Introduction
Cervical cancer remains the fourth leading cause of
cancer-related incidence and mortality, with 660,000 new cases and
350,000 deaths among women worldwide in 2022 (1). Despite ongoing advancements in disease
management, patient prognosis is influenced by multiple individual
factors, highlighting the urgent need for personalized therapeutic
strategies guided by reliable prognostic and predictive biomarkers
(2–4).
In this context, the cancer cell surface proteome
(surfaceome) has emerged as a critical area of study due to its
role in tumor progression and treatment response. Defined as the
complete set of plasma membrane proteins with at least one amino
acid residue exposed to the extracellular space (5), the surfaceome facilitates the
selective movement of substances and serves as an anchoring base
for the cytoskeleton, and its structure influences crucial
signaling processes triggered by interactions between cells and the
extracellular matrix. The surfaceome includes receptors,
transporters, channels, cell-adhesion proteins and enzymes, making
it a rich source of potential diagnostic biomarkers and important
targets for therapeutic interventions.
Interferon-induced transmembrane proteins (IFITMs)
are small Dispanin proteins containing two (trans) membrane helices
that localize in the plasma membrane and endosomal/lysosomal
membranes (6). Immunity-related
IFITMs (IFITM1, IFITM2 and IFITM3) are highly homologous, but they
differ in their subcellular localization and function, as
demonstrated in recent years (7,8).
IFITM1 differs from IFITM2 and IFITM3 by a 21-amino acid truncation
at the N-terminus and a 13-amino acid extension at the C-terminus.
Unlike IFITM2 and IFITM3, which are predominantly found in
endosomal membranes, IFITM1 is localized primarily in the plasma
membrane on the cell surface.
Immunity-related IFITMs are strongly induced by type
I and type II interferons (IFNs) and function mainly as restriction
factors that block viral infections. They also play an important
role in cancer progression (6,9–11).
Although the exact mechanisms of their function remain unknown,
IFITMs influence membrane structure and function (12–14),
the intracellular trafficking of endocytosed cargo (15), and pH regulation within the
vesicular environment (16,17). These processes are crucial for
regulating surface protein expression, trafficking and recycling,
leading us to hypothesize that IFITM1 could modulate surfaceome
composition.
The aim of the present study was to elucidate the
mechanisms by which IFITMs contribute to tumor progression. Initial
investigations revealed that the suppression of IFITM1 in cervical
cancer cells affects the levels of members of the IFN-related DNA
damage resistance signature (IRDS) (18), including major histocompatibility
complex class I (MHC-I), suggesting a possible explanation for the
inverse correlation between metastasis formation and IFITM1/3
expression in cervical cancer tissues. Building on these findings,
the authors focused on the cell surface proteome and, for the first
time to the best of our knowledge, it was demonstrated how IFITM1
levels impact the cervical cancer cell surfaceome and associated
cell phenotype.
Materials and methods
Cell culture
SiHa cell line [cat. no. HTB-35; American Type
Culture Collection (ATCC)] was cultured in RPMI-1640 medium (cat.
no. R4130; MilliporeSigma) supplemented with 10% fetal bovine serum
(cat. no. S-FBS-SA-015; Serana Europe GmbH) and
penicillin-streptomycin (cat. no. XC-A4122; Biosera) at 37°C with
5% CO2. The cells were passaged every two days using
Accutase (cat. no. A6964; MilliporeSigma). Cell line authentication
(STR profiling) and mycoplasma testing were performed
regularly.
The lentiviral CRISPR/Cas9 system was used to
generate single- and double-IFITM1- and IFITM3-knockout (KO) cells.
The generation and characterization of IFITM1- and IFITM3-KO SiHa
clones were described previously (18). SiHa wild-type (wt) and SiHa
scrambled cells served as controls. SiHa scrambled control was
generated using the CRISPR/Cas method with sgRNA that lacks a
complementary sequence in the genome. For all methods used to
evaluate the effect of IFNγ, the cells were treated with human IFNγ
recombinant protein (cat. no. PHC4031; Gibco™; Thermo Fisher
Scientific, Inc.) at a concentration of 100 ng/ml for 24 h. SiHa
cell line exhibits baseline IFN pathway activity, and the IFNγ
concentration used ensures effective and measurable exogenous
pathway stimulation, as confirmed by the detection of
IFN-stimulated protein levels following treatment with increasing
IFNγ concentrations.
Pulse stable isotope labeling with
amino acids in cell culture (SILAC), biotinylation and isolation of
cell surface proteins
For pulse SILAC, SiHa wt and IFITM1 KO cells (clones
no. I, II and IV) were cultured in biological triplicates for 24 h
on 10-cm plates in R6K4 RPMI SILAC medium (cat. no. SM022; Gemini
Biosciences) containing 13C-labeled arginine and 2D labeled lysine
supplemented with dialyzed FBS (cat. no. A3382001; Gibco; Thermo
Fisher Scientific, Inc.) and penicillin-streptomycin. Three
independently created clones (no. I, II and IV) were used as
biological replicates of IFITM1 KO cells to minimize the effect of
clonal selection. The cells from 3×10 cm-well plates were harvested
with Accutase, rinsed in ice-cold PBS and incubated with 0.8 mM
cell impermeable EZ-Link Sulfo-NHS-SS-Biotin (cat. no. 21331;
Thermo Fisher Scientific, Inc.) in PBS for 10 min. After
centrifugation at 200 × g/4°C/5 min, 50 mM Tris (pH 7.5) was added
to quench the excess Sulfo-NHS-SS Biotin. After washing in PBS, the
cell pellets were suspended in lysis buffer (2% NP-40, 0.2% SDS, 10
mM EDTA, 108 mM oxidized glutathione, 1X protease inhibitor mix)
and incubated on ice for 30 min. Biotinylated proteins were
incubated with Pierce high-capacity streptavidin agarose (cat. no.
20359; Thermo Fisher Scientific, Inc.) for 1 h; washed 3 times in
Buffer A (1% NP-40, 0.1% SDS, 5 mM oxidized glutathione), 2 times
in Buffer B (10% SDS, 5 mM oxidized glutathione), 2 times in Buffer
C (2 M NaCl, 0.1% SDS, 5 mM oxidized glutathione, heated to 40°C)
and 4 times in Buffer D (50 mM Tris (pH 8), 5 mM oxidized
glutathione); and then eluted with 200 mM DTT reducing agent in 125
mM Tris (pH 6.8) for 5 min at 85°C.
MS analysis
Peptide generation using FASP
Proteins were digested using a filter-aided sample
preparation protocol (FASP) (19).
Briefly, whole material obtained from enriched cell surface
proteins was loaded on Microcon Ultracel-30 spin filter columns
with a 30-kDa cutoff (MilliporeSigma). Urea buffer (8 M urea in 0.1
M Tris, pH 8.5) was added to the filter column, which was
subsequently centrifuged at 14,000 × g/20°C/15 min. Protein
reduction was performed via the addition of 100 mM Tris
(2-carboxyethyl) phosphine hydrochloride (MilliporeSigma) in urea
buffer at 37°C/30 min/600 rpm in a thermomixer. The column was
centrifuged at 14,000 × g/20°C/15 min, and free sulfhydryl groups
were alkylated using 300 mM iodoacetamide in urea buffer
(MilliporeSigma). Protein alkylation was performed in the dark for
20 min, followed by another centrifugation at 14,000 × g/15 min.
Ammonium bicarbonate was added to the column (final concentration
of 10 µM), followed by centrifugation at 14,000 × g/20 min. Trypsin
(Promega Corporation) was added at a 1:100 ratio, and the proteins
were digested overnight at 37°C. The columns were centrifuged at
14,000 × g/15 min, and the peptides were desalted on C18 Micro spin
columns (Harvard Apparatus).
LC-MS/MS analyses
The samples were separated using an UltiMate 3000
rapid separation liquid chromatography (RSLC) nano chromatograph
(Thermo Fisher Scientific, Inc.). Peptides were loaded on a
precolumn (µ-precolumn, 30 m i.d., 5 mm length, C18 PepMap 100, 5
µm particle size, 100 Å pore size) and further separated on an
Acclaim PepMap RSLC column (75 µm i.d., length 250 mm, C18,
particle size 2 µm, pore size 100 Å) with a flow rate of 300 nl/min
using a linear gradient of B (80% acetonitrile in 0.08% aq. formic
acid) in A (0.1% aq. formic acid): 2% B for 4 min, 2–40% B for 64
min, 40–98% B for 2 min, with A (0.1% aq. formic acid) and B (80%
acetonitrile in 0.08% aq. formic acid). Peptides eluted from the
column were introduced into an Orbitrap Elite mass spectrometer
(Thermo Fisher Scientific, Inc.) operating in the Top20
data-dependent acquisition mode. A survey scan of 400–2,000 m/z was
performed at 120,000 resolution with an AGC target of
1×106 and a 200 ms injection time, followed by 20
data-dependent MS2 scans performed in the LTQ linear ion trap with
1 microscan, a 10 ms injection time and 10,000 AGC.
Database searching and data
evaluation
The mass spectrometry data were processed using
Proteome Discoverer (Thermo Fisher Scientific, Inc.), version 2.2,
for SILAC-labeled samples. For the raw files of each triplicate
sample set, a consensus workflow was applied, consisting of the
following sequential nodes: the Minora feature detector, the
Precursor Ion Quantifier, and the Feature Mapper. Data processing
was performed using the Sequest HT engine with the following search
settings: database Swiss-Prot TaxID=9606, 2017-10-25, sequences
42252, taxonomy: Homo sapiens (updated September 2019) and database
with contaminants: PD_Contaminants_2015_5fasta; enzyme trypsin; 2
missed cleavage sites; precursor mass tolerance 10 ppm; fragment
mass tolerance 0.6 Da; static modification carbamidomethyl [+57.02
Da, (C)], label 13C(6) [+6.020 Da
(R)] and label 2H(4) [+4.025 Da
(K)]; dynamic modifications oxidation [+15.995 Da (M)], Met-loss +
Acetyl [protein terminus, −89.030 Da (M)]. The results were further
validated using the Percolator algorithm (version integrated in
Proteome Discoverer 2.2), which applies semi-supervised machine
learning for peptide-spectrum match validation and false discovery
rate (FDR) estimation, with a FDR threshold of 1%. For the protein
quantification and statistical assessment of the biological
triplicates, the Precursor Ions Quantifier was applied with the
following parameters: using only unique peptides and razor
peptides, normalization on total peptide amount, pairwise ratio
calculation and ANOVA (background-based) hypothesis test. The
relative quantification value is represented as the relative peak
area of the peptides with heavy isotope labels in SiHa wt cells
compared with IFITM1 KO cells or cells treated with IFNγ compared
with control SiHa cells without any treatment (Appendix S1). The normalized peptide
abundances were used to calculate the statistical significance of
differences between groups (wt vs. IFITM1 KO; untreated vs.
IFNγ-treated samples) using an unpaired two-tailed t-test.
Gene Ontology (GO) analysis was performed using
DAVID Bioinformatics Resources 6.8 (20), and protein-protein interactions were
mapped using STRING database (21).
The mass spectrometry proteomics data have been deposited in the
ProteomeXchange Consortium via the PRIDE repository (22) with the dataset identifier PXD053951
and the following access login credentials: Username: reviewer_pxd053951@ebi.ac.uk;
Password: ouMaYQI4Qg6T.
Flow cytometry
The cells were harvested using Accutase, washed in
1% BSA (cat. no. A3059; MilliporeSigma) in PBS, and incubated with
PE-conjugated anti-CD166 monoclonal antibody (cat. no. 12-1668-42)
or FITC-conjugated anti-CD40 monoclonal antibody (cat. no.
11-0409-42; both from Invitrogen; Thermo Fisher Scientific, Inc.)
for 30 min on ice in the dark. After a second wash, the cells were
resuspended in 1% BSA in PBS and measured with a FACSVerse
instrument (BD Biosciences). The results were analyzed using
FACSSuite (BD Biosciences). Statistical analysis was performed
using an unpaired t-test with data from three biological
replicates.
Protein extraction, PAGE and western
blotting
The cells were scraped into 1X LDS lysis buffer on
ice: 105 mM Tris hydrochloride, 140 mM Tris base, 75 mM LDS, 0.5 mM
EDTA, 10% glycerol, 1X protease inhibitor cocktail (cat. no. 78429;
Thermo Fisher Scientific, Inc.), and 0.1 mM PMSF (Thermo Fisher
Scientific, Inc.). The samples were frozen after lysis. The cell
lysates were sonicated and cleared by centrifugation (20,000 × g/30
min/4°C). The total protein concentration was measured using a DC
protein assay (cat. no. 5000112; Bio-Rad Laboratories, Inc.).
Appropriate amounts of cell lysates were mixed with DTT
(MilliporeSigma) to a final concentration of 100 mM and 4X NuPAGE
LDS Sample Buffer (cat. no. NP0008; Invitrogen; Thermo Fisher
Scientific, Inc.) and then heated at 95°C for 10 min. Samples (20
µg/lane) were loaded into 10% polyacrylamide gels, separated by
molecular weight, and transferred onto nitrocellulose membranes
using the Mini-PROTEAN system (Bio-Rad Laboratories, Inc.). The
membranes were blocked in 5% non-fat milk in phosphate-buffered
saline with 0.1% Tween 20 (PBST) at room temperature for 1 h and
incubated with primary antibodies diluted in milk buffer overnight
at 4°C. After washing in PBST, the membranes were incubated at room
temperature for 1 h with HRP-linked secondary antibodies. After
washing in PBST and PBS, the chemiluminescent signal was visualized
using enhanced chemiluminescence (ECL) reagents and a G:BOX Chemi
XRQ image analysis system and software (Syngene). Fiji ImageJ
software v. 2.14.0/1.54f (National Institutes of Health) (23) was used to quantify the signal
intensity of individual proteins relative to the intensity of the
loading control β-actin.
The primary antibodies and their dilutions used were
as follows: anti-IFITM1 (1:1,000; 5B5E2; cat. no. 60074-1-Ig;
Proteintech Group, Inc.), anti-IFITM2 (1:500; 3D5F7; cat. no.
66137-1-Ig; Proteintech Group, Inc.), anti-IFITM3 (1:1,000; D8E8G;
cat. no. 59212; Cell Signaling Technology, Inc.), anti-CD166
(1:1,000; EPR2759(2); cat. no. ab109215; Abcam) and anti-β-actin
(1:5,000; cat. no. MA1-140; Invitrogen; Thermo Fisher Scientific,
Inc.).
The secondary antibodies used were as follows:
anti-rabbit IgG (1:2,000; cat. no. 7074; Cell Signaling Technology,
Inc.) and anti-mouse IgG (1:5,000; Jackson ImmunoResearch
Laboratories, Inc.).
Migration and invasion assays
For the wound healing assay, cells
(7×105) were seeded into 12-well plates and incubated in
complete growth medium for 48 h. To inhibit cell proliferation and
avoid serum-free starvation conditions, the cells were treated with
mitomycin C (10 µg/ml; M5353; MilliporeSigma) and incubated for 2 h
at 37°C (24). The cells were then
scratched and washed twice with PBS. To observe cell migration, the
cells were cultivated in complete growth medium, and images were
captured at time 0 (t0) and after 24 h (t24)
using an Eclipse Ti-E inverted microscope (Nikon Corporation) at
×100 magnification. Fiji ImageJ software (23) with the Wound healing size tool
plugin (25) was used for scratch
area quantification. The migration rate (%) was calculated as
[(scratch area t0-scratch area t24)/scratch
area t0)]*100. The assay was repeated in biological
triplicate (each in technical duplicate), and statistical analysis
was conducted using an unpaired t-test.
For the Transwell migration assay, a
1×105 cell suspension in serum-free RPMI was placed in
the top chamber of each insert (cat. no. 140620; Thermo Fisher
Scientific, Inc.). Complete RPMI medium was added to the lower
chambers. The cells were incubated at 37°C for 24 h. After
incubation, the cells on the upper surface of the inserts were
removed, and the membranes were fixed with 70% ethanol at room
temperature for 15 min. The migratory cells were stained with 0.5%
crystal violet for 15 min. Images of the stained cells were
captured using an Eclipse Ti-S microscope (Nikon Corporation). The
number of migratory cells was calculated as an average of 5 fields
of view (100× magnification) in each of the independent
triplicates, and statistical analysis was conducted using an
unpaired t test.
To evaluate the invasion potential of the cells, a
3D Matrigel drop invasion assay was performed as established by
Aslan et al (26). Images
were captured 6 days after drop formation using an EVOS™ FL Digital
Inverted Microscope (Invitrogen™; objective magnification, ×4), and
the invasion area (mm2) per field of view was calculated
using Fiji ImageJ software (23).
Two independent fields of view were evaluated for each drop. The
assay was repeated in independent biological duplicates, each in
technical triplicates, and statistical analysis was conducted using
an unpaired t-test.
Cervicosphere formation assay
Cells were seeded in ultralow attachment plates
(Corning, Inc.) at a density of 10,000 viable cells per well using
a FACSAria III. The cells were cultured in K-SFM (Gibco; Thermo
Fisher Scientific, Inc.) supplemented with B27 (Gibco; Thermo
Fisher Scientific, Inc.), 10 ng/ml human epidermal growth factor
(MilliporeSigma) and 10 ng/ml bFGF (Gibco; Thermo Fisher
Scientific, Inc.). The cells were then cultivated for 10 days.
Images of the formed spheres were captured using an EVOS™ FL
Digital Inverted Microscope (Invitrogen™; Thermo Fisher Scientific,
Inc.; objective magnification, ×4), and the total number of spheres
per well was determined. The sphere size was evaluated using Fiji
ImageJ software (23) with a lower
threshold of 6,000 µm2. The assay was repeated in
independent biological triplicates, and statistical analysis was
conducted using an unpaired t test.
Reverse transcription-quantitative PCR
(RT-qPCR)
Cells (1×106) were seeded into 60 mm cell
culture dishes and incubated overnight in complete RPMI. The cells
were then treated with IFNγ (100 ng/ml; cat. no. PHC4031; Gibco;
Thermo Fisher Scientific, Inc.) and further incubated for 24 h. RNA
was isolated using the RNeasy Mini Kit (cat. no. 74104; Qiagen) and
transcribed into cDNA using the High-Capacity cDNA Reverse
Transcription Kit (cat. no. 4368814, Applied Biosystems™; Thermo
Fisher Scientific, Inc.) following the manufacturer's protocol.
Luna® Universal qPCR Master Mix (NEB) was used with 20 ng of cDNA
per 10 µl reaction in a 384-well plate, along with specific primers
(sequences below). Primers specific to ACTB were used as an
endogenous control. The thermocycling conditions recommended by the
master mix manufacturer were applied. qPCR was performed, and the
results were analyzed using the QuantStudio 5 system (Applied
Biosystems™; Thermo Fisher Scientific, Inc.). Relative gene
expression was quantified using the ΔΔCq method (2−ΔΔCq)
(27) from biological triplicates,
each with technical triplicates. The SiHa wt control was chosen as
the reference sample. Statistical analysis was conducted using the
unpaired t-test to compare IFITM1 KO samples vs. scrambled ctrl
(significance levels indicated by asterisks) and untreated (CTRL)
vs. IFNγ-treated (IFNγ) samples (indicated by hashtags) separately.
Primer sequences were as follows: IFITM1 forward,
5′-AGCACCATCCTTCCAAGGTCC-3′ and reverse,
5′-TAACAGGATGAATCCAATGGTC-3′; HLA-A forward,
5′-AAGTGGGAGGCGGCCCATGA-3′ and reverse,
5′-ATGTGTCTTGGGGGGGTCCGT-3′; HLA-B forward,
5′-ACTGAGCTTGTGGAGACCAGA-3′ and reverse, 5′-GCAGCCCCTCATGCTGT-3′;
HLA-C forward, 5′-ATCGTTGCTGGCCTGGCTGTCCT-3′ and reverse,
5′-TCATCAGAGCCCTGGGCACTGTT-3′; IGF2R forward,
5′-TTGAGTGGCGAACGCAGTATGC-3′ and reverse,
5′-CAGTGATGGCTTCCCAGTTGTC-3′; M6PR forward,
5′-CTCCAGTCCCTCATCTCACC-3′ and reverse, 5′-AGCCTGCTACACCATTTTGC-3′;
sortilin 1 (SORT1) forward, 5′-AGTTTCAGTGACCCACGTCAG and reverse,
5′-AGTAGGTCAGGTAACAAAGTCCAGT-3′; CD166/ALCAM forward,
5′-GAACACGATGAGGCAGACGA-3′ and reverse,
5′-TTCCATATTACCGAGGTCCTTGT-3′; CD40 forward,
5′-CTGGTCTCACCTCGCTATGG-3′ and reverse, 5′-GCAGTGGGTGGTTCTGGAT-3′;
CD276 forward, 5′-GGAGAATGCAGGAGCTGAGG-3′ and reverse,
5′-GCCAGAGGGTAGGAGCTGTA-3′; and ACTB forward,
5′-GGAACGGTGAAGGTGACAGC-3′ and reverse,
5′-ACCTCCCCTGTGTGGACTTG-3′.
Natural killer (NK) cytotoxicity
assay
The NK-92 cell line purchased from ATCC (cat. no.
CRL-2407) was cultured in Alpha Minimum Essential Medium without
ribonucleosides and deoxyribonucleosides (cat. no. 22561021; Gibco;
Thermo Fisher Scientific, Inc.) supplemented with 0.2 mM inositol
(cat. no. I7508; MilliporeSigma), 0.1 mM 2-mercaptoethanol (cat.
no. 21985023; Gibco; Thermo Fisher Scientific, Inc.), 0.02 mM folic
acid (cat. no. F8758; MilliporeSigma), 200 U/ml recombinant IL-2
(cat. no. PHC0026; Gibco; Thermo Fisher Scientific, Inc.), 25% FBS
and penicillin-streptomycin at 37°C with 5% CO2. The
cells were passaged regularly every 2 days at a concentration of
2–3×105 viable cells/ml. SiHa cells intended for
cocultivation were washed and resuspended in PBS to a concentration
of 1×106 cells/ml. The cells were stained with CFSE
solution (cat. no. 565082; BD Biosciences) at a final concentration
of 2 µM in a 37°C water bath for 10 min. The cells were then washed
thoroughly in PBS and complete RPMI. A total of 6×104
cells per well were seeded in 24-well plates and allowed to grow
overnight. The following day, complete RPMI was added to an NK-92
cell culture at a 1:1 ratio, and NK-92 cells were added at the
indicated ratios to the CFSE-stained SiHa cells. The cell lines
alone without coculture were used as controls. After 4 h of
co-cultivation at 37°C with 5% CO2, SiHa cells were
harvested with Accutase and mixed with collected medium containing
NK-92 and dead SiHa cells. Dead cells stained with 7-AAD (cat. no.
559925; BD Biosciences) for 10 min were analyzed by FACSVerse (BD
Biosciences). The dead SiHa cells were gated as the
CFSE+ 7-AAD+ population (Figs. S1 and S3). Statistical analysis was performed
using the unpaired t-test with data from three biological
replicates.
Statistical analysis
Statistical differences between two groups were
analyzed using an unpaired two-tailed t-test. For proteomic data
analysis, one-way ANOVA with background-based hypothesis testing
(as implemented in Proteome Discoverer 2.2) was applied, followed
by unpaired two-tailed t-tests for specific pairwise comparisons.
Data is presented as the mean ± standard error of the mean (SEM).
Statistical analysis and data visualization were performed using
GraphPad Prism version 8.0.1 (GraphPad Software; Dotmatics).
P<0.05 was considered to indicate a statistically significant
difference.
Results
IFITM1 modulates the cervical cancer
cell surfaceome in both IFNγ-dependent and IFNγ-independent
manners
To investigate the IFITM1-regulated cancer cell
surfaceome, we used the SiHa cervical cancer cell line, which
expresses relatively high levels of IFITM1 protein even without IFN
stimulation. In a previous study, it was demonstrated by the
authors that IFITM1 regulates the surface expression of human
leukocyte antigen B (HLA-B) in this cell line (18). IFITM1 KO SiHa cells were generated
using CRISPR/Cas9 gene editing, and their preparation and
characterization were performed as described previously (18).
To identify and quantify cell surface proteins whose
levels differ between SiHa wt and IFITM1 KO cells, a proteomic
approach was employed, combining SILAC, membrane protein isolation,
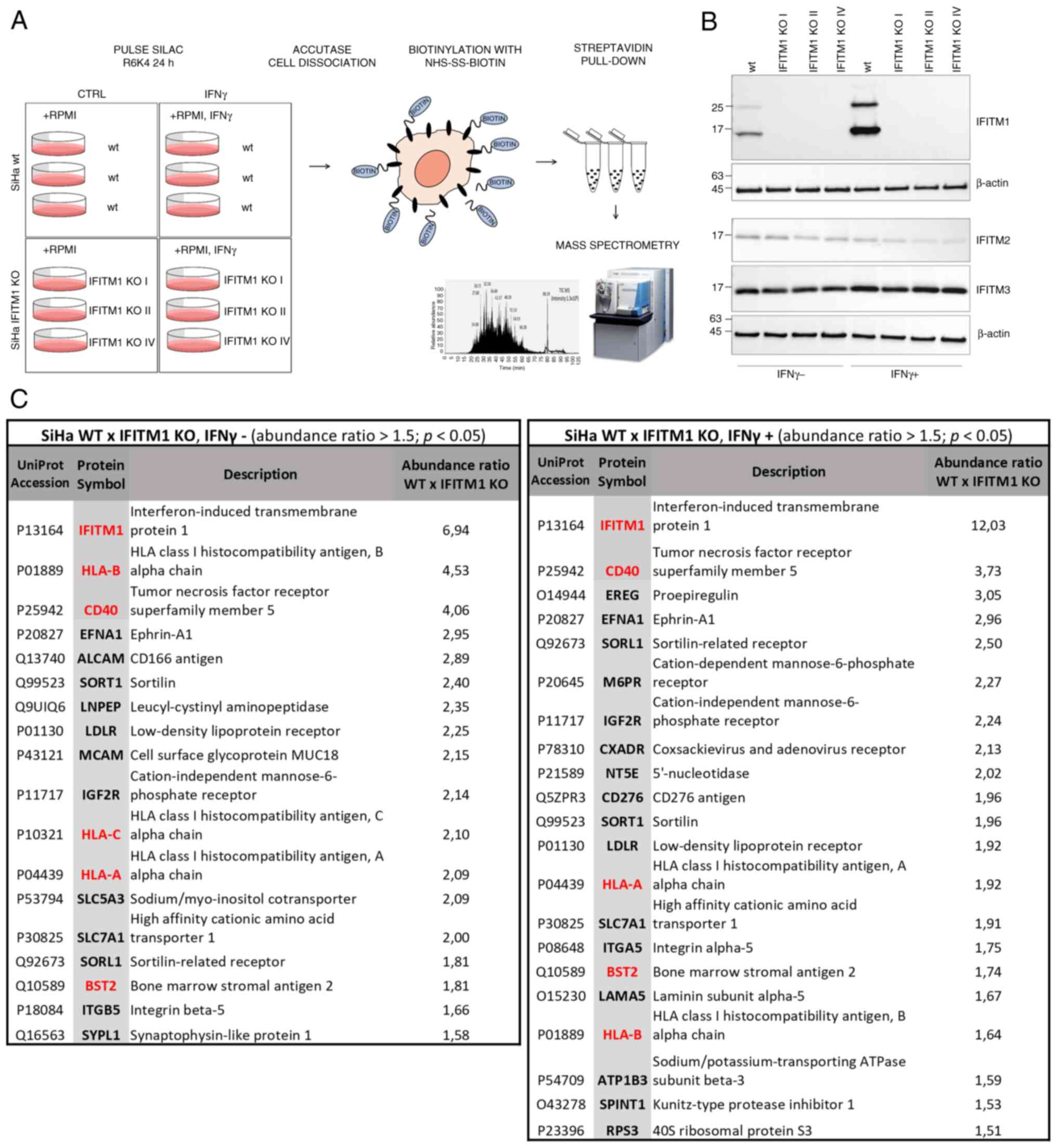
and MS analysis (Fig. 1A). To
capture proteins regulated only after IFN induction of IFITM1,
cells were treated with IFNγ alongside SILAC medium, allowing
protein turnover analysis. To minimize clonal selection bias, three
independently generated IFITM1 KO cell clones were used as
biological replicates (Fig. 1B).
Western blot analysis of IFITM1 levels revealed an additional
higher-molecular-weight band (~25 kDa) alongside the expected
15-kDa band. This band likely represents an IFITM1 dimer or a
post-translationally modified form of IFITM1, as previously
reported (6,28). In addition to IFITM1, the expression
of the related proteins IFITM2 and IFITM3 was examined, as they are
often affected in KO studies due to their high sequence similarity.
Western blot analysis confirmed that IFITM2 and IFITM3 expression
was preserved in IFITM1 KO clones. However, a slight decrease in
the protein levels is evident, particularly in the presence of
IFNγ. This may be a consequence of the tight co-regulation within
the IFITM family (6).
A set of proteins that were significantly
downregulated in IFITM1 KO cells with a fold change ≥1.5 and a
significance level of P<0.05 were identified and quantified: 18
proteins without IFNγ treatment and 21 proteins after IFNγ
treatment (Fig. 1C, Appendix S1). As expected, IFITM1 was the
protein with the greatest difference in expression and was also
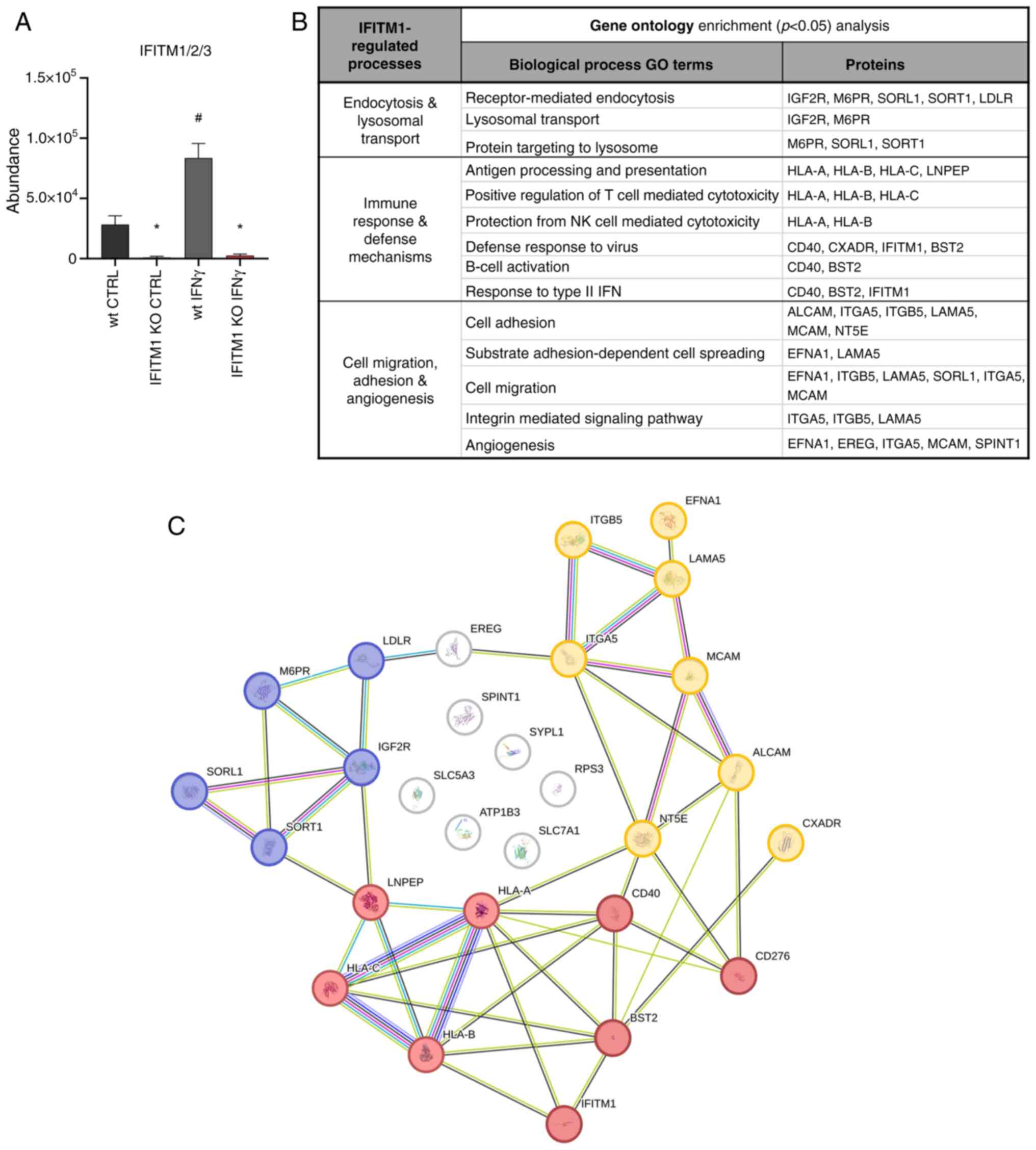
induced by IFNγ (Fig. 2A). GO
analysis using the DAVID bioinformatics tool (20) revealed that IFITM1-regulated
proteins are involved primarily in receptor-mediated endocytosis,
lysosomal transport of proteins, antiviral responses, antigen
processing and presentation, regulation of immune system
activity/function, and cell adhesion and migration (Fig. 2B). Using the STRING database of
known and predicted protein-protein interactions (21), three major groups of interacting
proteins that were significantly downregulated in IFITM1 KO cells
were identified (Fig. 2C).
IFITM1 protein regulates transcript
abundance and modulates protein dynamics
The first group of proteins consists of proteins
essential for antigen presentation and the immune response
(Fig. 2C, red). IFITM1 KO cells
presented reduced surface expression of all three major HLA class I
molecules [HLA-A, HLA-B and HLA-C (Fig.
3A)] along with decreased levels of antigen-trimming leucyl and
cystinyl aminopeptidase (LNPEP; Fig.
S2). Although IFNγ treatment increased HLA abundance in both wt
and IFITM1 KO cells, the levels remained significantly lower in
IFITM1 KO cells. Gene expression analysis confirmed the increased
transcriptional activity of HLA genes after IFNγ treatment
in both cell lines, with no significant differences between them.
These findings suggested that IFITM1 regulates HLA molecules at the
post-translational level (Fig. 3B),
which is consistent with the authors' previous findings concerning
HLA-B in IFITM1/3 double-KO SiHa cells (29).
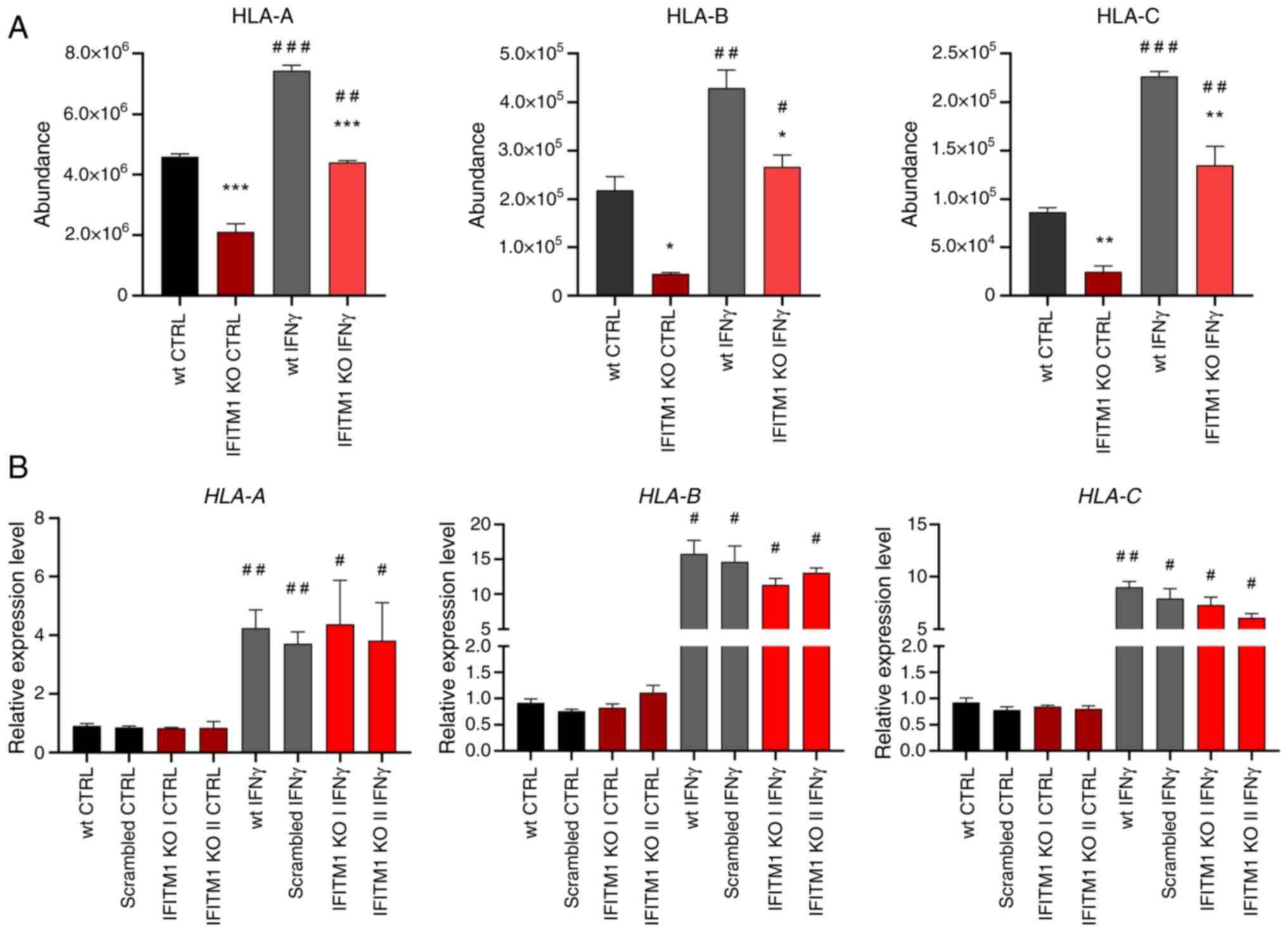 | Figure 3.Membrane protein levels and
associated gene expression of HLA class I molecules. (A) Peptide
abundances determined by mass spectrometry analysis as a measure of
HLA protein levels. (B) Determination of HLA gene expression
by reverse transcription-quantitative PCR. ACTB was used as
an endogenous control. The level of gene expression is relative to
the value of the wt control (reference sample). The error bars
represent the mean ± SEM from biological replicates. *P<0.05,
**P<0.01, and ***P<0.001 when comparing wt and IFITM1 KO;
#P<0.05, ##P<0.01, and
###P<0.001 when comparing untreated CTRL and
respective IFNγ-treated samples. HLA, human leukocyte antigen; wt,
wild-type; KO, knockout; CTRL, control; IFN, interferon; IFITM,
interferon-induced transmembrane protein. |
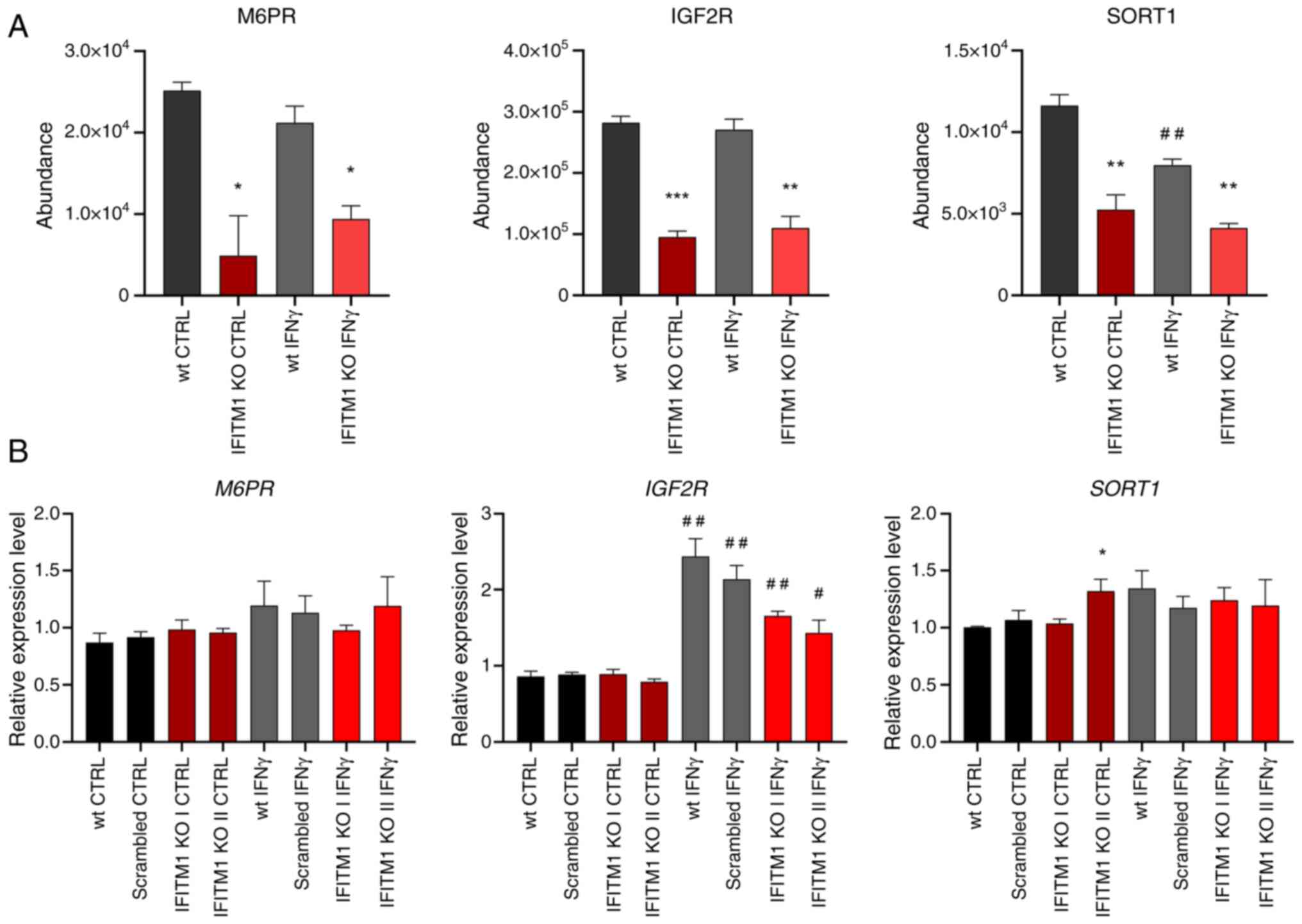
The second group of proteins contains proteins
pertaining to endocytosis, lysosomal transport, and lysosome
function (Fig. 2C, blue), including
mannose 6-phosphate (M6P) receptors (IGF2R and M6PR) and other
M6P-independent sorting receptors [SORT1, sortilin related receptor
1 (SORL1) and LDLR] that are critical for protein trafficking from
the Golgi apparatus to lysosomes and endosomes. IFNγ did not affect
the plasma membrane levels of these proteins (Figs. 4A and S2). Interestingly, SORT1 levels were
reduced upon IFNγ stimulation (Fig.
4A). Gene expression analysis of M6PR and IGF2R
revealed no significant differences between wt and IFITM1 KO cells
(Fig. 4B). Although surfaceome
analysis revealed stable IGF2R membrane levels after IFNγ
treatment, IGF2R gene expression was induced, which is
consistent with reports in microglia (30). Conversely, SORT1 gene
expression remained unchanged under IFNγ treatment (Fig. 4B), highlighting cell-specific
regulatory differences (31).
Variability among IFITM1 KO clones was observed, with one clone
displaying higher SORT1 expression, which did not persist in
the IFN-treated samples and was not reflected in the surfaceome
analysis.
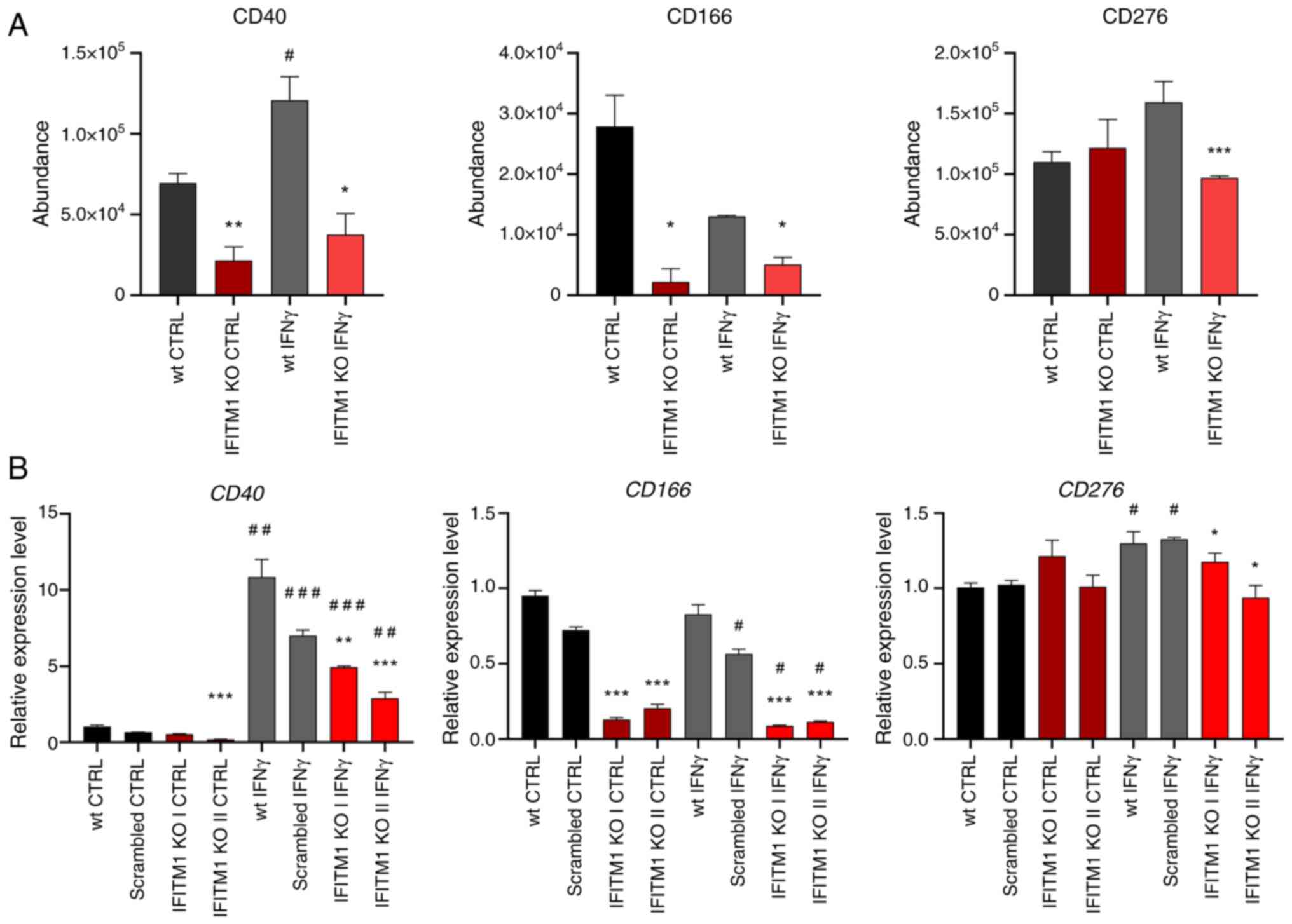
The third group consists of cell adhesion molecules
(CAMs) that mediate cell-cell and cell-extracellular matrix
interactions (Fig. 2C, yellow).
These multifunctional molecules often act as cell surface
receptors, are overexpressed in cancer, and represent potential
immunotherapy targets [for example, ALCAM-CD166; melanoma CAM
(MCAM)-CD146/Mucin 18, CXADR] (32–34).
Additionally, CD166, CD146 and CD40 play crucial roles in
activating immune responses in tumor tissues. IFITM1-regulated CAMs
include IFNγ-inducible (CD40, CD276) and non-IFNγ-inducible (CD166,
MCAM, EFNA1, CXADR) proteins (Figs.
1C, 5A and S2). Changes in CD40 and CD166 membrane
levels were found to be associated with their respective expression
data (Fig. 5B), confirming that
IFITM1 plays a significant role in regulating genes associated with
tumor progression. Interestingly, IFITM1 depletion alone does not
affect the expression or surface localization of CD276, an immune
checkpoint molecule. However, upon IFNγ treatment, CD276 expression
was upregulated at the RNA level in SiHa wt cells but not in IFITM1
KO cells. A similar trend was observed at the protein surface
level; however, the induction of CD276 in SiHa wt cells did not
reach statistical significance (P=0.06). These findings suggested
that IFITM1 plays an important role in the regulation of CD276 by
IFNγ and provide further evidence that IFITM1 acts as a master
regulator not only for IRDS proteins but also for cell surface
receptors.
Functional analyses confirm the
involvement of IFITM1 and IFITM3 in the mechanisms driving cancer
progression
Following the analysis of the data obtained from MS,
the effects of the IFITM1 protein were further validated by
performing functional assays focused on cancer cell adhesion,
migration, invasion and sphere formation-processes driven by
changes in surfaceome composition. To verify the impact of the
related IFITM3 protein and its possible relationship with IFITM1,
independent IFITM3 KO and double-knockout IFITM1+3 KO SiHa
derivatives, whose preparation has been previously described, were
also tested (18).
As expected, IFITM1 KO cells migrated more slowly
compared with SiHa wt cells, as shown by the results of the wound
healing (scratch) and Transwell migration assays, which were
evaluated after 24 h of incubation (Fig. 6A and B). Interestingly, IFITM3 KO
cells exhibited significantly reduced migration potential in the
scratch assay, although the Transwell assay did not confirm this
phenotype. When double-knockout IFITM1+3 KO cells were evaluated, a
phenotype with low migratory activity was observed, similar to that
of the IFITM1 KO clones.
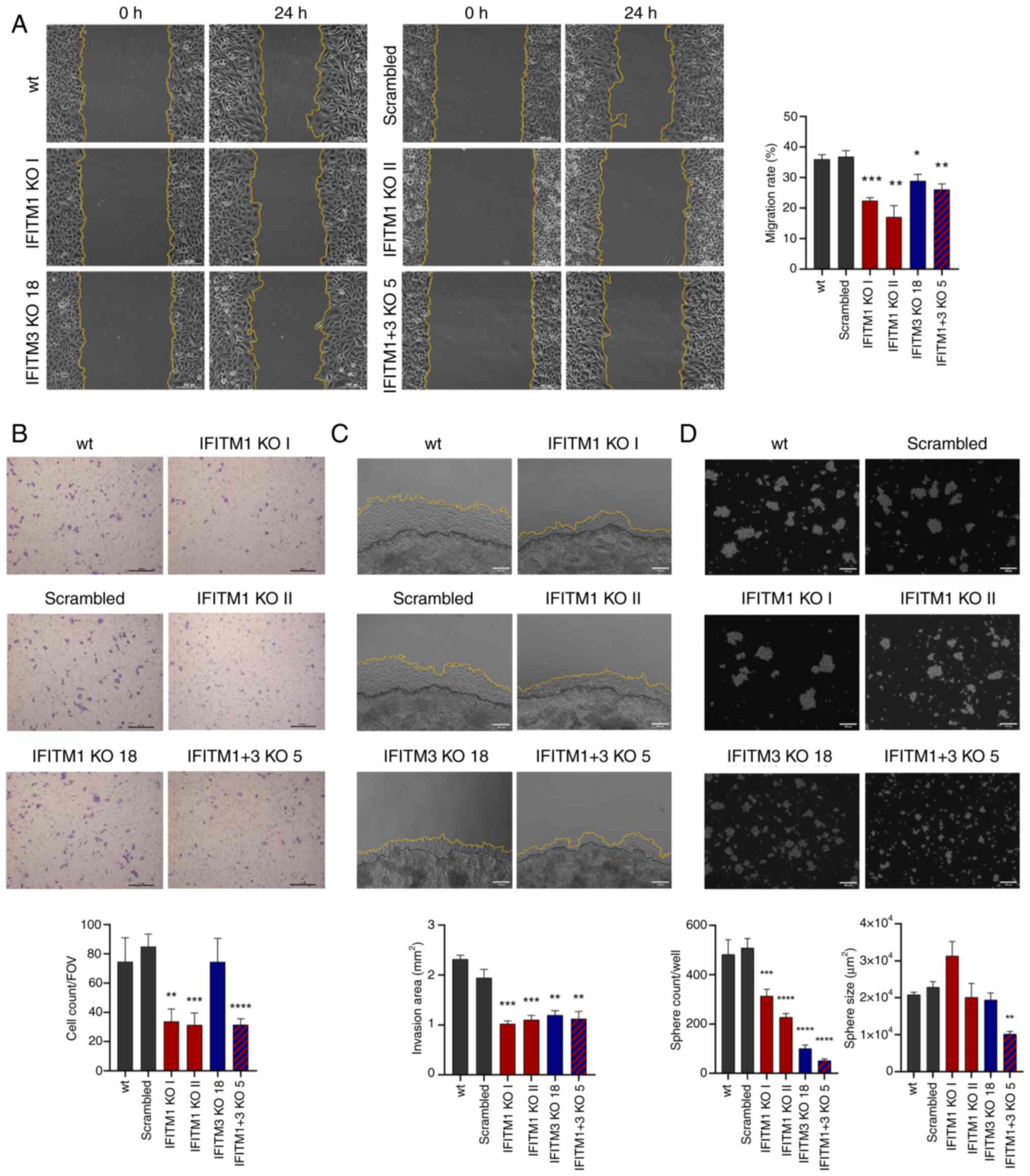 | Figure 6.Evaluation of SiHa IFITM KO cell
adhesion, migration and invasion abilities. (A) Representative
images (magnification, ×100) of the wound healing (scratch) assay,
with the borders of the scratch highlighted with yellow lines. The
scratch area was determined at 0 and 24 h using Fiji ImageJ, and
the values were used to calculate the migration rate (%). The
average migration rates of IFITM1 KO, IFITM3 KO and IFITM1+3 KO
cells in comparison with those of control cells (wt, scrambled) are
shown in the graph. (B) Representative images of migratory cells
after 24 h of Transwell assay (scale bar, 200 µm). The graph shows
the average number of cells per field of view (FOV) for individual
IFITM KO clones and their controls. (C) Representative images of
cells invading (yellow line) from 3D Matrigel drops (black line)
taken after 6 days (scale bar, 400 µm). The invasion area
(mm2) was determined by Fiji ImageJ, and the average
values for individual IFITM KO clones and controls are shown in the
graph. (D) Representative images of 10-day cervicopheres formed by
IFITM KO and control cells (scale bar, 400 µm). The average numbers
of all spheres formed per well are compared in the left graph. The
graph on the right shows the average sphere size (µm2)
that was determined using Fiji ImageJ. The error bars represent the
means from biological replicates ± SEM. *P<0.05, **P<0.01,
***P<0.001 and ****P<0.0001. IFITM, interferon-induced
transmembrane protein; KO, knockout; wt, wild-type. |
To assess invasive ability, a 3D Matrigel drop assay
was performed, which quantifies invading cells embedded in a
Matrigel drop (26). This method
confirmed a reduced capacity to invade through the Matrigel matrix
in all tested IFITM KO clones, on the basis of the extent of the
invasion area evaluated 6 days after Matrigel drop formation
(Fig. 6C).
To evaluate the role of IFITM proteins in cell-cell
adhesion and anchorage-independent growth, the ability of wt and
IFITM1 KO and/or IFITM3 KO cells to form cervicospheres was
compared. All the tested IFITM KO clones exhibited a significantly
reduced capacity to grow as spheres, suggesting that the IFITM1 and
IFITM3 proteins influence intercellular interactions and
cell-extracellular matrix adhesion (Fig. 6D). Notably, compared with IFITM1 KO
cells, IFITM3 KO cells displayed a greater reduction in sphere
formation. However, the most pronounced reduction in both the
number and size of spheres was observed in IFITM1+3 KO cells.
IFITM1 and IFITM3 KO cells show
reduced expression of immunity-related CD markers and increased
resistance to NK cell killing
Given the identified group of IFITM1-regulated
proteins involved in immune responses, two selected proteins were
further validated, CD40 and CD166, which are critical factors in
tumor-immune cell interactions.
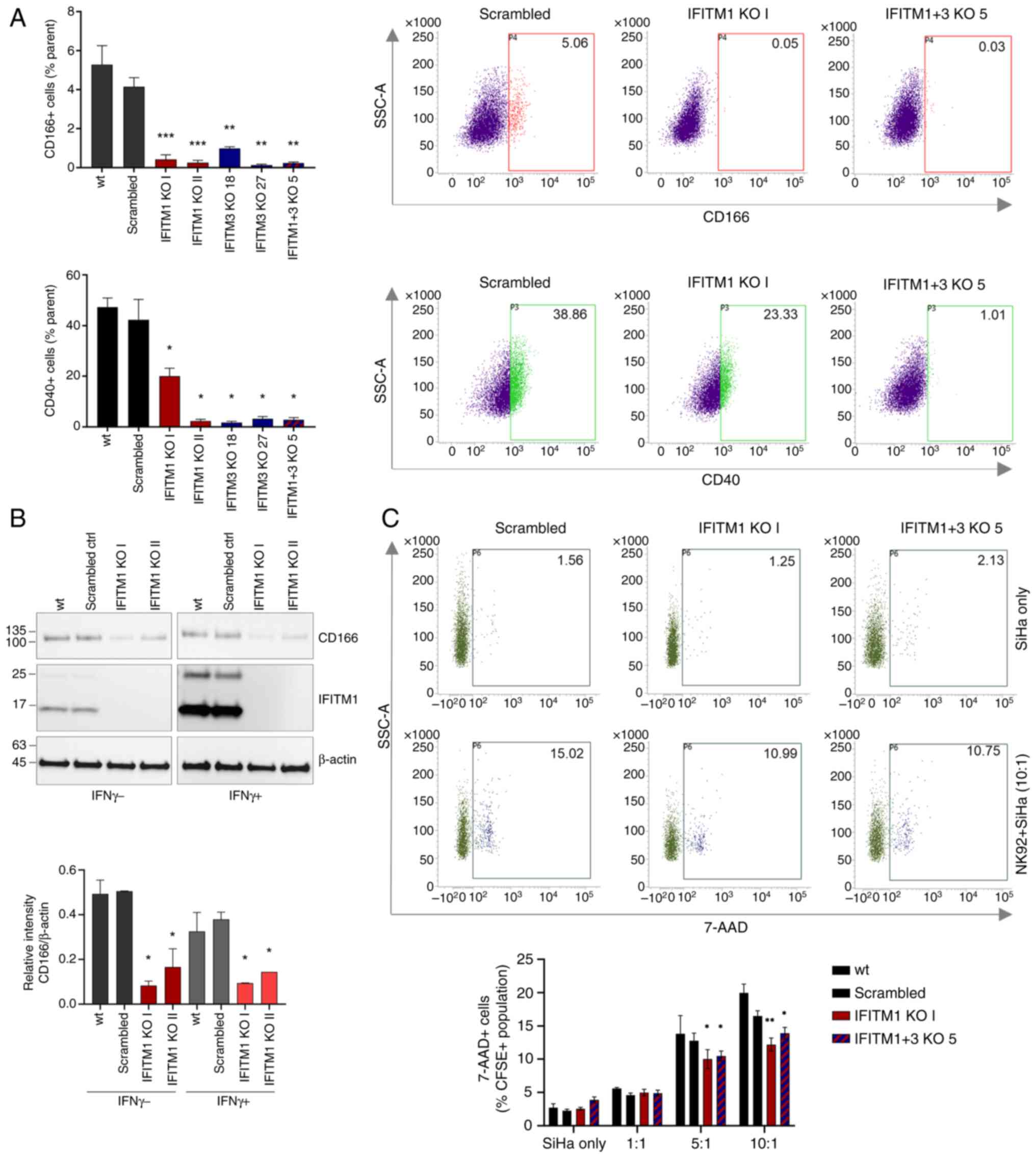
FACS analysis confirmed reduced CD40 and CD166
levels in all the tested IFITM KO cell clones (Figs. 7A and S3A). Immunoblotting also revealed a
decreased CD166 expression at the total protein level (Fig. 7B). Consistent with the MS and qPCR
findings, immunoblotting of SiHa cell lysates revealed that CD166
expression was not responsive to IFNγ stimulation (Fig. 7B).
Using the NK cytotoxicity assay, which involves the
co-cultivation of tumor and NK cells followed by an analysis of
tumor cell viability, the influence of IFITM proteins on
intercellular communication and the ability of NK cells to
recognize and eliminate tumor cells were examined. This assay
demonstrated that both IFITM1 KO and IFITM1+3 KO cells are
significantly more resistant to NK cell-mediated killing, as
evidenced by a lower percentage of dead (7-AAD+) tumor
cells after 4 h of co-cultivation (Figs. 7C and S3B).
Discussion
Despite advancements in prevention, detection and
treatment, cervical cancer remains a leading cause of
cancer-related mortality in women. Treatment is stage-dependent
(TNM, International Federation of Gynecology and Obstetrics
classification), with advanced cases requiring a combination of
surgery, radiotherapy, chemotherapy, or targeted therapy (2). Cisplatin remains the cornerstone of
chemotherapy, used alone or in combination, including as
neoadjuvant therapy to reduce tumor mass before surgery (35). Significant progress has been made in
targeted therapies, with bevacizumab (anti-vascular endothelial
growth factor) and pembrolizumab (anti-PD-1 immunotherapy) approved
for recurrent and metastatic cervical cancer (36,37).
Ongoing clinical trials are investigating additional approaches,
such as poly (ADP-ribose) polymerase inhibitors, mammalian target
of rapamycin inhibitors and therapeutic HPV vaccines (38).
Despite these advances, the prognosis for
advanced-stage disease remains poor due to tumor heterogeneity and
therapy resistance. The shift toward precision medicine underscores
the need for prognostic and predictive biomarkers to improve
patient stratification and treatment outcomes (3,4). In
this context, surfaceome analysis represents a powerful tool for
identifying therapeutic targets by profiling cell surface
proteins.
IFITM1 was originally identified as a leukocyte
membrane surface antigen and is now known primarily for its
significant antiviral activity (39). Over the past decade, increasing
evidence has shown that IFITM1 is overexpressed in numerous human
solid tumors (6,40). Aberrant expression of IFITM1
promotes tumor cell proliferation, inhibits cell death, stimulates
invasion and metastasis, contributes to cancer cell resistance to
endocrine therapy, chemotherapy and radiotherapy, and has
prognostic value for patient outcomes (6,11,40).
IFITM1 is also part of a subset of IFN-responsive genes that
contribute to the IRDS signature. The authors have demonstrated
that the suppression of IFITM1 via siRNA targeting can impair the
IRDS network (18,41). Current knowledge indicates that
IFITM proteins affect membrane structure and function,
intracellular trafficking, and the turnover of cell surface
receptors (15). IFITM1 has also
been reported to be a tight junction protein (42,43).
It was previously demonstrated that knocking out the IFITM1 and
IFITM3 proteins led to the downregulation of HLA-B expression on
the surface of cervical cancer cells, suggesting an explanation for
the inverse correlation between IFITM1/3 expression and metastasis
formation (18). In the present
study, it was confirmed that, in addition to HLA-B, IFITM1
regulates a whole set of surface proteins in cervical cancer cells,
both with and without dependence on IFNγ. Furthermore, IFITM1
knockout affects the regulation of specific protein surface
expression, either at the level of membrane dynamics or gene
transcription.
Most published studies focus on either one IFITM
family member or the combined effect of several IFITMs without
distinguishing individual members. This is because of the high
homology between IFITM proteins (6), which makes specific targeting
difficult. However, recent studies have overcome these obstacles
and are now describing the different functions of IFITM proteins
(44–47). This phenomenon is particularly
intriguing among such homologous proteins. Therefore, in addition
to focusing on IFITM1, validation and functional assays were
enriched with IFITM3 KO cells, as well as ‘double KO’ cells
depleted of both the IFITM1 and IFITM3 proteins, by the CRISPR/Cas9
gene-editing method to reveal their synergistic effects.
SILAC-based quantitative MS is a widely used
proteomic approach that enables the quantification and evaluation
of protein dynamics within living cells. An additional step
involving surface-protein biotinylation allowed us to enrich
samples with surface/membrane proteins, whose detection in total
lysates is challenging owing to their hydrophobic nature and the
abundance of intracellular proteins. Given the location of IFITM1
in the plasma membrane and its known role in altering membrane
properties during antiviral responses, the surfaceomes of IFITM1 KO
cells and parental SiHa wt cells were measured with or without IFNγ
treatment. The SILAC approach allowed us to evaluate the changes in
protein levels induced by the treatment and compare the membrane
protein levels between IFITM1 KO and SiHa wt cells, including
effects that were dependent on or independent of the presence of
IFNγ.
A total of 28 proteins that were differentially
expressed in wt and IFITM1 KO cells with a fold change of ≥1.5.
Among these proteins, 9 were significantly stimulated by IFNγ. The
present results suggested that IFITM1 affects surfaceome
composition even without IFNγ stimulation, at least in SiHa cell
line, which has a relatively high basal level of IFITM1.
Furthermore, the changes upon IFNγ stimulation indicate the
possible involvement of IFITM1 in the IFN-dependent regulation of
differentially expressed proteins or possible changes in the
functional properties of IFITM1 itself in the presence of IFN. A
detailed understanding of how IFITM1 contributes to IFN signaling
and the subsequent expression of IFN-inducible proteins deserves
further study.
As expected, the protein with the greatest
difference in expression between wt and IFITM1 KO cells was IFITM1.
Only one IFITM peptide was detected by MS, and this peptide is
common to the IFITM1, IFITM2 and IFITM3 proteins, thus
distinguishing between them was not possible. Western blot analysis
confirmed that IFITM1 expression was specifically ablated in IFITM1
KO cells, while IFITM2 and IFITM3 expression was preserved, albeit
with a slight decrease, particularly in the presence of IFNγ. This
reduction may result from the tight co-regulation of these proteins
due to shared regulatory elements, the stabilizing effects of
hetero-oligomer formation within cellular membranes, and
alterations in IFN response dynamics caused by IFITM1 KO, which may
further influence the expression of IFN-stimulated genes, including
IFITMs, through feedback regulation (6,18).
Together with the predominant localization of IFITM1 on the cell
surface (in contrast to IFITM2 and IFITM3, which are mostly located
in endosomes/lysosomes), it was hypothesized that the identified
peptide originated from IFITM1, with a minor contribution from
IFITM2/3, which can be temporarily found on the cell surface before
their internalization into endosomes (48). This also explains the presence of
small amounts of the IFITM peptide in the IFITM1 KO samples.
Despite the advantages of quantitative proteomics,
limitations in peptide quantification and reproducibility may lead
to the exclusion of relevant targets. Nonetheless, this approach
enabled us to identify entire functional protein groups, which led
to the discovery of IFITM1-regulated processes central to our
study. The proteins, whose levels in the plasma membrane were
downregulated by IFITM1 knockout, can be classified into three main
groups according to their function: Endocytosis and lysosomal
transport, antigen processing and presentation together with
regulation of the immune response, and cell adhesion and migration.
The regulation of the rate and manner of endocytosis controls the
exposure and abundance of membrane proteins at the cell surface.
Once internalized, proteins undergo recycling or degradation in
lysosomes through autophagy. IFITM proteins are known to modulate
membrane rigidity and curvature, cholesterol content, trafficking
of endocytosed cargo, and endosomal/lysosomal pH, thereby affecting
viral entry into cells (12,13,15–17,49).
According to our MS data, IFITM1 affects several proteins linked to
receptor-mediated endocytosis and endosomal/lysosomal trafficking
(LDLR, IGF2R, M6PR, SORT1 and SORL1). On the basis of the unchanged
gene expression levels of selected representatives of this
functional group (IGF2R, M6PR and SORT1) in IFITM1 KO cells, it was
hypothesized that, along with effects on the biophysical properties
of membranes, IFITM1 affects the exposure and turnover of numerous
membrane proteins, which may be another explanation for its broad
antiviral properties and underscores its significant role in tumor
progression. A more detailed description of how IFITM proteins
regulate the stability and localization of other proteins is the
subject of our further investigation.
The dysregulation of cell migration, adhesion and
invasion regulators, such as MCAM/CD146, ALCAM/CD166, CAR/CXADR,
EFNA1, LAMA5, CD73/NT5E and integrins, contributes to cancer
progression and metastasis (50–55).
The importance of IFITM1 in these processes is supported by
functional assays showing a decreased ability of IFITM1 KO cells to
migrate (toward the scratch or across the porous membrane) and
invade through Matrigel, which represents the extracellular matrix.
These results are consistent with findings in various tumor types
reported in the literature (56–58).
However, little is known about IFITM proteins in cervical cancer.
In contrast, Zheng et al (59) reported that migration was inhibited
after the transfection of HeLa cells with an IFITM1 recombinant
construct. While IFITM1 KO clones showed consistent results across
migration assays, the IFITM3 KO 18 clone exhibited a discrepancy.
IFITM3 KO cells migrated similarly to controls in the Transwell
assay, but their migratory ability was significantly reduced in the
scratch assay. This may be due to the distinct principles of each
assay: the Transwell assay measures individual cell migration,
where cytoskeletal components are crucial, while the scratch assay
focuses on collective migration, which relies on cell-cell adhesion
molecules. Both IFITM1 and IFITM3 are associated with membrane
structure and dynamics, which are closely linked to cytoskeletal
remodeling. However, their functions may differ due to differences
in their localization and expression patterns (6).
To evaluate adhesion properties, the cells were
allowed to proliferate in low-attachment plates, forcing them to
adhere to each other and form cervicospheres. IFITM1 and IFITM3 KO
clones showed a diminished ability to form spheres, as evaluated on
the basis of sphere counts. Compared with single KO, the knockout
of both IFITM1 and IFITM3 had a synergistic effect. Interestingly,
when evaluating the size of the formed spheres, clones deficient in
one of the IFITM proteins did not differ from the controls, whereas
deficiency in both proteins led to significantly smaller cell
aggregates. Sphere formation assays are typically used to evaluate
stem cell properties. However, changes in stem cell markers among
IFITM KO clones (data not shown) were not observed, indicating that
the observed differences are due to alterations in adhesion
proteins.
Surfaceome analysis confirmed our previous findings
that IFITM proteins affect the exposure of MHC-I molecules at the
surface of cervical cancer cells (18), which might be important for cancer
cell interactions with the immune system, supporting tumor immune
escape (60). These findings were
recently confirmed by She et al (61), who described the effect of IFITM1
expression on the brain colonization of human lung cancer cells
through the regulation of the membrane localization of MHC-I and
the enhancement of CD8+ T-cell cytolytic activity.
Interestingly, it was also shown that IFITM1 deficiency does not
affect the expression level of HLA genes, suggesting that
IFITM1 instead affects the dynamics of proteins within the
membrane. This finding is consistent with our previous results on
the HLA-B gene (29).
Additionally, IFN-independent LNPEP aminopeptidase, which is
important for antigen trimming before MHC-I-mediated presentation
(62), was identified among the
downregulated molecules in IFITM1 KO cells. Other proteins
regulated by IFITM1 that could be involved in antitumor immune
responses include CD40 (63–65),
CD166 (32,66), MCAM/CD146 (67,68)
and CXADR (52). Interestingly,
differences were observed in the involvement of the IFNγ signaling
pathway in the regulation of CD markers. The membrane levels of the
IFN-inducible CD40 and CD276 proteins correlated with the gene
expression levels. IFITM1 knockout leads to CD40 downregulation
irrespective of the presence of IFNγ and abolishes IFN-induced
CD276 expression, which is also reflected in the membrane exposure
of the protein. Furthermore, IFITM1 decreases the expression of
CD166, which is unresponsive to IFNγ treatment, as confirmed by
SILAC, qPCR and western blot analysis. The surface levels of the
CD40 and CD166 proteins were further validated and significant
downregulation of the IFITM1 and IFITM3 KO SiHa derivatives was
confirmed. Consistent with the differential levels of
immune-related proteins, IFITM1 KO and IFITM1+3 KO clones escaped
the effects of NK cells, as demonstrated by the NK cytotoxicity
assay. Other studies that link IFITM proteins with innate and
adaptive immunity are emerging, highlighting the potential role of
these proteins in tumor immunosurveillance (69–71).
Uncovering the exact mechanism by which IFITMs regulate these
proteins and thereby hinder antitumor immunity will be the subject
of our further investigations.
In conclusion, the importance of the IFITM1 and
IFITM3 proteins in processes that depend on the composition of
proteins in the plasma membrane, such as cell adhesion, migration,
invasion and interaction with immune cells was confirmed. The
presented results confirm the authors' hypothesis that IFITM
proteins regulate the expression and membrane exposure of proteins,
thereby facilitating tumor progression while also supporting tumor
immunosurveillance. The precise mechanism by which IFITM1 modulates
the membrane presentation of identified proteins remains the
subject of ongoing research.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Czech Science Foundation
(grant no. 23-06884S), the project SALVAGE (P JAC; grant no.
CZ.02.01.01/00/22_008/0004644) funded by the European Union and by
the State Budget of the Czech Republic, and MH CZ-DRO (MMCI; grant
no. 00209805).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author. The data generated in the
present study may be found in the PRIDE database under accession
number PXD053951 or at the following URL: https://www.ebi.ac.uk/pride/archive/projects/PXD053951.
Authors' contributions
NF and MN designed the present study, performed the
experiments and data analysis, and drafted the manuscript. LB
designed the study and conducted the experiments. LD and LU
performed proteomic measurements. LH designed the proteomic
methodology and data analysis. TRH and BV provided key revisions of
the manuscript and conducted data interpretation. NF and MN confirm
the authenticity of all the raw data. All authors read and approved
the final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
7-AAD
|
7-aminoactinomycin D
|
|
ALCAM
|
activated leukocyte cell adhesion
molecule
|
|
CAM
|
cell adhesion molecules
|
|
CFSE
|
carboxy-fluorescein succinimidyl
ester
|
|
CXADR
|
coxsackie virus and adenovirus
receptor
|
|
ECL
|
enhanced chemiluminescence
|
|
EFNA1
|
ephrin A1
|
|
FASP
|
filter-aided sample preparation
protocol
|
|
FDR
|
false discovery rate
|
|
GO
|
Gene Ontology
|
|
HLA
|
human leukocyte antigen
|
|
IFITM
|
interferon-induced transmembrane
protein
|
|
IFN
|
interferon
|
|
IGF2R
|
insulin-like growth factor 2
receptor
|
|
IRDS
|
IFN-related DNA damage resistance
signature
|
|
KO
|
knockout
|
|
LDLR
|
low-density lipoprotein receptor
|
|
LNPEP
|
leucyl and cystinyl
aminopeptidase
|
|
M6P
|
mannose-6-phosphate
|
|
M6PR
|
M6P receptor
|
|
MCAM
|
melanoma CAM
|
|
MHC-I
|
major histocompatibility complex
class I
|
|
PBST
|
phosphate-buffered saline with Tween
20
|
|
RSLC
|
rapid separation liquid
chromatography
|
|
SILAC
|
stable isotope labeling with amino
acids in cell culture
|
|
SORL1
|
sortilin related receptor 1
|
|
SORT1
|
sortilin 1
|
References
|
1
|
Bray F, Laversanne M, Sung H, Ferlay J,
Siegel RL, Soerjomataram I and Jemal A: Global cancer statistics
2022: GLOBOCAN estimates of incidence and mortality worldwide for
36 cancers in 185 countries. CA Cancer J Clin. 74:229–263. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
D'Augè TG, Donato VD and Giannini A:
Strategic approaches in management of early-stage cervical cancer:
A comprehensive editorial. Clin Exp Obstet Gynecol. 51:2352024.
View Article : Google Scholar
|
|
3
|
Garg P, Krishna M, Subbalakshmi AR,
Ramisetty S, Mohanty A, Kulkarni P, Horne D, Salgia R and Singhal
SS: Emerging biomarkers and molecular targets for precision
medicine in cervical cancer. Biochim Biophys Acta Rev Cancer.
1879:1891062024. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
D'Oria O, Bogani G, Cuccu I, D'Auge TG, Di
Donato V, Caserta D and Giannini A: Pharmacotherapy for the
treatment of recurrent cervical cancer: An update of the
literature. Expert Opin Pharmacother. 25:55–65. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bausch-Fluck D, Goldmann U, Müller S, van
Oostrum M, Müller M, Schubert OT and Wollscheid B: The in silico
human surfaceome. Proc Natl Acad Sci USA. 115:E10988–E10997. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Friedlová N, Zavadil Kokáš F, Hupp TR,
Vojtěšek B and Nekulová M: IFITM protein regulation and functions:
Far beyond the fight against viruses. Front Immunol.
13:10423682022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Narayana SK, Helbig KJ, McCartney EM, Eyre
NS, Bull RA, Eltahla A, Lloyd AR and Beard MR: The
interferon-induced transmembrane proteins, IFITM1, IFITM2, and
IFITM3 inhibit hepatitis C virus entry. J Biol Chem.
290:25946–25959. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Prelli Bozzo C, Nchioua R, Volcic M,
Koepke L, Krüger J, Schütz D, Heller S, Stürzel CM, Kmiec D,
Conzelmann C, et al: IFITM proteins promote SARS-CoV-2 infection
and are targets for virus inhibition in vitro. Nat Commun.
12:45842021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ren L, Du S, Xu W, Li T, Wu S, Jin N and
Li C: Current progress on host antiviral factor IFITMs. Front
Immunol. 11:5434442020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liao Y, Goraya MU, Yuan X, Zhang B, Chiu
SH and Chen JL: Functional involvement of interferon-inducible
trans-membrane proteins in antiviral immunity. Front Microbiol.
10:10972019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gómez-Herranz M, Taylor J and Sloan RD:
IFITM proteins: Understanding their diverse roles in viral
infection, cancer, and immunity. J Biol Chem. 299:1027412023.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li K, Markosyan RM, Zheng YM, Golfetto O,
Bungart B, Li M, Ding S, He Y, Liang C, Lee JC, et al: IFITM
proteins restrict viral membrane hemifusion. PLoS Pathog.
9:e10031242013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Amini-Bavil-Olyaee S, Choi YJ, Lee JH, Shi
M, Huang IC, Farzan M and Jung JU: The antiviral effector IFITM3
disrupts intracellular cholesterol homeostasis to block viral
entry. Cell Host Microbe. 13:452–464. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rahman K, Datta SAK, Beaven AH, Jolley AA,
Sodt AJ and Compton AA: Cholesterol binds the amphipathic helix of
IFITM3 and regulates antiviral activity. J Mol Biol.
434:1677592022. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Spence JS, He R, Hoffmann HH, Das T,
Thinon E, Rice CM, Peng T, Chandran K and Hang HC: IFITM3 directly
engages and shuttles incoming virus particles to lysosomes. Nat
Chem Biol. 15:259–268. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gerlach T, Hensen L, Matrosovich T,
Bergmann J, Winkler M, Peteranderl C, Klenk HD, Weber F, Herold S,
Pöhlmann S and Matrosovich M: pH optimum of hemagglutinin-mediated
membrane fusion determines sensitivity of influenza A viruses to
the interferon-induced antiviral state and IFITMs. J Virol.
91:e00246–17. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wee YS, Roundy KM, Weis JJ and Weis JH:
Interferon-inducible transmembrane proteins of the innate immune
response act as membrane organizers by influencing clathrin and
v-ATPase localization and function. Innate Immun. 18:834–845. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gómez-Herranz M, Nekulova M, Faktor J,
Hernychova L, Kote S, Sinclair EH, Nenutil R, Vojtesek B, Ball KL
and Hupp TR: The effects of IFITM1 and IFITM3 gene deletion on IFNγ
stimulated protein synthesis. Cell Signal. 60:39–56. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wiśniewski JR, Zougman A, Nagaraj N and
Mann M: Universal sample preparation method for proteome analysis.
Nat Methods. 6:359–362. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sherman BT, Hao M, Qiu J, Jiao X, Baseler
MW, Lane HC, Imamichi T and Chang W: DAVID: A web server for
functional enrichment analysis and functional annotation of gene
lists (2021 update). Nucleic Acids Res. 50:W216–W221. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Szklarczyk D, Gable AL, Nastou KC, Lyon D,
Kirsch R, Pyysalo S, Doncheva NT, Legeay M, Fang T, Bork P, et al:
The STRING database in 2021: Customizable protein-protein networks,
and functional characterization of user-uploaded gene/measurement
sets. Nucleic Acids Res. 49:D605–D612. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Perez-Riverol Y, Csordas A, Bai J,
Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J,
Mayer G, Eisenacher M, et al: The PRIDE database and related tools
and resources in 2019: Improving support for quantification data.
Nucleic Acids Res. 47:D442–D450. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Schindelin J, Arganda-Carreras I, Frise E,
Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S,
Schmid B, et al: Fiji: An open-source platform for biological-image
analysis. Nat Methods. 9:676–682. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Grada A, Otero-Vinas M, Prieto-Castrillo
F, Obagi Z and Falanga V: Research techniques made simple: Analysis
of collective cell migration using the wound healing assay. J
Invest Dermatol. 137:e11–e16. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Suarez-Arnedo A, Figueroa FT, Clavijo C,
Arbeláez P, Cruz JC and Muñoz-Camargo C: An image J plugin for the
high throughput image analysis of in vitro scratch wound healing
assays. PLoS One. 15:e02325652020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Aslan M, Hsu EC, Liu S and Stoyanova T:
Quantifying the invasion and migration ability of cancer cells with
a 3D Matrigel drop invasion assay. Biol Methods Protoc.
6:bpab0142021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nekulová M, Wyszkowska M, Friedlová N,
Uhrík L, Zavadil Kokáš F, Hrabal V, Hernychová L, Vojtěšek B, Hupp
TR and Szymański MR: Biochemical evidence for conformational
variants in the anti-viral and pro-metastatic protein IFITM1. Biol
Chem. 405:311–324. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gómez-Herranz M, Faktor J, Yébenes
Mayordomo M, Pilch M, Nekulova M, Hernychova L, Ball KL, Vojtesek
B, Hupp TR and Kote S: Emergent role of IFITM1/3 towards splicing
factor (SRSF1) and antigen-presenting molecule (HLA-B) in cervical
cancer. Biomolecules. 12:10902022. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Suh HS, Cosenza-Nashat M, Choi N, Zhao ML,
Li JF, Pollard JW, Jirtle RL, Goldstein H and Lee SC: Insulin-like
growth factor 2 receptor is an IFNgamma-inducible microglial
protein that facilitates intracellular HIV replication:
Implications for HIV-induced neurocognitive disorders. Am J Pathol.
177:2446–2458. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pirault J, Polyzos KA, Petri MH, Ketelhuth
DFJ, Bäck M and Hansson GK: The inflammatory cytokine
interferon-gamma inhibits sortilin-1 expression in hepatocytes via
the JAK/STAT pathway. Eur J Immunol. 47:1918–1924. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ferragut F, Vachetta VS, Troncoso MF,
Rabinovich GA and Elola MT: ALCAM/CD166: A pleiotropic mediator of
cell adhesion, stemness and cancer progression. Cytokine Growth
Factor Rev. 61:27–37. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wu Z, Wu Z, Li J, Yang X, Wang Y, Yu Y, Ye
J, Xu C, Qin W and Zhang Z: MCAM is a novel metastasis marker and
regulates spreading, apoptosis and invasion of ovarian cancer
cells. Tumour Biol. 33:1619–1628. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kawada M, Inoue H, Kajikawa M, Sugiura M,
Sakamoto S, Urano S, Karasawa C, Usami I, Futakuchi M and Masuda T:
A novel monoclonal antibody targeting coxsackie virus and
adenovirus receptor inhibits tumor growth in vivo. Sci Rep.
7:404002017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li J, Li Y, Wang H, Shen L, Wang Q, Shao
S, Shen Y, Xu H, Liu H, Cai R and Feng W: Neoadjuvant chemotherapy
with weekly cisplatin and paclitaxel followed by chemoradiation for
locally advanced cervical cancer. BMC Cancer. 23:512023. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tewari KS, Colombo N, Monk BJ, Dubot C,
Cáceres MV, Hasegawa K, Shapira-Frommer R, Salman P, Yañez E, Gümüs
M, et al: Pembrolizumab or placebo plus chemotherapy with or
without bevacizumab for persistent, recurrent, or metastatic
cervical cancer: Subgroup analyses from the KEYNOTE-826 randomized
clinical trial. JAMA Oncol. 10:1852024. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Monk BJ, Enomoto T, Kast WM, McCormack M,
Tan DSP, Wu X and González-Martín A: Integration of immunotherapy
into treatment of cervical cancer: Recent data and ongoing trials.
Cancer Treat Rev. 106:1023852022. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kumar Kore R, Shirbhate E, Singh V, Mishra
A, Veerasamy R and Rajak H: New investigational drug's targeting
various molecular pathways for treatment of cervical cancer:
Current status and future prospects. Cancer Invest. 42:627–642.
2024. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Bailey CC, Zhong G, Huang IC and Farzan M:
IFITM-family proteins: The cell's first line of antiviral defense.
Annu Rev Virol. 1:261–283. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liang R, Li X and Zhu X: Deciphering the
roles of IFITM1 in tumors. Mol Diagn Ther. 24:433–441. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Weichselbaum RR, Ishwaran H, Yoon T,
Nuyten DS, Baker SW, Khodarev N, Su AW, Shaikh AY, Roach P, Kreike
B, et al: An interferon-related gene signature for DNA damage
resistance is a predictive marker for chemotherapy and radiation
for breast cancer. Proc Natl Acad Sci USA. 105:18490–18495. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Popson SA, Ziegler ME, Chen X, Holderfield
MT, Shaaban CI, Fong AH, Welch-Reardon KM, Papkoff J and Hughes CC:
Interferon-induced transmembrane protein 1 regulates endothelial
lumen formation during angiogenesis. Arterioscler Thromb Vasc Biol.
34:1011–1019. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wilkins C, Woodward J, Lau DTY, Barnes A,
Joyce M, McFarlane N, McKeating JA, Tyrrell DL and Gale M Jr:
IFITM1 is a tight junction protein that inhibits hepatitis C virus
entry. Hepatology. 57:461–469. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Buchrieser J, Dufloo J, Hubert M, Monel B,
Planas D, Rajah MM, Planchais C, Porrot F, Guivel-Benhassine F, Van
der Werf S, et al: Syncytia formation by SARS-CoV-2-infected cells.
EMBO J. 39:e1062672020. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chmielewska AM, Gómez-Herranz M, Gach P,
Nekulova M, Bagnucka MA, Lipińska AD, Rychłowski M, Hoffmann W,
Król E, Vojtesek B, et al: The role of IFITM proteins in tick-borne
encephalitis virus infection. J Virol. 96:e01130212022. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Basile A, Zannella C, De Marco M, Sanna G,
Franci G, Galdiero M, Manzin A, De Laurenzi V, Chetta M, Rosati A,
et al: Spike-mediated viral membrane fusion is inhibited by a
specific anti-IFITM2 monoclonal antibody. Antiviral Res.
211:1055462023. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Meischel T, Fritzlar S, Villalon-Letelier
F, Tessema MB, Brooks AG, Reading PC and Londrigan SL: IFITM
proteins that restrict the early stages of respiratory virus
infection do not influence late-stage replication. J Virol.
95:e00837212021. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Jia R, Xu F, Qian J, Yao Y, Miao C, Zheng
YM, Liu SL, Guo F, Geng Y, Qiao W and Liang C: Identification of an
endocytic signal essential for the antiviral action of IFITM3. Cell
Microbiol. 16:1080–1093. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Ishikawa-Sasaki K, Murata T and Sasaki J:
IFITM1 enhances nonenveloped viral RNA replication by facilitating
cholesterol transport to the Golgi. PLoS Pathog. 19:e10113832023.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
von Lersner A, Droesen L and Zijlstra A:
Modulation of cell adhesion and migration through regulation of the
immunoglobulin superfamily member ALCAM/CD166. Clin Exp Metastasis.
36:87–95. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Shen X, Li M, Lei Y, Lu S, Wang S, Liu Z,
Wang C, Zhao Y, Wang A, Bi C and Zhu G: An integrated analysis of
single-cell and bulk transcriptomics reveals EFNA1 as a novel
prognostic biomarker for cervical cancer. Hum Cell. 35:705–720.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Owczarek C, Elmasry Y and Parsons M:
Contributions of coxsackievirus adenovirus receptor to
tumorigenesis. Biochem Soc Trans. 51:1143–1155. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Liu F, Wu Q, Dong Z and Liu K: Integrins
in cancer: Emerging mechanisms and therapeutic opportunities.
Pharmacol Ther. 247:1084582023. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Diao B, Sun C, Yu P, Zhao Z and Yang P:
LAMA5 promotes cell proliferation and migration in ovarian cancer
by activating Notch signaling pathway. FASEB J. 37:e231092023.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wang L, Zhou X, Zhou T, Ma D, Chen S, Zhi
X, Yin L, Shao Z, Ou Z and Zhou P: Ecto-5′-nucleotidase promotes
invasion, migration and adhesion of human breast cancer cells. J
Cancer Res Clin Oncol. 134:365–372. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Yang Y, Koh YW, Sari IN, Jun N, Lee S, Phi
LTH, Kim KS, Wijaya YT, Lee SH, Baek MJ, et al: Interferon-induced
transmembrane protein 1-mediated EGFR/SOX2 signaling axis is
essential for progression of non-small cell lung cancer. Int J
Cancer. 144:2020–2032. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Hatano H, Kudo Y, Ogawa I, Tsunematsu T,
Kikuchi A, Abiko Y and Takata T: IFN-induced transmembrane protein
1 promotes invasion at early stage of head and neck cancer
progression. Clin Cancer Res. 14:6097–6105. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Liang Y, Li E, Min J, Gong C, Gao J, Ai J,
Liao W and Wu L: MiR-29a suppresses the growth and metastasis of
hepatocellular carcinoma through IFITM3. Oncol Rep. 40:3261–3272.
2018.PubMed/NCBI
|
|
59
|
Zheng W, Zhao Z, Yi X, Zuo Q, Li H, Guo X,
Li D, He H, Pan Z, Fan P, et al: Down-regulation of IFITM1 and its
growth inhibitory role in cervical squamous cell carcinoma. Cancer
Cell Int. 17:882017. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Taylor BC and Balko JM: Mechanisms of
MHC-I downregulation and role in immunotherapy response. Front
Immunol. 13:8448662022. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
She X, Shen S, Chen G, Gao Y, Ma J, Gao Y,
Liu Y, Gao G, Zhao Y, Wang C, et al: Immune surveillance of brain
metastatic cancer cells is mediated by IFITM1. EMBO J.
42:e1111122023. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Saveanu L and van Endert P: The role of
insulin-regulated aminopeptidase in MHC class I antigen
presentation. Front Immunol. 3:572012. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Elgueta R, Benson MJ, de Vries VC, Wasiuk
A, Guo Y and Noelle RJ: Molecular mechanism and function of
CD40/CD40L engagement in the immune system. Immunol Rev.
229:152–172. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Grewal IS, Foellmer HG, Grewal KD, Xu J,
Hardardottir F, Baron JL, Janeway CA Jr and Flavell RA: Requirement
for CD40 ligand in costimulation induction, T cell activation, and
experimental allergic encephalomyelitis. Science. 273:1864–1867.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Djureinovic D, Wang M and Kluger HM:
Agonistic CD40 antibodies in cancer treatment. Cancers (Basel).
13:13022021. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Yang Y, Sanders AJ, Ruge F, Dong X, Cui Y,
Dou QP, Jia S, Hao C, Ji J and Jiang WG: Activated leukocyte cell
adhesion molecule (ALCAM)/CD166 in pancreatic cancer, a pivotal
link to clinical outcome and vascular embolism. Am J Cancer Res.
11:5917–5932. 2021.PubMed/NCBI
|
|
67
|
Jing L, An Y, Cai T, Xiang J, Li B, Guo J,
Ma X, Wei L, Tian Y, Cheng X, et al: A subpopulation of
CD146+ macrophages enhances antitumor immunity by
activating the NLRP3 inflammasome. Cell Mol Immunol. 20:908–923.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Duan H, Jing L, Jiang X, Ma Y, Wang D,
Xiang J, Chen X, Wu Z, Yan H, Jia J, et al: CD146 bound to LCK
promotes T cell receptor signaling and antitumor immune responses
in mice. J Clin Invest. 131:e1485682021. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Yánez DC, Ross S and Crompton T: The IFITM
protein family in adaptive immunity. Immunology. 159:365–372. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Cai Y, Ji W, Sun C, Xu R, Chen X, Deng Y,
Pan J, Yang J, Zhu H and Mei J: Interferon-induced transmembrane
protein 3 shapes an inflamed tumor microenvironment and identifies
immuno-hot tumors. Front Immunol. 12:7049652021. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Shen C, Li YJ, Yin QQ, Jiao WW, Li QJ,
Xiao J, Sun L, Xu F, Li JQ, Qi H and Shen AD: Identification of
differentially expressed transcripts targeted by the knockdown of
endogenous IFITM3. Mol Med Rep. 14:4367–4373. 2016. View Article : Google Scholar : PubMed/NCBI
|