Introduction
Lung cancer is the leading cause of cancer-related
deaths (18%) worldwide, characterized by fast growth, high
mortality and poor prognosis (1).
Lung cancer is generally classified into non-small-cell lung
carcinoma (NSCLC) and small-cell lung carcinoma based on
histological morphology. Lung adenocarcinoma (LUAD) is the major
type of NSCLC, ~50% of all lung cancers (2). Advanced LUAD patients have a poor
prognosis, and the average five-year survival rate is only 15%
(3). Currently, with the
development of biological therapy and immunotherapy, the clinical
treatment of LUAD has broken the traditional treatment methods
including surgery, radiotherapy and chemotherapy (4,5). For
patients with LUAD, prompt surgical intervention can significantly
lower death and postoperative recurrence rates (6). Therefore, more research into new LUAD
therapeutic targets is required. With the advancement of
bioinformatics, new cancer-related genes can be recognized.
Previous studies have successfully found some LUAD-related genes,
including C10rf74 (7), CLDN18
(8), and HMGB2 (9). The identification of potential targets
is conducive to the development of LUAD therapy.
Nucleophosmin (NPM1) is a multifunctional
phosphoprotein, expressed ubiquitously in all tissues (10). NPM1 serves as a molecular chaperone
for both nucleic acids and proteins with shuttling activity between
the cytoplasm and nucleus (11).
NPM1 participated in multiple cellular functions, including cell
cycle regulation and DNA repair (12). A previous study has revealed that
NPM1 regulates cell cycle progression and centrosome replication,
which is essential for cell growth and proliferation (13). NPM1 overexpression frequently
predicts poor prognosis in patients with breast cancer (14), oral squamous cell carcinomas
(15), and other cancers. Liu et
al (16) reported that NPM1 was
overexpressed in LUAD. However, its specific function and mechanism
were rarely explored.
The EGFR/MAPK signaling pathway, a conserved
receptor tyrosine kinase pathway, regulates cell survival, growth,
proliferation and differentiation (17). EGFR activation regulates the levels
of proteins associated with cancer cell growth, differentiation and
migration via the MAPK pathway (18). EGFR dimerizes and
auto-phosphorylates after binding to its ligand, triggering
downstream intracellular signaling through RAS/RAF/MEK/ERK
phosphorylation (19). EGFR/MAPK
signaling pathway is involved in multiple tumor progression; for
instance, gastric (20), ovarian
(21) and pancreatic cancer
(22). A recent study has revealed
that EGFR/MAPK signaling pathway was activated in LUAD to promote
tumor growth and metastasis (19).
Additionally, NPM1 could specifically activate EGFR/MAPK pathway in
prostate cancer (23). Whether NPM1
promotes LUAD progression through the EGFR/MAPK signaling pathway
remains unknown.
Therefore, in the present study, it was assessed
whether NPM1 is a valid target for LUAD prognosis via The Cancer
Genome Atlas (TCGA) database. The effects of NPM1 and EGFR/MAPK
signaling pathways on LUAD progression were explored in
vitro and in vivo. The present study aimed to prove that
NPM1 is a novel promising target in LUAD therapy through the
EGFR/MAPK signaling pathway.
Materials and methods
Data download and pre-processing
LUAD-related gene expression was acquired from the
TCGA database (https://tcga-data.nci.nih.gov/) via R software 3.6.5
(http://r-project.org). A total of 510 LUAD
samples and 58 healthy samples were collected. The mRNA expression
was obtained using the HUGO Gene Nomenclature Committee mRNA gene
annotation file (24). To ensure
high confidence in the results, the identification data were
standardized by localization probability >0.75.
Differentially expressed genes (DEGs)
and protein-protein interaction network (PPI) analysis
The GEO2R was used to identify DEGs between LUAD and
normal lung tissues. Significant criteria of DEGs were a |log fold
change (log2FC)|>1 and P<0.05. DEGs were put into
the STRING database (25) to get
their PPI networks. The networks were then identified and
visualized through Cytoscape software version 3.8.0 (26). Hub genes were obtained using the
cytoHubba plugin.
Diagnostic analysis
cBioPortal for Cancer Genomics is a common resource
for the interactive exploration of cancer genomics datasets
(27). Hub genes were put into
cBioPortal for mutation analysis with lung cancer. Next, the
receiver operating characteristic (ROC) curve was performed to
evaluate the diagnostic effect of hub genes. ROC curve was acquired
using survival ROC package 1.0.3. The GEPIA2 tool (28) was further used to validate survival
biomarkers, and Logrank P<0.01 was considered statistically
significant.
Cell culture and treatment
A total of three LUAD cell lines (A549, PC9 and
H1299) and human normal lung epithelial cell line BEAS-2B were
provided by the Chinese Academy of Sciences (Beijing, China). These
cells were cultured in Roswell Park Memorial Institute-1640
(RPMI-1640) medium with 10% fetal bovine serum (Gibco; Thermo
Fisher Scientific, Inc.) at 37°C with 5% CO2.
A549 cells (2×105 ml/well) were
inoculated in six-well plates and cultured to a confluence of
70–90% at 37°C with 5% CO2. Then, 1×108 TU/ml
short hairpin (sh)-negative control (NC; 40 µl/well) and sh-NPM1
(20 µl/well) were transfected to A549 cells using HighGene
transfection agent (ABclonal Biotech Co., Ltd.). After incubation
for 48 h, RPMI-1640 medium with 2.5 µg/ml puromycin was utilized to
select stably transfected cells. Subsequently, part of sh-NPM1
transfected A549 cells received 100 ng/ml EGF treatment for 10 min,
which has shown favorable efficiency in the activation of EGFR
(29). The sh-NPM1 sequence (Human)
was supplied by the Designer of Small Interfering RNA website
(http://biodev.cea.fr/DSIR/DSIR.html),
as follows: SS sequence, CCAAGAATGTGTTGTCCAA; AS sequence,
TTGGACAACACATTCTTGG.
Cell counting kit-8 (CCK-8) assay
A549 cell suspension was inoculated to a 96-well
plate (100 µl/well) and cultured at 37°C with 5% CO2.
After 10 µl CCK-8 solution (Beyotime Institute of Biotechnology)
was added into the well at 24 and 48 h, the plate continued to be
cultured for 2 h at 37°C. Subsequently, absorbance at 450 nm was
measured through a microplate reader (Molecular Devices, LLC).
Wound healing assay
Overall, A549 cells were treated with 0.25% trypsin
and cultured in six-well plates (5×105 cells/well). A
uniform scratch on the cell layer was created via a sterile pipette
tip. Cell migration images were captured at 0 and 24 h through a
light microscope (Olympus Corporation).
Transwell invasion assay
First, 50 mg/l Matrigel (Beijing Solarbio Science
& Technology Co., Ltd.) was diluted at 1:4, and 50 µl diluent
was then placed into the upper chamber for 4 h at 37°C. A549 cells
were diluted into 1×105 cell/ml suspension. Then, 200 µl
cell suspension was added into the upper chamber, with 600 µl
RPMI-1640 medium containing 10% FBS into the lower one. After being
cultured for 24 h and cleaned with phosphate-buffered saline (PBS;
Beyotime Institute of Biotechnology), cells were fixed with
methanol (Sinopharm Group Co., Ltd.) for 30 min and stained with
0.5% crystal violet (Beyotime Institute of Biotechnology) for 20
min. A total of three fields were randomly chosen and images were
captured, and the cell number was determined using ImageJ software
1.8.0 (National Institutes of Health).
Animal experiments
The animal experiments were approved (approval no.
kmmu20230850) by the Animal Research Ethics Committee of The First
Affiliated Hospital of Kunming Medical University (Kunming, China)
and conformed with the guidelines for the use of laboratory
animals. A total of 15 BALB/c male nude mice (age, 5 weeks-old;
weight, 18–22 g) were purchased from GemPharmatech Co. Ltd. All
mice were acclimatized in individually ventilated cages
(specific-pathogen-free conditions) at 22°C with 12/12 h light/dark
cycle, fed and watered ad libitum for 1 week.
Non-transfected or sh-NPM1/sh-NC-transfected A549 cells
(5×106 cells/100 µl) were subcutaneously injected into
mice, respectively, to establish the LUAD model.
The size of the tumor xenografts was measured
weekly, and tumor volume was calculated by the following formula:
Tumor volume (mm3)=(1/2 × length) × width2.
After 5 weeks of model construction, tumor weight was measured. The
mice were anesthetized by intraperitoneal injection of 50 mg/kg
sodium pentobarbital. At the end of the modeling, mice were then
sacrificed with 200 mg/kg sodium pentobarbital, and their death was
indicated by respiratory failure and cardiac arrest.
Western blotting
Using RIPA lysis buffer (Beijing Solarbio Science
& Technology Co., Ltd.), total proteins were extracted from
A549 cells and tumor tissues. Protein concentration was determined
by a BCA kit (Beyotime Institute of Biotechnology). The proteins
(25 µg) were separated in 10% SDS-PAGE gels and then transferred to
PVDF membranes, which were blocked with 5% non-fat dry milk for 1 h
at room temperature. After incubation with specific primary
antibodies (all purchased from Abcam) at 4°C overnight, the
membranes were washed thrice with 1X 0.05% TBST for 10 min and
incubated with goat-anti-rabbit IgG H&L secondary antibodies
(1:2,000; cat no. ab6721; Abcam) at room temperature for 1 h.
Protein bands were visualized by an ECL reagent (Applygen
Technologies, Inc.). The primary antibodies were as follows: NPM1
(1:400; cat no. ab183340), phosphorylated (p)-EGFR (1:1,000; cat
no. ab40815), EGFR (1:5,000; cat no. ab52894), p-MEK (1:2,500; cat
no. ab96379), MEK (1:2,500; cat no. ab32091), p-ERK (1:1,000; cat
no. ab201015), ERK (1:10,000; cat no. ab184699), GAPDH (1:2,500;
cat no. ab9485). The protein bands were visualized using an ECL kit
(Applygen Technologies, Inc.), and the relative protein levels were
quantified using ImageJ software 1.8.0 (National Institutes of
Health).
Statistical analyses
Each experiment was repeated at least thrice, and
data were analyzed by GraphPad Prism 8.0 (Dotmatics). Survival
analysis was performed using the ‘survival’ R package. The data
were expressed as the mean values ± standard deviation. Data from
multiple groups were analyzed by one-way ANOVA followed by Tukey's
post hoc test, and those from two groups were analyzed by Tukey's
test. P<0.05 was considered to indicate a statistically
significant difference.
Results
NPM1 is upregulated and related to
poor prognosis in LUAD
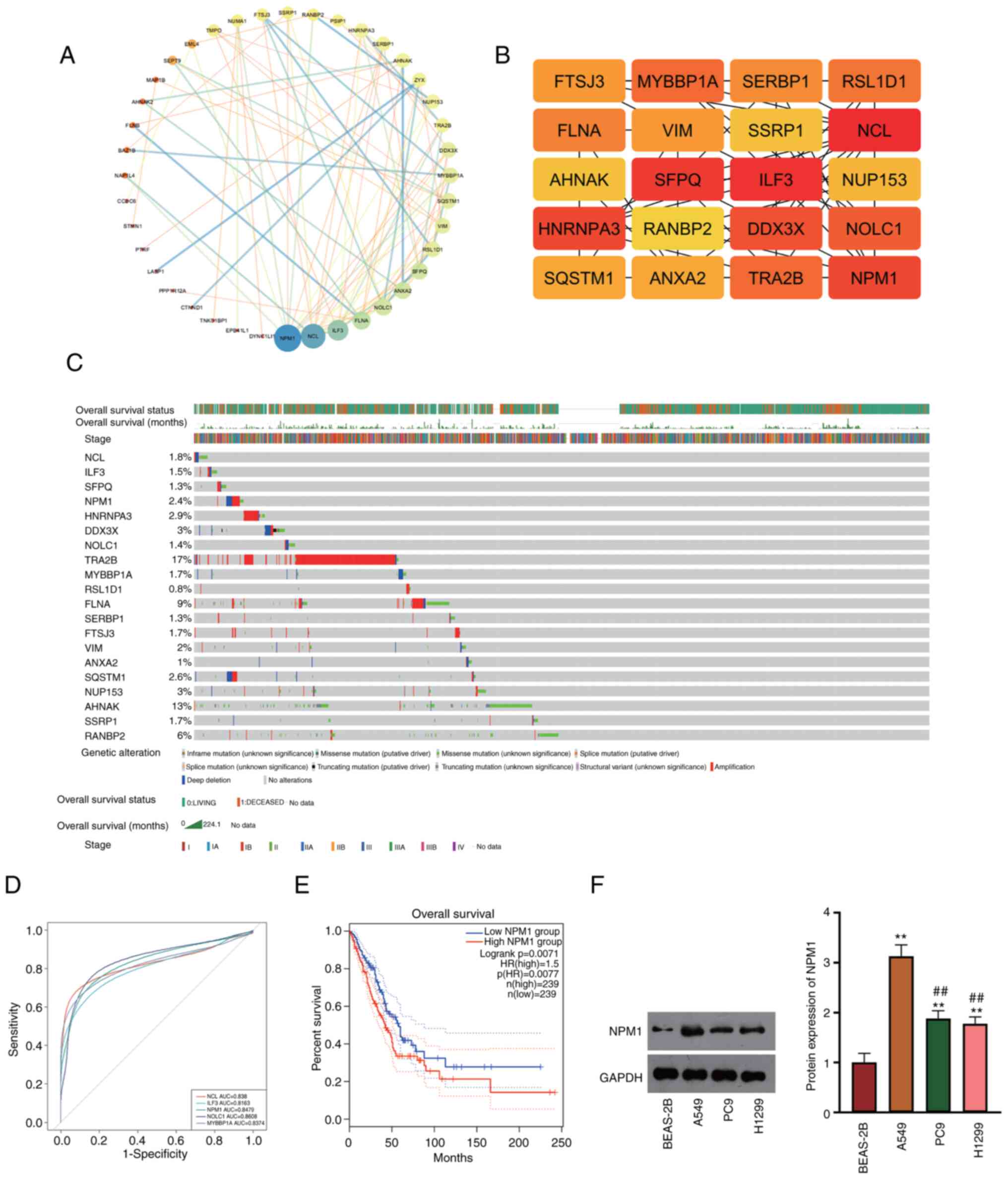
The PPI network of LUAD-related DEGs extracted by
GEO2R was obtained from the STRING database (Fig. 1A). The cytohubba plug-in was used to
obtain the top 20 hub genes, and NPM1 showed high relevance with
LUAD (Fig. 1B). Hub genes were then
inserted into GEPIA2 for survival analysis associated with LUAD.
The results revealed that NPM1 demonstrated a favorable prognostic
effect on overall survival with 2.4% of alteration rate, and the
major alteration type of NPM1 was amplification (Fig. 1C). Next, single gene ROC analysis of
hub genes was performed to evaluate the prognosis for patients with
LUAD. It was found that NPM1, NOLC1 and NCL showed favorable
diagnostic effects in LUAD (AUC >0.8, Fig. 1D). To investigate the effects of
NPM1 on survival and prognosis, patients with LUAD in TCGA-LUAD
were grouped into high- and low-NPM1 expression. The result
revealed that the survival time of patients in the high-NPM1 group
was shorter (Logrank P<0.01; Fig.
1E). Additionally, western blotting verified that NPM1 was
upregulated in LUAD cells (A549, PC9 and H1299) compared with
BEAS-2B, in which A549 cells showed the most obvious significance
(P<0.01, Fig. 1F). Therefore,
A549 cells were chosen for subsequent functional verification
experiments.
NPM1 silencing inhibits the
proliferation, migration and invasion of LUAD cells
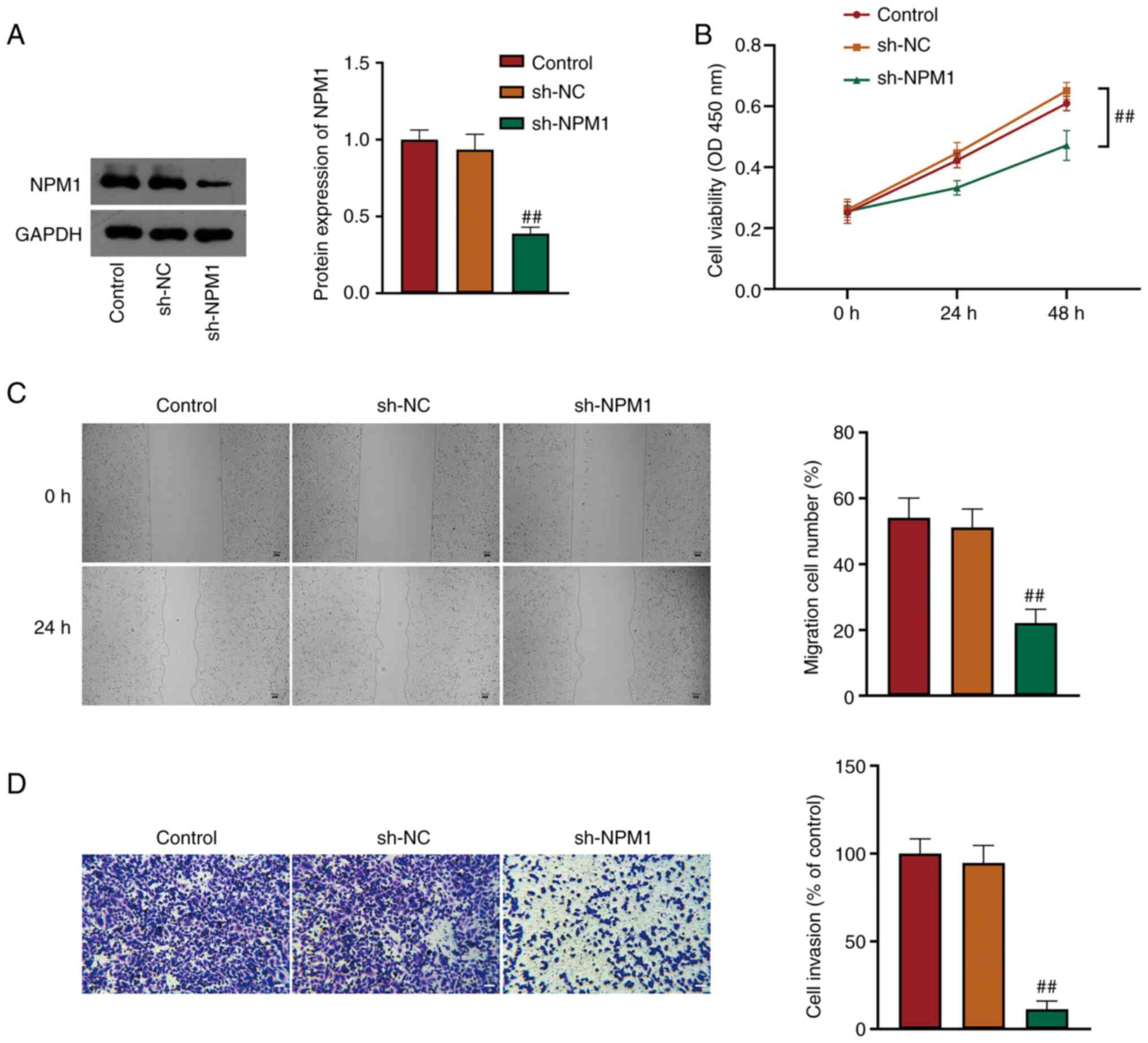
A549 cells were transfected with sh-NC/sh-NPM1 to
investigate the function of NPM1 in LUAD. NPM1 expression in A549
cells was detected. It was revealed that NPM1 protein expression
was significantly reduced in sh-NPM1 cells compared with sh-NC
cells (P<0.01, Fig. 2A), which
revealed successful transfection. CCK-8 assay indicated that NPM1
silencing suppressed A549 cell viability (P<0.01, Fig. 2B). As demonstrated by wound healing
and Transwell assays, A549 cell migration and invasion were
restrained by NPM1 knockdown (P<0.01, Fig. 2C and D).
NPM1 silencing suppresses malignant
characteristics by inhibiting the EGFR/MAPK signaling pathway
The EGFR/MAPK signaling pathway is abnormally
activated in LUAD, and NPM1 was related to signaling by EGFR in
cancer. To investigate whether NPM1 functions on LUAD malignant
characteristics via the EGFR/MAPK pathway, the protein expression
of p-EGFR, EGFR, p-MEK, MEK, p-ERK and ERK, which are the EGFR/MAPK
pathway-related proteins, were determined using western blot
analysis. The results demonstrated that protein levels of
p-EGFR/EGFR, p-MEK/MEK and p-ERK/ERK were all decreased in A549
cells after NPM1 knockdown (P<0.05, Fig. 3A). This result indicated that the
inhibitory effect of NPM1 silencing on LUAD malignancy may be
involved in suppressing the EGFR/MAPK pathway. Subsequently, EGF,
an activator of EGFR, was used to perform feedback experiments. It
was found that cell proliferation, migration and invasion were
increased in sh-NPM1 + EGF cells compared with sh-NPM1 cells
(P<0.01, Fig. 3B-D).
Furthermore, in comparison with sh-NPM1 cells, levels of
p-EGFR/EGFR, p-MEK/MEK, and p-ERK/ERK were successfully increased
in sh-NPM1 + EGF cells (P<0.05, Fig.
3E).
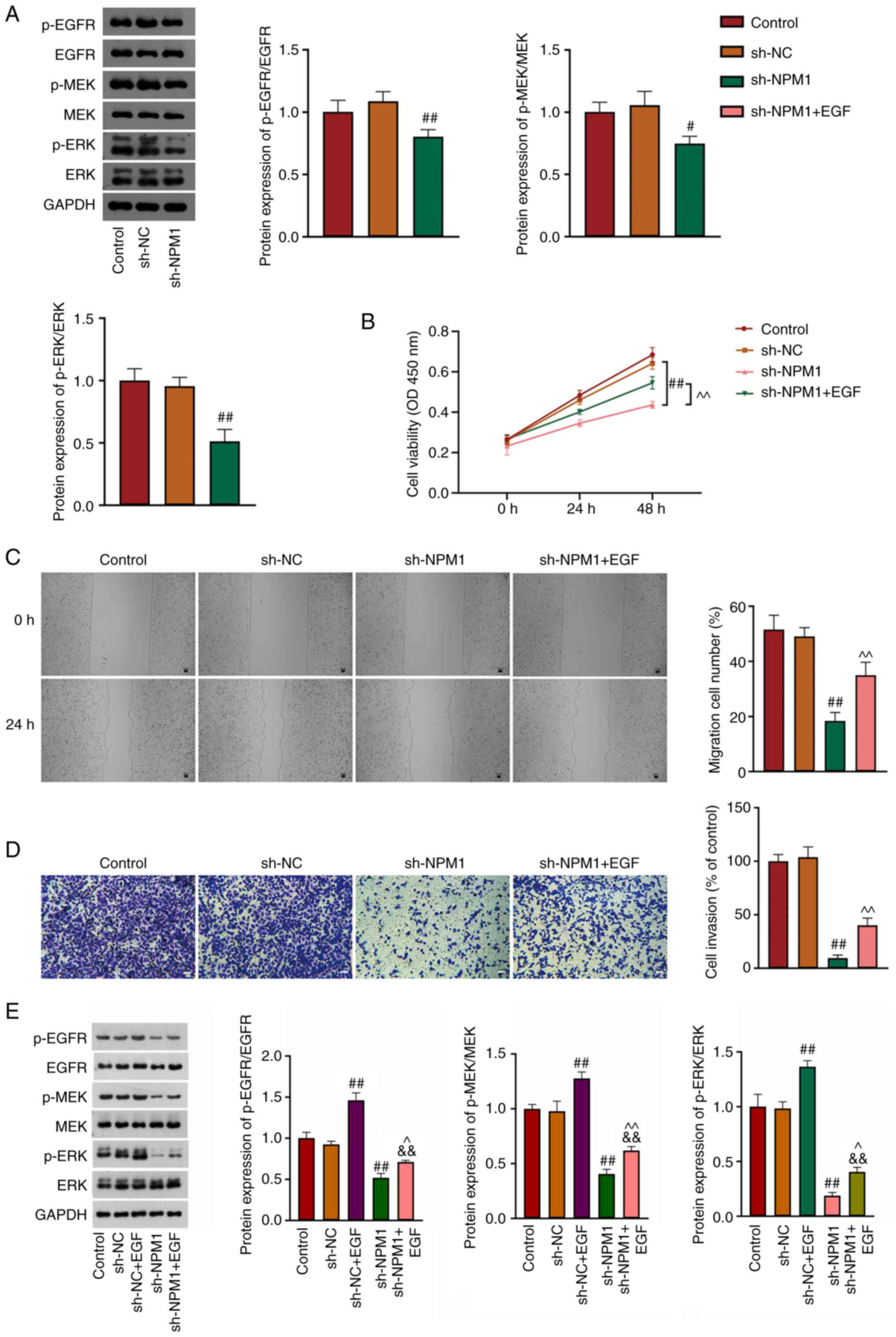 | Figure 3.NPM1 silencing suppresses malignant
characteristics by inhibiting the EGFR/MAPK signaling pathway. (A)
The expression of p-EGFR/EGFR, p-MEK/MEK and p-ERK/ERK was detected
using western blotting. (B) Cell viability was evaluated by Cell
Counting Kit-8 assay. (C) Cell migration at 0 and 24 h was
determined using wound healing assay. Scale bar, 50 µm. (D) Cell
invasion was detected by Transwell assay. Scale bar, 50 µm. (E) The
expression of p-EGFR/EGFR, p-MEK/MEK and p-ERK/ERK was evaluated
using western blotting. sh-NPM1 transfected A549 cells were treated
with EGF. #P<0.05 and ##P<0.01 vs. sh-NC,
&&P<0.01 vs. sh-NC + EGF, ^P<0.05 and
^^P<0.01 vs. sh-NPM1. NPM1, nucleophosmin; p-,
phosphorylated; sh-, short hairpin; NC, negative control. |
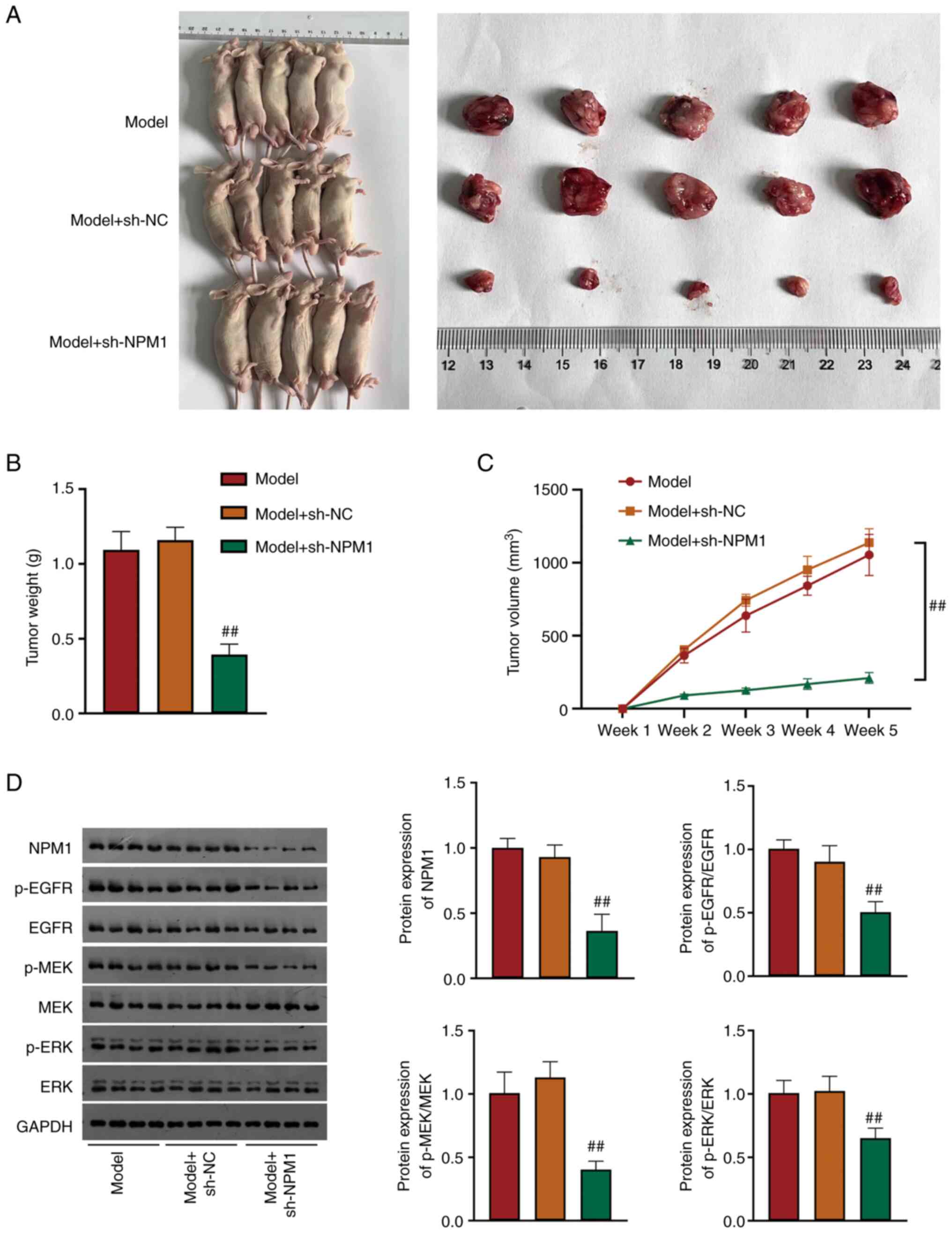
NPM1 silencing inhibits LUAD tumor
growth by restraining the EGFR/MAPK signaling pathway
A xenograft tumor mice model was conducted to deeply
explore the NPM1 effect on LUAD. Compared with model mice, the
tumor weight and volume were significantly smaller in
NPM1-knockdown mice (P<0.01, Fig.
4A-C). Additionally, protein levels of NPM1, p-EGFR/EGFR,
p-MEK/MEK and p-ERK/ERK were found to be significantly reduced in
NPM1-knockdown mice (P<0.01, Fig.
4D).
Discussion
LUAD with a poor prognosis is one of the leading
causes of global cancer-related mortalities (30). Clinical LUAD outcomes using current
treatments are not satisfactory (31). Identification of potential
biomarkers is important for predicting prognosis and guiding
individualized treatments. NPM1 is a highly abundant protein
crucial for multiple cellular functions. The present study revealed
that NPM1 is a potential target and is upregulated in LUAD. NPM1
prompted LUAD progression by activating the EGFR/MAPK signaling
pathway.
NPM1 is a well-known nucleocytoplasmic shuttling
protein required for normal cellular function. NPM1 is involved in
the maintenance of genomic stability, chromatin remodeling,
ribosome biogenesis, cell cycle progression and apoptosis (32). Overexpression, mutation,
translocation, function loss, or sporadic deletion of NPM1 would
result in cancer and tumorigenesis (33,34).
NPM1 overexpression is associated with high-grade malignancies and
poor prognosis. In those with pancreatic cancer, NPM1 promotes
aerobic glycolysis and tumor progression (35), and NPM1 promotes cell proliferation
by targeting PRDX6 in colorectal cancer (36). Previous studies indicated that NPM1
expression could predict the prognosis of prostate (37) and gastric cancers (38). The present bioinformatics analysis
showed that NPM1 was the hub gene in LUAD, and high expression of
NPM1 may indicate poor prognosis of LUAD patients. The current data
demonstrated that NPM1 was upregulated in LUAD cells. NPM1
overexpression could enhance the growth and proliferation of
various tumors (36,39,40).
Same as in previous studies, NPM1 silencing inhibited the
proliferation, migration and invasion of LUAD cells and suppressed
tumor growth in the present study.
Accumulating evidence revealed that the EGFR/MAPK
pathway is involved in NSCLC progression (19,41).
EGFR overexpression results in dimerization, auto-phosphorylation
and downstream activation of the PI3K, STAT and MAPK pathways.
These pathways mediate the transcription of genes that are
associated with transformation, proliferation, migration and
invasion (42). Phosphorylation and
activation of EGFR/MAPK signaling cascade are regarded as key
pathways in various cancers, for example, pancreatic cancer
(43), clear cell renal cell
carcinoma (44) and glioma
(45). EGFR is expressed in 85% of
NSCLC cells and is related to a poor prognosis (42). Activation of the EGFR/MAPK pathway
promotes cyclin D1 expression in NSCLC (42). The mechanism of NPM1 on LUAD
malignancy was further explored. The results indicated that
EGFR/MAPK pathway-related proteins were downregulated in LUAD after
NPM1 knockdown. A recent study revealed that NPM1 was related to
signaling by EGFR in cancer via GSEA analysis (16). In the present study, NPM1 knockdown
inhibited the malignant progression of LUAD by restraining the
EGFR/MAPK pathway. However, EGF (an activator of the EGFR/MAPK
pathway) reversed the effect of NPM1 silencing. These indicated
that NPM1 promotes the progression of LUAD by activating the
EGFR/MAPK signaling pathway.
In summary, the results of the present study
suggested that NPM1 is upregulated and associated with poor
prognosis in LUAD. Furthermore, it was identified that NPM1
promoted cell proliferation, migration, invasion and tumor growth
via the EGFR/MAPK pathway in LUAD. NPM1 is a potential therapeutic
target in LUAD. Although the current study improved our
understanding of NPM1 in LUAD, there were certain limitations.
Firstly, the clinical significance of NPM1 in the LUAD progression
requires further investigation. Secondly, the specific mechanism of
NPM1 on the EGFR/MAPK pathway needs to be explored.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science
Foundation of China (Study on anticancer effects of natural
bioactive compound C18H17NO6 and its mechanism of promoting
apoptosis on lung cancer cells; grant no. 81960545), the general
projects of Yunnan basic research plan (Mechanism of natural
anticancer compound M activating MAPKs signaling pathway and
inducing apoptosis of non-small cell lung cancer cells; grant no.
202001AT070026), the Joint special project of Yunnan Provincial
Department of Science and Technology (The role and mechanism of
natural active compound M in lung cancer metastasis; grant no.
202201AY070001-055) and the Talent Project of First Affiliated
Hospital of Kunming Medical University (grant no. 2022535D01).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ML and ZL conceptualized the present study. LW and
RW curated data and conducted investigation. RW, DZ, MC, SL, RL, CD
and MM conducted formal analysis and developed methodology. ZL
supervise the study and acquired funding. RW, ML and ZL conducted
project administration. RW, CD and ZL provided resources. SL, RL
and MM performed software analysis. RW, HZ, CD, SZ and ZL validated
data. RW and ML performed visualization. RW, CD and HZ wrote the
original draft. ML, SZ, CD and ZL wrote, reviewed and edited the
manuscript. RW, HZ, CD, SZ and ZL confirm the authenticity of all
the raw data. All authors reviewed the results, read and approved
the final version of the manuscript.
Ethics approval and consent to
participate
The animal experiments were approved (approval no.
kmmu20230850) by the Animal Research Ethics Committee of The First
Affiliated Hospital of Kunming Medical University (Kunming,
China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
NSCLC
|
non-small-cell lung carcinoma
|
|
LUAD
|
lung adenocarcinoma
|
|
NPM1
|
nucleophosmin
|
|
DEG
|
differentially expressed gene
|
|
PPI
|
protein-protein interaction
network
|
|
NC
|
negative control
|
|
ROC
|
receiver operating characteristic
|
|
CCK-8
|
Cell Counting Kit-8
|
References
|
1
|
Wu Z, Bai X, Lu Z, Liu S and Jiang H:
LINC01094/SPI1/CCL7 axis promotes macrophage accumulation in lung
adenocarcinoma and tumor cell dissemination. J Immunol Res.
2022:64507212022. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhao X, Chen Y, Sun X, He Z, Wu T, Wu C,
Chen J, Wang J, Diao K and Liu XS: Oncogenic EFNA4 amplification
promotes lung adenocarcinoma lymph node metastasis. Cancers
(Basel). 14:42262022. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bai J, Li H, Chen X, Chen L, Hu Y, Liu L,
Zhao Y, Zuo W, Zhang B and Yin C: LncRNA-AC009948.5 promotes
invasion and metastasis of lung adenocarcinoma by binding to
miR-186-5p. Front Oncol. 12:9499512022. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li R, Mu C, Cao Y and Fan Y: METTL7B
serves as a prognostic biomarker and promotes metastasis of lung
adenocarcinoma cells. Ann Transl Med. 10:8952022. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ma K, Jin Q, Wang M, Li X and Zhang Y:
Research progress and clinical application of predictive biomarker
for immune checkpoint inhibitors. Expert Rev Mol Diagn. 19:517–529.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Miller KD, Siegel RL, Lin CC, Mariotto AB,
Kramer JL, Rowland JH, Stein KD, Alteri R and Jemal A: Cancer
treatment and survivorship statistics, 2016. CA Cancer J Clin.
66:271–289. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Guo J, Li A, Guo R, He Q, Wu Y, Gou Y, Jin
J and Huang G: C1orf74 positively regulates the EGFR/AKT/mTORC1
signaling in lung adenocarcinoma cells. PeerJ. 10:e139082022.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu J, Yang H, Yin D, Jia Y, Li S and Liu
Y: Expression and prognostic analysis of CLDN18 and Claudin18.2 in
lung adenocarcinoma. Pathol Res Pract. 238:Aug 10–2022.(Epub ahead
of print). View Article : Google Scholar
|
|
9
|
Qiu X, Liu W, Zheng Y, Zeng K, Wang H, Sun
H and Dai J: Identification of HMGB2 associated with proliferation,
invasion and prognosis in lung adenocarcinoma via weighted gene
co-expression network analysis. BMC Pulm Med. 22:3102022.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
La Manna S, Florio D, Di Natale C, Lagreca
E, Sibillano T, Giannini C and Marasco D: Type C mutation of
nucleophosmin 1 acute myeloid leukemia: Consequences of intrinsic
disorder. Biochim Biophys Acta Gen Subj. 1866:1301732022.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Venanzi A, Rossi R, Martino G, Annibali O,
Avvisati G, Mameli MG, Sportoletti P, Tiacci E, Falini B and
Martelli MP: A curious novel combination of nucleophosmin (NPM1)
gene mutations leading to aberrant cytoplasmic dislocation of NPM1
in acute myeloid leukemia (AML). Genes (Basel). 12:14262021.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nemeckova S, Alexova-Zurkova K, Hainz P,
Krystofova J, Mackova J, Roubalova K, Stastna-Markova M, Vrana M
and Vydra J: Non-mutated nucleophosmin 1 is recognized by the CD8+
T lymphocytes of an AML patient after the transplantation of
hematopoietic stem cells from an HLA-haploidentical donor. Curr
Oncol. 29:2928–2934. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhou Y, Fang Y, Zhou J, Liu Y, Wu S and Xu
B: NPM1 is a novel therapeutic target and prognostic biomarker for
ewing sarcoma. Front Genet. 12:7712532021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zeng D, Xiao Y, Zhu J, Peng C, Liang W and
Lin H: Knockdown of nucleophosmin 1 suppresses proliferation of
triple-negative breast cancer cells through activating
CDH1/Skp2/p27kip1 pathway. Cancer Manag Res. 11:143–156. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Peng HH, Ko HH, Chi NC, Wang YP, Lee HC,
Pan PY, Kuo MY and Cheng SJ: Upregulated NPM1 is an independent
biomarker to predict progression and prognosis of oral squamous
cell carcinomas in Taiwan. Head Neck. 42:5–13. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu XS, Zhou LM, Yuan LL, Gao Y, Kui XY,
Liu XY and Pei ZJ: NPM1 is a prognostic biomarker involved in
immune infiltration of lung adenocarcinoma and associated with m6A
modification and glycolysis. Front Immunol. 12:7247412021.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Greenspan LJ, de Cuevas M, Le KH, Viveiros
JM and Matunis EL: Activation of the EGFR/MAPK pathway drives
transdifferentiation of quiescent niche cells to stem cells in the
Drosophila testis niche. Elife. 11:e708102022. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lim WC, Choi HK, Kim KT and Lim TG: Rose
(Rosa gallica) petal extract suppress proliferation, migration, and
invasion of human lung adenocarcinoma A549 cells through via the
EGFR signaling pathway. Molecules. 25:51192020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xu H, Yang X, Xuan X, Wu D, Zhang J, Xu X,
Zhao Y, Ma C and Li D: STAMBP promotes lung adenocarcinoma
metastasis by regulating the EGFR/MAPK signaling pathway.
Neoplasia. 23:607–623. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou Z, Wang W, Xie X, Song Y, Dang C and
Zhang H: Methylation-induced silencing of SPG20 facilitates gastric
cancer cell proliferation by activating the EGFR/MAPK pathway.
Biochem Biophys Res Commun. 500:411–417. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ji J, Li C, Wang J, Wang L, Huang H, Li Y
and Fang J: Hsa_circ_0001756 promotes ovarian cancer progression
through regulating IGF2BP2-mediated RAB5A expression and the
EGFR/MAPK signaling pathway. Cell Cycle. 21:685–696. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Huang J, Liu J and Qiu L: Transient
receptor potential vanilloid 1 promotes EGFR ubiquitination and
modulates EGFR/MAPK signalling in pancreatic cancer cells. Cell
Biochem Funct. 38:401–408. 2020. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Loubeau G, Boudra R, Maquaire S,
Lours-Calet C, Beaudoin C, Verrelle P and Morel L: NPM1 silencing
reduces tumour growth and MAPK signalling in prostate cancer cells.
PLoS One. 9:e962932014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gray KA, Seal RL, Tweedie S, Wright MW and
Bruford EA: A review of the new HGNC gene family resource. Hum
Genomics. 10:62016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
von Mering C, Huynen M, Jaeggi D, Schmidt
S, Bork P and Snel B: STRING: A database of predicted functional
associations between proteins. Nucleic Acids Res. 31:258–261. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tang Z, Kang B, Li C, Chen T and Zhang Z:
GEPIA2: An enhanced web server for large-scale expression profiling
and interactive analysis. Nucleic Acids Res. 47((W1)): W556–W560.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhou F, Yu T, Xiao F, Wang B, Tian W, Xu
R, Zhao X, Zeng A, Liu N, Wang Y, et al: Periostin promotes EMT via
inhibition of RIN1-mediated endocytosis of EGFR in gliomas. Holist
Integr Oncol. 1:192022. View Article : Google Scholar
|
|
30
|
Luo J and Du X: A promising prognostic
signature for lung adenocarcinoma (LUAD) patients basing on 6
hypoxia-related genes. Medicine (Baltimore). 100:e282372021.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yu Y, Wang Z, Zheng Q and Li J: GREB1L
overexpression correlates with prognosis and immune cell
infiltration in lung adenocarcinoma. Sci Rep. 11:132812021.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Box JK, Paquet N, Adams MN, Boucher D,
Bolderson E, O'Byrne KJ and Richard DJ: Nucleophosmin: From
structure and function to disease development. BMC Mol Biol.
17:192016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Karimi Dermani F, Gholamzadeh Khoei S,
Afshar S and Amini R: The potential role of nucleophosmin (NPM1) in
the development of cancer. J Cell Physiol. 236:7832–7852. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ruan Y, Xu H and Ji X: High expression of
NPM1 via the Wnt/β-catenin signalling pathway might predict poor
prognosis for patients with prostate adenocarcinoma. Clin Exp
Pharmacol Physiol. 49:525–535. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhu Y, Shi M, Chen H, Gu J, Zhang J, Shen
B, Deng X, Xie J, Zhan X and Peng C: NPM1 activates metabolic
changes by inhibiting FBP1 while promoting the tumorigenicity of
pancreatic cancer cells. Oncotarget. 6:21443–21451. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang D, Li Y, Liu Y, Cheng S, Liu F, Zuo
R, Ding C, Shi S and Liu G: NPM1 promotes cell proliferation by
targeting PRDX6 in colorectal cancer. Int J Biochem Cell Biol.
147:1062332022. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Dai L, Li J, Xing M, Sanchez TW, Casiano
CA and Zhang JY: Using serological proteome analysis to identify
serum anti-nucleophosmin 1 autoantibody as a potential biomarker in
European-American and African-American patients with prostate
cancer. Prostate. 76:1375–1386. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Guo CA, Su XL, Wang WJ, Xia TH, Cao XM,
Yuan SB, Wang WA, Zhang A and Liu HB: NPM1 is a diagnostic and
prognostic biomarker associated with the clinicopathological
characteristics of gastric cancer. Neoplasma. 69:965–975. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jing Y, Jiang X, Lei L, Peng M, Ren J,
Xiao Q, Tao Y, Tao Y, Huang J, Wang L, et al: Mutant NPM1-regulated
lncRNA HOTAIRM1 promotes leukemia cell autophagy and proliferation
by targeting EGR1 and ULK3. J Exp Clin Cancer Res. 40:3122021.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liu Q, Liu N, Shangguan Q, Zhang F, Chai
W, Tong X, Zhao X, Li Z, Qi D and Ye X: LncRNA SAMD12-AS1 promotes
cell proliferation and inhibits apoptosis by interacting with NPM1.
Sci Rep. 9:115932019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ishola AA, Chien CS, Yang YP, Chien Y,
Yarmishyn AA, Tsai PH, Chen JC, Hsu PK, Luo YH, Chen YM, et al:
Oncogenic circRNA C190 promotes non-small cell lung cancer via
modulation of the EGFR/ERK pathway. Cancer Res. 82:75–89. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Fu H, Gao H, Qi X, Zhao L, Wu D, Bai Y, Li
H, Liu X, Hu J and Shao S: Aldolase A promotes proliferation and
G1/S transition via the EGFR/MAPK pathway in non-small
cell lung cancer. Cancer Commun (Lond). 38:182018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wang J, Zhang Y, Wang Q, Wang L and Zhang
P: Study on the potential molecular mechanism of xihuang pill in
the treatment of pancreatic cancer based on network pharmacology
and bioinformatics. Evid Based Complement Alternat Med.
2022:46514322022.PubMed/NCBI
|
|
44
|
Ke X, Zeng X, Wei X, Shen Y, Gan J, Tang H
and Hu Z: MiR-514a-3p inhibits cell proliferation and
epithelial-mesenchymal transition by targeting EGFR in clear cell
renal cell carcinoma. Am J Transl Res. 9:5332–5346. 2017.PubMed/NCBI
|
|
45
|
Wang M, Zhao Y, Yu ZY, Zhang RD, Li SA,
Zhang P, Shan TK, Liu XY, Wang ZM, Zhao PC and Sun HW: Glioma
exosomal microRNA-148a-3p promotes tumor angiogenesis through
activating the EGFR/MAPK signaling pathway via inhibiting ERRFI1.
Cancer Cell Int. 20:5182020. View Article : Google Scholar : PubMed/NCBI
|