudero F, Knudsen AB, Zauber AG and Lansdorp-Vogelaar
I: Comparing the cost-effectiveness of innovative colorectal cancer
screening tests. J Natl Cancer Inst. 113:154–161. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Komor MA, Bosch LJ, Bounova G, Bolijn AS,
Delis-van DP, Rausch C, Hoogstrate Y, Stubbs AP, de Jong M, Jenster
G, et al: Consensus molecular subtype classification of colorectal
adenomas. J Pathol. 246:266–276. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Locke WJ, Guanzon D, Ma C, Liew YJ,
Duesing KR, Fung K and Ross JP: DNA methylation cancer biomarkers:
Translation to the clinic. Front Genet. 10:11502019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tse JWT, Jenkins LJ, Chionh F and
Mariadason JM: Aberrant DNA methylation in colorectal cancer: What
should we target? Trends Cancer. 3:698–712. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Carethers JM: Fecal DNA testing for
colorectal cancer screening. Annu Rev Med. 71:59–69. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Iyer GR and Hasan Q: Alteration of
methylation status in archival DNA samples: A qualitative
assessment by methylation specific polymerase chain reaction.
Environ Mol Mutagen. 61:837–842. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fu T, Sharmab A, Xie F, Liu Y, Li K, Wan
W, Baylin SB, Wolfgang CL and Ahuja N: Methylation of MGMT is
associated with poor prognosis in patients with stage III duodenal
adenocarcinoma. PLoS One. 11:e1629292016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Azuara D, Ausso S, Rodriguez-Moranta F,
Guardiola J, Sanjuan X, Lobaton T, Boadas J, Piqueras M, Monfort D,
Guino E, et al: New methylation biomarker panel for early diagnosis
of dysplasia or cancer in high-risk inflammatory bowel disease
patients. Inflamm Bowel Dis. 24:2555–2564. 2018.PubMed/NCBI
|
|
18
|
Pichon F, Shen Y, Busato F, P JS,
Jacquelin B, Grand RL, Deleuze JF, Muller-Trutwin M and Tost J:
Analysis and annotation of DNA methylation in two nonhuman primate
species using the Infinium Human Methylation 450K and EPIC
BeadChips. Epigenomics. 13:169–186. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li D, Guo J, Wang S, Zhu L and Shen Z:
Identification of novel methylated targets in colorectal cancer by
microarray analysis and construction of co-expression network.
Oncol Lett. 14:2643–2648. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wagner I and Capesius I: Determination of
5-methylcytosine from plant DNA by high-performance liquid
chromatography. Biochim Biophys Acta. 654:52–56. 1981. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li S and Tollefsbol TO: DNA methylation
methods: Global DNA methylation and methylomic analyses. Methods.
187:28–43. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kang B, Lee HS, Jeon SW, Park SY, Choi GS,
Lee WK, Heo S, Lee DH and Kim DS: Progressive alteration of DNA
methylation of Alu, MGMT, MINT2, and TFPI2 genes in colonic mucosa
during colorectal cancer development. Cancer Biomark. Jun
5–2021.(Epub ahead of print). doi: 10.3233/CBM-203259. View Article : Google Scholar
|
|
23
|
Naini MA, Kavousipour S, Hasanzarini M,
Nasrollah A, Monabati A and Mokarram P: O6-Methyguanine-DNA Methyl
Transferase (MGMT) promoter methylation in serum DNA of Iranian
patients with colorectal cancer. Asian Pac J Cancer Prev.
19:1223–1227. 2018.PubMed/NCBI
|
|
24
|
Kerachian MA and Kerachian M: Long
interspersed nucleotide element-1 (LINE-1) methylation in
colorectal cancer. Clin Chim Acta. 488:209–214. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang Y, Chen PM and Liu RB: Advance in
plasma SEPT9 gene methylation assay for colorectal cancer early
detection. World J Gastrointest Oncol. 10:15–22. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sun J, Zheng M, Li Y and Zhang S:
Structure and function of Septin 9 and its role in human malignant
tumors. World J Gastro Oncol. 12:619–631. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tóth K, Galamb O, Spisák S, Wichmann B,
Sipos F, Valcz G, Leiszter K, Molnár B and Tulassay Z: The
influence of methylated septin 9 gene on RNA and protein level in
colorectal cancer. Pathol Oncol Res. 17:503–509. 2011. View Article : Google Scholar
|
|
28
|
Wasserkort R, Kalmar A, Valcz G, Spisak S,
Krispin M, Toth K, Tulassay Z, Sledziewski AZ and Molnar B:
Aberrant septin 9 DNA methylation in colorectal cancer is
restricted to a single CpG island. BMC Cancer. 13:3982013.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hu J, Hu B, Gui YC, Tan ZB and Xu JW:
Diagnostic value and clinical significance of methylated SEPT9 for
colorectal cancer: A meta-analysis. Med Sci Monit. 25:5813–5822.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Church TR, Wandell M, Lofton-Day C, Mongin
SJ, Burger M, Payne SR, Castaños-Vélez E, Blumenstein BA, Rosch T,
Osborn N, et al: Prospective evaluation of methylated SEPT9 in
plasma for detection of asymptomatic colorectal cancer. Gut.
63:317–325. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Song L, Li Y, Jia J, Zhou G, Wang J, Kang
Q, Jin P, Sheng J, Cai G, Cai S and Han X: Algorithm Optimization
in Methylation Detection with Multiple RT-qPCR. PLoS One.
11:e1633332016. View Article : Google Scholar
|
|
32
|
Choi Y, Kim H, Chung H, Hwang JS, Shin JA,
Han IO and Oh ES: Syndecan-2 regulates cell migration in colon
cancer cells through Tiam1-mediated Rac activation. Biochem Biophys
Res Commun. 391:921–925. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhao G, Ma Y, Li H, Li S, Zhu Y, Liu X,
Xiong S, Liu Y, Miao J, Fei S, et al: A novel plasma based early
colorectal cancer screening assay base on methylated SDC2 and
SFRP2. Clin Chim Acta. 503:84–89. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Han YD, Oh TJ, Chung TH, Jang HW, Kim YN,
An S and Kim NK: Early detection of colorectal cancer based on
presence of methylated syndecan-2 (SDC2) in stool DNA. Clin
Epigenetics. 11:512019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Oh TJ, Oh HI, Seo YY, Jeong D, Kim C, Kang
HW, Han YD, Chung HC, Kim NK and An S: Feasibility of quantifying
SDC2 methylation in stool DNA for early detection of colorectal
cancer. Clin Epigenetics. 9:1262017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Su WC, Kao WY, Chang TK, Tsai HL, Huang
CW, Chen YC, Li CC, Hsieh YC, Yeh HJ, Chang CC, et al: Stool DNA
test targeting methylated syndecan-2 (SDC2) as a noninvasive
screening method for colorectal cancer. Biosci Rep. 29(41):
BSR202019302021. View Article : Google Scholar
|
|
37
|
Kim CW, Kim H, Kim HR, Kye BH, Kim HJ, Min
BS, Oh TJ, An S and Lee SH: Colorectal cancer screening using a
stool DNA-based SDC2 methylation test: A multicenter, prospective
trial. BMC Gastroenterol. 21:1732021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhao G, Liu X, Liu Y, Ma Y, Yang J, Li H,
Xiong S, Fei S, Zheng M and Zhao X: MethylatedSFRP2 and SDC2 in
stool specimens for Colorectal Cancer early detection: A
cost-effective strategy for Chinese population. J Cancer.
12:2665–2672. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Esteller M, Toyota M, Sanchez-Cespedes M,
Capella G, Peinado MA, Watkins DN, Issa JP, Sidransky D, Baylin SB
and Herman JG: Inactivation of the DNA repair gene
O6-methylguanine-DNA methyltransferase by promoter hypermethylation
is associated with G to A mutations in K-ras in colorectal
tumorigenesis. Cancer Res. 60:2368–2371. 2000.PubMed/NCBI
|
|
40
|
Shima K, Morikawa T, Baba Y, Nosho K,
Suzuki M, Yamauchi M, Hayashi M, Giovannucci E, Fuchs CS and Ogino
S: MGMT promoter methylation, loss of expression and prognosis in
855 colorectal cancers. Cancer Causes Control. 22:301–309. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Sartore-Bianchi A, Amatu A, Bonazzina E,
Stabile S, Giannetta L, Cerea G, Schiavetto I, Bencardino K,
Funaioli C, Ricotta R, et al: Pooled analysis of clinical outcome
of patients with chemorefractory metastatic colorectal cancer
treated within phase I/II clinical studies based on individual
biomarkers of susceptibility: A single-institution experience.
Target Oncol. 12:525–533. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Freitas M, Ferreira F, Carvalho S, Silva
F, Lopes P, Antunes L, Salta S, Diniz F, Santos LL, Videira JF, et
al: A novel DNA methylation panel accurately detects colorectal
cancer independently of molecular pathway. J Transl Med. 16:452018.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Qu X, Zhai Y, Wei H, Zhang C, Xing G, Yu Y
and He F: Characterization and expression of three novel
differentiation-related genes belong to the human NDRG gene family.
Mol Cell Biochem. 229:35–44. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Vaes N, Schonkeren SL, Rademakers G,
Holland AM, Koch A, Gijbels MJ, Keulers TG, de Wit M, Moonen L, Van
der Meer J, et al: Loss of enteric neuronal Ndrg4 promotes
colorectal cancer via increased release of Nid1 and Fbln2. EMBO
Rep. 22:e519132021. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Imperiale TF, Ransohoff DF, Itzkowitz SH,
Levin TR, Lavin P, Lidgard GP, Ahlquist DA and Berger BM:
Multitarget stool DNA testing for colorectal-cancer screening. N
Engl J Med. 370:1287–1297. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Bagheri H, Mosallaei M, Bagherpour B,
Khosravi S, Salehi AR and Salehi R: TFPI2 and NDRG4 gene promoter
methylation analysis in peripheral blood mononuclear cells are
novel epigenetic noninvasive biomarkers for colorectal cancer
diagnosis. J Gene Med. 22:e31892020. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Rademakers G, Massen M, Koch A, Draht MX,
Buekers N, Wouters K, Vaes N, De Meyer T, Carvalho B, Meijer GA, et
al: Identification of DNA methylation markers for early detection
of CRC indicates a role for nervous system-related genes in CRC.
Clin Epigenetics. 13:802021. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Mojtabanezhad Shariatpanahi A, Yassi M,
Nouraie M, Sahebkar A, Varshoee Tabrizi F and Kerachian MA: The
importance of stool DNA methylation in colorectal cancer diagnosis:
A meta-analysis. PLoS One. 13:e2007352018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lu H, Huang S, Zhang X, Wang D, Zhang X,
Yuan X, Zhang Q and Huang Z: DNA methylation analysis of SFRP2,
GATA4/5, NDRG4 and VIM for the detection of colorectal cancer in
fecal DNA. Oncol Lett. 8:1751–1756. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Xiao W, Zhao H, Dong W, Li Q, Zhu J, Li G,
Zhang S and Ye M: Quantitative detection of methylated NDRG4 gene
as a candidate biomarker for diagnosis of colorectal cancer. Oncol
Lett. 9:1383–1387. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wang C, Ouyang C, Cho M, Ji J, Sandhu J,
Goel A, Kahn M and Fakih M: Wild-type APC is associated with poor
survival in metastatic microsatellite stable colorectal cancer.
Oncologist. 26:208–214. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Matthaios D, Balgkouranidou I,
Karayiannakis A, Bolanaki H, Xenidis N, Amarantidis K, Chelis L,
Romanidis K, Chatzaki A, Lianidou E, et al: Methylation status of
the APC and RASSF1A promoter in cell-free circulating DNA and its
prognostic role in patients with colorectal cancer. Oncol Lett.
12:748–756. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Aitchison A, Hakkaart C, Day RC, Morrin
HR, Frizelle FA and Keenan JI: APC mutations are not confined to
hotspot regions in early-onset colorectal cancer. Cancers.
12:38292020. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Nunes S, Moreira-Barbosa C, Salta S, Palma
De Sousa S, Pousa I, Oliveira J, Soares M, Rego L, Dias T,
Rodrigues J, et al: Cell-free DNA methylation of selected genes
allows for early detection of the major cancers in women. Cancers.
10:3572018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Liang T, Wang H, Zheng Y, Cao Y, Wu X,
Zhou X and Dong S: APC hypermethylation for early diagnosis of
colorectal cancer: A meta-analysis and literature review.
Oncotarget. 8:46468–46479. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Loh K, Chia JA, Greco S, Cozzi SJ,
Buttenshaw RL, Bond CE, Simms LA, Pike T, Young JP, Jass JR, et al:
Bone morphogenic protein 3 inactivation is an early and frequent
event in colorectal cancer development. Genes Chromosomes Cancer.
47:449–460. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Houshmand M, Abbaszadegan MR and Kerachian
MA: Assessment of bone morphogenetic protein 3 methylation in
Iranian patients with colorectal cancer. Middle East J Dig Dis.
9:158–163. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Rokni P, Shariatpanahi AM, Sakhinia E and
Kerachian MA: BMP3 promoter hypermethylation in plasma-derived
cell-free DNA in colorectal cancer patients. Genes Genom.
40:423–428. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Kisiel JB, Yab TC, Nazer HF, Taylor WR,
Garrity-Park MM, Sandborn WJ, Loftus EV, Wolff BG, Smyrk TC,
Itzkowitz SH, et al: Stool DNA testing for the detection of
colorectal neoplasia in patients with inflammatory bowel disease.
Aliment Pharmacol Ther. 37:546–554. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Redwood DG, Asay ED, Blake ID, Sacco PE,
Christensen CM, Sacco FD, Tiesinga JJ, Devens ME, Alberts SR,
Mahoney DW, et al: Stool DNA testing for screening detection of
colorectal neoplasia in alaska native people. Mayo Clin Proc.
91:61–70. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
El BK, Tariq K, Himri I, Jaafari A, Smaili
W, Kandhro AH, Gouri A and Ghazi B: Decoding colorectal cancer
epigenomics. Cancer Genet. 220:49–76. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Baek YH, Chang E, Kim YJ, Kim BK, Sohn JH
and Park DI: Stool methylation-specific polymerase chain reaction
assay for the detection of colorectal neoplasia in Korean patients.
Dis Colon Rectum. 52:1452–1459, 1459-1463. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Grady WM, Yu M and Markowitz SD:
Epigenetic alterations in the gastrointestinal tract: Current and
emerging use for biomarkers of cancer. Gastroenterology.
160:690–709. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Laugsand EA, Brenne SS and Skorpen F: DNA
methylation markers detected in blood, stool, urine, and tissue in
colorectal cancer: A systematic review of paired samples. Int J
Colorectal Dis. 36:239–251. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Musher BL, Melson JE, Amato G, Chan D,
Hill M, Khan I, Kochuparambil ST, Lyons SE, Orsini JJ Jr, Pedersen
SK, et al: Evaluation of circulating tumor DNA for methylated BCAT1
and IKZF1 to detect recurrence of stage II/stage III colorectal
cancer (CRC). Cancer Epidemiol Biomarkers Prev. 29:2702–2709. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Rasmussen SL, Krarup HB, Sunesen KG,
Johansen MB, Stender MT, Pedersen IS, Madsen PH and
Thorlacius-Ussing O: Hypermethylated DNA, a circulating biomarker
for colorectal cancer detection. PLoS One. 12:e1808092017.
View Article : Google Scholar
|
|
67
|
Yu J, Xie Y, Li M, Zhou F, Zhong Z, Liu Y,
Wang F and Qi J: Association between SFRP promoter hypermethylation
and different types of cancer: A systematic review and
meta-analysis. Oncol Lett. 18:3481–3492. 2019.PubMed/NCBI
|
|
68
|
Hu H, Wang T, Pan R, Yang Y, Li B, Zhou C,
Zhao J, Huang Y and Duan S: Hypermethylated promoters of secreted
frizzled-related protein genes are associated with colorectal
cancer. Pathol Oncol Res. 25:567–575. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Hattori N, Sako M, Kimura K, Iida N,
Takeshima H, Nakata Y, Kono Y and Ushijima T: Novel prodrugs of
decitabine with greater metabolic stability and less toxicity. Clin
Epigenetics. 11:1112019. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Boughanem H, Cabrera-Mulero A,
Hernandez-Alonso P, Clemente-Postigo M, Casanueva FF, Tinahones FJ,
Morcillo S, Crujeiras AB and Macias-Gonzalez M: Association between
variation of circulating 25-OH vitamin D and methylation of
secreted frizzled-related protein 2 in colorectal cancer. Clin
Epigenetics. 12:832020. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Kumar A, Gosipatala SB, Pandey A and Singh
P: Prognostic relevance of SFRP1 gene promoter methylation in
colorectal carcinoma. Asian Pac J Cancer Prev. 20:1571–1577. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Bagci B, Sari M, Karadayi K, Turan M,
Ozdemir O and Bagci G: KRAS, BRAF oncogene mutations and tissue
specific promoter hypermethylation of tumor suppressor SFRP2,
DAPK1, MGMT, HIC1 and p16 genes in colorectal cancer patients.
Cancer Biomark. 17:133–143. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
van Loon K, Huijbers E and Griffioen AW:
Secreted frizzled-related protein 2: A key player in noncanonical
Wnt signaling and tumor angiogenesis. Cancer Metastasis Rev.
40:191–203. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Karam RA, Zidan HE, Abd Elrahman TM, Badr
SA and Amer SA: Study of p16 promoter methylation in Egyptian
colorectal cancer patients. J Cell Biochem. 120:8581–8587. 2018.
View Article : Google Scholar
|
|
75
|
Ye X, Mo M, Xu S, Yang Q, Wu M, Zhang J,
Chen B, Li J, Zhong Y, Huang Q and Cai C: The hypermethylation of
p16 gene exon 1 and exon 2: Potential biomarkers for colorectal
cancer and are associated with cancer pathological staging. BMC
Cancer. 18:10232018. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Lee M, Sup HW, Kyoung KO, Hee SS, Sun CM,
Lee SN and Koo H: Prognostic value of p16INK4a and p14ARF gene
hypermethylation in human colon cancer. Pathol Res Pract.
202:415–424. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Kim SH, Park KH, Shin SJ, Lee KY, Kim TI,
Kim NK, Rha SY, Roh JK and Ahn JB: p16 hypermethylation and KRAS
mutation are independent predictors of cetuximab plus FOLFIRI
chemotherapy in patients with metastatic colorectal cancer. Cancer
Res Treat. 48:208–215. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Shademan M, Zare K, Zahedi M, Mosannen MH,
Bagheri HH, Ghaffarzadegan K, Goshayeshi L and Dehghani H: Promoter
methylation, transcription, and retrotransposition of LINE-1 in
colorectal adenomas and adenocarcinomas. Cancer Cell Int.
20:4262020. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Schauer SN, Carreira PE, Shukla R,
Gerhardt DJ, Gerdes P, Sanchez-Luque FJ, Nicoli P, Kindlova M,
Ghisletti S, Santos AD, et al: L1 retrotransposition is a common
feature of mammalian hepatocarcinogenesis. Genome Res. 28:639–653.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Baba Y, Yagi T, Sawayama H, Hiyoshi Y,
Ishimoto T, Iwatsuki M, Miyamoto Y, Yoshida N and Baba H: Long
interspersed element-1 methylation level as a prognostic biomarker
in gastrointestinal cancers. Digestion. 97:26–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Jiang AC, Buckingham L, Bishehsari F,
Sutherland S, Ma K and Melson JE: Correlation of LINE-1
hypomethylation with size and pathologic extent of dysplasia in
colorectal tubular adenomas. Clin Transl Gastroenterol.
12:e003692021. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Barchitta M, Quattrocchi A, Maugeri A,
Vinciguerra M, Agodi A and Katoh M: LINE-1 hypomethylation in blood
and tissue samples as an epigenetic marker for cancer risk: A
systematic review and meta-analysis. PLoS One. 9:e1094782014.
View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Ye D, Jiang D, Li Y, Jin M and Chen K: The
role of LINE-1 methylation in predicting survival among colorectal
cancer patients: A meta-analysis. Int J Clin Oncol. 22:749–757.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Boughanem H, Martin-Nunez GM, Torres E,
Arranz-Salas I, Alcaide J, Morcillo S, Tinahones FJ, Crujeiras AB
and Macias-Gonzalez M: Impact of tumor LINE-1 methylation level and
neoadjuvant treatment and its association with colorectal cancer
survival. J Pers Med. 10:2192020. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Nagai Y, Sunami E, Yamamoto Y, Hata K,
Okada S, Murono K, Yasuda K, Otani K, Nishikawa T, Tanaka T, et al:
LINE-1 hypomethylation status of circulating cell-free DNA in
plasma as a biomarker for colorectal cancer. Oncotarget.
8:11906–11916. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Symonds EL, Pedersen SK, Murray DH, Jedi
M, Byrne SE, Rabbitt P, Baker RT, Bastin D and Young GP:
Circulating tumour DNA for monitoring colorectal cancer-a
prospective cohort study to assess relationship to tissue
methylation, cancer characteristics and surgical resection. Clin
Epigenetics. 10:632018. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Javierre BM, Rodriguez-Ubreva J,
Al-Shahrour F, Corominas M, Grana O, Ciudad L, Agirre X, Pisano DG,
Valencia A, Roman-Gomez J, et al: Long-range epigenetic silencing
associates with deregulation of Ikaros targets in colorectal cancer
cells. Mol Cancer Res. 9:1139–1151. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Mitchell S, Ho T, Brown G, Baker R, Thomas
M, McEvoy A, Xu Z, Ross J, Lockett T, Young G, et al: Evaluation of
methylation biomarkers for detection of circulating tumor DNA and
application to colorectal cancer. Genes (Basel). 7:1252016.
View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Pedersen SK, Symonds EL, Baker RT, Murray
DH, McEvoy A, Van Doorn SC, Mundt MW, Cole SR, Gopalsamy G, Mangira
D, et al: Evaluation of an assay for methylated BCAT1 and IKZF1 in
plasma for detection of colorectal neoplasia. BMC Cancer.
15:6542015. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Symonds EL, Pedersen SK, Baker RT, Murray
DH, Gaur S, Cole SR, Gopalsamy G, Mangira D, LaPointe LC and Young
GP: A blood test for methylated BCAT1 and IKZF1 vs. a fecal
immunochemical test for detection of colorectal neoplasia. Clin
Transl Gastroen. 7:e1372016. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Symonds EL, Pedersen SK, Murray D, Byrne
SE, Roy A, Karapetis C, Hollington P, Rabbitt P, Jones FS, LaPointe
L, et al: Circulating epigenetic biomarkers for detection of
recurrent colorectal cancer. Cancer. 126:1460–1469. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Murray DH, Symonds EL, Young GP, Byrne S,
Rabbitt P, Roy A, Cornthwaite K, Karapetis CS and Pedersen SK:
Relationship between post-surgery detection of methylated
circulating tumor DNA with risk of residual disease and
recurrence-free survival. J Cancer Res Clin. 144:1741–1750. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Pasha HF, Mohamed RH and Radwan MI:
RASSF1A and SOCS1 genes methylation status as a noninvasive marker
for hepatocellular carcinoma. Cancer Biomark. 24:241–247. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Blanchard TG, Czinn SJ, Banerjee V, Sharda
N, Bafford AC, Mubariz F, Morozov D, Passaniti A, Ahmed H and
Banerjee A: Identification of cross talk between FoxM1 and RASSF1A
as a therapeutic target of colon cancer. Cancers (Basel).
11:1992019. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Sanduleanu S and Siersema PD: Laterally
spreading tumor through the magnifying glass: We only see what we
know. Endoscopy. 48:421–423. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Ni HB, Wang FY, Xu J, He XJ, Chen J, Wu Q,
Wu JF and Sun YS: Screening and identification of a tumor specific
methylation phenotype in the colorectal laterally spreading tumor.
Eur Rev Med Pharmacol Sci. 21:2611–2616. 2017.PubMed/NCBI
|
|
97
|
Sun X, Yuan W, Hao F and Zhuang W:
Promoter methylation of RASSF1A indicates prognosis for patients
with stage II and III colorectal cancer treated with
oxaliplatin-based chemotherapy. Med Sci Monitor. 23:5389–5395.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Hu F, Chen L, Bi MY, Zheng L, He JX, Huang
YZ, Zhang Y, Zhang XL, Guo Q, Luo Y, et al: Potential of RASSF1A
promoter methylation as a biomarker for colorectal cancer:
Meta-analysis and TCGA analysis. Pathol Res Pract. 216:1530092020.
View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Shi B, Chu J, Gao Q and Tian T: Promoter
methylation of human mutL homolog 1 and colorectal cancer risk: A
meta-analysis. J Cancer Res Ther. 14:851–855. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Sun SY, Hu XT, Yu XF, Zhang YY, Liu XH,
Liu YH, Wu SH, Li YY, Cui SX and Qu XJ: Nuclear translocation of
ATG5 induces DNA mismatch repair deficiency (MMR-D)/microsatellite
instability (MSI) via interacting with Mis18alpha in colorectal
cancer. Br J Pharmacol. 178:2351–2369. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Zhang H, Lu Y, Xie Z and Wang K:
Relationship between human mutL Homolog 1 (hMLH1) hypermethylation
and colorectal cancer: A meta-analysis. Med Sci Monitor.
23:3026–3038. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Chung C: Predictive and prognostic
biomarkers with therapeutic targets in colorectal cancer: A 2021
update on current development, evidence, and recommendation. J
Oncol Pharm Pract. 107815522110055252021.PubMed/NCBI
|
|
103
|
Fu T, Liu Y, Li K, Wan W, Pappou EP,
Iacobuzio-Donahue CA, Kerner Z, Baylin SB, Wolfgang CL and Ahuja N:
Tumors with unmethylated MLH1 and the CpG island methylator
phenotype are associated with a poor prognosis in stage II
colorectal cancer patients. Oncotarget. 7:86480–86489. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Kuan JC, Wu CC, Sun CA, Chu CM, Lin FG,
Hsu CH, Kan PC, Lin SC, Yang T and Chou YC: DNA methylation
combinations in adjacent normal colon tissue predict cancer
recurrence: Evidence from a clinical cohort study. PLoS One.
10:e1233962015. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Maier S, Dahlstroem C, Haefliger C, Plum A
and Piepenbrock C: Identifying DNA methylation biomarkers of cancer
drug response. Am J Pharmacogenomics. 5:223–232. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Jover R, Nguyen TP, Perez-Carbonell L,
Zapater P, Paya A, Alenda C, Rojas E, Cubiella J, Balaguer F,
Morillas JD, et al: 5-Fluorouracil adjuvant chemotherapy does not
increase survival in patients with CpG island methylator phenotype
colorectal cancer. Gastroenterology. 140:1174–1181. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Oliver JA, Ortiz R, Jimenez-Luna C, Cabeza
L, Perazzoli G, Caba O, Mesas C, Melguizo C and Prados J:
MMR-proficient and MMR-deficient colorectal cancer cells:
5-Fluorouracil treatment response and correlation to CD133 and MGMT
expression. J Biosci. 45:1212020. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Cheng X, Xu X, Chen D, Zhao F and Wang W:
Therapeutic potential of targeting the Wnt/beta-catenin signaling
pathway in colorectal cancer. Biomed Pharmacother. 110:473–481.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Gujral TS, Chan M, Peshkin L, Sorger PK,
Kirschner MW and MacBeath G: A noncanonical Frizzled2 pathway
regulates epithelial-mesenchymal transition and metastasis. Cell.
159:844–856. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Qi L, Chen J, Zhou B, Xu K, Wang K, Fang
Z, Shao Y, Yuan Y, Zheng S and Hu W: HomeoboxC6 promotes metastasis
by orchestrating the DKK1/Wnt/β-catenin axis in right-sided colon
cancer. Cell Death Dis. 12:3372021. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Chen X, Liu HL, Zhao FH, Jiao ZX, Wang JS
and Dang YM: Wnt5a plays controversial roles in cancer progression.
Chin Med Sci J. 35:357–365. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Kim SH, Park KH, Shin SJ, Lee KY, Kim TI,
Kim NK, Rha SY and Ahn JB: CpG island methylator phenotype and
methylation of Wnt pathway genes together predict survival in
patients with colorectal cancer. Yonsei Med J. 59:5882018.
View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Hibi K, Mizukami H, Goto T, Kitamura Y,
Sakata M, Saito M, Ishibashi K, Kigawa G, Nemoto H and Sanada Y:
WNT5A gene is aberrantly methylated from the early stages of
colorectal cancers. Hepatogastroenterology. 56:1007–1009.
2009.PubMed/NCBI
|
|
114
|
Jiang G, Lin J, Wang W, Sun M, Chen K and
Wang F: WNT5A promoter methylation is associated with better
responses and longer progression-free survival in colorectal cancer
patients treated with 5-fluorouracil-based chemotherapy. Genet Test
Mol Bioma. 21:74–79. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Draht M, Goudkade D, Koch A, Grabsch HI,
Weijenberg MP, van Engeland M, Melotte V and Smits KM: Prognostic
DNA methylation markers for sporadic colorectal cancer: A
systematic review. Clin Epigenetics. 10:352018. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Nagaraju GP, Kasa P, Dariya B, Surepalli
N, Peela S and Ahmad S: Epigenetics and therapeutic targets in
gastrointestinal malignancies. Drug Discov Today.
S1359-6446(21)00202-6. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Jung G, Hernández-Illán E, Moreira L,
Balaguer F and Goel A: Epigenetics of colorectal cancer: Biomarker
and therapeutic potential. Nat Rev Gastro Hepat. 17:111–130. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Cervena K, Siskova A, Buchler T, Vodicka P
and Vymetalkova V: Methylation-based therapies for colorectal
cancer. Cells-Basel. 9:15402020. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Singh M, Kumar V, Sehrawat N, Yadav M,
Chaudhary M, Upadhyay SK, Kumar S, Sharma V, Kumar S, Dilbaghi N
and Sharma AK: Current paradigms in epigenetic anticancer
therapeutics and future challenges. Semin Cancer Biol.
S1044-579X(21)00063-8. 2021. View Article : Google Scholar
|
|
120
|
Wei T, Lin Y, Tang S, Luo C, Tsai C, Shun
C and Chen C: Metabolic targeting of HIF-1α potentiates the
therapeutic efficacy of oxaliplatin in colorectal cancer. Oncogene.
39:414–427. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Li J, Su X, Dai L, Chen N, Fang C, Dong Z,
Fu J, Yu Y, Wang W, Zhang H, et al: Temporal DNA methylation
pattern and targeted therapy in colitis-associated cancer.
Carcinogenesis. 41:235–244. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Pechalrieu D, Etievant C and Arimondo PB:
DNA methyltransferase inhibitors in cancer: From pharmacology to
translational studies. Biochem Pharmacol. 129:1–13. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Kisiel JB, Klepp P, Allawi HT, Taylor WR,
Giakoumopoulos M, Sander T, Yab TC, Moum BA, Lidgard GP, Brackmann
S, et al: Analysis of DNA methylation at specific loci in stool
samples detects colorectal cancer and high-grade dysplasia in
patients with inflammatory bowel disease. Clin Gastroenterol
Hepatol. 17:914–921.e5. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Chen J, Sun H, Tang W, Zhou L, Xie X, Qu
Z, Chen M, Wang S, Yang T, Dai Y, et al: DNA methylation biomarkers
in stool for early screening of colorectal cancer. J Cancer.
10:5264–5271. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Pan Y, Liu G, Zhou F, Su B and Li Y: DNA
methylation profiles in cancer diagnosis and therapeutics. Clin Exp
Med. 18:1–14. 2018. View Article : Google Scholar : PubMed/NCBI
|















