Introduction
RNA in living organisms can be divided into 2 groups
according to their coding potential for proteins: coding and
non-coding RNAs (ncRNA) (1–3). NcRNAs account for ~98% of RNAs in
mammals including humans (1–3). The discovery of small ncRNAs (sncRNAs)
has created a new research field and caused an upsurge in RNA
studies (1–3). SncRNAs are a large group of RNAs that do
not code for proteins and are less than 200 nucleotides in length,
usually ~20-30 nt (1,2). The role of sncRNAs in life goes far
beyond previous cognition (4–7).
SncRNAs have various family members and form complex
regulatory networks in cells (3–5). Among
which the most studied are small nucleolar RNA (3,4), small
interfering RNAs (5), microRNAs
(6) and PIWI-interacting RNAs
(piRNAs) (7) (Table I). SncRNAs are involved in the
regulation of various biological functions, including organism
immunity, growth, development, organ formation, cell proliferation,
cell differentiation and cell death (1–7).
Abnormalities in sncRNA expression and functions lead to numerous
functional disorders and health problems including long-term memory
disorder (2) and cancer (8–10). This
review discusses the biological function of piRNAs and the current
research progress made with regard to piRNAs in cancer.
 | Table I.Classification of small non-coding
RNA. |
Table I.
Classification of small non-coding
RNA.
| Classification | Length(nt) | Features |
|---|
| Small nucleolar
RNAs | 60-300 | Biosynthesis of
ribosomal RNA and guider of RNA modification |
| Small interfering
RNAs | ~21-22 | Gene regulation,
transposition and virus defense |
| microRNAs | ~21-22 | Transcriptional
regulation |
| PIWI-interacting
RNAs | ~24-32 | Gene silencing
regulation; degradation of mRNA transcripts; maintenance of
germline and stem cell function |
Discovery and characteristics of
piRNAs
piRNAs are a new and diverse group of sncRNAs and
the human genome consists of >30,000 piRNA species (8). In 2006, scientists discovered a new type
of small ncRNA in the testes of mice (9). Such small ncRNAs could interact with the
PIWI subfamily proteins of the AGO (Argonaut) family to serve
important biological roles, hence they are named PIWI-interacting
RNA (piRNA) (10).
piRNAs are generally derived from genomic sequences
(11,12). These macromolecules originate from 3
main genome regions: the intergenic region containing a large
number of transposition fragments and repetitive sequences, the
long-chain non-coding gene region and the 3′-UTR region of the mRNA
(11,12). PiRNAs produced from the intergenic
region are distributed as clusters called piRNA clusters (11,12). Some
piRNA clusters can be bidirectionally transcribed and consequently
generate 2 piRNAs (13,14). Among which, the antisense piRNA is
complementary to the DNA sequence template. The production of piRNA
does not require the involvement of the Dicer enzyme in RNase III.
The length of piRNA is ~24-32 nucleotides (13,14). In
piRNAs the first base at the 5′ end has a strong uracil bias and
the 3′ end is modified by methylation (13,14).
piRNAs are tissue-specific and distributed mainly in mammalian germ
cells and embryonic stem cells (11–14).
Biological functions of piRNAs
piRNAs are crucial for the
maintainance of genomic integrity and stability
Transposons are a type of mobile DNA elements
(15–17). Most of the active transposons in
organisms are RNA transposons, which are also known as reverse
transcriptional transposons that occupy a large proportion of the
genome (15–17). During the development of germ cells
and embryos, epigenetic reprogramming is activated and a large
number of transcriptional transposon RNA and transposon activity is
enhanced (15–17). Retrotransposons can move within the
chromosomes or between different chromosomes, hence increasing the
probability of structural and functional changes in the genome and
leading to serious diseases, such as cancer (15–17).
During germ cell development, piRNA combines with the PIWIL2 gene
(member 2 of the PIWI-like subfamily also known as HILI) to form a
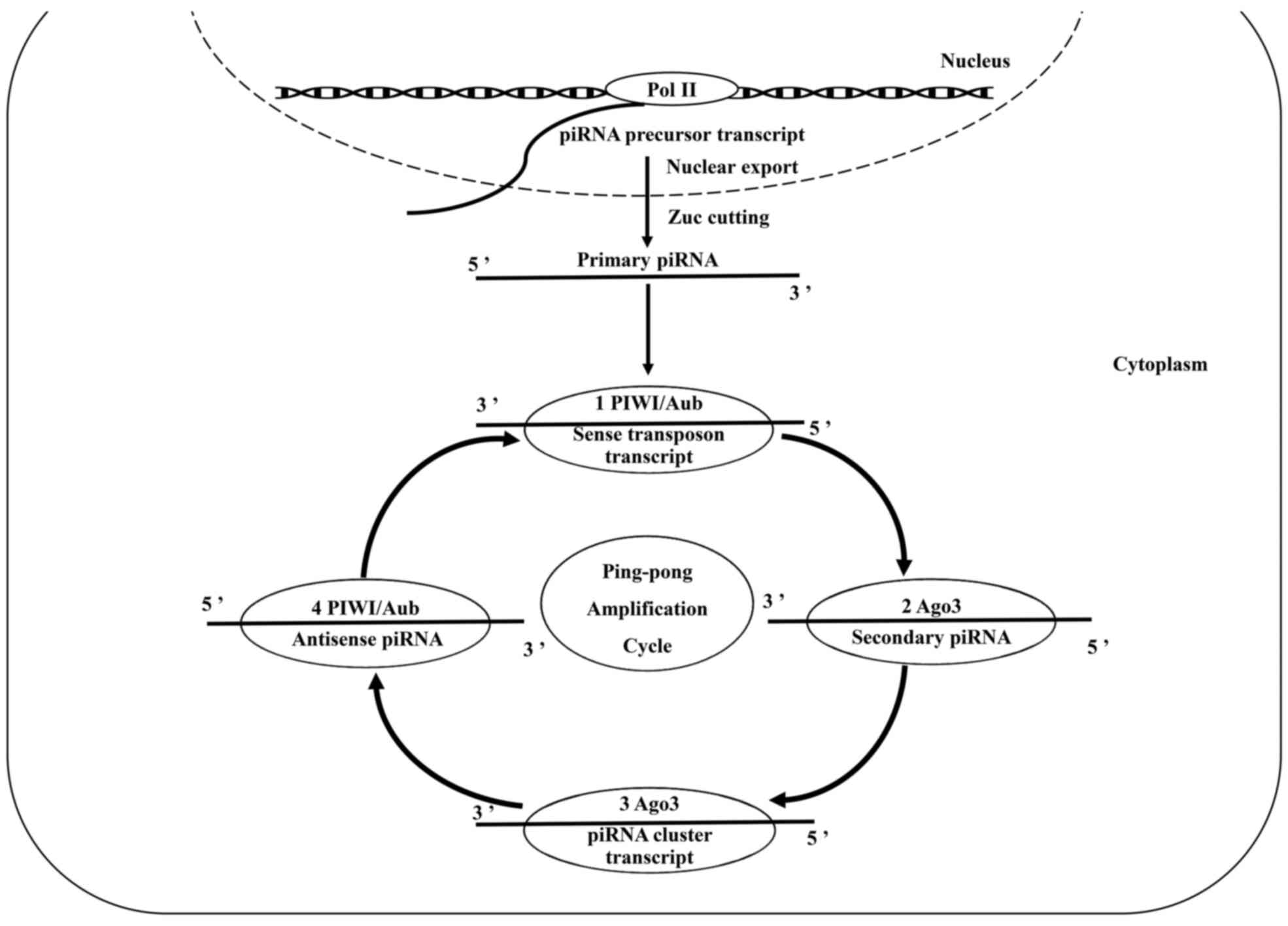
piRNA-induced gene silencing complex (pi-RISC) (18). In piRNA generation, the primary
production pathway enters the secondary generation pathway to form
a piRNA generation cycle, a process also known as the ‘ping-pong
cycle’ (19). The pi-RISC complex
uses active transposon transcripts as precursors and amplifies
piRNA in large quantities through the ping-pong cycle mechanism
(Fig. 1). piRNA binds with
transposons sequence by base complementation, hence completing
transcriptome cutting and consuming a large number of transposon
transcripts (20,21) (Fig. 1).
This process directly silences the ‘gene parasite’ transposition
element and protects germ cell genes from destruction (20,21). Thus,
piRNAs maintain genomic integrity and stability.
piRNAs regulates the degradation of
mRNA transcripts
PIWI/piRNA pathway mediates the degradation of a
large number of mRNA transcripts during mouse sperm formation,
particularly at the round spermatid stage (22,23). CAF1,
a subunit of the CCR4-NOT complex, is a magnesium dependent
deadenylase enzyme, which removes the poly(A) tail of mRNA
(22). piRNA recognizes the 3′-UTR in
mRNA sequences and its complementary sequence inhibits the activity
of mRNA deadenylase CAF1 (22). As a
result, the adenosine residues of mRNA become acidic and decay is
initiated by the piRNA binding with the PIWI protein (22,23).
Hence, piRNAs have an important regulatory role in the formation of
mice germ cells (22–25). Similar mechanisms have also been
observed in Drosophila germ cells (26).
piRNAs maintain germline and stem cell
function
piRNAs maintain the DNA integrity of germline stem
cells (24–28). Mouse PIWI homologs MIWI, MILI and
MIWI2 are highly expressed in mouse testes (24). The piRNA-dependent clearance of MIWI
via the anaphase-promoting complex/cyclosome (APC/C)-26S, mediated
ubiquitin proteome pathway is essential for mRNA stabilization and
proper sperm maturation during the late stages of sperm
development, indicating that the stage-specific regulation of
MIWI/piRNA is essential in male germ cell development (25). In model organisms, such as fruit
flies, piRNA pathways are involved in the decay of maternal
messenger RNA and the inhibition of translation in early embryos,
implying their direct regulatory role for genes, development such
as the embryonic posterior morphogen Nanos associated with embryo
(26). In addition, human HIWI
protein has 52% homology with Drosophila PIWI at the amino
acid level (27). HIWI genes are
expressed in developing fetal and adult tissues, including
primitive hematopoietic cells that are negatively regulated by HIWI
(27). The PIWI protein is a positive
modulator of adult stem cell generation and is required for
regeneration and tissue homeostasis during wound healing in lower
organisms, such as the planarian Schmidtea mediterranea
(28).
Progress of piRNAs in cancer research
piRNAs are associated with cancer development
(29). Given the abnormal expression
of piRNAs in different types of cancer, their functions cannot be
ignored (29). A number of piRNAs
with their binding partner, PIWI proteins, regulate the occurrence
and development of cancer (30,31).
piRNAs are differentially expressed in cancer and non-cancer
tissues and hence, can be used to distinguish them and provide new
cancer biomarkers (30,32). This section of the review mainly
summarizes the abnormal expression and relationship of piRNAs in
different human cancers including hepatocellular carcinoma (HCC),
gastric cancer (GC), colorectal cancer (CRC), osteosarcoma, lung
cancer, breast cancer (BC), prostate cancer, renal cell cancer
(RCC) etc.
piRNA and PIWILs in HCC
HCC is one of the most common malignant tumors and
among the top 5 cancers with the highest incidence and
cancer-related deaths in China and worldwide, ~906,000 new cases
and 830,000 deaths were reported worldwide in 2020 according to
Global Cancer Statistics 2020 (33).
In HCC, hundreds of piRNAs are differentially expressed according
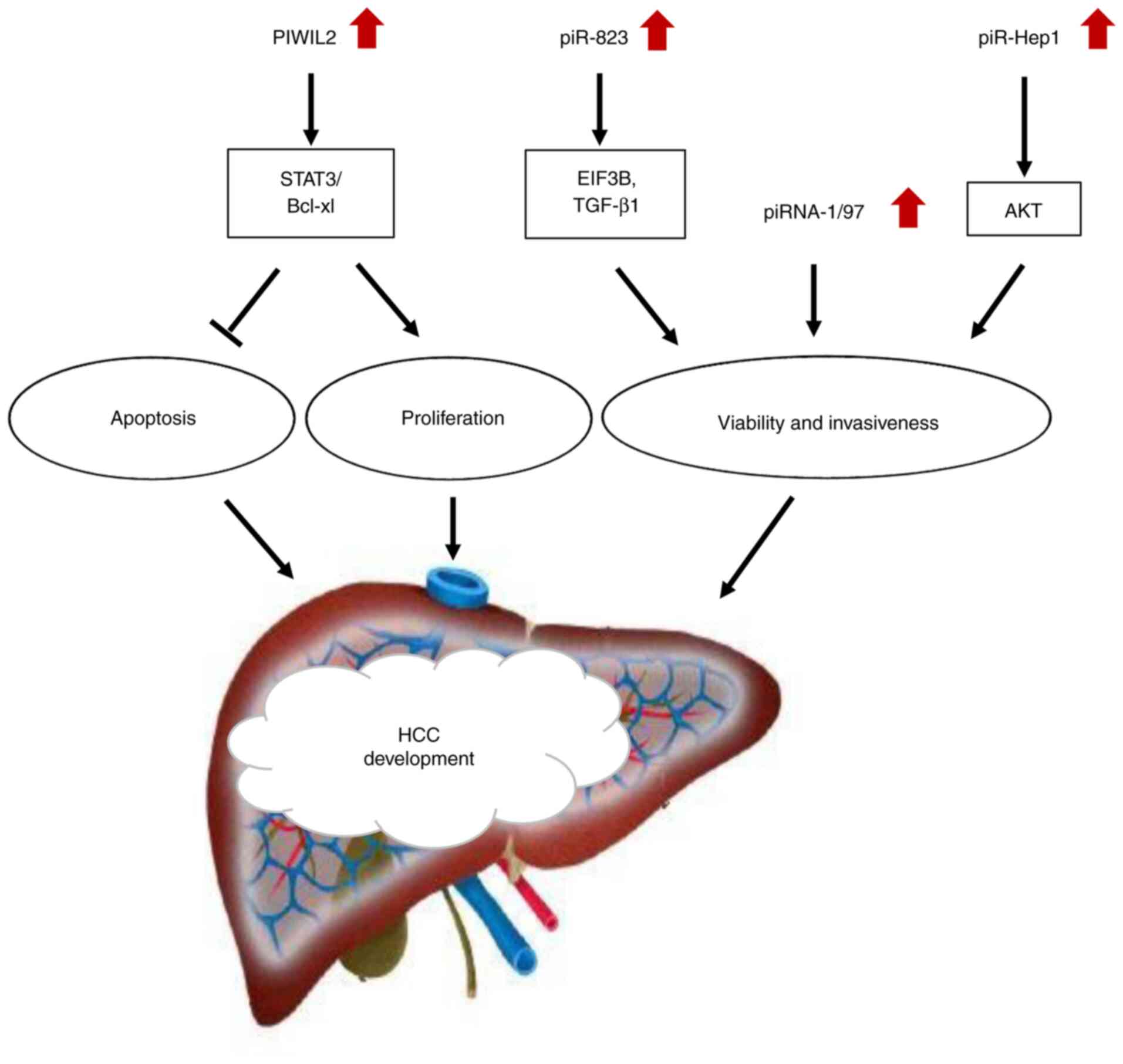
to small-RNA sequencing studies (34). Among which, piRNA-1/97 can promote the
migration and metastasis of hepatoma cells (34) (Table
II; Fig. 2). Several piRNAs
including piR-Hep1 (30), piR-823
(35) and piR-651 (36) have been identified in the
pathophysiology of HCC. piR-Hepl (30) is found to be upregulated in HCC tumors
compared with adjacent non-tumoral liver tissues and its silencing
inhibits cell survival, motility and tumor invasiveness mainly
through reducing the level of phosphorylated AKT (Table II; Fig.
2). In addition, the relative expression of PIWIL2 mRNA is
higher in HCC tissues compared with adjacent normal liver tissues
(37). A positive correlation was
found between PIWIL2 expression and piR-Hep1 level according to
Pearson's correlation analysis (30).
PIWIL2 acts as an oncogene by activating the STAT3/Bcl-xl cell
signaling pathway through endogenous RNAi mechanism, hence
inhibiting cell apoptosis and promoting cell proliferation
(31) (Table II; Fig.
2). AKT, the downstream target of the PIWIL2-activated
signaling pathway, promotes the key carcinogenic pathway of liver
cancer (30,31). Hence, the interaction between piR-Hep1
and PIWIL2 is crucial in the occurrence and development of tumors
(30) (Table II). Rizzo et al (38) found the specific expression pattern of
24 piRNAs, including piR-823 in dysplastic nodules and HCC. The
level of piR-823 is high during the progression from cirrhosis to
low- and high-grade proliferative nodules in HCC and is upregulated
in hepatic stellate cells (HSCs), which are responsible for liver
fibrogenesis (35). piR-823 also
activates HSCs and promotes extracellular matrix expression by
binding with eukaryotic initiation factor 3B (EIF3B) and
upregulating the expression of transforming growth factor β-1
(TGFβ-1) (35) (Table II; Fig.
2).
 | Table II.Summary of the abnormally expressed
piRNA and PIWIL in cancers of digestive system. |
Table II.
Summary of the abnormally expressed
piRNA and PIWIL in cancers of digestive system.
| Author(s),
year | Cancer type | piRNA or PIWIL | Expression | Tumor promoter or
suppressor | Molecular
mechanisms | Possible
applications | (Refs.) |
|---|
| Miao et al,
2018 | HCC | piRNA1/97 | Upregulated | Promoter | Promoted the
migration and metastasis of hepatoma cells | Potential markers
for monitoring metastasis | (34) |
| Law et al,
2013 |
| piR-Hep1 | Upregulated | Promoter | Promoted cell
viability, motility and invasiveness by activating AKT signaling
pathway | Potential
diagnostic biomarker or therapeutic target | (30) |
| Lee et al,
2006 |
| PIWIL2 | Upregulated | Promoter | Acted as an
oncogene by inhibition of apoptosis and promotion of proliferation
via STAT3/Bcl-xl signaling pathway | Potential
diagnostic biomarker or therapeutic target | (31) |
| Tang et al,
2018 |
| piR-823 | Upregulated | Promoter | Activated HSC and
promoted ECM expression through binding with EIF3B and upregulation
of TGF-β1 protein expression | Potential markers
for monitoring the occurrence of HCC | (35) |
| Cheng et al,
2011 |
| piR-651 | Upregulated | Promoter | Acted as an
oncogene diagnosis | Potential signs of
cancer | (36) |
| Cheng et al,
2011 | GC | piR-651 | Upregulated | Promoter | Promoted cell
cycle; associated with TNM stage | Diagnostic
biomarkers or treatment target | (36) |
| Cheng et al,
2012 |
| piR-823 | Downregulated | Suppressor | Inhibited tumor
growth in a dose-dependent manner | Diagnostic
biomarkers or treatment target | (32) |
| Ge et al,
2020 |
| piR-004918 and
piR-019308 (serum exosomes) | Upregulated | – | Abnormal expression
compared with unmetastatic GC | Potential markers
for monitoring GC metastasis | (44) |
| Cheng et al,
2011 | Colon cancer | piR-651 | Upregulated | – | Acted as an
oncogene | Diagnostic
biomarkers | (36) |
|
Vychytilova-Faltejskova et al,
2018 |
| piR-5937 and
piR-28876 (serum) | Downregulated | – | Abnormally low
expression in the serum of patients | Prognostic
molecular markers | (54) |
| Yin et al,
2017 | CRC | piR-823 | Upregulated | Promoter | Interacted with
HSF-1 and increased phosphorylation of HSF-1 at Ser326 | Diagnostic
biomarkers or therapeutic target | (47) |
| Weng et al,
2018 |
| piR-1245 | Upregulated | Promoter | Promoted tumor
progression by pi-RISC and its ability to repression of RNAs of
several tumor suppressor genes including ATF3, BTG1, DUSP1, FAS,
NFKBIA, UPP1, SESN2, TP53INP1 and MDX1 | Prognostic
molecular markers or therapeutic target | (48) |
| Chu et al,
2015 |
| piR-015551 | Upregulated | – | Positively
correlated with the expression level of lnc00964-3 | Potential markers
for monitoring development of CRC | (49) |
| Qu et al,
2019 |
| piR-017724
(serum) | Downregulated | – | Associated with the
poor overall survival and progression-free survival of
patients | Prognostic
molecular markers | (50) |
| Mai et al,
2018 |
| piR-54265 | Upregulated | Promoter | Promoted cell
proliferation, metastasis and chemoresistance of CRC cells by
binding to PIWIL2 and activation of the STAT3 signaling
pathway | Prognostic
molecular markers or therapeutic target | (51) |
| Tosar et al,
2021 |
| piR-54265
(serum) | Upregulated | Promoter | Significantly
elevated in serum of patients with primary and relapsed CRC | A valuable
biomarker for CRC screening, early detection and clinical
surveillance | (52) |
| Yin et al,
2019 |
| piR-18849 | Upregulated | Promoter | Positively
correlated with lymph node metastasis potential and negatively
correlated with tumor differentiation | Prognostic
molecular markers | (53) |
| Yin et al,
2019 |
| piR-19521 | Upregulated | Promoter | Negatively
correlated with the degree of tumor differentiation | Prognostic
molecular markers | (53) |
| Wang et al,
2020 |
| piR-020619 and
piR-020450 (serum) | Upregulated | – |
Abnormally high in the serum
of patients with CRC | Early biomarkers of
detection | (55) |
| Litwin et
al, 2018 |
| HIWI(PIWIL1) | Upregulated | – | Positively
correlated with OCT4 mRNA levels | Prognostic
molecular markers or treatment target | (56) |
| Litwin et
al, 2018 |
| HILI(PIWIL2) | Downregulated | – | Positively
correlated with SOX2 | Prognostic
molecular markers or treatment target | (56) |
Given the crucial role of piRNA-1/97 in the
migration and metastasis of liver cancer cells (34) (Table
II; Fig. 2), further experimental
studies may identify new pathways and molecular targets for the
clinical diagnosis and treatment of HCC. The abnormal expression
and involvement of piR-Hep1, PIWIL2, piRNA-1/97 and piR-823 in the
occurrence of HCC (30,31,37)
(Table II) and their close
relationship with the key downstream carcinogenic pathway of HCC
(Fig. 2) provide primary evidence for
the implication of piRNAs/PIWIL in the clinical detection and
treatment of liver cancer. Besides, abnormal piR-651 expression may
be a potential indicator of the development and progression of HCC
(36) and piR-823 accelerates
oncogenic processes during HCC (38)
(Table II). All the aforementioned
studies suggest that piRNA/PIWIL may be potential markers for liver
cancer detection.
piRNAs and GC
Although its incidence currently has a downward
trend, GC is still the third leading cause of cancer-related deaths
worldwide (39). In 2020,
>1,000,000 new cases and an estimated 769,000 deaths were
reported worldwide, with a substantial increase in Asian countries,
such as Mongolia and Japan where the GC incidence is twice as high
in men compared with women (33). GC
is asymptomatic in the early stage. A number of patients are
diagnosed at the middle or late stages of disease (39–42). piRNA
expression is often closely related to the malignant degree of GC
(40,41). In numerous GC cases, the expression
levels of piRNAs in gastric carcinoma tissues are highly altered
compared with those in normal gastric mucosa tissues (42).
piRNAs, such as piR-651 and piR-823 are
differentially expressed in GC tissues (43). Among them, piR-651 is notably
upregulated in several cancers including GC, CRC, lung cancer and
BC and in cancer cell lines, such as HepG2 (liver cancer), HeLa
(cervical cancer), BCAP-37 (BC), MSTO-211H (mesothelioma), NCI-H446
(lung cancer), MGC-803 and SGC-7901 (GC) (36). piR-651 inhibitor (antagonist) can
inhibit cell cycle and growth in GC cells, including MGC-803 and
SGC-7901 (36). In particular,
piR-651 antagomir arrests MGC-803 cells in the G2/M
phase (36). The aforementioned
studies suggested that piR-651 has an oncogenic role in development
of GC (Table II). piR-823 is
downregulated in GC tissues compared with non-cancerous tissues and
its mimics can inhibit the growth of MGC-803 and SGC-7901 cells
(32) (Table II). In addition, the levels of
piR-651 and piR-823 in the circulation of patients with GC were
lower compared with those in normal people (43). The positive detection rates of piR-651
(74.07%) and piR-823 (88.88%) are highly sensitive (43) and therefore, can be used as biomarkers
for GC diagnosis. Hence, the levels of piR-651 or piR-823 may be
useful to detect GC incidence and therapeutic manipulation of these
piRNAs could effectively inhibit the occurrence and development of
GC.
The expression levels of piR-004918 and piR-019308
in the serum exosomes of patients with metastatic GC were
significantly higher compared with patients with GC without
metastasis (44). So, serum
piR-004918 and piR-019308 could be used as potential markers to
monitor GC metastasis (44) (Table II).
piRNAs and PIWILs in CRC and colon
cancer
Early diagnosis of CRC is crucial for patient
survival (45,46). Although the overall molecular
mechanism of CRC has not been fully elucidated, piRNAs and PIWILs
are crucial for the early diagnosis of this disease (45,46).
piR-651 expression is higher in colon cancer tissues
compared with corresponding noncancerous normal tissues (36) (Table
II). However, the role of piR-651 in the development of colon
cancer remains largely unknown (36).
piR-823 interacts with heat shock transcription factor-1 and
increases its phosphorylation at Ser326, which in turn promotes its
transcriptional activity and increases oncogene expression
(47). The antagonist of piR-823
blocks the cell cycle at the G1 phase and increases the
apoptosis of CRC cells, such as HCT116 and DLD-1 (47) (Table
II). These findings indicate that piR-823 may serve as a
potential therapeutic target for CRC.
piR-1245 is also upregulated in CRC and its
expression is associated with late-stage and metastatic CRC
(48). However, an extremely high
piR-1245 expression affects the prognosis of patients with CRC
(48). Patients with a high
expression of piR-1245 have shorter overall survival time compared
with those with a low level of piR-1245 (48). Mechanistic studies found that a
specific pi-RISC formed by piR-1245 induces tumor progression
through its ability to repress the RNAs of several tumor suppressor
genes including activating transcription factor 3, B-cell
translocation gene 1, dual-specificity phosphatase-1, FAS, NFKBIA
(encoding nuclear factor of κ-light polypeptide gene enhancer in
B-cells inhibitor-α), uridine phosphorylase 1, sestrin2 (a highly
conserved and stress-inducible protein), tumor protein 53-induced
nuclear protein 1 and MAX dimerization protein 1, which are
potential targets complementary to piR-1245 (48). In CRC, the expression levels of these
targets are negatively associated with that of piR-1245 (48) (Table
II). piR-1245 also exerts oncogenic function in CRC by
promoting cell survival, migration and invasion and suppressing
apoptosis (48). Hence, piR-1245 may
be a potential diagnostic, prognostic, and/or therapeutic target
for CRC.
piR-015551, generated from long non-coding RNA (lnc)
00964-3, is positively correlated with lnc00964-3 according to
Pearson's correlation analysis and piR-015551 reduces the
expression of lnc00964-3 in CRC tissues (49) (Table
II). This finding suggests an interaction between lncRNAs and
piRNAs during the development and progression of CRC.
A total of 5 piRNAs (piR-001311, piR-004153,
piR-017723, piR-017724, and piR-020365) are differentially
expressed in the circulation of patients with CRC (50). The reduction of serum level of
piR-017724 is associated with patient survival rate and thus, may
be an independent prognostic factor for CRC detection (50). However, further study is needed to
increase its specificity (50)
(Table II). In addition, piR-54265
expression is substantially higher in CRC and its expression level
is associated with poor prognosis and poor overall survival time of
patients (51,52). piR-54265 promotes CRC cell invasion
and metastasis by binding to PIWIL2 and activating the STAT3
signaling pathway (51,52) (Table
II). Thus, piRNA-54265 may be an oncogenic RNA in CRC and can
be used as a therapeutic target.
In CRC tissues, piR-18849, piR-19521 and piR-17724
levels are increased (53). piR-18849
overexpression is positively correlated with lymph node metastasis
potential, but negatively correlated with tumor differentiation
degree, while piR-19521 expression is only negatively correlated
with the degree of tumor differentiation according to Spearman's
correlation analysis (53) (Table II). These findings revealed that
targeting piR-18849 and piR-19521 may be effective in blocking the
metastasis and differentiation of CRC. The differential expression
of these piRNAs may be useful in CRC detection.
The serum levels of piR-5937 and piR-28876 may be
applied to detect colon cancer with higher sensitivity and
specificity compared with the biomarkers carcinoembryonic antigen
and carbohydrate antigen 19-9 in patients with stage I colon cancer
(54) (Table II). In addition, the combined
monitoring of piR-020619 and piR-020450 can effectively distinguish
colon cancer tissues from normal tissues and is highly specific for
early colon cancer detection (55)
(Table II).
Human PIWI proteins, such as HIWI (PIWIL1) and HILI
(PIWIL2) act together in the occurrence of CRC (56). High HIWI and low HILI mRNA levels are
detected in CRC tissues, which are positively correlated with CRC
stem cell markers, OCT4 (a transcription factor of the POU protein
family) and SOX2 (a marker of embryonic stem cell pluripotency),
respectively, according to Spearman's correlation analysis
(56). This finding suggests that the
differential expression of HIWI, HILI and some cancer stem cell
markers in CRC may have a prognostic value and could provide a new
diagnostic and therapeutic approach for CRC treatment. This finding
also indicates the complexity of CRC and provides new avenues for
developing therapeutics against this disease.
piRNAs and osteosarcoma
Osteosarcoma is one of the most common primary
osteoblastic tumors (bone tumors) and according to data from
1984–2013 in the SEER (Surveillance, Epidemiology and End Results)
database of USA, the incidence of osteosarcoma between 0–29 years
of age remained relatively stable for the past 30 years (57). The survival rate of osteosarcoma
following surgery is only 15–20%, while its functional recovery
after amputation is poor and its disability and metastasis rates
are high (57,58). A total of 80–90% of patients with
osteosarcoma die of distant metastasis, such as lung or bone
metastases (58). Despite the unclear
etiology and pathogenesis mechanisms of osteosarcoma, recent
studies suggested that piRNAs serve an important role in the
development of osteosarcoma (59).
piR-39980 overexpression with piR-39980 mimic in 2
human osteosarcoma cell lines (143B and HOS) promoted cell
proliferation, migration and invasion via targeting serpin family B
member 1 (SERPINB1) and activating matrix metalloproteinase-2
(59). Inhibiting piRNA-39980
upregulated SERPINB1, promoted chromatin condensation and induced
y-H2AX accumulation and cell death (59). This finding revealed that piRNAs can
accelerate the metastatic potential of osteosarcoma by negatively
regulating tumor suppressors, such as SERPINB1 (59) (Table
III). Hence, piR-39980 may be a prognostic marker and
therapeutic target for osteosarcoma.
 | Table III.Summary of the abnormally expressed
piRNA and PIWIL in other types of cancers. |
Table III.
Summary of the abnormally expressed
piRNA and PIWIL in other types of cancers.
| Author(s),
year | Cancer type | piRNA or PIWIL | Expression | Tumor promoter or
suppressor | Molecular
mechanisms | Possible
applications | (Refs.) |
|---|
| Das et al,
2020 | Osteosarcoma | piR-39980 | Upregulated | Promoter | Promoted cell
migration and invasion by negatively regulating tumor suppressors,
such as SERPINB1 | Potential
diagnostic biomarker or therapeutic target | (59) |
| Peng et al,
2016 | Lung cancer | piR-55490 | Downregulated | Suppressor | Induced mTOR
degradation by binding to the 3′-UTR of mTOR mRNA and resulted in
inactivation of mTOR/AKT pathway | Potential
diagnostic biomarker or therapeutic target | (61) |
| Reeves et
al, 2017 |
| piR-34871,
piR-52200 | Upregulated | Promoter | Promoted
proliferation of lung cancer cell lines (A549 and H1299) by RASSF1C
regulating piRNA expression and inhibiting the AMPK pathway | Potential mechanism
molecules for future lung cancer research | (64) |
| Reeves et
al, 2017 |
| piR-35127,
piR-46545 | Downregulated | Suppressor | Blocked
proliferation of lung cancer cell lines (A549 and H1299) by RASSF1C
regulating piRNA expression and activating the AMPK pathway | Potential mechanism
molecules for future lung cancer research | (64) |
| Reeves et
al, 2012 |
| PIWIL1 | Upregulated | Promoter | Involved in the
initiation and progression of lung cancer through the MEK-ERK1/2
pathway | Potential mechanism
molecules for future lung cancer research | (65) |
| Xie et al,
2018 | Lung
adenocarcinoma | PIWIL1 | Upregulated | Promoter | Promoted the
proliferation, invasion and metastasis of lung adenocarcinoma
cells | Potential markers
for monitoring the development process | (66) |
| Li et al,
2016 | NSCLC | piR-651 | Upregulated | Promoter | Promoted cell
proliferation through cyclin D1 and CDK4 pathways | Potential tool for
the clinical diagnosis and treatment of NSCLC | (68) |
| Zhang et al,
2018 |
| piR-651 | Upregulated | Promoter | Promoted cell
proliferation, migration and invasion and inhibited cell
apoptosis | Potential biomarker
for the clinical diagnosis and treatment of NSCLC | (69) |
| Huang et al,
2013 | BC | piR-4987 | Upregulated | Promoter | Associated with
positive lymph nodes | Potential molecular
targets | (72) |
| Huang et al,
2013 |
| piR-20365,
piR-20485, piR-20582 | Upregulated | Promoter | Involved in the
occurrence and development of breast cancer | Potential molecular
targets | (72) |
| Oner et al,
2016 |
| piR-651,
piR-823 | Upregulated | Promoter | Promoted malignant
cell proliferation biomarkers or therapeutic targets | Potential
diagnostic | (74) |
| Fu et al,
2015 |
| piR-021285 | Upregulated | Promoter | Inhibited
methylation of CpG sites at 5′-UTR of first exon of ARHGAP11A mRNA,
which led to increased expression of ARHGAP11A and invasiveness of
breast cancer cells | Potential
diagnostic biomarkers or therapeutic targets | (75) |
| Maleki Dana P et
al, 2020 |
| piR-36712 | Downregulated | Suppressor | Promotes the
proliferation, invasion and migration of cancer cells;
Downregulated piR-36712 which led to SEPW1 mediated suppression of
P53, p21 and E-cadherin and upregulation of SLUG | Potential
prognostic molecular markers | (77) |
| Lee et al,
2010 |
| PIWIL2 | Upregulated | Promoter | Promoted breast
cancer cell survival by activating the STAT3/Bcl-xl pathway | Potential molecular
target for the clinical prognostic treatment | (80) |
| Zhang et al,
2013 |
| PIWIL2 |
Upregulated | Promoter | Acted as a positive
modulator of EMT in breast cancer stem cells by methylating Latexin
and suppressing its expression | Potential molecular
target for the clinical treatment | (81) |
| Krishnan et
al, 2016 |
| PIWIL3 | Upregulated | – | Related to overall
survival periods and recurrence free survival periods | Potential
independent prognostic marker | (78) |
| Heng et al,
2018 |
| PIWIL4 | Upregulated | Promoter | Triggered ER
pathway by upregulating the canonical ER signaling molecules, such
as Greb1, Tff1, Calcr and Ccnd1 | New regulators and
potential biological targets | (79) |
| Zuo et al,
2019 | Prostate
cancer | piR-000627,
piR-005553, piR-019346 | Upregulated | – | Associated with
biochemical recurrence (BCR) of prostate cancer | Potential
prognostic marker during treatment | (82) |
| Zhang et al,
2020 |
| piR-001773,
piR-017184 | Downregulated | Suppressor | Bound to 3′-UTR
site of PCDH9 and post-transcriptionally regulates PCDH9 | Potential
biological targets for future prostate cancer research | (83) |
| Chu et al,
2015 | Bladder cancer | piRABC | Downregulated | Suppressor | Suppressed the
development of bladder cancer by forming HIWI-piRABC complex and
targeting 3′-UTR of tumor necrosis factor superfamily member 4
(TNFSF4) mRNA which increases cell death resistance | Potential mechanism
molecules for future bladder cancer research | (84) |
| Ravo et al,
2015 | Endometrial
cancer | piR-020829,
piR-019914, piR-016735 | Upregulated | – | – | New biomarkers that
can be used to study early endometrial cancer | (85) |
| Ravo et al,
2015 |
| piR-020496 | Upregulated | Promoter | Targeted a
transcriptional co-repressor known as TLE4
(Transduction-protein-like enhancers of fragment 4) | New biomarkers that
can be used to study early endometrial cancer | (85) |
| Li et al,
2020 | PDAC | PIWIL1 | Upregulated | Promoter | Enhanced the
metastatic potential of PDAC by functioning as a co-activator of
anaphase-promoting complex/cyclosome (APC/C) E3 complex to
facilitate the degradation of pinin | Potential target of
PDAC | (86) |
| Lim et al,
2014 | Ovarian cancer | PIWIL1 | Upregulated | Suppressor | Reduced the
aggression of SKOV3 (ovarian cancer cell lines) | Potential mechanism
molecules for future ovarian cancer research | (87) |
| Busch et al,
2015 | ccRCC | piR-30924,
piR-38756 | Upregulated | Promoter | Related to tumor
recurrence and overall survival of clinical patients with
ccRCC | Independent
potential prognostic biomarker | (91) |
| Busch et al,
2015 |
| piR-57125 | Downregulated | Suppressor | Related to tumor
recurrence and overall survival of clinical patients with
ccRCC | Independent
potential prognostic biomarker | (91) |
| Zhao et al,
2019 |
| piR-34536,
piR-51810 | Downregulated | Suppressor | Found in ccRCC
mitochondria | Potentially new
prognostic biomarkers | (92) |
| Li et al,
2015 |
| piR-32051,
piR-39894, piR-43607 | Upregulated | Promoter | Associated with
metastasis molecules for future renal clear cell carcinoma
research | Potential
mechanism | (93) |
| Iliev et al,
2016 | RCC | piR-823
(urine) | Upregulated | – | – | Potential
diagnostic biomarker or therapeutic target for RCC | (94) |
| Stöhr et al,
2019 |
| PIWI-like 1 | – | Promoter | Associated with
shorter cancer-specific survival | Potential
indicators for the prognosis of patients with RCC | (96) |
piRNA and PIWILs in lung cancer
Lung cancer is one of the most common malignancies
worldwide (33). With an estimated
2.2 million new cancer cases and 1.8 million new deaths, lung
cancer is the second most commonly diagnosed cancer and the leading
cause of cancer-related death in 2020 (33). piRNAs are associated with the
progression of lung cancer (60–70).
In lung cancer, piRNAs, such as piR-010894-3 and
piR-001168-4 are upregulated in non-smoking patients with lung
tumors compared with patients who are smokers with lung tumors
(60). These constitutive and
differentially expressed piRNAs may be potential targets for
improving the diagnosis and treatment of patients with lung cancer
(60). A reduced piR-55490 expression
is negatively correlated with patients' overall survival with lung
cancer according to Spearman's correlation analysis (61). Restoring piR-55490 level inhibits the
proliferation of lung cancer cells mainly by binding to the 3′-UTR
of mTOR mRNA to induce its degradation and inactivate the mTOR/AKT
pathway (61) (Table III).
Ras association domain family 1C (RASSF1C) is one of
the two main subtypes of the RASSF1 gene, which has an
anti-apoptotic effect and promotes cell proliferation in BC cells,
such as breast cancer cell line T47D and lung cancer cells, such as
A549 and NCI-H1299) (62,63). In stably overexpressing RASSF1C lung
cancer cells, 4 piRNAs are differentially expressed (upregulated
piR-34871 and piR-52200 and downregulated piR-35127 and piR-46545)
(64). RASSF1C overexpression reduces
p-AMPK, p21 and p27 protein levels, implying that RASSF1C mediates
the regulation of piRNA expression by inhibiting the AMPK pathway
and thereby modulating the level of its target genes (64). In tumor tissues, piR-35127 is
negatively associated with RASSF1C (64). Silencing piR-34871 and piR-52200 or
overexpressing piR-35127 and piR-46545 can block the proliferation
of lung cancer cell lines (A549 and NCI-H1299) (64) (Table
III), indicating that these piRNAs are involved in the
regulation of lung cancer cells' transformation and tumorigenesis.
In addition, RASSF1C promotes the expression of PIWIL1 through the
MEK-ERK1/2 pathway and RASSF1C-PIWIL1 might be involved in the
initiation and progression of lung cancer (65) (Table
III). These studies revealed that RASSF1C is closely associated
with piRNA- and PIWIL1-mediated oncogenic processes in lung cancer
cells.
PIWIL1, a binding partner of piRNAs, is upregulated
in lung adenocarcinoma and promotes the proliferation, invasion,
and metastasis of lung adenocarcinoma cells (66). PIWIL1 expression is closely related to
the shortened overall survival time of patients with lung
adenocarcinoma (66) (Table III). Hence, the tumorigenic
processes in patients with lung cancer can be determined by
detecting the expression level of PIWIL1.
piR-651 is upregulated in numerous cancer tissues
and cell lines including GC, osteosarcoma, lung cancer, HCC, GC and
CRC (36,43,67). In a
highly metastatic human lung cancer cell line named 95-D, piR-651
enhances the carcinogenic potential by promoting cell
proliferation, migration, invasion and inhibiting apoptosis
(67).
In non-small cell lung cancer (NSCLC), piR-651
promotes cell proliferation through the cyclin D1 and
cyclin-dependent kinase 4 pathways (68) (Table
III). Inhibiting piR-651 reduces cell proliferation and
invasion and induces apoptosis in NSCLC (69) (Table
III). These findings indicate that piR-651 may be a potential
tool for the clinical diagnosis and treatment of lung cancer. The
differential expression of piRNA/piRNA-L has been observed in NSCLC
cell lines, such as H157, H226, H596, SK-MES-1, H522, H1437, H1792
and H1944 (70). piRNA/piRNA-L
regulates lung carcinogenesis by directly interacting with proteins
involved in the occurrence of lung tumors (70) and thus, may be a new tool for the
diagnosis and treatment of lung cancer. However, potential
applications of piRNAs in patients with lung cancer require further
confirmation.
piRNAs and PIWILs in BC
BC has surpassed lung cancer as the leading cause of
global cancer incidence in 2020, with an estimated 2.3 million new
cases and is the fifth leading cause of cancer mortality worldwide
(33). piRNAs are involved in the
occurrence of BC (71–81).
Compared with matched non-tumor tissues, a total of
4 piRNAs (piR-4987, piR-20365, piR-20485 and piR-20582) were
upregulated in breast cancer tissues and the increased piR-4987
expression was substantially associated with lymph node metastasis
(72) (Table III). The abnormal expression of
these piRNAs and the association of piR-4987 with lymph node
metastasis suggests their potential as important therapeutic
targets for BC.
In BC cells, the expression levels of DQ596670,
DQ598183, DQ597341, DQ598252, and DQ596311 are reduced, whereas
those of DQ598677, DQ597960, and DQ570994 are increased (73). A search for mRNAs targeted by the BC
piRNome revealed that these 8 piRNAs are involved in hormone
signaling, cell transformation, growth inhibition, and/or cell
cycle (73). These 8 piRNAs have
specific expression patterns in breast tumors and target several
key cancer cell pathways, such as Janus kinase-1, AKT3 etc.
(73). These findings suggest that
piRNAs may be a new class of primary regulators of BC development
and may be useful for the detection of this disease.
The expression level of piR-651 and piR-823 are
associated with hormone changes in gonadal development and BC
(74). piR-651 and piR-823
overexpression promotes cell proliferation in BC and prostate
cancer cell lines (74) (Table III). piRNAs, such as piR-021285 can
epigenetically control cancer-related genes in BC cells (75). In MCF cells, piRNA-021285 alters the
methylation status of several cancer-associated genes including the
ARHGAP11A gene (75). piRNA-021285
inhibits the methylation of CpG sites at 5′-UTR of the first exon
of ARHGAP11A mRNA, thus increasing ARHGAP11A level and the
invasiveness of BC cells (75)
(Table III). These studies reveal
that these piRNAs regulate the expression of oncogenes in BC.
Targeting piRNA-021285 may be therapeutically beneficial in BC
treatment.
piR-36712 functions as a tumor suppressor and its
low level is associated with poor clinical prognosis in patients
with BC. piRNA-36712 interacts with the RNAs generated by the
pseudogene of SEPW1 (SEPW1P) and inhibits SEPW1 expression by
competing with SEPW1P, microRNA-7 and microRNA-324 (77). In BC cells, downregulating piR-36712
leads to the SEPW1-mediated suppression of P53, p21 and E-cadherin
and the upregulation of Snail family transcriptional repressor 2
(snail2 or SLUG) (77). This event
promotes the proliferation, invasion, and migration of cancer cells
(76,77) (Table
III). Thus, by accelerating cancer development, SEPW1 may
worsen the prognosis of patients with BC.
In addition to piRNAs, PIWI genes are promising
prognostic markers for BC (78). In
breast tumor tissues, PIWIL1 and PIWIL3 are upregulated, whereas
PIWIL2 and PIWIL4 are downregulated. A total of 2 piRNAs
(piR-009051 and piR-021032) and PIWIL3 in tumor tissues were found
to be important for overall survival and recurrence free survival
periods in patients with BC (78). A
multivariate analysis confirmed that PIWIL3 is a potential
independent prognostic marker for BC (78). Given that the PIWIL proteins are
involved in piRNA biogenesis and the abnormal expression of these
genes in BC could lead to abnormal piRNA expression, these proteins
may be potential candidates for the prognosis of BC (78) (Table
III). In human BC cells, the expression level of PIWIL4 is
relatively high as it is required for cell growth, migration, and
invasion (79) (Table III). Given that estrogen receptor
(ER) signaling is involved in BC growth, an interaction occurs
between PIWIL4 and ER signaling pathway (79). PIWIL4 expression can be induced by ER
signaling, and PIWIL4 triggers ER pathway by upregulating the
canonical ER signaling molecules, such as growth regulation by
estrogen in breast cancer 1, trefoil factor 1, calcitonin receptor,
and cyclin D1 (79). These findings
suggest that PIWIL4 is a novel modulator of ER-dependent BC growth
and targeting this protein can inhibit the growth and migration of
these cancer cells (79) (Table III).
PIWIL2 is also abnormally expressed in BC cells and
promotes cell survival by activating the STAT3/Bcl-xl pathway
(80) (Table III). PIWIL2 specifically recognizes
the 3′ terminus of piR-932 and forms pi-RISC, which acts as a
positive modulator of epithelial-mesenchymal transition in BC stem
cells by methylating latexin and suppressing its expression
(81) (Table III). All these studies reveal that
the differentially expressed PIWIL genes in BC potentially
influence the tumorigenic processes and thus, can be used as
targets for the diagnosis and treatment of this disease.
piRNAs and prostate cancer
piRNAs serve an important role in numerous types of
cancer such as BC, lung cancer, prostate cancer, etc. (61,72,82). In
prostate cancer, the expression of certain piRNAs is associated
with the biochemical recurrence (BCR) of prostate cancer and thus
can be used to distinguish high-risk BCR patients from low-risk
patients (82). A total of 3 piRNAs
(hsa-piR-000627, hsa-piR-005553 and hsa-piR-019346) are associated
with prostate cancer BCR (82). Among
them, hsa-piR-000627 and hsa-piR-005553 have 343 common targeting
genes, 2 of which are mainly related to nucleoplasm and
intracellular transport (82)
(Table III). These studies reveal
that piRNAs regulate oncogenic processes during prostate cancer
development.
PCDH9 (member 9 of the protocadherin family) is a
tumor suppressor that is downregulated in prostate cancer and is a
potential target of a number of piRNAs including piR-001773 and
piR-017184. piR-001773/piR-017184 can directly bind to the 3′-UTR
sites of PCDH9 and post-transcriptionally regulate the expression
of PCDH9. The downregulation of piR-001773 and piR-017184 inhibits
tumor growth in vitro and in vivo (83) (Table
III). Given that piR-001773 and piR-017184 target PCDH9,
therapeutically suppressing these piRNAs may block tumor growth in
the prostate.
The targeted relationship between piRNA and tumor
suppressors provides important clues for prostate cancer mechanisms
and lays an important foundation for future prognosis monitoring
and therapeutical strategies of prostate cancer (82,83).
piRNAs and bladder cancer
In bladder cancer, DQ585569 is highly upregulated
and DQ594040 (piRABC) is downregulated. piRABC serves a
tumor-suppressive function by regulating cell proliferation, colony
formation and apoptosis in bladder cancer (84). piRABC can also suppress the
development of bladder cancer by forming the HIWI-piRABC complex
and targeting the 3′-UTR of tumor necrosis factor superfamily
member 4 mRNA, which increases cell death resistance (84) (Table
III).
piRNAs and endometrial cancer
Expression levels of has-piR-020829,
hsa-piR-019914, and hsa-piR-016735 are increased in endometrial
carcinoma and piR-020496 participates in endometrial cancer by
targeting a transcriptional co-repressor known as
transduction-protein-like enhancers of fragment 4 (85) (Table
III). Hence, these newly identified piRNAs may be used as novel
biomarkers for the early detection of endometrial cancer.
PIWIL and pancreatic duct cancer
(PDAC)
Without piRNA ligand, PIWIL1 activates anaphase by
functioning as a co-activator of APC/C E3 complex in human PDAC
(86). These complexes target and
facilitate the degradation of pinin, a key cell adhesion-related
protein and enhance the metastatic potential of PDAC (86) (Table
III). This phenomenon is opposite to the APC/C-mediated removal
of PIWIL1 during spermatogenesis (86). Hence, PIWILs could also function as
co-activator in malignant cells and PIWIL1 has an oncogenic
function in PDAC.
PIWIL and ovarian cancer
Abnormal expression of piRNA pathway genes, such as
PIWIL1 is accompanied by the upregulation of Maelstrom, a known
testis cancer gene in epithelial ovarian cancer and benign ovarian
tumors (87). However, their
expression reduces the aggressiveness and invasive potential of
ovarian cancer cell line SKOV3 (87)
(Table III). This study reveals
that PIWIL has a differential function in ovarian cancer depending
on the types of cells and tissues surrounding the cancer cells.
piRNAs and PIWILs in RCC
RCC is one of the deadliest malignancies of the
urinary system and represented 2.4% of all adult malignancies
worldwide in 2012 (88). Early
detection of kidney cancer is difficult due to its asymptomatic
nature and setting the early diagnostic markers and treatment in
patients with RCC remains challenging (89,90).
piRNAs, PIWIs and PIWILs serve important roles in the pathogenesis
of RCC (91–96).
The most common pathological and histological
subtype of RCC is renal clear cell carcinoma (ccRCC), which
accounts for ~70-80% of RCC cases (91). Numerous piRNAs including piR-30924,
piR-57125 and piR-38756 are abnormally expressed in primary
non-metastatic and metastatic ccRCC tissues (91). Metastatic primary tumors have higher
expression of piR-30924 and piR-38756 and lower piR-57125
expression compared with non-metastatic tumors (91). Hence, piR-30924 and piR-57125 can be
independent potential prognostic biomarkers (91) (Table
III). In addition, all these piRNAs are associated with tumor
recurrence and overall survival time and are likely to improve
prognostic information in patients with ccRCC (92). A total of 2 mitochondrial-derived
piRNAs, namely piR-34536 and piR-51810, have downregulated
expression in ccRCC tissues, but not in the serum (92). Hence, their levels could serve as
independent predictive markers to detect ccRCC progression, cancer
specificity and overall survival span of patients with ccRCC, thus
providing new ways to optimize individualized treatment specific to
RCC stages and ultimately improve patient survival (92) (Table
III). In addition, the abnormal expression levels of piR-32051,
piR-39894, and piR-43607 originating from the same piRNA cluster on
chromosome 17 are highly associated with ccRCC metastasis (93) (Table
III). In serum and urine, piR-823 expression is high in
patients with RCC and its level is positively associated with
adverse cancer outcomes (94). Hence,
urinary piR-823 may have an important diagnostic value in patients
with RCC (94) (Table III).
PIWIL1, PIWIL2, and PIWIL4 are downregulated in RCC
and their levels are associated with the clinical stage of tumor
and associated with poor survival in patients with RCC (95). These studies indicate that PIWIL1,
PIWIL2, and PIWIL4 may be useful prognostic biomarkers in patients
with RCC; however, due to their complex functioning mechanisms,
further confirmation studies are required (95). In addition, PIWI-like proteins serve
an important role in the pathogenesis of RCC (96). PIWI-like 1 expression is associated
with tumor staging and distant metastasis. The positivity of
PIWI-like 1 is associated with shorter cancer-specific survival.
Hence, the role and expression levels of PIWI-like proteins render
them as potential prognostic markers in patients with RCC (96) (Table
III).
Conclusion and future perspective
Knowledge about PIWIL and piRNA-associated pathways
and their biological functions has progressively increased
(15–28). Despite their highly altered levels in
numerous types of cancer, such as liver cancer, GC, CRC,
osteosarcoma, BC, lung cancer, prostate cancer, etc, the mechanisms
of PIWIL/piRNAs dependent regulation of cancer development are
largely unknown (44,50,54,55,85,94).
The differential expression of piRNAs in an organ-specific manner
in cancer tissues highlights that PIWIL/piRNAs may be useful in
specific diagnosis, prognosis, and molecular targeted therapy based
on the specific type and stage of cancer (34–36,48–53,80–85).
Therapeutic manipulation for some piRNAs can block/halt tumor
progression. For example, the antagonists of piRNAs such as piR-651
(36) and piR-823 (47) (Table
II) can inhibit cell cycle progression and thereby increase
tumor apoptosis and suppress tumor growth. Similarly, PIWIL2
inhibits tumor apoptosis and promotes cell proliferation by
activating the STAT3/Bcl-xl cell signaling pathway (36,80)
(Tables II and III). piR-651 promotes tumor cell
proliferation through the cyclin D1 and cyclin-dependent 4 pathways
(68) (Table III). Inhibiting PIWIL2 and piRNA-651
using siRNA/antagonists effectively attenuates cancer cell growth
(36,80). In addition, pi-RISC formed by piRNA
accelerates oncogenic processes by inhibiting the expression of
multiple tumor-suppressive genes (48) (Table
II). piRNA-021285 induces the methylation of cancer-associated
genes in BC cells. In particular, piR-021285 can modulate the
invasiveness of BC cells by methylating CpG sites at the first exon
of 5′-UTR on ARHGAP11A mRNA (75).
However, the variation of SNPs in piR-021285 leads to the increased
expression of ARHGAP11A and the enhancement of migration and
invasion of breast tumor cells (75)
(Table II). Similarly, piR-55490
binding to 3′-UTR of mTOR mRNA induces the degradation and
inactivation of the mTOR/AKT pathway, thus suppressing lung cancer
cell proliferation (61) (Table III). In addition, the oncogenic
piRNAs, piR-001773 and piR-017184 inhibit the expression of a tumor
suppressor PCDH9 in prostate cancer (83). piR-001773 and piR-017184 mediate the
post-transcriptional suppression of PCDH9 to accelerate prostate
tumor growth (83) (Table III). In contrast, oncogenic genes
such as RASSF1C can regulate the expression of piRNAs including
piR-35127 by inhibiting the AMPK pathway, which participates in
cancer progression (64) (Table III). RASSFIC promotes PIWIL1
expression through the MEK-ERK1/2 pathway, which participates in
lung cancer development (65)
(Table III). Although the functions
of PIWIL/piRNAs and associated pathways in various forms of cancer
are largely unknown, currently available reports provide a vital
clue that piRNAs and PIWIL are important contributors to the
development and regulation of various cancers. Interestingly,
PIWILs and numerous piRNAs have malignant cell type- and
tumor-specific functions, implying that they could be efficient
markers and effective/promising targets for the treatment of a
particular type of cancer.
piRNAs are abnormally expressed in tumors and may
represent potentially relevant tumor biomarkers (30–32,34,35).
This article reviews the importance of piRNAs in tumorigenesis,
proliferation, migration and metastasis of various tumors (30,34).
However, mechanisms for abnormal piRNAs/PIWI expression in various
cancers have not been clarified in most studies and applications of
piRNAs/PIWI in targeted therapy are only mentioned in a few studies
(47,53,83).
An in-depth understanding of the carcinogenic/tumor
suppressive mechanisms of PIWIL/piRNAs would provide a new avenue
in the therapeutic approach for cancer diagnosis and treatment. The
present review will provide new research ideas for future
piRNAs/PIWI research and more research will reveal in detail the
specific mechanisms between piRNAs and cancer and their potential
as cancer biomarkers and therapeutic agents.
Acknowledgements
Not applicable.
Funding
This work was funded by the National Natural
Science Foundation of China (grant no. 32000495), National Natural
Science Foundation of Shandong Province (grant nos. ZR2020MH202 and
ZR2020MH250),Key Research and Development Project of Shandong
Province (grant no. 2019GSF108100) and A Project of Shandong
Province Higher Educational Science and Technology Program (grant
no. J18KA290) and A Project Funding approved by the National
Medical Degree Postgraduate Education Steering Committee (grant no.
C-YX20190201-09).
Availability of data and materials
Data sharing is not applicable to this article, as
no datasets were generated or analyzed during the current
study.
Authors' contributions
CY and HQ wrote the manuscript. YC and MP were
responsible for figures and tables. ZL designed and edited the
manuscript. MP edited the tables and critically revised the
manuscript. Data authentication is not applicable. All authors have
read and approved the final version.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
piRNAs
|
PIWI-interacting RNA
|
|
sncRNAs
|
small ncRNAs
|
|
PIWIL
|
PIWI-like
|
|
HCC
|
hepatocellular carcinoma
|
|
HSC
|
hepatic stellate cell
|
|
GC
|
gastric cancer
|
|
CRC
|
cololectal cancer
|
|
NSCLC
|
non-small cell lung cancer
|
|
BC
|
breast cancer
|
|
EMT
|
epithelial-mesenchymal transition
|
|
PDAC
|
human pancreatic duct cancer
|
|
ccRCC
|
renal clear cell carcinoma
|
|
RCC
|
renal cell cancer
|
|
RASSF1C
|
Ras association domain family 1C
|
|
ER
|
estrogen receptor
|
|
BCR
|
biochemical recurrence
|
|
PCDH9
|
protocadherin family member 9
|
References
|
1
|
Romano G, Veneziano D, Acunzo M and Croce
CM: Small non-coding RNA and cancer. Carcinogenesis. 38:485–491.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Huang Y, Zhang JL, Yu XL, Xu TS, Wang ZB
and Cheng XC: Molecular functions of small regulatory noncoding
RNA. Biochemistry (Mosc). 78:221–230. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ohtani M: Transcriptional regulation of
snRNAs and its significance for plant development. J Plant Res.
130:57–66. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Scott MS and Ono M: From snoRNA to miRNA:
Dual function regulatory non-coding RNAs. Biochimie. 93:1987–1992.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Carthew RW and Sontheimer EJ: Origins and
mechanisms of miRNAs and siRNAs. Cell. 136:642–655. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu X, Luo G, Bai X and Wang XJ:
Bioinformatic analysis of microRNA biogenesis and function related
proteins in eleven animal genomes. J Genet Genomics. 36:591–601.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chalbatani GM, Dana H, Memari F,
Gharagozlou E, Ashjaei S, Kheirandish P, Marmari V, Mahmoudzadeh H,
Mozayani F, Maleki AR, et al: Biological function and molecular
mechanism of piRNA in cancer. Pract Lab Med. 13:e001132019.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cabral GF, Pinheiro JA, Vidal AF, Santos S
and Ribeiro-Dos-Santos A: piRNAs in gastric cancer: A new approach
towards translational research. Int J Mol Sci. 21:21262020.
View Article : Google Scholar
|
|
9
|
Vagin VV, Sigova A, Li C, Seitz H, Gvozdev
V and Zamore PD: A distinct small RNA pathway silences selfish
genetic elements in the germline. Science. 313:320–324. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Aravin A, Gaidatzis D, Pfeffer S,
Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ,
Kuramochi-Miyagawa S, Nakano T, et al: A novel class of small RNAs
bind to MILI protein in mouse testes. Nature. 442:203–207. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Han BW and Zamore PD: piRNAs. Curr Biol.
24:R730–R733. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rosenkranz D: piRNA cluster database: A
web resource for piRNA producing loci. Nucleic Acids Res.
44:D223–D230. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gunawardane LS, Saito K, Nishida KM,
Miyoshi K, Kawamura Y, Nagami T, Siomi H and Siomi MC: A
slicer-mediated mechanism for repeat-associated siRNA 5′ end
formation in Drosophila. Science. 315:1587–1590. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Klattenhoff C and Theurkauf W: Biogenesis
and germline functions of piRNAs. Development. 135:3–9. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ernst C, Odom DT and Kutter C: The
emergence of piRNAs against transposon invasion to preserve
mammalian genome integrity. Nat Commun. 8:14112017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Han YN, Li Y, Xia SQ, Zhang YY, Zheng JH
and Li W: PIWI proteins and PIWI-interacting RNA: Emerging roles in
cancer. Cell Physiol Biochem. 44:1–20. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Morrish TA, Gilbert N, Myers JS, Vincent
BJ, Stamato TD, Taccioli GE, Batzer MA and Moran JV: DNA repair
mediated by endonuclease-independent LINE-1 retrotransposition. Nat
Genet. 31:159–165. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lu Y, Li C, Zhang K, Sun H, Tao D, Liu Y,
Zhang S and Ma Y: Identification of piRNAs in hela cells by massive
parallel sequencing. BMB Rep. 43:635–641. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Brennecke J, Aravin AA, Stark A, Dus M,
Kellis M, Sachidanandam R and Hannon GJ: Discrete small
RNA-generating loci as master regulators of transposon activity in
Drosophila. Cell. 128:1089–1103. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Klenov MS, Lavrov SA, Stolyarenko AD,
Ryazansky SS, Aravin AA, Tuschl T and Gvozdev VA: Repeat-associated
siRNAs cause chromatin silencing of retrotransposons in the
drosophila melanogaster germline. Nucleic Acids Res. 35:5430–5438.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang SH and Elgin SC: Drosophila PIWI
functions downstream of piRNA production mediating a
chromatin-based transposon silencing mechanism in female germ line.
Proc Natl Acad Sci USA. 108:21164–21169. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gou LT, Dai P, Yang JH, Xue Y, Hu YP, Zhou
Y, Kang JY, Wang X, Li H, Hua MM, et al: Pachytene piRNAs instruct
massive mRNA elimination during late spermiogenesis. Cell Res.
25:2662015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Watanabe T, Cheng Ec, Zhong M and Lin H:
Retrotransposons and pseudogenes regulate mRNAs and lncRNAs via the
piRNA pathway in the germline. Genome Res. 25:368–380. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kuramochi-Miyagawa S, Kimura T, Ijiri TW,
Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, et
al: Mili, a mammalian member of piwi family gene, is essential for
spermatogenesis. Development. 131:839–849. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhao S, Gou LT, Zhang M, Zu LD, Hua MM,
Hua Y, Shi HJ, Li Y, Li J, Li D, et al: piRNA-triggered MIWI
ubiquitination and removal by APC/C in late spermatogenesis. Dev
Cell. 24:13–25. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rouget C, Papin C, Boureux A, Meunier AC,
Franco B, Robine N, Lai EC, Pelisson A and Simonelig M: Maternal
mRNA deadenylation and decay by the piRNA pathway in the early
Drosophila embryo. Nature. 467:1128–1132. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sharma AK, Nelson MC, Brandt JE, Wessman
M, Mahmud N, Weller KP and Hoffman R: Human CD34(+) stem cells
express the hiwi gene, a human homologue of the drosophila gene
piwi. Blood. 97:426–434. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Reddien PW, Oviedo NJ, Jennings JR, Jenkin
JC and Alvarado AS: SMEDWI-2 is a PIWI-like protein that regulates
planarian stem cells. Science. 310:1327–1330. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu Y: MicroRNAs and PIWI-interacting RNAs
in oncology. Oncol Lett. 12:2289–2292. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Law PT, Qin H, Ching AK, Lai KP, Co NN, He
M, Lung RW, Chan AW, Chan TF and Wong N: Deep sequencing of small
RNA transcriptome reveals novel non-coding RNAs in hepatocellular
carcinoma. J Hepatol. 58:1165–1173. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lee JH, Schütte D, Wulf G, Füzesi L,
Radzun HJ, Schweyer S, Engel W and Nayernia K: Stem-cell protein
Piwil2 is widely expressed in tumors and inhibits apoptosis through
activation of Stat3/Bcl-XL pathway. Hum Mol Genet. 15:201–211.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cheng J, Deng H, Xiao B, Zhou H, Zhou F,
Shen Z and Guo J: piR-823, a novel non-coding small RNA,
demonstrates in vitro and in vivo tumor suppressive activity in
human gastric cancer cells. Cancer Lett. 315:12–17. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Miao PZ, Yang Y, Chen EB, Zhu GQ, Wang B
and Dai Z: Differential expressions analysis of piwi-interacting
RNAs in hepatocellular carcinoma. Zhonghua Gan Zang Bing Za Zhi.
26:842–846. 2018.PubMed/NCBI
|
|
35
|
Tang X, Xie X, Wang X, Wang Y, Jiang X and
Jiang H: The combination of piR-823 and eukaryotic initiation
factor 3 B (EIF3B) activates hepatic stellate cells via
upregulating TGF-β1 in liver fibrogenesis. Med Sci Monit.
24:9151–9165. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cheng J, Guo JM, Xiao BX, Miao Y, Jiang Z,
Zhou H and Li QN: piRNA, the new non-coding RNA, is aberrantly
expressed in human cancer cells. Clin Chim Acta. 412:1621–1625.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zeng G, Zhang D, Liu X, Kang Q, Fu Y, Tang
B, Guo W, Zhang Y, Wei G and He D: Co-expression of Piwil2/Piwil4
in nucleus indicates poor prognosis of hepatocellular carcinoma.
Oncotarget. 8:4607–4617. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Rizzo F, Rinaldi A, Marchese G, Coviello
E, Sellitto A, Cordella A, Giurato G, Nassa G, Ravo M, Tarallo R,
et al: Specific patterns of PIWI-interacting small noncoding RNA
expression in dysplastic liver nodules and hepatocellular
carcinoma. Oncotarget. 7:54650–54661. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Martinez VD, Enfield KSS, Rowbotham DA and
Lam WL: An atlas of gastric PIWI-interacting RNA transcriptomes and
their utility for identifying signatures of gastric cancer
recurrence. Gastric Cancer. 19:660–665. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lin X, Xia Y, Hu D, Mao Q, Yu Z, Zhang H,
Li C, Chen G, Liu F, Zhu W, et al: Transcriptomewide piRNA
profiling in human gastric cancer. Oncol Rep. 41:3089–3099.
2019.PubMed/NCBI
|
|
42
|
Wang DW, Wang ZH, Wang LL, Song Y and
Zhang GZ: Overexpression of hiwi promotes growth of human breast
cancer cells. Asian Pac J Cancer Prev. 15:7553–7558. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Cui L, Lou Y, Zhang X, Zhou H, Deng H,
Song H, Yu X, Xiao B, Wang W and Guo J: Detection of circulating
tumor cells in peripheral blood from patients with gastric cancer
using piRNAs as markers. Clin Biochem. 44:1050–1057. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ge L, Zhang N, Li D, Wu Y, Wang H and Wang
J: Circulating exosomal small RNAs are promising non-invasive
diagnostic biomarkers for gastric cancer. J Cell Mol Med.
24:14502–14513. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Jing Z, Xi Y, Yin J and Shuwen H:
Biological roles of piRNAs in colorectal cancer. Gene.
769:1450632021. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Sarraf JS, Puty TC, da Silva EM, Allen
TSR, Sarraf YS, de Carvalho LEW, Adami F and de Oliveira EHC:
Noncoding RNAs and colorectal cancer: A general overview. Microrna.
9:336–345. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yin J, Jiang XY, Qi W, Ji CG, Xie XL,
Zhang DX, Cui ZJ, Wang CK, Bai Y, Wang J and Jiang HQ: piR-823
contributes to colorectal tumorigenesis by enhancing the
transcriptional activity of HSF1. Cancer Sci. 108:1746–1756. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Weng W, Liu N, Toiyama Y, Kusunoki M,
Nagasaka T, Fujiwara T, Wei Q, Qin H, Lin H, Ma Y and Goel A: Novel
evidence for a PIWI-interacting RNA (piRNA) as an oncogenic
mediator of disease progression, and a potential prognostic
biomarker in colorectal cancer. Mol Cancer. 17:162018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Chu H, Xia L, Qiu X, Gu D, Zhu L, Jin J,
Hui G, Hua Q, Du M, Tong N, et al: Genetic variants in noncoding
PIWI-interacting RNA and colorectal cancer risk. Cancer.
121:2044–2052. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Qu A, Wang W, Yang Y, Zhang X, Dong Y,
Zheng G, Wu Q, Zou M, Du L, Wang Y and Wang C: A serum piRNA
signature as promising non-invasive diagnostic and prognostic
biomarkers for colorectal cancer. Cancer Manag Res. 11:3703–3720.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Mai D, Ding P, Tan L, Zhang J, Pan Z, Bai
R, Li C, Li M, Zhou Y, Tan W, et al: PIWI-interacting RNA-54265 is
oncogenic and a potential therapeutic target in colorectal
adenocarcinoma. Theranostics. 8:5213–5230. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Mai D, Zheng Y, Guo H, Ding P, Bai R, Li
M, Ye Y, Zhang J, Huang X, Liu D, et al: Serum piRNA-54265 is a new
biomarker for early detection and clinical surveillance of human
colorectal cancer. Theranostics. 10:8468–8478. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Yin J, Qi W, Ji CG, Zhang DX, Xie XL, Ding
Q, Jiang XY, Han J and Jiang HQ: Small RNA sequencing revealed
aberrant piRNA expression profiles in colorectal cancer. Oncol Rep.
42:263–272. 2019.PubMed/NCBI
|
|
54
|
Vychytilova-Faltejskova P, Stitkovcova K,
Radova L, Sachlova M, Kosarova Z, Slaba K, Kala Z, Svoboda M, Kiss
I, Vyzula R, et al: Circulating PIWI-interacting RNAs piR-5937 and
piR-28876 are promising diagnostic biomarkers of colon cancer.
Cancer Epidemiol Biomarkers Prev. 27:1019–1028. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wang Z, Yang H, Ma D, Mu Y, Tan X, Hao Q,
Feng L, Liang J, Xin W, Chen Y, et al: Serum PIWI-interacting RNAs
piR-020619 and piR-020450 are promising novel biomarkers for early
detection of colorectal cancer. Cancer Epidemiol Biomarkers Prev.
29:990–998. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Litwin M, Dubis J, Arczyńska K, Piotrowska
A, Frydlewicz A, Karczewski M, Dzięgiel P and Witkiewicz W:
Correlation of HIWI and HILI expression with cancer stem cell
markers in colorectal cancer. Anticancer Res. 35:3317–3324.
2015.PubMed/NCBI
|
|
57
|
Wu J, Sun H, Li J, Guo Y, Zhang K, Lang C,
Zou C and Ma H: Increased survival of patients aged 0–29 years with
osteosarcoma: A period analysis, 1984–2013. Cancer Med.
7:3652–3661. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Vijayamurugan N and Bakhshi S: Review of
management issues in relapsed osteosarcoma. Expert Rev Anticancer
Ther. 14:151–161. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Das B, Jain N and Mallick B: piR-39980
promotes cell proliferation, migration and invasion, and inhibits
apoptosis via repression of SERPINB1 in human osteosarcoma. Biol
Cell. 112:73–91. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Nogueira Jorge NA, Wajnberg G, Ferreira
CG, de Sa Carvalho B and Passetti F: snoRNA and piRNA expression
levels modified by tobacco use in women with lung adenocarcinoma.
PLoS One. 12:e01834102017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Peng L, Song L, Liu C, Lv X, Li X, Jie J,
Zhao D and Li D: piR-55490 inhibits the growth of lung carcinoma by
suppressing mTOR signaling. Tumour Biol. 37:2749–2756. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Dammann R, Li C, Yoon JH, Chin PL, Bates S
and Pfeifer GP: Epigenetic inactivation of a RAS association domain
family protein from the lung tumour suppressor locus 3p21.3. Nat
Genet. 25:315–319. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
63
|
Reeves ME, Firek M, Chen ST and Amaar Y:
The RASSF1 gene and the opposing effects of the RASSF1A and RASSF1C
isoforms on cell proliferation and apoptosis. Mol Biol Int.
2013:1450962013. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Reeves ME, Firek M, Jliedi A and Amaar YG:
Identification and characterization of RASSF1C piRNA target genes
in lung cancer cells. Oncotarget. 8:34268–34282. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Reeves ME, Baldwin ML, Aragon R, Baldwin
S, Chen ST, Li X, Mohan S and Amaar YG: RASSF1C modulates the
expression of a stem cell renewal gene, PIWIL1. BMC Res Notes.
5:2392012. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Xie K, Zhang K, Kong J, Wang C, Gu Y,
Liang C, Jiang T, Qin N, Liu J, Guo X, et al: Cancer-testis gene
PIWIL1 promotes cell proliferation, migration, and invasion in lung
adenocarcinoma. Cancer Med. 7:157–166. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Yao J, Wang YW, Fang BB, Zhang SJ and
Cheng BL: piR-651 and its function in 95-D lung cancer cells.
Biomed Rep. 4:546–550. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Li D, Luo Y, Gao Y and Yang Y, Wang Y, Xu
Y, Tan S, Zhang Y, Duan J and Yang Y: piR-651 promotes tumor
formation in non-small cell lung carcinoma through the upregulation
of cyclin D1 and CDK4. Int J Mol Med. 38:927–936. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Zhang SJ, Yao J, Shen BZ, Li GB, Kong SS,
Bi DD, Pan SH and Cheng BL: Role of piwi-interacting RNA-651 in the
carcinogenesis of non-small cell lung cancer. Oncol Lett.
15:940–946. 2018.PubMed/NCBI
|
|
70
|
Mei Y, Wang Y, Kumari P, Shetty AC, Clark
D, Gable T, MacKerell AD, Ma MZ, Weber DJ, Yang AJ, et al: A
piRNA-like small RNA interacts with and modulates p-ERM proteins in
human somatic cells. Nat Commun. 6:73162015. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Guo B, Li D, Du L and Zhu X: piRNAs:
Biogenesis and their potential roles in cancer. Cancer Metastasis
Rev. 39:567–575. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Huang G, Hu H, Xue X, Shen S, Gao E, Guo
G, Shen X and Zhang X: Altered expression of piRNAs and their
relation with clinicopathologic features of breast cancer. Clin
Transl Oncol. 15:563–568. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Hashim A, Rizzo F, Marchese G, Ravo M,
Tarallo R, Nassa G, Giurato G, Santamaria G, Cordella A, Cantarella
C and Weisz A: RNA sequencing identifies specific PIWI-interacting
small non-coding RNA expression patterns in breast cancer.
Oncotarget. 5:9901–9910. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Öner C, Öner DT and Çolak E: Estrogen and
androgen hormone levels modulate the expression of PIWI interacting
RNA in prostate and breast cancer. PLoS One. 11:e01590442016.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Fu A, Jacobs DI, Hoffman AE, Zheng T and
Zhu Y: PIWI-interacting RNA 021285 is involved in breast
tumorigenesis possibly by remodeling the cancer epigenome.
Carcinogenesis. 36:1094–1102. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Dana PM, Mansournia MA and Mirhashemi SM:
PIWI-interacting RNAs: New biomarkers for diagnosis and treatment
of breast cancer. Cell Biosci. 10:442020. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Tan L, Mai D, Zhang B, Jiang X, Zhang J,
Bai R, Ye Y, Li M, Pan L, Su J, et al: PIWI-interacting RNA-36712
restrains breast cancer progression and chemoresistance by
interaction with SEPW1 pseudogene SEPW1P RNA. Mol Cancer. 18:92019.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Krishnan P, Ghosh S, Graham K, Mackey JR,
Kovalchuk O and Damaraju S: Piwi-interacting RNAs and PIWI genes as
novel prognostic markers for breast cancer. Oncotarget.
7:37944–37956. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Heng ZSL, Lee JY, Subhramanyam CS, Wang C,
Thanga LZ and Hu Q: The role of 17β-estradiol-induced upregulation
of piwi-like 4 in modulating gene expression and motility in breast
cancer cells. Oncol Rep. 40:2525–2535. 2018.PubMed/NCBI
|
|
80
|
Lee JH, Jung C, Javadian-Elyaderani P,
Schweyer S, Schütte D, Shoukier M, Karimi-Busheri F, Weinfeld M,
Rasouli-Nia A, Hengstler JG, et al: Pathways of proliferation and
antiapoptosis driven in breast cancer stem cells by stem cell
protein piwil2. Cancer Res. 70:4569–4579. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Zhang H, Ren Y, Xu H, Pang D, Duan C and
Liu C: The expression of stem cell protein Piwil2 and piR-932 in
breast cancer. Surg Oncol. 22:217–223. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Zuo Y, Liang Y, Zhang J, Hao Y, Li M, Wen
Z and Zhao Y: Transcriptome analysis identifies piwi-interacting
RNAs as prognostic markers for recurrence of prostate cancer. Front
Genet. 10:10182019. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Zhang L, Meng X, Li D and Han X:
piR-001773 and piR-017184 promote prostate cancer progression by
interacting with PCDH9. Cell Signal. 76:1097802020. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Chu H, Hui G, Yuan L, Shi D, Wang Y, Du M,
Zhong D, Ma L, Tong N, Qin C, et al: Identification of novel piRNAs
in bladder cancer. Cancer Lett. 356:561–567. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Ravo M, Cordella A, Rinaldi A, Bruno G,
Alexandrova E, Saggese P, Nassa G, Giurato G, Tarallo R, Marchese
G, et al: Small non-coding RNA deregulation in endometrial
carcinogenesis. Oncotarget. 6:4677–4691. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Li F, Yuan P, Rao M, Jin CH, Tang W, Rong
YF, Hu YP, Zhang F, Wei T, Yin Q, et al: piRNA-independent function
of PIWIL1 as a co-activator for anaphase promoting
complex/cyclosome to drive pancreatic cancer metastasis. Nat Cell
Biol. 22:425–438. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Lim SL, Ricciardelli C, Oehler MK, Tan IM,
Russell D and Grützner F: Overexpression of piRNA pathway genes in
epithelial ovarian cancer. PLoS One. 9:e996872014. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Capitanio U, Bensalah K, Bex A, Boorjian
SA, Bray F, Coleman J, Gore JL, Sun M, Wood C and Russo P:
Epidemiology of renal cell carcinoma. Eur Urol. 75:74–84. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Owens B: Kidney cancer. Nature.
537:S972016. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Busch J, Ralla B, Jung M, Wotschofsky Z,
Trujillo-Arribas E, Schwabe P, Kilic E, Fendler A and Jung K:
Piwi-interacting RNAs as novel prognostic markers in clear cell
renal cell carcinomas. J Exp Clin Cancer Res. 34:612015. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Zhao C, Tolkach Y, Schmidt D, Toma M,
Muders MH, Kristiansen G, Müller SC and Ellinger J: Mitochondrial
PIWI-interacting RNAs are novel biomarkers for clear cell renal
cell carcinoma. World J Urol. 37:1639–1647. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Li Y, Wu X, Gao H, Jin JM, Li AX, Kim YS,
Pal SK, Nelson RA, Lau CM, Guo C, et al: Piwi-interacting RNAs
(piRNAs) are dysregulated in renal cell carcinoma and associated
with tumor metastasis and cancer-specific survival. Mol Med.
21:381–388. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Iliev R, Fedorko M, Machackova T,
Mlcochova H, Svoboda M, Pacik D, Dolezel J, Stanik M and Slaby O:
Expression levels of PIWI-interacting RNA, piR-823, are deregulated
in tumor tissue, blood serum and urine of patients with renal cell
carcinoma. Anticancer Res. 36:6419–6423. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Iliev R, Stanik M, Fedorko M, Poprach A,
Vychytilova-Faltejskova P, Slaba K, Svoboda M, Fabian P, Pacik D,
Dolezel J and Slaby O: Decreased expression levels of PIWIL1,
PIWIL2, and PIWIL4 are associated with worse survival in renal cell
carcinoma patients. Onco Targets Ther. 9:217–222. 2016.PubMed/NCBI
|
|
96
|
Stöhr CG, Steffens S, Polifka I, Jung R,
Kahlmeyer A, Ivanyi P, Weber F, Hartmann A, Wullich B, Wach S and
Taubert H: Piwi-like 1 protein expression is a prognostic factor
for renal cell carcinoma patients. Sci Rep. 9:17412019. View Article : Google Scholar : PubMed/NCBI
|