Introduction
Colorectal cancer (CRC) remains the third leading
cause of cancer-related mortality worldwide regardless of the
availability of improved diagnostic and therapeutic strategies
(1). Surgical resection in
combination with radiotherapy and chemotherapy, depending on cancer
stage, offers the best chances of survival for patients and
represents the standard of clinical care (2). Removal of the primary tumor carries a
risk of subsequent metastatic tumor spread, due to the potential
release of tumor cells into the circulation. In addition, surgical
trauma to adjacent healthy tissues can support metastasis, since
neoangiogenesis increases during wound healing (3,4).
Perioperative factors such as the choice of anesthesia may have an
influence on carcinogenesis, metastasis, recurrence, and the final
clinical outcome; however these effects have not been fully
elucidated (5,6). Surgery for CRC is performed under
general anesthesia. For the maintenance of anesthesia, two
anesthetics are most commonly used: Either total intravenous
anesthesia (TIVA) using propofol, or a volatile anesthetic gas
(VAG), with sevoflurane being the mostly commonly administered. For
TIVA, a reduced intraoperative inflammatory reaction has been
observed in several studies (7–9). In
a retrospective study on CRC patients who received propofol or
desflurane (another VAG), the use of propofol led to a
significantly longer and higher overall survival with reduced
metastasis occurrence (10–13).
However, the mechanisms underlying these observations remain
largely unknown. In contrast to TIVA, studies investigating VAG in
this context have demonstrated no consistent trend concerning tumor
cell growth, metastasis or protective effects (11,14–17).
Recently, the identification of regulatory non-coding RNAs,
including microRNAs (miRNAs/miRs) and long non-coding RNAs
(lncRNAs), has increased the complexity of the molecular landscape
(18,19).
Numerous investigations have characterized the
important function of miRNAs in tumor biology. A recent study by
the authors demonstrated anesthetic-specific effects on expression
levels of miRNAs co-precipitating with extracellular vesicles when
the VAG sevoflurane was compared to TIVA. In silico target
analyses of microRNA expression patterns in this study indicated an
inhibitory effect of propofol on crucial carcinoma-related
pathways, including proliferation, migration and enhanced apoptosis
(19). In contrast to the
well-established role of miRNAs, lncRNAs and their impact on cancer
is a relatively new research field, both being specifiable on their
size and function. LncRNAs are not translated, consist of minimum
200 nucleotides up to kilobases (20–22)
and have been attributed important functions in transcription,
translation and post-translational modification (23,24).
In the field of tumor biology, lncRNAs can either inhibit tumor
growth or act as oncogenes, promoting cell proliferation and
migration (25–27); however, the potential effect of
anesthetic agents on lncRNAs has not been investigated previously,
to the best of our knowledge.
The hypothesis that the present study was based on
is that anesthetic-specific lncRNA expression changes are induced
in the blood circulation and may play a crucial role in tumor
outcome.
The primary aim of the present study was to perform
a proof-of-concept study, in order to demonstrate the differential
effects of TIVA (propofol) on blood-derived lncRNA and mRNA
profiles, in comparison to VAG (sevoflurane), during CRC resection.
Secondly, the present study was designed to provide preliminary
evidence of cancer relevant signaling changes induced by
lncRNA-mRNA regulatory effects. It was designed as a prospective,
matched-case, non-randomized pilot study, including patients
undergoing colorectal cancer surgery at the LMU Hospital Munich,
anesthetized with TIVA or VAG and fulfilling the predefined
inclusion criteria. To obtain holistic lncRNA and mRNA expression
profiles, a high-throughput sequencing approach with subsequent
quantitative PCR confirmation was applied. Differential gene
expression (DGE) analysis of mRNA and lncRNA sequencing data
permitted the validation of in silico identified
target-mRNAs and to confirm significant changes in target-mRNA
expression. Consequently, it was feasible to identify
differentially regulated lncRNAs, which possibly modulate
target-mRNAs and play a crucial role in cancer relevant signaling
pathways. This type of differentially expressed and
anesthetic-specific lncRNAs and their identified target-mRNAs
represent candidates for the still unexplained mechanism that may
influence long-term survival of cancer patients following the
surgical removal of the tumor.
Materials and methods
Patient identification and
selection
Patient recruitment and matching were performed as
previously described (19). The
Ethics Committee of the Medical Faculty of the
Ludwig-Maximilians-University (LMU) Munich, Germany (to which the
Institute of Human Genetics and the Department of Anesthesiology
are assigned) approved the present study (protocol-no. 232-16). The
present study was performed in accordance with the Declaration of
Helsinki. Written informed consent was obtained and study samples
were pseudonymized. Inclusion and exclusion criteria were applied
for both the TIVA and VAG group. The inclusion criteria were the
presence of a primary colorectal cancer scheduled for operative
therapy and consent to the study. The exclusion criteria were the
following: i) Denied consent; ii) an age <18 years; iii)
pregnancy; iv) the simultaneous occurrence of CRC with another
primary tumor; v) severe organ dysfunction (liver, kidney); vi)
chronic inflammatory or autoimmune disorder (e.g., rheumatoid
arthritis); vii) the use of immunosuppressive medication; and viii)
contraindications for epidural anesthesia. The final study cohort
consisted of 12 patients receiving TIVA and 10 patients receiving
VAG. All patients underwent open surgery procedure for CRC
resection and were recruited at the University Hospital of Munich
from 04/2017 to 06/2018. Statistical power calculation for sample
size assessment was not performed, since to the best of our
knowledge there are no studies available dealing with the effect of
anesthetic agents on blood-derived lncRNA expression profiles. The
patient cohorts were comparable with regard to demographic
variables, comorbidities and perioperatively administered
medication. The mean age was 71.90 years with a standard deviation
(SD) of 10.13 years for the VAG group, and 61.58 years with a SD of
10.95 years for the TIVA group. The sex distribution was 7 males/5
females in the TIVA group, and 8 males/2 females in the VAG group.
In particular, the cohorts did not differ significantly in the
primary matching goals of tumor stage (P=0.162) and localization
(P=0.595) (Table I).
 | Table I.Overview of the patient
characteristics divided according to the anesthetic agent used. |
Table I.
Overview of the patient
characteristics divided according to the anesthetic agent used.
| Parameter | TIVA | VAG | P-value |
|---|
| Sex |
|
|
|
|
Male | 7 | 8 | 0.531 |
|
Female | 5 | 2 |
|
| Age in
yearsa (age
range) | 59.5
(54.8-66.0) | 71.0
(66.2-80.8) | 0.016 |
| BMI
(kg/m2)a | 25.1
(21.3-26.9) | 26.7
(24.1-36.6) | 0.069 |
| ASA
classificationb
(3/2) |
|
|
|
| 3 | 10 | 9 | 0.865 |
| 2 | 2 | 1 |
|
| Tumor location |
|
|
|
|
Rectal | 6 | 7 | 0.595 |
| Right
colon | 3 | 3 |
|
| Left
colon | 1 | 0 |
|
|
Sigmoid | 2 | 0 |
|
| UICC
stagec |
|
| 0.162 |
| I | 1 | 3 |
|
| II | 3 | 2 |
|
|
III | 8 | 3 |
|
| IV | 0 | 2 |
|
| Coronary heart
disease |
|
|
|
| No | 12 | 7 | 0.156 |
|
Yes | 0 | 3 |
|
| Arterial
hypertension |
|
|
|
| No | 8 | 4 | 0.412 |
|
Yes | 4 | 6 |
|
| Diabetes |
|
|
|
| No | 12 | 6 | 0.062 |
|
Yes | 0 | 4 |
|
| Kidney
insufficiency |
|
|
|
| No | 12 | 9 | 0.926 |
|
Yes | 0 | 1 |
|
| Atrial
fibrillation |
|
|
|
| No | 12 | 7 | 0.156 |
|
Yes | 0 | 3 |
|
| Preoperative
radiation |
|
|
|
| No | 7 | 5 | 0.969 |
|
Yes | 5 | 5 |
|
| Preoperative
chemotherapy |
|
|
|
| No | 7 | 7 | 0.903 |
|
Yes | 5 | 3 |
|
| Epidural
anesthesia |
|
|
|
| No | 2 | 1 | 0.865 |
|
Yes | 10 | 9 |
|
| Duration of
anesthesia, in min; value (range)a | 390.0
(286.2-545.5) | 337.5
(313.2-418.8) | 0.5 |
| Maximum dosage of
noradrenaline in mg/h; value (range)a | 0.4 (0.3-0.6) | 0.6 (0.5-1.0) | 0.016 |
| Maximum MAC; value
(range)a,d | 0.0 (0.0-0.0) | 1.2 (1.0-1.4) | <0.001 |
| Fluids
(liters)a | 3.5 (3.0-5.25) | 4.25
(3.63-4.9) | 0.264 |
| Duration of
surgery, in min; value (range)a | 282.5
(191.8-403.0) | 234.0
(204.2-291.8) | 0.288 |
| Cumulative dosage
of ropivacained in
mg; value (range)a | 190.0
(155.0-241.2) | 165.0
(152.5-180.0) | 0.113 |
| Cumulative dosage
of propofol during anesthesia in mg/kg of body weight (value
range)a | 32.0
(23.5-44.9) | 5.6 (1.7-7.6) | <0.001 |
| Cumulative dosage
of sufentanil in mg/kg of body weight; value (range)a | 1.0 (0.6-1.2) | 0.9 (0.7-1.0) | 0.334 |
Anesthesiologic procedures and sample
collection
Standard balanced anesthesia consisting of an
epidural in combination with general anesthesia was planned for all
patients. For 3 patients, epidural anesthesia was not possible. due
to anatomical reasons. General anesthesia was induced with propofol
in both groups and subsequently maintained with propofol (TIVA) or
sevoflurane (VAG).
A maximum of 44.2 ml of blood was collected per
patient. Venous blood was drawn through intravascular catheters at
the preoperative time point and after termination of surgery during
wound closure (post-operative time point). For collection and
stabilization of the 2.5 ml whole blood samples, RNA tubes
(PAXgene; Qiagen GmbH; cat. no. 762165) were used according to the
manufacturer's protocol and stored at −80°C until further
processing.
RNA extraction, depletion, library
preparation and sequencing
Total blood RNA containing the red and the white
blood cell fraction (ratios of lymphocytes and macrophages as
physiologically present in human blood) was extracted using the
PAXgene system with the PAXgene blood miRNA kit (Qiagen GmbH; cat.
no. 763134). To achieve higher concentrations, the samples were
evaporated and eluted in a smaller volume of 28 µl. The integrity
of total blood cell derived RNA was assessed using capillary
electrophoresis, using the RNA 6000 Nano kit (Agilent Technologies
GmbH; cat. no. 5067-1511) on the Bioanalyzer 2100 (Agilent
Technologies GmbH). To remove highly abundant RNA like cytoplasmic
and mitochondrial rRNA and globin mRNA the QIAseq®
FastSelect -rRNA/Globin kit (Qiagen GmbH; cat. no. 335376) was
utilized. QIAseqTM Stranded Total RNA Library kit
(Qiagen GmbH; cat. no. 180743) was used to generate RNA sequencing
libraries from 800 ng total RNA input. Due to a moderate sample
quality an insert size of ~150–250 bp was generated by a ten-minute
incubation at 95°C. To assess the size distribution and
concentration of final libraries the High Sensitivity DNA kit
(Agilent Technologies GmbH; cat. no. 5067-4626) on Bioanalyzer 2100
(Agilent Technologies GmbH) was performed. All kit protocols were
applied according to the manufacturer's instructions. Samples were
divided into four sequencing runs-two runs with 12 samples each and
two with 10 samples each. In total, 50 cycles of single-end
sequencing on the HiSeq2500 (Illumina GmbH) were performed, using
the HiSeq Rapid SBS and SR Cluster kits (Illumina GmbH; Cat. nos.
FC-402-4022 and GD-402-4002).
Bioinformatic RNA sequencing
analysis
3′ adaptor sequences were excluded from the raw data
sequences using Cutadapt (v2.8) (28) and reads with <16 nucleotides
remaining were omitted. Trimmed reads were analyzed with the Fast
QC software v.0.11.9 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/)
to evaluate phred scores and read lengths before aligning them
against the human genome (GRCh38) with STAR (v2.7.3a) (29). Aligned reads were then annotated
using RSEM (v1.3.1) (30), to
count reads mapping to coding sequences.
All samples were included for subsequent DGE
analyses. DGE analyses for the identification of differentially
expressed genes between the two time points, preoperatively and
after the termination of the surgery during wound closure
(postoperative) within each anesthetic group and between the TIVA-
and VAG-groups, were performed using DESeq2 (v1.30.1) (31). Technical variation introduced by
multiple sequencing runs was accounted for in the model.
Benjamini-Hochberg method controlling the false discovery rate was
applied to reduce type I error accumulation.
For lncRNA and mRNA analysis, transcripts that
fulfilled the following filter criteria were selected: i) Mean
expression (BaseMean) ≥50 normalized reads; ii) absolute value of
log2 fold change (|log2FC|) ≥1 for lncRNA,
respectively |log2FC| ≥0.5 for mRNA transcripts; and
iii) an adjusted P-value (padj) ≤0.1.
Target and pathway analysis
Using the software tool RNAInter v.4.0, target-mRNAs
of significantly regulated lncRNAs were identified (32). Filter criteria for RNAInter were
set to interaction type ‘RNA-RNA interactions’, species ‘Homo
sapiens’ and a confidence score >0.5. The resulting target-mRNAs
were compared with the mRNA DGE data. For lncRNAs without results
(n=18 VAG, n=29 TIVA) in RNAInter, target-mRNAs were identified
using a lncRNA identification and functional annotation tool called
LncADeep v.1.0 (33), which among
others identified the interaction with Kyoto Encyclopedia of Genes
and Genomes (KEGG) pathways (34).
However, only few pathways were unique (n=1 for VAG, n=14 for TIVA)
and had no clinical relevance for CRC or anesthesia-related effects
(Table SI). Ingenuity Pathway
Analysis (IPA® version 81348237, Qiagen Digital
Insights, a subsidary of Qiagen Inc.) was used for in silico
analysis. Target-mRNAs were entered into IPA®, and only
experimentally confirmed relationships were considered for
signaling pathways identification and regulatory effect
characterization.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) validation
A technical validation via RT-qPCR (StepOnePlus™
Real-Time PCR System; Applied Biosystems™; Thermo Fisher
Scientific, Inc.) was performed from the same RNA sample utilized
for RNA sequencing libraries. A total RNA quantity of 400 ng was
used for reverse transcription, applying the QuantiTect Reverse
Transcription kit (Qiagen GmbH; Cat. no. 205313). The resulting
cDNA was diluted 1:20 for RT-qPCR using the SsoAdvanced Universal
SYBR-Green Supermix Kit (Bio-Rad Laboratories GmbH; Cat. no.
175272) and PrimePCR (lncRNA) SYBR-Green Assays (Bio-Rad
Laboratories GmbH). Additionally, QuantiNova LNA PCR Assays for
lncRNAs, HELLPAR_2464189 and TSIX_1589209 (Qiagen GmbH) and
QuantiTect Primer Assay for lncRNA CCDC26 (https://geneglobe.qiagen.com/) were used for repeating
the lncRNA HELLPAR, TSIX and CCDC26 quantification (Table II). Stable reference gene
candidates to normalize relative gene expression levels were
selected from RNA sequencing data utilizing the software tools
geNorm and NormFinder (NormqPCR v1.44.0) (35–37).
geNorm determines expression stability by assessing the pairwise
variation as the standard deviation of the logarithmically
transformed expression ratios of a particular gene to all other
genes (termed gene-stability measure M). Lower M values indicate a
more stable expression and genes with higher M values are
iteratively discarded to reevaluate the remaining genes to find
well suited references for normalization. Similarly, NormFinder
estimates variances on logarithmically transformed measured gene
expression to define a stability value for each gene, additionally
attempting to minimize the estimated intra- and inter-group
variation as well in a model-based approach. ZNF207, CAPZB and
CORO1A were identified as reference candidates for the TIVA cohort
among the top 15 in both tools with an M value threshold <0.5,
and VIM, VMP1, RASSF2 and DENND3 as reference candidates for the
VAG cohort (mean M value over all genes for TIVA 0.76 and for VAG
0.83). The RT-qPCR cycling conditions were as follows: 2 min at
95°C once for activation, followed by 40 cycles of 5 sec at 95°C
and 30 sec at 60°C. Melt curve steps were 65°C to 95°C at 0.5°C
increments for 5 sec/step. Following RT-qPCR, data were normalized
with the geometric mean of the selected stable reference genes
(ZNF207, CORO1A for the TIVA cohort, and VIM, VMP1, RASSF2, DENND3
for the VAG cohort. Relative quantification was performed applying
the 2−ΔΔCq method (38).
 | Table II.RT-qPCR validation of selected
lncRNAs and mRNAs. |
Table II.
RT-qPCR validation of selected
lncRNAs and mRNAs.
Statistical analysis
Comparison of patients' demographical data and
clinical parameters were performed with the non-parametic
Mann-Whitney U test. Fisher's exact test was performed for two
categorical variables, tumor location and stage according to Union
for International Cancer Control (UICC). The Chi-squared test was
performed for all other categorical variables. Differential gene
expression data was corrected for false discovery according to the
Benjamini-Hochberg correction (padj). IPA® (version
60467501; Qiagen, Inc.) was used for statistical data analyses. A
P-value (adjusted P-value for NGS data) ≤0.1 was considered to
indicate a statistically significant difference.
Results
RNA extraction and sequencing quality
and quantity
The RNA integrity number of the extracted RNA
determined by bioanalyzer was 8.033±1.333. Sequencing-quality
control was performed using Fast QC software. Sequencing libraries
of the runs were reasonably homogenous in size and composition
(Figs. 1 and 2, and Table III). Run 1 and run 2 contained
two VAG and three TIVA patients, and run 3 and run 4 contained 3
patients each.
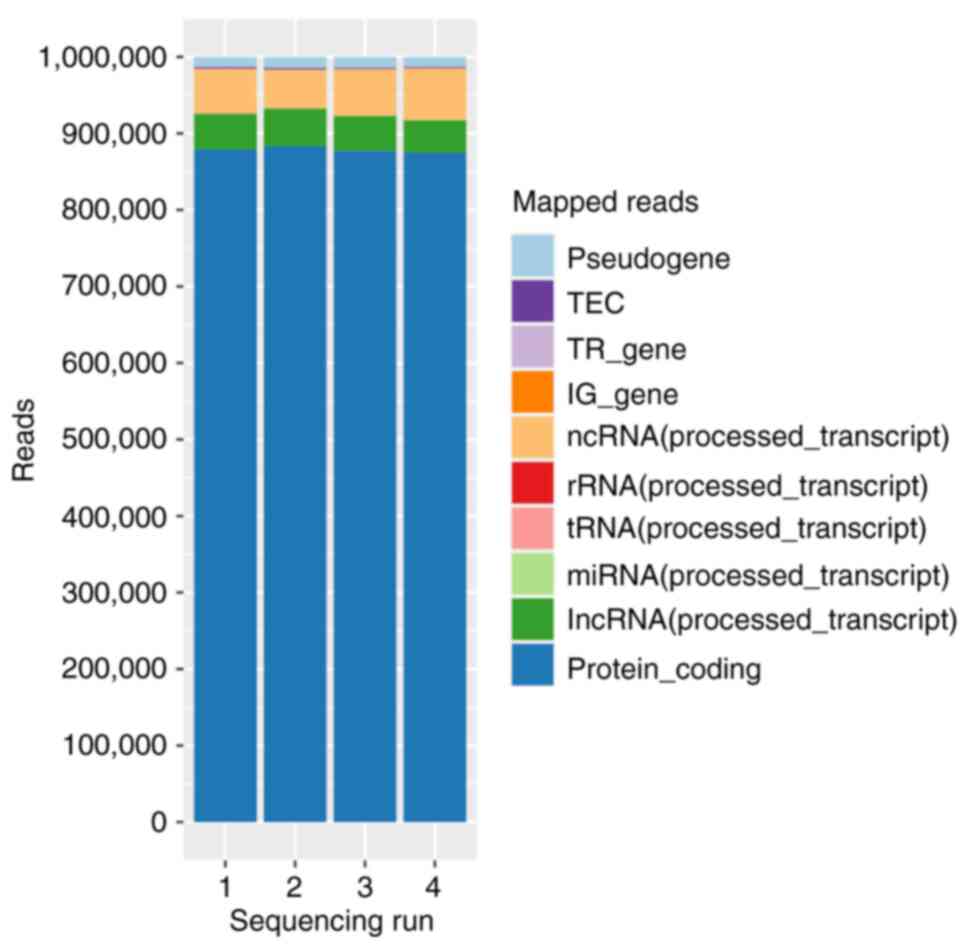 | Figure 1.Mean annotation distribution for each
of four sequencing runs. Within a run, both time points of patients
from the TIVA and VAG groups were included. TIVA, total intravenous
anesthesia; VAG, volatile anesthetic gas; RPM, reads per million;
miRNA, micro RNA; tRNA, transfer-RNA, rRNA, ribosomal RNA, ncRNA,
non-coding RNA, IG_gene, immunoglobulins_gene; TR_gene, thyroid
hormone receptor_gene; TEC, tyrosine-protein kinase. |
 | Table III.RNA sequencing quality and
quantity. |
Table III.
RNA sequencing quality and
quantity.
| Sequencing
category | Mean library size
(reads) | Mapped reads
(%) | lncRNA (%) | mRNA (%) |
|---|
| run 1 | 4.11E6±1.25E6 | 79.70±19.32 | 4.61±5.05 | 87.47±4.84 |
| run 2 | 3.54E6±9.60E5 | 75.38±19.87 | 4.81±5.01 | 88.25±6.86 |
| run 3 | 3.43E6±6.10E5 | 78.60±12.01 | 4.54±2.38 | 87.72±2.63 |
| run 4 | 2.76E6±5.85E5 | 78.10±14.33 | 4.18±3.89 | 87.51±4.66 |
| preTIVA | 3.22E6±8.83E5 | 73.46±18.82 | 4.06±2.93 | 86.89±3.90 |
| postTIVA | 3.85E6±8.31E5 | 81.77±14.03 | 4.74±3.08 | 88.81±2.26 |
| preVAG | 3.08E6±1.10E6 | 70.65±26.92 | 4.66±4.71 | 86.28±5.74 |
| postVAG | 3.27E6±9.75E5 | 81.05±20.04 | 4.69±6.23 | 88.14±8.04 |
Anesthesia-induced changes in lncRNA
profiles
An intra-group comparison (paired analysis) was
performed. DGE analysis considered the two time points within each
of the two groups. In total, 35 differentially regulated lncRNAs in
the TIVA-group (all upregulated) and 25 in the VAG-group (24
upregulated, one downregulated) were identified. A total of eight
lncRNAs (RP6-159A1.4, MIR646HG, LINC00694, ST3GAL4-AS1, LINC00937,
CTB-131B5.2, PLBD1-AS1, AC091878.1) were upregulated in both groups
(Table IV). Several of these
lncRNAs have already been shown to be associated with cancer:
PLBD1-AS1 (39), TTN-AS1 (40–42),
LINC01001 (43), RP11-701P16.5
(44), CTB-31N19.3/METTL9
(45), ST20-AS1 (46), HELLPAR (47), CCDC26 (48–50),
LINC00511 (51–53), SNHG23 (MEG8) (54), TSIX (55), LINC01127 (56).
 | Table IV.lncRNAs regulated postoperatively
compared with preoperative time point. |
Table IV.
lncRNAs regulated postoperatively
compared with preoperative time point.
| Regulation | lncRNA (Refs.) | baseMean |
log2FC | padj |
|---|
| TIVA
upregulated | RP6-159A1.4 | 3882.13 | −1.73 | 2.76E-06 |
|
| MIR646HG | 313.00 | −1.54 | 6.02E-04 |
|
| LINC00694 | 241.00 | −1.77 | 8.83E-04 |
|
| ST3GAL4-AS1 | 201.26 | −1.48 | 3.15E-03 |
|
| LINC00937 | 161.64 | −1.53 | 2.13E-04 |
|
| CTB-131B5.2 | 144.24 | −2.19 | 2.36E-04 |
|
| PLBD1-AS1 (40) | 123.81 | −1.18 | 2.64E-02 |
|
| AC091878.1 | 68.23 | −1.31 | 3.81E-02 |
|
| FAM157C | 1792.54 | −1.25 | 2.04E-03 |
|
| FAM157A | 1274.90 | −1.00 | 1.39E-02 |
|
| TTN-AS1
(41–43) | 434.98 | −1.03 | 2.05E-03 |
|
| FAM157B | 432.07 | −1.39 | 1.44E-03 |
|
|
RP11-81A1.6 | 422.04 | −1.18 | 1.92E-04 |
|
| LINC01001
(44) | 360.38 | −1.22 | 1.39E-02 |
|
|
RP11-83A24.2 | 280.34 | −1.02 | 5.03E-03 |
|
|
RP11-563J2.2 | 271.15 | −1.54 | 7.84E-05 |
|
|
RP4-669L17.10 | 269.11 | −1.02 | 3.92E-03 |
|
|
AC138035.2 | 129.84 | −1.14 | 4.87E-04 |
|
|
RP3-368A4.6 | 120.61 | −1.16 | 1.46E-02 |
|
|
LINC00211 | 119.63 | −1.45 | 4.93E-08 |
|
|
AC003104.1 | 116.80 | −1.10 | 1.97E-02 |
|
|
AC007278.2 | 112.79 | −2.57 | 6.14E-06 |
|
|
AC007278.3 | 112.72 | −2.52 | 9.08E-07 |
|
|
RP11-296O14.3 | 111.83 | −1.05 | 5.20E-03 |
|
|
RP11-563J2.3 | 104.44 | −1.62 | 5.95E-06 |
|
|
RP11-65L3.2 | 94.53 | −1.32 | 5.42E-04 |
|
|
CTC-490G23.2 | 78.78 | −2.34 | 2.32E-04 |
|
|
RP11-561P12.5 | 73.08 | −1.41 | 1.92E-02 |
|
|
RP11-212I21.4 | 72.92 | −1.01 | 4.70E-02 |
|
|
RP11-701P16.5 (45) | 65.57 | −1.77 | 3.22E-03 |
|
|
CTB-31N19.3/METTL9 (46) | 64.83 | −1.88 | 2.00E-06 |
|
|
RP11-981G7.1 | 61.78 | −1.11 | 2.66E-02 |
|
|
CTD-2530H12.2 | 56.48 | −1.44 | 1.30E-03 |
|
| ST20-AS1
(47) | 67.92 | −1.05 | 5.36E-02 |
|
|
RP11-242C19.2 | 52.96 | −1.50 | 3.92E-03 |
| VAG
upregulated | RP6-159A1.4 | 2051.03 | −1.42 | 9.55E-05 |
|
| MIR646HG | 161.36 | −1.45 | 1.46E-07 |
|
| LINC00694 | 129.39 | −1.90 | 3.06E-06 |
|
| ST3GAL4-AS1 | 94.53 | −1.70 | 2.75E-05 |
|
| LINC00937 | 82.99 | −1.54 | 1.99E-04 |
|
| CTB-131B5.2 | 65.08 | −2.12 | 4.52E-06 |
|
| PLBD1-AS1 | 67.11 | −1.53 | 5.30E-05 |
|
| AC091878.1 | 60.44 | −1.67 | 1.41E-03 |
|
| HELLPAR
(48) | 343.21 | −1.23 | 3.26E-02 |
|
|
RP11-76E17.3 | 156.81 | −1.95 | 4.15E-07 |
|
|
RP11-638I8.1 | 155.68 | −1.07 | 1.94E-03 |
|
|
RP13-580B18.4 | 113.59 | −1.76 | 2.05E-03 |
|
|
RP11-191L9.4 | 104.61 | −1.67 | 1.28E-03 |
|
|
CTA-212A2.3 | 82.30 | −1.47 | 4.25E-02 |
|
|
RP3-412A9.17 | 68.59 | −1.52 | 4.33E-02 |
|
| CCDC26
(49–51) | 57.83 | −1.43 | 4.52E-02 |
|
| LINC00511
(52–54) | 56.07 | −1.50 | 1.52E-02 |
|
| SNHG23
(55) | 51.90 | −1.97 | 9.79E-03 |
|
| TSIX
(56) | 51.72 | −1.42 | 3.10E-02 |
|
| LINC01127
(57) | 63.81 | −1.07 | 5.85E-02 |
|
|
RP11-989E6.10 | 82.68 | −1.29 | 6.24E-02 |
|
|
CTA-228A9.4 | 61.44 | −1.57 | 7.35E-02 |
|
|
RP3-394A18.1 | 114.24 | −1.19 | 5.04E-02 |
|
|
CH507-528H12.1 | 846.35 | −1.13 | 7.58E-02 |
| VAG
downregulated | AATBC | 58.95 | 1.34 | 5.29E-04 |
Anesthesia-induced changes in mRNA
profiles
In addition to lncRNAs, DGE analysis of mRNAs
between the pre- and postoperative time points revealed 1595 mRNAs
in the TIVA-group and 947 in the VAG-group, respectively. Of these,
1047 mRNAs (TIVA) and 399 mRNAs (VAG) were agent-specific (Table SII).
In silico identification of
lncRNA-associated target-mRNAs
Overall, 16 possible target-mRNAs were identified in
relation to six lncRNAs in the TIVA-group. Concerning seven lncRNA
from the VAG-group, 252 target-mRNAs were related. HELLPAR targeted
207 mRNAs, TSIX 36 and CCDC26 one, respectively. For 29 (TIVA) and
18 (VAG) lncRNAs, no target-mRNAs could be identified. The in
silico identified target-mRNAs were compared with the mRNAs
from the DGE analysis. In total, 12.5% (2 out of 16) of the in
silico identified target-mRNAs for the TIVA-group, 9.13% (23
out of 252) for the VAG-group, respectively, satisfied the cut-off
requirements for of the DGE analysis. Those remaining significantly
regulated target mRNAs are presented in Table V and were subsequently analyzed
with IPA®.
 | Table V.Target-mRNAs identified in
silico and verified in the DGE of the intra-group
comparison. |
Table V.
Target-mRNAs identified in
silico and verified in the DGE of the intra-group
comparison.
|
|
| Target mRNA |
|---|
|
|
|
|
|---|
| Group | lncRNA | mRNA | baseMean |
log2FC | padj |
|---|
| TIVA | FAM157A | HIPK2 | 789.76 | −0.78 | 4.21E-02 |
|
| ST20-AS1 | BCL9L | 249.43 | 0.80 | 5.85E-02 |
| VAG | CCDC26 | BCL2 | 136.61 | 1.15 | 1.10E-06 |
|
| TSIX | TYW5 | 64.77 | −1.01 | 8.06E-04 |
|
|
| PHACTR2 | 133.80 | −0.56 | 2.51E-02 |
|
| HELLPAR | TMEM170B | 199.91 | −0.67 | 2.57E-02 |
|
|
| SSBP2 | 72.63 | 0.67 | 8.54E-02 |
|
|
| SPN
(CD43) | 190.04 | 0.67 | 2.44E-02 |
|
|
| SPATA13 | 443.65 | −0.62 | 6.28E-02 |
|
|
| SLC46A3 | 53.37 | −0.69 | 5.91E-02 |
|
|
| RNF165 | 54.38 | −0.96 | 5.25E-02 |
|
|
| P2RX7
(P2X7) | 52.66 | 0.81 | 4.69E-02 |
|
|
| MUC4 | 105.31 | −1.65 | 3.31E-02 |
|
|
| MDFIC | 73.85 | 0.71 | 5.31E-02 |
|
|
| KLRD1 | 243.76 | −0.94 | 8.29E-02 |
|
|
| GPR27 | 151.79 | −1.14 | 9.44E-08 |
|
|
| EXOC6 | 166.52 | −0.88 | 7.74E-03 |
|
|
| EVL | 303.50 | 0.87 | 5.84E-02 |
|
|
| DGKH | 95.39 | −1.02 | 7.76E-03 |
|
|
| CTSC | 248.17 | 0.89 | 2.57E-04 |
|
|
| BEST1 | 377.61 | −0.59 | 7.12E-02 |
|
|
| AUTS2 | 101.90 | −0.85 | 6.26E-02 |
|
|
| ASPH | 364.13 | −1.52 | 1.34E-05 |
|
|
| ALDH5A1 | 110.39 | 0.60 | 6.15E-02 |
|
|
| AACS | 60.32 | −1.17 | 3.74E-02 |
IPA® analysis
To determine relevant biological functions and
signaling pathways of differentially expressed target-mRNAs, the
target-mRNAs were uploaded into IPA® for comparative
analyses (Fig. S1).
In the TIVA group, four canonical pathways were
determined, all of which involved the serine/threonine protein
kinase, homeodomain interacting protein kinase 2 (HIPK2), which is
upregulated by the lncRNA FAM157A.
For the VAG-group, IPA analysis revealed eleven
signaling pathways. In seven pathways the anti-apoptotic protein
BCL2, downregulated by lncRNA CCDC26, were predicted to be
involved. In the four other pathways, sialophorin (SPN; CD43), a
highly glycosylated transmembrane protein, purinergic receptor P2X
7 (P2RX7), a purine receptor for ATP, and aldehyde dehydrogenase 5
family member A1 (ALDH5A1), a succinate semialdehyde dehydrogenase,
all target-mRNAs of HELLPAR, were involved.
Additionally, target-mRNAs were also categorized to
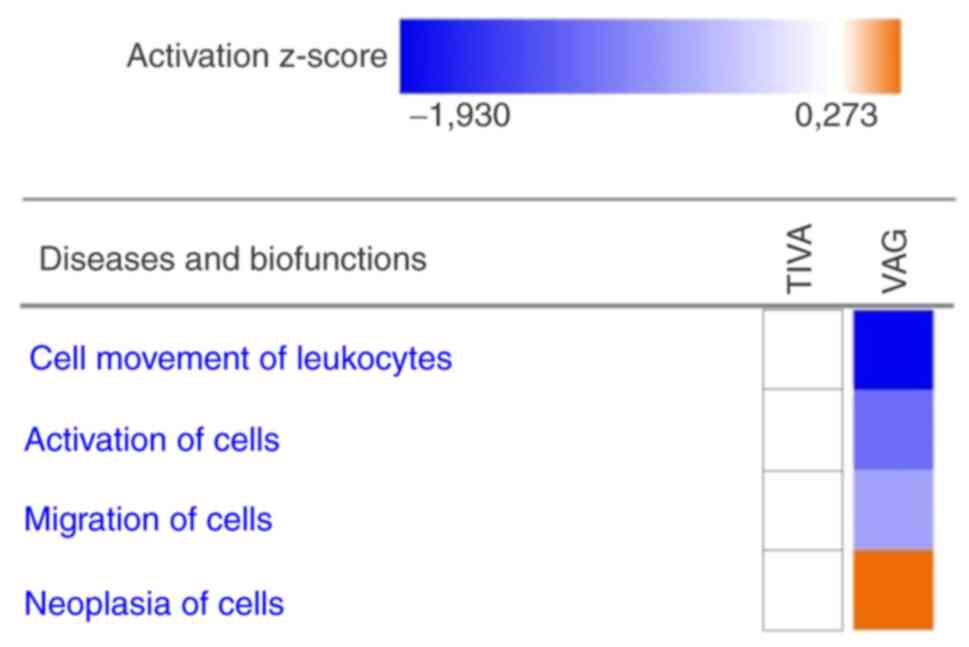
related diseases and biofunctions. The target-mRNAs of VAG
demonstrated a predicted downregulation in ‘Cell movement of
leukocytes’, ‘Activation of cells’ and ‘Migration of cells’. For
‘Neoplasia of cells’ an upregulation was predicted. For TIVA, no
change was predicted (Fig. 3).
Technical validation
The regulation of selected lncRNAs and mRNAs were
validated by using RT-qPCR. The results of the RT-qPCR were
compared with the corresponding results of the RNA sequencing
(Table V). In contrast to mRNA
assays, assays for lncRNAs have not yet been well established.
Nevertheless, the results for FAM157A and ST20-AS1 were obtained
that confirmed the previous results. For HELLPAR, CCDC26 and TSIX,
although different assays have been used (Bio-Rad Laboratories,
Inc. and Qiagen, Inc.), no results could be obtained.
Discussion
The primary aim of the present proof-of-concept
study was to analyze possible anesthesia-induced lncRNA and mRNA
expression changes in blood. Furthermore, a main focus of the
present study was to decipher the potential role of lncRNAs and
their target-mRNAs as molecular players in the observed beneficial
effect of TIVA on cancer outcome. In a previously performed study
by the authors, anesthesia-specific miRNA expression changes were
detected in circulating extracellular vesicles of CRC patients
undergoing tumor resection (19).
The present study extended earlier observations by the authors and
concentrated on lncRNAs derived from blood, since lncRNAs represent
a highly cancer-related class of non-coding RNAs (57). Blood samples obtained prior to the
induction of anesthesia and following the wound closure of patients
receiving either TIVA or VAG were compared and a comprehensive
lncRNA and mRNA expression profiling analysis was performed.
Overall, 35 differentially regulated lncRNAs for the TIVA-group and
25 for the VAG-group were identified (Table IV). With the exclusion of one
lncRNA in the VAG-group, all were upregulated. These results
demonstrated that lncRNA expression changes can be specific for the
anesthetic regime. In addition, 1,595 differentially regulated
mRNAs in the TIVA- and 947 in the VAG-intra-group comparison were
detected, of which 1047 mRNAs (TIVA) and 399 mRNAs (VAG) were
anesthetic-specific (Table
SII).
Several lncRNAs from Table IV had already been shown to be
associated with tumor growth (41,43–46),
cancer cell proliferation, migration and invasion (58–61).
However, the majority of lncRNAs identified in the present study
have not been previously investigated. Thus far, a limited amount
of research data linking lncRNAs from blood and type of anesthetic
used to cancer is available. As a consequence of limited data on
specific lncRNAs, knowledge on their target-mRNAs and association
to pathway regulations is even more scarce. To date, mainly cancer
tissues or cell lines have been examined directly. For example, in
human CRC cell lines, it was demonstrated that propofol promotes
apoptosis, proliferation and inhibits invasion, in which the
lncRNAs HOTAIR and HOXA11-AS were largely involved (62,63).
However, these lncRNAs, which have already been frequently
associated with cancer, could not be confirmed in the present
study. Instead, the combined sequencing and in silico
analyses of the present study revealed an association of the lncRNA
FAM157A (via target-mRNA HIPK2) for the TIVA group, respectively of
the lncRNAs CCDC26 (via target-mRNA BCL2) and HELLPAR (via
target-mRNA SPN and P2RX7) for the VAG group with cancer-related
pathways. Concerning FAM157A the analyses of the present study
provided novel evidence for an association of this lncRNA and its
identified target HIPK2 to several pivotal cancer signaling
pathways including ‘p53 Signaling’, ‘Cell Cycle: G2/M DNA damage
checkpoint regulation’, ‘Molecular Mechanisms of Cancer’ and the
‘Senescence Pathway’. These canonical pathways were sourced from
the Knowledge Base item underlying the IPA®
software and used for the causal analytic tools ‘Mechanistic
Networks’, ‘Causal Network Analysis’ and ‘Downstream Effects
Analysis’ implemented in the software (64). The IPA Knowledge Base is
created by millions of manually curated data obtained from
scientific journals, publicly available molecular content
databases, textbooks and more and allows the query, visualization
and computation across the Knowlegde Base in relationship to
miRNA and mRNA findings entered into IPA®. Previous
in vitro and in vivo studies suggested that HIPK2 is
a potential tumor suppressor and DNA damage-responsive kinase that
activates the apoptotic program in a phosphorylation-dependent
manner by targeting different downstream targets (65–67).
Upregulated HIPK2 has been demonstrated to interact with the tumor
suppressor p53, leading to cell cycle arrest of cancer cells during
surgery (68). The results of the
present study not only corroborate the effect of HIPK2 on the tumor
suppressor protein p53, but also give further insight on HIPK2
upstream regulatory mechanism and a possible explanation for the
anti-tumor effect of TIVA.
Another finding that emerged from the analyses is
the importance of the lncRNA HELLPAR that is upregulated after VAG
anesthesia. From a CRC study comparing patients with or without
metastases, it is known that HELLPAR is involved in the regulation
of proliferation and invasive ability of tumor cells, with higher
expression in patients with metastases than without (47).
In the in silico analyses of the present
study, it was demonstrated that HELLPAR regulates 20 out of 25
target-mRNAs in different directions. Two of these target-mRNAs,
SPN (CD43) and P2RX7 (P2X7), both downregulated, were included in
the ‘B-Cell Development’-signaling pathway and the ‘inflammasome
pathway’ derived from the aforementioned IPA Knowledge Base
in colon carcinoma cells. Anti-adhesive function of SPN (CD43)
expression has been associated with inhibition of adhesion to the
extracellular matrix, which has implications for tumorigenesis and
metastasis of CRC cells (69). The
upregulation of HELLPAR with the resulting downregulation of CD43
may therefore lead to an increased adhesion of cancer cells and
thus to increased metastasis. High P2RX7 expression correlated with
tumor size, metastasis and poor overall survival (70). The downregulation observed in the
present study could indicate a protective effect.
Another interesting finding was the association of
lncRNA CCDC26 upregulation with its downregulated target-mRNA BCL2
involved in the IPA Knowledge Base canonical pathways ‘p53
signaling’, ‘Autophagy’, ‘Interferon Signaling’, ‘Neuroinflammation
Signaling Pathway’, ‘Cytotoxic T lymphocyte-mediated apoptosis of
target cells’, ‘Docosahexaenoic Acid (DHA) Signaling’ and ‘Myc
Mediated Apoptosis Signaling’. Decreased overall survival as well
as tumor growth and apoptosis have already been linked to high
CCDC26 expression (50,71). In pancreatic cancer cell cultures,
CCDC26 knockdown decreased BCL2 mRNA and protein levels, while in
cancer tissue CCDC26 and BCL2 were upregulated (72). Downregulation of the anti-apoptotic
BCL2 ensures apoptosis in the mitochondria via release of
cytochrome c and also in the nucleus, via the p53 signaling pathway
(72–75). This could be explained by the
VAG-induced conversion of the positive correlation between CCDC26
and BCL2 into negative correlation, thus exerting a protective
effect.
An interesting conclusion derived from the results
of the present study based on the mRNA data from Table V is that by using the
abovementioned IPA® Knowlegde Base tool for
interpretation of these transcription data, remarkable inhibitory
effects were identified on the causal networks ‘cell movement of
leukocytes’, ‘activation of cells’ and ‘migration of cells’ in the
patients’ group anesthetized by VAG. These signaling networks are
crucial for the effectiveness of immune cells against tumor cells
and metastasis. Constraining these cellular actions culminated in a
downgraded immunological response against cancer cells. The
importance of an intact immune response for tumor elimination has
been proven by numerous studies (76–78).
Limitations of the present study are the relatively
small sample size and that the results are not complemented by
functional studies but were created by a combined sequencing and
in silico analysis of observational data. In addition, the
lncRNA and mRNA profile data are based on whole blood samples, with
the profile changes deriving from the white cell fraction which is
hypothesized by the authors that may be the blood fraction
primarily involved in fighting cancer. It is therefore very likely
that the anesthetic-related lncRNA response observed in the present
study may reflect a global host response, which could in turn have
effects on tumor outcome. These findings could provide evidence of
a novel lncRNA mediated mechanism of anesthetic effect on
immunologic and inflammatory signaling. However, an exact
delineation of the relationship between anesthetic action, the
immune system and cancer was not the primary aim of the present
study. Drawing blood after surgery termination before wound closure
was chosen as second time point, in order to ensure a sufficiently
long-time interval for possible anesthesia-induced effects on
intracellular RNA expression. Certainly, it should be subsequently
hypothesized that the consequences of including RNA expression
changes that might be induced by the surgical procedure, at least
partly, or by stress response. However, this type of confounding
effects may affect both patient groups. Further limiting factors
that may have an impact on the results are demographic and clinical
differences present in the two cohorts. For elderly patients and
patients with serious comorbidities, VAG was the choice of
anesthetic in the present study. Thus, an age difference between
the two groups with a P=0.016 was present and was hardly avoidable.
Concerning UICC stages, stage 4 was present in two patients of the
VAG group and none in the TIVA group. The influence of tumor stage
on lncRNA profile is, however, beyond the scope of this pilot study
and should be considered for larger cohort future studies.
In summary, the results of the present study
demonstrated that general anesthesia is able to orchestrate lncRNA
expression in blood, with differential and specific effects of the
anesthetic agent. The analyses identified target-mRNAs for more
than 17.1% (TIVA) and 28.0% (VAG) of these lncRNAs and classified
them in the context of cancer-relevant signaling pathways.
Canonical pathways identified for TIVA were cancer-specific and
suggestive of anti-tumor effects, whereas the possible influence of
VAG on tumor progression was less clear. The analysis of
target-mRNA of VAG revealed a markedly worsened immunological
response against cancer. These results provided preliminary
evidence for the presence of a novel lncRNA-mediated mechanism of
anesthetic action that, in addition to other immunoregulatory
effects, may influence tumor outcome. The lncRNA results of the
present study may only serve as a possible mechanistic explanation
for observations in several larger cohorts in retrospective
studies. Furthermore, the present study demonstrated novel effects,
to the best of our knowledge, of anesthetic agents on lncRNAs with
immunologic consequences not previously investigated.
According to the study design, the feasibility of
detecting anesthesia specific expression changes in blood-derived
lncRNA and mRNA profiles was demonstrated. Secondly, the results of
the present study, which included a combined high-throughput
sequencing and bioinformatic analysis, possibly indicated a
non-negligible role of lncRNAs as molecular players for the
proposed negative outcome of CRC patients anesthetized with VAG, in
comparison to TIVA anesthetization. In addition to studies on
circulating cell-free lncRNA profiles, the blood-derived lncRNA
landscape may provide significant insights for different research
inquiries on solid cancers and thus it appears that the further
elucidation of its potential may be be a promising future study
aim.
Supplementary Material
Supporting Data
Supporting Data
Supporting Data
Acknowledgements
The authors would like to thank Franz Jansen,
Institute of Human Genetics, University Hospital, LMU Munich for
his excellent technical assistance for RT-qPCR assays.
Funding
The present study was funded by the Monika Kutzner Stiftung
Grant, Rechtsfähige Stiftung des privaten Rechts zur Förderung der
Krebsforschung Bayerische Straße 8, 10707 Berlin.
Availability of data and materials
All data generated in the present study may be
acquired from the European Nucleotide Archive (ENA) under the
accession number, PRJEB56067 (https://www.ebi.ac.uk/ena/browser/view/PRJEB56067).
All other datasets used and/or analyzed during the current study
are available from the corresponding author on reasonable
request.
Authors' contributions
MR, GS, MWP and OKS contributed to the study
conception and design. Patient recruitment, blood sampling and
patient data acquisition were performed by FB, ASM and MB.
Molecular analysis was performed by AL and MR. Bioinformatic
analysis was performed by BK, ASM and GS. AL and MR drafted the
manuscript. OKS, GS and MWP prepared the manuscript. BK and MWP
confirm the authenticity of all the raw data. All authors have
commented on previous versions of the manuscript and have read and
approved the final manuscript.
Ethics approval and consent to
participate
The Ethics Committee of the Medical Faculty of the
Ludwig-Maximilians-University (LMU) Munich, Germany (to which the
Institute of Human Genetics and the Department of Anesthesiology
are assigned) approved the present study (protocol-no. 232-16). The
present study was performed in accordance with the Declaration of
Helsinki. Written informed consent was obtained and study samples
were pseudonymized.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CRC
|
colorectal cancer
|
|
DGE
|
differential gene expression
|
|
FC
|
fold change
|
|
padj
|
adjusted P-value
|
|
PCA
|
principal component analysis
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
TIVA
|
total intravenous anesthesia
|
|
VAG
|
volatile anesthetic gas
|
|
HIPK2
|
homeodomain interacting protein kinase
2
|
|
SPN
|
sialophorin
|
|
P2RX7
|
purinergic receptor P2X 7
|
|
ALDH5A1
|
aldehyde dehydrogenase 5 family member
A1
|
|
lncRNA
|
long non-coding RNA
|
References
|
1
|
Araghi M, Soerjomataram I, Jenkins M,
Brierley J, Morris E, Bray F and Arnold M: Global trends in
colorectal cancer mortality: Projections to the year 2035. Int J
Cancer. 144:2992–3000. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Werner J and Heinemann V: Standards and
challenges of care for colorectal cancer today. Visc Med.
32:156–157. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Demicheli R, Retsky MW, Hrushesky WJ, Baum
M and Gukas ID: The effects of surgery on tumor growth: A century
of investigations. Ann Oncol. 19:1821–1828. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tohme S, Simmons RL and Tsung A: Surgery
for cancer: A trigger for metastases. Cancer Res. 77:1548–1552.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jin Z, Li R, Liu J and Lin J: Long-term
prognosis after cancer surgery with inhalational anesthesia and
total intravenous anesthesia: A systematic review and
meta-analysis. Int J Physiol Pathophysiol Pharmacol. 11:83–94.
2019.PubMed/NCBI
|
|
6
|
Makito K, Matsui H, Fushimi K and Yasunaga
H: Volatile versus total intravenous anesthesia for cancer
prognosis in patients having digestive cancer surgery.
Anesthesiology. 133:764–773. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Guerrero Orriach JL, Raigon Ponferrada A,
Malo Manso A, Herrera Imbroda B, Escalona Belmonte JJ, Ramirez
Aliaga M, Ramirez Fernandez A, Diaz Crespo J, Soriano Perez AM,
Fontaneda Heredia A, et al: Anesthesia in combination with propofol
increases disease-free survival in bladder cancer patients who
undergo radical tumor cystectomy as compared to inhalational
anesthetics and opiate-based analgesia. Oncology. 98:161–167. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jun IJ, Jo JY, Kim JI, Chin JH, Kim WJ,
Kim HR, Lee EH and Choi IC: Impact of anesthetic agents on overall
and recurrence-free survival in patients undergoing esophageal
cancer surgery: A retrospective observational study. Sci Rep.
7:140202017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zheng X, Wang Y, Dong L, Zhao S, Wang L,
Chen H, Xu Y and Wang G: Effects of propofol-based total
intravenous anesthesia on gastric cancer: A retrospective study.
Onco Targets Ther. 11:1141–1148. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wu ZF, Lee MS, Wong CS, Lu CH, Huang YS,
Lin KT, Lou YS, Lin C, Chang YC and Lai HC: Propofol-based total
intravenous anesthesia is associated with better survival than
desflurane anesthesia in colon cancer surgery. Anesthesiology.
129:932–941. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang T, Fan Y, Liu K and Wang Y: Effects
of different general anaesthetic techniques on immune responses in
patients undergoing surgery for tongue cancer. Anaesth Intensive
Care. 42:220–227. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yan T, Zhang GH, Wang BN, Sun L and Zheng
H: Effects of propofol/remifentanil-based total intravenous
anesthesia versus sevoflurane-based inhalational anesthesia on the
release of VEGF-C and TGF-β and prognosis after breast cancer
surgery: A prospective, randomized and controlled study. BMC
Anesthesiol. 18:1312018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Abel F, Giebel B and Frey UH: Agony of
choice: How Anesthetics affect the composition and function of
extracellular vesicles. Adv Drug Deliv Rev. 175:1138132021.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ecimovic P, McHugh B, Murray D, Doran P
and Buggy DJ: Effects of sevoflurane on breast cancer cell function
in vitro. Anticancer Res. 33:4255–4260. 2013.PubMed/NCBI
|
|
15
|
Iwasaki M, Zhao H, Jaffer T, Unwith S,
Benzonana L, Lian Q, Sakamoto A and Ma D: Volatile anaesthetics
enhance the metastasis related cellular signalling including CXCR2
of ovarian cancer cells. Oncotarget. 7:26042–26056. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kvolik S, Glavas-Obrovac L, Bares V and
Karner I: Effects of inhalation anesthetics halothane, sevoflurane,
and isoflurane on human cell lines. Life Sci. 77:2369–2383. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Müller-Edenborn B, Roth-Z'graggen B,
Bartnicka K, Borgeat A, Hoos A, Borsig L and Beck-Schimmer B:
Volatile anesthetics reduce invasion of colorectal cancer cells
through down-regulation of matrix metalloproteinase-9.
Anesthesiology. 117:293–301. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Forrest ME, Saiakhova A, Beard L, Buchner
DA, Scacheri PC, LaFramboise T, Markowitz S and Khalil AM: Colon
cancer-upregulated long non-coding RNA lincDUSP Regulates cell
cycle genes and potentiates resistance to apoptosis. Sci Rep.
8:73242018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Buschmann D, Brandes F, Lindemann A,
Maerte M, Ganschow P, Chouker A, Schelling G, Pfaffl MW and
Reithmair M: Propofol and sevoflurane differentially impact
MicroRNAs in circulating extracellular vesicles during colorectal
cancer resection: A pilot study. Anesthesiology. 132:107–120. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Dinger ME, Pang KC, Mercer TR and Mattick
JS: Differentiating protein-coding and noncoding RNA: Challenges
and ambiguities. PLoS Comput Biol. 4:e10001762008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bánfai B, Jia H, Khatun J, Wood E, Risk B,
Gundling WE Jr, Kundaje A, Gunawardena HP, Yu Y, Xie L, et al: Long
noncoding RNAs are rarely translated in two human cell lines.
Genome Res. 22:1646–1657. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Guttman M, Amit I, Garber M, French C, Lin
MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al:
Chromatin signature reveals over a thousand highly conserved large
non-coding RNAs in mammals. Nature. 458:223–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ji Q, Zhang L, Liu X, Zhou L, Wang W, Han
Z, Sui H, Tang Y, Wang Y, Liu N, et al: Long non-coding RNA MALAT1
promotes tumour growth and metastasis in colorectal cancer through
binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2
complex. Br J Cancer. 111:736–748. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Siddiqui H, Al-Ghafari A, Choudhry H and
Al Doghaither H: Roles of long non-coding RNAs in colorectal cancer
tumorigenesis: A review. Mol Clin Oncol. 11:167–172.
2019.PubMed/NCBI
|
|
27
|
Thiele JA, Hosek P, Kralovcova E, Ostasov
P, Liska V, Bruha J, Vycital O, Rosendorf J, Opattova A, Horak J,
et al: lncRNAs in Non-Malignant tissue have prognostic value in
colorectal cancer. Int J Mol Sci. 19:26722018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Martin M: Cutadapt removes adapter
sequences from high-throughput sequencing reads. EMBnet. J.
17:10–12. 2011.
|
|
29
|
Dobin A, Davis CA, Schlesinger F, Drenkow
J, Zaleski C, Jha S, Batut P, Chaisson M and Gingeras TR: STAR:
Ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li B and Dewey CN: RSEM: Accurate
transcript quantification from RNA-Seq data with or without a
reference genome. BMC Bioinformatics. 12:3232011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15:5502014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lin Y, Liu T, Cui T, Wang Z, Zhang Y, Tan
P, Huang Y, Yu J and Wang D: RNAInter in 2020: RNA interactome
repository with increased coverage and annotation. Nucleic Acids
Res. 48(D1): D189–D197. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yang C, Yang L, Zhou M, Xie H, Zhang C,
Wang MD and Zhu H: LncADeep: An ab initio lncRNA identification and
functional annotation tool based on deep learning. Bioinformatics.
34:3825–3834. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kanehisa M and Sato Y: KEGG Mapper for
inferring cellular functions from protein sequences. Protein Sci.
29:28–35. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Vandesompele J, De Preter K, Pattyn F,
Poppe B, Van Roy N, De Paepe A and Speleman F: Accurate
normalization of real-time quantitative RT-PCR data by geometric
averaging of multiple internal control genes. Genome Biol.
3:RESEARCH00342002. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Andersen CL, Jensen JL and Ørntoft TF:
Normalization of real-time quantitative reverse transcription-PCR
data: A model-based variance estimation approach to identify genes
suited for normalization, applied to bladder and colon cancer data
sets. Cancer Res. 64:5245–5250. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Perkins JR, Dawes JM, McMahon SB, Bennett
DL, Orengo C and Kohl M: ReadqPCR and NormqPCR: R packages for the
reading, quality checking and normalisation of RT-qPCR
quantification cycle (Cq) data. BMC Genomics. 13:2962012.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Deng X, Bi Q, Chen S, Chen X, Li S, Zhong
Z, Guo W, Li X, Deng Y and Yang Y: Identification of a
Five-Autophagy-Related-lncRNA signature as a novel prognostic
biomarker for hepatocellular carcinoma. Front Mol Biosci.
7:6116262021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lin C, Zhang S, Wang Y, Wang Y, Nice E,
Guo C, Zhang E, Yu L, Li M, Liu C, et al: Functional role of a
novel long noncoding RNA TTN-AS1 in esophageal squamous cell
carcinoma progression and metastasis. Clin Cancer Res. 24:486–498.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang Y, Li D, Lu J, Chen L, Zhang S, Qi W,
Li W and Xu H: Long noncoding RNA TTN-AS1 facilitates tumorigenesis
and metastasis by maintaining TTN expression in skin cutaneous
melanoma. Cell Death Dis. 11:6642020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Jia Y, Duan Y, Liu T, Wang X, Lv W, Wang
M, Wang J and Liu L: LncRNA TTN-AS1 promotes migration, invasion,
and epithelial mesenchymal transition of lung adenocarcinoma via
sponging miR-142-5p to regulate CDK5. Cell Death Dis. 10:5732019.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hong MEI, Changen LI, Liang Y and Yingfei
GAO: Expression of lncRNA LINC01001 in breast cancer and its effect
on proliferation of MCF-7 cells. Chin J Cancer Biother. 25:158–162.
2018.
|
|
44
|
Zhou M, Guo M, He D, Wang X, Cui Y, Yang
H, Hao D and Sun J: A potential signature of eight long non-coding
RNAs predicts survival in patients with non-small cell lung cancer.
J Transl Med. 13:2312015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Lv M, Cao D, Zhang L, Hu C, Li S, Zhang P,
Zhu L, Yi X, Li C, Yan A, et al: METTL9 regulates N1-histidine
methylation of zinc transporters to promote tumor growth. bioRxiv.
2021.2004.2020.440582. 2021.
|
|
46
|
Wang W, Zhao Z, Yang F, Wang H, Wu F,
Liang T, Yan X, Li J, Lan Q, Wang J and Zhao J: An immune-related
lncRNA signature for patients with anaplastic gliomas. J
Neurooncol. 136:263–271. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kutilin DS, Gusareva MA, Kosheleva NG,
Zinkovich MS, Gvaramiya AK, Gappoeva MA, Fatkina NB, Krokhmal JN,
Vasilieva EO, Karnauhova EA, et al: Differential expression of long
noncoding RNAs in patients with metastatic and nonmetastatic
colorectal cancer. J Clin Oncol. 39 (15_suppl):e150232021.
View Article : Google Scholar
|
|
48
|
Gourvest M, Brousset P and Bousquet M:
Long noncoding RNAs in acute myeloid leukemia: Functional
characterization and clinical relevance. Cancers (Basel).
11:16382019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Chen C, Wang P, Mo W, Zhang Y, Zhou W,
Deng T, Zhou M, Chen X, Wang S and Wang C: lncRNA-CCDC26, as a
novel biomarker, predicts prognosis in acute myeloid leukemia.
Oncol Lett. 18:2203–2211. 2019.PubMed/NCBI
|
|
50
|
Ma X, Li Y, Song Y and Xu G: Long
Noncoding RNA CCDC26 promotes thyroid cancer malignant progression
via miR-422a/EZH2/Sirt6 Axis. Onco Targets Ther. 14:3083–3094.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Hu Y, Zhang Y, Ding M and Xu R: LncRNA
LINC00511 Acts as an oncogene in colorectal cancer via sponging
miR-29c-3p to upregulate NFIA. Onco Targets Ther. 13:13413–13424.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhang J, Sui S, Wu H, Zhang J, Zhang X, Xu
S and Pang D: The transcriptional landscape of lncRNAs reveals the
oncogenic function of LINC00511 in ER-negative breast cancer. Cell
Death Dis. 10:5992019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Lu G, Li Y, Ma Y, Lu J, Chen Y, Jiang Q,
Qin Q, Zhao L, Huang Q, Luo Z, et al: Long noncoding RNA LINC00511
contributes to breast cancer tumourigenesis and stemness by
inducing the miR-185-3p/E2F1/Nanog axis. J Exp Clin Cancer Res.
37:2892018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Terashima M, Ishimura A, Wanna-Udom S and
Suzuki T: MEG8 long noncoding RNA contributes to epigenetic
progression of the epithelial-mesenchymal transition of lung and
pancreatic cancer cells. J Biol Chem. 293:18016–18030. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Salama EA, Adbeltawab RE and El Tayebi HM:
XIST and TSIX: Novel cancer immune biomarkers in
PD-L1-Overexpressing breast cancer patients. Front Oncol.
9:14592020. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Jing L, Gong M, Lu X, Jiang Y, Li H and
Cheng W: LINC01127 promotes the development of ovarian tumors by
regulating the cell cycle. Am J Transl Res. 11:406–417.
2019.PubMed/NCBI
|
|
57
|
Fang Y and Fullwood MJ: Roles, functions,
and mechanisms of long Non-coding RNAs in cancer. Genomics
Proteomics Bioinformatics. 14:42–54. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Feng H, Zhang X, Lai W and Wang J: Long
non-coding RNA SLC16A1-AS1: Its multiple tumorigenesis features and
regulatory role in cell cycle in oral squamous cell carcinoma. Cell
Cycle. 19:1641–1653. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Li F, Li Q and Wu X: Construction and
analysis for differentially expressed long non-coding RNAs and
MicroRNAs mediated competing endogenous RNA network in colon
cancer. PLoS One. 13:e01924942018. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Zhao X, Fan Y, Lu C, Li H, Zhou N, Sun G
and Fan H: PCAT1 is a poor prognostic factor in endometrial
carcinoma and associated with cancer cell proliferation, migration
and invasion. Bosn J Basic Med Sci. 19:274–281. 2019.PubMed/NCBI
|
|
61
|
Zhang Q, Ding Z, Wan L, Tong W, Mao J, Li
L, Hu J, Yang M, Liu B and Qian X: Comprehensive analysis of the
long noncoding RNA expression profile and construction of the
lncRNA-mRNA co-expression network in colorectal cancer. Cancer Biol
Ther. 21:157–169. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Zhang YF, Li CS, Zhou Y and Lu XH: Effects
of propofol on colon cancer metastasis through STAT3/HOTAIR axis by
activating WIF-1 and suppressing Wnt pathway. Cancer Med.
9:1842–1854. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Ren YL and Zhang W: Propofol promotes
apoptosis of colorectal cancer cells via alleviating the
suppression of lncRNA HOXA11-AS on miRNA let-7i. Biochem Cell Biol.
98:90–98. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Krämer A, Green J, Pollard J Jr and
Tugendreich S: Causal analysis approaches in ingenuity pathway
analysis. Bioinformatics. 30:523–530. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Sombroek D and Hofmann TG: How cells
switch HIPK2 on and off. Cell Death Differ. 16:187–194. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Hofmann TG, Möller A, Sirma H, Zentgraf H,
Taya Y, Dröge W, Will H and Schmitz ML: Regulation of p53 activity
by its interaction with homeodomain-interacting protein kinase-2.
Nat Cell Biol. 4:1–10. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
67
|
Ozaki T and Nakagawara A: Role of p53 in
cell death and human cancers. Cancers (Basel). 3:994–1013. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Puca R, Nardinocchi L, Givol D and D'Orazi
G: Regulation of p53 activity by HIPK2: Molecular mechanisms and
therapeutical implications in human cancer cells. Oncogene.
29:4378–4387. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Park WS, Kim HJ, Lee GK, Son HS and Bae Y:
Anti-adhesive functions of CD43 expressed on colon carcinoma cells
through the modulation of integrins. Exp Mol Pathol. 92:82–89.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Lara R, Adinolfi E, Harwood CA, Philpott
M, Barden JA, Di Virgilio F and McNulty S: P2X7 in cancer: From
molecular mechanisms to therapeutics. Front Pharmacol. 11:7932020.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Hirano T, Yoshikawa R, Harada H, Harada Y,
Ishida A and Yamazaki T: Long noncoding RNA, CCDC26, controls
myeloid leukemia cell growth through regulation of KIT expression.
Mol Cancer. 14:902015. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Peng W and Jiang A: Long noncoding RNA
CCDC26 as a potential predictor biomarker contributes to
tumorigenesis in pancreatic cancer. Biomed Pharmacother.
83:712–717. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Hardwick JM and Soane L: Multiple
functions of BCL-2 family proteins. Cold Spring Harb Perspect Biol.
5:a0087222013. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Tsujimoto Y: Role of Bcl-2 family proteins
in apoptosis: Apoptosomes or mitochondria? Genes Cells. 3:697–707.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Chiou SK, Rao L and White E: Bcl-2 blocks
p53-dependent apoptosis. Mol Cell Biol. 14:2556–2563. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Blagih J, Buck MD and Vousden KH: p53,
cancer and the immune response. J Cell Sci. 133:jcs2374532020.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Bronte V, Cingarlini S, Marigo I, De Santo
C, Gallina G, Dolcetti L, Ugel S, Peranzoni E, Mandruzzato S and
Zanovello P: Leukocyte infiltration in cancer creates an
unfavorable environment for antitumor immune responses: A novel
target for therapeutic intervention. Immunol Invest. 35:327–357.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Garziera M and Toffoli G: Inhibition of
host immune response in colorectal cancer: Human leukocyte
antigen-G and beyond. World J Gastroenterol. 20:3778–3794. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Brierley JD, Gospodarowicz MK and
Wittekind C: TNM Classification of Malignant Tumours. 8th Edition.
Wiley-Blackwell; New Jersey: pp. pp2722016
|