Introduction
Lung cancer has the highest rates of morbidity and
mortality among cancers worldwide (1). Lung adenocarcinoma (LUAD) is the most
common pathologic type and a highly invasive and lethal type of
cancer. Early-stage LUAD can easily invade blood vessels and
lymphatic vessels. Clinically, patients with early-stage cancer do
not have obvious symptoms, often, the tumor has metastasized prior
to causing the corresponding symptoms. Therefore, most patients
with LUAD miss the optimal time for treatment. In recent years,
great improvements have been achieved in the diagnosis and
treatment of LUAD. However, the long-term survival rate of patients
with LUAD remains low (2), and the
local progression, recurrence, and metastasis of tumors are the
main reasons for poor prognosis. Therefore, it is of great
significance to further investigate biomarkers related to the
diagnosis and prognosis of LUAD in order to improve the overall
efficacy of treatment against lung cancer (3).
Excision repair cross-complementation group 6 like
(ERCC6L) is a newly discovered DNA helicase (4), also termed polo kinase 1 interaction
checkpoint helicase (PICH). It has been demonstrated that ERCC6L is
a member of the ERCC6 subfamily, SNF2 (5). Previous research has revealed that
the ERCC6L protein is related to embryonic development, indicating
that ERCC6L may play an important role in maintaining growth
(6). In addition, Baumann et
al revealed that ERCC6L binds to a mitogen-regulated kinase
[polo-like kinase 1 (PLK1)] and is involved in remodeling
centromeric chromatin (7). PLK1
regulates cell division and proliferation, and is considered a
genetic marker for tumor development (8,9).
Dysfunction of ERCC6L induces DNA damage that affects the cell
division and cycle, potentially promoting the development of cancer
(10,11). In recent years, continuous research
has shown that ERCC6L appears to play a critical role in
tumorigenesis and progression (12). For example, ERCC6L is overexpressed
in breast and kidney cancers and is significantly associated with
poor prognosis. ERCC6L was found to be upregulated in
hepatocellular carcinoma, neuroblastoma, gastric cancer and
colorectal cancer, and silencing of ERCC6L inhibited cancer cell
growth, invasion, metastasis and the EMT process. Furthermore,
aberrant expression of ERCC6L promotes tumor progression via the
PI3K/AKT and NF-κB pathways. This evidence has demonstrated that
ERCC6L may be a valuable biomarker of tumors and may act as an
important mediator in the malignant biological behavior of tumors.
However, the role of ERCC6L in the development of LUAD and its
underlying molecular mechanism remain unknown. In the present
study, the expressions profiles and clinical significance of ERCC6L
in LUAD were investigated. In addition, the biological functions
and molecular mechanism of ERCC6L, involved in the tumor
progression of LUAD were explored in vitro and in
vivo.
Materials and methods
Materials
Paraffin tissue specimens were obtained from the
People's Hospital of Guangxi Zhuang Autonomous Region (Nanning,
China) from October 2010 to September 2012. The clinicopathological
data of 85 patients with LUAD were obtained from the Department of
Pathology of the hospital. The extracted information included age,
sex, tumor size, lymph node metastasis, TNM stage, pathological
grade and life status. The histological staging was based on the
2018 edition of the World Health Organization (WHO), and the tumor
staging was based on the 8th edition of the American Joint
Committee on Cancer (AJCC) (13).
Patients with LUAD included in the present study did not receive
any antitumor treatment preoperatively, develop infectious
diseases, or expire within one month after surgery. Follow-up data,
including outpatient follow-up and telephone follow-up, were
collected from the electronic follow-up system established by the
department; data collection was initiated one month after surgical
treatment of the patient and ended in mid-2019. Cancer tissue was
defined as the solid non-necrotizing tissue of the tumor; normal
lung tissue adjacent to the cancer was defined as the lung tissue 5
cm away from the edge of the tumor. The tissue samples were fixed
with 10% neutral formalin for 24 h at room temperature (RT),
embedded in paraffin, and then serially sectioned at 5 µm. All
sections were preserved by the scientific research laboratory
following standard procedures. The present study was approved
(approval no. KY-KJT-2021-125) by the Ethics
Committee/Institutional Review Committee of the People's Hospital
of Guangxi Zhuang Autonomous Region and all participants provided
written informed consent in accordance with the Declaration of
Helsinki. The histopathological results were reviewed by two
experienced pathologists.
Methods
Immunohistochemical assay
Grilled paraffin slices at 60°C for 2 h; For
dewaxing and hydration, the sections were placed in xylene lotion
and soaked for 20 min in 100, 95, 90, 85, and 80% ethanol. For
antigen repair, 1X citric acid repair solution (pH 6.0) was boiled
in a pressure cooker for 3 min, and cooled with water for 10 min.
To eliminate endogenous peroxidase activity, 3%
H2O2 (50 µl) was added, and the sections were
incubated at RT for 10 min. The sections were blocked in goat serum
(1:10; product code SL038; Beijing Solarbio Science &
Technology Co., Ltd.) and incubated at RT for 15–20 min. Following
washing with phosphate-buffered saline (PBS), the sections were
incubated with 50 µl of rabbit anti-human ERCC6L polyclonal
antibody (1:200; cat. no. PA5-62199; Invitrogen; Thermo Fisher
Scientidic, Inc.) at 4°C overnight. To display antigen
distribution, the sections were incubated with 50 µl of ready-use
Max Vision™ HRP-polymer anti-rabbit reagent (cat. no. KIT-5001;
Fuzhou Maixin Biotech Co., Ltd.) for 15 min at RT, closed with a
warm box, and rinsed with PBS. Subsequently, freshly prepared
3,3′-diaminobenzidine solution was added for color development. The
sections were then incubated in hematoxylin dye for 2 min at RT,
and excess dye was removed by rinsing. Subsequently, the sections
were differentiated in hydrochloric acid for 5 sec, submerged in
ammonia solution for 5 min, and rinsed with water. The slices were
dehydrated in gradient alcohol (80, 85, 90,95, and 100%) for 10 min
and soaked in xylene lotion for 10 min. After sealing with neutral
gum, the sections were observed and photographed under a light
microscope(magnification, ×200; Olympus MX51; Olympus
Corporation).
Clinical sample analysis
To identify potential biomarkers of LUAD, the
association between the expression of ERCC6L, malignant
pathological features, and overall survival (OS) in 85 clinical
LUAD samples were analyzed. The clinical samples were divided into
two groups, namely high- and low-ERCC6L expression groups,
according to the median expression of ERCC6L mRNA.
Data mining
Normal samples from the tumor database of The Cancer
Genome Atlas (TCGA; http://cancergenome.nih.gov/) were integrated to
identify the expression characteristics of ERCC6L mRNA. From TCGA
database, 514 cases of primary LUAD and 59 cases of normal controls
were collected. ERCC6L mRNA expression and clinicopathological
characteristics were analyzed. Of the 514 patients, 505 had
complete clinical and follow-up information. The role of ERCC6L
mRNA expression in patients with LUAD was determined through
analysis of TCGA clinical data of those 505 patients. The extracted
data included age, sex, smoking history, tumor size, lymph node
infiltration, distant metastasis, TNM stage, residual tumor,
recurrence status, life status, OS, progression-free interval
(PFI), disease-free interval (DFI) and disease-specific survival
(DSS).
Two lung cancer gene microarray datasets (GSE31210
and GSE30219) (14,15) were downloaded from the Gene
Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/), to validate the
expression and clinical significance of ERCC6L.
Genetic and epigenetic analysis
To investigate the mechanism of ERCC6L dysregulation
in LUAD, its gene-level threshold Gistic2.0-processed copy number
variation (CNV) (16) (n=511) and
DNA methylation status (n=453, methylation 450K) were concurrently
collected from TCGA. Data on ERCC6L mRNA expression, copy number,
and DNA methylation in LUAD cell lines were obtained from the
Cancer Cell Line Encyclopedia (CCLE) (https://dashela.broad institute.org/ccle) (17,18).
This is an online database that provides public access, analysis,
and visualization of genomic data (e.g., gene expression, gene
methylation, and mutation data) for >1,100 cell lines. UALCAN
Network (http://ualcan.path.uab.edu) (19) was used to analyze the level of
ECRR6L promoter methylation in tumor tissue and normal tissue.
Identification of differentially
expressed genes associated with ERCC6L in LUAD
LinkedOmics (http://www.linkedomics.org) (20) is a new open portal offering
multiple omics data analysis of all 32 types of cancer included in
TCGA. In the present study, it was used to identify differentially
expressed genes related to ERCC6L in LUAD. A Benjamini-Hochberg
false discovery rate (FDR) <0.01 and P-value <0.05 were set
as critical criteria. The top three factors that were positively
correlated with ERCC6L were selected for further analysis.
Pearson's correlation was conducted to analyze the expression
between ERCC6L and selected genes. Gene Expression Profiling
Interactive Analysis 2 (GEPIA2; http://gepia2.cancer-pku.cn) (21) is an online interactive web server
that obtains RNA sequencing data from tumor and normal samples in
TCGA and the Genotype-Tissue Expression Project for analysis of the
expression profile and prognostic value of selected genes. Patients
with LUAD were grouped according to the median mRNA expression.
P-values <0.05 denoted statistically significant
differences.
Functional and pathway enrichment
analysis
Gene set enrichment analysis (GSEA) using the
LinkInterpreter module of LinkedOmics (22) was performed for ERCC6L and its
co-expressed genes using TCGA RNA-sequencing (RNA-seq) data.
Analysis of Kyoto Encyclopedia of Genes and Genomes (KEGG)
(23) and Gene Ontology (GO)
(24) pathways (e.g., cellular
components, molecular functions, and biological processes)
identified potential pathways of ERCC6L. FDR <0.05 and P-value
<0.05 denoted statistically significant differences.
Single-cell level analysis
The cancer single-cell state atlas (CancerSEA;
http://biocc.hrbmu.edu.cn/CancerSEA/)
(25) was used to investigate the
function of cancer cells of different states at the single-cell
level. This analysis was conducted to reveal significant
differences in enrichment using data from the Molecular Signatures
Database (MSigDB) (https://www.gsea-msigdb.org/gsea/msigdb) and analyze
the function of ERCC6L. For each single-cell dataset derived from
circulating tumor cells, the Spearman rank correlation test was
used to analyze the significant correlation between gene expression
and functional status; multiple comparisons were corrected for
FDRs.
Cell culture
Human LUAD cell lines (A549, PC9, and H1975) and
293T cells were obtained from the Typical Culture Preservation
Committee of the Chinese Academy of Sciences (Shanghai, China). The
culture conditions of the three cell lines were: 10% Fetal bovine
serum, 1% streptomycin, and RPMI-1640 medium. The cells were
cultured at 37°C in 5% CO2. The reagents were purchased
from Thermo Fisher Scientific, Inc. (GIBCO-BRL; Invitrogen; Thermo
Fisher Scientific, Inc.).
Lentivirus design and cell
transfection
Suzhou GenePharma Co., Ltd. was commissioned to
design and construct a lentiviral vector for the target gene. The
lentivirus RNAi vector was LV-3 of a four-plasmid system. The LV3
(H1/GFP-Puro) vector was synthesized for the lentivirus-short
hairpin-ERCC6L (Lv-shRNA-ERCC6L). RNA interference target
(sh-ERCC6L) was designed according to the sequence of the ERCC6L
gene and a GV248-GFP-lentiviral vector was used as a negative
control (sh-NC). The sh-ERCC6L sequence was
5′-GGTGGTGTCGGTTTAACATTA-3′, while that of the sh-NC sequence was
5′-TTCTCCGAACGTGTCACGGT-3′. A total of three lentivirus packaging
plasmids were used to transfect into 293T cells. The released virus
was harvested and concentrated by ultracentrifugation at 40,000 g
for 2 h at 4°C. The titer of the lentivirus was 5×108
TU/ml. A549 and PC9 cells were transfected with the lentivirus at a
multiplicity of infection (MOI) of 10 with 5 µg/ml polybrene
(product no. H9268; Sigma-Aldrich; Merck KGaA). The enhanced GFP
gene was integrated into the transfected plasmid to distinguish the
successful transfection of cells. The stable expressing cells were
screened by treating with 4 µg/ml puromycin (product no. P8833;
Sigma-Aldrich; Merck KGaA) for 2 weeks.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Collected transfected A549 and PC9 cells were added
with an appropriate amount of TRIzol reagent (Beijing Solarbio
Science & Technology Co., Ltd.). Total RNA extraction was
performed according to the instructions provided by the
manufacturer. After the RNA was diluted with RNase-free water, 1 µl
was added to the NanoDrop-2000 micro-nucleic acid detector to
determine the absorbance, and the samples were stored at −80°C.
cDNA was synthesized according to the manufacturer's instructions
using a reverse transcription kit [PrimeScript™ RT reagent Kit with
gDNA Eraser (Perfect Real Time), cat. no. RR047A; Takara Bio,
Inc.]. Real-time PCR was then performed using SYBR-Green qPCR
Master Mix (Takara Bio, Inc.) on an ABI 7500 real-time quantitative
fluorescence PCR system (Applied BioSystems; Thermo Fisher
Scientific, Inc.). Briefly, the qPCR cycling condition were an
initial denaturation step at 95°C for 30 sec, followed by an
annealing with 40 cycles at a melting temperature of 95°C for 5
sec, and an extension with 40 cycles at a temperature of 60°C for
34 sec. The RT-qPCR primers for ERCC6L and GAPDH were purchased
from Sangon Biotech Co., Ltd. The specific primers used were as
follows: ERCC6L forward, 5′-tcctcctcacaggaacccca-3′ and reverse,
5′-ccagcagggacccttgacaa-3′; GAPDH forward,
5′-CATGAGAAGTATGACAACAGCCT-3′ and reverse,
5′-AGTCCTTCCACGATACCAAG-3′. GAPDH was used as the internal control,
and the results were analyzed using the 2−ΔΔCq method
(26).
Western blot analysis
Radioimmunoprecipitation (RIPA) assay protein
extraction reagent and 10 g/l benzene methyl sulfonyl fluoride
(both from Beyotime Institute of Biotechnology) were used to lyse
H1975, PC9, and A549 cells. A BCA protein detection kit was used to
determine the protein concentration (Beyotime Institute of
Biotechnology). Appropriate samples and molecular weight standard
marker were absorbed and slowly added into the glue hole. An equal
volume of 1X loading buffer was added to the swimming lanes on both
sides of the sample. Electrophoresis was performed at 80 V for
approximately 20 min and at 120 V for approximately 90 min. The
proteins (10 µg) were separated using 10% SDS-PAGE and transferred
to polyvinylidene difluoride membranes. The membranes were blocked
with 5% skim milk (high-protein skim high-calcium milk powder; Yili
Co., Ltd., Inner Mongolia, China) for 2 h at RT. Subsequently, the
membrane was incubated with Tris-buffered saline containing 0.5%
Tween-20 (TBST) (Beyotime Institute of Biotechnology) solution
containing anti-ERCC6L antibody (1:1,000; cat. no. 15688-1-AP),
neural-cadherin (N-cadherin; 1:2,000; cat. no. 22018-1-AP),
epithelial cadherin (E-cadherin; 1:5,000; cat. no. 20874-1-AP),
vimentin (1:2,000; cat. no. 10366-1-AP), snail family
transcriptional repressor 1 (Snai1; 1:500; cat. no. 13099-1-AP),
snail family transcriptional repressor 1 (Snai2; 1:500; cat. no.
12129-1-AP), notch receptor 3 (Notch 3; 1:500; cat. no.
55114-1-AP), β-catenin (1:5,000; cat. no. 51067-2-AP), GAPDH
(1:5,000; cat. no. 10494-1-AP) at 4°C for 12 h. All antibodies were
purchased from Wuhan Sanying Biotechnology. Subsequently, after
washing with TBST, the membrane was incubated with horseradish
peroxidase-labeled anti-rabbit antibodies (1:5,000; bs-80295G-HRP;
BIOSS) at RT for 2 h. The proteins were visualized using the
Odyssey FC imaging system (LI-COR Biosciences).
Cell proliferation assay
Transfected A549 and PC9 cells were cultured in
96-well plates (1,500 cells/well) and transfected for 0, 24, 48, 72
and 96 h, respectively. A total of 20 µl Cell Counting Kit-8
(CCK-8) reagent (Dojindo Laboratories, Inc., Japan) was added to
part of the cells in darkness. Following incubation for 2 h, the
optical density value for each well at 450 nm was detected using a
microplate instrument (Thermo Fisher Scientific, Inc.).
Colony formation assay
Suspended transfected A549 and PC9 cells were seeded
into a six-well plate at a density of 500 cells/ml. The culture was
terminated when white clones in the plate were visually observed.
The cells were cultured at 37°C and 5% CO2 for 14 days.
Next, 1 ml of 0.5% crystal violet solution (Beijing Solarbio
Science & Technology Co., Ltd.) was added to each well after
cells were fixed by pure methanol at RT, and then the cells were
stained for 30 min at RT. Subsequently, the number of clones was
recorded. The number of cells per clone consisted of >50
cells.
Apoptosis and cell cycle assay
In this study, the Annexin V-fluorescein
isothiocyanate/propidium iodide (Annexin V-FITC/PI; Nanjing KeyGen
Biotech Co., Ltd.) method was used to detect cell apoptosis by flow
cytometry. Transfected A549 and PC9 cells were collected, and
3–5×105/ml resuspended cells were placed in a centrifuge
tube. Following centrifugation at 300 × g for 5 min at RT, binding
buffer (500 µl) was added to resuspend the cells. After adding 5 µl
of Annexin V-allophycocyanin, the cells were incubated for 5–10
min. Next, PI dye solution (5 µl) was added and mixed. The solution
was incubated at RT for 5–15 min in the dark. Flow cytometry (BD
FACSCanto™ II) was performed, and the data were analyzed using
FlowJo VX software (FlowJo LLC).
Similarly, transfected A549 and PC9 cells were
collected and fixed with anhydrous ethanol until the final
concentration was 75% at 4°C overnight. The cells were then
incubated overnight at 4°C. The following day, PI/RNase Staining
Buffer (0.5 ml; BD Biosciences) was added. Following incubation at
RT for 15 min in the dark, the cells were fully mixed and examined
using flow cytometry (BD FACSCanto™ II). The results of the cell
cycle were analyzed using the Modfit LT 3.1 software (Verity
Software House, Inc.).
Wound healing assay
Approximately 8×105 transfected A549 and
PC9 cells were added into each well of a six-well plate, allowed to
reach a density >90%, and then cultured in serum-free medium.
Horizontal lines were marked evenly at the back of the six-well
plate (every 0.5-1 cm), crossing the wells. At least five lines
crossed each well. Images were captured at 0, 24 and 48 h using a
fluorescence microscope (magnification, ×200; Olympus IX71; Olympus
Corporation). The ImageJ software (version 1.8.0; National
Institutes of Health) was used to analyze the wound healing. The
experiment was performed thrice.
Cell migration assay
Transfected A549 and PC9 cells were collected and
their density was adjusted to 2–3×105/ml. The cell
migration assay was carried out using an 8.0-µm pore size Transwell
chamber (product number 3422; Corning, Inc.). In the lower chamber,
complete medium containing 15% fetal bovine serum (600 µl) was
added. In the upper chamber, cell suspension (100 µl) was added.
Following incubation with 5% CO2 at 37°C for 18–24 h,
the compartment was removed using tweezers. The fluid in the upper
compartment was drained, and the cells on the surface of the
basement membrane of the upper compartment were gently removed
using a wet cotton swab. The chamber was moved to the well
containing 600 µl of methanol; the cells were fixed at RT for 30
min and stained with 600 µl of 0.1% crystal violet dye for 20 min
at RT. The bottom membrane of the chamber was gently cut with a
razor blade and allowed to dry. Following transfer to a slide, one
drop of neutral resin was added, and the slide was covered with a
cover glass. Five visual fields were randomly selected for counting
under a fluorescence microscope (magnification, ×200; Olympus
IX71).
Cell invasion assay
A 24-well plate with a small chamber containing an
8.0-µm pore size Matrigel-coated membrane (product number 354480;
BioCoat™ Matrigel®; BD Biosciences) was used for the
A549 and PC9 cells invasion assay. The BioCoat Matrigel invasion
chambers were rehydrated with warm (37°C) serum-free medium for 30
min at RT according to the manufacturer's instructions. Following
rehydration, the medium was carefully removed. Cells were collected
and their density was adjusted to 2–3×105 cells/ml.
Serum-free medium (200 µl) was added to the upper chamber of the
24-well plates for activation for 30 min; complete medium with 15%
serum (600 µl) was added to the lower chamber. Subsequently, cell
suspension (100 µl) was added to the upper chamber, and the cells
were incubated with 5% CO2 for 30–34 h at RT. The upper
compartment fluid was blotted, and the cells on the surface of the
upper compartment substratum were gently removed using a wet cotton
swab. The subsequent operations were analogous to the cell
migration assay.
Establishment of a mouse xenograft
model of LUAD
A mouse xenograft model of LUAD was performed under
the guidance of the Ethics Committee, combining methods previously
described (27–29). The animal experiment of this study
was approved (approval no. 202103009) by The Animal Care and
Welfare Committee of Guangxi Medical University (Nanning, China).
The BALB/c nude mice were raised at the Experimental Animal Center
of Guangxi Medical University under specific-pathogen free (SPF)
conditions, with a constant temperature (20–24°C) and humidity
(45–65%) and a 12-h light-dark cycle. All nude mice were provided
with food and sterile water ad libitum. A total of 14 female
mice (four weeks old; weight, 10–12 g) were divided into a
sh-ERCC6L group and a sh-NC group (n=7 mice per group). A total of
2×106 cells transfected with either sh-ERCC6L or sh-NC
were subcutaneously injected into the right armpit of each mouse.
The mice were monitored daily and tumor volumes were recorded every
4 days. After 15 days of inoculation, the nude mice were
anesthetized by intraperitoneal injection of 4% chloral hydrate
(200 mg/ kg) and intramuscular injection of ketorolac (1mg/kg;
painkiller), and then euthanized by cervical dislocation. The final
volume and weight of the tumors were measured. The tumor volume was
calculated as: (Length × width2)/2.
Data processing and statistical
analysis
The SPSS Statistics 20.0 software (IBM Corp.) was
used for statistical analysis. Quantitative data were presented as
the mean ± standard deviation. Welch's t-test (for unpaired
samples) and χ2 test were performed to determine the
association between the mRNA levels of ERCC6L and
clinicopathological features. For patients with duplicated ERCC6L
expression data, the median values were selected for analysis. The
receiver operating characteristic (ROC) curve was used to determine
the diagnostic and prognostic value of ERCC6L expression in LUAD.
Kaplan-Meier curves for OS, PFI, DFI, and DSS were generated using
the GraphPad Prism 8.0.2 (GraphPad Software, Inc.) software. The
significant difference between the survival curves was identified
by log-rank testing. Univariate and multivariate Cox regression
analyses were performed to determine the independent prognostic
factors. P-values <0.05 were considered to indicate
statistically significant differences.
Results
Expression profiles of ERCC6L in
pan-cancer and LUAD tissues
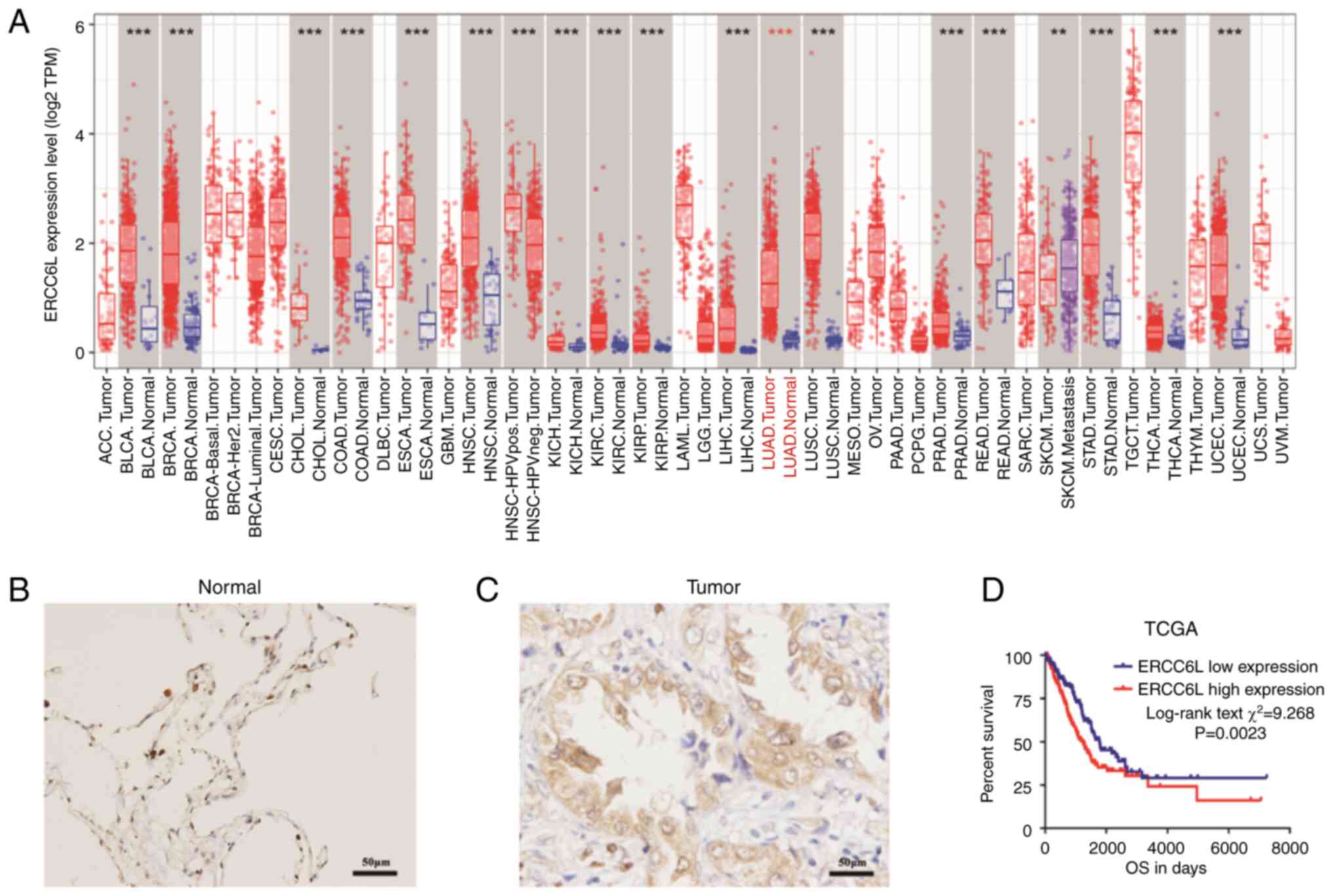
The pan-cancer RNA-seq data of ERCC6L was first
extracted from TCGA database, and the mRNA expression levels of
ERCC6L were analyzed in primary tumors and normal solid tissues
with a sample size of ≥3. The results revealed that ERCC6L mRNA was
upregulated in most human malignant solid tumors including LUAD,
and it may be involved in the occurrence and development of cancers
(Fig. 1A). The expression of
ERCC6L protein in paraffin tissue samples of LUAD from our hospital
was then detected by immunohistochemistry. The results showed that
the alveolar epithelial cells of normal lung tissue exhibited
negative or weak positive staining (Fig. 1B), while the tumor tissue exhibited
strong positive expression (brown color) (Fig. 1C). Collectively it was demonstrated
that the expression of ERCC6L protein was significantly higher in
LUAD tissues.
Association between ERCC6L protein
expression and clinicopathological features in patients with
LUAD
The expression of ERCC6L in patients with LUAD
divided into the high- and low-expression groups according to the
median, and the association between the expression of ERCC6L and
the clinical characteristics of patients was analyzed (Table I). The results showed that ERCC6L
protein expression was significantly higher in tumor tissues of
patients with positive nodal invasion (N1/2/3; P=0.017) or advanced
TNM stage (III/IV; P=0.024), and those who expired (P=0.009).
However, high ERCC6L expression was not associated with age, sex,
tumor size, or pathological grade. To investigate whether ERCC6L
expression was associated with patient prognosis, clinical
follow-up information was extracted, and OS was used as the
endpoint. Kaplan-Meier analysis revealed that patients with high
ERCC6L expression in lung glands had poor OS (P=0.0023; Fig. 1D). Subsequently, Cox regression
analysis was conducted to evaluate the prognostic value of ERCC6L
in patients with LUAD (Table II).
Univariate regression analysis showed that OS was significantly
associated with high ERCC6L expression (P=0.002), nodal invasion
(P<0.001), and tumor size (P=0.025) in LUAD. Multivariate
regression analysis further confirmed that high ERCC6L expression
(P=0.035) and nodal invasion (P=0.005) were independent prognostic
factors of poor OS in patients with LUAD.
 | Table I.Association of the
clinicopathological parameters between high and low ERCC6L
expression groups in 85 patients with LUAD. |
Table I.
Association of the
clinicopathological parameters between high and low ERCC6L
expression groups in 85 patients with LUAD.
|
| ERCC6L
expression |
|
|
|---|
|
|
|
|
|
|---|
| Parameters | Low (n=46) | High (n=39) |
χ2/t | P-value |
|---|
| Age (years) |
|
| <0.001 | 0.995 |
|
≤65 | 33 | 28 |
|
|
|
>65 | 13 | 11 |
|
|
| Sex |
|
| 1.708 | 0.191 |
|
Male | 23 | 25 |
|
|
|
Female | 23 | 14 |
|
|
| Tumor size |
|
| 0.805 | 0.369 |
|
T1-2 | 12 | 7 |
|
|
|
T3-4 | 34 | 32 |
|
|
| Nodal invasion |
|
| 5.662 | 0.017a |
| No | 26 | 12 |
|
|
|
Yes | 20 | 27 |
|
|
| Clinical stage |
|
| 5.108 | 0.024a |
|
I/II | 29 | 15 |
|
|
|
III/IV | 17 | 24 |
|
|
| Pathology
grade |
|
| 2.419 | 0.120 |
|
I/II | 33 | 23 |
|
|
|
III | 11 | 16 |
|
|
| Living status |
|
| 6.830 | 0.009a |
|
Alive | 19 | 6 |
|
|
|
Dead | 27 | 33 |
|
|
 | Table II.Univariate and multivariate analysis
of overall survival in 85 patients with LUAD. |
Table II.
Univariate and multivariate analysis
of overall survival in 85 patients with LUAD.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Parameters | HR | 95% CI
(lower/upper) | P-value | HR | 95% CI
(lower/upper) | P-value |
|---|
| Age (years) ≤65 vs.
>65 | 1.186 | 0.688 | 2.045 | 0.539 |
|
|
|
|
| Sex Female vs.
Male | 0.888 | 0.532 | 1.480 | 0.648 |
|
|
|
|
| Tumor size T1/2 vs.
T3/4 | 2.258 | 1.107 | 4.605 | 0.025a | 1.506 | 0.708 | 3.205 | 0.288 |
| Nodal invasion Yes
vs. No | 2.991 | 1.719 | 5.205 |
<0.001a | 2.347 | 1.292 | 4.263 | 0.005a |
| Pathology grade
I/II vs. III | 1.144 | 0.669 | 1.959 | 0.623 |
|
|
|
|
| ERCC6L expression
High vs. low | 2.245 | 1.340 | 3.761 | 0.002a | 1.771 | 1.041 | 3.013 | 0.035a |
Association between ERCC6L mRNA
expression and clinicopathological features in patients with LUAD
based on TCGA database
RNA-seq data of LUAD was extracted from the TCGA
database for further analysis with a larger sample size. The
association between the mRNA expression levels of ERCC6L and
clinicopathological parameters in 514 LUAD tissues and 59 normal
lung tissues was analyzed. The results suggested that ERCC6L mRNA
expression was significantly upregulated in LUAD tissues (Fig. 2A). Owing to individual differences
between patients, the mRNA expression of ERCC6L in cancer tissues
and normal tissues of the same patient was compared (Fig. 2B). This comparison confirmed the
overexpression of ERCC6L mRNA in LUAD tissues. As revealed in
Fig. 2C-I, the data analysis
revealed that the mRNA expression of ERCC6L was significantly
upregulated in patients with smoking history, positive nodal
invasion and advanced clinical staging, as well as those who
expired, irrespective of sex, tumor size and distal metastasis. In
addition, although the expression levels of ERCC6L mRNA appeared to
be higher in residual and recurrent tumors, no significant
differences were observed. (Figs.
2J and S1).
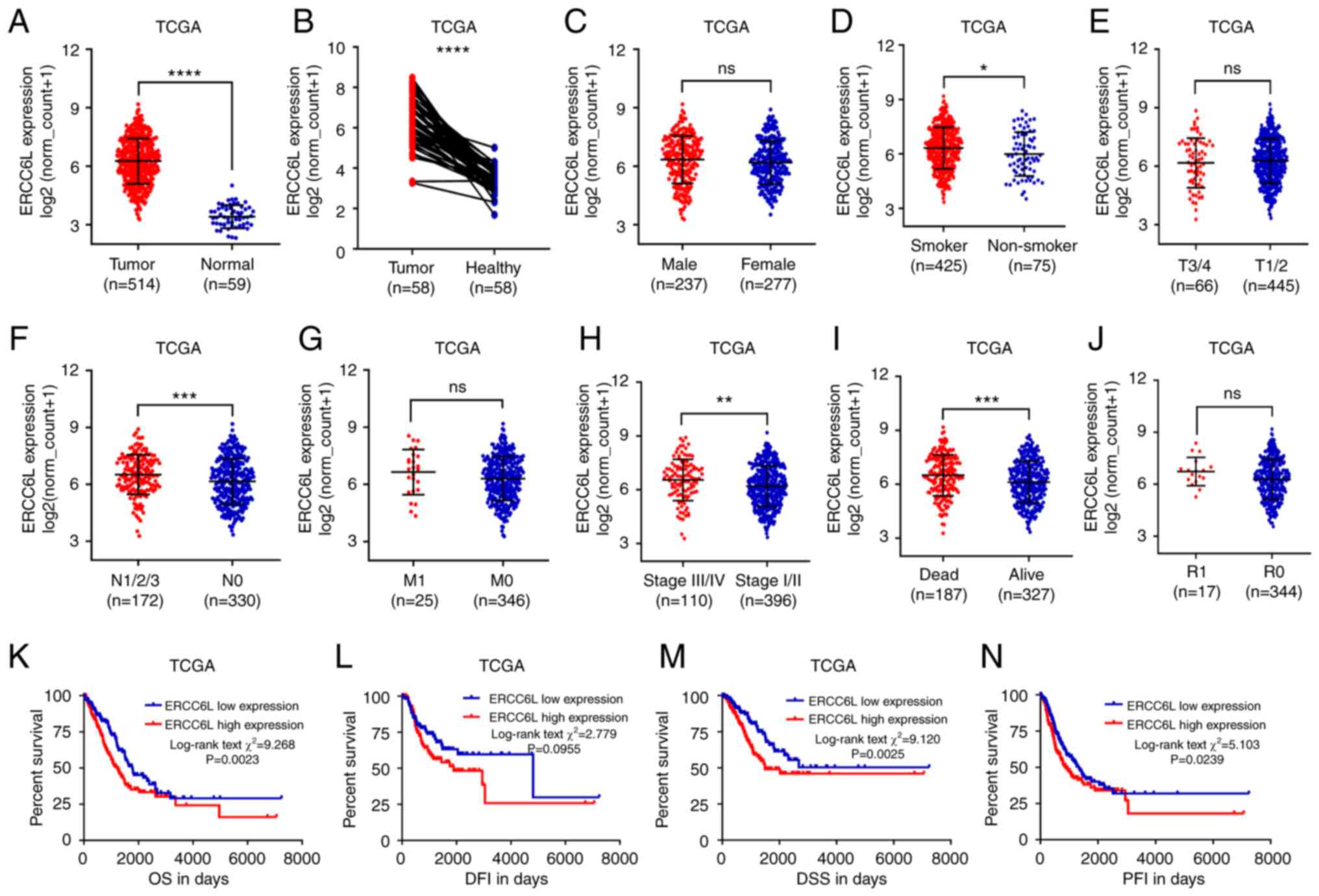 | Figure 2.Association of ERCC6L mRNA expression
with clinicopathological features and clinical outcomes in patients
with LUAD based on the TCGA database. (A) Plot chart showing the
mRNA expression levels of ERCC6L in LUAD tissues and normal lung
tissues. (B) Plot chart showing the mRNA expression of ERCC6L in
LUAD and paired normal tissues. (C-J) Plot chart showing ERCC6L
mRNA expression between (C) male and female, (D) smoker and
non-smoker, (E) cases with T3/4 and T1/2, (F) cases with or without
nodal invasion, (G) cases with or without distant metastasis, (H)
cases with stages III/IV and I/II, (I) live and dead cases, and (J)
cases with or without residual tumor. (K-N) High ERCC6L RNA
expression was associated with unfavorable outcomes in patients
with LUAD. Kaplan-Meier curves of (A) OS, (B) DFI, (C) DSS and (D)
PFI in cases with LUAD. *P<0.05, **P<0.01, ***P<0.001 and
****P<0.0001. ERCC6L, excision repair cross-complementation
group 6 like; LUAD, lung adenocarcinoma; TCGA, The Cancer Genome
Atlas; OS, overall survival; DFI, disease-free interval; DSS,
disease-specific survival; PFI, progression-free interval; ns, not
significant. |
Association between ERCC6L mRNA
expression and outcome of patients with LUAD
To investigate the prognostic value of ERCC6L mRNA
expression in LUAD, the OS, PFI, DFI, and DSS was analyzed in the
TCGA cohort. Kaplan-Meier analysis demonstrated that patients with
LUAD with high ERCC6L mRNA expression in the TCGA-LUAD cohort had
poor OS, DFI, DSS, and PFI (Fig.
2K-N). Subsequently, Cox regression analysis was performed to
further determine whether ERCC6L was an independent risk factor in
patients with LUAD. As revealed in Table III, in terms of OS, univariate
analysis showed that clinical stage, residual tumor, and ERCC6L
mRNA expression were associated with OS (P<0.001, P<0.001,
and P=0.003, respectively). In contrast, age, sex and smoking
history were not associated with OS (P>0.05). Multivariate
analysis further identified clinical stage, residual tumor and
ERCC6L mRNA expression as independent risk factors of poor OS in
patients with LUAD (P<0.001, P=0.001, and P=0.035,
respectively). Moreover, univariate analysis showed that residual
tumor (P=0.017) was associated with DFI, whereas age, sex, smoking
history, and ERCC6L mRNA expression were not (P>0.05).
Multivariate analysis showed that residual tumor (P=0.035) was an
independent risk factor of poor DFI in patients with LUAD. In terms
of DSS, univariate analysis showed that clinical stage, residual
tumor and high ERCC6L mRNA expression were associated with DSS
(P<0.001, P<0.001, and P=0.003, respectively). Multivariate
analysis further identified clinical stage, residual tumor and
ERCC6L mRNA expression as independent risk factors of poor DSS in
patients with LUAD (P<0.001, P=0.001, and P=0.011,
respectively). Univariate analysis revealed that clinical stage,
residual tumor, and ERCC6L mRNA expression were associated with PFI
(P=0.003, P<0.001, and P=0.029, respectively), whereas age, sex
and smoking history were not (P>0.05). Multivariate analysis
further confirmed that clinical stage, residual tumor, and ERCC6L
mRNA expression were independent risk factors of poor PFI in
patients with LUAD (P=0.036, P=0.003, and P=0.047, respectively).
The aforementioned results indicated that the upregulation of
ERCC6L mRNA expression in TCGA-LUAD was negatively correlated with
OS, DSS, and PFI.
 | Table III.Univariate and multivariate analysis
of OS, DFI, DSS and PFI in patients with LUAD in the TCGA
database. |
Table III.
Univariate and multivariate analysis
of OS, DFI, DSS and PFI in patients with LUAD in the TCGA
database.
|
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|
|---|
|
| Parameters | HR | 95% CI
(lower/upper) | P-value | HR | 95% CI
(lower/upper) | P-value |
|---|
| OS | Age
(Continuous) | 1.008 | 0.992 | 1.023 | 0.330 |
|
|
|
|
|
| Sex Female vs.
Male | 0.938 | 0.702 | 1.255 | 0.669 |
|
|
|
|
|
| Smoking history
2/3/4/5 vs. 1 | 0.912 | 0.604 | 1.377 | 0.661 |
|
|
|
|
|
| Clinical stage
III/IV vs. I/II | 2.651 | 1.945 | 3.613 |
<0.001a | 2.400 | 1.748 | 3.295 |
<0.001a |
|
| Residual tumors Yes
vs. No | 3.937 | 2.204 | 7.033 |
<0.001a | 2.753 | 1.524 | 4.973 | 0.001a |
|
| ERCC6L expression
High vs. low | 1.551 | 1.156 | 2.081 | 0.003a | 1.771 | 1.041 | 3.013 | 0.035a |
| DFI | Age
(Continuous) | 1.016 | 0.994 | 1.039 | 0.158 |
|
|
|
|
|
| Gender Female vs.
Male | 0.814 | 0.536 | 1.237 | 0.335 |
|
|
|
|
|
| Smoking history
2/3/4/5 vs. 1 | 0.742 | 0.436 | 1.263 | 0.272 |
|
|
|
|
|
| Clinical stage
III/IV vs. I/II | 0.803 | 0.371 | 1.740 | 0.578 |
|
|
|
|
|
| Residual tumors Yes
vs. No | 4.108 | 1.281 | 13.170 | 0.017a | 3.571 | 1.097 | 11.627 | 0.035a |
|
| ERCC6L expression
High vs. low | 1.434 | 0.944 | 2.178 | 0.091 | 1.337 | 0.873 | 2.048 | 0.181 |
| DSS | Age
(Continuous) | 0.998 | 0.970 | 1.007 | 0.206 |
|
|
|
|
|
| Sex Female vs.
Male | 1.040 | 0.716 | 1.512 | 0.836 |
|
|
|
|
|
| Smoking history
2/3/4/5 vs. 1 | 1.037 | 0.599 | 1.795 | 0.896 |
|
|
|
|
|
| Clinical stage
III/IV vs. I/II | 2.472 | 1.662 | 3.676 |
<0.001a | 2.131 | 1.420 | 3.199 |
<0.001a |
|
| Residual tumors Yes
vs. No | 5.025 | 2.489 | 10.142 |
<0.001a | 3.557 | 1.738 | 7.279 | 0.001a |
|
| ERCC6L expression
High vs. low | 1.801 | 1.229 | 2.637 | 0.003a | 1.670 | 1.125 | 2.480 | 0.011a |
| PFI | Age
(Continuous) | 0.998 | 0.984 | 1.012 | 0.733 |
|
|
|
|
|
| Sex Female vs.
Male | 0.929 | 0.706 | 1.223 | 0.601 |
|
|
|
|
|
| Smoking history
2/3/4/5 vs. 1 | 0.970 | 0.654 | 1.440 | 0.881 |
|
|
|
|
|
| Clinical stage
III/IV vs. I/II | 1.622 | 1.180 | 2.230 | 0.003a | 1.424 | 1.023 | 1.981 | 0.036a |
|
| Residual tumors Yes
vs. No | 3.312 | 1.780 | 6.162 |
<0.001a | 2.620 | 1.379 | 4.976 | 0.003a |
|
| ERCC6L expression
High vs. low | 1.359 | 1.033 | 1.788 | 0.029a | 1.330 | 1.004 | 1.762 | 0.047a |
Validation of the expression and
clinical significance of ERCC6L in LUAD based on the GEO
database
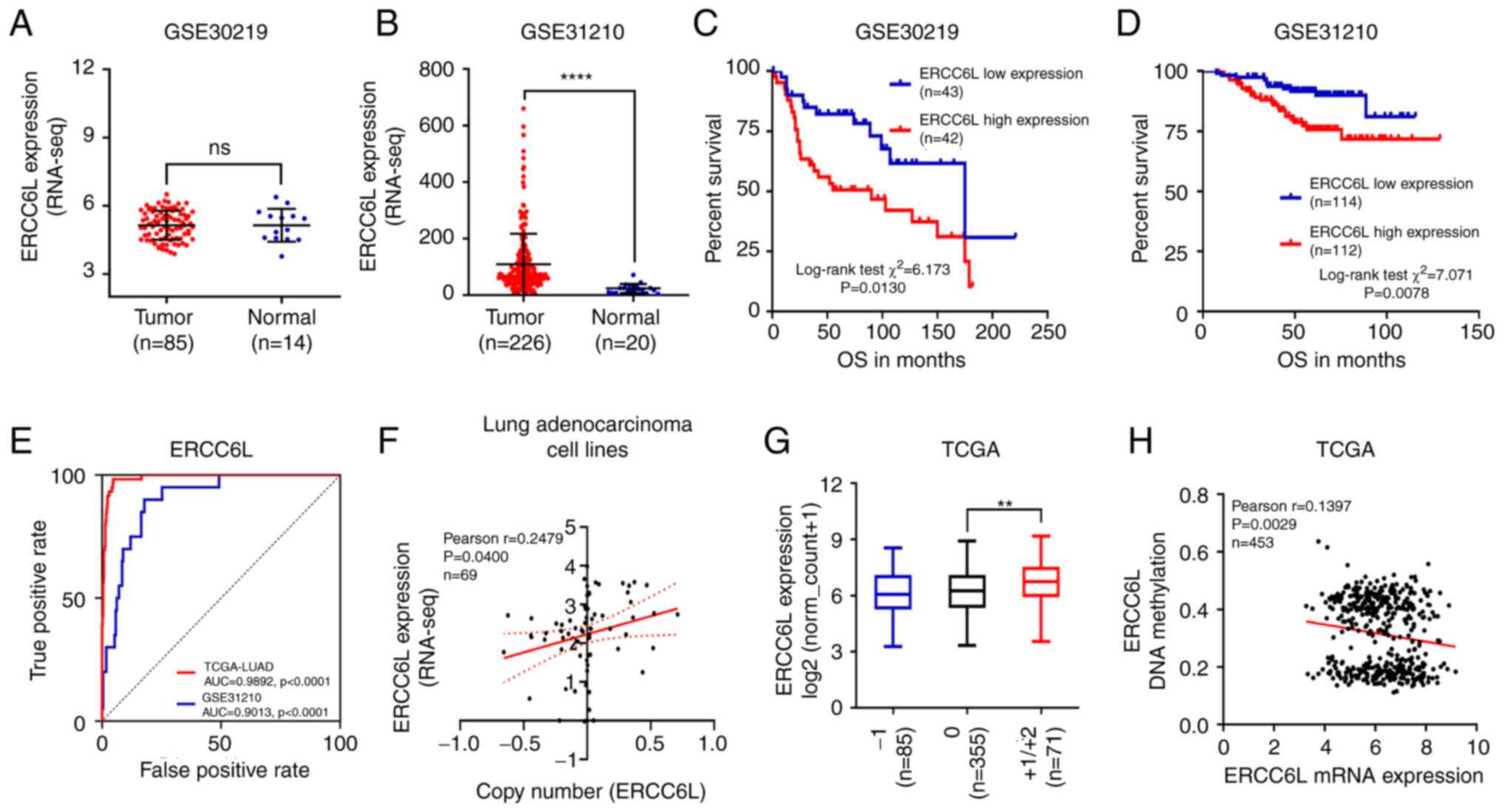
Data from the GEO database were used to validate the
aforementioned results, and the independent array sets GSE30219 and
GSE31210 were selected for analysis. In the GSE30219 dataset, there
was no significant change in the mRNA expression levels of ERCC6L
(P>0.05; Fig. 3A). However, in
the GSE31210 microarray dataset, the mRNA expression levels of
ERCC6L were significantly increased in LUAD tissue compared with
normal lung tissue (P<0.001; Fig.
3B). LUAD tissues were divided into high- and low-expression
groups according to the median mRNA expression of ERCC6L.
Kaplan-Meier curve showed that, in the GSE30219 (Fig. 3C) and GSE31210 (Fig. 3D) datasets, the OS was
significantly shorter in patients with high ERCC6L mRNA expression
than in those with low ERCC6L mRNA expression. In addition, the
area under the curve value of ERCC6L expression for the diagnosis
of LUAD in the TCGA cohort was 0.9892 (P<0.0001). Meanwhile, in
the GSE31210 array set, the area under the curve value of ERCC6L in
LUAD was 0.9013 (P<0.0001; Fig.
3E).
Regulation of ERCC6L gene expression
at DNA amplification and methylation levels
Deep sequencing data extracted from the TCGA
database was used to investigate the potential mechanism involved
in the dysregulation of the ERCC6L gene in LUAD. The association
between ERCC6L mRNA expression and copy number data was examined in
69 LUAD cell lines from the Encyclopedia of Cancer Cell Lines. The
results showed that the mRNA expression of ERCC6L was positively
correlated with the DNA copy number (Fig. 3F). In addition, based on TCGA
database, 71 patients (13.9%) had low or high DNA amplification,
while 85 patients (16.6%) had DNA deletion (Fig. 3G). Furthermore, the correlation
between ERCC6L mRNA expression and DNA methylation was examined in
453 patients with primary LUAD. The results showed that the mRNA
expression of ERCC6L was negatively associated with DNA methylation
in LUAD (Fig. 3H).
GSEA of genes co-expressed with ERCC6L
in LUAD
To explore the potential functions and pathways of
ERCC6L involvement in LUAD, RNA-seq data was analyzed from 515
patients with LUAD from TCGA database using LinkedOmics. The
LinkFinder module of LinkedOmics was used to identify genes
differentially expressed in correlation with ERCC6L in LUAD. The
threshold was FDR <0.01 A volcano plot revealed differentially
expressed genes positively (red dots) and negatively (green dots)
correlated with ERCC6L in LUAD (Fig.
4A). The heat map showed the top 50 significant genes that were
positively or negatively correlated with ERCC6L, respectively
(Fig. 4B and C). As revealed in
Fig. 4D, a very strong positive
correlation was observed between ERCC6L expression and CDCA5
(r=0.8344), KIF4A (r=0.8743) and TPX2 (r=0.8464). Consistent with
the ERCC6L gene, these three genes were upregulated in tumors and
significantly associated with poor OS in patients with LUAD vs.
normal controls (Fig. 4E and
F).
Moreover, the LinkInterpreter module of LinkedOmics
was used to conduct GSEA of genes co-expressed with ERCC6L in LUAD.
The top 10 significant GO terms and signaling pathway enrichment
results are presented in Fig. 5A and
B. The results suggested that the genes positively co-expressed
with ERCC6L were mainly located in the chromosomal regions of
condensation, ‘chromosomal region’, ‘spindle’, and ‘replication
fork’. These genes are generally involved in ‘chromosome
segregation’, ‘DNA replication’, ‘chromatin assembly or
disassembly’, ‘spindle organization’ and ‘cell cycle checkpoint’.
Molecular function ontology analysis found that genes were mainly
enriched in GO terms, such as ‘catalytic activity acting on DNA’,
‘DNA helicase activity’, ‘single-stranded DNA binding’ and ‘DNA
replication’. In addition, the KEGG pathway analysis showed that
significant enrichment pathways were ‘cell cycle’, ‘DNA
replication’, ‘homologous recombination’, ‘oocyte meiosis’,
‘proteasome’, ‘spliceosome’ and the ‘P53 signaling pathway’.
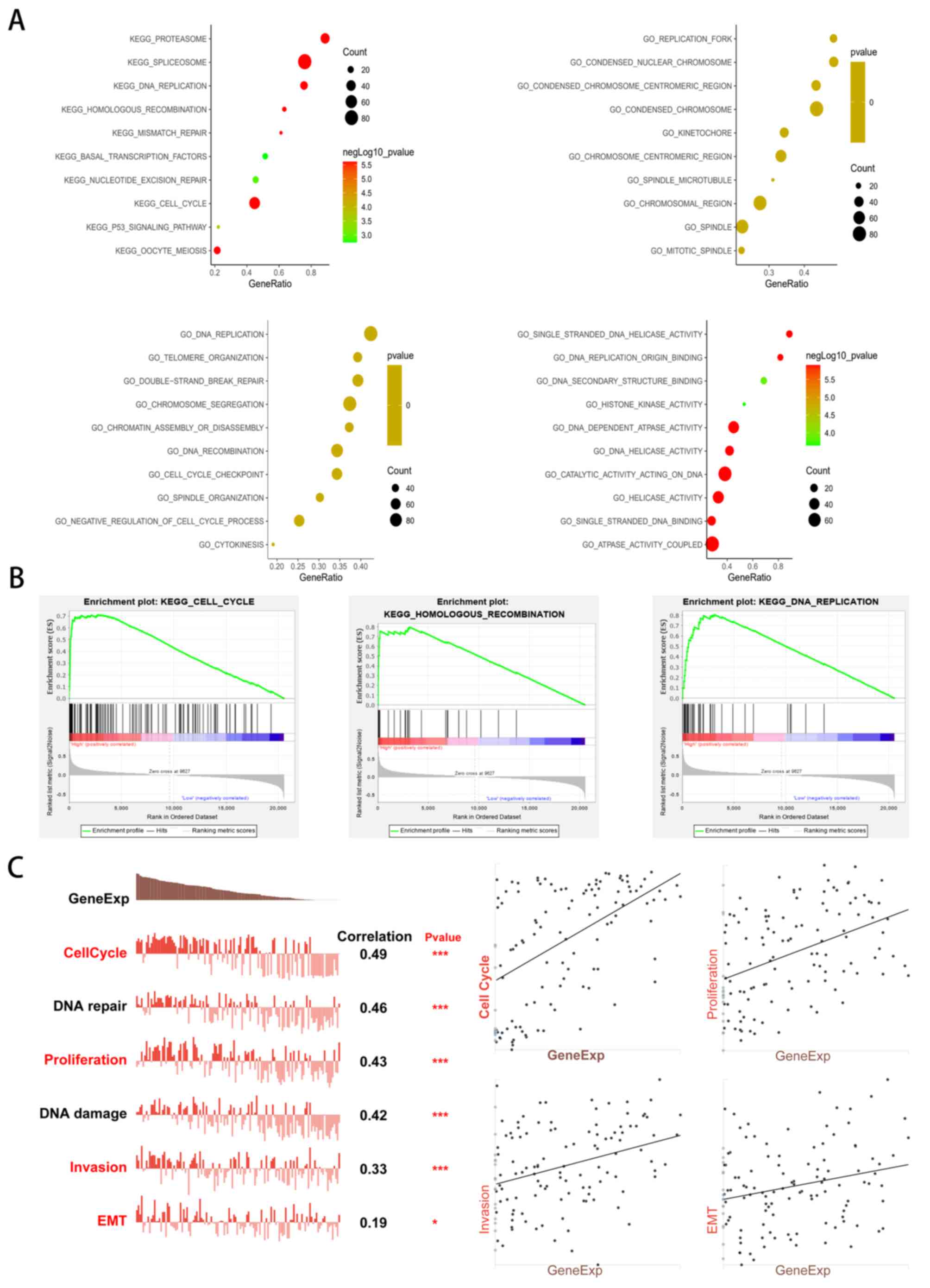 | Figure 5.GSEA analysis and single-cell gene
functional analysis of ERCC6L in LUAD. (A) Bubble plots showing the
top 10 GO and KEGG pathway terms of genes co-expressed with ERCC6L
in LUAD. (B) The top 3 KEGG pathways of genes positively
co-expressed with ERCC6L in LUAD. ERCC6L upregulation was
associated with ‘cell cycle’, ‘homologous recombination’ and ‘DNA
replication’. (C) Gene functional analysis of ERCC6L in LUAD at the
single-cell level. Gene expression of ERCC6L was significantly
correlated with cell cycle, DNA repair, proliferation, DNA damage,
invasion, and EMT. Spearman's correlation was conducted. *P<0.05
and ***P<0.001. GSEA, gene set enrichment analysis; ERCC6L,
excision repair cross-complementation group 6 like; LUAD, lung
adenocarcinoma; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of
Genes and Genomes; EMT, epithelial-mesenchymal transition. |
Gene functional analysis of ERCC6L in
LUAD at the single-cell level
Considering the heterogeneity among cancer cells,
the biological functions of genes were further investigated at the
single-cell level. The CancerSEA database was used to identify the
main biological functions of ERCC6L in LUAD. As revealed in
Fig. 5C, gene expression of ERCC6L
was significantly correlated with the cell cycle, DNA repair,
proliferation, DNA damage, invasion and EMT [P=0.49, 0.46, 0.43,
0.42, 0.33 (P<0.001) and 0.19 (P<0.05), respectively].
Construction of the ERCC6L
gene-silencing LUAD cell line
The endogenous expression levels of ERCC6L mRNA and
protein were examined in three LUAD cell lines, namely A549, H1975,
and PC9. The results of qPCR and western blot analysis showed that
ERCC6L was expressed in all three cell lines (Fig. 6A). Subsequently, the expression of
endogenous ERCC6L in LUAD cell lines was knocked down using
lentivirus-mediated shRNA. In this study, A549 and PC9 cells were
selected for transfection with lentivirus, and stably transfected
cells were constructed. The efficiency of interference with the
ERCC6L gene was determined by qPCR and western blotting. The
results showed that the efficiency of ERCC6L inhibition was 89 and
85% in A549 and PC9 cells, respectively (Fig. 6B). Knockdown of ERCC6L expression
also inhibited ERCC6L protein expression in A549 and PC9 cells,
respectively (Fig. 6C).
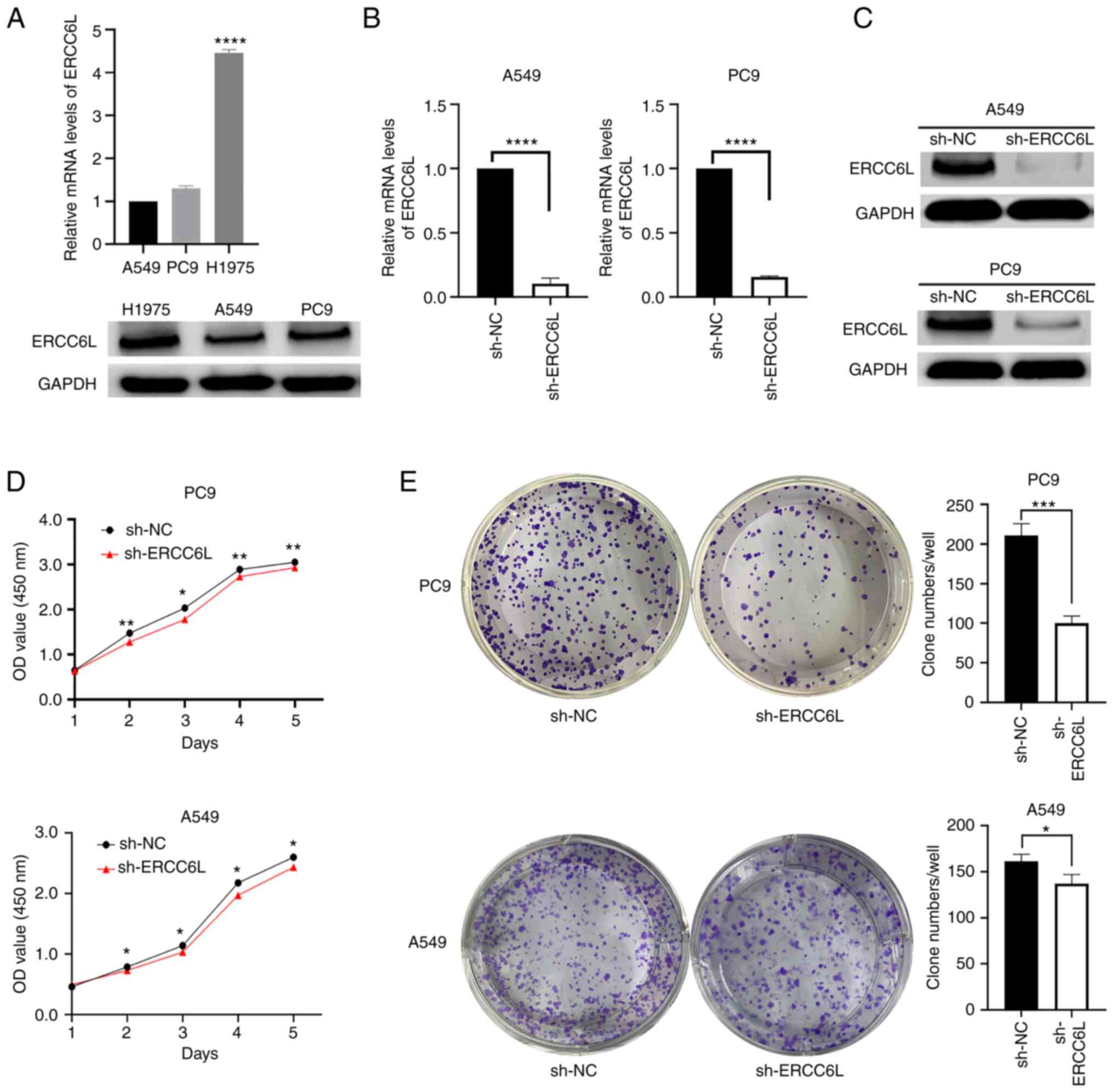 | Figure 6.Knockdown of ERCC6L inhibits cell
proliferation in LUAD in vitro. (A) The endogenous
expression of ERCC6L mRNA and protein in A549, H1975 and PC9 cells
was detected by qPCR and western blot analysis, respectively. (B)
The transfection efficiency of lentivirus-mediated shRNA was
verified by qPCR assay. Following knockdown of ERCC6L, the mRNA
expression level was decreased in A549 and PC9 cells as detected by
qPCR assay. (C) Following knockdown of ERCC6L, the protein
expression level was downregulated in A549 and PC9 cells as
detected by western blot analysis. (D) CCK-8 assay was performed to
detect the proliferative activity of PC9 and A549 cells after
silencing of ERCC6L. (E) Knockdown of ERCC6L inhibited the ability
of plate clone formation in A549 and PC9 cells. *P<0.05,
**P<0.01, ***P<0.001 and ****P<0.0001 compared with sh-NC.
ERCC6L, excision repair cross-complementation group 6 like; LUAD,
lung adenocarcinoma; qPCR, quantitative polymerase chain reaction;
sh-, short hairpin; NC, negative control. |
Effect of ERCC6L on the proliferation
of LUAD cells
CCK-8 assay and plate clone formation assay were
performed to assess the effect of ERCC6L on cell proliferation in
LUAD. The results revealed that the downregulation of ERCC6L
expression in A549 and PC9 cells decreased cell viability; the
difference was statistically significant compared with the sh-NC
group (Fig. 6D). In addition,
knockdown of ERCC6L reduced the number and volume of A549 and PC9
cell clones and decreased the proliferation of these cells
(Fig. 6E).
Effect of ERCC6L on cell apoptosis and
the cell cycle of LUAD cells
Flow cytometric analysis was performed to explore
the effects of ERCC6L on cell apoptosis and the cell cycle in LUAD.
As revealed in Fig. 7A, the cell
apoptotic rate of PC9 and A549 cells transfected with sh-ERCC6L was
significantly higher than that of the control. In addition,
following the downregulation of ERCC6L expression, the proportions
of A549 and PC9 cells in the G1 phase decreased, while those in the
S phase increased (Fig. 7B).
Effect of ERCC6L on cell migration and
invasion of LUAD cells
Wound-healing and Transwell assays were performed to
assess the effect of ERCC6L on cell migration and invasion in LUAD.
The result of the wound-healing experiment showed that, compared
with the sh-NC group, A549 and PC9 cells with ERCC6L knockdown
showed significantly reduced migratory ability 24 and 48 h after
the wound, as well as reduced wound-healing ability and relatively
wide wound spacing (Fig. 8A). For
the cell migration assay, the results revealed that the number of
cells penetrating the membrane in the A549-sh-ERCC6L and
PC9-sh-ERCC6L groups was significantly lower than that recorded in
the NC group (P<0.0001 for both; Fig. 8B). Knockdown of ERCC6L could weaken
the migratory ability of LUAD cells in vitro. For the cell
invasion assay, compared with the sh-NC group, the number of A549
and PC9 cells transfected with sh-ERCC6L that penetrated the
membrane was significantly decreased (P<0.001 for both; Fig. 8C). Collectively, these results
suggested that knockdown of ERCC6L could reduce the invasive
ability of LUAD cells in vitro.
 | Figure 8.Effect of ERCC6L expression on cell
migration, invasion, the EMT process and subcutaneous tumorigenesis
of A549 and PC9 cells. (A) Representative images of A549 and PC9
cells transfected with sh-ERCC6L as detected by scratch assay. (B)
Following knockdown of ERCC6L, Transwell migration assays were used
to detect the migration ability of A549 and PC9 cells. (C) The
BioCoat Matrigel invasion assay was used to examine the effect of
ERCC6L on the invasion ability of A549 and PC9 cells. (D) Effect of
ERCC6L expression on EMT marker molecules and nuclear transcription
factors in LUAD cells. (E) Knockdown of ERCC6L inhibited growth of
xenografted tumor in nude mice. (E-1) Representative image of
subcutaneous xenografts of nude mice in sh-ERCC6L and sh-NC groups.
(E-2,3) The tumor sizes and tumor weights of PC9 cells subcutaneous
xenograft were compared between sh-ERCC6L and sh-NC groups. (E-4)
Representative immunohistochemical staining of ERCC6L in
subcutaneous tumors of nude mice (magnification, ×400).
***P<0.001 and ****P<0.0001. ERCC6L, excision repair
cross-complementation group 6 like; EMT, epithelial-mesenchymal
transition; sh-, short haiprin; LUAD, lung adenocarcinoma; NC,
negative control. |
Association between ERCC6L and EMT, as
well as the Wnt/β-catenin signaling pathway
To explore the potential molecular mechanism by
which ERCC6L is involved in the progression of LUAD, the expression
of EMT-related biomarkers and key molecules of the Wnt/β-catenin
signaling pathway was examined. As revealed in Fig. 8D, compared with sh-NC, sh-ERCC6L
decreased the protein expression levels of N-cadherin, Snai1 and
Snai2 in A549 and PC9 cells, while it upregulated the protein
expression levels of E-cadherin. Furthermore, knockdown of ERCC6L
downregulated the expression levels of Notch 3 and β-catenin. These
results suggested that ERCC6L may co-regulate EMT through the
Wnt/β-catenin and Wnt/Notch 3 signaling pathways, thus affecting
the occurrence and development of LUAD.
Effect of knocking down ERCC6L
expression on the proliferation of LUAD cells in vivo
The aforementioned results showed that knockdown of
ERCC6L inhibited the proliferation of LUAD cells in vitro.
To verify if these results are reproducible in vivo, a LUAD
xenograft tumor model in nude mice was established. Tumorigenesis
could be observed 4–7 days after inoculation. The state of the
mice, including food intake, appeared to be normal throughout the
experiment. As revealed in Fig.
8E, mice injected with sh-ERCC6L-transfected LUAD PC9 cells had
reduced tumorigenicity, tumor volume and tumor weight compared with
the control mice (P<0.05). In line with the in vitro
findings, these results suggested that the low-expression of ERCC6L
inhibited the proliferation of LUAD cells in vivo.
Discussion
The development of new genome-wide sequencing tests,
as well as molecular targeted drugs and antibodies, has improved
the overall efficacy of treatment against cancer. However, LUAD (a
common type of lung cancer) is associated with poor prognosis and
short survival. Therefore, it is necessary to investigate the
mechanism involved in the tumorigenesis and progression of LUAD and
further establish effective treatment strategies (30,31).
In recent years, it has been consistently reported that the
expression levels of SNF2 family members, ERCC6 and ERCC6L, are
significantly associated with various types of cancers. For
example, Zhao et al (32)
reported that high expression of ERCC6 was associated with poor OS
in patients with colorectal cancer who received or did not receive
treatment with 5-fluorouracil. Luo et al (33) revealed that ERCC6 may be a
biomarker for the prognosis of gastric cancer. Pu et al
(34) demonstrated that the
expression of ERCC6L was significantly correlated with the clinical
survival of patients with breast and gastric cancer. Yu et
al (35) revealed that liver
cancer patients with low ERCC6L expression had significantly longer
OS. Huang et al (36) also
demonstrated the clinical potential of interfering with ERCC6L
expression to improve the treatment of high-risk patients with
triple-negative breast cancer. These studies suggested that ERCC6L
is a valuable prognostic marker for patients with cancer and may be
a prognostic biomarker. Nevertheless, little is known about the
expression profile and function of ERCC6L in LUAD.
In this study, TCGA-RNA-seq data was initially used
to demonstrate that ERCC6L is upregulated in most types of cancer,
including LUAD. The immunohistochemical results based on paraffin
sections revealed that ERCC6L was overexpressed in the tumor
tissues of patients with LUAD. Subsequently, the clinical
significance of ERCC6L expression in LUAD was further analyzed by
integrating clinical samples with TCGA-LUAD data. The results
showed that high ERCC6L expression was significantly associated
with lymph node metastasis, TNM stage, and survival status.
Therefore, ERCC6L may be involved in the carcinogenesis and
progression of LUAD.
In clinical prognostic studies, objective OS events
are typically considered the end point. This may affect clinical
judgment due to the longer follow-up time for OS. For LUAD (a
highly aggressive tumor type), using more accurate DFI, PFI, and
DSS results with relatively short follow-up times as reference is
of great importance. Because patients may relapse or deteriorate in
a short period of time, this approach has practical guiding
significance for biological research, such as tumor invasion and
evaluation of clinical treatment effects. In the present study,
high expression of ERCC6L in both LUAD clinical samples and the
TCGA-LUAD dataset was significantly associated with shorter OS;
this observation was consistent with the analysis of two GEO
independent datasets. Cox regression analysis confirmed that ERCC6L
expression was an independent risk factor affecting the prognosis
of LUAD. Furthermore, in terms of PFI and DSS, ERCC6L was also
indicated as an independent predicator in LUAD. In addition, the
ROC analysis suggested that ERCC6L mRNA expression has certain
diagnostic value. Collectively, these results suggest that ERCC6L
is a promising biomarker for the diagnosis and prognosis of LUAD.
This evidence also laid a foundation for further investigation on
the function and mechanism of ERCC6L in LUAD.
Bioinformatics analysis can improve the
reproducibility and effectiveness of diagnostic markers of cancer
at different levels (e.g., molecular and pathway). In the present
study, transcriptome data of LUAD was collected and epigenetic
analysis, pathway analysis, and single-cell level analysis were
performed to demonstrate that ERCC6L expression may affect the
biological behavior of LUAD. To the best of our knowledge, the
underlying molecular mechanisms of ERCC6L dysregulation in tumors
are not yet fully understood. Considering the potential diagnostic
and prognostic value of ERCC6L in LUAD, it is important to explore
the potential mechanism of ERCC6L dysregulation in LUAD. Genetic
and epigenetic changes (e.g., CNVs, DNA methylation, and somatic
mutations) often cause abnormalities in gene expression and cancer
cell behavior. CNVs can regulate the expression of specific genes
through a dose-effect relationship (37,38).
In this study, DNA amplification was the main type that caused
changes in ERCC6L and was positively correlated with upregulation
of this gene. This association between CNVs and ERCC6L mRNA
expression was also confirmed in LUAD cell lines. Therefore, it was
hypothesized that the aberrant expression and dysregulation of
ERCC6L in LUAD may be due to the alteration of chromosomal
structure. In addition, ERCC6L mRNA expression was negatively
correlated with methylation levels, indicating that ERCC6L
expression may be modulated by DNA methylation in LUAD.
By identifying genes co-expressed with ERCC6L in
LUAD, the possible biological roles and mechanisms of ERCC6L were
further speculated from the perspective of gene enrichment
analysis. The most significant positively correlated genes were
CDCA5, KIF4A, and TPX2, which were overexpressed in tumor tissues
of LUAD. These genes were positively correlated with ERCC6L
expression and associated with poor OS. By GSEA analysis, these
genes were revealed to be mainly concentrated in chromosomes,
spindles, and microtubules and were primarily involved in cell
cycle-related activities and regulation (e.g., mitotic cell cycle
phase transition, chromosome separation, cell cycle checkpoint
activity, etc.). Similarly, the cell cycle was the most significant
signaling pathway for the enriched genes co-expressed with ERCC6L
in LUAD. The cell cycle plays an important role in tumorigenesis
(39). Dysregulation of the cell
cycle can lead to cancer cell proliferation and tumor growth
(40). Zhang et al
(41) demonstrated that ERCC6L
could enhance cell viability in vitro and promote tumor
growth in vivo by regulating the mitogen-activated protein
kinase (MAPK) signal transduction pathway after knockout of ERCC6L
expression in 786-O cells. Chen et al (42) reported that ERCC6L promotes the
development of liver cancer by activating the PI3K/AKT and nuclear
factor-κB (NF-κB) signaling pathways. Nevertheless, whether ERCC6L
affects the cell cycle of LUAD warrants further investigation.
Heterogeneity among cancer cells poses a major
challenge to the diagnosis and treatment of cancer. Single-cell
sequencing technology provides an unprecedented opportunity to
accurately decipher the functional status of individual cancer
cells (43). Functional analysis
of ERCC6L in individual LUAD cells showed that overexpression of
ERCC6L was positively correlated with the cell cycle, DNA repair,
proliferation, DNA damage, invasion and EMT. The regulation of the
cell cycle by ERCC6L overexpression was consistent with the results
of the GSEA using data from the TCGA-LUAD dataset. In general, the
results of the cancer cell line and single-cell analyses support
the notion that the regulation of cell cycle progression may be the
primary function of ERCC6L in LUAD cells.
Fernandez-Cuesta et al (43) showed that lung cancer types are not
the result of early progenitor cell lesions of highly aggressive
lung neuroendocrine tumors, instead, they arise through independent
cellular mechanisms. Inactivation of chromatin remodeling genes is
sufficient to drive lung carcinoid transformation. In fact, it has
been shown that ERCC6L is involved in the remodeling of centromere
chromatin, and its binding to PLK1 can enhance the function of
ERCC6L in the consolidation of mitotic chromosomes (44–46).
Notably, PLK1 plays an important role in cell division and
proliferative life activities (36), indicating that the dysregulation of
ERCC6L expression can lead to dysregulation of mitosis. In turn,
this process affects the proliferative ability of cells. It has
been reported that inactivation of ERCC6L gene results in p53
activation, DNA damage, and apoptosis in mouse embryos (47). It was hypothesized that ERCC6L
plays an important role in the development of LUAD. Furthermore,
continuous studies have shown that high expression of ERCC6L can
promote abnormal cell proliferation and inhibit cell apoptosis in
various malignant tumors, such as breast, gastric, and liver
cancer. For example, knockdown of ERCC6L could significantly
inhibit the proliferation of breast cancer cells and induce
apoptosis in vitro (48).
Pu et al (34) found that
ERCC6L regulated the cell cycle through the RAB31/MAPK/cyclin
dependent kinase 2 (CDK2) pathway and promoted the proliferation of
cancer cells. Hence, it was hypothesized that ERCC6L may have a
similar biological function in LUAD.
In the present study, a shRNA lentivirus was used to
construct stable LUAD cell lines in which ERCC6L was knocked down.
In cell function experiments, this method can achieve a relatively
stable silencing effect of ERCC6L gene expression. Knockdown of
ERCC6L inhibited the proliferation of A549 and PC9 cells in
vivo and in vitro. Downregulation of ERCC6L expression
lead to cell cycle arrest in the S phase and promoted apoptosis.
These results indicated that the dysregulation of ERCC6L expression
could affect the proliferative ability, apoptosis level, and cycle
distribution of LUAD cells.
EMT is a process in which fully differentiated
epithelial cells are transformed into mesenchymal phenotypes and
plays an important role in embryonic development, wound healing and
tumor invasion (49,50). EMT is a reversible process
initiated by several factors, including TGF-β, Notch, Wnt,
fibroblast growth factor (FGF), and epidermal growth factor (EGF)
(51). In addition, the process is
mediated by EMT-induced transcription factors, such as Snai1/2,
Slug, Twist, zinc finger E-box binding homeobox 1 (ZEB1), and ZEB2.
E-cadherin, N-cadherin, β-catenin, and Snai are important molecules
involved in cell adhesion and EMT, and play important roles in
organ formation, tissue homeostasis, and maintenance of epithelial
integrity and polarity (52–54).
Loss of E-cadherin function is associated with poor prognosis and
survival in patients with various types of cancer (55). β-Catenin (a membrane adhesion
protein complex) is an important component of the Wnt signaling
pathway. Numerous studies have shown that mutations or
dysregulation of components of the Wnt signaling pathway are
associated with cancer in humans (56). The expression levels of E-cadherin
and β-catenin play an important role in the occurrence of EMT in
ovarian cancer cells (57,58). E-cadherin expression is
downregulated in numerous cancer cell lines with enhanced invasion
and migration phenotypes (59).
The Wnt/β-catenin pathway directly or indirectly upregulates the
expression of key transcription factors regulating E-cadherin
(60). The Notch signaling pathway
plays a key role in cell development, affecting cell processes
(e.g., differentiation, proliferation and migration) and
participating in the occurrence and progression of cancer (61,62).
Activation of EMT signaling in cancer cells is widely thought to
contribute to metastasis, recurrence, or resistance to therapy;
hence, molecules that regulate EMT are also considered drug targets
(63). Changes in the occurrence
and metastasis of lung cancer can be judged by changes in the
levels of related EMT markers. Studies have shown that ERCC6L
promotes the growth and invasion of colorectal cancer cells
(64). Similarly, the present
study demonstrated that inhibition of ERCC6L expression attenuated
the migratory and invasive activities of A549 and PC9 cells.
Following the downregulation of ERCC6L expression, the protein
expression levels of N-cadherin, Snai1, Snai2, Notch 3, and
β-catenin were decreased, whereas those of E-cadherin were
increased. These results suggested that ERCC6L may influence the
EMT of LUAD through the Wnt/β-catenin and Wnt/Notch 3 signaling
pathways.
Clinically, LUAD is often diagnosed at an advanced
stage; thus, it is difficult to determine the earliest time-point
of EMT initiation. Currently, the effects of the expression of
related genes on cell location remain unknown. Moreover, it is
impossible to determine its influence on the treatment of tumors
and their recurrence and metastasis after treatment. Therefore,
discovering specific targets of epigenetic silencing during EMT is
of great importance. The present study showed that ERCC6L is
involved in regulating the changes in EMT, providing a new
potential research target for the mechanism of EMT in LUAD.
In summary, ERCC6L was overexpressed in LUAD tissues
compared with normal lung tissues. Increased expression of ERCC6L
was significantly correlated with nodal invasion and advanced TNM
staging, and acted as an independent risk factor for prognosis in
patients with LUAD. DNA amplification and hypomethylation may
contribute to ERCC6L dysregulation in LUAD. Knockdown of KIF18A
inhibited LUAD cell proliferation in vitro and in
vivo, attenuated cell migration and invasion, induced apoptosis
and S-phase arrest. ERCC6L may regulate EMT through the
Wnt/β-catenin and Wnt/Notch 3 signaling pathways, leading to
malignant biological behavior (i.e., metastasis and invasion) of
LUAD. Therefore, ERCC6L may be a potential therapeutic target for
lung cancer. Nevertheless, there were several limitations in the
present study. For example, only a few case samples were included
in the studies on the association between the expression of ERCC6L
and clinicopathological characteristics of patients; this is
because fresh tissue samples need to be collected during surgery.
Hence, the source of samples was limited, and the number of samples
obtained was relatively small. Moreover, since the topic of the
present study is that ERCC6L overexpression confers malignant
phenotypes of LUAD, the effect of ERCC6L on normal cell lines
(e.g., immortalized bronchial cells) should be further examined.
Finally, the in-depth molecular mechanism involved in the genetic
and epigenetic levels (e.g. DNA copy number and methylation), and
in the promotion of lymph node metastasis and invasion of LUAD by
ERCC6L remains to be fully elucidated.
Supplementary Material
Supporting Data
Acknowledgements
The present study was jointly accomplished by The
People's Hospital of Guangxi Zhuang Autonomous Region (also known
as Guangxi Academy of Medical Sciences, Nanning, China) and Guangxi
University. We are grateful to the Experimental Animal Center of
Guangxi Medical University (Nanning, China) for the support of
animal experiments. We would also like to thank the Research and
Experiment Center of The People's Hospital of Guangxi Zhuang
Autonomous Region for the support of cell and molecular biology
experiments. We would also like to thank Dr Jiao Lan and Dr
Mingzheng Mo from The People's Hospital of Guangxi Zhuang
Autonomous Region for their excellent technical assistance.
Funding
The present study was supported by Guangxi Natural Science
Foundation (grant no. 2018GXNSFBA281058), the Basic Ability
Improvement Project of Young and Middle-aged Teachers in Guangxi
Universities (grant no. 2018KY0050) and the National Natural
Science Foundation of China (grant no. 82060078).
Availability of data and materials
The processed data required to reproduce these
findings cannot be shared at this time as the data also form part
of an ongoing study. Requests to access the datasets should be
directed to the corresponding author YZ (yl.zhong@whu.edu.cn).
Authors' contributions
SL, LJ, XH, MY, HL, BL, ZW and YZ conceived and
designed the study. XH, LJ, SL, ZW and YZ performed data curation.
SL, HL, BL, MY and XH performed formal analysis of the data. XH,
SL, LJ and YZ wrote the original draft of the manuscript. XH, SL,
LJ, BL, HL, MY, and ZW wrote, reviewed and edited the final
manuscript. SL, LJ and XH contributed equally to this work. SL, XH
and YZ confirmed the authenticity of all the raw data. All authors
contributed to the article and read and approved the submitted
final version.
Ethics approval and consent to
participate
The present study was approved (approval no.
KY-KJT-2021-125) by the Ethics Committee of the People's Hospital
of Guangxi Zhuang Autonomous Region (Nanning, China). All
participants provided written informed consent in accordance with
the Declaration of Helsinki. The animal experiment of this study
was approved (approval no. 202103009) by The Animal Care and
Welfare Committee of Guangxi Medical University (Nanning,
China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that the research was conducted
in the absence of any commercial or financial relationships that
could be construed as potential competing interests.
References
|
1
|
Ferlay J, Colombet M, Soerjomataram I,
Mathers C, Parkin DM, Piñeros M, Znaor A and Bray F: Estimating the
global cancer incidence and mortality in 2018: GLOBOCAN sources and
methods. Int J Cancer. 144:1941–1953. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Quintanal-Villalonga Á and Molina-Pinelo
S: Epigenetics of lung cancer: A translational perspective. Cell
Oncol (Dordr). 42:739–756. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kim D, Lee YS, Kim DH and Bae SC: Lung
cancer staging and associated genetic and epigenetic events. Mol
Cells. 43:1–9. 2020.PubMed/NCBI
|
|
4
|
Santamaria A, Neef R, Eberspächer U, Eis
K, Husemann M, Mumberg D, Prechtl S, Schulze V, Siemeister G,
Wortmann L, et al: Use of the novel Plk1 inhibitor
ZK-thiazolidinone to elucidate functions of Plk1 in early and late
stages of mitosis. Mol Biol Cell. 18:4024–4036. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Xu Y, Chen X and Li Y: ERCC6L, a gene of
SNF2 family, may play a role in the teratogenic action of alcohol.
Toxicol Lett. 157:233–239. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yin Y, Tang L, Zhang J, Tang B and Li Z:
Molecular Cloning and Gene Expression Analysis of ERCC6L in Sika
Deer (Cervus nippon hortulorum). PLoS One. 6:e209292011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Baumann C, Körner R, Hofmann K and Nigg
EA: PICH, a centromere-associated SNF2 family ATPase, is regulated
by Plk1 and required for the spindle checkpoint. Cell. 128:101–114.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bhola NE, Jansen VM, Bafna S, Giltnane JM,
Balko JM, Estrada MV, Meszoely I, Mayer I, Abramson V, Ye F, et al:
Correction: Kinome-wide functional screen identifies role of PLK1
in Hormone-independent, ER-positive breast cancer. Cancer Res.
79:8762019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Helmke C, Becker S and Strebhardt K: The
role of Plk3 in oncogenesis. Oncogene. 35:135–147. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Abbasi R, Ramroth H, Becher H, Dietz A,
Schmezer P and Popanda O: Laryngeal cancer risk associated with
smoking and alcohol consumption is modified by genetic
polymorphisms in ERCC5, ERCC6 and RAD23B but not by polymorphisms
in five other nucleotide excision repair genes. Int J Cancer.
125:1431–1439. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hübner NC, Wang LH, Kaulich M, Descombes
P, Poser I and Nigg EA: Re-examination of SiRNA specificity
questions role of PICH and Tao1 in the spindle checkpoint and
identifies Mad2 as A sensitive target for small RNAs. Chromosoma.
119:149–165. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang G, Ma J, Xiong J, Huang X, Han X, Yu
X and Jiang X: Upregulation of excision repair
cross-complementation group 6-Like (ERCC6L) promotes tumor growth
in hepatocellular carcinoma. Dig Dis Sci. 66:1097–1109. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rami-Porta R, Asamura H, Travis WD and
Rusch VW: Lung cancer-major changes in the American Joint Committee
on Cancer eighth edition cancer staging manual. CA Cancer J Clin.
67:138–155. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yamauchi M, Yamaguchi R, Nakata A, Kohno
T, Nagasaki M, Shimamura T, Imoto S, Saito A, Ueno K, Hatanaka Y,
et al: Epidermal growth factor receptor tyrosine kinase defines
critical prognostic genes of stage I lung adenocarcinoma. PLoS One.
7:e439232012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rousseaux S, Debernardi A, Jacquiau B,
Vitte AL, Vesin A, Nagy-Mignotte H, Moro-Sibilot D, Brichon PY,
Lantuejoul S, Hainaut P, et al: Ectopic activation of germline and
placental genes identifies aggressive metastasis-prone lung
cancers. Sci Transl Med. 5:186ra662013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mermel CH, Schumacher SE, Hill B, Meyerson
ML, Beroukhim R and Getz G: GISTIC2. 0 facilitates sensitive and
confident localization of the targets of focal somatic copy-number
alteration in human cancers. Genome Biol. 12:R412011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nusinow DP, Szpyt J, Ghandi M, Rose CM,
McDonald ER III, Kalocsay M, Jané-Valbuena J, Gelfand E, Schweppe
DK, Jedrychowski M, et al: Quantitative proteomics of the cancer
cell line encyclopedia. Cell. 180:387–402.e16. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Barretina J, Caponigro G, Stransky N,
Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehár J, Kryukov GV,
Sonkin D, et al: The cancer cell line encyclopedia enables
predictive modelling of anticancer drug sensitivity. Nature.
483:603–607. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Vasaikar SV, Straub P, Wang J and Zhang B:
LinkedOmics: Analyzing multi-omics data within and across 32 cancer
types. Nucleic Acids Res. 46:D956–D963. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tang Z, Kang B, Li C, Chen T and Zhang Z:
GEPIA2: An enhanced web server for large-scale expression profiling
and interactive analysis. Nucleic Acids Res. 47:W556–W560. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yuan H, Yan M, Zhang G, Liu W, Deng C,
Liao G, Xu L, Luo T, Yan H, Long Z, et al: CancerSEA: A cancer
single-cell state atlas. Nucleic Acids Res. 47:D900–D908. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gong HX, He L, Li XP, Wang YD, Li Y, Huang
JJ, Wang Z, Xie D, Kung HF and Peng Y: Effective antitumor immunity
against murine gliomas using dendritic cells transduced with
hTERTC27 recombinant adenovirus. Oncol Rep. 27:1163–1169. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang Y, Guo Y, Tan S, Ke B, Tao J, Liu H,
Jiang J, Chen J, Chen G and Wu B: β-Arrestin1 enhances
hepatocellular carcinogenesis through inflammation-mediated Akt
signalling. Nat Commun. 6:73692015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lin CH, Wang CH, Hsu SL, Liao LY, Lin TA
and Hsueh CM: Molecular mechanisms responsible for neuron-derived
conditioned medium (NCM)-mediated protection of ischemic brain.
PLoS One. 11:e01466922016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhong Y, Jiang L, Lin H, Li X, Long X,
Zhou Y, Li B and Li Z: Overexpression of KIF18A promotes cell
proliferation, inhibits apoptosis, and independently predicts
unfavorable prognosis in lung adenocarcinoma. IUBMB Life.
71:942–955. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhong Y, Jiang L, Long X, Zhou Y, Deng S,
Lin H and Li X: Clinical significance and integrative analysis of
kinesin family member 18B in lung adenocarcinoma. OncoTargets Ther.
12:9249–9264. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhao Z, Zhang G and Li W: Elevated
Expression of ERCC6 confers resistance to 5-fluorouracil and is
associated with poor patient survival in colorectal cancer. DNA
Cell Biol. 36:781–786. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Luo SS, Liao XW, Zhu XD and Doetsch PW:
Prognostic value of excision repair cross-complementing mRNA
expression in gastric cancer. Biomed Res Int. 2018:62046842018.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Pu SY, Yu Q, Wu H, Jiang JJ, Chen XQ, He
YH and Kong QP: ERCC6L, A DNA helicase, is involved in cell
proliferation and associated with survival and progress in breast
and kidney cancers. Oncotarget. 8:42116–42124. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yu B, Liang H, Ye Q and Wang Y:
Upregulation of ERCC6L is associated with tumor progression and
unfavorable prognosis in hepatocellular carcinoma. J Gastrointest
Oncol. 11:1009–1023. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Huang Y, Li W, Yan W, Wu J, Chen L, Yao X,
Gu F, Lv L, Zhao J, Zhao M, et al: Loss of PICH promotes chromosome
instability and cell death in triple-negative breast cancer. Cell
Death Dis. 10:4282019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gamazon ER and Stranger BE: The impact of
human copy number variation on gene expression. Brief Funct
Genomics. 14:352–357. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Gao Y and Teschendorff AE: Epigenetic and
genetic deregulation in cancer target distinct signaling pathway
domains. Nucleic Acids Res. 45:583–596. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ingham M and Schwartz GK: Cell-cycle
therapeutics come of age. J Clin Oncol. 35:2949–2959. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Malumbres M and Barbacid M: Cell cycle,
CDKs and cancer: A changing paradigm. Nat Rev Cancer. 9:153–166.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang G, Yu Z, Fu S, Lv C, Dong Q, Fu C,
Kong C and Zeng Y: ERCC6L That Is Up-regulated in high grade of
renal cell carcinoma enhances cell viability in vitro and promotes
tumor growth in vivo potentially through modulating MAPK signalling
pathway. Cancer Gene Ther. 26:323–333. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chen H, Wang H, Yu X, Zhou S, Zhang Y and
Wang Z, Huang S and Wang Z: ERCC6L Promotes the progression of
hepatocellular carcinoma through activating PI3K/AKT and NF-κB
signaling pathway. BMC Cancer. 20:8532020. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Fernandez-Cuesta L, Peifer M, Lu X, Sun R,
Ozretić L, Seidal D, Zander T, Leenders F, George J, Müller C, et
al: Frequent mutations in chromatin-remodelling genes in pulmonary
carcinoids. Nature Commun. 5:35182014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kurasawa Y and Yu-Lee LY: PICH and
cotargeted Plk1 coordinately maintain prometaphase chromosome arm
architecture. Mol Biol Cell. 21:1188–1199. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Rouzeau S, Cordelières FP,
Buhagiar-Labarchède G, Hurbain I, Onclercq-Delic R, Gemble S,
Magnaghi-Jaulin L, Jaulin C and Amor-Guéret M: Bloom's Syndrome and
PICH Helicases cooperate with topoisomerase IIα in centromere
disjunction before anaphase. PLoS One. 7:e339052012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Leng M, Besusso D, Jung SY, Wang Y and Qin
J: Targeting Plk1 to chromosome arms and regulating chromosome
compaction by the PICH ATPase. Cell Cycle. 7:1480–1489. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Albers E, Sbroggiò M, Pladevall-Morera D,
Bizard AH, Avram A, Gonzalez P, Martin-Gonzalez J, Hickson ID and
Lopez-Contreras AJ: Loss of PICH results in chromosomal
instability, p53 activation, and embryonic lethality. Cell Rep.
24:3274–3284. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Liu J, Sun J, Zhang Q and Zeng Z: shRNA
knockdown of DNA Helicase ERCC6L expression inhibits human breast
cancer growth. Mol Med Rep. 18:3490–3496. 2018.PubMed/NCBI
|
|
49
|
Greenburg G and Hay ED: Epithelia
suspended in collagen gels can lose polarity and express
characteristics of migrating mesenchymal cells. J Cell Biol.
95:333–339. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Nawshad A, LaGamba D, Polad A and Hay ED:
Transforming growth factor-beta signaling during
epithelial-mesenchymal transformation: Implications for
embryogenesis and tumor metastasis. Cells Tissues Organs.
179:11–23. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Otsuki Y, Saya H and Arima Y: Prospects
for new lung cancer treatments that target EMT signaling. Dev Dyn.
247:462–472. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Vu T and Datta PK: Regulation of EMT in
colorectal cancer: A culprit in metastasis. Cancers (Basel).
9:1712017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Kaszak I, Witkowska-Piłaszewicz O,
Niewiadomska Z, Dworecka-Kaszak B, Ngosa Toka F and Jurka P: Role
of cadherins in cancer-a review. Int J Mol Sci. 21:76242020.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Shook D and Keller R: Mechanisms,
mechanics and function of epithelial-mesenchymal transitions in
early development. Mech Dev. 120:1351–1383. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wong SHM, Fang CM, Chuah LH, Leong CO and
Ngai SC: E-cadherin: Its dysregulation in carcinogenesis and
clinical implications. Crit Rev Oncol Hematol. 121:11–22. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Tafrihi M and Nakhaei Sistani R:
E-Cadherin/β-catenin complex: A target for anticancer and
antimetastasis plants/plant-derived compounds. Nutr Cancer.
69:702–722. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Al-Alem L and Curry TE Jr: Ovarian cancer:
Involvement of the matrix metalloproteinases. Reproduction.
150:R55–R64. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Kenny HA and Lengyel E: MMP-2 functions as
an early response protein in ovarian cancer metastasis. Cell Cycle.
8:683–688. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Mendonsa AM, Na TY and Gumbiner BM:
E-cadherin in contact inhibition and cancer. Oncogene.
37:4769–4780. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Nguyen VHL, Hough R, Bernaudo S and Peng
C: Wnt/β-catenin signalling in ovarian cancer: Insights into its
hyperactivation and function in tumorigenesis. J Ovarian Res.
12:1222019. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Artavanis-Tsakonas S, Rand MD and Lake RJ:
Notch signaling: Cell fate control and signal integration in
development. Science. 284:770–776. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Siebel C and Lendahl U: Notch signaling in
development, tissue homeostasis, and disease. Physiol Rev.
97:1235–1294. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
O'Leary K, Shia A and Schmid P: Epigenetic
regulation of EMT in non-small cell lung cancer. Curr Cancer Drug
Targets. 18:89–96. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Xie Y, Yu J, Wang F, Li M, Qiu X, Liu Y
and Qi J: ERCC6L Promotes Cell Growth and Invasion in Human
Colorectal Cancer. Oncol Lett. 18:237–246. 2019.PubMed/NCBI
|