Introduction
Acute myeloid leukemia (AML), a heterogeneous
disease of, but not restricted to, the bone marrow is characterized
by abnormal clonal augmentation of immature myoblasts and presents
with a poor prognosis due to rapid progress (1,2). AML
accounts for more than a half of leukemia-associated mortality,
with a 5-year survival rate of ~27% (3). Especially, the elderly are more
sensitive to the development of AML; most cases present in
individuals aged >65 years, which adds the difficulty of
management due to relatively poor physical condition (4). Although novel therapies have been
introduced for AML treatment over past decades, limited progress
has been made in enhancing patient survival profile (5,6).
Hence, to develop novel treatment options for AML, its associated
molecule mechanisms, such as driver genes and pathways, have been
extensively studied (7,8).
Kinesins are a class of motor proteins that are key
for intracellular transport of multiple materials, such as
adenovirus, and participate in almost all important cellular
functions (9–11). Kinesin family member 2A (KIF2A), a member of the
Kinesin-13 family, regulates mitosis via supporting the formation
of the bipolar spindle (12).
KIF2A is promising target in the exploration of cancer pathology,
as indicated by reports of its function and potential clinical
utility in multiple types of cancer, including nasopharyngeal
carcinoma, non-small cell lung cancer and AML (13–15). Previously,
KIF2A has been shown to be correlated with clinical features of
patients with AML and regulate AML cell proliferation and
apoptosis, indicating that KIF2A is involved in the regulation of
AML pathology (15). However, the
mechanistic role of KIF2A in AML has not been fully identified.
The present study aimed to detect the effect of
KIF2A on AML cell survival and chemosensitivity, as well as the
pathways by which it exerts these effects.
Materials and methods
Subjects
The present study was approved by the Institutional
Review Board of First Affiliated Hospital of Anhui Medical
University. A total of 58 newly diagnosed patients with AML and 30
healthy subjects (56.7% males and 43.3% females) with a mean age of
53.2±6.6 years (range, 33 to 79 years) from First Affiliated
Hospital of Anhui Medical University (Anhui, China) were enrolled
from January 2015 to December 2018. All patients were newly
diagnosed with AML rather than acute promyelocytic leukemia and
were aged >18 years, without a history of other hematological
malignancy or cancer or radiotherapy or chemotherapy before
enrollment. The healthy subjects were all aged >18 years,
recruited during scheduled bone marrow donation and blood routine
indexes, tumor markers and blood biochemistry tests were normal.
All subjects provided written informed consent.
Data and sample collection
After recording the clinical characteristics of
patients with AML (Table I), bone
marrow samples were collected before starting induction therapy.
Bone marrow samples were also collected from healthy donors.
Immediately following collection, bone marrow mononuclear cells
(BMMCs) were isolated from bone marrow by gradient centrifugation
(at 600 × g for 15 min at 25°C) using Percoll (Sigma-Aldrich; Merck
KGaA), followed by reverse transcription quantitative (RT-q)PCR
assay for determination of KIF2A expression levels. For patients
with AML, remission status was documented following completion of
the response evaluation for induction therapy. Clinical follow-up
was conducted until December 31, 2019. Event-free survival (EFS)
and overall survival (OS) were estimated for analysis of survival
prognosis; in addition, patients with AML were divided into high-
and low KIF2A expression groups according to the median value of
KIF2A expression (2.638).
 | Table I.Clinical features of patients with
AML. |
Table I.
Clinical features of patients with
AML.
| Characteristic | Patients
(n=58) |
|---|
| Age, years |
|
| Mean ±
SD | 57.9±11.6 |
| Median
(IQR) | 60.0
(48.8-68.0) |
| Sex (%) |
|
|
Male | 34.0 (58.6) |
|
Female | 24.0 (41.4) |
| WBC
(×109/l) |
|
| Mean ±
SD | 17.2±16.1 |
| Median
(IQR) | 11.6
(4.6-25.1) |
| BM blasts, % |
|
| Mean ±
SD | 73.8±13.5 |
| Median
(IQR) | 75.0
(61.8-85.0) |
| FAB classification
(%) |
|
| M1 | 3.0 (5.2) |
| M2 | 16.0 (27.6) |
| M4 | 17.0 (29.3) |
| M5 | 22.0 (37.9) |
| Cytogenetic
abnormality (%) |
|
| NK | 29.0 (50.0) |
| CK | 7.0 (12.1) |
|
inv(16) or t(16;16) | 4.0 (6.9) |
| MK | 2.0 (3.4) |
| +8 | 2.0 (3.4) |
|
t(9;11) | 2.0 (3.4) |
| -7 or
7q- | 1.0 (1.7) |
| -5 or
5q- | 1.0 (1.7) |
|
t(8;21) | 1.0 (1.7) |
| Other
non-defined | 11.0 (19.0) |
| Molecular
abnormality (%) |
|
| Mutated
NPM1 | 17.0 (29.3) |
| Mutated
FLT3-ITD | 15.0 (25.9) |
| Mutated
WT1 | 7.0 (12.1) |
| Mutated
CEBPA | 6.0 (10.3) |
| Risk classification
(%) |
|
|
Favorable | 11.0 (19.0) |
|
Intermediate | 31.0 (53.4) |
|
Poor | 16.0 (27.6) |
Cell line culture and KIF2A expression
detection
Human AML cell lines, including KG-1 [American Type
Culture Collection (ATCC)], Kasumi-1 (ATCC), MOLM-13 [Deutsche
Sammlung von Mikroorganismen und Zellkulturen (DSMZ)] and OC-AML2
(DSMZ), were cultured with 90% RPMI-1640 supplemented with 10%
fetal bovine serum (FBS; both Hyclone; Cytiva) according to the
manufacturer's instructions. The cells were cultured in 5%
CO2 at 37°C. The expression of KIF2A in AML cell lines
was determined by RT-qPCR and western blotting using BMMCs from
healthy subjects as the control.
KIF2A small interfering RNA (siRNA)
transfection
The transfection of 50 nM negative control (NC) or
KIF2A siRNA (both Shanghai GenePharma Co., Ltd.) into AML cells
(KG-1 and Kasumi-1) was performed using HillyMax reagent (Dojindo
Molecular Technologies, Inc.) at 37°C for 6 h. Untreated cells were
labeled as Untreated cells. At 48 h after transfection, RT-qPCR,
western blotting and Annexin V/propidium iodide (AV/PI) and
chemosensitivity assay were performed. At 0, 24, 48 and 72 h after
transfection, Cell Counting Kit-8 (CCK-8) assay was performed. The
target sequence of KIF2A siRNA was 5′-GGCAAAGAGAUUGACCUGG-3′ and
the scrambled sequence used for NC siRNA was
5′-AAGAACAACACAAAAGAACAG-3′.
740 Y-P incubation
The concentration of 740 Y-P (Selleck Chemicals)
incubated with AML cells (KG-1 and Kasumi-1) was 25 µg/ml, and the
cells were incubated for 48 h at 37°C. The transfection of NC or
KIF2A siRNA was performed as aforementioned at the same time as 740
Y-P incubation. Cells were divided into five groups as follows: i)
Untreated cells; ii) NC siRNA, cells transfected with NC siRNA;
iii) KIF2A siRNA, cells transfected with KIF2A siRNA; iv) NC siRNA
+ 740 Y-P, cells were transfected with NC siRNA and incubated with
740 Y-P and v) KIF2A siRNA + 740 Y-P, cells were transfected with
KIF2A siRNA and incubated with 740 Y-P. Following incubation,
RT-qPCR, western blotting, CCK-8 and AV/PI and chemosensitivity
assay were performed.
Ras homolog family member A (RhoA)
plasmid transfection
The sequence of RhoA was obtained from the National
Center of Biotechnology Information (reference no. NM_001664.4).
The sequence used for NC was 5′-AACACCGAACGAGACAGGATT-3′. NC or
RhoA overexpression plasmids were constructed using pcDNA3.1 vector
(Shanghai GenePharma Co., Ltd.). Then, 50 nM NC or RhoA
overexpression plasmid was co-transfected with 50 nM NC or KIF2A
siRNA into AML cells (KG-1 and Kasumi-1) using HilyMax (Dojindo
Molecular Technologies, Inc.) at 37°C for 6 h. Cells were divided
into groups as follows: i) Untreated cells; ii) NC siRNA + NC
plasmid, cells transfected with NC siRNA and NC overexpression
plasmid; iii) KIF2A siRNA + NC plasmid, cells transfected with
KIF2A siRNA and NC overexpression plasmid; iv) NC siRNA + RhoA
plasmid, cells transfected with NC siRNA and RhoA overexpression
plasmid and v) KIF2A siRNA + RhoA plasmid, cells transfected with
KIF2A siRNA and RhoA overexpression plasmid. RT-qPCR, western
blotting and CCK-8, AV/PI and chemosensitivity assay were performed
48 h after transfection.
CCK-8 assay
CCK-8 (Sigma-Aldrich; Merck KGaA) assay was
performed. The KG-1and Kasumi-1 cells (3×103) were
plated, and 10 µl reagent was added for 3 h. Then, the optical
density value was read using a microplate reader (BioTek
Instruments, Inc.) at 450 nm.
AV/PI assay
AV/PI assay was performed using an Annexin V
Apoptosis Detection kit I (BD Biosciences). The KG-1and Kasumi-1
cells (1×105; 100 µl) were stained with AV (5 µl) and PI
(5 µl) for 15 min at room temperature after being digested. A
FACSCalibur flow cytometer (BD Biosciences) was used to analyze the
cells. The data was analyzed by FlowJo 7.6 (BD Biosciences).
RT-qPCR
The extraction of total RNA from BMMCs from healthy
subjects was performed using RNeasy Mini kit (Qiagen GmbH) in
accordance with the manufacturer's protocol. The complementary DNA
was generated using qPCR RT Master Mix (Toyobo Life Science) and 1
µg RNA. Realtime PCR Master Mix (Toyobo Life Science) was used to
perform qPCR. The thermocycling conditions were as follows: 95°C
for 60 sec, followed by 40 cycles of 95°C for 15 sec and 61°C for
30 sec. Relative expression was calculated via the
2−ΔΔCq method (16).
The primers for KIF2A and β-actin (internal reference) were as
follows: KIF2A forward, 5′-GCCGAATACATCAAGCAAT-3′ and reverse,
5′-CTCTCCAGGTCAATCTCTT-3′ and β-actin forward,
5′-TCGTGCGTGACATTAAGGAGAAG-3′ and reverse,
5′-AGGAAGGAAGGCTGGAAGAGTG-3′.
Western blot analysis
RIPA lysis buffer (Beyotime Institute of
Biotechnology) was used to isolate total protein from AML cells and
BMMCs from healthy subjects. The quantification of total protein
was performed via BCA Protein Assay kit (Thermo Fisher Scientific,
Inc.). Then, the total protein (20 µg) was separated by 4–20%
SDS-PAGE (Beyotime Institute of Biotechnology) and transferred to
nitrocellulose membranes (Pall Life Sciences). The membranes were
then blocked with 5% BSA (Beyotime, China) at 37°C for 1 h.
Afterwards, the diluted primary antibodies (4°C, overnight) and
secondary antibody (37°C, 1 h) were then incubated with the
membrane. Protein bands were visualized using ECL western blotting
substrate (Thermo Fisher Scientific, Inc.) and quantified with
ImageJ V 1.52v (National Institutes of Health). The antibodies are
listed in Table SI.
Chemosensitivity assay
Following incubation as aforementioned, the KG-1and
Kasumi-1 cells (5×103) were treated with different
concentrations of Adriamycin (ADR; 0.0, 0.1, 0.2, 0.4, 0.8 and 1.6
µM) (Selleck Chemicals) or Arabinofuranosyl Cytidine (AraC; 0, 10,
20, 40, 80 and 160 nM) in a 96-well plate for 48 h at 37°C. Then,
CCK-8 assay was performed as aforementioned.
Statistical analysis
Statistical analysis was performed using GraphPad
Prism 7.02 (GraphPad Software Inc.). Data are presented as the mean
± standard deviation or median and interquartile range. Differences
in expression levels between groups were determined by
Kruskal-Wallis (with post hoc Dunn's multiple comparisons test) or
Mann-Whitney U test. Correlation analysis was performed using
Spearman's test. Receiver operating characteristic (ROC) curve
analysis was used to estimate the value of KIF2A in distinguishing
patients with AML from healthy donors as well as CR from non-CR
AML. EFS and OS are shown as Kaplan-Meier curves and were analyzed
by Log-rank test. Comparation among groups was assessed by one-way
ANOVA, subsequently, the multiple comparisons were assessed by
Dunnett or Tukey's test. A two-side P<0.05 was considered to
indicate a statistically significant difference.
Results
Correlation of KIF2A with risk,
feature and prognosis of AML
A total of patients with 58 AML (58.6% males and
41.4% females) were enrolled, with a mean age of 57.9±11.6 years
(Table I). A total of 30 healthy
donors were also recruited. KIF2A was upregulated in patients with
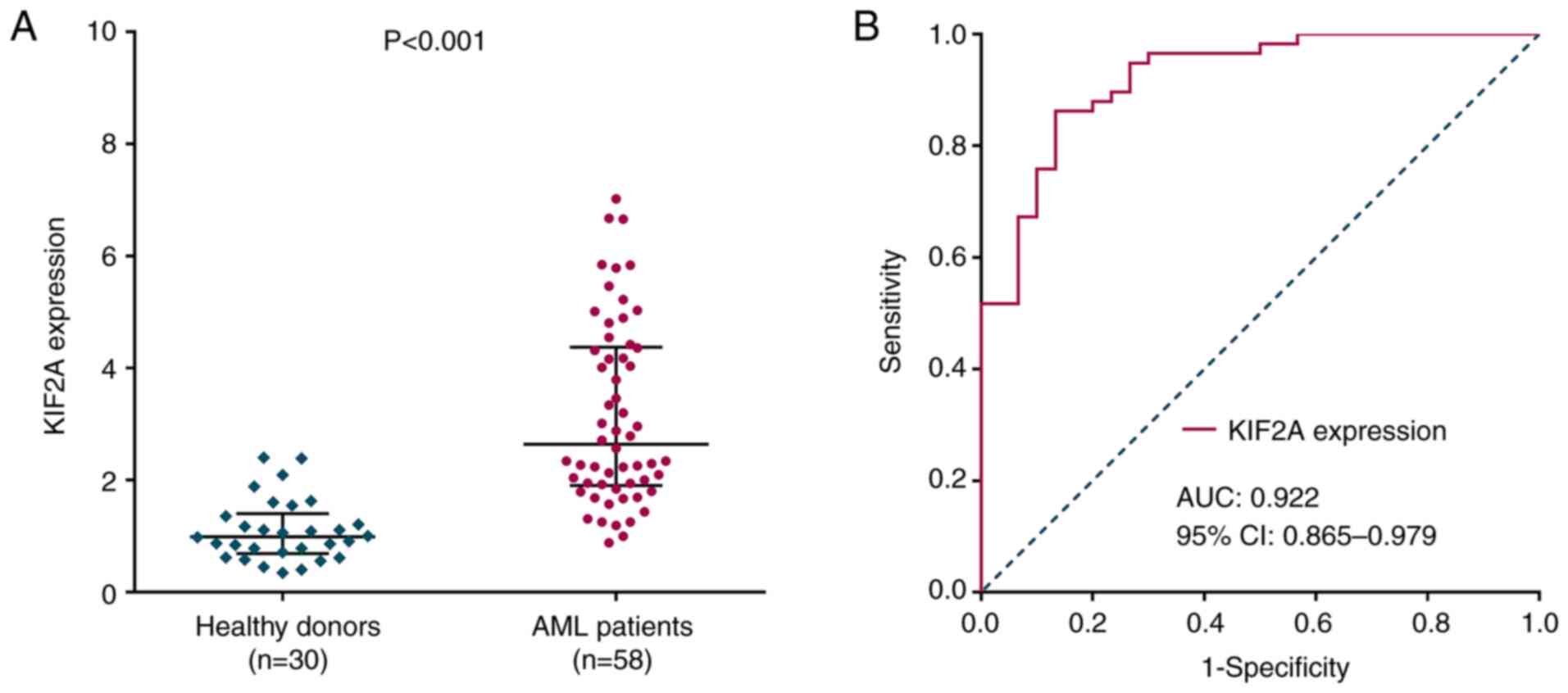
AML compared with healthy donors (P<0.001; Fig. 1A). ROC analysis revealed the
ability of KIF2A to differentiate between patients with AML and
healthy donors, with the area under curve (AUC) of 0.922 (95% CI,
0.865-0.979; Fig. 1B).
Furthermore, KIF2A was correlated with increased bone marrow blasts
(P=0.018; Fig. 2A) and poor
National Comprehensive Cancer Network risk classification (P=0.016;
Fig. 2C) but was not correlated
with FAB classification (P=0.528; Fig.
2B) in patients with AML. In addition, KIF2A was downregulated
in patients with complete response (CR) compared with non-CR
patients (P=0.029; Fig. 2D). ROC
analysis demonstrated that KIF2A differentiated between CR and
non-CR patients, with an AUC of 0.695 (95% CI, 0.529-0.861;
Fig. 2E). EFS (P=0.026; Fig. 2F) exhibited a less favorable
association whereas OS (P=0.092; Fig.
2G) was not significantly different between patients with high
and low KIF2A expression.
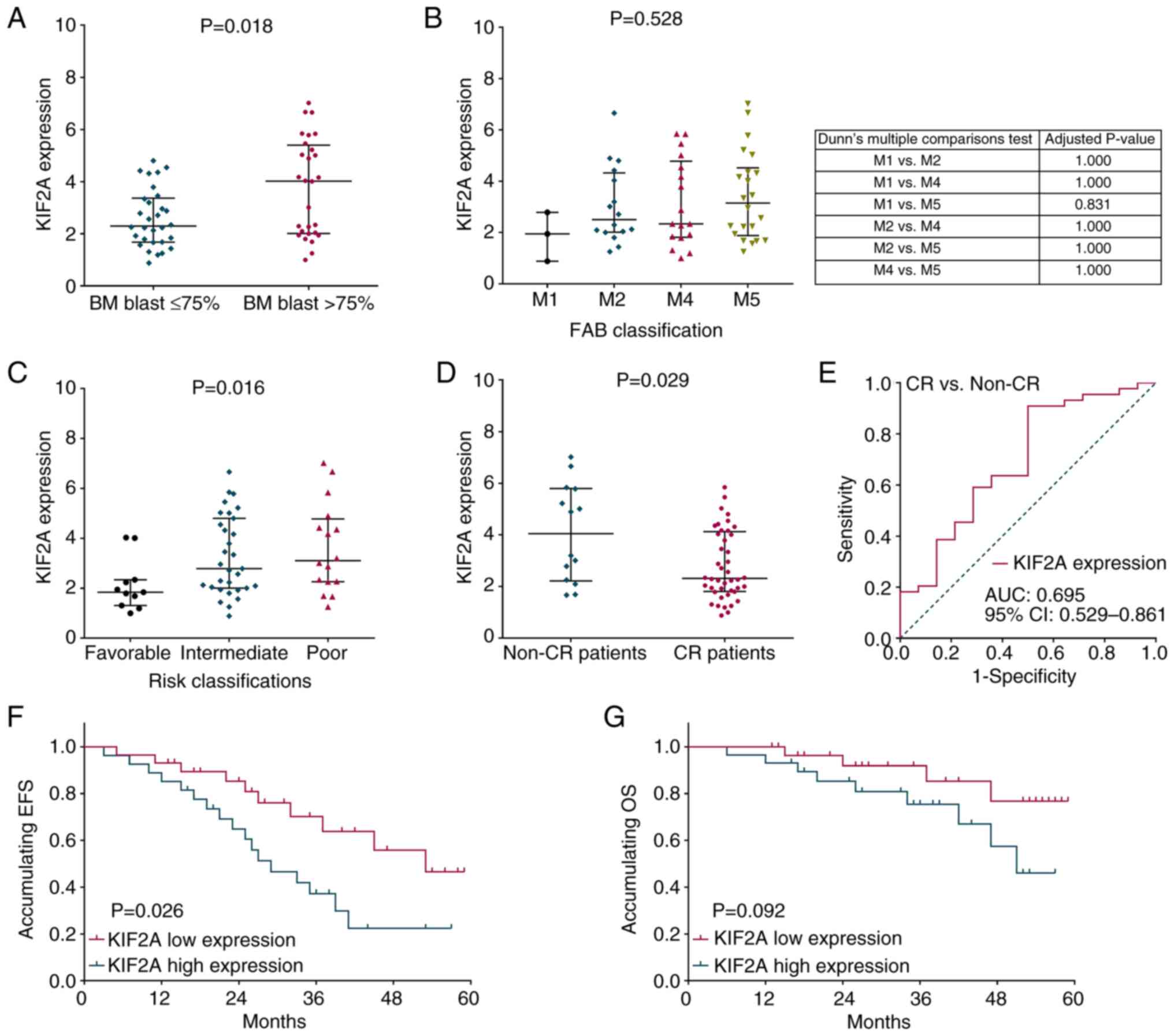 | Figure 2.Correlation of KIF2A expression with
clinical features and efficacy in patients with AML. The
correlation of KIF2A expression with (A) BM blasts, (B) FAB
classification, (C) risk classification and (D) CR. (E) KIF2A
expression can differentiate between patients with and without CR.
Correlation of KIF2A expression with (F) EFS and (G) OS. KIF2A,
kinesin family member 2A; AML, acute myeloid leukemia; BM, bone
marrow; FAB, French-American-British; CR, complete response; EFS,
event-free survival; OS, overall survival; AUC, area under
curve. |
KIF2A expression in AML cell
lines
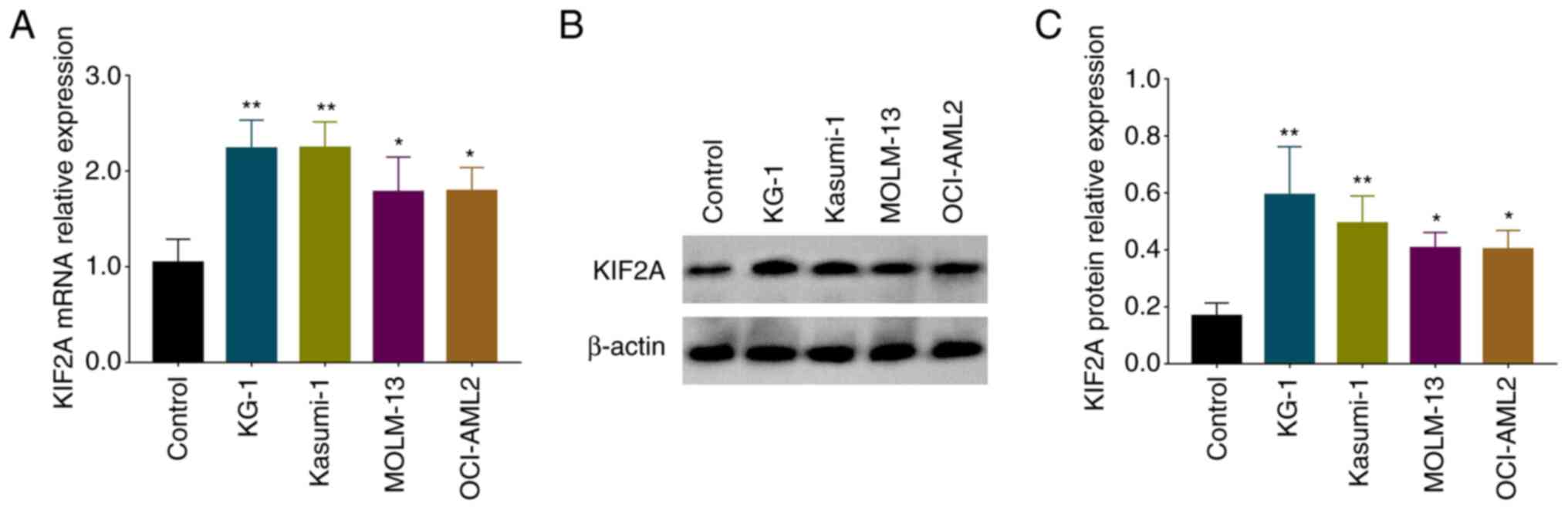
KIF2A mRNA expression levels were increased in AML
cell lines KG-1 (P<0.01), Kasumi-1 (P<0.01), MOLM-13
(P<0.05) and OCI-AML2 (P<0.05) compared with NC cells
(Fig. 3A). Protein expression of
KIF2A was also elevated in KG-1 (P<0.01), Kasumi-1 (P<0.01),
MOLM-13 (P<0.05) and OCI-AML2 (P<0.05) cell lines compared
with NC cells (Fig. 3B and C).
Expression of KIF2A mRNA and protein levels was higher in KG-1 and
Kasumi-1 cell lines, thus, KG-1 and Kasumi-1 cell lines were
selected for subsequent molecular experiments.
Effect of KIF2A inhibition on AML cell
proliferation, apoptosis and chemosensitivity
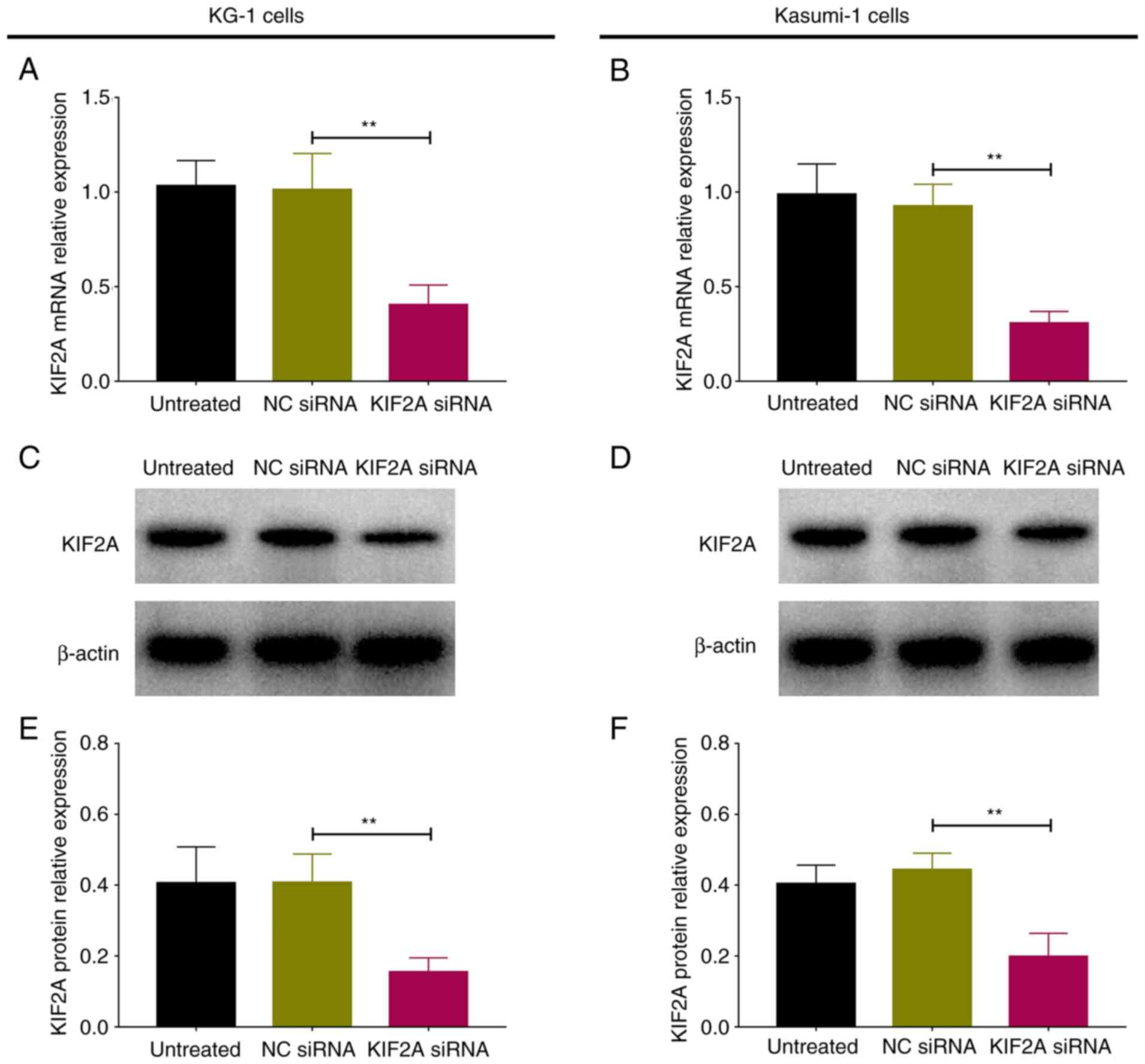
Following transfection, KIF2A mRNA and protein
relative expression levels were downregulated in KIF2A siRNA
compared with NC siRNA group in KG-1 and Kasumi-1 cells (all
P<0.01; Fig. 4). These data
indicated that transfection of KIF2A siRNA was successful in both
cell lines.
Following transfection, in KG-1 cells, KIF2A siRNA
inhibited proliferation but promoted cell apoptosis (both
P<0.01; Fig. 5A-C).
Chemosensitivity to ADR was enhanced by KIF2A siRNA at 0.2
(P<0.05), 0.4 (P<0.01), 0.8 (P<0.05) and 1.6 µM
(P<0.05; Fig. 5D).
Chemosensitivity to AraC was enhanced by KIF2A siRNA at 20
(P<0.05), 40 (P<0.01), 80 (P<0.05) and 160 µM (P<0.05;
Fig. 5E). In Kasumi-1 cells, KIF2A
also decreased cell proliferation (P<0.05; Fig. 5F), but increased cell apoptosis
(P<0.01; Fig. 5G and H). In
addition, chemosensitivity to ADR was promoted by KIF2A siRNA at
0.4 (P<0.01), 0.8 (P<0.05) and 1.6 µM (P<0.05; Fig. 5I); chemosensitivity to AraC was
enhanced by KIF2A siRNA at 20 (P<0.05), 40 (P<0.01), 80
(P<0.01) and 160 µM (P<0.05; Fig. 5J).
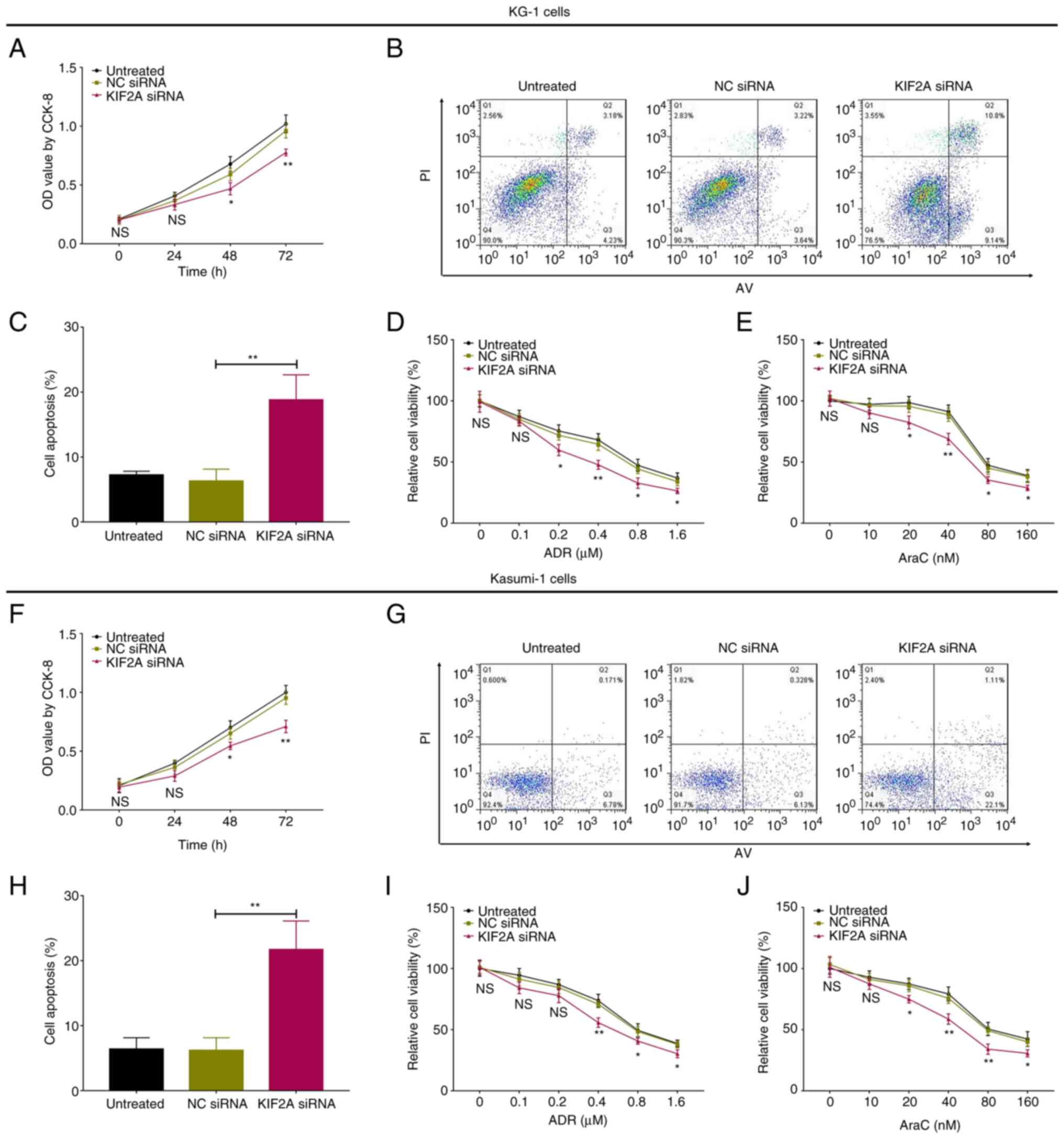 | Figure 5.KIF2A siRNA regulates proliferation,
apoptosis and chemosensitivity of AML cells. Cell (A)
proliferation, (B and C) cell apoptosis and chemosensitivity to (D)
ADR and (E) AraC in KG-1 cells transfected with KIF2A siRNA. Cell
(F) proliferation, (G and H) apoptosis and chemosensitivity to (I)
ADR and (J) AraC in Kasumi-1 cells transfected with KIF2A siRNA.
*P<0.05, **P<0.01 KIF2A vs. NC siRNA. KIF2A, kinesin family
member 2A; AML, acute myeloid leukemia; NC, negative control; ADR,
Adriamycin; AraC, Arabinofuranosyl Cytidine; si, small interfering;
OD, optical density; CCK-8, Cell Counting Kit-8. |
PI3K/AKT and RhoA/rho-associated
coiled-coil containing protein kinase (ROCK) signaling pathways are
inhibited by KIF2A inhibition in AML cells
In KG-1 cells, phosphorylated (p-)PI3K (P<0.01),
p-AKT (P<0.05), RhoA (P<0.05) and ROCK1 (P<0.05) protein
expression levels were inhibited by KIF2A siRNA (Fig. 6A and C). Similarly, in Kasumi-1
cells, KIF2A siRNA suppressed p-PI3K (P<0.01), p-AKT
(P<0.01), RhoA (P<0.01) and ROCK1 (P<0.05) protein
expression levels.
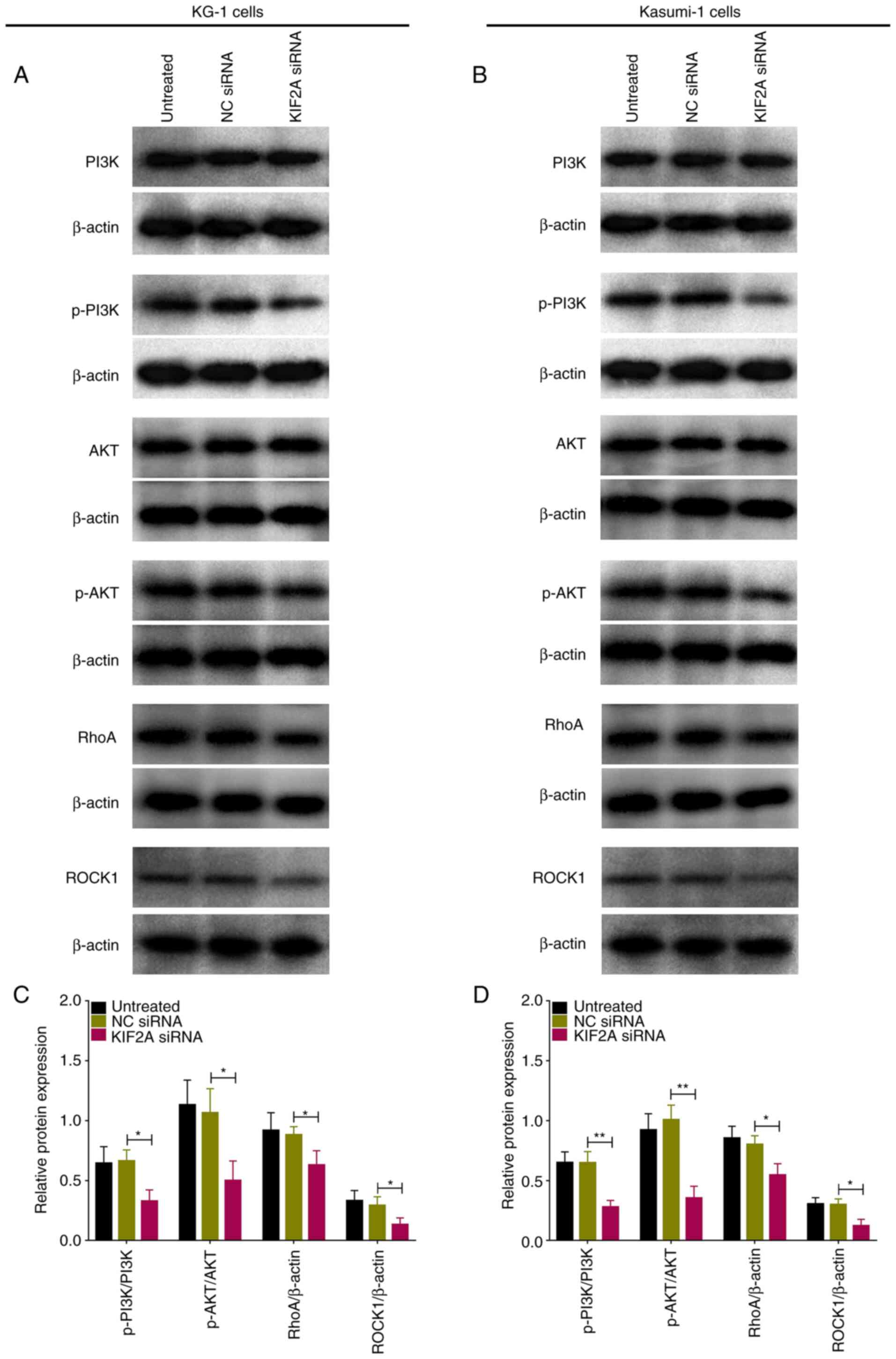 | Figure 6.KIF2A siRNA regulates PI3K/AKT and
RhoA/ROCK pathways in AML cells. Western blot analysis of (A) KG-1
and (B) Kasumi-1 cells following transfection with KIF2A siRNA.
Relative PI3K, p-PI3K, AKT, p-AKT, RhoA and ROCK1 protein
expression levels in (C) KG-1 and (D) Kasumi-1 cells transfected
with KIF2A siRNA. *P<0.05, **P<0.01. KIF2A, kinesin family
member 2A; PI3K, phosphatidylinositol 3-kinase; AKT, protein kinase
B; RhoA, ras homolog family member A; ROCK1, Rho associated
coiled-coil containing protein kinase 1; AML, acute myeloid
leukemia; NC, negative control; si, small interfering; p-,
phosphorylated. |
PI3K/AKT pathway activation regulates
proliferation, apoptosis and chemosensitivity in KIF2A-knockdown
AML cells
In rescue experiments, the PI3K/AKT pathway
activator (740 Y-P) elevated p-PI3K and p-AKT expression levels in
both wild-type (P<0.05 and P<0.01) and KIF2A-knockdown KG-1
cells (both P<0.01; Fig. 7A and
C). Additionally, PI3K/AKT pathway activator also elevated
p-PI3K and p-AKT expression in both wild-type and KIF2A-knockdown
Kasumi-1 cells (all P<0.01; Fig. 7B
and D).
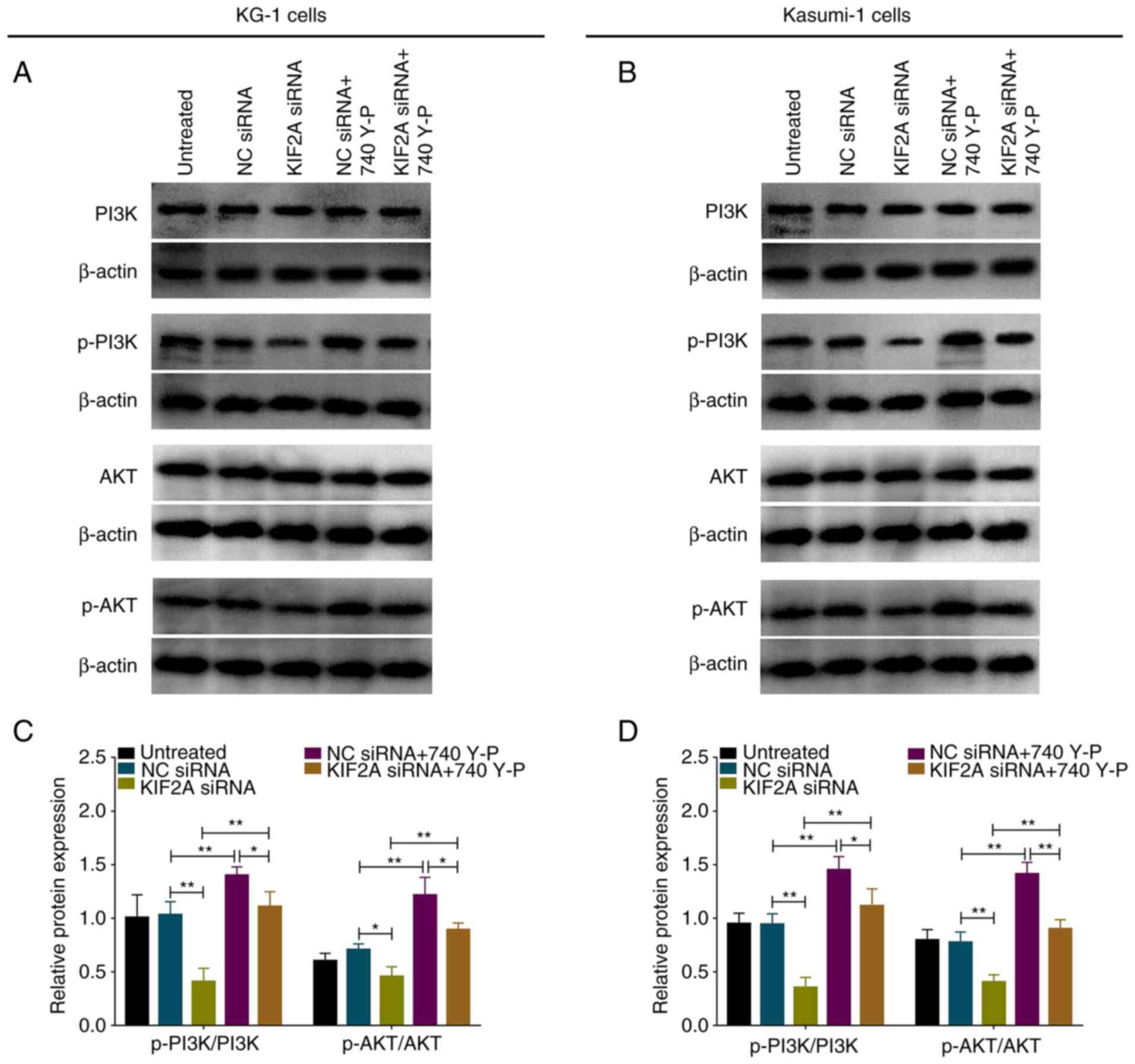 | Figure 7.PI3K/AKT activator treatment in AML
cells. Western blot analysis of (A) KG-1 and (B) Kasumi-1 cells.
Relative PI3K, p-PI3K, AKT and p-AKT protein expression levels in
(C) KG-1 and (D) Kasumi-1 cells (B and D). *P<0.05, **P<0.01.
PI3K, phosphatidylinositol 3-kinase; AKT, protein kinase B; AML,
acute myeloid leukemia; p-, phosphorylated; NC, negative control;
KIF2A, kinesin family member 2A; si, small interfering. |
In KG-1 cells, the PI3K/AKT pathway activator
increased cell proliferation but inhibited cell apoptosis and
chemosensitivity to ADR and AraC (all P<0.05; Fig. 8A-E); furthermore, PI3K/AKT
activation also reversed the effect of KIF2A siRNA on cell
proliferation, apoptosis and chemosensitivity (all P<0.05). In
Kasumi-1 cells, the PI3K/AKT pathway activator exhibited similar
effects as in KG-1 cells (Fig.
8F-J).
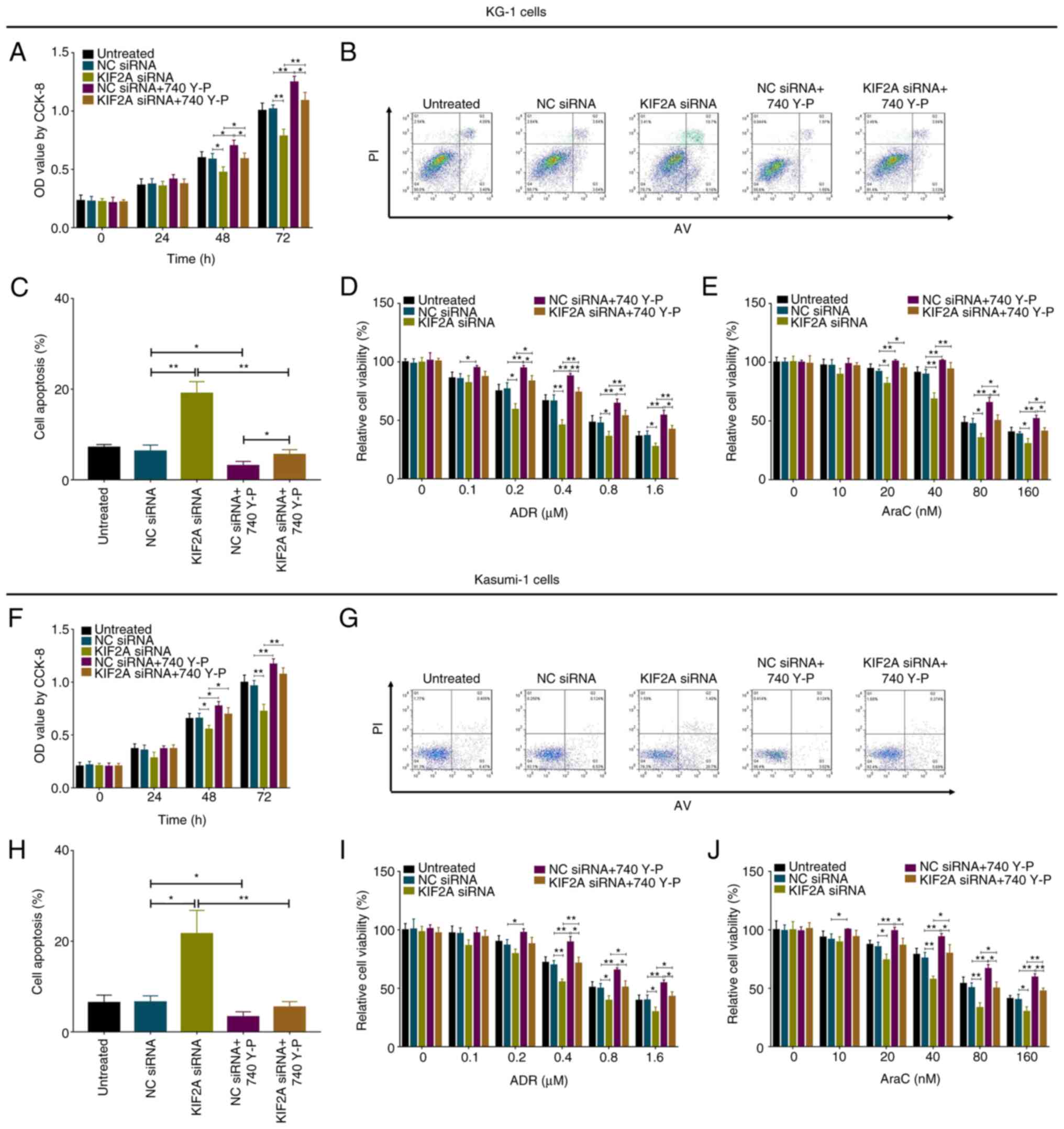 | Figure 8.PI3K/AKT activator rescues the effect
of KIF2A siRNA on AML cells. (A) Proliferation, (B and C) apoptosis
and chemosensitivity to (D) ADR and (E) AraC in KG-1 cells. (F)
Proliferation, (G and H) apoptosis and chemosensitivity to (I) ADR
and (J) AraC in Kasumi-1 cells. *P<0.05, **P<0.01. PI3K,
phosphatidylinositol 3-kinase; AKT, protein kinase B; KIF2A,
kinesin family member 2A; AML, acute myeloid leukemia; NC, negative
control; si, small interfering; OD, optical density; CCK-8, Cell
Counting Kit-8. |
RhoA/ROCK pathway activation regulates
proliferation, apoptosis and chemosensitivity in KIF2A-knockdown
AML cells
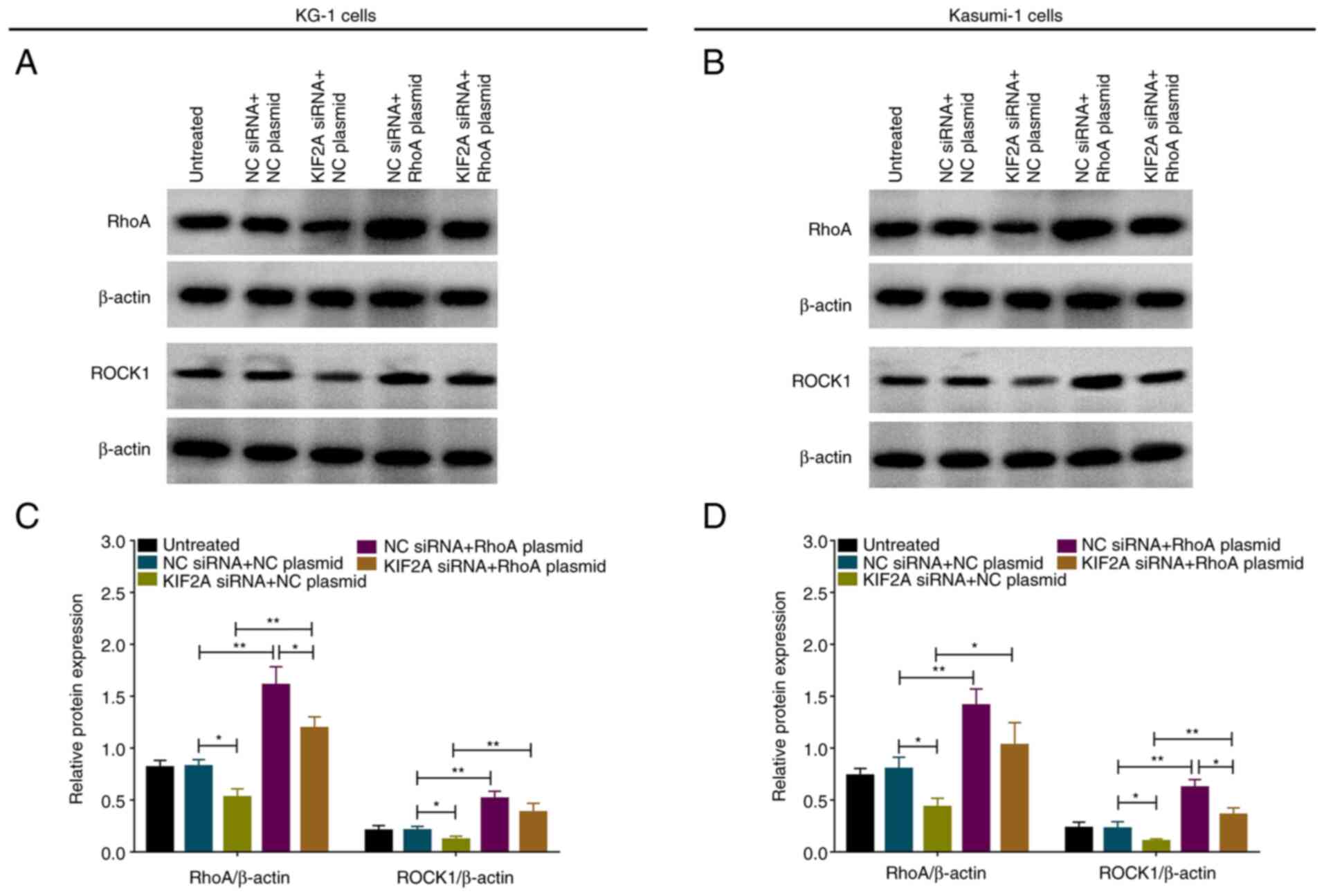
In rescue experiments, RhoA overexpression plasmid
elevated RhoA and ROCK1 expression levels in both wild-type and
KIF2A-knockdown KG-1 and Kasumi-1 cells (all P<0.05; Fig. 9A-D).
In both KG-1 (Fig.
S1A) and Kasumi-1 cells (Fig.
S1B), RhoA expression was elevated in RhoA plasmid compared
with NC plasmid group, indicating successful transfection (both
P<0.001). Moreover, in KG-1 cells, RhoA overexpression plasmid
enhanced cell proliferation, but inhibited cell apoptosis and
chemosensitivity to ADR and AraC (all P<0.05); it also
eliminated the effect of KIF2A siRNA on KG-1 cell proliferation,
apoptosis and chemosensitivity (P<0.05; Fig. 10A-E). In Kasumi-1 cells, RhoA
overexpression plasmid showed similar effects as in KG-1 cells
(Fig. 10F-J).
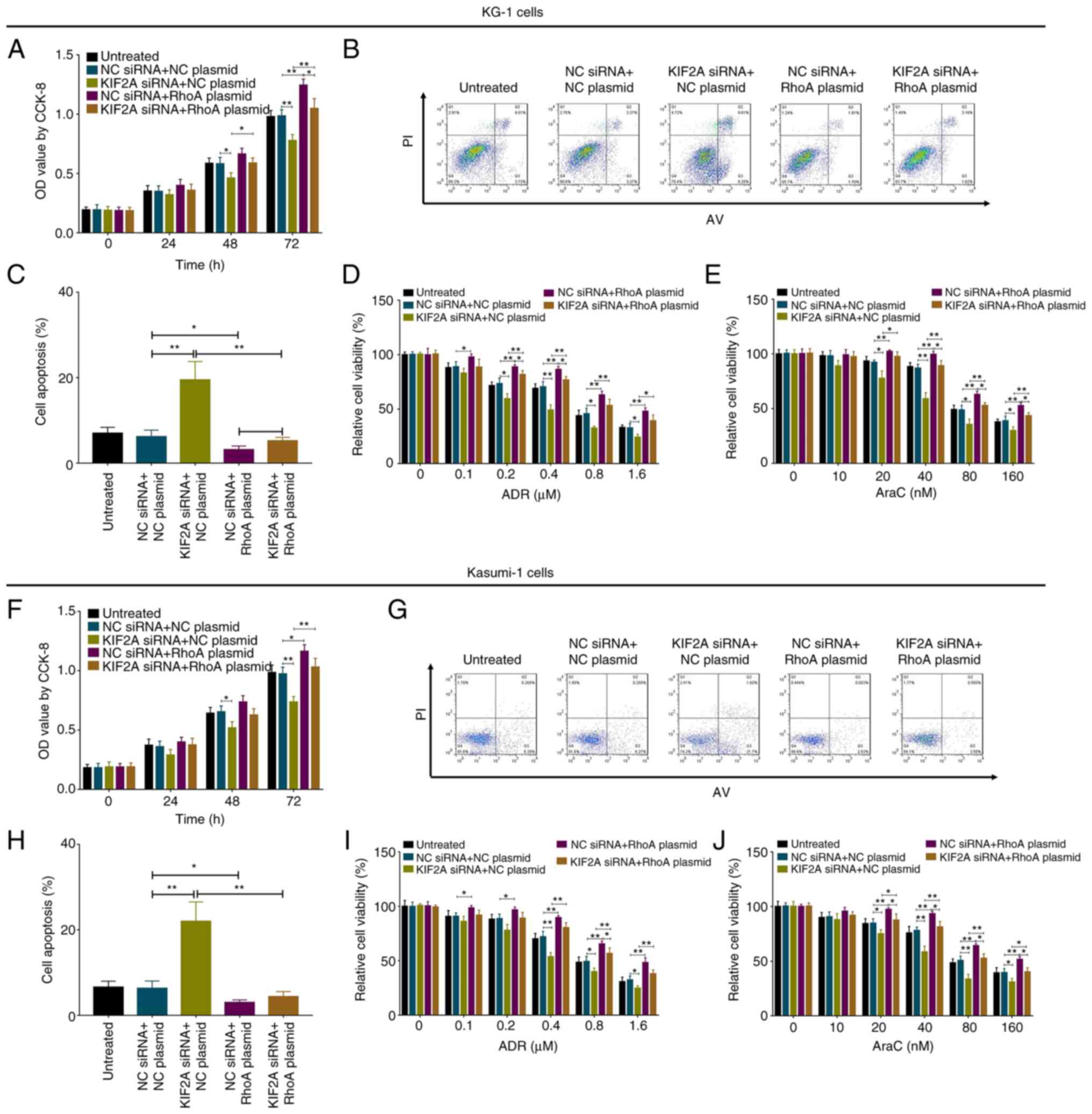 | Figure 10.RhoA overexpression plasmid decreases
the effect of KIF2A siRNA on AML cells. (A) Proliferation, (B and
C) apoptosis and chemosensitivity to (D) ADR and (E) AraC in KG-1
cells following RhoA overexpression. (F) Proliferation, (G and H)
apoptosis and chemosensitivity to (I) ADR and (J) AraC in Kasumi-1
cells following RhoA overexpression. *P<0.05, **P<0.01. RhoA,
ras homolog family member A; KIF2A, kinesin family member 2A; AML,
acute myeloid leukemia; ADR, Adriamycin; AraC, Arabinofuranosyl
Cytidine; NC, negative control; si, small interfering; OD, optical
density; CCK-8, Cell Counting Kit-8. |
Discussion
Similarly to other kinesins, KIF2A is essential for
cell mitosis. Moreover, KIF2A may serve as a biomarker or
therapeutic target in patients with cancer, including AML (17,18).
To the best of our knowledge, there is limited evidence to support
the potential clinical use of KIF2A in AML. However, KIF2A has been
found to be correlated with clinical features or prognosis in other
types of cancer. For example, a previous study based on The Cancer
Genome Atlas database demonstrated that KIF2A is upregulated in
esophageal squamous cell carcinoma tissue and its expression is
correlated with worse disease-free survival in patients (19). Another study observed increased
expression of KIF2A in patients with gastric cancer, and KIF2A
expression is associated with worse histological type, higher TNM
stage, lymph node metastasis and decreased 5-year survival rate; in
addition, KIF2A is also an independent predicting factor for worse
prognosis in patients with gastric cancer (20). These studies indicated that KIF2A
may be a prognostic biomarker for patients with cancer. However,
further studies of the clinical value of KIF2A in cancer are
required.
In view of the mechanistic role of KIF2A in cancer,
a study revealed that KIF2A short hairpin RNA notably suppresses
tumor cell proliferation, migration and invasion, and also inhibits
tumor growth and metastasis in osteosarcoma mice (21). In addition, KIF2A expression is
elevated in gastric cancer cells; inhibiting KIF2A expression in
gastric cancer cells suppresses cell invasion via downregulating
membrane type 1-matrix metalloproteinase (22). A previous study demonstrated that
KIF2A presents with higher expression in grade III–IV glioma tissue
compared with grade I–II glioma tissue, and KIF2A inhibition
results in suppression of glioma cell proliferation, migration,
invasion and promotion of apoptosis (23). A previous study revealed that in
lung adenocarcinoma, KIF2A suppression decreases migration and
proliferation but promotes apoptosis in A549 cells (24). These previous studies suggest that
KIF2A participates in cancer pathology primarily by regulating cell
function. In the present study, KIF2A was upregulated in patients
with AML and positively correlated with BM blast percentage and
risk classification but negatively correlated with treatment
response and survival profile. It was hypothesized that KIF2A
exerted its clinical effect in the pathology of AML by modulating
cancer cell functions via interaction with multiple factors, as
indicated by previous studies (21–24). Here, KIF2A siRNA inhibited
proliferation but enhanced apoptosis, chemosensitivity to ADR and
AraC and expression levels of mRNA/proteins associated with
PI3K/AKT and RhoA/ROCK pathway in AML cells. These data suggested a
regulatory role of KIF2A in AML.
The PI3K/AKT and RhoA/ROCK signaling pathways are
involved in the development of AML, especially the PI3K/AKT
signaling pathway, which has been widely studied (25,26).
To the best of our knowledge, however, the role of RhoA/ROCK is
less reported in AML. A previous study demonstrated that an
inhibitor of both PI3K and histone deacetylase elevates the
antitumor activity of venetoclax in preclinical AML mice (27). Additionally, in AML cells,
Metrine® notably suppresses cell viability and enhances
cell apoptosis in a time- and dose-dependent manner by
downregulating the PI3K/AKT/mTOR signaling pathway (28). Another study reported that ISC-4, a
PI3K/AKT signaling pathway inhibitor, decreases cell survival and
clonogenicity but promotes apoptosis and sensitivity to cytarabine
in AML cells; it also enhances disease progression in a mouse model
of preclinical AML (29). With
regard to the RhoA/ROCK signaling pathway, a study demonstrated
that ROCK1 is targeted by microRNA (miR)-592 to suppress cell
proliferation, migration and invasion and enhance apoptosis in AML
cells (30). Moreover, a long
intergenic non-coding RNA LINC00662 increases proliferation but
decreases apoptosis in AML cells by activating ROCK1 via sponging
of miR-340-5p (31). These
previous findings identified the role of the PI3K/AKT and RhoA/ROCK
signaling pathways in AML by enhancing disease progression disease
via modulation of cell function, most of which are accomplished by
interacting with other factors.
To the best of our knowledge, there are few reports
on the regulation of PI3K/AKT and RhoA/ROCK signaling pathways by
KIF2A. A previous study revealed that KIF2A inhibition enhance
squamous cell carcinoma of the oral tongue (SCCOT) cell apoptosis
by downregulating the PI3K/AKT signaling pathway, which indicates
that KIF2A acts as a tumor promotor by activating this pathway in
SCCOT (32). Another study
demonstrated that silencing of KIF2A notably elevates apoptosis but
decreases proliferation, migration and invasion of gastric cancer
cells by mediating AKT signaling (17). To the best of our knowledge,
however, no studies have reported regulation of RhoA/ROCK signaling
pathway by KIF2A in AML or other types of cancer. The present study
identified a role of KIF2A in the regulation of AML pathology and
demonstrated that KIF2A silencing inhibited proliferation, enhanced
apoptosis and chemosensitivity; it also found that KIF2A regulated
the PI3K/AKT and RhoA/ROCK signaling pathways to affect the
malignant behavior of AML cells. These data enriched understanding
of the mechanism underlying the progression of AML and provided a
basis for investigation of novel targeted therapy.
There was a limitation of the present study: Few
people were willing to give bone marrow, thus, it the sample size
of controls was smaller than that of patients with AML.
In conclusion, KIF2A was correlated with worse
clinical features and survival profile in patients with AML; its
knockdown suppressed proliferation but promoted apoptosis and
chemosensitivity via inactivating PI3K/AKT and RhoA/ROCK signaling
pathways in AML cells. These data suggested the potential of KIF2A
as a prognostic marker and treatment target for AML management.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
RX designed the experiments. XL analyzed the data
and drafted the manuscript. XL and RX confirm the authenticity of
all the raw data. XL and RX revised the manuscript. Both authors
read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Institutional
Review Board of First Affiliated Hospital of Anhui Medical
University (approval no. PJ2020-12-39). All subjects provided
written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
KIF2A
|
kinesin family member 2A
|
|
AML
|
acute myeloid leukemia
|
|
ADR
|
Adriamycin
|
|
RT-q
|
reverse transcription-quantitative
|
|
EFS
|
Event-free survival
|
|
OS
|
overall survival
|
|
NC
|
negative control
|
|
CCK-8
|
Cell Counting Kit-8
|
|
AV/PI
|
Annexin V/propidium iodide
|
|
siRNA
|
small interfering RNA
|
References
|
1
|
Kuykendall A, Duployez N, Boissel N,
Lancet JE and Welch JS: Acute myeloid leukemia: The good, the bad,
and the ugly. Am Soc Clin Oncol Educ Book. 38:555–573. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jung J, Cho BS, Kim HJ, Han E, Jang W, Han
K, Lee JW, Chung NG, Cho B, Kim M and Kim Y: Reclassification of
acute myeloid leukemia according to the 2016 WHO classification.
Ann Lab Med. 39:311–316. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
National Cancer Institute: SEER Cancer
stat facts: Acute myeloid leukemia (AML). https://seer.cancer.gov/statfacts/html/amyl.html/December
17–2018.
|
|
4
|
National Cancer Institute: SEER Cancer
Statistics Review, 1975-2016. https://seer.cancer.gov/csr/1975_2016/April
18–2019
|
|
5
|
Jetani H, Navarro-Bailón A, Maucher M,
Frenz S, Verbruggen CM, Yeguas A, Vidriales MB, González M,
Saborido JR, Kraus S, et al: Siglec-6 is a novel target for CAR
T-cell therapy in acute myeloid leukemia (AML). Blood. Jul
21–2021.(Epub ahead of print). doi: 10.1182/blood.2020009192.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Michelozzi IM, Kirtsios E and Giustacchini
A: Driving CAR T stem cell targeting in acute myeloid leukemia: The
roads to success. Cancers (Basel). 13:28162021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Song MK, Park BB and Uhm JE: Targeted
therapeutic approach based on understanding of aberrant molecular
pathways leading to leukemic proliferation in patients with acute
myeloid leukemia. Int J Mol Sci. 22:57892021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhao M, Wang J, Qu M, Zhao Y, Wang H, Ke
Y, Liu Y, Lei ZN, Liu HM, Hu Z, et al: OGP46 induces
differentiation of acute myeloid leukemia cells via different
optimal signaling pathways. Front Cell Dev Biol. 9:6529722021.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lu W and Gelfand VI: Moonlighting motors:
Kinesin, dynein, and cell polarity. Trends Cell Biol. 27:505–514.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen K, Nam W and Epureanu BI: Collective
intracellular cargo transport by multiple kinesins on multiple
microtubules. Phys Rev E. 101:0524132020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Scherer J, Yi J and Vallee RB: Role of
cytoplasmic dynein and kinesins in adenovirus transport. FEBS Lett.
594:1838–1847. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ganem NJ and Compton DA: The KinI kinesin
Kif2a is required for bipolar spindle assembly through a functional
relationship with MCAK. J Cell Biol. 166:473–478. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang Q, Lu D, Liu W, Ye S, Guo H, Liao T
and Chen C: Effects of KIF2A on the prognosis of nasopharyngeal
carcinoma and nasopharyngeal carcinoma cells. Oncol Lett.
18:2718–2723. 2019.PubMed/NCBI
|
|
14
|
Wang G, Wang Z and Yu H: Kinesin family
member 2A high expression correlates with advanced tumor stages and
worse prognosis in non-small cell lung cancer patients. J Clin Lab
Anal. 34:e231352020.PubMed/NCBI
|
|
15
|
Ding T, Li J, Sun J, Fan X, Shi C, Zhou D
and Deng R: Association of kinesin family member 2A with increased
disease risk, deteriorative clinical characteristics, and shorter
survival profiles in acute myeloid leukemia. Braz J Med Biol Res.
54:e91732020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang X, Wang Y, Liu X, Zhao A, Yang Z,
Kong F, Sun L, Yu Y and Jiang L: KIF2A promotes the progression via
AKT signaling pathway and is upregulated by transcription factor
ETV4 in human gastric cancer. Biomed Pharmacother. 125:1098402020.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li X, Shu K, Wang Z and Ding D: Prognostic
significance of KIF2A and KIF20A expression in human cancer: A
systematic review and meta-analysis. Medicine (Baltimore).
98:e180402019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li D, Sun H, Meng L and Li D: The
Overexpression of kinesin superfamily protein 2A (KIF2A) was
associated with the proliferation and prognosis of esophageal
squamous cell carcinoma. Cancer Manag Res. 12:3731–3739. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang S, Huang F, Wang Y, Song Q, Yang X
and Wu H: KIF2A overexpression and its association with
clinicopathologic characteristics and poor prognoses in patients
with gastric cancer. Dis Markers. 2016:74845162016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang ZX, Ren SC, Chang ZS and Ren J:
Identification of kinesin family member 2A (KIF2A) as a promising
therapeutic target for osteosarcoma. Biomed Res Int.
2020:71027572020.PubMed/NCBI
|
|
22
|
Zhao P, Lan F, Zhang H, Zeng G and Liu D:
Down-regulation of KIF2A inhibits gastric cancer cell invasion via
suppressing MT1-MMP. Clin Exp Pharmacol Physiol. 45:1010–1018.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang X, Ma C, Wang Q, Liu J, Tian M, Yuan
Y, Li X and Qu X: Role of KIF2A in the progression and metastasis
of human glioma. Mol Med Rep. 13:1781–1787. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xie T, Li X, Ye F, Lu C, Huang H, Wang F,
Cao X and Zhong C: High KIF2A expression promotes proliferation,
migration and predicts poor prognosis in lung adenocarcinoma.
Biochem Biophys Res Commun. 497:65–72. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Darici S, Alkhaldi H, Horne G, Jørgensen
HG, Marmiroli S and Huang X: Targeting PI3K/Akt/mTOR in AML:
Rationale and clinical evidence. J Clin Med. 9:29342020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nepstad I, Hatfield KJ, Grønningsæter IS
and Reikvam H: The PI3K-Akt-mTOR signaling pathway in human acute
myeloid leukemia (AML) cells. Int J Mol Sci. 21:29072020.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li X, Su Y, Hege K, Madlambayan G, Edwards
H, Knight T, Polin L, Kushner J, Dzinic SH, White K, et al: The
HDAC and PI3K dual inhibitor CUDC-907 synergistically enhances the
antileukemic activity of venetoclax in preclinical models of acute
myeloid leukemia. Haematologica. 106:1262–1277. 2021.PubMed/NCBI
|
|
28
|
Hao Y, Zhang N, Wei N, Yin H, Zhang Y, Xu
H, Zhu C and Li D: Matrine induces apoptosis in acute myeloid
leukemia cells by inhibiting the PI3K/Akt/mTOR signaling pathway.
Oncol Lett. 18:2891–2896. 2019.PubMed/NCBI
|
|
29
|
Annageldiyev C, Tan SF, Thakur S,
Dhanyamraju PK, Ramisetti SR, Bhadauria P, Schick J, Zeng Z, Sharma
V, Dunton W, et al: The PI3K/AKT pathway inhibitor ISC-4 induces
apoptosis and inhibits growth of leukemia in preclinical models of
acute myeloid leukemia. Front Oncol. 10:3932020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xu Y, Li K, Wang SB and Yang SG: MiR-592
functions as a tumor suppressor in acute myeloid leukemia by
targeting ROCK1 and predicts patients' prognosis. Eur Rev Med
Pharmacol Sci. 23:1610–1619. 2019.PubMed/NCBI
|
|
31
|
Liu Y, Gao X and Tian X: High expression
of long intergenic non-coding RNA LINC00662 contributes to
malignant growth of acute myeloid leukemia cells by upregulating
ROCK1 via sponging microRNA-340-5p. Eur J Pharmacol.
859:1725352019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang K, Lin C, Wang C, Shao Q, Gao W, Song
B, Wang L, Song X, Qu X and Wei F: Silencing Kif2a induces
apoptosis in squamous cell carcinoma of the oral tongue through
inhibition of the PI3K/Akt signaling pathway. Mol Med Rep.
9:273–278. 2014. View Article : Google Scholar : PubMed/NCBI
|