Introduction
Lymphoma is a type of malignancy originating from
the lymphatic-hematopoietic system, which mainly includes Hodgkin's
lymphoma and non-Hodgkin's lymphoma, the latter accounting for
80–90% of all lymphoma cases (1).
Non-Hodgkin's lymphoma is derived from natural killer cells, or
B/T-lymphocytes. Diffuse large B-cell lymphoma (DLBCL) is the most
common type, and accounts for 30–40% of non-Hodgkin's lymphomacases
(2). DLBCL primarily occurs in the
lymph nodes, and also in extranodal tissues, including the
gastrointestinal tract, bone and central nervous system (3). Due to its heterogeneity, there are
differences in the clinical presentation of DLBCL. The diagnosis of
DLBCL cases is generally made from the pathological determination
of excisional lymph nodes (2,4).
Therefore, the development of sensitive and specific markers for
the clinical diagnosis of DLBCL would be beneficial.
Calponin 3 (CNN3) is a thin filament-associated
protein, implicated in the modulation and regulation of smooth
muscle contraction. Initially, CNN3 was revealed to inhibit the Mg
ATPase activity of smooth muscle myosin (5,6).
Subsequent evidence demonstrated that CNN3 induced actin
polymerization and hindered depolymerization of actin filaments
(7). Furthermore, CNN3 is also
expressed in the brain, in order to regulate the dendritic spine
plasticity of hippocampal neurons, with the systemic knockout of
CNN3 resulting in embryonic and neonatal lethality, due to
developmental defects of the central nervous system (8,9).
Additionally, CNN3 regulates the actin cytoskeleton rearrangement
of choriocarcinoma cells (10),
suggesting its effects on tumorigenesis. Recent reports have
demonstrated the cancer-promoting roles of CNN3. For instance, the
ectopic expression of CNN3 promotes the growth and metastasis of
cervical cancer cells (11). CNN3
enhances the invasion and drug resistance of digestive tract cancer
cells, including colon cancer and gastric cancer cells (12,13).
However, the effects of CNN3 on hematopoietic malignancies remain
unknown. In the present study, gain- and loss-of-function assays
were performed to confirm the roles of CNN3 in DLBCL cells.
Additionally, forkhead box O3 (FOXO3) has been
reported to function as a tumor suppressor in various cancer types,
including gastric (14), prostate
(15) and lung cancer (16). A recent study reported that the
silencing of FOXO3 promoted the proliferation and inhibited the
apoptosis of DLBCL cells (17). As
a transcription factor, FOXO3 simultaneously functions as a
transcription enhancer and suppressor. For instance, FOXO3 promotes
the transcription of BH3 interacting domain death agonist and
cathepsin L (18,19), and inhibits the transcription of
integrin β1 and interleukin 10 (20,21).
The data of the present study demonstrated that the
expression of CNN3 was downregulated by FOXO3, suggesting that
FOXO3 may function by regulating CNN3 in DLBCL cells. To verify
this hypothesis, the effects of FOXO3 and CNN3 were evaluated in
DLBCL cells, and rescue experiments were also performed in the
present study.
Materials and methods
Cell culture and treatment
The human DLBCL cell lines, DB (cat. no. CL-0645,
Procell Life Science & Technology Co., Ltd.) and SU-DHL-4 (cat.
no. CC1606, Guangzhou Cellcook Biotech Co., Ltd.), were cultured
with RPMI-1640 (Beijing Solarbio Science & Technology Co.,
Ltd.) containing 10% fetal bovine serum (FBS) at 37°C with 5%
CO2, and 293T cells (cat. no. ZQ0033, Shanghai Zhong
Qiao Xin Zhou Biotechnology Co., Ltd.) were cultured with DMEM
(Wuhan Servicebio Biotechnology Co., Ltd.) supplemented with 10%
FBS.
Cell transfection
To control the expression of CNN3 and FOXO3, the
coding sequence of CNN3 or FOXO3 was cloned into pcDNA3.1 vector
(Shaanxi Youbio Technology Co., Ltd.). The short interference RNA
(siRNA), short hairpin RNA (shRNA) targeting CNN3 and their
negative control (NC) sequences were synthesized by General Biology
Co., Ltd., and shRNA was inserted into pRNA-H1.1 (General Biology
Co., Ltd.). All siRNA and shRNA sequences are presented in Table I. The DB or SU-DHL-4 cells were
transfected with plasmid or RNA (original concentration: Plasmid,
0.1 µg/µl; siRNA, 0.01 nmol/µl) using Lipo8000 reagent (Beyotime
Institute of Biotechnology) 2.5 µg plasmid or 100 pmol siRNA for
2–3×105 cells in 200 µl solution) for 4–6 h.
Subsequently, 20 h following transfection, the cells were used in
subsequent analyses. The siRNA and shRNA sequences are presented in
Table I.
 | Table I.Sequence information of siRNAs and
shRNAs. |
Table I.
Sequence information of siRNAs and
shRNAs.
| Name | Sequence
(5′-3′) |
|---|
| CNN3 siRNA-a
sense |
GCAAGUAUAUGAUCCCAAATT |
| CNN3 siRNA-a
anti-sense |
UUUGGGAUCAUAUACUUGCTT |
| CNN3 siRNA-b
sense |
GCUCAGUGAAGAAGGUCAATT |
| CNN3 siRNA-b
anti-sense |
UUGACCUUCUUCACUGAGCTT |
| CNN3 siRNA-c
sense |
GGAGUUAAGUAUGCAGAAATT |
| CNN3 siRNA-c
anti-sense |
UUUCUGCAUACUUAACUCCTT |
| CNN3 siRNA-d
sense |
CGGCCGAAGUCAAGAACAATT |
| CNN3 siRNA-d
anti-sense |
UUGUUCUUGACUUCGGCCGTT |
| siRNA NC sense |
UUCUCCGAACGUGUCACGUTT |
| siRNA NC
anti-sense |
ACGUGACACGUUCGGAGAATT |
| CNN3 shRNA-1 |
GGCAAGTATATGATCCCAAATTCAAGAGATTTGGGATCATATACTTGCTTTTT |
| CNN3 shRNA-2 |
GGCTCAGTGAAGAAGGTCAATTCAAGAGATTGACCTTCTTCACTGAGCTTTTT |
| shRNA NC |
GTTCTCCGAACGTGTCACGTTTCAAGAGAACGTGACACGTTCGGAGAATTTTT |
It was observed that the DB cells grew slightly more
rapidly than the SU-DHL-4 cells; thus, the DB cells were used in
the xenograft experiment. In order to obtain cells stably
overexpressing CNN3 or cells in which CNN3 was silenced, DB cells
transfected with overexpression or knockdown vector were treated
with G418 (200 µg/ml; Beijing Solarbio Science & Technology
Co., Ltd.) for ~6 weeks. The cells remaining alive were considered
as CNN3-stably-overexpressing or -silenced DLBCL cells.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA extractionwas performed using TRIpure
lysis buffer (Beijing BioTeke Corporation Co., Ltd.) and
trichloromethane (Shanghai GenePharma Co., Ltd.), and purified with
isopropanol and 75% ethanol (Shanghai GenePharma Co., Ltd.).
Following quantification with a NANO2000 spectrophotometer (Thermo
Fisher Scientific, Inc.), the RNA was reverse transcribed into cDNA
using BeyoRT II M-MLV reverse transcriptase (Beyotime Institute of
Biotechnology). Subsequently, qPCR was performed to determine the
mRNA levels of FOXO3 and CNN3. The PCR procedure was performed as
follows: Pre-denaturation at 94°C for 5 min 10 sec, annealing at
60°C for 20 sec, extension at 72°C for 30 sec, followed with 40
cycles of 72°C for 2 min and 30 sec, 40°C for 1 min 30 sec, melting
60–94°C every 1°C for 1 sec, and 25°C for 1–2 min. The primers were
synthesized by General Biology Co., Ltd., and all primer
information is presented in Table
II. The PCR data were analyzed using the 2−ΔΔCq
method (22).
 | Table II.PCR primer sequences. |
Table II.
PCR primer sequences.
| Gene ID | Name | Temperature
(°C) | Sequence | Length of
amplification (bp) |
|---|
| NM_001455.4 | FOXO3 forward | 52.9 |
5′-TGACGACAGTCCCTCCC-3′ | 112 |
|
| FOXO3 reverse | 53.2 |
5′-GCTGGCGTTAGAATTGGT-3′ |
|
| NM_001839.5 | CNN3 forward | 46.4 |
5′-ATCATCCTCTGCGAACT-3′ | 136 |
|
| CNN3 reverse | 45.2 |
5′-CCATAAGCCTGAATAGC-3′ |
|
|
| ChIP CNN3
forward-1 | 44.4 |
5′-AAATCCTGCACTCCTTA-3′ | 138 |
|
| ChIP CNN3
reverse-1 | 44.1 |
5′-CTGACTGCTCCTGTTGT-3′ |
|
|
| ChIP CNN3
forward-2 | 46.8 |
5′-ACGGTTCAACTATGATGTT-3′ | 166 |
|
| ChIP CNN3
reverse-2 | 47.6 |
5′-CTCTTGCCACCACTTTC-3′ |
|
Western blot analysis
Protein was extracted from the DB or SU-DHL-4 cells
using RIPA lysis buffer (Beijing Solarbio Science & Technology
Co., Ltd.) supplemented with 1% phenylmethylsulfonyl fluoride
(Beijing Solarbio Science & Technology Co., Ltd.). The protein
concentration was determined using a BCA protein assay kit (Beijing
Solarbio Science & Technology Co., Ltd.). After mixing with 6X
loading buffer, the protein was denatured in boiling. A total of 20
µl of protein sample (10–20 µg protein) was loaded onto sodium
lauryl sulfate-polyacrylamide gel, and subjected to
electrophoresis. The gel concentration was 5–12% depending on the
protein size. Subsequently, the proteins were transferred onto PVDF
membranes, blocked with skim milk (Sangon Biotech Co., Ltd.) at
room temperature for 30 min, and incubated with the following
antibodies at a 1:1,000 dilution at 4°C overnight: Rabbit
proliferation cell nuclear antigen (PCNA; cat. no. 24036-1-AP;
Wuhan Sanying Biotechnology), rabbit cyclin D1 (cat. no. AF0931;
Affinity Biosciences), rabbit cyclin-dependent kinase (CDK)6 (cat.
no. A0106, ABclonal Biotech Co., Ltd.), rabbit CDK2 (cat. no.
A0294; ABclonal Biotech Co., Ltd.), rabbit phosphorylated
retinoblastoma transcription co-repressor (pRB; Ser807/811; cat.
no. AP0484; ABclonal Biotech Co., Ltd.), rabbit cleaved caspase-3
(cat. no. AF7022, Affinity Biosciences), rabbit cleaved poly
ADP-ribose polymerase-1 (PARP-1; cat. no. AF7023, ABclonal Biotech
Co., Ltd.), cleaved caspase-9 (cat. no. 20750; Cell Signaling
Technology, Inc.), rabbit FOXO3 (cat. no. A0102; ABclonal Biotech
Co., Ltd.), rabbit CNN3 (cat. no. DF9323; Affinity Biosciences) or
mouse GAPDH (cat. no. 60004-1-Ig; Wuhan Sanying Biotechnology).
After rinsing with 0.15% TBST buffer, the protein was incubated
with goat anti-rabbit IgG labeled with horseradish peroxidase (HRP;
cat. no. SE134; Beijing Solarbio Science & Technology Co.,
Ltd.) or goat anti-mouse IgG labeled with HRP (cat. no. SE131;
Beijing Solarbio Science & Technology Co., Ltd.) at a 1:3,000
dilution, and reacted with ECL reagent (Beijing Solarbio Science
& Technology Co., Ltd.), followed by signal exposure in the
dark. The optical density of the bands was analyzed using
Gel-Pro-Analyzer 4.0 software (Media Cybernetics, Inc.).
Cell counting kit (CCK)-8 assay
The DB or SU-DHL-4 cells were cultured in 96-well
plates. Following culture for 0 h, 24 h, 48 h or 72 h, the CCK-8
reagent (Biosharp Life Sciences) was added with 10 µl per well to
treat the cells at 37°C. After 2 h later, the optical density was
measured at 450 nm (OD450) of the supernatant with a microplate
reader (BioTek Instruments, Inc.).
EdU assay
The DB or SU-DHL-4 cells were stained with 10 µM EdU
reagent (Nanjing KeyGen Biotech. Co., Ltd.) at 37°C for 2 h, and
fixed with 4% paraformaldehyde for 15 min. Subsequently, all cells
were washed with PBS containing 3% bovine serum albumin (BSA),
incubated with 0.5% Triton X-100 for 20 min and incubated with
Click-iT reagent (Nanjing KeyGen Biotech. Co., Ltd.) at room
temperature for 30 min in the dark. Finally, the cells were stained
with DAPI reagent (Beijing Solarbio Science & Technology Co.,
Ltd.) at room temperature for 5 min, and photographed at a ×400
magnification.
Flow cytometry
Flow cytometry was conducted in order to analyze
cell cycle and apoptosis of DB and SU-DHL-4 cells. For apoptosis
determination, the cells were incubated with Annexin V-FITC (cat.
no. BL110A, Biosharp Life Sciences) for 10 min and propidium iodide
(PI) (cat. no. BL110A, Biosharp Life Sciences) for 5 min at room
temperature in the dark. Subsequently, the Annexin
V+PI− cells were counted as early apoptotic
cells, and Annexin V+PI+ cells were counted
as late apoptotic cells. For cell cycle analysis, the cells were
fixed with 70% ethanol at 4°C for 2 h, stained with PI at 37°C for
30 min in the dark, and detected using a flow cytometer (ACEA
Bioscience, Inc.). The apoptosis analysis were performed with
NovoExpress 1.4.1 software (ACEA Bioscience, Inc.).
Dual-luciferase reporter assay
In order to confirm the binding between FOXO3 and
the promoter sequence of CNN3, dual-luciferase reporter assay was
executed in 293T cells. The cells were transfected with pGL3 vector
(Shaanxi Youbio Technology Co., Ltd.) containing CNN3 promoter
fragment and FOXO3 overexpression plasmid (Shaanxi Youbio
Technology Co., Ltd.) at 37°C in the presence of Lipo8000 reagent
(Beyotime Institute of Biotechnology) for 4–6 h. Following
transfection for 48 h, the cells were collected and treated with a
dual-luciferase reporter assay kit (Nanjing KeyGen Biotech. Co.,
Ltd.), and the Firefly and Renilla value was determined
using a microplate reader (BioTek Instruments, Inc.).
Chromatin immunoprecipitation
(ChIP)
The ChIP assay was performed to confirm the binding
between FOXO3 and the promoter sequence of CNN3 using a ChIP kit
(cat. no. P2078, Beyotime Institute of Biotechnology) according to
the manufacturer's instruction. Briefly, the cells were transfected
with FOXO3 overexpression plasmid (FOXO3 coding sequence cloned
into pcDNA3.1 vector) (Shaanxi Youbio Technology Co., Ltd.). After
24 h, the cells were treated with 1% formaldehyde for crosslinking
for 10 min, and terminated with adding of glycine (0.125 M).
Following ultrasonication at 300 W for 10 sec for 12 times using an
ultrasonic homogenizer (Ningbo Scientz Biotechnology Co., Ltd.) and
centrifugation at 10,000 × g at 4°C for 20 min, the supernatant was
isolated. Cell lysates (100 µl) were added into 900 µl dilution
buffer, and incubated with agarose beads (60 µl Protein A/G per 1
ml reaction solution) bound with antibody (IgG, cat. no. A7016,
Beyotime Institute of Biotechnology; anti-FOXO3, cat. no.
NBP2-16521, Novus Biologicals, LLC) at 4°C for ~2 h. Thereafter,
the DNA-protein complex was eluted from beads with centrifugation
at room temperature at 1,000 × g for 1 min twice, and the
combination was broken by de-crosslinking in NaCl solution at
65°Covernight. The protein was degraded with proteinase K (cat. no.
ST535, Beyotime Institute of Biotechnology), and the DNA was
collected for PCR and electrophoresis to detect the promoter
sequence of CNN3. Primer sequences are presented in Table II. The cell lysate before the
incubation with beads served as input control.
Xenograft model
A total of 30 healthy BALB/C nude mice (4–6 weeks
old, weighing 20–22 g) were kept in a sterile environment (12/12 h
light/dark cycle at 22±1°C with a humidity of 45–55%) with free
access to water and food. The mice were randomly divided into five
groups as follows: The vector, CNN3, shNC, shCNN3-1 and shCNN3-2
groups (n=6 per group). The DB cells stably overexpressing CNN3 or
cells in whichCNN3 was silenced were mixed with an equal volume of
Matrigel (Corning, Inc.), and subcutaneously injected into mice
(106 cells per mouse), following anesthetization with
inhalant of 2–3% isoflurane (3% for induction and 2% for
maintenance). At 1 week post-inoculation, the tumor volume was
determined every 3 days. The maximum allowable tumor size was 15
mm. Following inoculation for 22 days, the diameter of the maximum
tumor was >14 mm. Therefore, the mice underwent euthanasia with
an intraperitoneal injection of pentobarbital sodium (200 mg/kg),
and the subcutaneous tumors were isolated for detections. The total
duration of the xenograft experiment was 22 days. The humane
endpoints were set according to Institutional Animal Care and Use
Committee Guidebook, and no animal was euthanized due to humane
endpoint sprior to the experimental endpoint. Animal death was
verified by the disappearance of a heartbeat. The health and
behaviors of animals were monitored daily.
The project design experimental procedure was in
line with the Guide for the Care and Use of Laboratory Animals (8th
Edition, NIH), and approved by the Ethics Committee of Shenyang
Medical College (approval no. SYYXY2021080101).
Hematoxylin and eosin (H&E)
staining
The tumor tissues were fixed with 4%
paraformaldehyde at room temperature overnight, washed with
non-sterile water, and dehydrated with ethanol and xylene. The
tissues were then embedded with paraffin, and cut into 5-µm-thick
sections, which were deparaffinized. Subsequently, the sections
were stained with hematoxylin (Beijing Solarbio Science &
Technology Co., Ltd.) for 5 min, soaked in 1% hydrochloric
acid/ethanol for 3 sec, and counterstained with eosin (Sangon
Biotech Co., Ltd.) for 3 min. The aforementioned incubations were
performed at room temperature. Finally, the sections were
re-dehydrated, and mounted with gum. Images were obtained using a
photon microscope (Olympus Corporation) at a ×400
magnification.
Immunohistochemical staining
The subcutaneous tumor tissues from mice were fixed
into paraffin sections, as described above. Following
deparaffinization, the sections reacted with antigen retrieval
reagent (containing 1.8 mM citric acid and 8.2 mM sodium citrate)
for 10 min. The sections were then blocked with 3%
H2O2 and 1% BSA respectively, and incubated
with antibody against PCNA (cat. no. 24036-1-AP, Wuhan Sanying
Biotechnology) or cyclin D1 (cat. no. AF0931, Jiangsu Affinity
Biosciences Biology Research Company) at a 1:100 dilution at 4°C
overnight in a humid box. After washing with PBS, the sections were
incubated with HRP-labeled IgG (cat. no. 31460, Thermo Fisher
Scientific, Inc.) at a 1:500 dilution at 37°Cfor 60 min, reacted
with DAB reagent for several seconds, and stained with hematoxylin
(Beijing Solarbio Science & Technology Co., Ltd.) at room
temperature for 3 min. Finally, the sections were dehydrated with
ethanol and xylene (75% ethanol for 2 min, 85% ethanol for 2 min,
95% ethanol for 2 min, 100% ethanol for 5 min twice, xylene for 10
min twice), and mounted with gum. Images were obtained using a
photon microscope (Olympus Corporation) at ×400 magnification.
Bioinformatics analysis
The medical bank Gene Expression Profiling
Interactive Analysis (GEPIA) (http://gepia.cancer-pku.cn/) [the dataset sources were
The Cancer Genome Atlas (TCGA) and GTEx] was used to analyze the
expression of CNN3 in DLBCL tissues. The expression pattern of CNN3
was analyzed by ggplot2 and circlize software, and GO enrichment
analysis was carried out with clusterProfiler software. An unpaired
t-test was used to compare the data between two groups. The online
bioinformatics website JASPAR (https://jaspar.genereg.net/) was used to predict the
binding between FOXO3 and promoter sequence of CNN3, and the
potential binding sites were identified.
Statistical analysis
The data in the present study are presented as the
mean ± SD, and analyzed by GraphPad Prism 7.0 software (GraphPad
Software, Inc.). The data from two independent groups were compared
using an unpaired Student's t-test. The data from multiple groups
were analyzed using one- or two-way ANOVA, followed by post hoc
Bonferroni comparisons. A value of P<0.05 was considered to
indicate a statistically significant difference.
Results
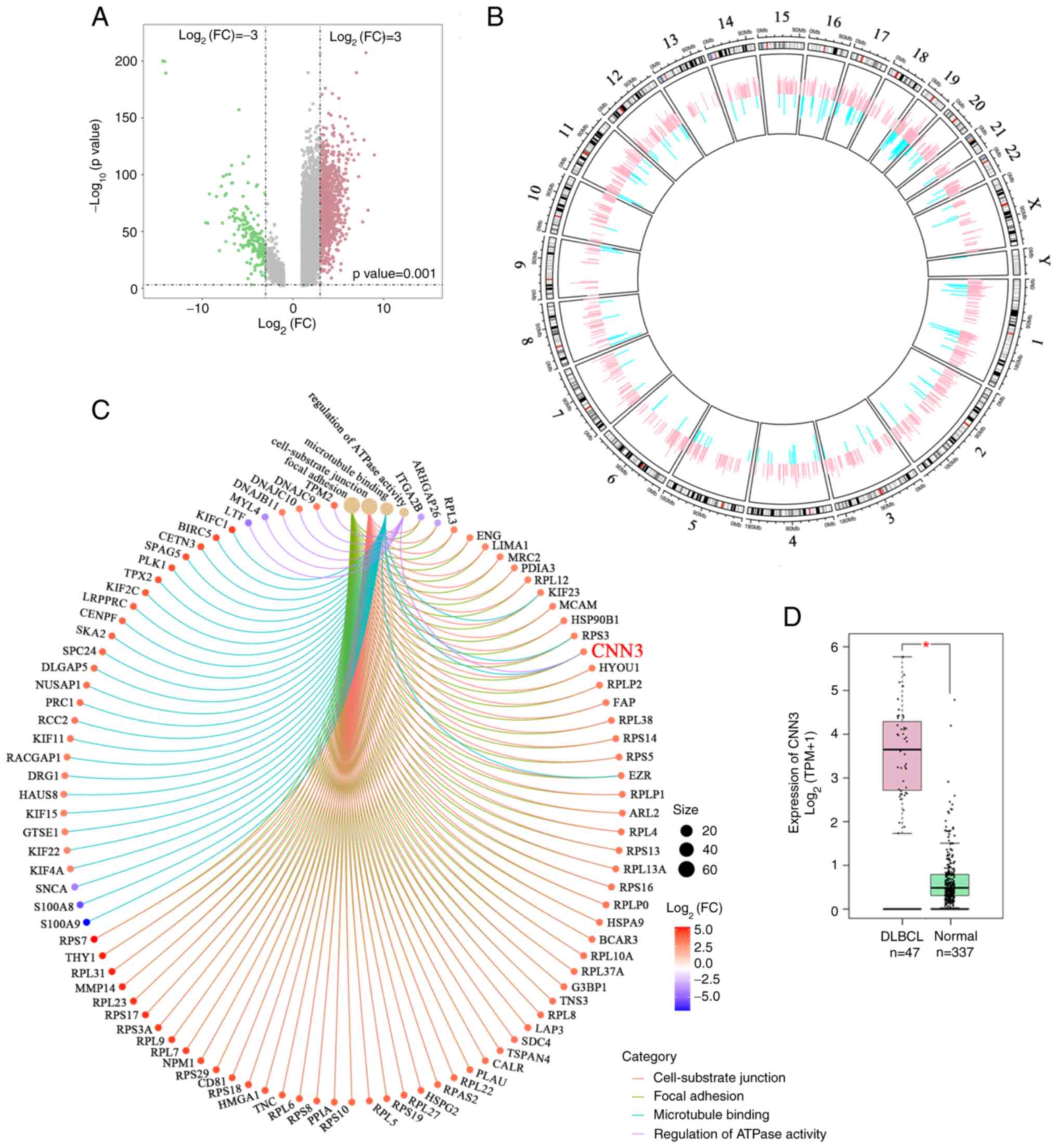
CNN3 is highly expressed in DLBCL
specimens
The dysregulated genes in DLBCL specimens from a
cancer databank GEPIA (23). Gene
Expression Profiling Interactive with values P<0.001 and
log2FC>3 or <-3 were analyzed. According to
Fig. 1A, 1,440 genes were highly
expressed, and 206 genes were expressed at low levels in DLBCL
specimens, as compared with the blood samples from healthy
controls. These abnormally expressed genes were distributed in all
chromosomes (Fig. 1B). Gene
Ontology (GO) enrichment analysis revealed that CNN3 was
simultaneously enriched in four pathways, namely focal adhesion,
cell-substrate junction, microtubule binding, and the regulation of
ATPase activity in DLBCL specimens (Fig. 1C). An additional analysis revealed
that CNN3 mRNA expression was significantly upregulated in DLBCL
specimens, as compared with normal blood samples (Fig. 1D).
CNN3 promotes the proliferation and
cell cycle transition of DLBCL cells
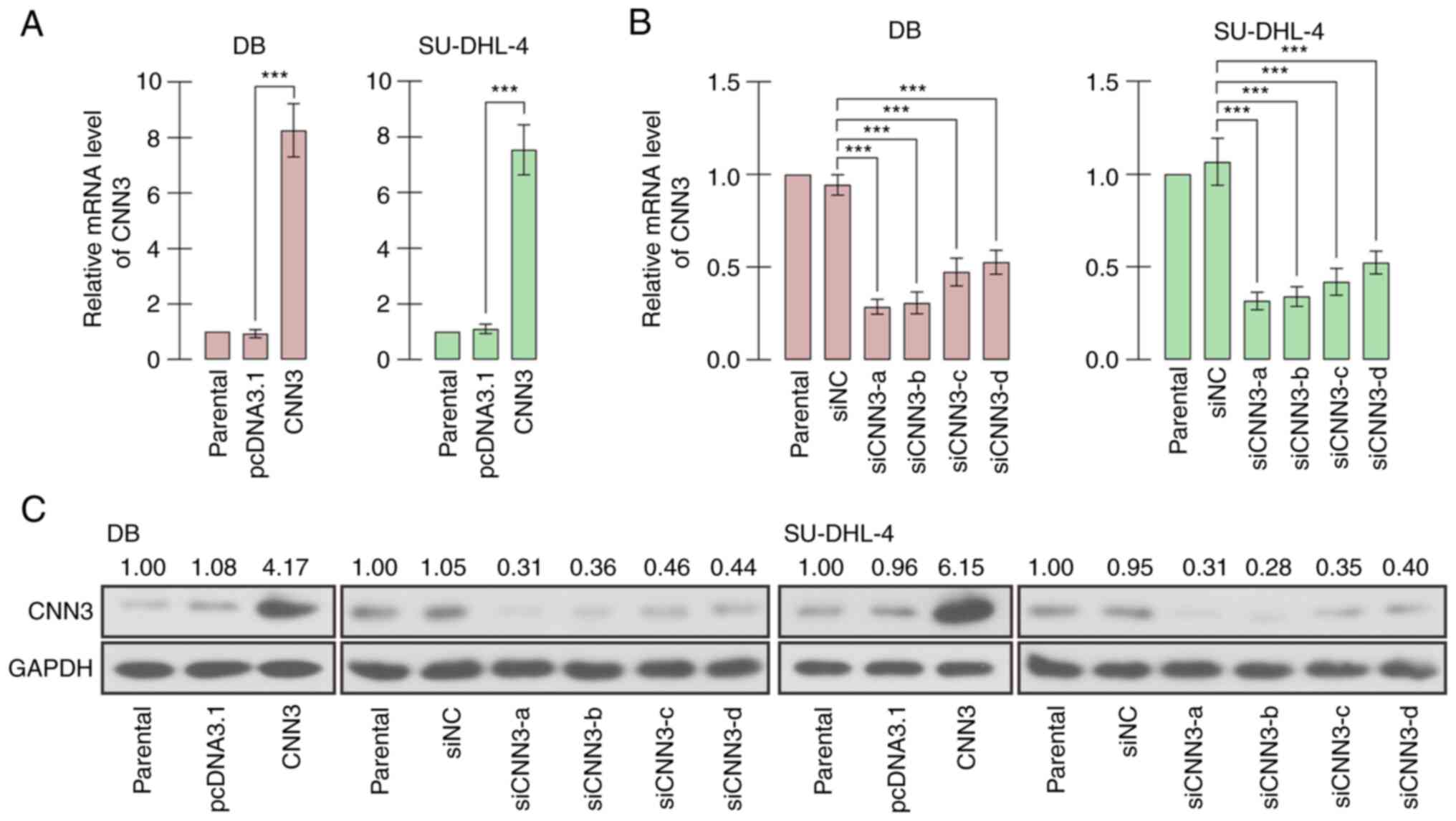
The overexpression vector and siRNAs targeting CNN3
were synthesized and transfected into DB and SU-DHL-4 cells in
order to regulate the expression of CNN3, and RT-qPCR and western
blot analysis confirmed the overexpression and silencing efficiency
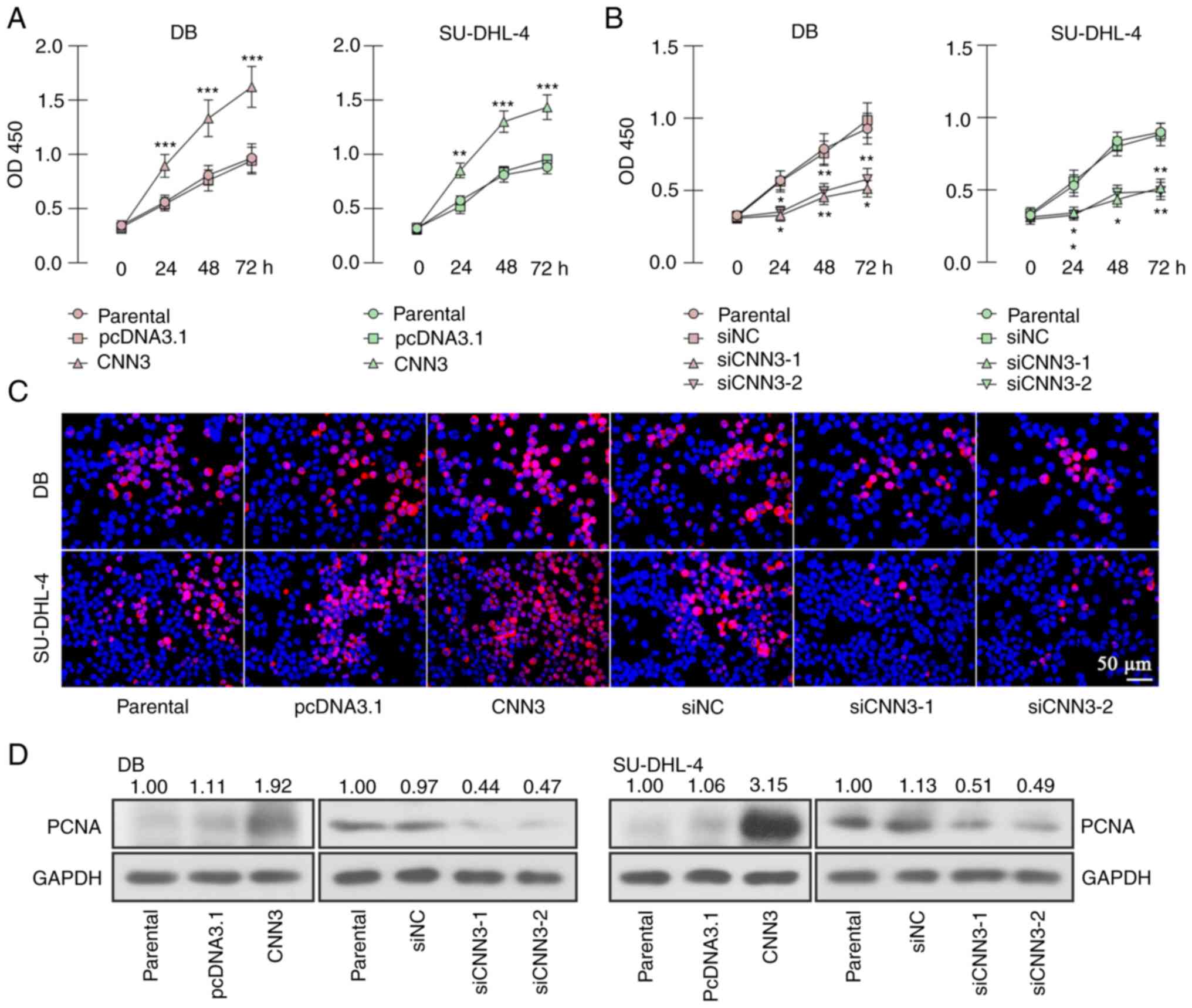
(Fig. 2). The results of CCK-8
assay revealed that the viability of CNN3-overexpressing DB and
SU-DHL-4 cells was increased and that of the CNN3-silenced cells
was decreased (Fig. 3A and B). EdU
staining revealed that the live cell numbers were increased
following the ectopic expression of CNN3, and decreased following
CNN3 knockdown, compared with the empty vector or siNC control
(Fig. 3C). Western blot analysis
revealed that the expression of the proliferation marker, PCNA, was
elevated following CNN3 overexpression and decreased following CNN3
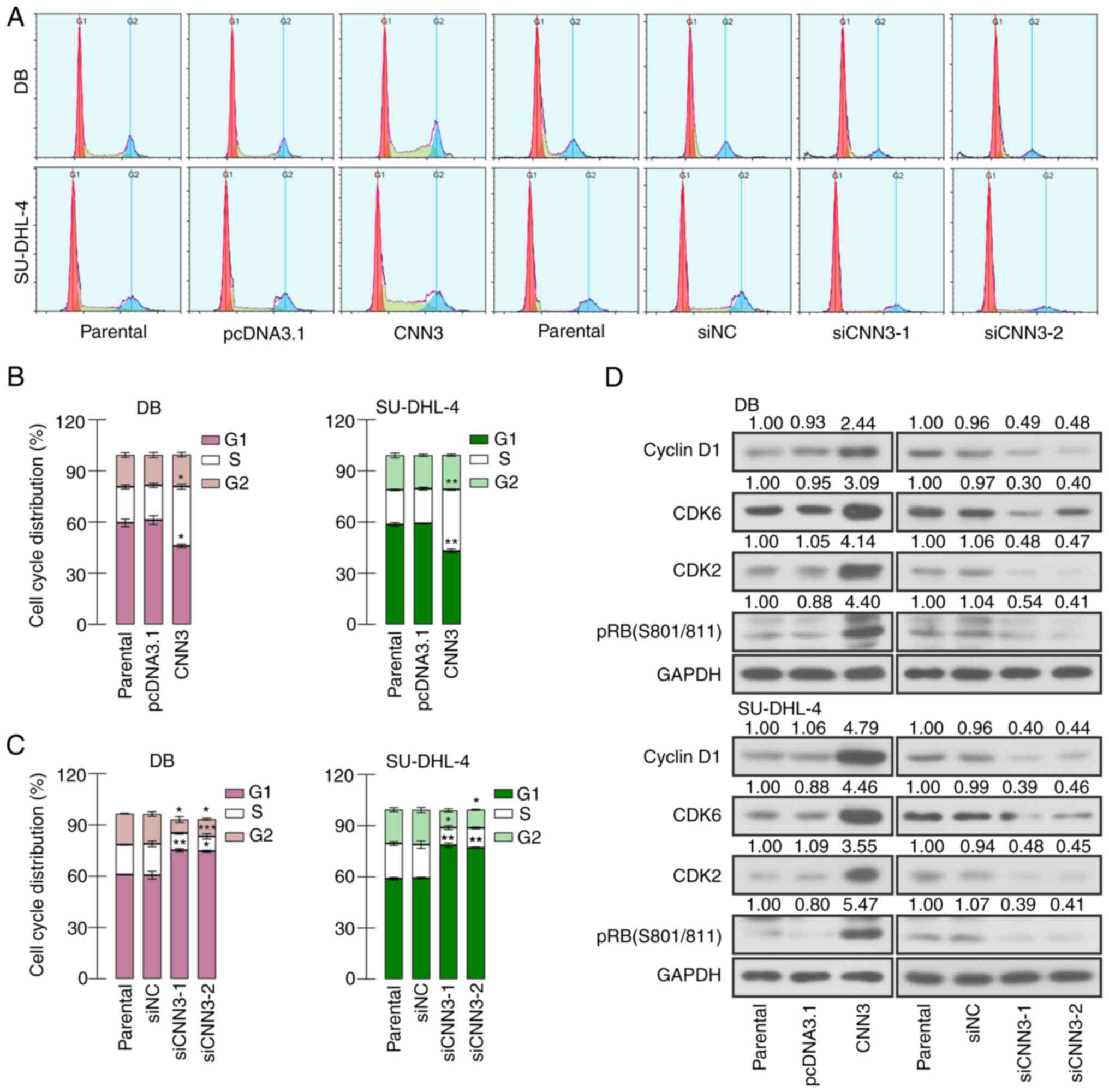
silencing (Fig. 3D). Subsequently,
the results of flow cytometry of the DB and SU-DHL-4 cells
demonstrated that CNN3 overexpression elevated the proportion of
cells in the S phase and reduced the proportion of cells in the G1
phase (Fig. 4A and B).
Additionally, the proportion of cells in the S phase was decreased
and that of cells in the G1 phase was increased following the
knockdown of CNN3 (Fig. 4A and C).
Thereafter, the expression of several cell cycle-related proteins
was measured using western blot analysis. The results revealed that
the levels of cyclin D1, CDK6, CDK2 and (Ser801/811) were increased
following the overexpression of CNN3 and decreased following CNN3
silencing (Fig. 4D). These results
suggested that CNN3 facilitated the proliferation and cell cycle
G1/S transition of DLBCL cells.
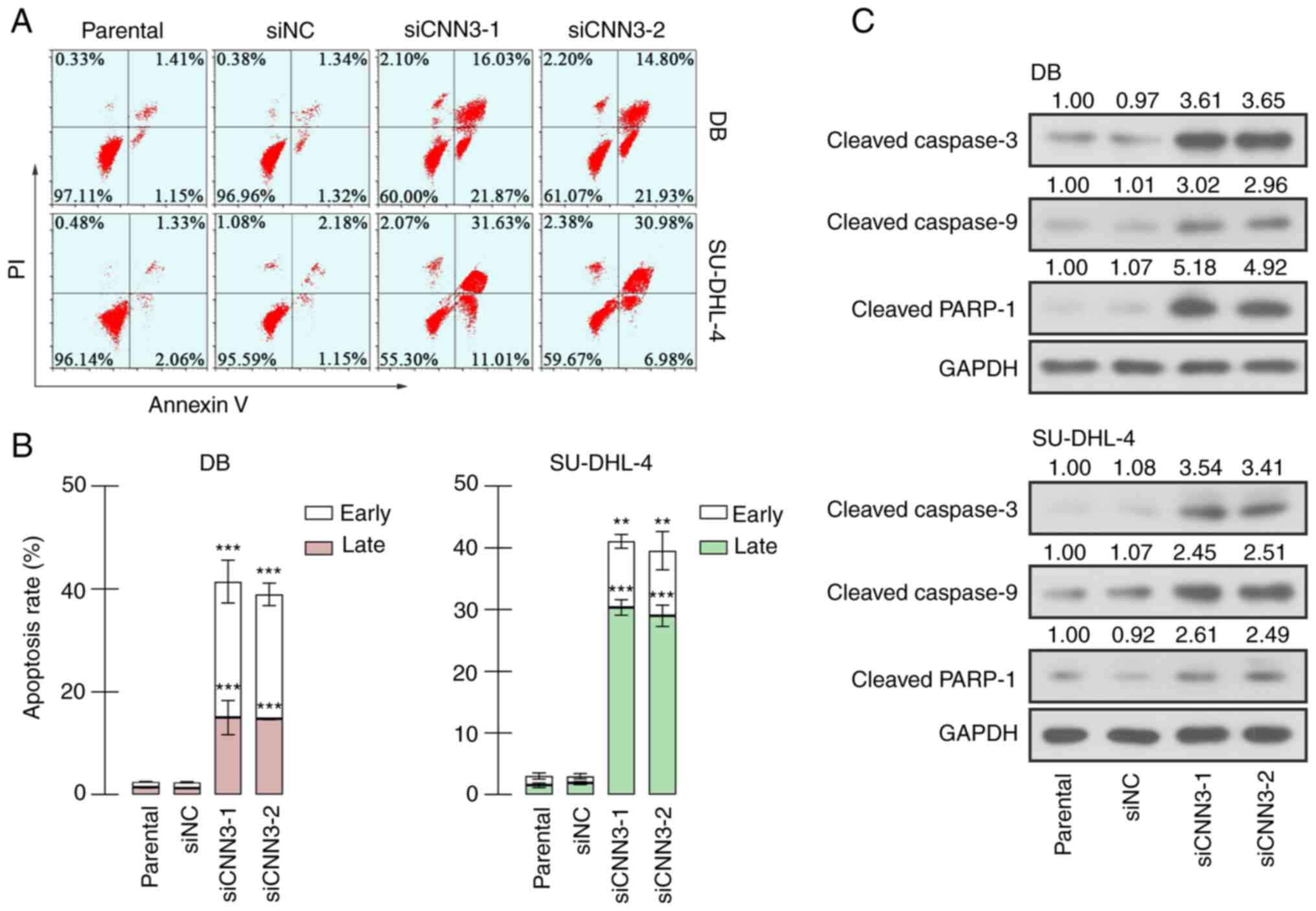
CNN3 silencing induces the apoptosis
of DLBCL cells
The apoptosis of the DB and SU-DHL-4 cells was
analyzed using flow cytometry. As shown in Fig. 5A and B, it was evident that CNN3
knockdown significantly induced the apoptosis of the DB and
SU-DHL-4 cells, including both early (Annexin
V+PI−) and late apoptosis (Annexin
V+PI+). The results of western blot analysis
also revealed that the levels of the apoptotic markers, cleaved
caspase-3/9 and cleaved PARP-1, were increased in the CNN3-silenced
DB and SU-DHL-4 cells (Fig. 5C). On
the whole, these results suggested that CNN3 knockdown resulted in
the increased apoptosis of DLBCL cells.
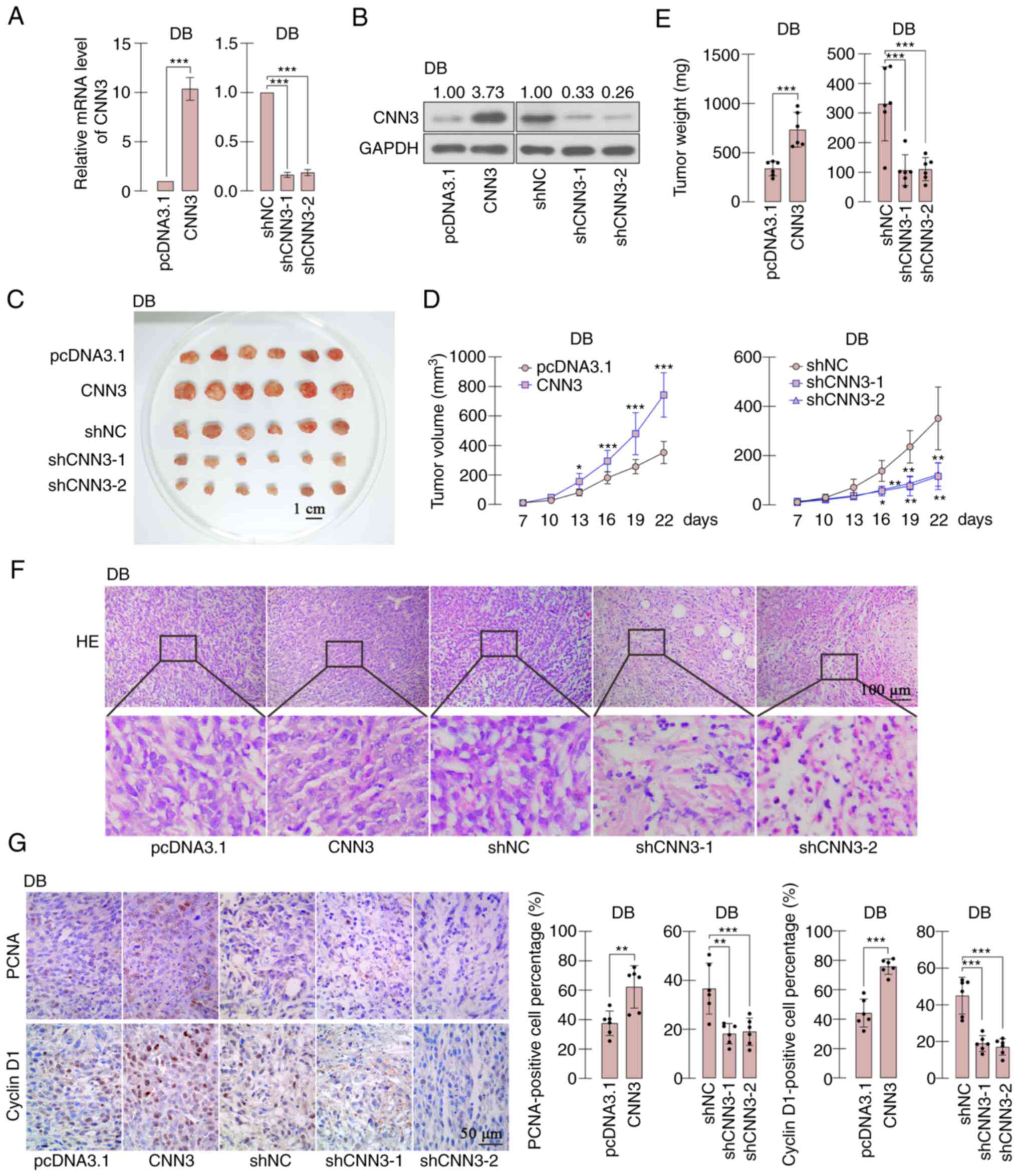
CNN3 enhances the growth of DLBCL
cells in vivo
A tumor xenograft experiment was carried out to
detect the effects of CNN3 on the growth of DLBCL cells in nude
mice. The DB cells with stably overexpressing CNN3 or cells in
which CNN3 was silenced were screened, and alterations in
expression were confirmed using RT-qPCR and western blot analysis
(Fig. 6A and B). The screened DB
cells were subcutaneously inoculated into mice for 22 days. The
results demonstrated that the growth of CNN3-overexpressing DB
cells was increased and that of the CNN3-silenced cells was
decreased, in comparison to the growth of the respective controls
(Fig. 6C-E). H&E staining
revealed nuclear shrinkage and intercellular cavity in the
CNN3-silenced tumors, whereas the CNN3-overexpressing tumors were
more compactly structured in comparison with the control (Fig. 6F). Immunohistochemical staining
revealed that the expression of PCNA and cyclin D1 was increased in
tumors overexpressing CNN3, suggesting that CNN3 facilitated the
growth of DB cells; by contrast, PCNA and cyclin D1 expression was
decreased in tumors derived fromCNN3-silenced cells, suggesting the
inhibition of tumor growth (Fig.
6G).
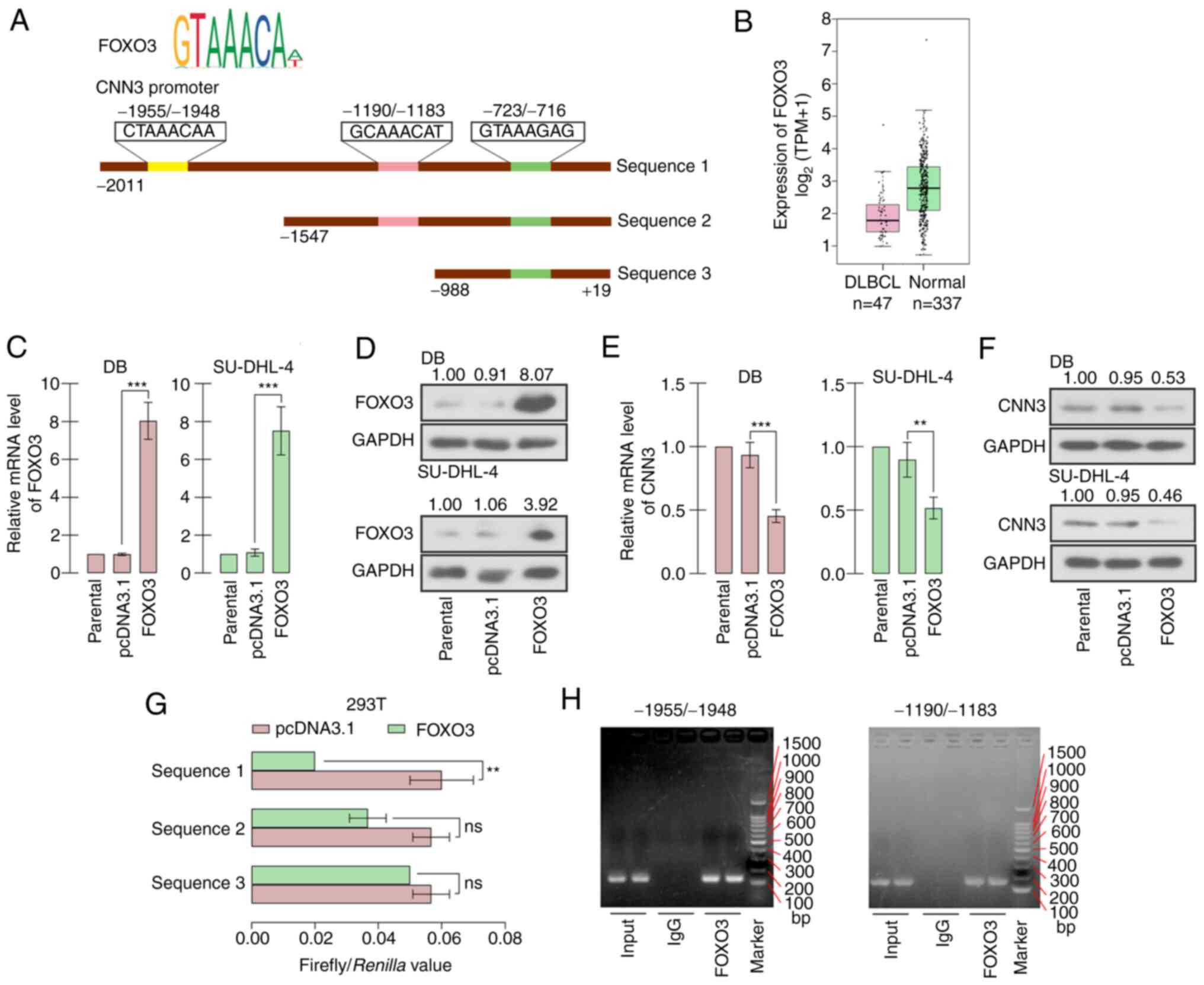
CNN3 expression is negatively
regulated by FOXO3
The analysis from bioinformatics websites
demonstrated that the promoter sequence of CNN3 was potentially
bound by FOXO3, and the latent binding sites are displayed in
Fig. 7A. The analysis from the
GEPIA database revealed that the FOXO3 expression was slightly
decreased in DLBCL specimens, in comparison with normal blood
samples (Fig. 7B). In order to
investigate the function of FOXO3, a FOXO3 overexpression vector
was constructed and its availability was verified at the
transcription and translation levels in DB and SU-DHL-4 cells
(Fig. 7C and D). The mRNA and
protein expression levels of CNN3 were simultaneously decreased by
~50% following FOXO3 overexpression (Fig. 7E and F). Dual-luciferase reporter
assay demonstrated that the suppression of FOXO3 luciferase
activity of the reporter vector containing the CNN3 promoter
sequence was significantly attenuated following the deletion of the
−1955/-1948 and −1190/-1183 sites, particularly the former
(Fig. 7G). ChIP assay revealed that
FOXO3 certainly bound to the −1955/-1948 and −1190/-1183 sites of
CNN3 promoter sequence. In addition, the binding affinity of FOXO3
to −1955/-1948 site appeared greater than that to the −1190/-1183
site (Fig. 7H).
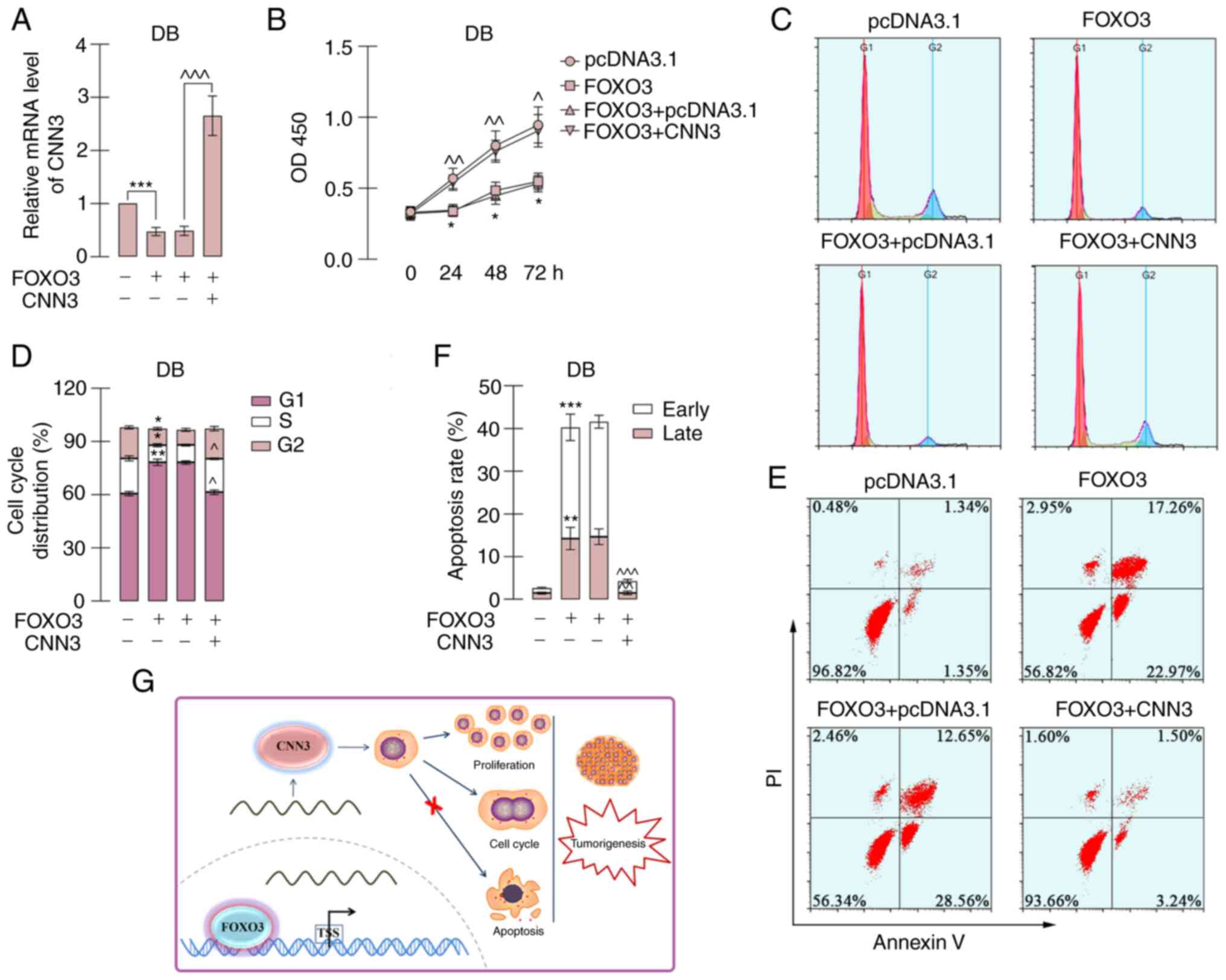
Thereafter, rescue experiments were performed by the
co-expression of FOXO3 and CNN3. The overexpression vector of CNN3
only contained its coding sequence, not the promoter sequence.
Thus, the ectopic expression of CNN3 was not regulated by FOXO3. As
demonstrated in Fig. 8A, the
transfection of the CNN3 overexpression vector reversed the
FOXO3-induced decrease in CNN3 expression. Moreover, the recovered
CNN3 expression abrogated the FOXO3-induced inhibition of the
proliferation and cell cycle transition, and the induction of
apoptosis of DB cells (Fig. 8B-F).
On the whole, these results suggested that FOXO3 bound to the
promoter of CNN3 and negatively regulated its tumor-promoting roles
in DLBCL cells (Fig. 8G).
Discussion
FOX is an evolutionarily conserved transcription
factor family, which has a deeply conserved forkhead winged
helix-turn-helix DNA binding domain (24). FOXO3, also known as FOXO3a or
forehead in rhabdomyosarcoma-like 1, belongs to the FOXO subfamily
(25). FOXO3 mediates multiple
pathological and physiological processes by controlling the
transcription of target genes involved in cell proliferation, cell
cycle transition, apoptosis, DNA damage and autophagy (26). It has been reported that FOXO3
inhibited tumorigenesis by suppressing the expression of
proliferation-related genes, including CCND1 (27) and CCNA1 (28), and inducing the expression of
apoptotic genes, including BIM (29) and NOXA (30). The loss of FOXO3 expression
significantly accelerates B lymphomagenesis in MYC-mutant mice
(31), and the silencing of FOXO3
inhibits the apoptosis of DLBCL cells (17). In the present study, the ectopic
expression of FOXO3 induced the apoptosis, and attenuated the
proliferation and cell cycle transition of DLBCL cells. It was also
demonstrated that FOXO3 bound to the promoter sequence of CNN3 and
suppressed its transcription, with the effects of FOXO3 being
significantly reversed by CNN3 expression.
The cancer-promoting roles of CNN3 have been
reported in several solid tumors, and its effect on lymphomagenesis
was first demonstrated in the present study, to the best of our
knowledge. CNN3 is an isoform of calponin, acting as an
actin-binding protein for the inhibition of myosin ATPase and the
stabilization of the actin cytoskeleton (32). CNN3 affects the motility, migration,
adhesion, differentiation, phagocytosis and fusion of multiple
cells by regulating the actin cytoskeleton. Its effects on
proliferation have been reported in recent years. For instance,
CNN3 knockdown impairs the proliferation of myoblasts and
fibroblasts (33,34). Increased expression of CNN3 promoted
the proliferation of osteosarcoma and cervical cancer cells
(11,35). By contrast, in a previous study by
Yang et al (36), CNN3 was
reported to suppress the proliferation of lung cancer cells. The
tumor databank analysis demonstrated that the expression of CNN3 in
tumor specimens was relatively different, compared with their
respective control normal tissues, suggesting that CNN3 may be
expressed with tissue specificity, resulting in its discrepant
functions in different organs and tissues. Nevertheless, this
speculation requires further verification in future studies. In the
present study, CNN3 was highly expressed in DLBCL specimens,
accompanied by promoting effects on the proliferation of DLBCL
cells. However, the clinical data from the tumor databank analyzed
in the present study included DLBCL specimens and normal blood
samples (the dataset sources are TCGA and GTEx). It was
hypothesized that it may be more reasonable to compare the
expression of CNN3 in DLBCL specimens and normal lymph node
tissues. Due to the difficulty of normal lymph node collection,
this comparison has not yet been realized. In future studies, the
authors aim to collect DLBCL specimens and normal lymph node
tissues to confirm the expression of CNN3.
The data of the present study demonstrated that CNN3
promoted the proliferation and cell cycle transition, and its
knockdown stimulated the apoptosis of DLBCL cells. In the majority
of adult tissues, cells are in a quiescent state (G0 phase), and
can be stimulated to re-enter the cell cycle by mitogenic signals.
Uncontrolled proliferation and abnormally activated cell cycle are
the primary characteristics of malignant cells (37). Cyclins interact with CDKs for the
regulation of the G0/G1, S, G2 and M phase progression in mammalian
cells (37). Cyclin D firstly
senses the mitogenic signals, binding to CDK4 and CDK6, and cells
inter the G1 phase to initiate DNA synthesis (38,39).
The activation of the cyclin D-CDK4/6 complex phosphorylates the
pocket protein including RB to enable E2F transcription factor to
induce the transcription of downstream genes involved in cell
cycle. This includes cyclin E, which binds to CDK2, with the formed
complex further phosphorylating RB to create a positive feedback
loop (37,38,40).
In the results of the present study, CNN3 induced the increased
levels of cyclin D1, CDK6, CDK2 and pRB in DLBCL cells, accompanied
by an accelerated G1/S transition. These data suggested that CNN3
facilitated the proliferation of DLBCL cells by accelerating cell
cycle G1/S transition.
Suppressed apoptosis is another characteristic of
malignant tumor cells. Apoptosis is a type of cell death actively
initiated by cells themselves, and helpful for the survival of the
whole organism. The apoptotic process in mammalian cells is
evolutionarily conserved and precisely regulated. The extrinsic
apoptotic signals are delivered by death receptors, including Fas
and tumor necrosis factor receptors (TNFRs), and their adapters,
including Fas-associated death domain and TNFR-associated death
domain to active caspase-8 (41,42).
Intrinsic apoptosis is initiated by the mitochondrial pathway. The
permeability of the mitochondrial membrane is increased and
cytochrome c is released into the cytoplasm to activate
caspase-9 (43). The intrinsic and
extrinsic apoptotic pathways merge to caspase-3, a well-known
executioner agent that is responsible for the cleavage of protein
kinase, DNA repair proteins and cytoskeletal proteins (44). PARPs are
poly-ADP-ribosyltransferases that mediate poly-ADP-ribosylation of
nuclear proteins involved in DNA repair. Among 18 members of the
PARP subfamily, PARP-1 has been the most extensively studied
(45). PARP-1 catalyzes the
synthesis of polymers of ADP-ribose with NAD+ as
substrates (46). In response to
DNA strand breakage, PARP-1 is cleaved and activated by caspase-3
(47); thus, the presence of
cleaved PARP-1 is generally considered as a marker of apoptosis
(46). In the present study, the
levels of cleaved caspase-3, caspase-9 and PARP-1 were all
increased following CNN3 knockdown, suggesting the activation of
the intrinsic apoptosis of DLBCL cells. Moreover, flow cytometry
was used to measure the apoptosis by Annexin V and PI staining. In
the early stage of apoptosis, the phosphatidylserine (PS) in the
inner layers of cytomembrane is flipped out to the outer layers to
be recognized for phagocytosis (41). As a phospholipid binding protein,
Annexin V specifically and strongly interacts with PS, and the
labeled Annexin V can indicate early apoptotic cells (48). In the middle and late stages of
apoptosis, PI can pass through the impaired cytomembrane to stain
DNA; thus, Annexin V+PI+ is considered as a
marker of late apoptosis. The present study also demonstrated that
CNN3 silencing concurrently triggered the early and late apoptosis
of DLBCL cells.
DLBCL is a malignant tumor originating from lymphoid
tissue. Due to its high heterogeneity, there are variations in the
treatment and outcomes of patients with DLBCL. Based on gene
expression profiling, DLBCL is divided into two distant subtypes:
Germinal center B-cell-like (GCB) and activated B-cell-like (ABC).
These two subtypes arise from different stages of lymphoid
differentiation with separate oncogenic mechanisms, and the
prognosis of patients with the ABC subtype is poorer (49–51).
For instance, B-cell receptor signaling and nuclear factor κB are
generally activated in the ABC subtype and B-cell leukemia/lymphoma
6 and enhancer of zeste 2 polycomb repressive complex 2 subunit
tend to be expressed in the GCB subtype (52). Both FOXO3 and CNN3 have not been
found to be associated with the classification of DLBCL. The
authors aim to investigate the outcomes of patients with DLBCL with
a high or low expression of FOXO3 or CNN3 and the association
between FOXO3 or CNN3 and the ABC/GCB subtype in future studies.
Nevertheless, the findings of the present study suggested that
FOXO3 and CNN3 may be considered as novel therapeutic targets for
DLBCL.
In conclusion, the present study demonstrated that
CNN3 promoted tumor growth in vivo and in vitro, and
cell cycle G1/S transition, and inhibited the apoptosis of DLBCL
cells. FOXO3 bound the promoter sequence of CNN3 and inhibited its
transcription. The lymphoma suppressive roles of FOXO3 were
antagonized by CNN3 overexpression. These findings may provide
novel insight into the diagnosis and treatment of patients with
DLBCL in clinical practice.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science
Foundation of China (grant no. U1908215), the General Program of
the National Natural Science Foundation of China (grant no.
62273330), and the Fundamental Research Funds for the Central
Universities, the Provincial and Ministerial Science Foundation
(Millions of Talents Project of Liaoning Province in 2019).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XX designed the study and coordinated the
experiments. ML, XW, QG and HW performed the experiments and
analyzed the data. The manuscript was drafted by XX. All authors
have read and approved the final manuscript. XX and ML confirmed
the authenticity of all the raw data.
Ethics approval and consent to
participate
The project design experimental procedure was in
line with Guide for the Care and Use of Laboratory Animals (8th
Edition, NIH), and approved by the Ethics Committee of Shenyang
Medical College (approval no. SYYXY2021080101).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Armitage JO, Gascoyne RD, Lunning MA and
Cavalli F: Non-Hodgkin lymphoma. Lancet. 390:298–310. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Guerard EJ and Bishop MR: Overview of
non-Hodgkin's lymphoma. Dis Mon. 58:208–218. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Martelli M, Ferreri AJ, Agostinelli C, Di
Rocco A, Pfreundschuh M and Pileri SA: Diffuse large B-cell
lymphoma. Crit Rev Oncol Hematol. 87:146–171. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liu Y and Barta SK: Diffuse large B-cell
lymphoma: 2019 Update on diagnosis, risk stratification, and
treatment. Am J Hematol. 94:604–616. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Winder SJ and Walsh MP: Smooth muscle
calponin. Inhibition of actomyosin MgATPase and regulation by
phosphorylation. J Biol Chem. 265:10148–10155. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Winder SJ, Walsh MP, Vasulka C and Johnson
JD: Calponin-calmodulin interaction: Properties and effects on
smooth and skeletal muscle actin binding and actomyosin ATPases.
Biochemistry. 32:13327–13333. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kake T, Kimura S, Takahashi K and Maruyama
K: Calponin induces actin polymerization at low ionic strength and
inhibits depolymerization of actin filaments. Biochem J.
312:587–592. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ferhat L, Esclapez M, Represa A, Fattoum
A, Shirao T and Ben-Ari Y: Increased levels of acidic calponin
during dendritic spine plasticity after pilocarpine-induced
seizures. Hippocampus. 13:845–858. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Flemming A, Huang QQ, Jin JP, Jumaa H and
Herzog S: A conditional knockout mouse model reveals that
calponin-3 is dispensable for early B cell development. PLoS
One. 10:e01283852015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shibukawa Y, Yamazaki N, Kumasawa K,
Daimon E, Tajiri M, Okada Y, Ikawa M and Wada Y: Calponin 3
regulates actin cytoskeleton rearrangement in trophoblastic cell
fusion. Mol Biol Cell. 21:3973–3984. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xia L, Yue Y, Li M, Zhang YN, Zhao L, Lu
W, Wang X and Xie X: CNN3 acts as a potential oncogene in cervical
cancer by affecting RPLP1 mRNA expression. Sci Rep. 10:24272020.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nair VA, Al-Khayyal NA, Sivaperumal S and
Abdel-Rahman WM: Calponin 3 promotes invasion and drug resistance
of colon cancer cells. World J Gastrointest Oncol. 11:971–982.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hong KS, Kim H, Kim SH, Kim M and Yoo J:
Calponin 3 regulates cell invasion and doxorubicin resistance in
gastric cancer. Gastroenterol Res Pract. 2019:30249702019.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tsuji T, Maeda Y, Kita K, Murakami K, Saya
H, Takemura H, Inaki N, Oshima M and Oshima H: FOXO3 is a latent
tumor suppressor for FOXO3-positive and cytoplasmic-type gastric
cancer cells. Oncogene. 40:3072–3086. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shukla S, Bhaskaran N, Maclennan GT and
Gupta S: Deregulation of FoxO3a accelerates prostate cancer
progression in TRAMP mice. Prostate. 73:1507–1517. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Blake DC Jr, Mikse OR, Freeman WM and
Herzog CR: FOXO3a elicits a pro-apoptotic transcription program and
cellular response to human lung carcinogen nicotine-derived
nitrosaminoketone (NNK). Lung Cancer. 67:37–47. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zheng X, Rui H, Liu Y and Dong J:
Proliferation and apoptosis of B-cell lymphoma cells under targeted
regulation of FOXO3 by miR-155. Mediterr J Hematol Infect Dis.
12:e20200732020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bi C and Wang G: LINC00472 suppressed by
ZEB1 regulates the miR-23a-3p/FOXO3/BID axis to inhibit the
progression of pancreatic cancer. J Cell Mol Med. 25:8312–8328.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yu S, Yu Y, Zhang W, Yuan W, Zhao N, Li Q,
Cui Y, Wang Y, Li W, Sun Y and Liu T: FOXO3a promotes gastric
cancer cell migration and invasion through the induction of
cathepsin L. Oncotarget. 7:34773–34784. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hu C, Ni Z, Li BS, Yong X, Yang X, Zhang
JW, Zhang D, Qin Y, Jie MM, Dong H, et al: hTERT promotes the
invasion of gastric cancer cells by enhancing FOXO3a ubiquitination
and subsequent ITGB1 upregulation. Gut. 66:31–42. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bouzeyen R, Haoues M, Barbouche MR, Singh
R and Essafi M: FOXO3 transcription factor regulates IL-10
expression in mycobacteria-infected macrophages, tuning their
polarization and the subsequent adaptive immune response. Front
Immunol. 10:29222019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Benayoun BA, Caburet S and Veitia RA:
Forkhead transcription factors: Key players in health and disease.
Trends Genet. 27:224–232. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Anderson MJ, Viars CS, Czekay S, Cavenee
WK and Arden KC: Cloning and characterization of three human
forkhead genes that comprise an FKHR-like gene subfamily. Genomics.
47:187–199. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu Y, Ao X, Ding W, Ponnusamy M, Wu W,
Hao X, Yu W, Wang Y, Li P and Wang J: Critical role of FOXO3a in
carcinogenesis. Mol Cancer. 17:1042018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ai B, Kong X, Wang X, Zhang K, Yang X,
Zhai J, Gao R, Qi Y, Wang J, Wang Z and Fang Y: LINC01355
suppresses breast cancer growth through FOXO3-mediated
transcriptional repression of CCND1. Cell Death Dis. 10:5022019.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Marlow LA, von Roemeling CA, Cooper SJ,
Zhang Y, Rohl SD, Arora S, Gonzales IM, Azorsa DO, Reddi HV, Tun
HW, et al: Foxo3a drives proliferation in anaplastic thyroid
carcinoma through transcriptional regulation of cyclin A1: A
paradigm shift that impacts current therapeutic strategies. J Cell
Sci. 125:4253–4263. 2012.PubMed/NCBI
|
|
29
|
Yamamura Y, Lee WL, Inoue K, Ida H and Ito
Y: RUNX3 cooperates with FoxO3a to induce apoptosis in gastric
cancer cells. J Biol Chem. 281:5267–5276. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Obexer P, Geiger K, Ambros PF, Meister B
and Ausserlechner MJ: FKHRL1-mediated expression of Noxa and Bim
induces apoptosis via the mitochondria in neuroblastoma cells. Cell
Death Differ. 14:534–547. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Vandenberg CJ, Motoyama N and Cory S:
FoxO3 suppresses Myc-driven lymphomagenesis. Cell Death Dis.
6:e20462016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liu R and Jin JP: Calponin isoforms CNN1,
CNN2 and CNN3: Regulators for actin cytoskeleton functions in
smooth muscle and non-muscle cells. Gene. 585:143–153. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
She Y, Li C, Jiang T, Lei S, Zhou S, Shi H
and Chen R: Knockdown of CNN3 impairs myoblast proliferation,
differentiation, and protein synthesis via the mTOR pathway. Front
Physiol. 12:6592722021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Daimon E, Shibukawa Y and Wada Y: Calponin
3 regulates stress fiber formation in dermal fibroblasts during
wound healing. Arch Dermatol Res. 305:571–584. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Dai F, Luo F, Zhou R, Zhou Q, Xu J, Zhang
Z, Xiao J and Song L: Calponin 3 is associated with poor prognosis
and regulates proliferation and metastasis in osteosarcoma. Aging
(Albany NY). 12:14037–14049. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yang C, Zhu S, Feng W and Chen X: Calponin
3 suppresses proliferation, migration and invasion of non-small
cell lung cancer cells. Oncol Lett. 22:6342021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Otto T and Sicinski P: Cell cycle proteins
as promising targets in cancer therapy. Nat Rev Cancer. 17:93–115.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Malumbres M and Barbacid M: Cell cycle,
CDKs and cancer: A changing paradigm. Nat Rev Cancer. 9:153–166.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Malumbres M and Barbacid M: To cycle or
not to cycle: A critical decision in cancer. Nat Rev Cancer.
1:222–231. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sherr CJ and Roberts JM: Living with or
without cyclins and cyclin-dependent kinases. Genes Dev.
18:2699–2711. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hengartner MO: Apoptosis: Corralling the
corpses. Cell. 104:325–328. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Schneider P and Tschopp J: Apoptosis
induced by death receptors. Pharm Acta Helv. 74:281–286. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Danial NN and Korsmeyer SJ: Cell death:
Critical control points. Cell. 116:205–219. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ghobrial IM, Witzig TE and Adjei AA:
Targeting apoptosis pathways in cancer therapy. CA Cancer J Clin.
55:178–194. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Amé JC, Spenlehauer C and de Murcia G: The
PARP superfamily. Bioessays. 26:882–893. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Koh DW, Dawson TM and Dawson VL: Mediation
of cell death by poly(ADP-ribose) polymerase-1. Pharmacol Res.
52:5–14. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Tewari M, Quan LT, O'Rourke K, Desnoyers
S, Zeng Z, Beidler DR, Poirier GG, Salvesen GS and Dixit VM:
Yama/CPP32 beta, a mammalian homolog of CED-3, is a
CrmA-inhibitable protease that cleaves the death substrate
poly(ADP-ribose) polymerase. Cell. 81:801–809. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
van Engeland M, Nieland LJ, Ramaekers FC,
Schutte B and Reutelingsperger CP: Annexin V-affinity assay: A
review on an apoptosis detection system based on phosphatidylserine
exposure. Cytometry. 31:1–9. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Rosenwald A, Wright G, Chan WC, Connors
JM, Campo E, Fisher RI, Gascoyne RD, Muller-Hermelink HK, Smeland
EB, Giltnane JM, et al: The use of molecular profiling to predict
survival after chemotherapy for diffuse large-B-cell lymphoma. N
Engl J Med. 346:1937–1947. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Lenz G, Wright G, Dave SS, Xiao W, Powell
J, Zhao H, Xu W, Tan B, Goldschmidt N, Iqbal J, et al: Stromal gene
signatures in large-B-cell lymphomas. N Engl J Med. 359:2313–2323.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Scott DW, Mottok A, Ennishi D, Wright GW,
Farinha P, Ben-Neriah S, Kridel R, Barry GS, Hother C, Abrisqueta
P, et al: Prognostic significance of diffuse large B-cell lymphoma
cell of origin determined by digital gene expression in
formalin-fixed paraffin-embedded tissue biopsies. J Clin Oncol.
33:2848–2856. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Sehn LH and Salles G: Diffuse large B-cell
lymphoma. N Engl J Med. 384:842–858. 2021. View Article : Google Scholar : PubMed/NCBI
|