Introduction
Cervical cancer is the fourth most common malignancy
in women (1,2). Although therapeutic methods and
treatment techniques for tumors have improved recently, the
outcomes for patients with cervical cancer remain poor (3). The main reasons for poor outcomes are
recurrence and metastasis, which remain huge challenges in the
treatment of cervical cancer. Therefore, it is necessary to
identify biomarkers for cervical cancer diagnosis and
treatment.
Plasminogen activator urokinase (PLAU), also known
as name urokinase-type plasminogen activator (uPA), exerts multiple
biological effects in various physiological and pathological
processes, such as keratinocyte proliferation (4), air-way inflammation (5) and rheumatoid arthritis (6). Additionally, PLAU is an important
modulator of tumorigenesis and progression. PLAU is overexpressed
in several tumors, including head and neck squamous cell carcinoma
(HNSCC) (7), gastric
adenocarcinoma (8) and breast
cancers (9). PLAU participates in
tumor progression by promoting tumor cell proliferation, cell
migration, invasion, and epithelial-mesenchymal transition
(10–12). Given the oncogenic role of PLAU, it
is considered a potential prognostic marker for tumors (7,13,14).
To date, PLAU has been reported to be involved in cervical cancer
progression. PLAU is upregulated and plays a vital role in the
invasion and metastasis of advanced cervical cancer (15–17).
Therefore, it is necessary to elucidate the mechanisms by which
PLAU expression is regulated.
PLAU is regulated by several factors. Certain miRNAs
have been reported to regulate PLAU expression: miR-23b-3p
upregulates PLAU, thereby affecting HNSCC progression (7); meanwhile, the lncRNA TRPM2-AS
promotes the progression of gastric adenocarcinoma by regulating
PLAU (8). In addition to miRNAs,
transcription factors are important for gene regulation.
Transcription factors are the main elements of transcription, which
is the process of RNA synthesis according to the genomic DNA
sequence (18). Transcription
factors can bind to specific sequences in the promoter of target
genes, thereby inducing the upregulation or downregulation of
target genes (18–20). Several transcription factors
regulate PLAU at the transcriptional level. Specificity protein 1
(Sp1) and Sp3 transcription factors can aid in the transcription of
PLAU (21,22). Transcription factors Ets1 and Ets2
can bind to the enhancer region of PLAU, thereby increasing
transcription (23). However, the
mechanisms underlying PLAU transcription remain unclear.
YinYang1 (YY1), a member of the GLI-Krüppel family
of zinc-finger DNA-binding proteins, is a transcription factor
widely expressed in various tissues. YY1 can activate the
transcription of several target genes, such as FOXE1, TNK2-AS1 and
LINC00466 (24–26). By regulating these target genes,
YY1 participates in various biological functions, including cell
proliferation (27,28), apoptosis (29), invasion and migration (30), radioresistance (31) and drug resistance (32). However, it is unknown whether YY1
is associated with PLAU transcription.
In the current study, the expression of PLAU in
cervical cancer was analyzed and the role of PLAU in cervical
cancer cell proliferation, migration and invasion, as well as the
location of the PLAU core promoter, were determined. An important
transcription factor, YY1, which is involved in PLAU transcription,
was identified.
Materials and methods
Clinical specimens
Cervical cancer tissues (16 pairs) and the matched
adjacent normal tissues were collected at Liaocheng People's
Hospital (Liaocheng, China) from February 2021 to November 2021.
All the cases have not received any treatment before hospitalized
and were diagnosed as cervical cancer by two pathologists. The
present study was approved (approval no. LC2021015) by the Ethics
Committee of Liaocheng People's Hospital (Liaocheng, China).
Written informed consent was provided by all patients.
Cell lines and cell culture
Both HeLa and HT3 cell lines were purchased from the
American Type Culture Collection. The cells were cultured in
Dulbecco's modified Eagle's medium (DMEM) containing 10% FBS (both
from Thermo Fisher Scientific, Inc.) in an incubator with 5%
CO2 at 37°C.
Analyzing the expression of PLAU in
cervical cancer based on UALCAN database
The UALCAN database (http://ualcan.path.uab.edu/) is a comprehensive web
resource for analyzing cancer OMICS data, which can be used for
performing pan-cancer gene expression analysis (33).
RNA extraction
For cervical cancer tissues and matched adjacent
normal tissues, tissues were placed into microtubes containing 1 ml
precooled TRIzol® (Thermo Fisher Scientific, Inc.) and
smashed in a tissue grinding machine. For cells cultured in
six-well plates, after the media were discarded, 1 ml TRIzol
reagent was added to each well. After the tissues or cells were
lysed using TRIzol reagent, the lysates were transferred to new
microtubes. The RNA was extracted by adding 500 µl chloroform to
each tube. The RNA was then separated by adding 200 µl isopropanol.
RNA concentration was measured using an ultramicro
spectrophotometer (Implen GmbH).
Reverse transcription-quantitative PCR
(RT-qPCR)
RNA (2 µg) was reverse-transcribed into cDNA using a
Titan One Tube RT-PCR kit (Roche Diagnostics) following the
manufacturer's instruction. YY1 and PLAU mRNA expression in
cervical cancer tissues and adjacent normal tissues were measured
with TaqMan Real-Time PCR Assays using a BeyoFast™ Probe
qPCR Mix kit (Beyotime Institute of Biotechnology) following the
manufacturer's protocol. The primers and probes used were listed in
Table SI. The thermocycling
conditions of TaqMan Real-Time PCR assays were as follows: 95°C, 10
min; 95°C, 15 sec; and 60°C, 60 sec for 40 cycles. The expression
of YY1, PLAU, MMP-9, E-cadherin, and Cyclin D1 in HeLa cells and
HT3 cells were measured with SYBR Green Real-Time PCR Assays using
a FastStart Universal SYBR Green Master (ROX) kit (Roche
Diagnostics) following the manufacturer's protocol with the
following conditions: 95°C, 10 min; 95°C, 15 sec; 60°C, 60 sec for
40 cycles, and the primers used were listed in Table SII. The expression of target genes
was quantified using the 2−ΔΔCq method, in accordance
with a previous study (34).
PLAU and YY1 knockdown by lentiviral
infection
The lentiviral vectors (Lv)-small hairpin RNA
(shRNA) based on a 3rd generation lentiviral system targeting PLAU
(Lv-shSPP1), Lv-shYY1, and negative control (Lv-shCon) were
purchased from Shanghai GeneChem Co., Ltd. Briefly, 1.5 µg shRNA
vector and 1.5 µg package mix plasmids were transfected into 293T
cells (cat. no. C6008; Beyotime Institute of Biotechnology) with
Lipofectamine 3000® reagent (cat. no. L3000001; Thermo
Fisher Scientific, Inc.) for 24 h at 37°C, and the medium was then
harvested and the virus was extracted by 80,000 ×
g-ultracentrifugation at 4°C for 2 h in a centrifuge (Beckman
Coulter, Inc.). For lentiviral transduction, the cells were seeded
in six-well plates at a density of 50,000 cells/well. The
lentiviral vector was added at a multiplicity of infection of 15,
with 5 µl polybrene (Sigma-Aldrich; Merck KGaA) per well. After 72
h, cells infected with the lentiviral vectors were selected using 1
µg/ml puromycin (Beyotime Institute of Biotechnology). Knockdown
efficiency was evaluated using RT-qPCR.
Transwell assay
Transwell migration assays were performed using
Transwell plates (Corning, Inc.) with 8 µm-wells in the membrane.
For the migration assay, cells in 200 µl DMEM without FBS were
seeded into the upper chamber of the plate at 100,000 cells/well.
In the lower chamber, 500 µl DMEM media with 15% FBS was added.
After 12 h, the cells remaining in the upper chamber were wiped
away and the cells transferred to the lower chamber were stained
with crystal violet (Beyotime) at 25°C for 3 min. Then the stained
cells were observed using a T2R inverted (T2R, Nikon Corporation)
fluorescence microscope at ×200 magnification under brigbtfield
counted in three randomly selected visual fields. The invasion
assay was similar to the transwell migration assay, except that the
upper chambers were pre-coated with 20 µg of ECM gel
(MilliporeSigma).
Cell Counting Kit-8 (CCK-8)
proliferation assay
HeLa and HT3 Cells were seeded in 96-well plate at
5,000 cells per-well and cultured in an incubator with 5%
CO2 at 37°C. After being cultured for 0, 24 and 48 h, 10
µl of CCK-8 reagents (cat. no. GB707; Dojindo, Laboratories, Inc.)
were added into each well, and incubated for 1 h at 37°C. Then the
optical density was measured at 450 nm in a spectrophotometer
(BioTek Instruments, Inc.,).
Construction of PLAU promoter
reporter
The sequence of the PLAU promoter was obtained from
NCBI (https://www.ncbi.nlm.nih.gov/nuccore/NG_011904.1?from=5001&to=11398&report=genbank).
The region of −1500/+18 region (transcription starting site was
considered as +1) was cloned from the genomic DNA of HeLa cells and
inserted into the pGL3-basic vector. The promoter-reporter was
confirmed by Sanger sequencing in an Illumina NextSeq 500
instrument (Illumina, Inc.) at Sangon Biotech Co., Ltd. and named
pGL3-PLAU.
Deletion mutation and site-directed
mutation
In accordance with a previous study (35), the deletion mutation was performed
by self-linking the fragment of pGL3-PLAU with the exception of the
region needing to be deleted. The fragment was obtained using a PCR
procedure (95°C, 1 min; 95°C, 15 sec; 55°C, 30 sec; 72°C, 5 min for
30 cycles; 72°C, 10 min) with pGL3-PLAU as the template. The
forward primer was designed based on the sequence at the downstream
of the deletion region of pGL3-PLAU, and the reverse primer was
designed based on the sequence at the upstream of the deletion
region of pGL3-PLAU. The primers were listed in Table SIII. Subsequently, the PCR
products were extracted using an Agarose Gel Extraction kit (cat.
no. D0056; Beyotime Institute of Biotechnology). Following
incubation with T4 polynucleotide kinase (New England BioLabs,
Inc.) at 37°C for 1 h, the product was self-linked using a T4 DNA
ligase (New England BioLabs, Inc.) at 37°C for 1 h. The deletion
mutants were confirmed by sequencing at Sangon Biotech Co., Ltd.
The primer used for DNA sequencing was:
5′-CTAGCAAAATAGGCTGTCCC-3′.
According to a previous study (35), site-directed mutagenesis was
performed using PCR procedure (95°C, 1 min; 95°C, 15 sec; 55°C, 30
sec; 72°C, 5 min for 30 cycles; 72°C, 10 min) with pGL3-PLAU
(−900/-1100) as the template; and the primers used are listed in
Table SIV. pGL3-PLAU-MTA and
pGL3-PLAU-MTB were constructed based on the pGL3-PLAU as template.
pGL3-PLAU-MTA were constructed based on the pGL3-PLAU-MTA as
template. After digestion with Dpn I-restricted enzyme (New England
BioLabs, Inc.), the product was transferred to the Top 10 bacterial
E. coli cells (Beyotime Institute of Biotechnology) for
amplification. The mutations were confirmed by Sanger
sequencing.
Luciferase activity assay
HeLa cells were seeded in 12-well plates at 50,000
cells/well. After the cells reached 80% confluence, 1 µg of
promoter-reporter and 1 µg pRL Renilla Luciferase Control
Reporter Vectors (negative control) were transfected into the cells
using Lipofectamine 3000 reagent (Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. After 24 h, the cells
were lysed in the lysis buffer of the Promega E1500 Luciferase
Assay System (Promega Corporation) and the cell lysates were
incubated with the luciferin of the Promega E1500 Luciferase Assay
System. The activity of Renilla luciferase was measured
using a Renilla-Glo® Luciferase Assay Kit (cat.
no. E2710; Promega Corporation). Luciferase activity was measured
using the Promega LumiPro instrument (Promega Corporation). The
promoter activity was normalized as follows: Promoter-reporter
activity/Renilla luciferase activity.
Statistical analysis
Transwell assay, CCK-8 assay, RT-qPCR and luciferase
activity assays were biological repeated three times and the data
are presented as the mean ± SD. Statistical analysis was performed
using the SPSS software (version 16.0 IBM). Differences of gene
expression between normal tissues and cervical cancer tissue of
TCGA were analyzed with unpaired Student's t-test. The significance
of the differences in the expression of YY1 and PLAU between the
cervical cancer tissues and the paired normal tissues was
determined using a paired t-test. Tukey's post hoc test following
one-way ANOVA was used to evaluate the significance of the
differences among multiple independent groups. The correlation
between PLAU expression and YY1 was analyzed using Spearman's rank
correlation coefficient analysis with Rho and P-values as
indicated. P<0.05 was considered to indicate a statistically
significant difference.
Results
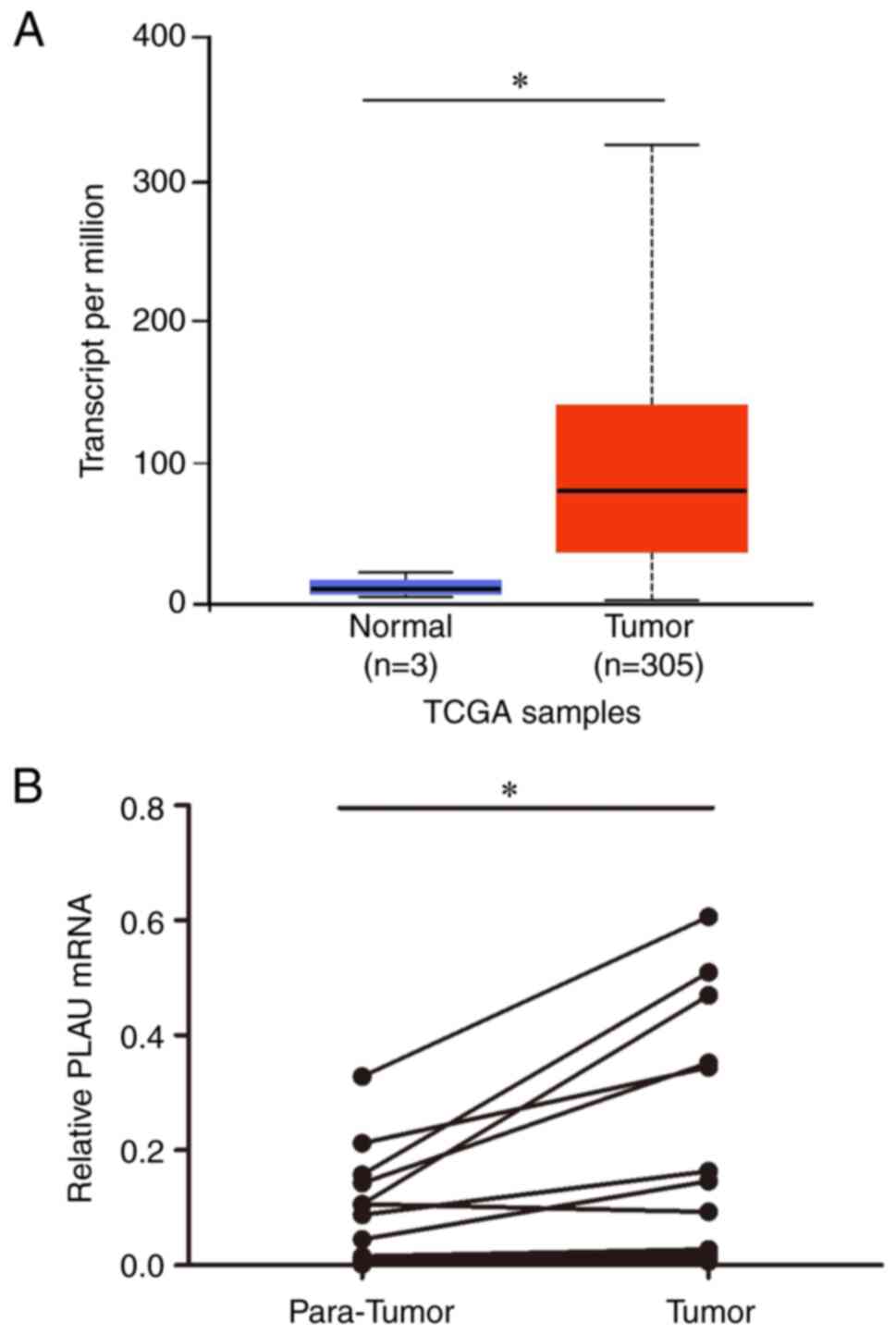
PLAU is overexpressed in cervical
cancer
The expression of cervical cancer in UALCAN
(http://ualcan.path.uab.edu/) was
analyzed based on The Cancer Genome Atlas (TCGA) database. As shown
in Fig. 1A, cervical cancer
tissues showed higher PLAU expression than normal cervical tissues.
To further verify the PLAU expression, PLAU expression was measured
and compared in 16 cervical cancer tissues and matched para-cancer
tumors using RT-qPCR. As revealed in Fig. 1B, PLAU was upregulated in cervical
cancer tissues compared with that in matched para-cancer normal
tissues.
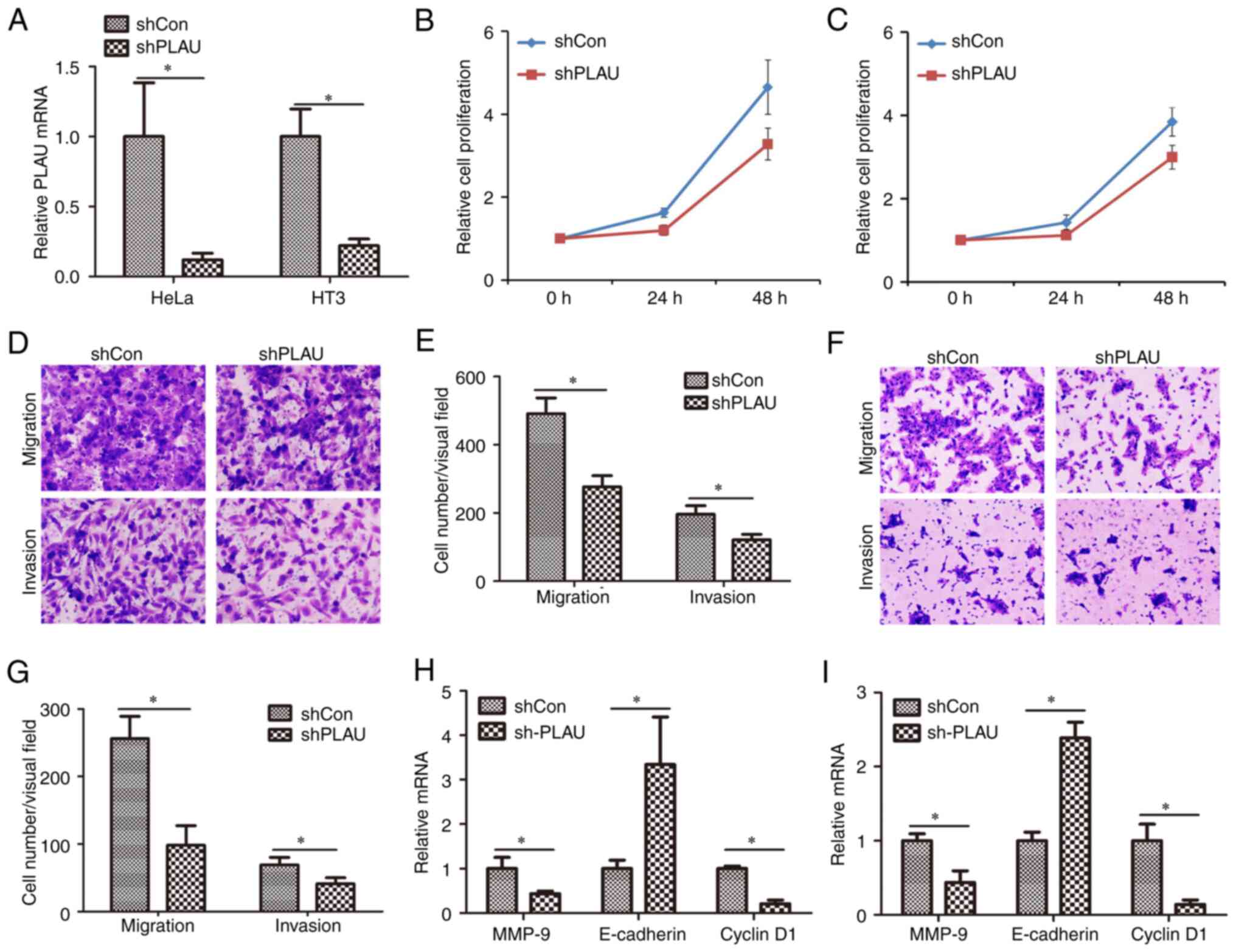
Knockdown of PLAU inhibits cell
proliferation, migration and invasion of cervical cancer
PLAU was knocked down in two cervical cancer cell
lines, HeLa and HT3, by lentiviral infection. As revealed in
Fig. 2A, PLAU expression was
significantly decreased in HeLa cells infected with Lv-shPLAU
compared with PLAU expression in those infected with Lv-shCon;
similarly, PLAU expression was significantly decreased in HT3 cells
infected with Lv-shPLAU compared with PLAU expression in those
infected with Lv-shCon. Cell proliferation was measured using CCK-8
assay (Fig. 2B and C). It was
demonstrated that proliferation of cells infected with Lv-shPLAU
was significantly inhibited compared with those infected with
Lv-shCon. HeLa cells infected with Lv-shCon or Lv-shPLAU were
subjected to Transwell assays to evaluate cell migration and
invasion capacities. As revealed in Fig. 2D and E, in HeLa cells, Lv-shPLAU
infection induced weaker migration and invasion capacities than
Lv-shCon infection. Similarly, Lv-shPLAU infection weakened the
migration and invasion capacities of HT3 cells compared with
Lv-shPLAU infection (Fig. 2F and
G). Furthermore, the mechanism of PLAU regulating cell
proliferation, migration and invasion was determined by measuring
the mRNA expression levels of several proliferation, migration and
invasion-associated genes. Knockdown of PLAU induced downregulation
of MMP-9 and cyclin D1 mRNA levels, as well as upregulation of
E-cadherin mRNA levels in HeLa cells (Fig. 2H) and HT3 cells (Fig. 2I). These results suggested that
PLAU knockdown inhibits the migration and invasion of cervical
cancer cells.
Identification of the PLAU core
promoter
Considering the overexpression and important role of
PLAU in cervical cancer, it is necessary to elucidate the
regulation of PLAU. A PLAU promoter-reporter, pGL3-PLAU, was
constructed; and based on that a series of deletion mutant types of
the PLAU promoter-reporter were constructed. The activity of these
reporters was compared and it was found that when the −1100/-901
region was deleted, the promoter activity significantly decreased
(Fig. 3A). The −1100/-901 region
was cloned into the pGL3-basic vector and the activities of
pGL3-PLAU and pGL3-PLAU (−1100/-901) were compared. As revealed in
Fig. 3B, until it was narrowed to
−1100/-901, the promoter still possessed 78.13% activity of the
full-length promoter, suggesting that the core promoter of PLAU is
located in the −901/-1100 regions.
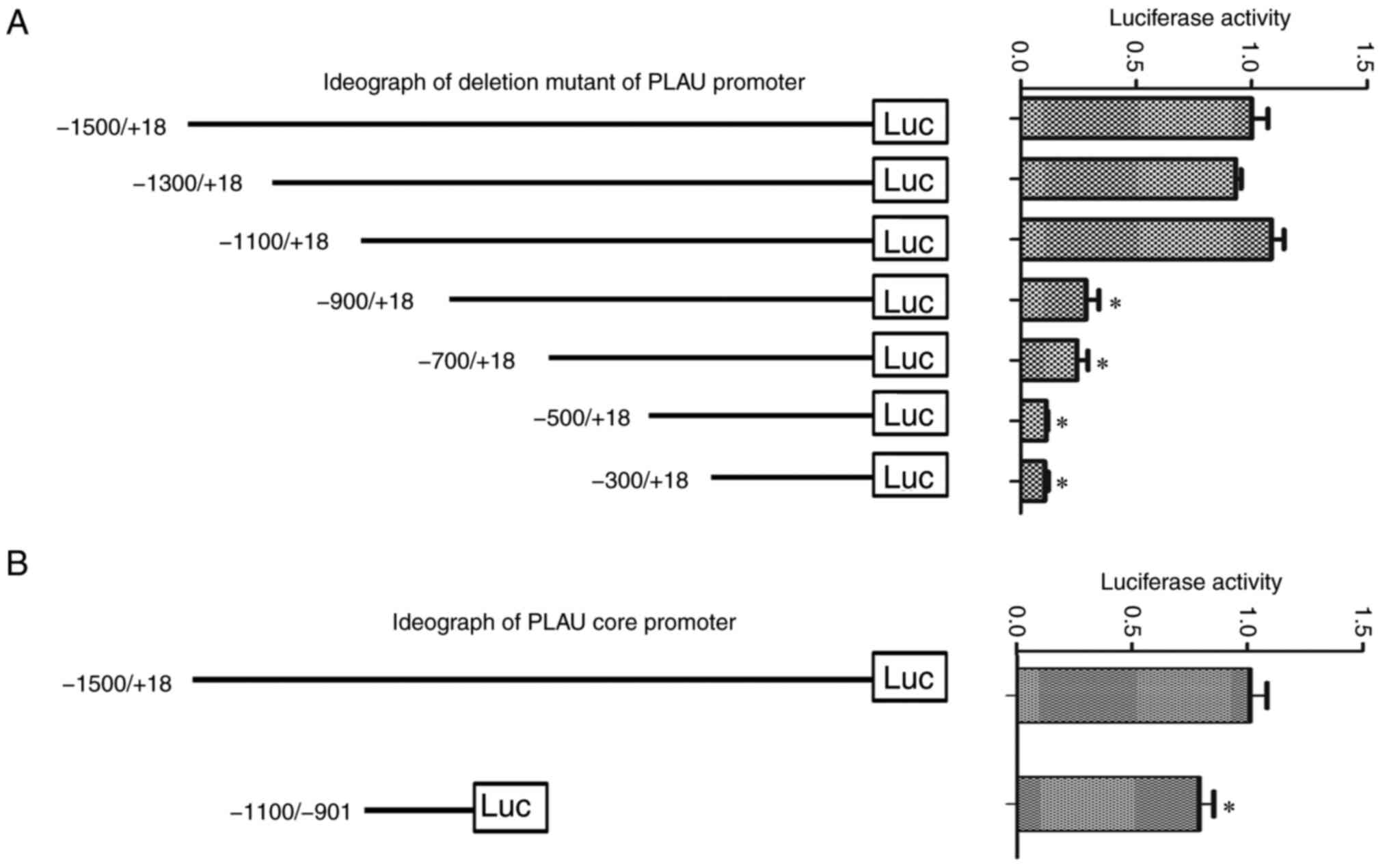 | Figure 3.Identification of the PLAU core
promoter. (A) Based on the pGL3-PLAU, a series of deletion mutants
of the PLAU promoter (−1500/+18, −1300/+18, −1100/+18, −900/+18,
−700/+18, −500/+18, −300/+18) were constructed and transfected into
HeLa cells to measure the luciferase activity. Left panel, the
ideograph of the deletion mutants of the PLAU promoter; right
panel, relative luciferase activity of each mutant. (B) Left panel,
ideograph of the PLAU core promoter; right panel, relative
luciferase activity. *P<0.05 vs. the −1500/+18. PLAU,
plasminogen activator urokinase; luc, luciferase. |
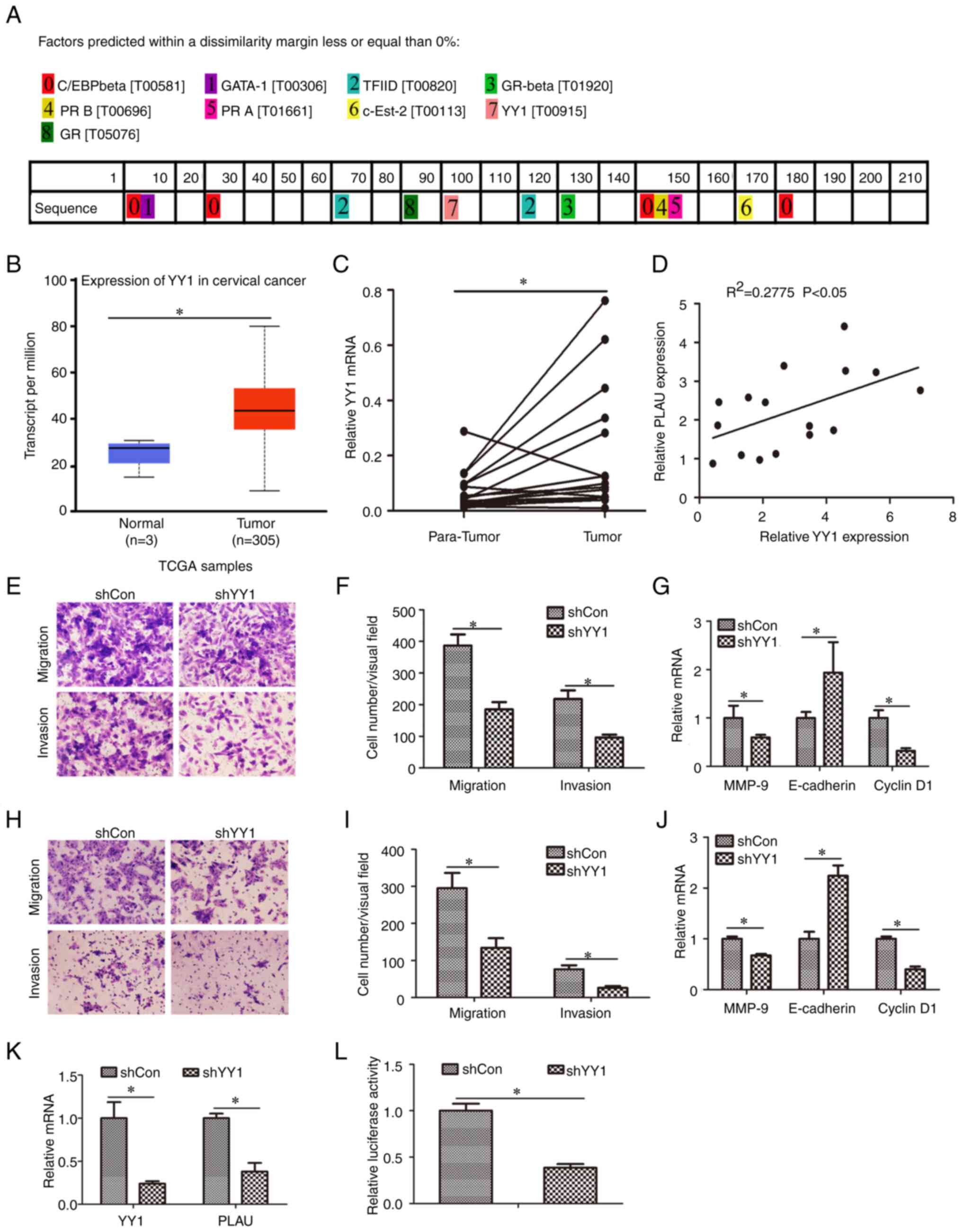
YY1 regulates PLAU expression at the
transcriptional level
The −1100/-901 sequence was submitted to the online
PROMO software (http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3).
Several transcription factors were predicted, including TF II D,
YY1, GATA1 and GR-β (Fig. 4A). The
expression of the predicted transfection factors was analyzed
(Fig. S1). The expression of
C/EBPβ, GATA1, TBP (encoding TBP, the key subunit of TF II D), PR
(encoding PRA and PRB), SULTIE1 (encoding c-Est-2) and GR (encoding
GR and GR-α) showed no statistically significant difference between
normal and cervical cancer tissues. However, the expression of YY1
was overexpressed in cervical cancer compared with normal tissues
(Fig. 4B). YY1 expression was
further evaluated in the collected cervical tissues. As revealed in
Fig. 4C, YY1 in cervical cancer
was higher than in adjacent normal tissues. In addition, it was
found that the expression of PLAU is positively correlated to YY1
in cervical cancer tissues (Fig.
4D). Furthermore, the role of YY1 in cervical cancer cell
migration and invasion was evaluated. Cervical cancer cells were
infected with the lentivirus Lv-shYY1 and it was observed that
knockdown of YY1 inhibited cell migration and invasion of HeLa
(Fig. 4E and F) and HT3 cells
(Fig. 4H and I). Besides,
knockdown of YY1 induced downregulation of MMP-9 and cyclin D1 mRNA
levels, as well as upregulation of E-cadherin mRNA levels in HeLa
(Fig. 2G) and HT3 cells (Fig. 2J). PLAU mRNA was also measured; as
demonstrated in Fig. 4K, Lv-shYY1
infection decreased PLAU mRNA, indicating that YY1 is a regulator
of PLAU. To evaluate whether YY1 regulates PLAU through
transcription, the PLAU core promoter-reporter was transfected into
cells with or without YY1 knockdown and promoter activity was
determined. The results showed that following Lv-shYY1 infection,
core promoter activity was downregulated in HeLa cells (Fig. 4L), indicating that YY1 regulates
the transcription of PLAU.
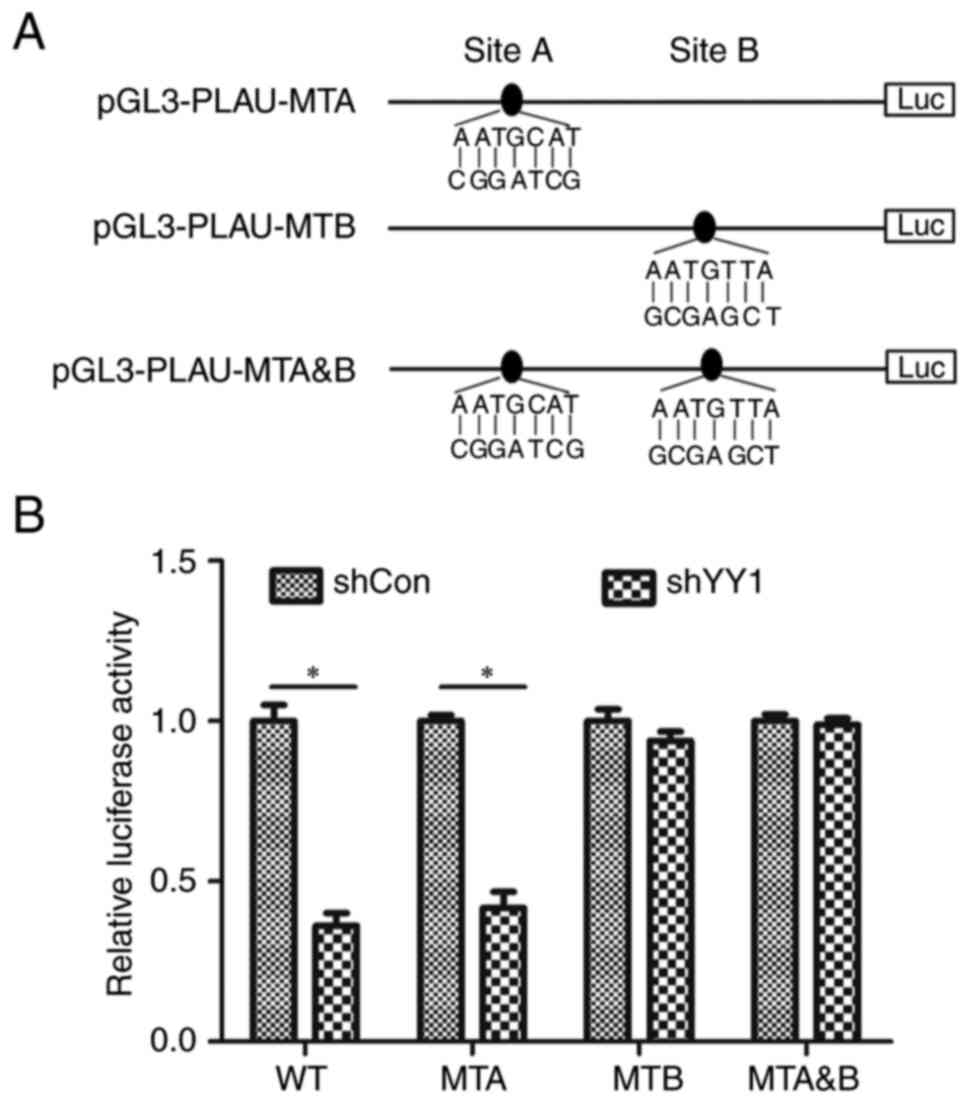
Identification of the YY1 binding site
on the PLAU promoter
According to PROMO software, there are two putative
binding sites in the core promoter region of PLAU: Site A is
located at −1024/-1030, while Site B is located at −974/-980. To
determine which site was responsible for YY1 regulating PLAU, Site
A and Site B were mutated separately and Site A and Site B were
double-mutated; they were named pGL3-PLAU-MTA, pGL3-PLAU-MTB and
pGL3-PLAU-MTA&B, respectively (Fig. 5A). pGL3-PLAU-MTA, pGL3-PLAU-MTB and
pGL3-PLAU-MTA&B were transfected into HeLa cells with or
without YY1 knockdown and promoter activity was measured. As
revealed in Fig. 5B, YY1 knockdown
induced a decrease in the activity of pGL3-PLAU and pGL3-PLAU-MTA,
instead of PGL3-PLAU-MTB and pGL3-PLAU-MTA&B, indicating that
Site B is the main site responsible for YY1 regulating PLAU.
Discussion
In the current study, it was found that PLAU was
overexpressed in cervical cancer and knockdown of PLAU weakened the
cell proliferation, migration and invasion capacities of cervical
cancer cells. The location of the PLAU core promoter and an
important transcription factor, YY1, which regulates PLAU promoter
activity, were also identified.
Using the UALCAN database and RT-qPCR assay, it was
found that PLAU was overexpressed in cervical cancer and promoted
cell, proliferation, migration and invasion, indicating that PLAU
promotes cervical cancer progression. Actually, upregulation of
PLAU, as well as its oncogenic role, has already been observed in
other tumor types, such as HNSCC and gastric cancer (7,8). In
cervical cancer, PLAU has been reported to regulate tumor invasion
and metastasis (15–17). Our study further revealed the
important role of PLAU in tumor progression. The present results
verified previous studies about higher expression of PLAU in
cervical cancer and its role in cervical cancer cell migration and
invasion (15–17); therefore, not only for HNSCC and
gastric cancer, but also for cervical cancer PLAU can serve as a
potential prognostic marker and therapeutic target, and this is
just the reason why the regulation of PLAU transcriptional
regulation was investigated in cervical cancer.
Transcription is the process by which RNA is
synthesized according to the genomic sequence of DNA. The
transcription process requires two key elements, the promoter and
transcription factor binding to the promoter (18,36,37).
Revealing the mechanisms by which an oncogene is regulated will
help to further understand the process of tumorigenesis and
progression, as well as to determine novel therapeutic targets
(35). In the present study, a
PLAU promoter-reporter was constructed and the core promoter
located in the region −901/-1100 was identified. Based on the
sequence of the core promoter, it was predicted that several
transcription factors putatively bind to the core promoter;
however, among these transcription factors, only the expression of
YY1 showed significant difference between normal tissues and
cervical cancers. YY1, a member of the GLI-Krüppel family of
zinc-finger DNA-binding proteins, is a ubiquitously expressed
transcription factor. YY1 has been reported to activate the
transcription of several target genes, such as FOXE1, TNK2-AS1, and
LINC00466 (24–26). In the current study, it was
demonstrated that YY1 knockdown induced downregulation of PLAU
promoter activity and mRNA expression, indicating that PLAU is a
newly identified target gene of YY1. YY1 plays a critical role in
tumorigenesis and progression by participating in various
biological processes, including cell proliferation (27,28),
apoptosis (29), invasion and
migration (30), radioresistance
(31), and drug resistance
(32).
It was observed that PLAU is correlated with YY1
expression in cervical cancer in the cervical cancer tissues
collected in Liaocheng People's Hospital. In addition, it was found
that knockdown of YY1 induced downregulation of PLAU mRNA and
promoter activity; therefore, the regulatory role of the
transcription factor YY1 on PLAU expression was confirmed. To the
best of our knowledge, this is the first study to demonstrate how
PLAU is regulated at the transcriptional level. Our finding that
YY1 regulates PLAU reveals a new mechanism of YY1-associated
cervical cancer progression, suggesting a strategy for targeting
PLAU, which is short of an applicable drug in cervical cancer
patients, through inhibiting YY1. Although the YY1-regulating PLAU
in cervical cancer was confirmed, whether this regulation was
unique in cervical cancer or a universal effect in different tumor
types is unknown.
As predicted by PROMO software, there are two
potential binding sites for YY1 in the PLAU core promoter. Using
site-directed mutation and luciferase activity assay, it was
revealed that mutation of Site B (−974/-980) not Site A
(−1024/-1030) abolished YY1′s effects on PLAU promoter activity,
suggesting that Site B is the main site responsible for
YY1-mediated regulation of PLAU. Blocking Site A may abolish the
regulatory effects of YY1 on PLAU and may be a promising target for
cervical cancer therapy. There are unclear details about YY1
regulating PLAU. YY1 can regulate target genes with various
co-factors, such as p300 and HDAC1 (38–40);
however, whether YY1 regulates PLAU expression needing such
co-factors remains unknown.
HPV infection is a risk factor of cervical cancer.
In the present study, the HPV was not in our concern, thus it was
not analyzed whether the samples were infected with HPV. It was
determined that YY1/PLAU influences the migration and invasion in
HeLa and HT-3 cells. HeLa cells are infected with HPV18, while HT-3
cells are not infected with HPV; therefore, the present results
suggested that YY1/PLAU regulates cell migration and invasion
independent of HPV infection.
There are several limitations to the present study.
Firstly, 16 pairs of cervical cancers and the matched adjacent
normal tissues were only collected, and extension the number of
specimen pairs will be helpful in further confirming the conclusion
of the present study. Secondly, the mRNA levels of YY1 and PLAU
were evaluated; results would provide an improved understanding if
protein levels are also detected. Nevertheless, it is considered
that the mRNA level, to a certain extent, can confirm the
expression of PLAU and YY1. Thirdly, our study detected the role of
PLAU in regulating cervical cancer cell migration and invasion and
explored the mechanism of how PLAU is regulated, but the mechanism
of how PLAU regulates cervical cancer cell migration and invasion
was not detected; our future studies will elucidate the mechanism
by which PLAU regulates cell migration and invasion.
In conclusion, it was demonstrated that PLAU is
overexpressed in cervical cancer and the promotion role of PLAU in
cervical cancer was observed. The core promoter of PLAU was
identified and it was determined that YY1 may be a crucial
transcription factor of PLAU. The findings of the present study
suggested that YY1/PLAU may be potential therapeutic targets for
cervical cancer treatment.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the Figshare repository (https://figshare.com/s/1966432ad77921b295cd).
Authors' contributions
CX designed the study. YG and PX performed the cell
culture and western blotting experiments, and drafted the
manuscript. HL and XM collected the tissue samples and performed
the RT-qPCR experiments, and the construction of PLAU
promoter-reporter. PX performed the statistical analysis, the
luciferase assay and participated in data analysis. YG and CX
confirm the authenticity of the raw data. All authors have read and
approved the final version of the manuscript, and agree to be
accountable for all aspects of the present research, ensuring that
the accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
The present study was approved (approval no.
LC2021015) by the Ethics Committee of Liaocheng People's Hospital
(Liaocheng, China). Written informed consent was provided by all
patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer statistics, 2022. CA Cancer J Clin. 72:7–33. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Benard VB, Watson M, Saraiya M, Harewood
R, Townsend JS, Stroup AM, Weir HK and Allemani C: Cervical cancer
survival in the United States by race and stage (2001–2009):
Findings from the CONCORD-2 study. Cancer. 123 (Suppl
24):S5119–S5137. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Corley SM, Mendoza-Reinoso V, Giles N,
Singer ES, Common JE, Wilkins MR and Beverdam A: Plau and Tgfbr3
are YAP-regulated genes that promote keratinocyte proliferation.
Cell Death Dis. 9:11062018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Peng W, Wu Y, Zhang G, Zhu W, Chang M,
Rouzi A, Jiang W, tong J, Wang Q, Liu J, et al: GLIPR1 protects
against cigarette smoke-induced airway inflammation via PLAU/EGFR
signaling. Int J Chron Obstruct Pulmon Dis. 16:2817–2832. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Buckley BJ, Ali U, Kelso MJ and Ranson M:
The urokinase plasminogen activation system in rheumatoid
arthritis: Pathophysiological roles and prospective therapeutic
targets. Curr Drug Targets. 20:970–981. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Huo Z, Li X, Zhou J, Fan Y, Wang Z and
Zhang Z: Hypomethylation and downregulation of miR-23b-3p are
associated with upregulated PLAU: A diagnostic and prognostic
biomarker in head and neck squamous cell carcinoma. Cancer Cell
Int. 21:5642021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sun J, Zhou F, Xue J, Ji C, Qu Y and Pan
Y: Long non-coding RNA TRPM2-AS regulates microRNA miR-138-5p and
PLAU (plasminogen activator, urokinase) to promote the progression
of gastric adenocarcinoma. Bioengineered. 12:9753–9765. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Moquet-Torcy G, Tolza C, Piechaczyk M and
Jariel-Encontre I: Transcriptional complexity and roles of
Fra-1/AP-1 at the uPA/Plau locus in aggressive breast cancer.
Nucleic Acids Res. 42:11011–11024. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen G, Sun J, Xie M, Yu S, Tang Q and
Chen L: PLAU promotes cell proliferation and epithelial-mesenchymal
transition in head and neck squamous cell carcinoma. Front Genet.
12:6518822021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang S, Jiang L, Han Y, Chew SH, Ohara Y,
Akatsuka S, Weng L, Kawaguchi K, Fukui T, Sekido Y, et al:
Urokinase-type plasminogen activator receptor promotes
proliferation and invasion with reduced cisplatin sensitivity in
malignant mesothelioma. Oncotarget. 7:69565–69578. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zou B, Li J, Xu K, Liu JL, Yuan DY, Meng Z
and Zhang B: Identification of key candidate genes and pathways in
oral squamous cell carcinoma by integrated bioinformatics analysis.
Exp Ther Med. 17:4089–4099. 2019.PubMed/NCBI
|
|
13
|
Wu M, Wei B, Duan SL, Liu M, Ou-Yang DJ,
Huang P and Chang S: Methylation-driven gene PLAU as a potential
prognostic marker for differential thyroid carcinoma. Front Cell
Dev Biol. 10:8194842022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang H, Lu L, Liang X and Chen Y:
Identification of prognostic genes in the pancreatic adenocarcinoma
immune microenvironment by integrated bioinformatics analysis.
Cancer Immunol Immunother. 71:1757–1769. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Daneri-Navarro A, Macias-Lopez G,
Oceguera-Villanueva A, Del Toro-Arreola S, Bravo-Cuellar A,
Perez-Montfort R and Orbach-Arbouys S: Urokinase-type plasminogen
activator and plasminogen activator inhibitors (PAI-1 and PAI-2) in
extracts of invasive cervical carcinoma and precursor lesions. Eur
J Cancer. 34:566–569. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kobayashi H, Fujishiro S and Terao T:
Impact of urokinase-type plasminogen activator and its inhibitor
type 1 on prognosis in cervical cancer of the uterus. Cancer Res.
54:6539–6548. 1994.PubMed/NCBI
|
|
17
|
Koelbl H, Kirchheimer JC, Tatra G, Christ
G and Binder BR: Increased plasma levels of urokinase-type
plasminogen activator with endometrial and cervical cancer. Obstet
Gynecol. 72:252–256. 1988.PubMed/NCBI
|
|
18
|
Kou XX, Hao T, Meng Z, Zhou YH and Gan YH:
Acetylated Sp1 inhibits PTEN expression through binding to PTEN
core promoter and recruitment of HDAC1 and promotes cancer cell
migration and invasion. Carcinogenesis. 34:58–67. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
de Jonge WJ, Patel HP, Meeussen JVW and
Lenstra TL: Following the tracks: How transcription factor binding
dynamics control transcription. Biophys J. 121:1583–1592. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hong JH and Zhang HG: Transcription
factors involved in the development and prognosis of cardiac
remodeling. Front Pharmacol. 13:8285492022. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Su SC, Lin CW, Yang WE, Fan WL and Yang
SF: The urokinase-type plasminogen activator (uPA) system as a
biomarker and therapeutic target in human malignancies. Expert Opin
Ther Targets. 20:551–566. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mahmood N, Mihalcioiu C and Rabbani SA:
Multifaceted role of the urokinase-type plasminogen activator (uPA)
and its receptor (uPAR): Diagnostic, prognostic, and therapeutic
applications. Front Oncol. 8:242018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nagamine Y, Medcalf RL and Muñoz-Cánoves
P: Transcriptional and posttranscriptional regulation of the
plasminogen activator system. Thromb Haemost. 93:661–675. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li S, Zhang Z, Peng H and Xiao X:
YY1-induced up-regulation of FOXE1 is negatively regulated by
miR-129-5p and contributes to the progression of papillary thyroid
microcarcinoma. Pathol Res Pract. 221:1533372021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yao W, Yan Q, Du X and Hou J: TNK2-AS1
upregulated by YY1 boosts the course of osteosarcoma through
targeting miR-4319/WDR1. Cancer Sci. 112:893–905. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li F, Shen ZZ, Xiao CM and Sha QK:
YY1-mediated up-regulation of lncRNA LINC00466 facilitates glioma
progression via miR-508/CHEK1. J Gene Med. 23:e32872021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang Q, Wan M, Shi J, Horita DA, Miller
LD, Kute TE, Kridel SJ, Kulik G and Sui G: Yin Yang 1 promotes
mTORC2-mediated AKT phosphorylation. J Mol Cell Biol. 8:232–243.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wu S, Wang H, Li Y, Xie Y, Huang C, Zhao
H, Miyagishi M and Kasim V: Transcription factor YY1 promotes cell
proliferation by directly activating the pentose phosphate pathway.
Cancer Res. 78:4549–4562. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhai Z, Ren Y, Shu C, Chen D, Liu X, Liang
Y, Li A and Zhou J: JAC1 targets YY1 mediated JWA/p38 MAPK
signaling to inhibit proliferation and induce apoptosis in TNBC.
Cell Death Discov. 8:1692022. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Guan C, Liu L, Zhao Y, Zhang X, Liu G,
Wang H, Gao X, Zhong X and Jiang X: YY1 and eIF4A3 are mediators of
the cell proliferation, migration and invasion in
cholangiocarcinoma promoted by circ-ZNF609 by targeting miR-432-5p
to regulate LRRC1. Aging (Albany NY). 13:25195–25212. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhao L, Li R, Qiu JZ, Yu JB, Cao Y and
Yuan RT: YY1-mediated PTEN dephosphorylation antagonizes IR-induced
DNA repair contributing to tongue squamous cell carcinoma
radiosensitization. Mol Cell Probes. 53:1015772020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhao L, Li R and Gan YH: Knockdown of Yin
Yang 1 enhances anticancer effects of cisplatin through protein
phosphatase 2A-mediated T308 dephosphorylation of AKT. Cell Death
Dis. 9:7472018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bai G, Wei N, Li F, Zhao P, Meng Z, Zou B,
Liu Y, Xu K, Li K, Yao C and Yang P: Function and transcriptional
regulation of TCTN1 in oral squamous cell carcinoma. Oncol Rep.
47:262022. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lynch TR, Xue M, Czerniak CW, Lee C and
Kimble J: Notch-dependent DNA cis-regulatory elements and their
dose-dependent control of C. elegans stem cell self-renewal.
Development. 149:dev2003322022. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yin S, NandyMazumdar M, Paranjapye A and
Harris A: Cross-talk between enhancers, structural elements and
activating transcription factors maintains the 3D architecture and
expression of the CFTR gene. Genomics. 114:1103502022.(Epub ahead
of print). View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Nakayama T, Fukutomi T, Terao Y and
Akagawa K: Syntaxin 1A gene is negatively regulated in a
cell/tissue specific manner by YY1 transcription factor, which
binds to the −183 to −137 promoter region together with gene
silencing factors including histone deacetylase. Biomolecules.
11:1462021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hou X, Li Y, Huang Y, Zhao H and Gui L:
Adenosine receptor A1-A2a heteromers regulate EAAT2 expression and
glutamate uptake via YY1-induced repression of PPARγ transcription.
PPAR Res. 2020:24102642020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wei X, Liu F, Jiang X, Xu X, Zhou T and
Kang C: YY1 promotes telomerase activity and laryngeal squamous
cell carcinoma progression through impairment of GAS5-mediated p53
stability. Front Oncol. 11:6924052021. View Article : Google Scholar : PubMed/NCBI
|