Introduction
Lung cancer is responsible for one fifth of
cancer-related deaths worldwide (1). Non-small cell lung cancer (NSCLC) is
the commonest type of lung cancer (2,3).
Some targeted therapies against epidermal growth factor receptor,
anaplastic lymphoma kinase and c-ros oncogene 1 receptor tyrosine
kinase have been confirmed to provide survival benefits to NSCLC
patients. However, a considerable proportion of NSCLC remains
incurable, mainly due to late diagnosis and drug resistance
(4–6). Therefore, it is necessary to
understand the pathogenesis of NSCLC for proper diagnosis and
development of new treatment modalities. Fanconi anemia (FA) is a
rare recessive disorder characterized by anemia and bone marrow
failure. It is caused by mutations in the FA family proteins
(7,8). The FA family consists of ~19 genes
associated with cell cycle progression, regulation and DNA damage
repair (9,10). FANCI is an FA family protein; its
gene is located on the long arm of chromosome 15. FANCI forms a
complex with its molecular chaperone FANCD2 to participate in DNA
repair and ribosome biosynthesis. The complex also regulates the
cell cycle in the S and G2 phases (11–13).
The FA core complex, which is an E3 ubiquitin ligase,
monoubiquitinates FANCD2 and FANCI when DNA is damaged and
stabilizes both proteins (14,15).
Recently, FANCI was reported to be involved in the occurrence and
development of several tumors. For example, FANCI is associated
with susceptibility to familial prostate cancer (16). It has also been used as a novel
marker for hepatitis B virus-associated hepatocellular carcinoma
(17). In lung adenocarcinoma
(LUAD), it co-operates with inosine monophosphate dehydrogenase
type II to promote tumor growth (18). However, the understanding of the
role of FANCI in tumor development and progression and the
corresponding mechanism involved is still very limited, especially
in the case of NSCLC.
Ubiquitin-conjugating enzyme E2T (UBE2T) is an E2
ubiquitin ligase involved in the FA pathway. It binds to FANCL and
facilitates the monoubiquitination of FANCD2/FANCI to promote DNA
interstrand cross-link (ICL) repair (19,20).
Increasing evidence demonstrates that UBE2T is involved in multiple
cancers. It is reported to decrease BRCA1 expression and promote
breast cancer progression (21).
It has been identified as a putative biomarker of bladder cancer
(22). In addition, UBE2T-mediated
monoubiquitination of H2AX induces hepatocellular carcinoma
radioresistance by activating checkpoint kinase 1 (23). Based on the cancer genome atlas
database, UBE2T was found to be associated with a poor prognosis in
NSCLC (24). It also promotes
epithelial-mesenchymal transition (EMT) and accelerates NSCLC cell
proliferation, migration and invasion (25). In the EMT, epithelial cells gain
migratory and invasive properties, owing to the loss of apico-basal
polarity and cell-cell adhesion, to become mesenchymal stem cells.
EMT is closely related to tumors, depending on the biological
processes of wound healing, cell migration and proliferation
(26–28). Although the aforementioned studies
have found that UBE2T is associated with NSCLC, the role of UBE2T
in the carcinogenesis of NSCLC is yet to be explored in depth.
In the current study, FANCI was found to be
overexpressed and to serve a critical role in NSCLC progression by
promoting EMT. The pathogenesis mechanism involved the
UBE2T-mediated stabilization and monoubiquitination of FANCI.
Materials and methods
Sample collection
A total of 32 pairs of fresh NSCLC and adjacent (~5
cm) non-cancerous lung tissue samples were collected from patients
(all >18 years old) who underwent surgery in Fujian Provincial
Hospital between January 2015 and January 2018. The clinical
information (shown in Table I) of
all the patients was collected by the informed consent. This study
was approved by the Medical Ethics Committee of Fujian Provincial
Hospital (approval no. K2018-015-11) and was conducted in
accordance with the Declaration of Helsinki. Written informed
consent was obtained from all participants.
 | Table I.Correlation of lncRNA FANCI
expression with clinical variables in NSCLC patients. |
Table I.
Correlation of lncRNA FANCI
expression with clinical variables in NSCLC patients.
|
|
| FANCI
expression |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
feature | n | High | Low | P-value |
|---|
| Sex |
|
|
|
|
|
Male | 22 | 10 | 12 | 0.7043 |
|
Female | 10 | 6 | 4 |
|
| Age (years) |
|
|
|
|
|
<60 | 19 | 11 | 8 | 0.4725 |
|
≥60 | 13 | 5 | 8 |
|
| Tumor size
(cm) |
|
|
|
|
|
<3 | 17 | 5 | 12 | 0.032 |
| ≥3 | 15 | 11 | 4 |
|
| Lymphatic
metastasis |
|
|
|
|
| N0 | 18 | 4 | 14 | 0.001 |
|
N1-N3 | 14 | 12 | 2 |
|
| Distant
metastasis |
|
|
|
|
| M0 | 17 | 5 | 12 | 0.032 |
| M1 | 15 | 11 | 4 |
|
Cell culture and transfection
A total of five NSCLC cell lines (H1650, H1975,
HCC827, A549 and H1299) and a human normal epithelial (HBE) cell
line were purchased from the Cell Bank of Type Culture Collection
of the Chinese Academy of Sciences. RPMI-1640 (cat. no. 11875093;
Thermo Fisher Scientific, Inc.) medium containing 10% fetal bovine
serum (cat. no. A3160901; Gibco; Thermo Fisher Scientific, Inc.)
was used to culture cells at 37°C with 5% CO2 in a
humidified chamber and were used for transfection when they reached
60–70% confluence. FANCI-targeting short hairpin (sh)RNA
(ATGTAAGCTCGGAGCTAATAT) and scrambled negative control (NC1
(sh-FANCI); ACGUGACACGUUGGAGAAT), sh-UBE2T (TTGTCTGGATGTTCTCAAATT)
and negative control (NC2 (sh-UBE2T); UUCUCCGAACGUGUCACGUTT), as
well as the pEX-UBE2T and pEX-WDR48 overexpression and the empty
vectors (pEX-1) were synthesized by Shanghai GenePharma Co., Ltd.
and 1 µg/µl was used for transfection with
Lipofectamine® 3000 (cat. no. L3000015; Invitrogen;
Thermo Fisher Scientific, Inc.) at 37°C for 24 h, according to the
manufacturer's protocol. Subsequent experiments were performed 24 h
after transfection.
Reverse transcription-quantitative
(RT-q) PCR
Total RNA was isolated from NSCLC tissues and
cultured cells (at 80% density) using TRIzol® reagent
(cat. no. 15596018; Thermo Fisher Scientific, Inc.) Nanodrop 2000
ultramicro spectrophotometer was applied for RNA purity and
quantification. The cDNA Synthesis SuperMix for qPCR (cat. no.
11141ES10; Shanghai Yeasen Biotechnology Co., Ltd.) was used to
synthesize cDNA according to the manufacturer's instructions. For
RT-qPCR, SYBR Premix Ex Taq II (cat. no. RR420A; Takara
Biotechnology Co., Ltd.) was used. RT-qPCR was conducted using a
Roche Light Cycler 480 system (Roche Diagnostics, GmbH), according
to the manufacturer's protocol. The following thermocycling
conditions were used for qPCR: initial denaturation at 95°C for 5
min; followed by 40 cycles at 95°C for 10 sec of denaturation, 60°C
for 20 sec of annealing and elongation; and 72°C for 20 sec of
final extension. The 2−ΔΔCq method was used to calculate
the relative expression of FANCI and UBE2T (29). β-Actin served as an internal
control. Primer sequences used in this study are listed in Table II.
 | Table II.Primer sequences (5′-3′) for
reverse-transcription quantitative PCR. |
Table II.
Primer sequences (5′-3′) for
reverse-transcription quantitative PCR.
| Gene | Primer sequences
(5′-3′) |
|---|
| FANCI F |
CCACCTTTGGTCTATCAGCTTC |
| FANCI R |
CAACATCCAATAGCTCGTCACC |
| UBE2T F |
TTGATTCTGCTGGAAGGATTTG |
| UBE2T R |
CAGTTGCGATGTTGAGGGAT |
| β-actin F |
AGGGGCCGGACTCGTCATACT |
| β-actin R |
GGCGGCACCACCATGTACCCT |
Western blotting (WB)
Precooled RIPA buffer (cat. no. P0013B; Beyotime
Institute of Biotechnology) was used to extract total protein from
cells and tissues. The BCA Protein Assay kit (cat. no. P0012S;
Beyotime Institute of Biotechnology) was used to determine protein
concentrations. Proteins (60 µg per lane) were separated on 10%
SDS-PAGE. Subsequently, the proteins were transferred onto
polyvinylidene fluoride membranes (cat. no. 32031602;
MilliporeSigma). Then, the membranes were blocked with 5% fat-free
milk at room temperature for 2 h. Following blocking, the membranes
were incubated with primary antibodies (1:1,000, anti-FANCI, cat.
no. ab74332; 1:1,000, anti-FANCD2, cat. no. ab108928; cat. no.
Abcam) 1:1,000, anti-UBE2T, cat. no. ab179802; 1:1,000, anti-WDR48,
cat. no. ab230645; 1:1,000, anti-E-cadherin, cat. no. ab40772;
1:1,000, anti-N-cadherin, cat. no. ab76011; and 1:1,000,
anti-Vimentin, cat. no. ab92547) purchased from Abcam at 4°C
overnight, followed by incubation with corresponding secondary
antibodies for 1 h at room temperature. β-actin (1:2,000, cat. no.
ab8226; Abcam) was used as an internal reference protein. The
membranes were visualized using ECL solution (E411-04, Vazyme
Biotech Co., Ltd.). Image Lab (version 3.0; Bio-Rad Laboratories,
Inc.) was used for densitometry.
Cell proliferation
Cell proliferation was detected using a Cell
counting kit-8 (CCK-8) assay (cat. no. CK04; Dojindo Laboratories,
Inc.). Cells (~4×103) were seeded into each well of a
96-well plate and maintained for the indicated times. Next, 10 µl
of CCK-8 reagent was added to each well for another 2 h of
incubation. Finally, the absorbance was measured at 450 nm.
Cell migration
Cell migration was tested using Transwell and wound
healing assays. For the Transwell assay, ~5×105 cells,
mixed in 200 µl basal medium, were added to the upper and lower
chambers of the Transwell. They were then supplemented with 500 µl
medium. After incubation for 24 h at 37°C, cells were fixed using
4% paraformaldehyde for 30 min, stained with 0.1% crystal violet
for 30 min at room temperature (cat. no. C0121; Beyotime Institute
of Biotechnology) and then observed under a light microscope
(Olympus Corporation). For the scratch assay (wound healing assay),
cells were first seeded into a 6-well plate and incubated for 24 h
at 37°C using RPMI-1640 (without FBS). Then, a scratch was created
with a 10 µl pipette tip in a cell monolayer. The images were
captured upon scratching and after 24 h of scratching.
Flow cytometry
Cell cycle and apoptosis were determined via flow
cytometry using the Annexin V-FITC/PE cell apoptosis and cell cycle
detection kit (cat. no. C1052; Beyotime Institute of Biotechnology)
at room temperature for 30 min according to the manufacturer's
protocols (30). The cell
apoptosis percentage was calculated as Q2+Q4. A flow cytometer
(BeamCyte; BEAMDIAG) was used for the flow cytometry. The flow
cytometric data were then analyzed with CytoSYS 1.0 software
(BEAMDIAG).
Immunoprecipitation (IP)
IP was performed using the Crosslink Magnetic
IP/Co-IP kit (cat. no. 88804; Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. Briefly, after cell
lysis, 10–20 µl of lysis solution was collected as an input for
further detection. The rest of the cell lysate was treated with an
anti-UBE2T antibody (cat. no. ab179802; 1:1,000; Abcam) at 4°C for
12–16 h, Then, protein A/G magnetic beads (cat. no. HY-K0202;
MedChemExpress) were added to the lysate. After incubation of ~4 h,
they were collected and washed thrice to obtain the co-precipitated
proteins. Subsequently, WB was conducted with anti-FANCI (1:200;
cat. no. ab74332; cat. no. Abcam) and anti-FANCD2 (1:200; cat. no.
ab108928; Abcam) antibodies, as aforementioned.
Mouse tumor xenografts
BALB/c nude mice (n=12; age, 6 weeks; female) were
purchased from GemPharmatech Co. Ltd. A549/sh-NC cells or
A549/sh-FANCI cells (~5×106) were injected into the mice
in the right flanks subcutaneously (n=6 per group). Tumor volume
was recorded every week. The mice were sacrificed by using
pentobarbital sodium (200 mg/kg) via injection into the caudal
vein. Mortality was confirmed by the stopping of the heart and
breathing rate (5 min), as well as the disappearance of the foot
withdrawal reflex. The tumor tissues were collected for subsequent
experiments. The xenograft mice in the present study survived until
to sacrifice (five weeks later). Appropriate humane end points were
set, including the tumor burden should be <10% of body weight;
tumors should be ≥20 mm; tumor rapid growth causing ulceration,
necrosis or infection, interference with eating or ability to walk.
The Animal Care and Use Committee of the Fujian Provincial Hospital
(approval no. A2018-016-12) approved the in vivo
experiments.
Immunohistochemistry (IHC)
IHC staining was performed on paraffin-embedded
mouse tissue sections. The sections were treated with primary
anti-FANCI (1:1,000; cat. no. ab74332; Abcam), anti-Ki67 (1:1,000;
cat. no. ab16667; Abcam) and anti-cleaved-Caspase-3 (1:1,000; cat.
no. 9661; CST) antibodies overnight at 4°C. After washing with PBS,
samples were incubated with corresponding secondary antibodies for
30 min at 37°C and DAPI for 10 min at room temperature. By
comparing the entire tissue area at ×10 magnification, the positive
staining score was defined as 0, 1, 2, 3 and 4 for 0, 1–25, 26–50,
51–75 and 76–100% staining, respectively. If the staining score was
≥ 3, the protein was considered to be highly expressed.
Statistical analysis
The data were analyzed using GraphPad Prism 6.01
software (GraphPad, USA). The differences between NSCLC tumor
tissues and paired adjacent tissues were assessed using paired
Student's t-test. The differences between other two groups were
assessed using unpaired Student's t-test and differences between
more than two groups were assessed using one-way ANOVA with a
Tukey's post hoc test. P<0.05 was considered to indicate a
statistically significant difference.
Results
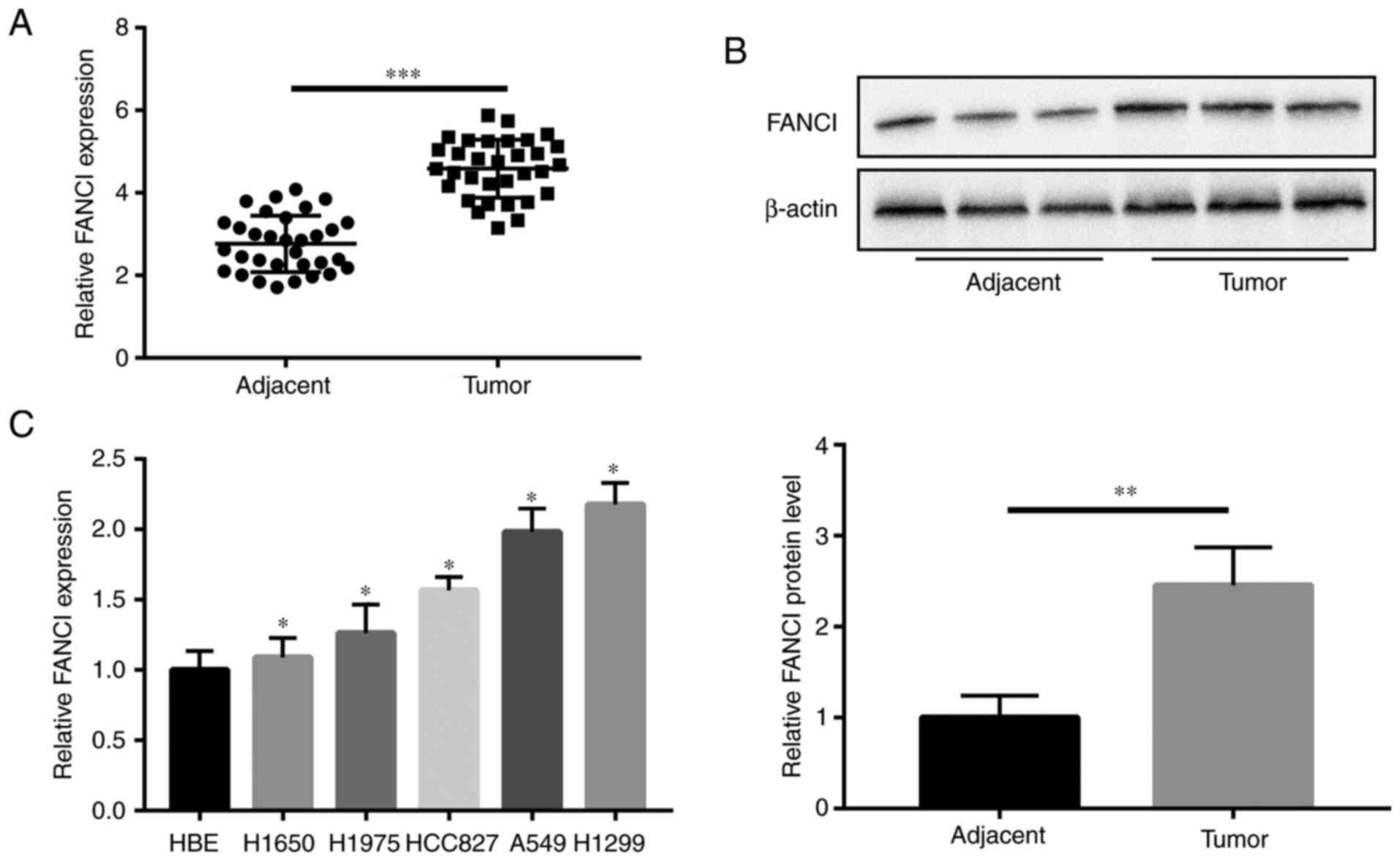
FANCI is highly expressed in NSCLC
tissues and cell lines
Compared with the expression of FANCI in paired
adjacent tissues, all 32 NSCLC tumor tissues showed higher
expression of FANCI, as per results of RT-qPCR (Fig. 1A). As shown in Table I, significant correlations were
found between high levels of FANCI expression and tumor size
(P=0.032), lymphatic metastasis (P=0.001) and distant metastasis
(P=0.032). WB revealed the upregulation of FANCI in the tumors
(Fig. 1B), as well as in the NSCLC
cell lines (H1650, H1975, HCC827, A549 and H1299), compared with
its expression in HBE cells. Among all NSCLC cell lines, A549 and
H1299 cells had the highest expression of FANCI (Fig. 1C). Therefore, H1299 and A549 cells
were chosen for subsequent experiments.
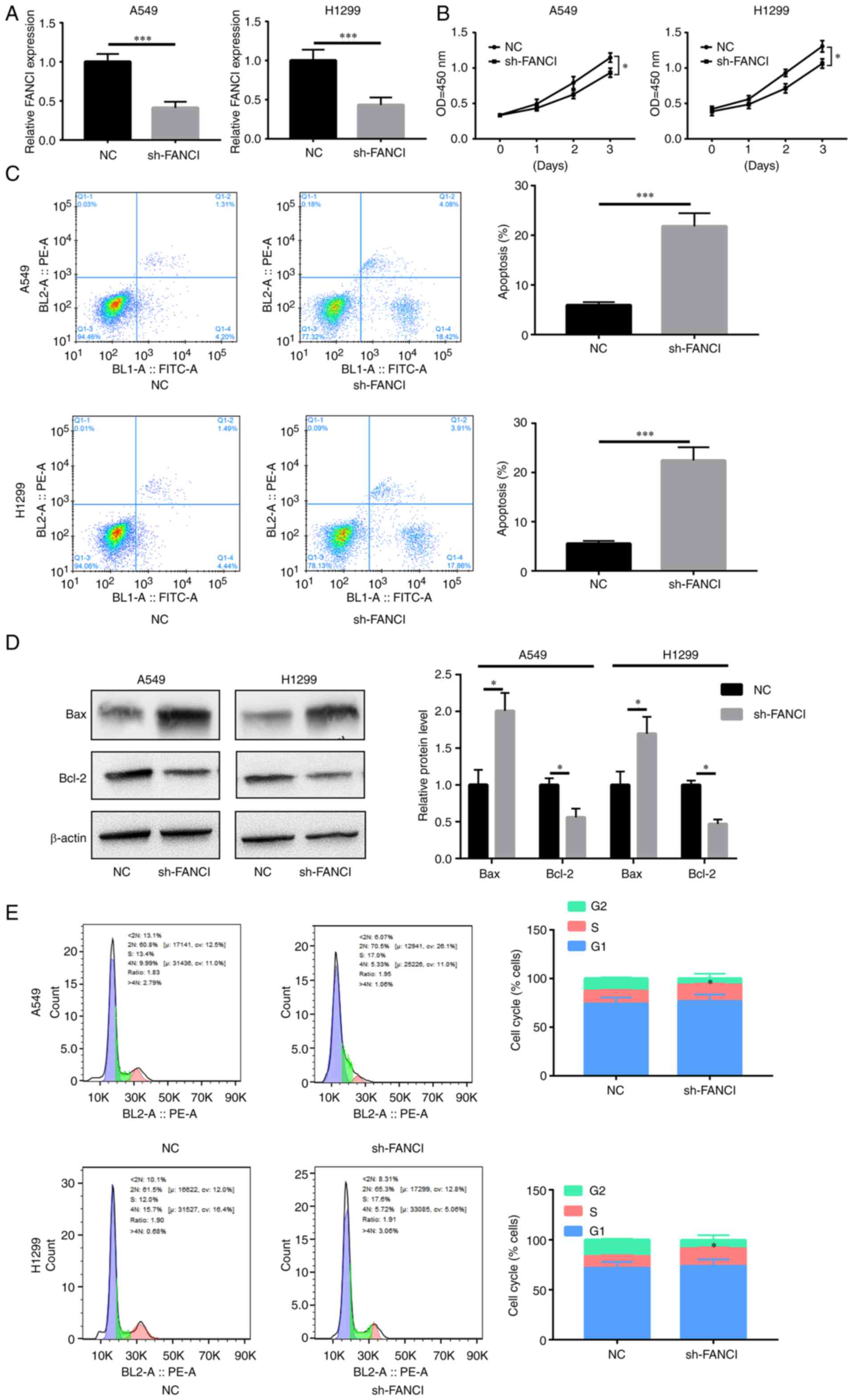
Downregulation of FANCI inhibits NSCLC
cell proliferation
FANCI was knocked down in A549 and H1299 cells to
evaluate its role in NSCLC. RT-qPCR showed that sh-FANCI
effectively decreased FANCI expression (Fig. 2A). The results of the CCK-8 assay
indicated that FANCI knockdown decreased proliferation of cancer
cells, compared with the control group (Fig. 2B). Flow cytometry analysis revealed
an elevated rate of apoptosis upon FANCI knockdown (Fig. 2C). The level of pro-apoptotic
protein Bax increased, while that of anti-apoptotic protein Bcl-2
decreased (Fig. 2D). Furthermore,
the cell cycle was blocked upon gene silencing (Fig. 2E). The above data showed that
knockdown of FANCI abated cell proliferation by promoting apoptosis
and cell cycle arrest in NSCLC cells.
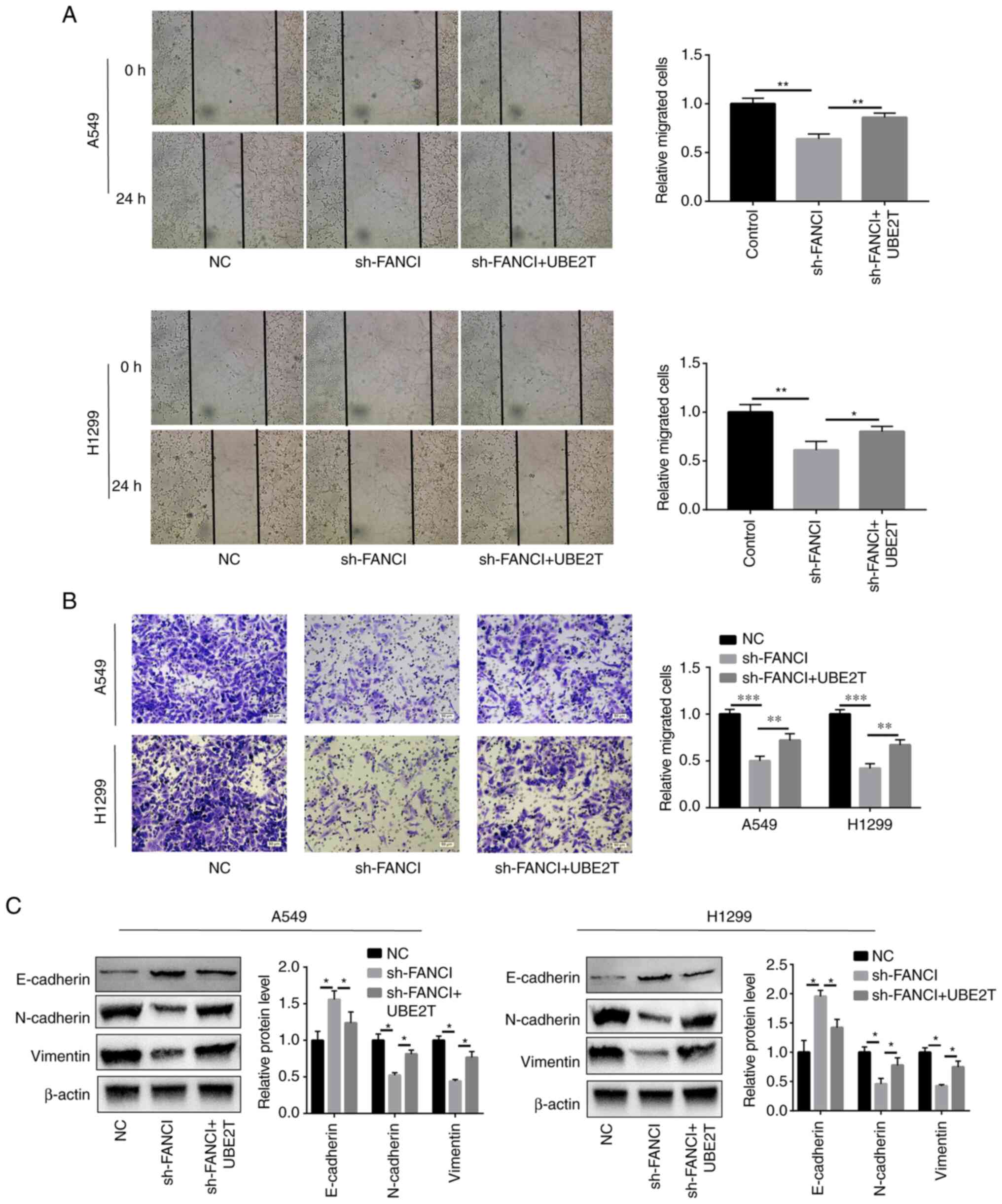
FANCI regulates migration and EMT in
NSCLC cells
Cell migration and invasion were attenuated,
compared with those of the scrambled NC group, when FANCI
expression was downregulated (Fig. 3A
and B). Furthermore, to determine the effect of FANCI on EMT,
the EMT markers including cadherin (E-cadherin and N-cadherin) and
vimentin were evaluated in A549 and H1299 cells. As the WB results
showed, when FANCI was knocked down, the E-cadherin expression
level was raised, but that of N-cadherin and vimentin was decreased
(Fig. 3C), indicating FANCI could
regulate the expression of cadherin. The above findings
demonstrated that FANCI affects EMT in NSCLC by regulating the
expression of cadherin and vimentin.
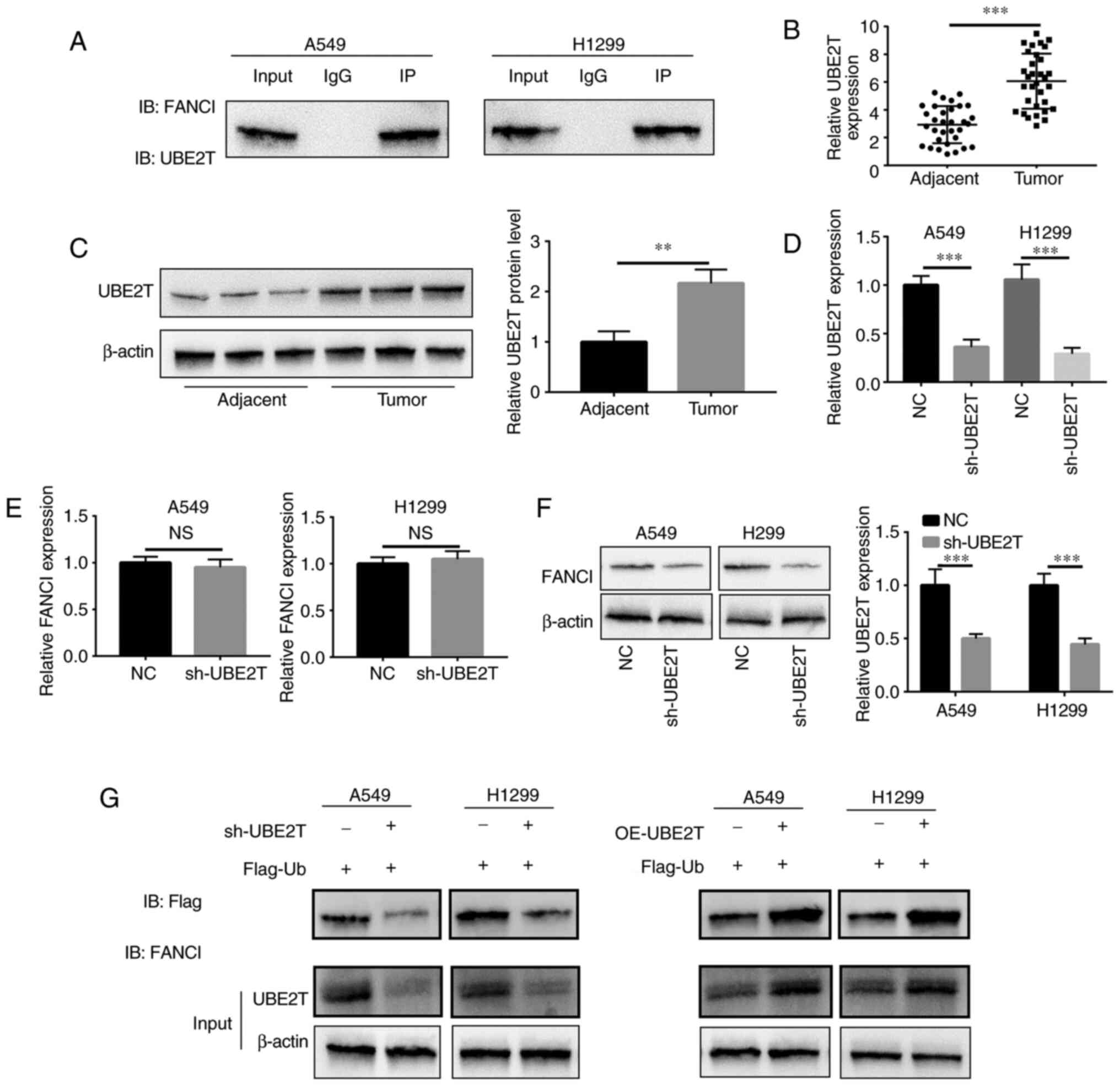
UBE2T contributes to
monoubiquitination of FANCI in NSCLC
UBE2T induced monoubiquitination of FANCI to
activate a downstream pathway. IP revealed that UBE2T bound to
FANCI in A549 and H1299 cells (Fig.
4A). In NSCLC, the mRNA and protein expression of UBE2T in
tumor tissues was higher compared with that in adjacent tissues
(Fig. 4B and C). sh-UBE2T-mediated
knockdown of UBE2Twas confirmed via RT-qPCR (Fig. 4D). Upon downregulation
ofUBE2Texpression, FANCI mRNA level did not change significantly
(Fig. 4E), but its protein level
decreased (Fig. 4F), suggesting
that UBE2T might regulate the stability of FANCI by binding to it.
To explore the monoubiquitination of FANCI by UBE2T, co-IP was
conducted and ubiquitin content was examined. The results showed
that the ubiquitin content in A549/sh-UBE2T and
H1299/sh-UBE2T cells was downregulated, compared with ubiquitin
content in A549/NC and H1299/NC cells (Fig. 4G). By contrast, the ubiquitin
content was higher inA549 and H1299/UBE2Tcells than in A549 and
H1299/Vector cells (Fig. 4G),
indicating that UBE2T mediated monoubiquitination of FANCI in
NSCLC. As FANCI is a paralog of FANCD2, the monoubiquitination of
FANCD2 by UBE2T was also detected. It was found that the knockdown
of UBE2T decreased the monoubiquitination of FANCD2, while
the overexpression of UBE2T increased it (Fig. S1). However, the knockdown of
FANCD2 did not have any effect on cell growth (Fig. S2).
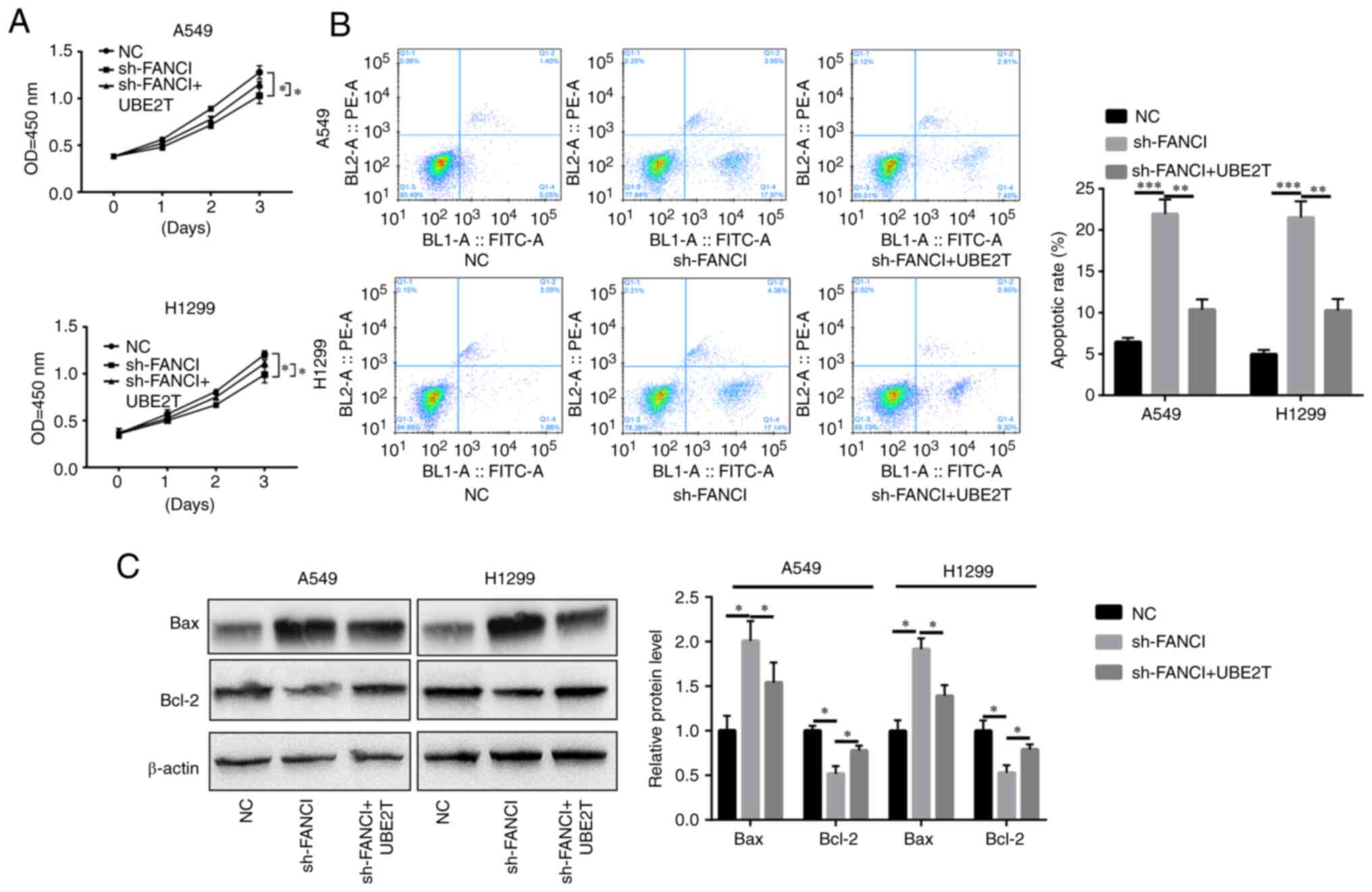
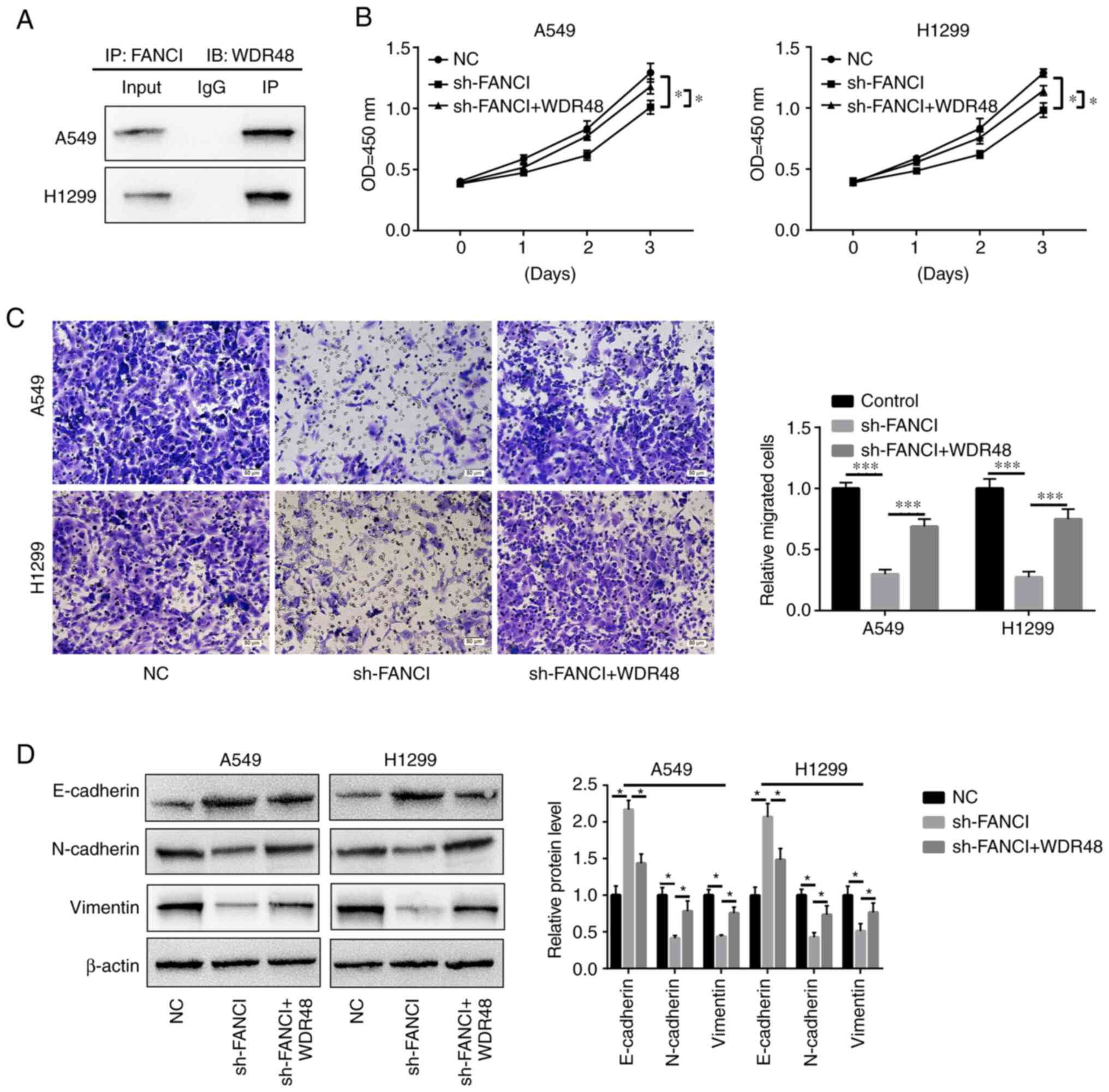
UBE2T mitigates the inhibitory effects
of FANCI downregulation
To explore whether UBE2T could alleviate the effects
of FANCI knockdown in NSCLC cells, UBE2T was overexpressed in
FANCI-knockdown cells. It was found that overexpression of
UBE2T promoted the proliferation of, and repressed apoptosis in,
A549 and H1299 cells when FANCI was knocked down (Fig. 5A-C). Furthermore, cell migration
and invasion were also enhanced upon upregulation of UBE2T
expression (Fig. 6A and B). The
levels of EMT markers cadherin and vimentin were quantified using
WB. Although FANCI silencing increased E-cadherin expression level
and decreased N-cadherin and vimentin expression levels, the
overexpression of UBE2T partly reversed this effect (Fig. 6C). The above results revealed that
the functions of FANCI in NSCLC are regulated by UBE2T.
FANCI functions via binding to
WDR48
Through in silico analysis (String:
http://cn.string-db.org/), it was found that
WDR48, which is related to EMT and cell growth (31), might be a downstream factor of
FANCI. IP assay confirmed that FANCI could bind to WDR48 in A549
and H1299 cells (Fig. 7A). The
upregulation of WDR48 reversed the inhibitory effects of FANCI
silencing on cell proliferation and migration (Fig. 7B and C) and EMT. It downregulated
E-cadherin expression and upregulated that of N-cadherin and
vimentin (Fig. 7D). The above
findings suggest that FANCI might function by binding to WDR48.
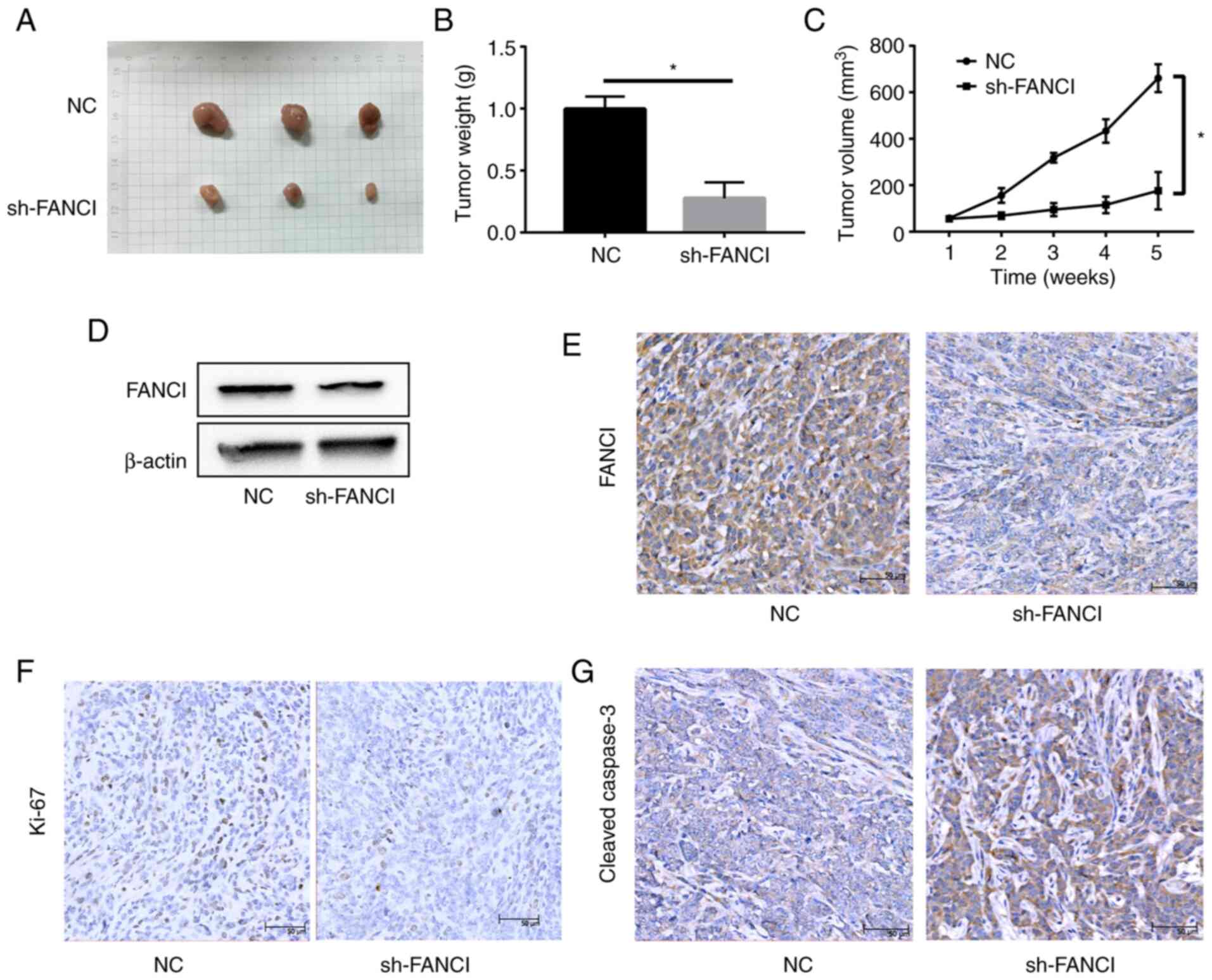
Knockdown of FANCI inhibits tumor
growth in vivo
A549/sh-NC and A549/sh-FANCI cells were injected
subcutaneously into nude mice to establish tumor xenografts. The
tumor size and weight significantly reduced in the sh-FANCI group,
compared with the sh-NC group. The tumor volume showed a similar
pattern (Fig. 8A-C). WB and IHC
revealed low FANCI expression in the sh-FANCI group (Fig. 8D and E). In addition, the
expression of proliferation marker Ki67 was lower, whereas that of
cleaved-caspase 3 was higher, in the sh-FANCI group (Fig. 8F and G). These data suggested that
knockdown of FANCI significantly inhibited tumor growth in
vivo.
Discussion
Despite remarkable efforts to treat NSCLC, such as
surgery, chemotherapy, radiotherapy and targeted therapy, the
5-year overall survival rate remains unsatisfactory (32,33).
Elucidating the underlying mechanisms of NSCLC tumorigenesis
requires intensive and consistent exploration. An ICL refers to the
covalent bond between complementary bases of double-stranded DNA,
which is one of the most toxic types of DNA damage (34). The FA pathway is responsible for
repairing ICLs in the S phase and maintaining genomic stability
(35). FANCL, an E3 ubiquitin
ligase, serves an irreplaceable role in FA pathway activation and
ICL repair. When ICLs occur on DNA, the FA core complex is
recruited to the stagnated replication fork. FANCL, the E3
ubiquitin ligase subunit in the FA-core complex, interacts
specifically with UBE2T and then, FANCL and UBE2T promote the
monoubiquitination of the FANCI-FANCD2 heterodimer. The
ubiquitinated FANCI-FANCD2 complex recruits downstream
endonucleases to cut DNA. Then, the downstream proteins undergo
cross-injury synthesis and homologous recombination repair to
finally complete the repair of ICLs (36). FANCI was also revealed as a
biomarker for the poor prognosis of LUAD (18); however, a previous study found a
tumor suppressive role of FANCI, wherein it acted as a negative
factor for the Akt pathway by regulating PHLPP phosphatases
(37). Therefore, the role of
FANCI in tumors remains unclear and further studies are needed to
explore it properly. The present study discovered that FANCI is
overexpressed in NSCLC tumor tissues. Knockdown of FANCI inhibited
EMT, proliferation, migration, invasion and tumor growth in NSCLC
cells. Although FANCI is a paralog of FANCD2 and UBE2T regulates
the monoubiquitination of FANCD2 in NSCLC cells, we found that
downregulation of FANCD2 did not influence cell growth. Based on
these results, we propose that FANCI, but not FANCD2, functions as
an oncogene in NSCLC. Thus, FANCI is a novel potential therapeutic
target for NSCLC.
UBE2T is an E2 enzyme that is widely dysregulated in
numerous cancers and functions as an oncoprotein (38,39).
Recently, UBE2T was reported to promote β-catenin nuclear
translocation in HCC through the MAPK/ERK axis (40). In addition, UBE2T promotes the
progression of LUAD by regulating autophagy through the
p53/AMPK/mTOR signaling pathway (41). As a member of the E2 family, UBE2T
enhances DNA crosslinking-induced damage repair by
monoubiquitinating the FANCI-FANCD2 complex (42). Some studies have indicated that
FANCD2 is a preferred substrate for ubiquitination in the
FANCI-FANCD2 complex (15,43), but it becomes a poor substrate for
ubiquitination when FANCI is absent (44). Compared with FANCD2, free FANCI is
more efficiently ubiquitinated by the FA-core complex (44,45).
It has also been reported that the abundance of FANCI in U2OS cells
is nearly 10 times more than that of FANCD2, indicating that FANCI
may also be ubiquitinated alone in cells to a certain degree
(46). The present study found
that UBE2T expression was upregulated in NSCLC tumor tissues.
Therefore, in terms of mechanism, it was hypothesized that FANCI
might be regulated by UBE2T during the development and progression
of NSCLC. According to the co-IP and WB assays, UBE2T interacted
with FANCI and stabilized the FANCI level. Ubiquitin-like protein 5
(UBL5) has also been reported to bind FANCI and promote its
stability (47). The present study
discovered that overexpression of UBE2T increased the
monoubiquitination level of FANCI in NSCLC cells. The foregoing
results suggested that the function of FANCI in NSCLC was probably
modulated by UBE2T-mediated monoubiquitination. However, it is
still not well known how the FA-core complex and UBE2T specifically
regulate FANCI monoubiquitination and this requires further
research. In addition, the rescue experiments showed that
overexpression of UBE2T partly reversed the FANCI knockdown-induced
inhibition of EMT, cell growth, migration and invasion, indicating
that UBE2T regulates the function of FANCI in NSCLC.
WDR48 is critical for EMT. The inhibition of
endogenous WDR48 expression is reported to significantly decrease
TGF-β-induced EMT, migration and invasion in tumor cells (47). The present study predicted the
binding of FANCI and WDR48 using in silico analysis and
confirmed it via IP assay. Furthermore, rescue experiments showed
that the upregulation of WDR48 also reversed the FANCI
knockdown-induced inhibition of EMT and cell growth. These findings
indicate the probable role of WDR48 in FANCI function in NSCLC.
In conclusion, the present study revealed the
oncogenic role of FANCI in promoting cell growth and EMT in NSCLC.
This function of FANCI is mediated by WDR48 and regulated by
UBE2T-induced monoubiquitination of FANCI. In the present study,
FANCI emerged as a potential target for NSCLC therapy that may also
serve as a biomarker for predicting its poor prognosis.
Supplementary Material
Supporting Data
Acknowledgments
Not applicable.
Funding
The present study was funded by the National Natural Science
Foundation of China (grant no. 82074189), Fujian Natural Science
Foundation (grant no. 2021J01380) and Science and Technology
Planning Project of Fujian Provincial Health Commission (grant no.
2021zylc31).
Availability of data and materials
The datasets in the present study are available from
the corresponding author on reasonable request.
Authors' contributions
JZ and XL designed the study. JWa and JZ performed
the experiments. XL and JW prepared the figures. JZ and XL confirm
the authenticity of all the raw data. JWu, JH and ZL contributed to
the drafting of the manuscript and the final approval of the
version to be published. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Medical Ethics
Committee of Fujian Provincial Hospital (approval no. K2018-015-11)
and written informed consent was obtained from all the
participants.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CCK-8
|
Cell Counting Kit-8
|
|
IP
|
immunoprecipitation
|
|
EMT
|
epithelial-mesenchymal transition
|
|
FANCD2
|
Fanconi anemia complementation group
D2
|
|
FANCI
|
Fanconi anemia complementation group
I
|
|
FANCL
|
Fanconi anemia complementation group
L
|
|
HBE
|
human bronchial epithelial
|
|
ICL
|
interstrand cross-link
|
|
IP
|
immunoprecipitation
|
|
LUAD
|
lung adenocarcinoma
|
|
NSCLC
|
non-small cell lung cancer
|
|
RT-qPCR
|
reverse-transcription quantitative
PCR
|
|
shRNA
|
short hairpin RNA
|
|
UBE2T
|
ubiquitin-conjugating enzyme E2T
|
|
WB
|
western blotting
|
|
WDR48
|
WD repeat domain 48
|
References
|
1
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer statistics, 2022. CA Cancer J Clin. 72:7–33. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hiley CT, Le Quesne J, Santis G, Sharpe R,
de Castro DG, Middleton G and Swanton C: Challenges in molecular
testing in non-small-cell lung cancer patients with advanced
disease. Lancet. 388:1002–1011. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Del Re M, Crucitta S, Gianfilippo G,
Passaro A, Petrini I, Restante G, Michelucci A, Fogli S, de Marinis
F, Porta C, et al: Understanding the mechanisms of resistance in
EGFR-positive NSCLC: From tissue to liquid biopsy to guide
treatment strategy. Int J Mol Sci. 20:39512019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Doval DC, Desai CJ and Sahoo TP:
Molecularly targeted therapies in non-small cell lung cancer: The
evolving role of tyrosine kinase inhibitors. Indian J Cancer. 56
(Suppl):S23–S30. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kelaidi C, Makis A, Petrikkos L, Antoniadi
K, Selenti N, Tzotzola V, Ioannidou ED, Tsitsikas K, Kitra V,
Kalpini-Mavrou A, et al: Bone marrow failure in Fanconi anemia:
Clinical and genetic spectrum in a cohort of 20 pediatric patients.
J Pediatr Hematol Oncol. 41:612–617. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Engel NW, Schliffke S, Schüller U, Frenzel
C, Bokemeyer C, Kubisch C and Lessel D: Fatal myelotoxicity
following palliative chemotherapy with cisplatin and gemcitabine in
a patient with stage IV cholangiocarcinoma linked to post mortem
diagnosis of Fanconi anemia. Front Oncol. 9:4202019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Katsuki Y and Takata M: Defects in
homologous recombination repair behind the human diseases: FA and
HBOC. Endocr Relat Cancer. 23:T19–T37. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dong H, Nebert DW, Bruford EA, Thompson
DC, Joenje H and Vasiliou V: Update of the human and mouse Fanconi
anemia genes. Hum Genomics. 9:322015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kottemann MC and Smogorzewska A: Fanconi
anaemia and the repair of Watson and Crick DNA crosslinks. Nature.
493:356–363. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Joo W, Xu G, Persky NS, Smogorzewska A,
Rudge DG, Buzovetsky O, Elledge SJ and Pavletich NP: Structure of
the FANCI-FANCD2 complex: Insights into the Fanconi anemia DNA
repair pathway. Science. 333:312–316. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sondalle SB, Longerich S, Ogawa LM, Sung P
and Baserga SJ: Fanconi anemia protein FANCI functions in ribosome
biogenesis. Proc Natl Acad Sci USA. 116:2561–2570. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Meetei AR, de Winter JP, Medhurst AL,
Wallisch M, Waisfisz Q, van de Vrugt HJ, Oostra AB, Yan Z, Ling C,
Bishop CE, et al: A novel ubiquitin ligase is deficient in Fanconi
anemia. Nat Genet. 35:165–170. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Smogorzewska A, Matsuoka S, Vinciguerra P,
McDonald ER III, Hurov KE, Luo J, Ballif BA, Gygi SP, Hofmann K,
D'Andrea AD and Elledge SJ: Identification of the FANCI protein, a
monoubiquitinated FANCD2 paralog required for DNA repair. Cell.
129:289–301. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Paulo P, Maia S, Pinto C, Pinto P,
Monteiro A, Peixoto A and Teixeira MR: Targeted next generation
sequencing identifies functionally deleterious germline mutations
in novel genes in early-onset/familial prostate cancer. PLoS Genet.
14:e10073552018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xie S, Jiang X, Zhang J, Xie S, Hua Y,
Wang R and Yang Y: Identification of significant gene and pathways
involved in HBV-related hepatocellular carcinoma by bioinformatics
analysis. PeerJ. 7:e74082019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zheng P and Li L: FANCI cooperates with
IMPDH2 to promote lung adenocarcinoma tumor growth via a
MEK/ERK/MMPs pathway. Onco Targets Ther. 13:451–463. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Machida YJ, Machida Y, Chen Y, Gurtan AM,
Kupfer GM, D'Andrea AD and Dutta A: UBE2T is the E2 in the Fanconi
anemia pathway and undergoes negative autoregulation. Mol Cell.
23:589–596. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Longerich S, San Filippo J, Liu D and Sung
P: FANCI binds branched DNA and is monoubiquitinated by
UBE2T-FANCL. J Biol Chem. 284:23182–23186. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ueki T, Park JH, Nishidate T, Kijima K,
Hirata K, Nakamura Y and Katagiri T: Ubiquitination and
downregulation of BRCA1 by ubiquitin-conjugating enzyme E2T
overexpression in human breast cancer cells. Cancer Res.
69:8752–8760. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhu X, Li T, Niu X, Chen L and Ge C:
Identification of UBE2T as an independent prognostic biomarker for
gallbladder cancer. Oncol Lett. 20:442020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sun J, Zhu Z, Li W, Shen M, Cao C, Sun Q,
Guo Z, Liu L and Wu D: UBE2T-regulated H2AX monoubiquitination
induces hepatocellular carcinoma radioresistance by facilitating
CHK1 activation. J Exp Clin Cancer Res. 39:2222020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wu ZH, Zhang YJ and Sun HY: High ubiquitin
conjugating enzyme E2 T mRNA expression and its prognostic
significance in lung adenocarcinoma: A study based on the TCGA
database. Medicine (Baltimore). 99:e185432020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yin H, Wang X, Zhang X, Zeng Y, Xu Q, Wang
W, Zhou F and Zhou Y: UBE2T promotes radiation resistance in
non-small cell lung cancer via inducing epithelial-mesenchymal
transition and the ubiquitination-mediated FOXO1 degradation.
Cancer Lett. 494:121–131. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hay ED: An overview of
epithelio-mesenchymal transformation. Acta Anat (Basel). 154:8–20.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Thiery JP: Epithelial-mesenchymal
transitions in development and pathologies. Curr Opin Cell Biol.
15:740–746. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhao L, Sun H, Kong H, Chen Z, Chen B and
Zhou M: The lncrna-TUG1/EZH2 axis promotes pancreatic cancer cell
proliferation, migration and EMT phenotype formation through
sponging Mir-382. Cell Physiol Biochem. 42:2145–2158. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang X, Yin H, Zhang H, Hu J, Lu H, Li C,
Cao M, Yan S and Cai L: NF-κB-driven improvement of EHD1
contributes to erlotinib resistance in EGFR-mutant lung cancers.
Cell Death Dis. 9:4182018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Han D, Wang L, Chen B, Zhao W, Liang Y, Li
Y, Zhang H, Liu Y, Wang X, Chen T, et al: USP1-WDR48 deubiquitinase
complex enhances TGF-β induced epithelial-mesenchymal transition of
TNBC cells via stabilizing TAK1. Cell Cycle. 20:320–331. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cao J, Yuan P, Wang Y, Xu J, Yuan X, Wang
Z, Lv W and Hu J: Survival rates after lobectomy, segmentectomy,
and wedge resection for non-small cell lung cancer. Ann Thorac
Surg. 105:1483–1491. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Giaj-Levra N, Borghetti P, Bruni A,
Ciammella P, Cuccia F, Fozza A, Franceschini D, Scotti V, Vagge S
and Alongi F: Current radiotherapy techniques in NSCLC: Challenges
and potential solutions. Expert Rev Anticancer Ther. 20:387–402.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Rozelle AL, Cheun Y, Vilas CK, Koag MC and
Lee S: DNA interstrand cross-links induced by the major oxidative
adenine lesion 7,8-dihydro-8-oxoadenine. Nat Commun. 12:18972021.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yang Y, Guo T, Liu R, Ke H, Xu W, Zhao S
and Qin Y: FANCL gene mutations in premature ovarian insufficiency.
Hum Mutat. 41:1033–1041. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ceccaldi R, Sarangi P and D'Andrea AD: The
Fanconi anaemia pathway: New players and new functions. Nat Rev Mol
Cell Biol. 17:337–349. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang X, Lu X, Akhter S, Georgescu MM and
Legerski RJ: FANCI is a negative regulator of Akt activation. Cell
Cycle. 15:1134–1143. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang W, Zhang Y, Yang Z, Liu X, Yang P,
Wang J, Hu K, He X, Zhang X and Jing H: High expression of UBE2T
predicts poor prognosis and survival in multiple myeloma. Cancer
Gene Ther. 26:347–355. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liu LP, Yang M, Peng QZ, Li MY, Zhang YS,
Guo YH, Chen Y and Bao SY: UBE2T promotes hepatocellular carcinoma
cell growth via ubiquitination of p53. Biochem Biophys Res Commun.
493:20–27. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lioulia E, Mokos P, Panteris E and Dafou
D: UBE2T promotes β-catenin nuclear translocation in hepatocellular
carcinoma through MAPK/ERK-dependent activation. Mol Oncol.
16:1694–1713. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhu J, Ao H, Liu M, Cao K and Ma J: UBE2T
promotes autophagy via the p53/AMPK/mTOR signaling pathway in lung
adenocarcinoma. J Transl Med. 19:3742021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Nepal M, Che R, Ma C, Zhang J and Fei P:
FANCD2 and DNA damage. Int J Mol Sci. 18:18042017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Sato K, Toda K, Ishiai M, Takata M and
Kurumizaka H: DNA robustly stimulates FANCD2 monoubiquitylation in
the complex with FANCI. Nucleic Acids Res. 40:4553–4561. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang S, Wang R, Peralta C, Yaseen A and
Pavletich NP: Structure of the FA core ubiquitin ligase closing the
ID clamp on DNA. Nat Struct Mol Biol. 28:300–309. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
van Twest S, Murphy VJ, Hodson C, Tan W,
Swuec P, O'Rourke JJ, Heierhorst J, Crismani W and Deans AJ:
Mechanism of ubiquitination and deubiquitination in the Fanconi
anemia pathway. Mol Cell. 65:247–259. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Beck M, Schmidt A, Malmstroem J, Claassen
M, Ori A, Szymborska A, Herzog F, Rinner O, Ellenberg J and
Aebersold R: The quantitative proteome of a human cell line. Mol
Syst Biol. 7:5492011. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Oka Y, Bekker-Jensen S and Mailand N:
Ubiquitin-like protein UBL5 promotes the functional integrity of
the Fanconi anemia pathway. EMBO J. 34:1385–1398. 2015. View Article : Google Scholar : PubMed/NCBI
|