Introduction
Lung cancer is a common malignant tumour
(approximately 11.4% of all cancers diagnosed), and its
cancer-related mortality (18.0% of the total cancer-related deaths)
ranks first among various types of tumours (1). There are two major pathological types of
lung cancer: Non-small cell lung cancer (NSCLC) and small-cell lung
cancer. NSCLC, as the main subtype, accounts for more than 75% of
cases (2). The standard treatment
option for NSCLC is surgical resection combined with adjuvant
radiotherapy/chemotherapy and molecular-targeted agents. However,
numerous patients with NSCLC are diagnosed at a late stage and are
not suitable for surgical treatment (3). Currently, the molecular mechanisms of
NSCLC progression remain incompletely elucidated, and the treatment
efficiency is far from what individuals anticipate. The 5-year
survival rate is <15% in individuals with NSCLC. Accordingly, it
is necessary to determine the oncogenic factors and/or tumour
suppressors in the progression of NSCLC to provide potential
treatment targets.
An important matter for tumour occurrence and
development is the overgrowth of cancer cells without limitation,
which is derived from the dysfunction of cell death. Thus,
regulators of cell apoptosis are regarded as tumour-related
molecules and inducing apoptosis in cancer cells is considered an
important method for mitigating tumour progression (4). Accumulating evidence has indicated that
a few transmembrane proteins (TMEMs) are up- or downregulated in
some types of tumours and participate in regulating tumour
progression and tumorigenesis (5–7). The
upregulation of TMEM16A has been reported to promote cancer
metastasis and be correlated with a poor prognosis in human gastric
cancer (8). Cytosolic TMEM88has been
revealed to increase the expression of Snail and promote the
metastasis of cancer cells in triple-negative breast and lung
cancer (9,10). TMEM100, belonging to the family of
transmembrane proteins, is well conserved in vertebrates. There are
two putative transmembrane domains in TMEM100 (11). The known cellular functions of TMEM100
mainly include arterial endothelial differentiation and vascular
morphogenesis. In mouse embryos, TMEM100 knockdown leads to
cardiovascular developmental disorders, heart defects, and a
failure of vascular remodelling (12). During atrioventricular canal cushion
formation, endothelial-mesenchymal transformation is impaired by
TMEM100 deficiency (13). Moreover,
TMEM100 mitigates the metastasis and proliferation of cancer cells
in hepatocellular carcinoma (14).
However, the pathophysiologic roles of TMEM100 in the survival of
NSCLC cells remain to be explored.
The present research aimed to determine the
regulatory roles of TMEM100 in the apoptosis of NSCLC cells and the
corresponding mechanisms involved to provide potential treatment
targets for this fatal disease.
Materials and methods
Cell culture
H358, H1650, H1299, A549, H1975, and H460 cells were
obtained from the American Type Culture Collection (ATCC). Cells
were cultured in cell culture flasks in DMEM (Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS;
Gibco; Thermo Fisher Scientific, Inc.). Cells were cultured in an
atmosphere containing 5% CO2 at 37°C. Serum deprivation
(SD) was utilized as an apoptotic model to induce cell apoptosis in
the appropriate experiments.
Cell transfection
Cells at the logarithmic growth phase were
trypsinized and plated in 6-well plates at a density of
~1×105 cells/well. After 24 h of incubation,
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) was
used to transfect negative control (NC; 50 nM) and miR-106b mimics
(miR-106b; 50 nM) or miR-106b inhibitor (anti-miR-106b; 50 nM) and
anti-NC (50 nM) or empty vector and recombinant survivin plasmid (1
µg), diluted with Opti-MEM according to the manufacturer's
instructions. After 6 h of transfection at 37°C, the culture medium
was renewed and then the cells were collected for subsequent
experiments. The sequences were as follows: NC forward,
5′-UCACAACCUCCUAGAAAGAGUAGA−3′ and reverse,
5′-UCUACUCUUUCUAGGAGGUUGUGA−3′; miR-106b mimics forward,
5′-UAAAGUGCUGACAGUGCAGAU−3′ and reverse,
5′-AUCUGCACUGUCAGCACUUUA−3′; anti-miR-106b
5′-AUCUGCACUGUCAGCACUUUA−3′; and anti-NC
5′-UCUACUCUUUCUAGGAGGUUGUGA−3′. The recombinant survivin plasmid
was obtained from Addgene, Inc. Both miR mimics and inhibitors were
purchased from Shanghai Biotend Biological Technology, Co., Ltd.
The lentiviruses (the lentiviral expression vector pLKO.1) of
TMEM100 overexpression and knockdown were purchased from Shanghai
GenePharma Co., Ltd. Briefly, pLKO.1/puro was used as the
lentiviral plasmid and lentiviruses were generated by the 2nd
generation system. Lentiviruses were produced by co-transfection of
293T cells (ATCC) with the lentiviral vector (1.5 µg) and packaging
vectors (0.5 µg psPAX2 and 0.5 µg pMD2.G). After 48 h of
transfection, the lentiviral supernatant was collected. The
lentiviruses were utilized to infect the target cells
(1×105) at a multiplicity of infection (MOI) of 20,
including H358, H460, H1299, and H1975, in the presence of 5 µg/ml
polybrene (Sigma-Aldrich; Merck KGaA). After 48 h of incubation,
the cells were cultured in DMEM with 10% FBS at 37°C in the
presence of 3 µg/ml puromycin. After a week, cells were then used
for the subsequent experimentations.
Western blot analysis
The proteins were extracted from cell lines (H358,
H460, H1299, and H1975) by using RIPA (P0013B; Beyotime Institute
of Biotechnology) and protein concentrations were determined by
bicinchoninic acid (BCA). Cell lysates in equal amounts (50 µg
protein) were loaded on sodium dodecyl sulfate-polyacrylamide gels
(12%), and then the gels were transferred to polyvinylidene
difluoride membranes (Roche Diagnostics). After 2 h of transfer,
the membranes were incubated with 5% skimmed milk for 1 h at 25°C
for blocking, and then the primary antibodies were added to the
membranes overnight at 4°C. Primary antibodies included TMEM100,
survivin and cleaved caspase-3 (1:500; product codes ab117973,
ab76424, and ab2302, respectively; all from Abcam), Bim and
cytochrome c (both 1:500; product nos. 2933 and 11940,
respectively), or β-actin (1:3,000; product no. 3700; all from Cell
Signaling Technology, Inc.). After extensive washing with TBST
(0.1% Tween) for ~40 min, the corresponding horseradish
peroxidase-conjugated secondary antibodies (1:5,000; product nos.
7074 and 7076; Cell Signaling Technology, Inc.) were added to the
membranes for 1 h at room temperature. After extensive washing with
TBST for ~40 min, the signal was next visualized with
chemiluminescence substrate (Pierce Biotechnology; Thermo Fisher
Scientific, Inc.) by chemiluminescence detection. Densitometric
analysis was performed using Image Lab software (version 4.1;
Bio-Rad Laboratories, Inc.).
Reverse transcription-quantitative
(RT-q)PCR
RT-qPCR was performed as previously described
(15). TRIzol reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) and a miRNA isolation kit (Ambion;
Thermo Fisher Scientific, Inc.) were used to extract total RNA
according to the protocol provided by the supplier. For miRNA
analysis, RT-qPCR TaqMan probes for miR-106b were obtained from
Applied Biosystems; Thermo Fisher Scientific, Inc., and TaqMan
premix (Takara Bio, Inc.) was used for RT-qPCR. For mRNA analysis,
a SYBR Green PCR system (Applied Biosystems; Thermo Fisher
Scientific, Inc.) was applied after reverse transcription. The
primers were designed and were as follows: TMEM100 forward,
5′-TGCTGTGGTTGTCTTCATCG-3′ and reverse,
5′-CTCTCCCGTCTCTTGGCTTTC−3′; and β-actin forward,
TTGTTACAGGAAGTCCCTTGCC and reverse, 5′-ATGCTATCACCTCCCCTGTGTG-3′.
Actin and snRNA U6 (cat. no. 4427975; Thermo Fisher Scientific,
Inc.) were utilized as normalization controls to quantify mRNA and
miRNA expression levels, respectively.
Cell viability assay
Cell Counting Kit-8 (CCK-8; Dojindo Molecular
Technologies, Inc.) was used to determine cell viability. Cells
were cultured in a 96-well plate at 3×103 cells/well at
37°C. After adding 10 µl of CCK-8 solution to each well for 2 h at
37°C, the absorbance was measured with a microplate reader at 450
nm.
Colony formation assay
Cells were cultured at a density of
0.8×103 cells (H358 cells) per well or at a density of
3×103 cells (H460 cells) per well in 6-cm Petri dishes.
A total of 15 days later, 500 µl of 4% paraformaldehyde was added
to each well for 10 min at 25°C. Then, 1% crystal violet
(Sigma-Aldrich; Merck KGaA) was utilized to stain the cells for 10
min at 25°C. Colonies (approximately >60 cells) could then be
observed directly with the unaided eye, and only clearly visible
colonies (megascopic cell colonies) observed with the unaided eye
were counted and analysed.
Caspase-3 activity assay
A caspase-3 assay kit (Invitrogen; Thermo Fisher
Scientific, Inc.) was utilized to detect the activity of caspase-3,
according to the manufacturer's instructions. Briefly, 50 µl of
cell lysis buffer from the kit was added to the harvested cells on
ice for 30 min. Then, the lysed cells were centrifuged at 2,795 × g
for 5 min at 4°C. After adding 50 µl of 2X substrate working
solution, the mixture was incubated at room temperature for 30 min.
The fluorescence was then measured using a microplate reader
(excitation, 342 nm and emission, 441 nm).
Analyses of GEO datasets
To determine the expression of mRNA or miRNA in
NSCLC tissues, GEO datasets (the public datasets, www.ncbi.nlm.nih.gov/gds) were analysed in our
study. GSE19804 and GSE27262 were used to examine the expression of
TMEM100 in NSCLC tissues and paired peritumoral tissues (16,17).
GSE3141 was utilised to investigate the clinical significance of
TMEM100 in NSCLC tissues (18). MiRNA
expression profiling of NSCLC tissues and non-tumor adjacent
tissues was acquired from the datasets (GSE63805 and GSE36681)
(19,20). Oncomine (a public database; www.oncomine.org) was utilized to examine the
expression of TMEM100 in different cancers.
Luciferase experiments
TargetScan (version 7.2; www.targetscan.org) and miRanda (www.microRNA.org) were utilized to identify the
potential miRNAs that target TMEM100 based on the sequence of
TMEM100 3′untranslated region (UTR) (21,22). As
previously described, a dual luciferase reporter gene assay was
conducted (15). Briefly, the
full-length human TMEM100 3′UTRs containing the wild-type (WT)
sites or mutant-type (MT) sites were cloned into the pGL3-Promoter
vector (Promega Corporation). By using Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.), control vectors,
pGL3-TMEM100 3′UTR and miRNA mimics (Life Technologies; Thermo
Fisher Scientific, Inc.) were transfected into 293 cells. According
to the protocol provided by the supplier (Promega Corporation), the
dual luciferase reporter assay was performed, and firefly
luciferase luminescence was recorded by a VICTOR multilabel counter
(Berthold Technologies).
TUNEL assay
A One Step TUNEL apoptosis assay kit was purchased
from Beyotime Institute of Biotechnology. TUNEL-positive cells were
determined according to the manufacturer's instructions. Briefly,
cells were cultured in a 6-well plate at 5×104
cells/well at 37°C for 12 h. Then, the plate was washed with PBS
for 5 min and fixed in 4% paraformaldehyde for 30 min at 25°C.
After washing with PBS for 5 min, PBS containing 0.3% Triton X-100
was added to the cells for 5 min at room temperature. Then, the
cells were washed twice with PBS for 5 min and treated with 100 µl
TUNEL test solution (10 µl TdT enzyme and 90 µl fluorescent
labelling solution) per sample at 37°C for 60 min. After adding 200
µl mounting medium (P0126; Beyotime Institute of Biotechnology),
the stained samples (without the nuclear stain) were observed under
a fluorescence microscope (magnification, ×200; BX61; Olympus
Corporation). A total of 5 visual fields were selected randomly for
each group.
Statistical analysis
All values are expressed as the mean ± standard
error of the mean (SEM) from at least three independent
experiments. Paired and unpaired Student's t-tests or one-way ANOVA
followed by Dunnett's post hoc test were performed to evaluate the
statistical significance. Survival analysis was carried out by
using Kaplan-Meier analysis and log-rank testing was selected to
compare patient survival between subgroups. Correlations were
calculated by Pearson's rank correlation coefficients. P<0.05
was considered to indicate a statistically significant difference.
GraphPad Prism version 5.0 (GraphPad Software, Inc.) was utilized
to perform the statistical analyses.
Results
Downregulation of TMEM100 is
associated with poor clinical outcomes in NSCLC patients
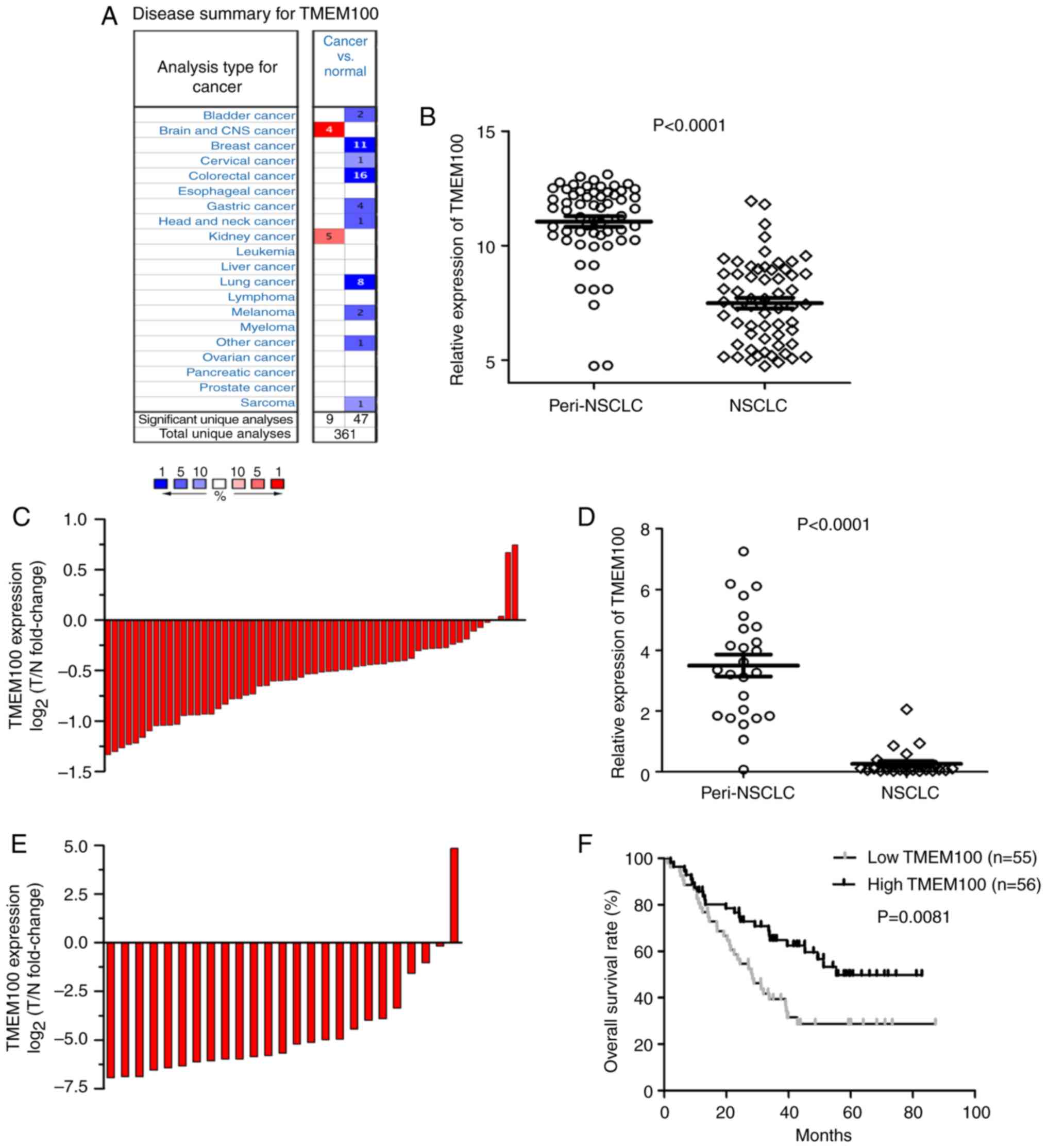
The expression of TMEM100 using Oncomine was
analysed, and it was revealed that TMEM100 expression was
downregulated in some types of cancer tissues, including breast,
colorectal and lung cancer (Fig. 1A).
To determine the role of TMEM100 in the progression of NSCLC,
TMEM100 expression in NSCLC tissues and paired peritumoral tissues
was firstly examined via two public data sets (GSE19804 and
GSE27262). As revealed in Fig. 1B and
C, the mRNA levels of TMEM100 were markedly decreased in 60
cases of tumor tissues compared with paired peritumoral tissues.
Consistent results were acquired in the other data set. The
expression of TMEM100 in tumour tissues was downregulated compared
with paired peritumoral tissues in 25 patients (Fig. 1D and E).
The clinical significance of TMEM100 in NSCLC
tissues (GSE3141) was then investigated. The patients were divided
into two groups based on the mRNA levels of TMEM100 (the
high-TMEM100 group and the low-TMEM100 group). As revealed in
Fig. 1F, our results indicated that
patients with low TMEM100 expression were associated with poorer
overall survival (median overall survival time, 28.4 months;
P<0.01) than patients with high TMEM100 expression (median
overall survival time, 55.4 months). Thus, these results indicated
that TMEM100 could be considered as a potential molecule for
predicting the prognosis in individuals with NSCLC.
Overexpression of TMEM100 suppresses
cell growth and elicits apoptosis in NSCLC cells
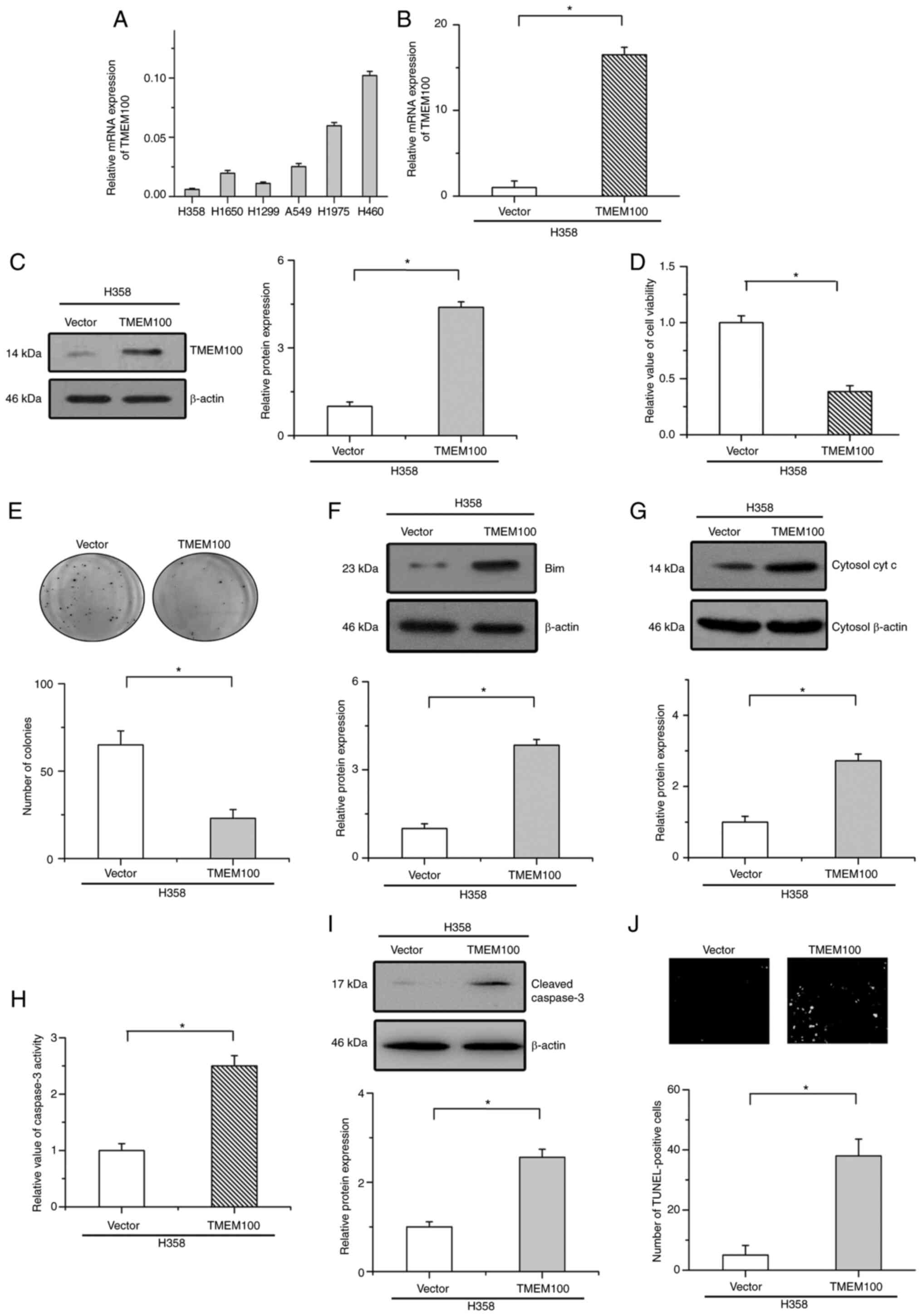
To investigate the cellular function of TMEM100 in
the progression of NSCLC, the endogenous expression of TMEM100 was
then detected in six NSCLC cell lines (Fig. 2A). Based on the endogenous expression
of TMEM100 in these cells, TMEM100 was stably overexpressed in H358
cells (low expression levels of endogenous TMEM100) via lentiviral
infection. RT-qPCR and western blot analyses were utilized to
confirm the overexpression efficiency (Fig. 2B and C). As revealed in Fig. 2D and E, the overexpression of TMEM100
decreased cell viability and inhibited colony formation in H358
cells. Moreover, the expression of key proapoptotic proteins (Bim
and cytochrome c) in mitochondrial apoptosis, the activity
and protein expression of cleaved caspase-3 and TUNEL assays were
examined to validate apoptosis in NSCLC cells. The results revealed
that the overexpression of TMEM100 induced the expression of Bim
and enhanced cytochrome c release into the cytoplasm
(Fig. 2F and G). Consistent with
these results, overexpression of TMEM100 significantly increased
the activity and protein expression of cleaved caspase-3 and the
number of TUNEL-positive cells (Fig.
2H-J). These results indicated that TMEM100 induced apoptosis
in NSCLC cells.
TMEM100 knockdown facilitates cell
survival in NSCLC
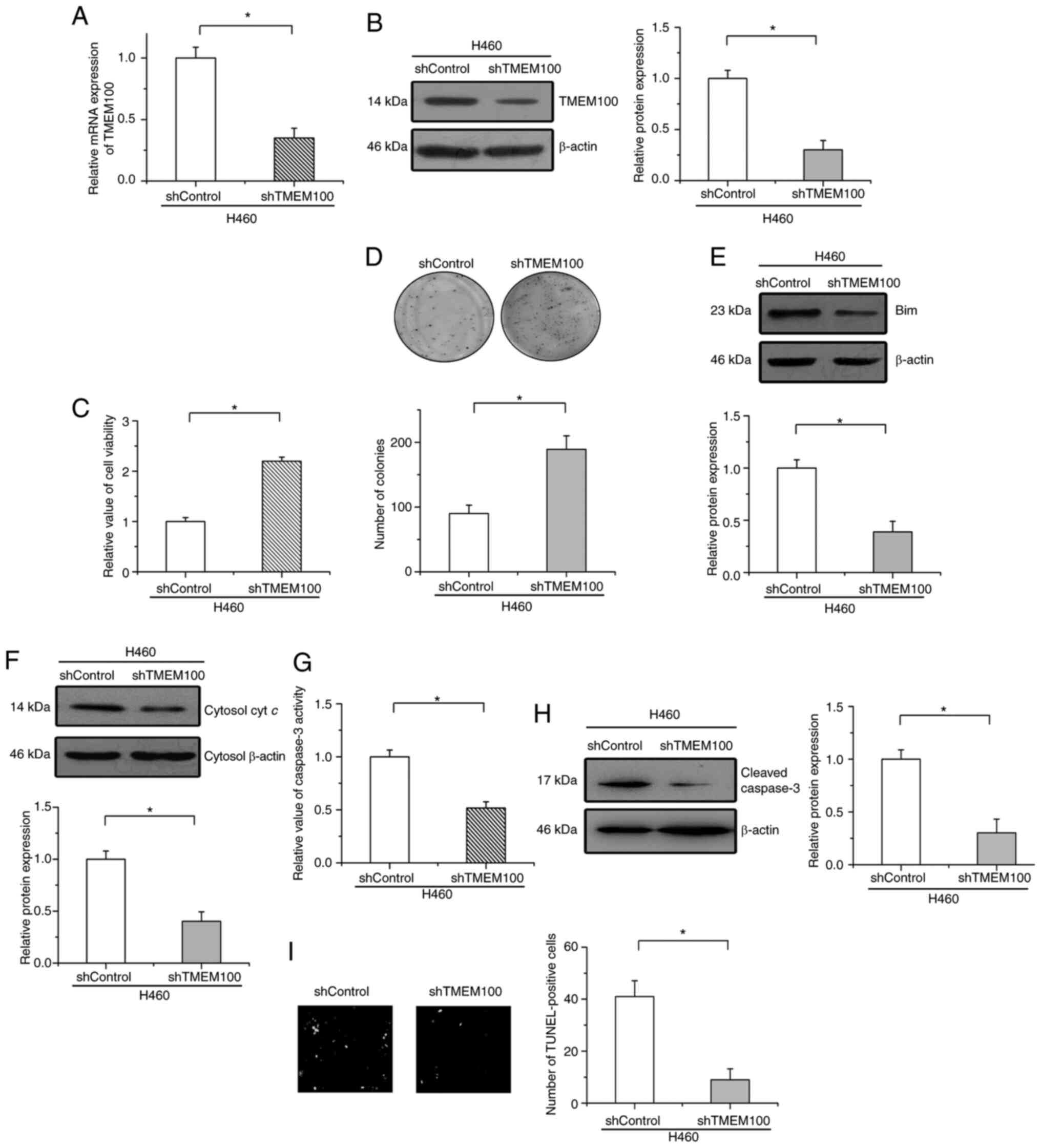
To represent the physiological functions of TMEM100
on cell survival in NSCLC, TMEM100 expression was knocked down with
a shRNA. As revealed in Fig. 3A and
B, the efficiency of knockdown was confirmed by RT-qPCR and
western blot analyses in H460 cells (high expression levels of
endogenous TMEM100). SD was then utilized as an apoptotic model to
examine the effects of TMEM100 on cell apoptosis. It was
established that the knockdown of TMEM100 significantly increased
cell viability and enhanced colony formation after treatment with
SD in H460 cells (Fig. 3C and D).
Moreover, the protein levels of Bim and cytochrome c in the
cytoplasm were both attenuated by TMEM100 knockdown in starved H460
cells (Fig. 3E and F). The knockdown
of TMEM100 suppressed the activation and expression of cleaved
caspase-3 and decreased the number of TUNEL-positive cells in
starved H460 cells (Fig. 3G-I). These
results indicated that TMEM100 knockdown promoted cell survival and
inhibited cell apoptosis in NSCLC.
TMEM100 negatively regulates cell
survival in NSCLC cells
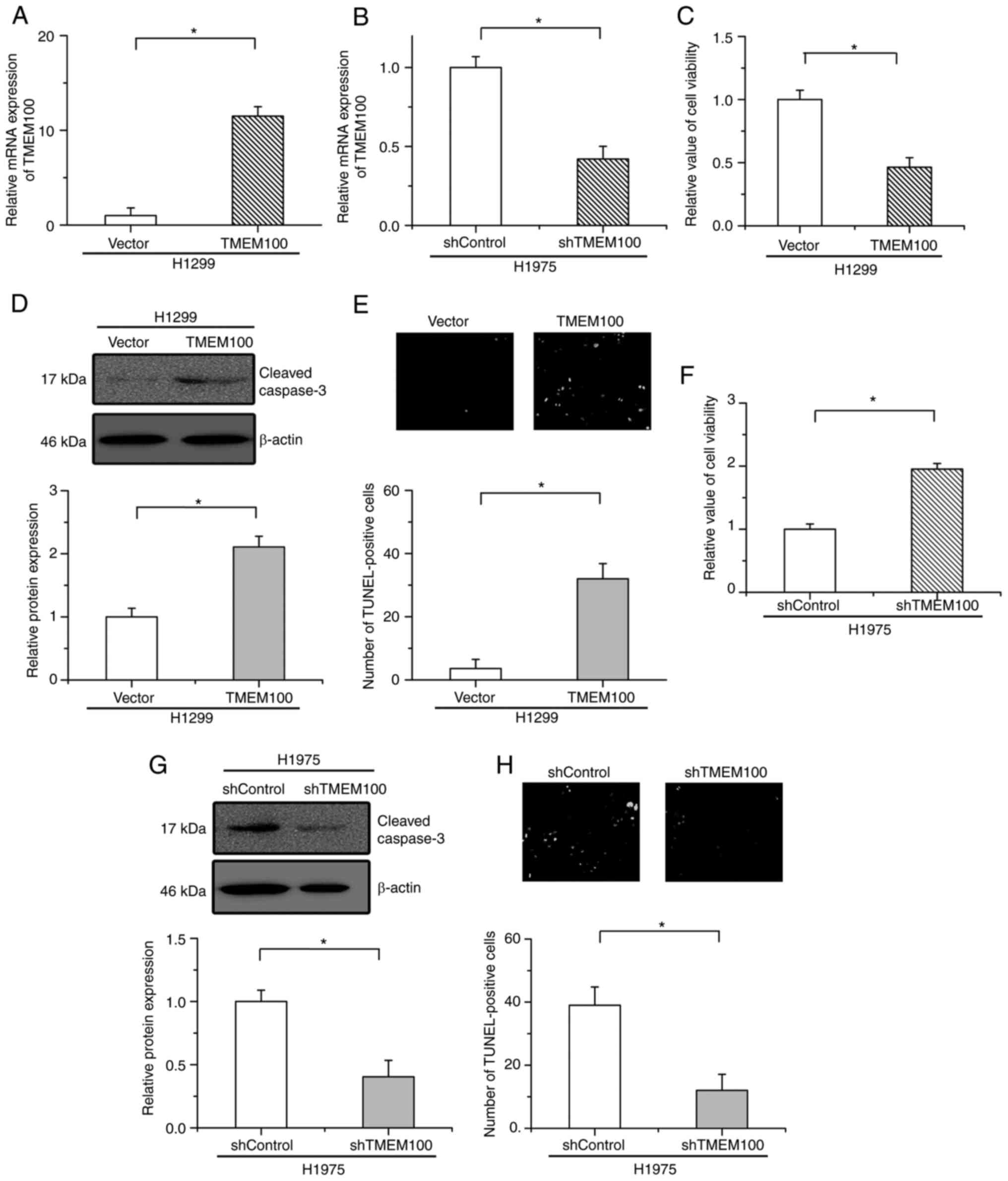
Furthermore, to determine whether the regulatory
effects of TMEM100 on cell apoptosis are general in other NSCLC
cells, H1299 and H1975 cells were then selected to establish
TMEM100 overexpression and TMEM100-knockdown cells, respectively.
The overexpression and knockdown efficiencies were verified by
RT-qPCR (Fig. 4A and B). As
demonstrated in Fig. 4C,
overexpression of TMEM100 significantly suppressed H1299 cell
viability. The expression of cleaved caspase-3 and the number of
TUNEL-positive cells were increased by TMEM100 overexpression
(Fig. 4D and E). Conversely,
knockdown of TMEM100 significantly increased cell viability and
mitigated the expression of cleaved caspase-3 and the number of
TUNEL-positive cells in starved H1975 cells (Fig. 4F-H). These results indicated that
TMEM100 facilitated cell apoptosis in NSCLC cells.
MiRNA-106b acts as an upstream
regulator of TMEM100 in NSCLC
MiRNAs, which can block the translation or initiate
the transcript degradation of target mRNAs, play a critical role in
reducing protein expression. To search for miRNAs that potentially
target TMEM100 and to identify the upstream regulator of TMEM100,
TargetScan and miRanda were utilized to determine miRNAs that
potentially target the 3′UTR of TMEM100 mRNA (21,22). Based
on the analysis results, 30 potential upstream miRNAs (including
miR-520e, miR-106b, miR-195, miR-497, miR-16 andmiR-15a) were
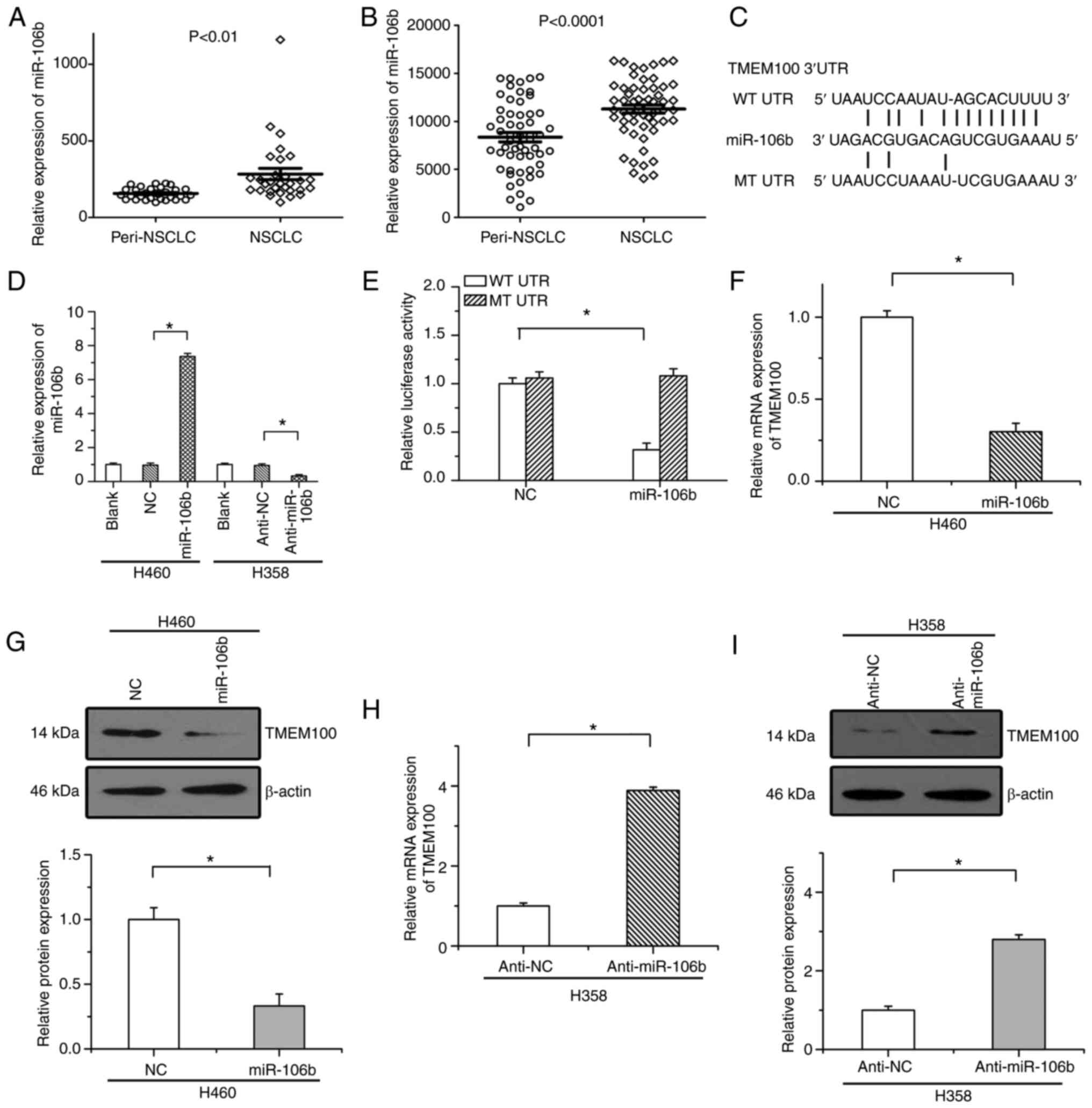
identified. The expression of these miRNAs in NSCLC tissues and
peritumoral tissues was then examined via two public datasets
(GSE63805 and GSE36681). Among these miRNAs, it was revealed that
only miR-106b was significantly upregulated in NSCLC tissues in
both datasets (as revealed in Fig. 5A and
B), indicating that miR-106b likely functions as an upstream
regulator of TMEM100 in NSCLC.
Then, it was studied whether TMEM100 was regulated
by miR-106b. As revealed in Fig. 5C,
there was a specific sequence conserved in the 3′UTR of TMEM100
mRNA, which was predicted to integrate with miR-106b. Results of
RT-qPCR revealed that expression of miR-106b was significantly
increased by miR-106b mimics (miR-106b) and decreased by a
miR-106b-specific inhibitor (anti-miR-106b) (Fig. 5D). A luciferase reporter assay was
utilized to demonstrate the predicted combination of miR-106b and
the 3′UTR of TMEM100 mRNA. Our results revealed that the luciferase
activity of the WT plasmid was significantly suppressed by
miR-106b, while the luciferase activity of the MT plasmid was not
affected by miR-106b (Fig. 5E).
Moreover, it was also demonstrated that the mRNA and protein
expression levels of TMEM100 were significantly downregulated by
miR-106b (Fig. 5F and G), whereas
anti-miR-106b led to the increased expression of TMEM100 (Fig. 5H and I). These results indicated that
miR-106b served as an upstream regulator of TMEM100 in NSCLC.
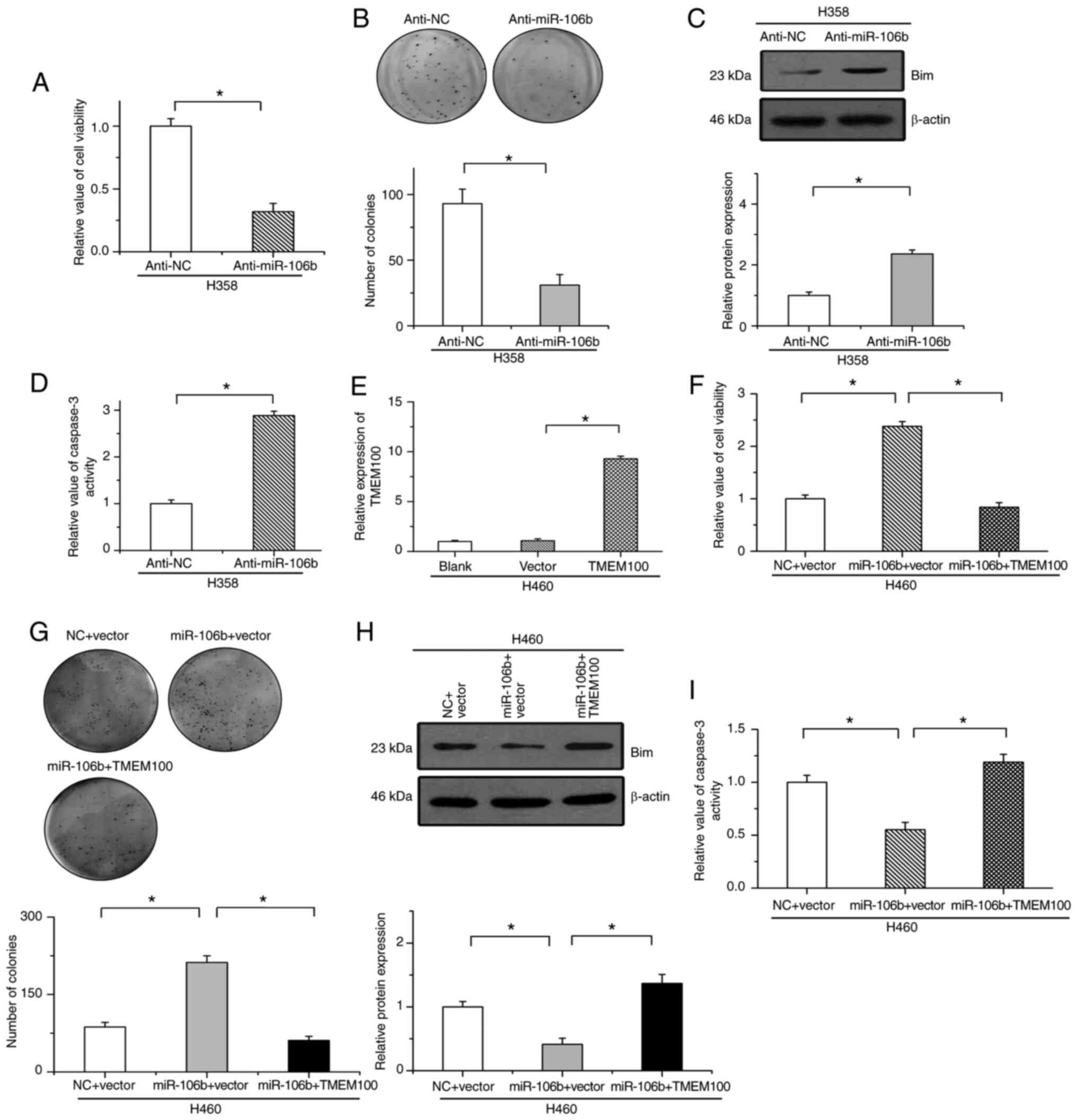
miR-106b facilitates cell survival by
serving as an oncogenic factor
The regulatory effects of miR-106b on cell survival
in NSCLC were then examined. Our results revealed that the
inhibition of miR-106b mitigated cell growth and suppressed colony
formation (Fig. 6A and B). The
protein levels of Bim and caspase-3 activity were significantly
increased by treatment with anti-miR-106b in H358 cells (Fig. 6C and D). Conversely, the efficiency of
TMEM100 overexpression was verified by RT-qPCR in H460 cells
(Fig. 6E). It was revealed that
miR-106b increased cell viability and promoted colony formation
after treatment with SD in H460 cells, which was attenuated by the
reintroduction of TMEM100 (Fig. 6F and
G). Additionally, miR-106b-inhibited Bim expression and
caspase-3 activation were eliminated by the restoration of TMEM100
in starved H460 cells (Fig. 6H and
I). These results indicated that miR-106b inhibited apoptosis
by reducing TMEM100 expression in NSCLC cells.
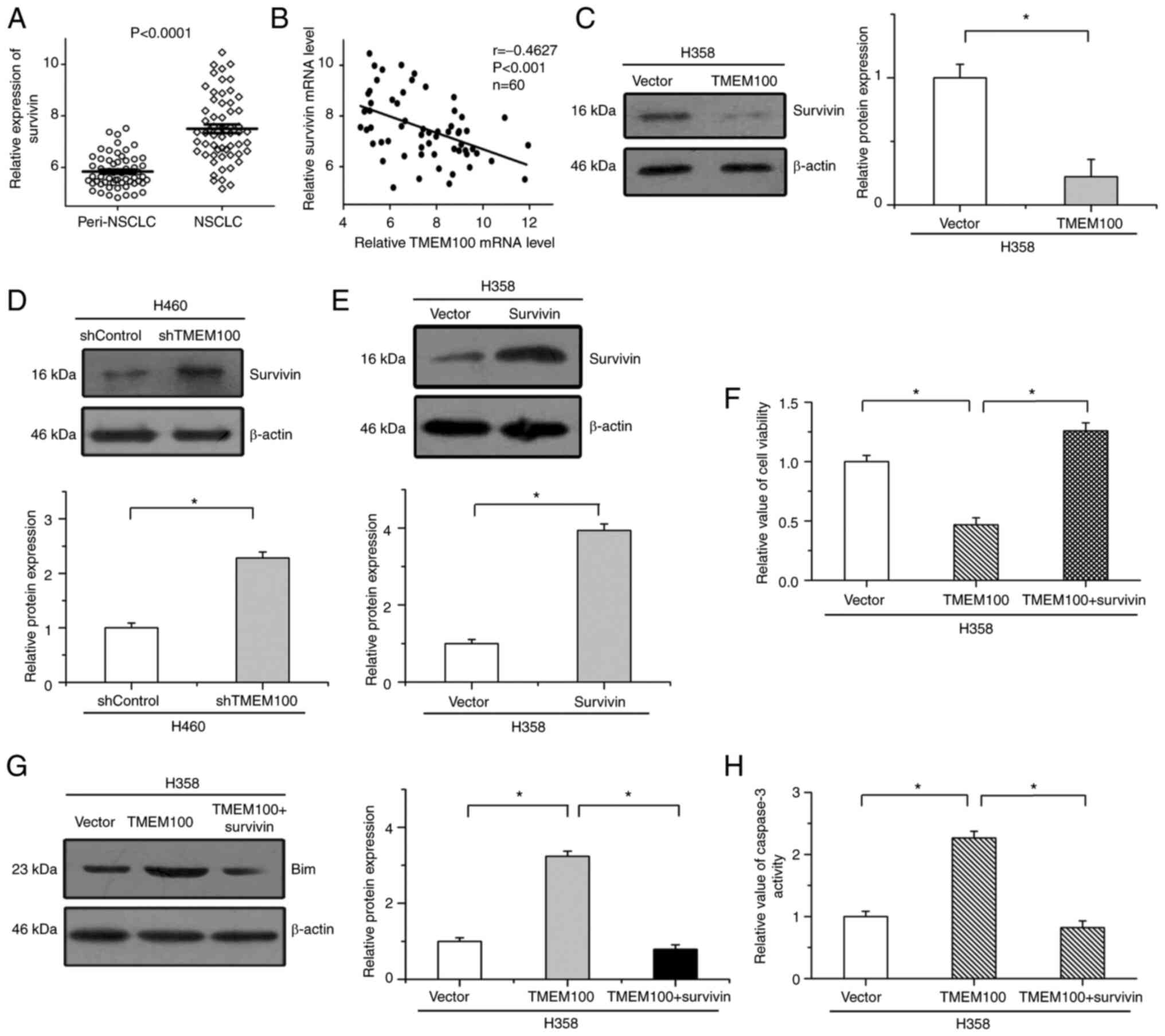
Effects of TMEM100 on cell apoptosis
are attenuated by the reintroduction of survivin
Survivin, a critical regulator of cell apoptosis,
has been reported to play important roles in the progression of
NSCLC (23). As revealed in Fig. 7A, the expression of survivin was
significantly upregulated in 60 NSCLC tissues (GSE19804) compared
with paired peritumoral tissues. There were highly negative
correlations between TMEM100 expression and survivin expression in
the same NSCLC tissues (r=−0.46, P<0.001) (Fig. 7B). It was then examined whether
survivin was involved in the inhibitory effects of TMEM100 on cell
survival. Our results revealed that the overexpression of TMEM100
mitigated the protein levels of survivin, while the expression of
survivin was increased by TMEM100 knockdown (Fig. 7C and D). Furthermore, to determine the
roles of survivin in TMEM100-regulated cell apoptosis, survivin
expression was increased by transfection with a recombinant plasmid
(Fig. 7E). It was revealed that the
inhibitory effects of TMEM100 on cell viability were mitigated by
the increased expression of survivin (Fig. 7F). In addition, TMEM100-enhanced Bim
expression and caspase-3 activation were antagonized by the
restoration of survivin (Fig. 7G and
H). These results indicated that the roles of TMEM100 in cell
survival were, at least in part, mediated by negatively regulating
survivin in NSCLC.
Discussion
Mounting evidence has indicated that transmembrane
proteins play important roles in the progression and development of
some malignancies (8,10,24).
Cytosolic TMEM88 has been revealed to stimulate the invasion and
metastasis of cancer cells. Higher expression of cytosolic TMEM88
was revealed to be closely associated with poorer differentiation,
higher TNM stage, and worse overall survival (10). Increased expression of TMEM16A has
been revealed in some tumours, including gastric cancer (8), prostate carcinoma (24), as well as head and neck squamous cell
carcinoma (25). Additionally, the
increased expression of TMEM16A was revealed to be positively
associated to tumour stage and negatively associated with the
overall survival of patients (8).
TMEM100, a member of the transmembrane protein family, has also
been revealed to participate in regulating the progression of some
cancers. A previous study revealed that TMEM100 inhibited
metastasis and proliferation, and the levels of TMEM100 were
correlated with tumour size, TNM stage, overall survival and
disease-free survival in hepatocellular carcinoma (14). Han et al revealed that
overexpression of TMEM100 suppressed cell proliferation and
mitigated the migration and invasion of NSCLC cells, whereas
TMEM100 knockdown promoted cell proliferation and migration
(26). However, the proliferation of
cancer cells is determined not only by cell proliferation but also
by cell death. Apoptosis, an important form of regulated cell
death, also plays a critical regulatory role in the progression of
cancer (4). At present, whether the
regulation of cell apoptosis is also involved in the inhibitory
effects of TMEM100 on NSCLC progression remains uncertain. In the
present study, new evidence was provided to indicate that TMEM100,
negatively regulated by miR-106b, induced cell apoptosis and
inhibited the survival of NSCLC cells by suppressing survivin
expression, consistent with a previous study revealing that TMEM100
served as a tumour suppressor in NSCLC (26).
Apoptosis acts as a negative regulator of cell
growth. Accumulating evidence has indicated that the inhibition of
cell apoptosis is commonly observed in some cancer tissues, and
genes involved in modulating cell apoptosis are regarded as a new
class of tumour-related genes (27,28).
Inducing cellular apoptosis is one of the most effective strategies
for relieving the progression of tumours. A previous study revealed
that mitochondrial dysfunction was an early-stage event that
triggered the intrinsic apoptotic pathway (29). Bim is a critical mediator of the
mitochondrial apoptotic pathway and participates in regulating the
function of mitochondria. Bim can cause a decrease in the
mitochondrial membrane potential and induce the opening of the
mitochondrial permeability transition pore either by directly
activating proapoptotic Bax/Bak or by antagonizing antiapoptotic
Bcl-2 (30). A disruption in the
mitochondrial membrane potential leads to the release of cytochrome
c from the mitochondria into the cytoplasm, which in turn
triggers the activation of caspase cascades and elicits cell
apoptosis (31). To study the role of
TMEM100 in the survival of NSCLC cells, the expression of Bim,
cytochrome c release and caspase-3 activation were examined
under conditions of TMEM100 overexpression or knockdown. Our
results revealed that the overexpression of TMEM100 induced the
expression of Bim, promoted the release of cytochrome c into
the cytoplasm, and increased the activity of caspase-3. Conversely,
TMEM100 knockdown had the opposite effects. These results indicated
that TMEM100 elicited cell apoptosis and inhibited the progression
of NSCLC by acting as a tumour suppressor.
An important finding of the present study was that
TMEM100 facilitated cell apoptosis by suppressing survivin
expression in NSCLC. Survivin belongs to the inhibitor of apoptosis
(IAP) gene family and plays important roles in the progression of
some human malignancies, including NSCLC (23,32). Its
roles in cell survival and apoptosis have been widely studied in
NSCLC. The inhibitory effects of nicotine on chemotherapeutic
drug-induced cell apoptosis have been reported to be mediated by
upregulating XIAP and survivin (33).
Ezponda et al reported that SF2/ASF promotes the stability
of survivin mRNA and that the downregulation of SF2/ASF induces
apoptosis by reducing the expression of survivin in NSCLC cells
(23). In LKB1-deficient lung
adenocarcinomas, survivin was responsible for promoting malignant
progression by acting as the downstream mediator of YAP (34). In the present study, our results
revealed that the expression of survivin was negatively correlated
with TMEM100 expression in NSCLC tissues. Moreover, the roles of
TMEM100 in cell apoptosis were attenuated by the reintroduction of
survivin. These results indicated that TMEM100 facilitated cell
apoptosis, at least in part, by inhibiting survivin expression in
NSCLC.
Another notable finding of the present study was
that TMEM100 was negatively regulated by miR-106b in NSCLC cells.
MiRNAs, a large family of short, noncoding endogenous RNAs, can
bind to a specific sequence conserved in the 3′UTR of their target
genes and post-transcriptionally regulate their expression
(35). Currently, a number of
microRNAs have been indicated to regulate various types of
pathological processes, including the initiation and development of
cancers (36–38). However, the effects of miR-106b on
different human cancers are not consistent. Previous studies have
demonstrated that miR-106b enhanced the metastasis and
proliferation of cancer cells and boosted tumorigenesis in
colorectal cancer (39),
hepatocellular carcinoma (40), and
glioma (41).Conversely, miR-106b, as
a tumour suppressor, has been reported to be significantly
downregulated in clinical samples of giant bone cell tumours and
inhibited metastasis and tumorigenesis in thyroid (42) and breast cancer (43). To date, the precise roles of miR-106b
in NSCLC have been inconclusive. In the present research, our
results revealed that miR-106b, which was markedly upregulated in
NSCLC tissues, interacted with the 3′UTR of TMEM100 mRNA and
suppressed its expression. Moreover, the inhibitory roles of
miR-106b in cell apoptosis were eliminated by the restoration of
TMEM100 in NSCLC. These results indicated that miR-106b-mitigated
TMEM100 expression promoted the progression of NSCLC. However,
further studies are still needed to validate the roles of TMEM100
in NSCLC in vivo. Although it was demonstrated that TMEM100
regulates cell survival by survivin in NSCLC, other parallel
pathways involved should be investigated in future studies.
In conclusion, our results revealed that TMEM100
induced cell apoptosis in NSCLC by acting as a tumour suppressor.
Moreover, miR-106b was responsible for the decreased expression of
TMEM100 in NSCLC. These results indicated an important underlying
regulatory mechanism and provided potential treatment targets for
NSCLC.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81700863, 81802626
and 81972854), the Shanghai Sailing Program (grant no.
16YF1401300), the Traditional Chinese Medicine and Western Medicine
of Shanghai Municipal Committee for Health and Family Planning
(grant no. ZHYY-ZXYJHZX-201610), the Shanghai Jiao Tong University
Medical Engineering Cross Fund (grant no. YG2016QN52) and the
Incubating Program for Clinical Research and Innovation of Renji
Hospital (grant no. PYII-17-006).
Availability of data and materials
All data generated and/or analyzed during the
present study are included in this published article.
Authors' contributions
JM and LZ conceived and designed the experiments.
JM, CM and TY performed the experiments. YB, MY and XM analyzed the
results. XM, CM and LZ wrote the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Péchoux CL, Sun A, Slotman BJ, De
Ruysscher D, Belderbos J and Gore EM: Prophylactic cranial
irradiation for patients with lung cancer. Lancet Oncol.
17:e277–e293. 2016. View Article : Google Scholar
|
|
3
|
Rosenzweig KE and Gomez JE: Concurrent
chemotherapy and radiation therapy for inoperable locally advanced
non-small-cell lung cancer. J Clin Oncol. 35:6–10. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Evan GI and Vousden KH: Proliferation,
cell cycle and apoptosis in cancer. Nature. 411:342–348. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sun HF, Yang XL, Zhao Y, Tian Q, Chen MT,
Zhao YY and Jin W: Loss of TMEM126A promotes extracellular matrix
remodeling, epithelial-to-mesenchymaltransition, and breast cancer
metastasis by regulating mitochondrial retrograde signaling. Cancer
Lett. 440-441:189–201. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Oktay MH and Jones JG: TMEM: A novel
breast cancer dissemination marker for the assessment of metastatic
risk. Biomark Med. 9:81–84. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wrzesiński T, Szelag M, Cieślikowski WA,
Ida A, Giles R, Zodro E, Szumska J, Poźniak J, Kwias Z, Bluyssen HA
and Wesoly J: Expression of pre-selected TMEMs with predicted ER
localization as potential classifiers of ccRCC tumors. BMC Cancer.
15:5182015. View Article : Google Scholar
|
|
8
|
Liu F, Cao QH, Lu DJ, Luo B, Lu XF, Luo RC
and Wang XG: TMEM16A overexpression contributes to tumor invasion
and poor prognosis of human gastric cancer through TGF-β signaling.
Oncotarget. 6:11585–11599. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yu X, Zhang X, Zhang Y, Jiang G, Mao X and
Jin F: Cytosolic TMEM88 promotes triple-negative breast cancer by
interacting with Dvl. Oncotarget. 6:25034–25045. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang X, Yu X, Jiang G, Miao Y, Wang L,
Zhang Y, Liu Y, Fan C, Lin X, Dong Q, et al: Cytosolic TMEM88
promotes invasion and metastasis in lung cancer cells by binding
DVLS. Cancer Res. 75:4527–4537. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Moon EH, Kim MJ, Ko KS, Kim YS, Seo J, Oh
SP and Lee YJ: Generation of mice with a conditional and reporter
allele for Tmem100. Genesis. 48:673–678. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Somekawa S, Imagawa K, Hayashi H, Sakabe
M, Ioka T, Sato GE, Inada K, Iwamoto T, Mori T, Uemura S, et al:
Tmem100, an ALK1 receptor signaling-dependent gene essential for
arterial endothelium differentiation and vascular morphogenesis.
Proc Natl Acad Sci USA. 109:12064–12069. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mizuta K, Sakabe M, Hashimoto A, Ioka T,
Sakai C, Okumura K, Hattammaru M, Fujita M, Araki M, Somekawa S, et
al: Impairment of endothelial-mesenchymal transformation during
atrioventricular cushion formation in TMEM100 null embryos. Dev
Dyn. 244:31–42. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ou D, Yang H, Hua D, Xiao S and Yang L:
Novel roles of TMEM100: Inhibition metastasis and proliferation of
hepatocellular carcinoma. Oncotarget. 6:17379–17390. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang L, Du Y, Xu S, Jiang Y, Yuan C, Zhou
L, Ma X, Bai Y, Lu J and Ma J: DEPDC1, negatively regulated by
miR-26b, facilitates cell proliferation via the up-regulation of
FOXM1 expression in TNBC. Cancer Lett. 442:242–251. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lu TP, Tsai MH, Lee JM, Hsu CP, Chen PC,
Lin CW, Shih JY, Yang PC, Hsiao CK, Lai LC and Chuang EY:
Identification of a novel biomarker, SEMA5A, for non-small cell
lung carcinoma in nonsmoking women. Cancer Epidemiol Biomarkers
Prev. 19:2590–2597. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wei TY, Juan CC, Hisa JY, Su LJ, Lee YC,
Chou HY, Chen JM, Wu YC, Chiu SC, Hsu CP, et al: Protein arginine
methyltransferase 5 is a potential oncoprotein that upregulates G1
cyclins/cyclin-dependent kinases and the phosphoinositide
3-kinase/AKT signaling cascade. Cancer Sci. 103:1640–1650. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bild AH, Yao G, Chang JT, Wang Q, Potti A,
Chasse D, Joshi MB, Harpole D, Lancaster JM, Berchuck A, et al:
Oncogenic pathway signatures in human cancers as a guide to
targeted therapies. Nature. 439:353–357. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Robles AI, Arai E, Mathé EA, Okayama H,
Schetter AJ, Brown D, Petersen D, Bowman ED, Noro R, Welsh JA, et
al: An integrated prognostic classifier for Stage I lung
adenocarcinoma based on mRNA, microRNA, and DNA methylation
biomarkers. J Thorac Oncol. 10:1037–1048. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jang JS, Jeon HS, Sun Z, Aubry MC, Tang H,
Park CH, Rakhshan F, Schultz DA, Kolbert CP, Lupu R, et al:
Increased miR-708 expression in NSCLC and its association with poor
survival in lung adenocarcinoma from never smokers. Clin Cancer
Res. 18:3658–3667. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
John B, Enright AJ, Aravin A, Tuschl T,
Sander C and Marks DS: Human MicroRNA targets. PLoS Biol.
2:e3632004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ezponda T, Pajares MJ, Agorreta J,
Echeveste JI, López-Picazo JM, Torre W, Pio R and Montuenga LM: The
oncoprotein SF2/ASF promotes non-small cell lung cancer survival by
enhancing survivinexpression. Clin Cancer Res. 16:4113–4125. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu W, Lu M, Liu B, Huang Y and Wang K:
Inhibition of Ca(2+)-activated Cl(−) channel ANO1/TMEM16A
expression suppresses tumor growth and invasiveness in human
prostate carcinoma. Cancer Lett. 326:41–51. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dixit R, Kemp C, Kulich S, Seethala R,
Chiosea S, Ling S, Ha PK and Duvvuri U: TMEM16A/ANO1 is
differentially expressed in HPV-negative versus HPV-positive head
and neck squamous cell carcinoma through promoter methylation. Sci
Rep. 5:166572015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Han Z, Wang T, Han S, Chen Y, Chen T, Jia
Q, Li B, Li B, Wang J, Chen G, et al: Low-expression of TMEM100 is
associated with poor prognosis in non-small-cell lung cancer. Am J
Transl Res. 9:2567–2578. 2017.PubMed/NCBI
|
|
27
|
Haag T, Herkt CE, Walesch SK, Richter AM
and Dammann RH: The apoptosis associated tyrosine kinase gene is
frequently hypermethylated in human cancer and is regulated by
epigenetic mechanisms. Genes Cancer. 5:365–374. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Taubert H, Würl P, Greither T, Kappler M,
Bache M, Bartel F, Kehlen A, Lautenschläger C, Harris LC, Kaushal
D, et al: Stem cell-associated genes are extremely poor prognostic
factors for soft-tissue sarcoma patients. Oncogene. 26:7170–7174.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yang M, Wang B, Gao J, Zhang Y, Xu W and
Tao L: Spinosad induces programmed cell death involves
mitochondrial dysfunction and cytochrome C release in Spodoptera
frugiperda Sf9 cells. Chemosphere. 169:155–161. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Brunelle JK and Letai A: Control of
mitochondrial apoptosis by the Bcl-2 family. J Cell Sci.
122:437–441. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu G, Sheng Y, Zhang M and Sun D: A
polysaccharide from the leaves of Aralia elata induces apoptosis in
U-2 OS cells viamitochondrial-dependent pathway. Int J Biol
Macromol. 93:418–425. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ambrosini G, Adida C and Altieri DC: A
novel anti-apoptosis gene, survivin, expressed in cancer and
lymphoma. Nat Med. 3:917–921. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Dasgupta P, Kinkade R, Joshi B, Decook C,
Haura E and Chellappan S: Nicotine inhibits apoptosis induced by
chemotherapeutic drugs by up-regulating XIAP and survivin. Proc
Natl Acad Sci USA. 103:6332–6337. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang W, Gao Y, Li F, Tong X, Ren Y, Han
X, Yao S, Long F, Yang Z, Fan H, et al: YAP promotes malignant
progression of Lkb1-deficient lung adenocarcinoma through
downstream regulation of survivin. Cancer Res. 75:4450–4457. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Latronico MV, Catalucci D and Condorelli
G: Emerging role of microRNAs in cardiovascular biology. Circ Res.
101:1225–1236. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Nicolas FE and Lopez-Martinez AF:
MicroRNAs in human diseases. Recent Pat DNA Gene Seq. 4:142–154.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Blitzblau RC and Weidhaas JB: MicroRNA
binding-site polymorphisms as potential biomarkers of cancer risk.
Mol Diagn Ther. 14:335–42. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sun HL, Cui R, Zhou J, Teng KY, Hsiao YH,
Nakanishi K, Fassan M, Luo Z, Shi G, Tili E, et al: ERK activation
globally downregulates miRNAs through phosphorylating Exportin-5.
Cancer Cell. 30:723–736. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang GJ, Li JS, Zhou H, Xiao HX, Li Y and
Zhou T: MicroRNA-106b promotes colorectal cancer cell migration and
invasion by directly targeting DLC1. J Exp Clin Cancer Res.
34:732015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yen CS, Su ZR, Lee YP, Liu IT and Yen CJ:
miR-106b promotes cancer progression in hepatitis B
virus-associated hepatocellular carcinoma. World J Gastroenterol.
22:5183–5192. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu F, Gong J, Huang W, Wang Z, Wang M,
Yang J, Wu C, Wu Z and Han B: MicroRNA-106b-5p boosts glioma
tumorigensis by targeting multiple tumor suppressor genes.
Oncogene. 33:4813–4822. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Carvalheira G, Nozima BH and Cerutti JM:
microRNA-106b-mediated down-regulation of C1orf24 expression
induces apoptosis and suppresses invasion of thyroid cancer.
Oncotarget. 6:28357–28370. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ni X, Xia T, Zhao Y, Zhou W, Wu N, Liu X,
Ding Q, Zha X, Sha J and Wang S: Downregulation of miR-106b induced
breast cancer cell invasion and motility in association with
overexpression of matrix metalloproteinase 2. Cancer Sci.
105:18–25. 2014. View Article : Google Scholar : PubMed/NCBI
|