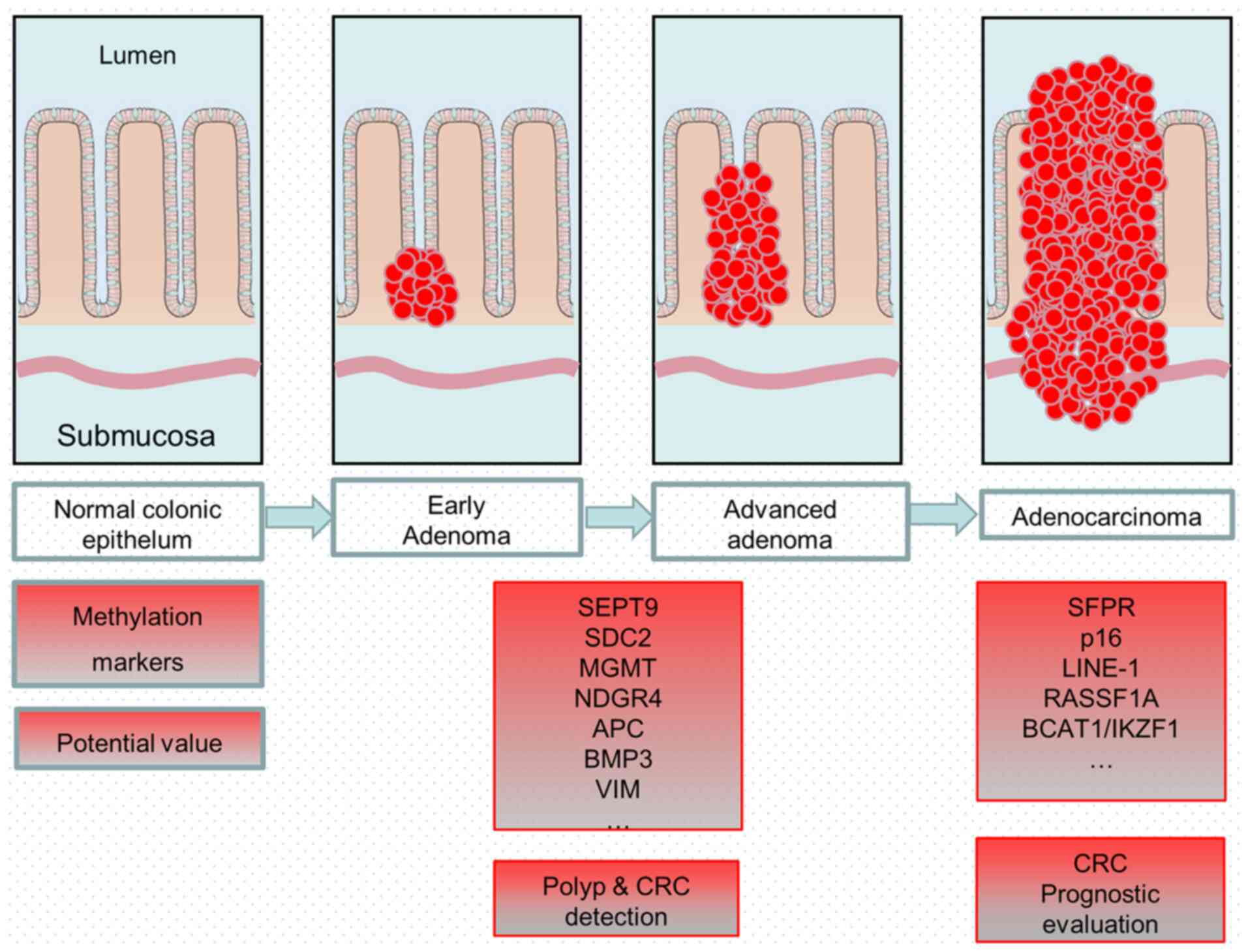
|
1
|
Liu Y, Guo F, Zhu X, Guo W, Fu T and Wang
W: Death domain-associated protein promotes colon cancer metastasis
through direct interaction with ZEB1. J Cancer. 11:750–758. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang C, Liu Y, Guo W, Zhu X, Ahuja N and
Fu T: MAPT promoter CpG island hypermethylation is associated with
poor prognosis in patients with stage II colorectal cancer. Cancer
Manag Res Volume. 11:7337–7343. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jones PA, Ohtani H, Chakravarthy A and De
Carvalho DD: Epigenetic therapy in immune-oncology. Nat Rev Cancer.
19:151–161. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bates SE: Epigenetic therapies for cancer.
N Engl J Med. 383:650–663. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Deans C and Maggert KA: What do you mean,
‘epigenetic’? Genetics. 199:887–896. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Al AN, Tupper C and Jialal I: Genetics,
epigenetic mechanism. 2021.PubMed/NCBI
|
|
7
|
Okugawa Y, Grady WM and Goel A: Epigenetic
alterations in colorectal cancer: Emerging biomarkers.
Gastroenterology. 149:1204–1225.e12. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
van der Pol Y and Mouliere F: Toward the
early detection of cancer by decoding the epigenetic and
environmental fingerprints of cell-free DNA. Cancer Cell.
36:350–368. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Luo H, Wei W, Ye Z, Zheng J and Xu RH:
Liquid biopsy of methylation biomarkers in cell-free DNA. Trends
Mol Med. 27:482–500. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Peterse E, Meester R, de Jonge L, Omidvari
AH, Alarid-Escudero F, Knudsen AB, Zauber AG and Lansdorp-Vogelaar
I: Comparing the cost-effectiveness of innovative colorectal cancer
screening tests. J Natl Cancer Inst. 113:154–161. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Komor MA, Bosch LJ, Bounova G, Bolijn AS,
Delis-van DP, Rausch C, Hoogstrate Y, Stubbs AP, de Jong M, Jenster
G, et al: Consensus molecular subtype classification of colorectal
adenomas. J Pathol. 246:266–276. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Locke WJ, Guanzon D, Ma C, Liew YJ,
Duesing KR, Fung K and Ross JP: DNA methylation cancer biomarkers:
Translation to the clinic. Front Genet. 10:11502019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tse JWT, Jenkins LJ, Chionh F and
Mariadason JM: Aberrant DNA methylation in colorectal cancer: What
should we target? Trends Cancer. 3:698–712. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Carethers JM: Fecal DNA testing for
colorectal cancer screening. Annu Rev Med. 71:59–69. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Iyer GR and Hasan Q: Alteration of
methylation status in archival DNA samples: A qualitative
assessment by methylation specific polymerase chain reaction.
Environ Mol Mutagen. 61:837–842. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fu T, Sharmab A, Xie F, Liu Y, Li K, Wan
W, Baylin SB, Wolfgang CL and Ahuja N: Methylation of MGMT is
associated with poor prognosis in patients with stage III duodenal
adenocarcinoma. PLoS One. 11:e1629292016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Azuara D, Ausso S, Rodriguez-Moranta F,
Guardiola J, Sanjuan X, Lobaton T, Boadas J, Piqueras M, Monfort D,
Guino E, et al: New methylation biomarker panel for early diagnosis
of dysplasia or cancer in high-risk inflammatory bowel disease
patients. Inflamm Bowel Dis. 24:2555–2564. 2018.PubMed/NCBI
|
|
18
|
Pichon F, Shen Y, Busato F, P JS,
Jacquelin B, Grand RL, Deleuze JF, Muller-Trutwin M and Tost J:
Analysis and annotation of DNA methylation in two nonhuman primate
species using the Infinium Human Methylation 450K and EPIC
BeadChips. Epigenomics. 13:169–186. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li D, Guo J, Wang S, Zhu L and Shen Z:
Identification of novel methylated targets in colorectal cancer by
microarray analysis and construction of co-expression network.
Oncol Lett. 14:2643–2648. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wagner I and Capesius I: Determination of
5-methylcytosine from plant DNA by high-performance liquid
chromatography. Biochim Biophys Acta. 654:52–56. 1981. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li S and Tollefsbol TO: DNA methylation
methods: Global DNA methylation and methylomic analyses. Methods.
187:28–43. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kang B, Lee HS, Jeon SW, Park SY, Choi GS,
Lee WK, Heo S, Lee DH and Kim DS: Progressive alteration of DNA
methylation of Alu, MGMT, MINT2, and TFPI2 genes in colonic mucosa
during colorectal cancer development. Cancer Biomark. Jun
5–2021.(Epub ahead of print). doi: 10.3233/CBM-203259. View Article : Google Scholar
|
|
23
|
Naini MA, Kavousipour S, Hasanzarini M,
Nasrollah A, Monabati A and Mokarram P: O6-Methyguanine-DNA Methyl
Transferase (MGMT) promoter methylation in serum DNA of Iranian
patients with colorectal cancer. Asian Pac J Cancer Prev.
19:1223–1227. 2018.PubMed/NCBI
|
|
24
|
Kerachian MA and Kerachian M: Long
interspersed nucleotide element-1 (LINE-1) methylation in
colorectal cancer. Clin Chim Acta. 488:209–214. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang Y, Chen PM and Liu RB: Advance in
plasma SEPT9 gene methylation assay for colorectal cancer early
detection. World J Gastrointest Oncol. 10:15–22. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sun J, Zheng M, Li Y and Zhang S:
Structure and function of Septin 9 and its role in human malignant
tumors. World J Gastro Oncol. 12:619–631. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tóth K, Galamb O, Spisák S, Wichmann B,
Sipos F, Valcz G, Leiszter K, Molnár B and Tulassay Z: The
influence of methylated septin 9 gene on RNA and protein level in
colorectal cancer. Pathol Oncol Res. 17:503–509. 2011. View Article : Google Scholar
|
|
28
|
Wasserkort R, Kalmar A, Valcz G, Spisak S,
Krispin M, Toth K, Tulassay Z, Sledziewski AZ and Molnar B:
Aberrant septin 9 DNA methylation in colorectal cancer is
restricted to a single CpG island. BMC Cancer. 13:3982013.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hu J, Hu B, Gui YC, Tan ZB and Xu JW:
Diagnostic value and clinical significance of methylated SEPT9 for
colorectal cancer: A meta-analysis. Med Sci Monit. 25:5813–5822.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Church TR, Wandell M, Lofton-Day C, Mongin
SJ, Burger M, Payne SR, Castaños-Vélez E, Blumenstein BA, Rosch T,
Osborn N, et al: Prospective evaluation of methylated SEPT9 in
plasma for detection of asymptomatic colorectal cancer. Gut.
63:317–325. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Song L, Li Y, Jia J, Zhou G, Wang J, Kang
Q, Jin P, Sheng J, Cai G, Cai S and Han X: Algorithm Optimization
in Methylation Detection with Multiple RT-qPCR. PLoS One.
11:e1633332016. View Article : Google Scholar
|
|
32
|
Choi Y, Kim H, Chung H, Hwang JS, Shin JA,
Han IO and Oh ES: Syndecan-2 regulates cell migration in colon
cancer cells through Tiam1-mediated Rac activation. Biochem Biophys
Res Commun. 391:921–925. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhao G, Ma Y, Li H, Li S, Zhu Y, Liu X,
Xiong S, Liu Y, Miao J, Fei S, et al: A novel plasma based early
colorectal cancer screening assay base on methylated SDC2 and
SFRP2. Clin Chim Acta. 503:84–89. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Han YD, Oh TJ, Chung TH, Jang HW, Kim YN,
An S and Kim NK: Early detection of colorectal cancer based on
presence of methylated syndecan-2 (SDC2) in stool DNA. Clin
Epigenetics. 11:512019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Oh TJ, Oh HI, Seo YY, Jeong D, Kim C, Kang
HW, Han YD, Chung HC, Kim NK and An S: Feasibility of quantifying
SDC2 methylation in stool DNA for early detection of colorectal
cancer. Clin Epigenetics. 9:1262017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Su WC, Kao WY, Chang TK, Tsai HL, Huang
CW, Chen YC, Li CC, Hsieh YC, Yeh HJ, Chang CC, et al: Stool DNA
test targeting methylated syndecan-2 (SDC2) as a noninvasive
screening method for colorectal cancer. Biosci Rep. 29(41):
BSR202019302021. View Article : Google Scholar
|
|
37
|
Kim CW, Kim H, Kim HR, Kye BH, Kim HJ, Min
BS, Oh TJ, An S and Lee SH: Colorectal cancer screening using a
stool DNA-based SDC2 methylation test: A multicenter, prospective
trial. BMC Gastroenterol. 21:1732021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhao G, Liu X, Liu Y, Ma Y, Yang J, Li H,
Xiong S, Fei S, Zheng M and Zhao X: MethylatedSFRP2 and SDC2 in
stool specimens for Colorectal Cancer early detection: A
cost-effective strategy for Chinese population. J Cancer.
12:2665–2672. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Esteller M, Toyota M, Sanchez-Cespedes M,
Capella G, Peinado MA, Watkins DN, Issa JP, Sidransky D, Baylin SB
and Herman JG: Inactivation of the DNA repair gene
O6-methylguanine-DNA methyltransferase by promoter hypermethylation
is associated with G to A mutations in K-ras in colorectal
tumorigenesis. Cancer Res. 60:2368–2371. 2000.PubMed/NCBI
|
|
40
|
Shima K, Morikawa T, Baba Y, Nosho K,
Suzuki M, Yamauchi M, Hayashi M, Giovannucci E, Fuchs CS and Ogino
S: MGMT promoter methylation, loss of expression and prognosis in
855 colorectal cancers. Cancer Causes Control. 22:301–309. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Sartore-Bianchi A, Amatu A, Bonazzina E,
Stabile S, Giannetta L, Cerea G, Schiavetto I, Bencardino K,
Funaioli C, Ricotta R, et al: Pooled analysis of clinical outcome
of patients with chemorefractory metastatic colorectal cancer
treated within phase I/II clinical studies based on individual
biomarkers of susceptibility: A single-institution experience.
Target Oncol. 12:525–533. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Freitas M, Ferreira F, Carvalho S, Silva
F, Lopes P, Antunes L, Salta S, Diniz F, Santos LL, Videira JF, et
al: A novel DNA methylation panel accurately detects colorectal
cancer independently of molecular pathway. J Transl Med. 16:452018.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Qu X, Zhai Y, Wei H, Zhang C, Xing G, Yu Y
and He F: Characterization and expression of three novel
differentiation-related genes belong to the human NDRG gene family.
Mol Cell Biochem. 229:35–44. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Vaes N, Schonkeren SL, Rademakers G,
Holland AM, Koch A, Gijbels MJ, Keulers TG, de Wit M, Moonen L, Van
der Meer J, et al: Loss of enteric neuronal Ndrg4 promotes
colorectal cancer via increased release of Nid1 and Fbln2. EMBO
Rep. 22:e519132021. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Imperiale TF, Ransohoff DF, Itzkowitz SH,
Levin TR, Lavin P, Lidgard GP, Ahlquist DA and Berger BM:
Multitarget stool DNA testing for colorectal-cancer screening. N
Engl J Med. 370:1287–1297. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Bagheri H, Mosallaei M, Bagherpour B,
Khosravi S, Salehi AR and Salehi R: TFPI2 and NDRG4 gene promoter
methylation analysis in peripheral blood mononuclear cells are
novel epigenetic noninvasive biomarkers for colorectal cancer
diagnosis. J Gene Med. 22:e31892020. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Rademakers G, Massen M, Koch A, Draht MX,
Buekers N, Wouters K, Vaes N, De Meyer T, Carvalho B, Meijer GA, et
al: Identification of DNA methylation markers for early detection
of CRC indicates a role for nervous system-related genes in CRC.
Clin Epigenetics. 13:802021. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Mojtabanezhad Shariatpanahi A, Yassi M,
Nouraie M, Sahebkar A, Varshoee Tabrizi F and Kerachian MA: The
importance of stool DNA methylation in colorectal cancer diagnosis:
A meta-analysis. PLoS One. 13:e2007352018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lu H, Huang S, Zhang X, Wang D, Zhang X,
Yuan X, Zhang Q and Huang Z: DNA methylation analysis of SFRP2,
GATA4/5, NDRG4 and VIM for the detection of colorectal cancer in
fecal DNA. Oncol Lett. 8:1751–1756. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Xiao W, Zhao H, Dong W, Li Q, Zhu J, Li G,
Zhang S and Ye M: Quantitative detection of methylated NDRG4 gene
as a candidate biomarker for diagnosis of colorectal cancer. Oncol
Lett. 9:1383–1387. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wang C, Ouyang C, Cho M, Ji J, Sandhu J,
Goel A, Kahn M and Fakih M: Wild-type APC is associated with poor
survival in metastatic microsatellite stable colorectal cancer.
Oncologist. 26:208–214. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Matthaios D, Balgkouranidou I,
Karayiannakis A, Bolanaki H, Xenidis N, Amarantidis K, Chelis L,
Romanidis K, Chatzaki A, Lianidou E, et al: Methylation status of
the APC and RASSF1A promoter in cell-free circulating DNA and its
prognostic role in patients with colorectal cancer. Oncol Lett.
12:748–756. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Aitchison A, Hakkaart C, Day RC, Morrin
HR, Frizelle FA and Keenan JI: APC mutations are not confined to
hotspot regions in early-onset colorectal cancer. Cancers.
12:38292020. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Nunes S, Moreira-Barbosa C, Salta S, Palma
De Sousa S, Pousa I, Oliveira J, Soares M, Rego L, Dias T,
Rodrigues J, et al: Cell-free DNA methylation of selected genes
allows for early detection of the major cancers in women. Cancers.
10:3572018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Liang T, Wang H, Zheng Y, Cao Y, Wu X,
Zhou X and Dong S: APC hypermethylation for early diagnosis of
colorectal cancer: A meta-analysis and literature review.
Oncotarget. 8:46468–46479. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Loh K, Chia JA, Greco S, Cozzi SJ,
Buttenshaw RL, Bond CE, Simms LA, Pike T, Young JP, Jass JR, et al:
Bone morphogenic protein 3 inactivation is an early and frequent
event in colorectal cancer development. Genes Chromosomes Cancer.
47:449–460. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Houshmand M, Abbaszadegan MR and Kerachian
MA: Assessment of bone morphogenetic protein 3 methylation in
Iranian patients with colorectal cancer. Middle East J Dig Dis.
9:158–163. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Rokni P, Shariatpanahi AM, Sakhinia E and
Kerachian MA: BMP3 promoter hypermethylation in plasma-derived
cell-free DNA in colorectal cancer patients. Genes Genom.
40:423–428. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Kisiel JB, Yab TC, Nazer HF, Taylor WR,
Garrity-Park MM, Sandborn WJ, Loftus EV, Wolff BG, Smyrk TC,
Itzkowitz SH, et al: Stool DNA testing for the detection of
colorectal neoplasia in patients with inflammatory bowel disease.
Aliment Pharmacol Ther. 37:546–554. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Redwood DG, Asay ED, Blake ID, Sacco PE,
Christensen CM, Sacco FD, Tiesinga JJ, Devens ME, Alberts SR,
Mahoney DW, et al: Stool DNA testing for screening detection of
colorectal neoplasia in alaska native people. Mayo Clin Proc.
91:61–70. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
El BK, Tariq K, Himri I, Jaafari A, Smaili
W, Kandhro AH, Gouri A and Ghazi B: Decoding colorectal cancer
epigenomics. Cancer Genet. 220:49–76. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Baek YH, Chang E, Kim YJ, Kim BK, Sohn JH
and Park DI: Stool methylation-specific polymerase chain reaction
assay for the detection of colorectal neoplasia in Korean patients.
Dis Colon Rectum. 52:1452–1459, 1459-1463. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Grady WM, Yu M and Markowitz SD:
Epigenetic alterations in the gastrointestinal tract: Current and
emerging use for biomarkers of cancer. Gastroenterology.
160:690–709. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Laugsand EA, Brenne SS and Skorpen F: DNA
methylation markers detected in blood, stool, urine, and tissue in
colorectal cancer: A systematic review of paired samples. Int J
Colorectal Dis. 36:239–251. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Musher BL, Melson JE, Amato G, Chan D,
Hill M, Khan I, Kochuparambil ST, Lyons SE, Orsini JJ Jr, Pedersen
SK, et al: Evaluation of circulating tumor DNA for methylated BCAT1
and IKZF1 to detect recurrence of stage II/stage III colorectal
cancer (CRC). Cancer Epidemiol Biomarkers Prev. 29:2702–2709. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Rasmussen SL, Krarup HB, Sunesen KG,
Johansen MB, Stender MT, Pedersen IS, Madsen PH and
Thorlacius-Ussing O: Hypermethylated DNA, a circulating biomarker
for colorectal cancer detection. PLoS One. 12:e1808092017.
View Article : Google Scholar
|
|
67
|
Yu J, Xie Y, Li M, Zhou F, Zhong Z, Liu Y,
Wang F and Qi J: Association between SFRP promoter hypermethylation
and different types of cancer: A systematic review and
meta-analysis. Oncol Lett. 18:3481–3492. 2019.PubMed/NCBI
|
|
68
|
Hu H, Wang T, Pan R, Yang Y, Li B, Zhou C,
Zhao J, Huang Y and Duan S: Hypermethylated promoters of secreted
frizzled-related protein genes are associated with colorectal
cancer. Pathol Oncol Res. 25:567–575. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Hattori N, Sako M, Kimura K, Iida N,
Takeshima H, Nakata Y, Kono Y and Ushijima T: Novel prodrugs of
decitabine with greater metabolic stability and less toxicity. Clin
Epigenetics. 11:1112019. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Boughanem H, Cabrera-Mulero A,
Hernandez-Alonso P, Clemente-Postigo M, Casanueva FF, Tinahones FJ,
Morcillo S, Crujeiras AB and Macias-Gonzalez M: Association between
variation of circulating 25-OH vitamin D and methylation of
secreted frizzled-related protein 2 in colorectal cancer. Clin
Epigenetics. 12:832020. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Kumar A, Gosipatala SB, Pandey A and Singh
P: Prognostic relevance of SFRP1 gene promoter methylation in
colorectal carcinoma. Asian Pac J Cancer Prev. 20:1571–1577. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Bagci B, Sari M, Karadayi K, Turan M,
Ozdemir O and Bagci G: KRAS, BRAF oncogene mutations and tissue
specific promoter hypermethylation of tumor suppressor SFRP2,
DAPK1, MGMT, HIC1 and p16 genes in colorectal cancer patients.
Cancer Biomark. 17:133–143. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
van Loon K, Huijbers E and Griffioen AW:
Secreted frizzled-related protein 2: A key player in noncanonical
Wnt signaling and tumor angiogenesis. Cancer Metastasis Rev.
40:191–203. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Karam RA, Zidan HE, Abd Elrahman TM, Badr
SA and Amer SA: Study of p16 promoter methylation in Egyptian
colorectal cancer patients. J Cell Biochem. 120:8581–8587. 2018.
View Article : Google Scholar
|
|
75
|
Ye X, Mo M, Xu S, Yang Q, Wu M, Zhang J,
Chen B, Li J, Zhong Y, Huang Q and Cai C: The hypermethylation of
p16 gene exon 1 and exon 2: Potential biomarkers for colorectal
cancer and are associated with cancer pathological staging. BMC
Cancer. 18:10232018. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Lee M, Sup HW, Kyoung KO, Hee SS, Sun CM,
Lee SN and Koo H: Prognostic value of p16INK4a and p14ARF gene
hypermethylation in human colon cancer. Pathol Res Pract.
202:415–424. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Kim SH, Park KH, Shin SJ, Lee KY, Kim TI,
Kim NK, Rha SY, Roh JK and Ahn JB: p16 hypermethylation and KRAS
mutation are independent predictors of cetuximab plus FOLFIRI
chemotherapy in patients with metastatic colorectal cancer. Cancer
Res Treat. 48:208–215. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Shademan M, Zare K, Zahedi M, Mosannen MH,
Bagheri HH, Ghaffarzadegan K, Goshayeshi L and Dehghani H: Promoter
methylation, transcription, and retrotransposition of LINE-1 in
colorectal adenomas and adenocarcinomas. Cancer Cell Int.
20:4262020. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Schauer SN, Carreira PE, Shukla R,
Gerhardt DJ, Gerdes P, Sanchez-Luque FJ, Nicoli P, Kindlova M,
Ghisletti S, Santos AD, et al: L1 retrotransposition is a common
feature of mammalian hepatocarcinogenesis. Genome Res. 28:639–653.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Baba Y, Yagi T, Sawayama H, Hiyoshi Y,
Ishimoto T, Iwatsuki M, Miyamoto Y, Yoshida N and Baba H: Long
interspersed element-1 methylation level as a prognostic biomarker
in gastrointestinal cancers. Digestion. 97:26–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Jiang AC, Buckingham L, Bishehsari F,
Sutherland S, Ma K and Melson JE: Correlation of LINE-1
hypomethylation with size and pathologic extent of dysplasia in
colorectal tubular adenomas. Clin Transl Gastroenterol.
12:e003692021. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Barchitta M, Quattrocchi A, Maugeri A,
Vinciguerra M, Agodi A and Katoh M: LINE-1 hypomethylation in blood
and tissue samples as an epigenetic marker for cancer risk: A
systematic review and meta-analysis. PLoS One. 9:e1094782014.
View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Ye D, Jiang D, Li Y, Jin M and Chen K: The
role of LINE-1 methylation in predicting survival among colorectal
cancer patients: A meta-analysis. Int J Clin Oncol. 22:749–757.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Boughanem H, Martin-Nunez GM, Torres E,
Arranz-Salas I, Alcaide J, Morcillo S, Tinahones FJ, Crujeiras AB
and Macias-Gonzalez M: Impact of tumor LINE-1 methylation level and
neoadjuvant treatment and its association with colorectal cancer
survival. J Pers Med. 10:2192020. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Nagai Y, Sunami E, Yamamoto Y, Hata K,
Okada S, Murono K, Yasuda K, Otani K, Nishikawa T, Tanaka T, et al:
LINE-1 hypomethylation status of circulating cell-free DNA in
plasma as a biomarker for colorectal cancer. Oncotarget.
8:11906–11916. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Symonds EL, Pedersen SK, Murray DH, Jedi
M, Byrne SE, Rabbitt P, Baker RT, Bastin D and Young GP:
Circulating tumour DNA for monitoring colorectal cancer-a
prospective cohort study to assess relationship to tissue
methylation, cancer characteristics and surgical resection. Clin
Epigenetics. 10:632018. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Javierre BM, Rodriguez-Ubreva J,
Al-Shahrour F, Corominas M, Grana O, Ciudad L, Agirre X, Pisano DG,
Valencia A, Roman-Gomez J, et al: Long-range epigenetic silencing
associates with deregulation of Ikaros targets in colorectal cancer
cells. Mol Cancer Res. 9:1139–1151. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Mitchell S, Ho T, Brown G, Baker R, Thomas
M, McEvoy A, Xu Z, Ross J, Lockett T, Young G, et al: Evaluation of
methylation biomarkers for detection of circulating tumor DNA and
application to colorectal cancer. Genes (Basel). 7:1252016.
View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Pedersen SK, Symonds EL, Baker RT, Murray
DH, McEvoy A, Van Doorn SC, Mundt MW, Cole SR, Gopalsamy G, Mangira
D, et al: Evaluation of an assay for methylated BCAT1 and IKZF1 in
plasma for detection of colorectal neoplasia. BMC Cancer.
15:6542015. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Symonds EL, Pedersen SK, Baker RT, Murray
DH, Gaur S, Cole SR, Gopalsamy G, Mangira D, LaPointe LC and Young
GP: A blood test for methylated BCAT1 and IKZF1 vs. a fecal
immunochemical test for detection of colorectal neoplasia. Clin
Transl Gastroen. 7:e1372016. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Symonds EL, Pedersen SK, Murray D, Byrne
SE, Roy A, Karapetis C, Hollington P, Rabbitt P, Jones FS, LaPointe
L, et al: Circulating epigenetic biomarkers for detection of
recurrent colorectal cancer. Cancer. 126:1460–1469. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Murray DH, Symonds EL, Young GP, Byrne S,
Rabbitt P, Roy A, Cornthwaite K, Karapetis CS and Pedersen SK:
Relationship between post-surgery detection of methylated
circulating tumor DNA with risk of residual disease and
recurrence-free survival. J Cancer Res Clin. 144:1741–1750. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Pasha HF, Mohamed RH and Radwan MI:
RASSF1A and SOCS1 genes methylation status as a noninvasive marker
for hepatocellular carcinoma. Cancer Biomark. 24:241–247. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Blanchard TG, Czinn SJ, Banerjee V, Sharda
N, Bafford AC, Mubariz F, Morozov D, Passaniti A, Ahmed H and
Banerjee A: Identification of cross talk between FoxM1 and RASSF1A
as a therapeutic target of colon cancer. Cancers (Basel).
11:1992019. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Sanduleanu S and Siersema PD: Laterally
spreading tumor through the magnifying glass: We only see what we
know. Endoscopy. 48:421–423. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Ni HB, Wang FY, Xu J, He XJ, Chen J, Wu Q,
Wu JF and Sun YS: Screening and identification of a tumor specific
methylation phenotype in the colorectal laterally spreading tumor.
Eur Rev Med Pharmacol Sci. 21:2611–2616. 2017.PubMed/NCBI
|
|
97
|
Sun X, Yuan W, Hao F and Zhuang W:
Promoter methylation of RASSF1A indicates prognosis for patients
with stage II and III colorectal cancer treated with
oxaliplatin-based chemotherapy. Med Sci Monitor. 23:5389–5395.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Hu F, Chen L, Bi MY, Zheng L, He JX, Huang
YZ, Zhang Y, Zhang XL, Guo Q, Luo Y, et al: Potential of RASSF1A
promoter methylation as a biomarker for colorectal cancer:
Meta-analysis and TCGA analysis. Pathol Res Pract. 216:1530092020.
View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Shi B, Chu J, Gao Q and Tian T: Promoter
methylation of human mutL homolog 1 and colorectal cancer risk: A
meta-analysis. J Cancer Res Ther. 14:851–855. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Sun SY, Hu XT, Yu XF, Zhang YY, Liu XH,
Liu YH, Wu SH, Li YY, Cui SX and Qu XJ: Nuclear translocation of
ATG5 induces DNA mismatch repair deficiency (MMR-D)/microsatellite
instability (MSI) via interacting with Mis18alpha in colorectal
cancer. Br J Pharmacol. 178:2351–2369. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Zhang H, Lu Y, Xie Z and Wang K:
Relationship between human mutL Homolog 1 (hMLH1) hypermethylation
and colorectal cancer: A meta-analysis. Med Sci Monitor.
23:3026–3038. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Chung C: Predictive and prognostic
biomarkers with therapeutic targets in colorectal cancer: A 2021
update on current development, evidence, and recommendation. J
Oncol Pharm Pract. 107815522110055252021.PubMed/NCBI
|
|
103
|
Fu T, Liu Y, Li K, Wan W, Pappou EP,
Iacobuzio-Donahue CA, Kerner Z, Baylin SB, Wolfgang CL and Ahuja N:
Tumors with unmethylated MLH1 and the CpG island methylator
phenotype are associated with a poor prognosis in stage II
colorectal cancer patients. Oncotarget. 7:86480–86489. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Kuan JC, Wu CC, Sun CA, Chu CM, Lin FG,
Hsu CH, Kan PC, Lin SC, Yang T and Chou YC: DNA methylation
combinations in adjacent normal colon tissue predict cancer
recurrence: Evidence from a clinical cohort study. PLoS One.
10:e1233962015. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Maier S, Dahlstroem C, Haefliger C, Plum A
and Piepenbrock C: Identifying DNA methylation biomarkers of cancer
drug response. Am J Pharmacogenomics. 5:223–232. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Jover R, Nguyen TP, Perez-Carbonell L,
Zapater P, Paya A, Alenda C, Rojas E, Cubiella J, Balaguer F,
Morillas JD, et al: 5-Fluorouracil adjuvant chemotherapy does not
increase survival in patients with CpG island methylator phenotype
colorectal cancer. Gastroenterology. 140:1174–1181. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Oliver JA, Ortiz R, Jimenez-Luna C, Cabeza
L, Perazzoli G, Caba O, Mesas C, Melguizo C and Prados J:
MMR-proficient and MMR-deficient colorectal cancer cells:
5-Fluorouracil treatment response and correlation to CD133 and MGMT
expression. J Biosci. 45:1212020. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Cheng X, Xu X, Chen D, Zhao F and Wang W:
Therapeutic potential of targeting the Wnt/beta-catenin signaling
pathway in colorectal cancer. Biomed Pharmacother. 110:473–481.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Gujral TS, Chan M, Peshkin L, Sorger PK,
Kirschner MW and MacBeath G: A noncanonical Frizzled2 pathway
regulates epithelial-mesenchymal transition and metastasis. Cell.
159:844–856. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Qi L, Chen J, Zhou B, Xu K, Wang K, Fang
Z, Shao Y, Yuan Y, Zheng S and Hu W: HomeoboxC6 promotes metastasis
by orchestrating the DKK1/Wnt/β-catenin axis in right-sided colon
cancer. Cell Death Dis. 12:3372021. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Chen X, Liu HL, Zhao FH, Jiao ZX, Wang JS
and Dang YM: Wnt5a plays controversial roles in cancer progression.
Chin Med Sci J. 35:357–365. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Kim SH, Park KH, Shin SJ, Lee KY, Kim TI,
Kim NK, Rha SY and Ahn JB: CpG island methylator phenotype and
methylation of Wnt pathway genes together predict survival in
patients with colorectal cancer. Yonsei Med J. 59:5882018.
View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Hibi K, Mizukami H, Goto T, Kitamura Y,
Sakata M, Saito M, Ishibashi K, Kigawa G, Nemoto H and Sanada Y:
WNT5A gene is aberrantly methylated from the early stages of
colorectal cancers. Hepatogastroenterology. 56:1007–1009.
2009.PubMed/NCBI
|
|
114
|
Jiang G, Lin J, Wang W, Sun M, Chen K and
Wang F: WNT5A promoter methylation is associated with better
responses and longer progression-free survival in colorectal cancer
patients treated with 5-fluorouracil-based chemotherapy. Genet Test
Mol Bioma. 21:74–79. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Draht M, Goudkade D, Koch A, Grabsch HI,
Weijenberg MP, van Engeland M, Melotte V and Smits KM: Prognostic
DNA methylation markers for sporadic colorectal cancer: A
systematic review. Clin Epigenetics. 10:352018. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Nagaraju GP, Kasa P, Dariya B, Surepalli
N, Peela S and Ahmad S: Epigenetics and therapeutic targets in
gastrointestinal malignancies. Drug Discov Today.
S1359-6446(21)00202-6. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Jung G, Hernández-Illán E, Moreira L,
Balaguer F and Goel A: Epigenetics of colorectal cancer: Biomarker
and therapeutic potential. Nat Rev Gastro Hepat. 17:111–130. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Cervena K, Siskova A, Buchler T, Vodicka P
and Vymetalkova V: Methylation-based therapies for colorectal
cancer. Cells-Basel. 9:15402020. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Singh M, Kumar V, Sehrawat N, Yadav M,
Chaudhary M, Upadhyay SK, Kumar S, Sharma V, Kumar S, Dilbaghi N
and Sharma AK: Current paradigms in epigenetic anticancer
therapeutics and future challenges. Semin Cancer Biol.
S1044-579X(21)00063-8. 2021. View Article : Google Scholar
|
|
120
|
Wei T, Lin Y, Tang S, Luo C, Tsai C, Shun
C and Chen C: Metabolic targeting of HIF-1α potentiates the
therapeutic efficacy of oxaliplatin in colorectal cancer. Oncogene.
39:414–427. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Li J, Su X, Dai L, Chen N, Fang C, Dong Z,
Fu J, Yu Y, Wang W, Zhang H, et al: Temporal DNA methylation
pattern and targeted therapy in colitis-associated cancer.
Carcinogenesis. 41:235–244. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Pechalrieu D, Etievant C and Arimondo PB:
DNA methyltransferase inhibitors in cancer: From pharmacology to
translational studies. Biochem Pharmacol. 129:1–13. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Kisiel JB, Klepp P, Allawi HT, Taylor WR,
Giakoumopoulos M, Sander T, Yab TC, Moum BA, Lidgard GP, Brackmann
S, et al: Analysis of DNA methylation at specific loci in stool
samples detects colorectal cancer and high-grade dysplasia in
patients with inflammatory bowel disease. Clin Gastroenterol
Hepatol. 17:914–921.e5. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Chen J, Sun H, Tang W, Zhou L, Xie X, Qu
Z, Chen M, Wang S, Yang T, Dai Y, et al: DNA methylation biomarkers
in stool for early screening of colorectal cancer. J Cancer.
10:5264–5271. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Pan Y, Liu G, Zhou F, Su B and Li Y: DNA
methylation profiles in cancer diagnosis and therapeutics. Clin Exp
Med. 18:1–14. 2018. View Article : Google Scholar : PubMed/NCBI
|