Introduction
Glucosamine is a natural amino sugar that is used as
one of the building blocks of the body, such as in cartilage and
tendon. Therefore, it has been used to treat patients with painful
knee osteoarthritis and in complementary and alternative medicine
as an osteoarthritis cure (1,2). In
addition, a number of studies have reported that glucosamine
inhibits cell proliferation and induces cell cycle arrest and
apoptosis in various types of cancer (3,4). Other
studies have described target proteins of glucosamine, such as
transglutaminase 2, p70S6K, hypoxia-inducible factor 1α (HIF-1α),
cyclooxygenase-2 (COX-2) and the insulin-like growth factor 1
receptor (IGF-1R)/Akt pathway (5–9). These
studies propose that glucosamine has potential value as an
antiproliferative drug.
Facilitative glucose transporters (GLUTs) are
integral membrane proteins that have 12 transmembrane domains and a
binding site for hexose substrates such as glucose, fructose and
glucosamine (10). A total of 13
GLUT isoforms with highly conserved amino acid sequences and 12
hydrophobic α-helical domains have been identified and
characterized in mammalian cells. Each GLUT isoform has a different
expression level in various tissues and organs and plays a specific
role in the energy-independent uptake of hexoses. For instance,
GLUT1, which is highly expressed in all tissues, is in charge of
basal glucose transport; GLUT2, which is abundant in liver,
pancreatic islet and retinal cells, has a higher affinity for
glucosamine than glucose (11).
Cancer cells undergo metabolic reprogramming to
support their rapid growth and proliferation. One hallmark of this
reprogramming is the Warburg effect, which describes the preference
of cancer cells for aerobic glycolysis, even in the presence of
sufficient oxygen (12). This
phenomenon allows cancer cells to generate ATP quickly while
producing metabolic intermediates essential for biosynthesis. A
number of previous studies have also demonstrated that GLUTs are
overexpressed in various types of cancer, facilitating increased
glucose uptake to sustain the heightened glycolytic flux. This
overexpression of GLUT proteins has been closely associated with
aggressive tumor behavior, including metastasis and poor prognosis
(13,14).
Once hexoses are transported into the cytoplasm,
they are catalyzed by hexokinases (HKs), which carry out the first
and rate-limiting step of the hexose metabolic pathway. In addition
to glycolysis, HK also performs the irreversible step in the
hexosamine pathway that converts hexose to hexose-6-phosphate,
using an ATP molecule to start the process (15). In humans and other mammals, four HK
isoforms have identified; HKI, II, III and IV (glucokinase). HKI,
II and III have a high affinity for glucose and a molecular mass of
~100 kDa, whereas HKIV has a molecular mass of ~50 kDa and can only
phosphorylate glucose, giving it a higher Km than the
other HKs (16). During cancer
progression, among those HKs, HKII, which has low affinity for
glucose and is only slightly expressed in normal tissues, is
markedly upregulated (17),
suggesting that HKII plays an important role in malignancy.
HIF-1, a heterodimer consisting of α and β subunits,
is stabilized under hypoxic conditions and plays a pivotal role in
the glycolytic phenotype in human cancers (18). It has been reported that HIF-1α acts
as a transcription factor and increases expressions of GLUTs and
HKs and thus, tumor cells which are in hypoxic conditions activate
HIF-1α to facilitate glucose uptake and glycolysis to satisfy their
energy demands (19). Furthermore,
since the metabolic reprogramming induced by HIF-1α helps cancer
cells to grow, cancer cells turn on HIF-1α even with enough
oxygens, a phenomenon called pseudohypoxia (20). These features make cancer cells more
resistant to glycolysis inhibitors, such as 2-deoxy-D-glucose
(2-DG), by increasing the number of glycolytic enzymes (21).
Malignant cancer cells have an elevated and
accelerated glucose metabolism due to increased requirements for
glucose, which is essential as an energy source (17). In addition, dysfunction of the Krebs
cycle caused by mitochondrial mutations leads to increased
dependence on glycolysis by cancer cells for ATP production
(22). Therefore, it is not
surprising that the increased aerobic glycolysis in cancer is
closely related to overexpression of GLUTs and HKII in various
types of cancer. A number of researchers have used glucose analogs
such as fluorodeoxyglucose and 2-DG and HKII inhibitors such as
3-bromopyruvate (3-BrPA) to study interference of glucose uptake
and glycolysis (15,23). As stated for the aforementioned
molecules, these results indicate that glucosamine also may be a
glycolysis inhibitor.
Based on these preliminary studies, the present
study sought to determine whether the antiproliferative activity of
glucosamine occurs because it is a glycolysis inhibitor in various
cancer cells and whether GLUTs, HK isoforms and HIF-1α affect the
sensitivity to glucosamine of cancer cells. It showed a new
mechanism of glucosamine to specifically inhibit the glycolytic
pathway to suppress cancer progression.
Materials and methods
Cell lines and agents
Human liver cancer cell lines (HepG2 and Hep3B),
non-small cell lung cancer cell lines (A549, H1299 and H460) and
breast cancer cell lines (MCF-7, MDA-MB-231 and T47D) were
purchased from the American Type Culture Collection and cultured in
RPMI 1640 (PAA Laboratories GmbH) supplemented with 10% fetal
bovine serum (FBS; PAA Laboratories GmbH) and 100 U/ml
penicillin-streptomycin (Gibco; Thermo Fisher Scientific, Inc.) at
37°C in a humidified atmosphere with 5% CO2. The
authenticity of all eight cell lines used in the present study was
validated by short tandem repeat (STR) DNA profiling, which was
performed using PowerPlex 18D system by commercial service (Cosmo
Genetech).
D-(+)-glucose, D-(+)-glucosamine hydrochloride,
cytochalasin B and phloretin were purchased from MilliporeSigma,
while 2-DG and 3-BrPA were purchased from Tokyo Chemical Industry.
In the competition experiment between glucose and glucosamine,
glucose-free RPMI 1640 medium (MilliporeSigma) was used and cells
were pre-incubated for 24 h with the indicated doses of glucose
prior to being treated with glucosamine.
Semiquantitative RT-PCR
Total RNA was isolated from cells using
TRIzol® reagent (Thermo Fisher Scientific, Inc.) and
chloroform extraction. First-strand cDNA was synthesized from 5 µg
of total RNA using Superscript II reverse transcriptase
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. Semiquantitative PCR was performed in
a thermal cycler (MyCycler; Bio-Rad Laboratories, Inc.) using
gene-specific primer sets and sequences of primers are listed in
the supplementary Table SI. The
thermocycling conditions were as follows: Initial denaturation at
95°C for 5 min, followed by 25–45 cycles of denaturation at 95°C,
annealing at 42–60°C for 30 sec (depending on the target gene), and
elongation at 72°C for 1 min, with a final extension at 72°C for 5
min. The reactions were stopped during the exponential phase to
ensure accurate comparisons. ACTB was used as a reference
gene. PCR products were electrophoresed on 1.2–1.5% agarose gel and
visualized using ethidium bromide.
siRNA transfection
A549 and HepG2 cells (2.5×105 cells/well)
were seeded in 6-well plates and transfected with double-stranded
small interfering RNA (siRNA). Target-specific siRNAs were designed
for knockdown of GLUT1, GLUT2 and HIF1A were from
Bioneer Corporation and MBiotech, respectively. A scrambled siRNA
(siSCR) was synthesized and validated by Bioneer Corporation. The
sense and anti-sense sequences of siRNAs are listed in the Table SII. Following 25 pmole of siRNA
transfection for 24 h at 37°C using Lipofectamine®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to
the manufacturer's instructions, subsequent experiments were
conducted. Knockdown of GLUT1, GLUT2 and HIF1A expression
were validated by semiquantitative RT-PCR. The thermocycling
conditions were as follows: Initial denaturation at 95°C for 5 min,
followed by 25–45 cycles of denaturation at 95°C, annealing at
42–60°C for 30 sec (depending on the target gene), and elongation
at 72°C for 1 min, with a final extension at 72°C for 5 min. The
reactions were stopped during the exponential phase to ensure
accurate comparisons. ACTB was used as a reference gene. PCR
products were electrophoresed on 1.2–1.5% agarose gel and
visualized using ethidium bromide.
Western blotting
Whole cell lysates were prepared using RIPA-B lysis
buffer containing 150 mM NaCl, 1 mM EDTA (pH 8.0), 20 mM Tris-Cl
(pH 7.4), 1% NP-40, 0.5% sodium deoxycholate, 1% Triton X-100, 1 mM
Na3VO4, 5 mM NaF, 10% glycerol and protease
inhibitor cocktail (Roche Diagnostics GmbH). Protein concentration
was determined using BCA protein assay kit (Thermo Fisher
Scientific, Inc.). Equivalent amounts of proteins (10–20 µg per
lane, depending on the target protein) were separated by
electrophoresis on 6, 8, 10 or 12% SDS-PAGE and then transferred to
a polyvinylidene fluoride membrane (Pall Corporation). Membranes
were blocked with 5% skimmed milk at room temperature for 1 h. The
membranes were then incubated overnight at 4°C with primary
antibodies against phosphorylated (p-)Akt (1:1,000, cat. no. 9271,
Cell Signaling Technology, Inc.), Akt (1:1,000, cat. no. 4691, Cell
Signaling Technology, Inc.), a/β-tubulin (1:1,000, cat. no. 2148,
Cell Signaling Technology, Inc.), IGF-1R (1:1,000; cat. no. sc-713,
Santa Cruz Biotechnology, Inc.) and COX-2 (1:1,000; cat. no.
sc-1745, Santa Cruz Biotechnology, Inc.). After washing with
Tris-buffered saline containing 0.1% Tween 20, the membranes were
incubated with horseradish peroxidase-conjugated secondary
antibodies (1:10,000; cat. no. AbC-5001 for anti-mouse antibody and
cat. no. AbC-5003 for anti-rabbit antibody, Abclon) for 1 h at room
temperature. Immunoreactive bands were detected using enhanced
chemiluminescence (AbSignal; Abclon).
MTT assay
Cells (3.0×103 cells/well) were seeded
into 96-well plates, incubated overnight at 37°C and treated with
glucosamine or other agents. Control groups were given dimethyl
sulfoxide (DMSO; 0.1% final concentration) or phosphate-buffered
saline (PBS) vehicle. Cell viability was measured by MTT assay. An
insoluble purple formazan precipitate was solubilized in DMSO and
the absorbance was determined by microplate spectrometer at 562
nm.
Cell cycle analysis
A549 and HepG2 cells (1.5×105) were
plated onto 60×15 mm dishes for 1 day and then cells were treated
with glucose (1, 10, 20, or 50 mM) and glucosamine (1, 2, or 5 mM)
at 37°C. Glucose was administered for 48 h and glucosamine was
co-administered with glucose during the last 24 h of that period.
After treatment, cells were washed once with PBS, trypsinized and
harvested. All the harvested cells were fixed in 70% ethanol for 1
h at 4°C. After centrifugation for 5 min. at 1,000 × g and 4°C,
cell pellets were washed twice with PBS and then stained with PI
containing RNase A (40 µg/ml) for 30 min at 4°C in the dark. Total
DNA contents were detected by BD FACSCalibur flow cytometer (BD
Biosciences), and analyzed using FlowJo 10.10.0 (FlowJo LLC).
Apoptosis analysis
A549 and HepG2 cells (1.5×105) were
plated onto 60×15 mm dishes for 1 day and after treated with
glucosamine for 48 h, cells were harvested and washed twice with
PBS on ice. Then, cells were stained using the FITC Annexin V
Apoptosis Detection Kit I (BD Pharmingen; BD Biosciences). Cells
were resuspended in 1× binding buffer containing 5 µl of
fluorescein isothiocyanate (FITC)-Annexin V and 5 µl of PI. After
staining, cells were detected by BD FACSCalibur flow cytometer (BD
Biosciences) at 488 and 633 nm, and analyzed using FlowJo 10.10.0
(FlowJo LLC). The apoptotic rate was calculated as the percentage
of early apoptotic cells and the percentage of late apoptotic
cells. All procedures were carried out according to the
manufacturer's instructions.
Measurement of glucosamine consumption
rate
Cells (2.5×105 cells/well) were seeded in
6-well plates and incubated under RPMI 1640 medium containing the
indicated concentrations of glucosamine. Glucosamine in harvested
culture medium was reacted using D-Glucosamine Assay Kits
(K-GAMINE; Megazyme) and optical density was measured with a
spectrophotometer at 340 nm. All procedures were carried out
according to the manufacturer's instructions.
Measurement of lactate production
The cellular glycolytic efficiency could be
estimated by measuring the cellular lactate generation rate. Cells
(2.5×105 cells/well) in the exponential growth phase
were plated in 6-well plates for 1 day and cells were washed and
treated with indicated concentrations of glucosamine for an
additional 24 h. Aliquots of the culture medium were withdrawn and
used for analysis of secreted lactic acid using the YSI 2300D STAT
Plus Glucose and Lactate Analyzer (YSI Inc.).
Whole genome gene expression
analysis
A549 cells were treated with DMSO or 5 mM of
glucosamine for 24 h at 37°C and RNA was isolated. From each
sample, 500 ng of total RNA was biotinylated and amplified using
the Illumina TotalPrep RNA Amplification Kit (Ambion; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions. The
cRNA yield was measured using RiboGreen RNA quantitation kit
(Invitrogen; Thermo Fisher Scientific, Inc. and 750 ng of the cRNA
sample was hybridized on a human HT-12 expression bead chip
(Illumina, Inc.) profiling 48,804 transcripts per sample. The chips
were stained with streptavidin and scanned using an Illumina
BeadArray Reader (Illumina, Inc.). BeadStudio v3 (Illumina, Inc.)
was used to quantile-normalize the data.
Gene ontology (GO) analysis was performed on genes
that were increased or decreased by more than 2-fold in the
glucosamine-treated sample compared with the control. The analysis
was conducted using the gseGO function in the clusterProfiler
package (ver. 4.14.3) (24) in R
software (ver. 4.4.1, R Core Team, 2024) (25), based on the Gene Set Enrichment
Analysis approach. GO terms which have adjust P-value <0.01 were
considered markedly regulated GO. Up- or downregulated GO terms
were featured by R software.
Kyoto Encyclopedia of Genes and Genomes (KEGG)
pathway enrichment analysis was performed on differentially
expressed genes using the enrichKEGG function from the
clusterProfiler package in R software. The result of glycolysis
pathway (hsa00010) was visualized using the Pathview package
(Release 3.20) (26).
Statistical analysis
All statistical analyses were conducted using
GraphPad Prism 8.0 (Dotmatics) and Microsoft Excel (Microsoft
Corporation). For comparisons involving more than three groups,
one-way ANOVA was performed, followed by Tukey's post hoc test. For
comparisons between two groups, the Student's t-test was employed.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Glucosamine regulates some gene sets
related to glycolysis
In 1953, J.H. Quastel and A. Cantero demonstrated,
for the first time, that D-glucosamine inhibits the tumor growth in
a xenograft mouse model (27).
After their findings, a number of studies have sought to discover
the mechanism and related molecules in this antiproliferative
effect. A number of studies have demonstrated that glucosamine
decreases the ATP level in cancer cells and inhibits oncogenes,
such as IGF-1R, HIF-1α and COX-2 (8,9,21).
However, the mechanism has not been described in detail. Therefore,
the present study used a microarray to screen genes that are
regulated directly or affected indirectly by glucosamine. Genes
induced by glucosamine more than two folds were categorized mainly
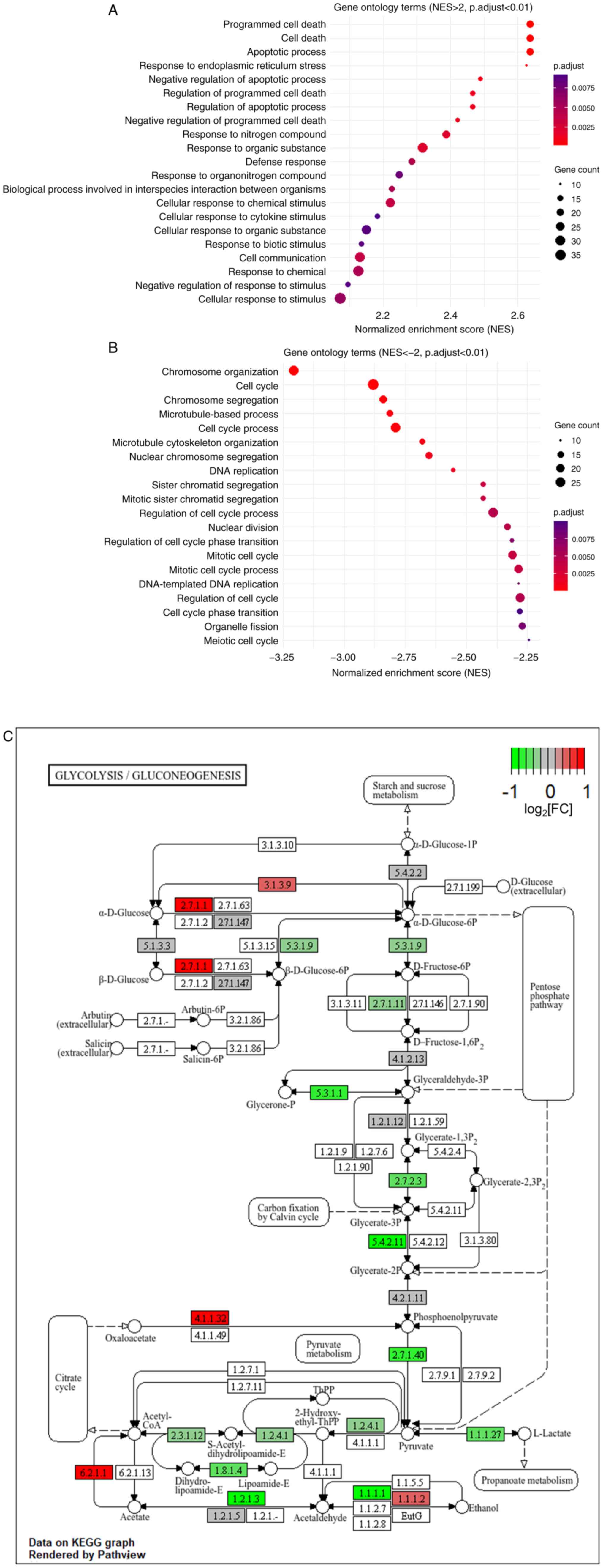
into pro-apoptosis and cell death (Fig.
1A), while genes downregulated by glucosamine more than two
folds mostly belonged to cell cycle and DNA damage responses
(Fig. 1B). Next, the present study
examined whether the genes involved in carbohydrate metabolism,
biomolecules biosynthesis and glycosylation, which pathways are
highly associated with glycolysis (28–30),
are regulated by glucosamine. Notably, as shown in Table SIII, several genes upregulated by
glucosamine more than twofold were involved in these pathways.
Also, pathway enrichment of differential genes was used and then
the KEGG pathway analysis results viewed to see whether the
differential genes are enriched in glycolysis. As a result,
differential genes are represented in the glycolysis pathway and
their expression is primarily downregulated by glucosamine
(Fig. 1C). With all these findings,
glucosamine's similar structure to glucose and its reduction of ATP
levels in cancer cells (4), as
demonstrated by previous studies, it was assumed that glycolysis is
the main target pathway of glucosamine.
Competitive relationship between
glucose and glucosamine constrains the inhibitory effects of
glucosamine in cancer cells
Other than its NH2 group, glucosamine has
a structure very similar to that of glucose. In fact, several
studies have demonstrated that glucosamine competes with glucose in
carbohydrate metabolism (31–33).
Based on this, whether its antiproliferative effect is linked to
this competitive relationship was investigated. A recent study has
shown that glucosamine exhibits antiproliferative effects on a
liver cancer cell line (34), while
our previous research demonstrated similar effects on lung cancer
cell lines (9). Therefore, the
present study initially conducted experiments on HepG2 liver cancer
cell line and A549 lung cancer cell line. Under low level of
glucose, glucosamine inhibited proliferation of A549 and HepG2
cancer cells, which anti-proliferative effects of glucosamine were
reversed by increasing glucose concentration (Fig. 2A). Glucose-treated cells also showed
a dose-dependent recovery of the G0/G1 arrest induced by
glucosamine (Fig. 2B). Since
previous studies showed that glucosamine inhibits the IGF-1R/Akt
pathway (9,35), the present study next investigated
the effect of glucose on these target molecules. As expected,
IGF-1R and p-Akt expression levels that had been reduced by
glucosamine were restored as concentrations of glucose increased
(Fig. 2C). To determine whether the
competitive effect of glucose and glucosamine occurs during
glycolysis in cancer cells, the level of lactate, which is produced
from pyruvate during glycolysis was investigated. Glucosamine
markedly suppressed lactate production, but this suppression was
nearly reversed with a high dose of glucose (Fig. 2D). These findings suggested that the
antiproliferative effect of glucosamine was closely related to
competition between glucose and glucosamine, which occurs during
glycolysis.
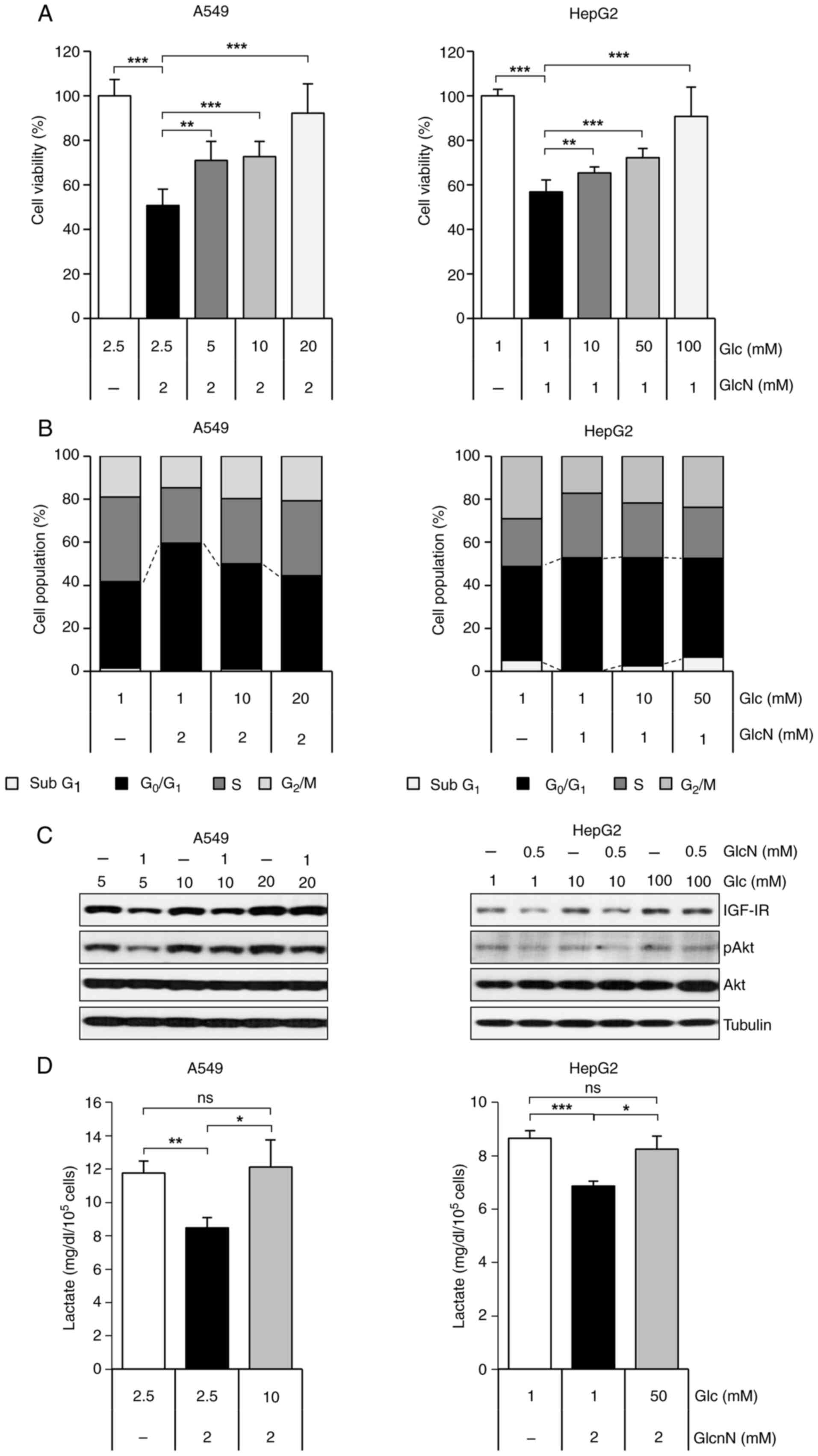 | Figure 2.GlcN-induced anticancer effect occurs
through competition with Glc. (A) Cell viability of A549 and HepG2
cells after treatment with Glc and GlcN for 48 h. Cell viability
was measured by MTT assay and results are presented as mean ±
standard deviation. (B) Cell population by cell cycle of A549 and
HepG2 cells after treated with Glc and GlcN. Cells were treated
with Glc for 24 h, followed by additional 24 h treatment with GlcN.
Cell cycle analysis was performed after propidium-iodide staining.
(C) Western blot analysis of IGF-1R, p-Akt and Akt expression
levels in A549 and HepG2 cells. Cells were treated with indicated
concentrations of Glc and GlcN for 24 h. (D) Lactate production in
A549 and HepG2 cell lines after treatment with Glc and GlcN for 24
h. Results are presented as mean ± standard deviation. *P<0.05,
**P<0.01, ***P<0.001, ns, no significance; Glc, glucose;
GlcN, glucosamine; p-, phosphorylated; IGF-1R, insulin-like growth
factor 1 receptor. |
Antiproliferative effect of
glucosamine is related to basal expression level of GLUT2 and basal
activity of glucosamine consumption
Glucosamine enters the cell mostly via GLUT2, along
with other isoforms of GLUT (11).
Although several studies have examined the kinetics of glucosamine
transport, there is no direct evidence on how glucosamine,
particularly its antiproliferative effects, influences these
kinetics. Thus, it was hypothesized that cancer cells with higher
expression levels of GLUT2 were more sensitive to the
antiproliferative effect of glucosamine. First, the expression
level of GLUT1 and GLUT2 in cancer cell lines
originating from various types of cancer including liver cancer,
lung cancer and breast cancer was screened. For this purpose, two
liver cancer cell lines (HepG2 and Hep3B), three lung cancer cell
lines (A549, H1299 and H460) and three breast cancer cell lines
(MCF-7, MDA-MB-231 and T47D) were used. Among them, liver cancer
cell lines showed higher levels of GLUT2 compared with the
others, while most of the cell lines showed similar levels of
GLUT1 (Fig. 3A). Then, to
assess the correlation between glucosamine uptake and GLUT2
expression levels, a glucosamine consumption assay was performed
and the results showed that liver cancer cell lines, which showed
highly expressed GLUT2, consumed glucosamine more actively
than others (Fig. 3B). Next,
whether the expression level of GLUT2 and the rate of
glucosamine uptake are related to the antiproliferative effect of
glucosamine was investigated. As a result, liver cancer cell lines,
which showed higher expressions of GLUT2 and actively
consumed glucosamine, were more sensitive to glucosamine than
others (Fig. 3C). Along with these
results, HepG2 and Hep3B cells showed greater decreases in IGF-1R
and p-Akt expression levels in the presence of glucosamine than
A549 cells, which were the most sensitive among the remaining cell
lines (Fig. 3D). Taken together,
these results implied that the antiproliferative effect of
glucosamine has a strong relationship with the expression level of
GLUT2 and the uptake rate of glucosamine.
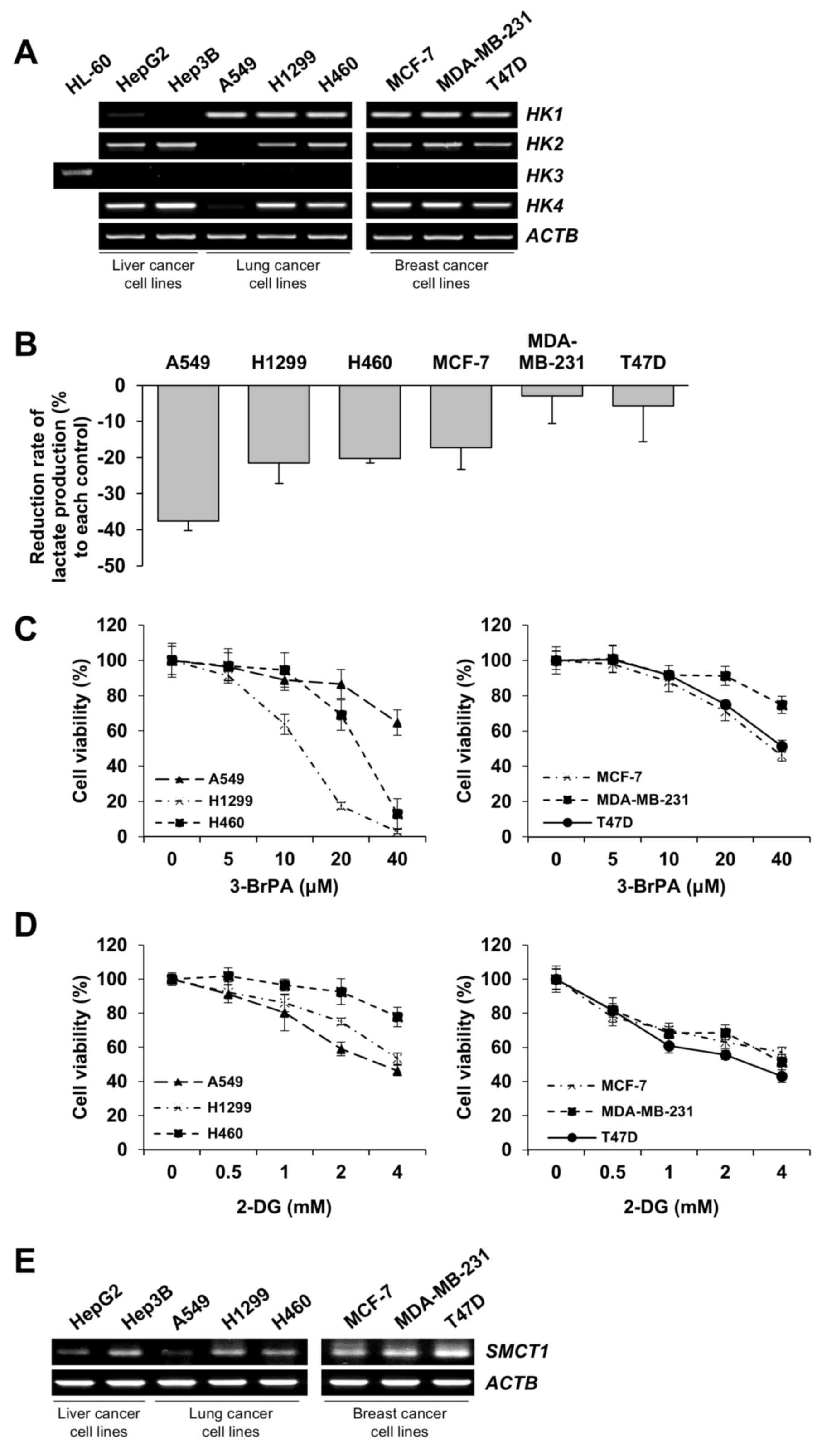
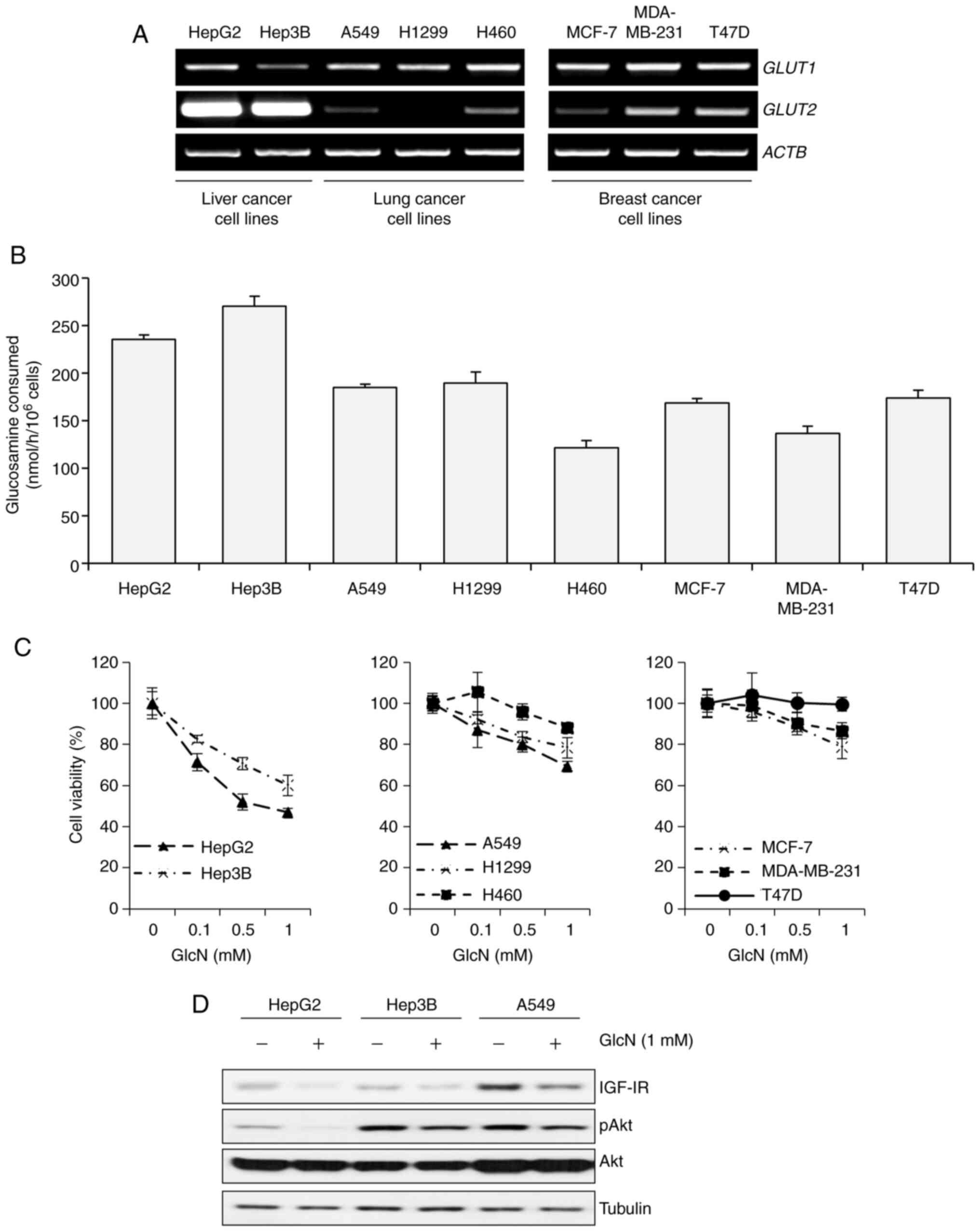 | Figure 3.GLUT2 basal level is associated with
GlcN sensitivity in liver, lung and breast cancer cell lines. (A)
Semiquantitative PCR of GLUT1 and GLUT2 in various
cancer cell lines. Total mRNA from the following cancer cell lines
was isolated and used for analysis: HepG2, Hep3B, A549, H1299,
H460, MCF-7, MDA-MB-231 and T47D. ACTB was included as a
loading control. (B) GlcN consumption rates in the eight cell
lines. Each cell line was incubated in Glc-free RPMI 1640
containing both 1 mM Glc and 5 mM GlcN for 6 h. Results are
presented as mean ± standard deviation. (C) Cell viability of the
eight cell lines after treated with GlcN for 48 h. Cell viability
was measured by MTT assay and results are presented as mean ±
standard deviation. (D) Western blot analysis of IGF-1R, p-Akt and
Akt expression levels in HepG2, Hep3B and A549 cells after 1mM of
glucosamine treatment for 12 h. GLUT, glucose transporter; GlcN,
glucosamine; IGF-1R, insulin-like growth factor 1 receptor. |
Antiproliferative effect of
glucosamine is dependent on the absorption process of glucosamine
into cancer cell lines through GLUT2, not GLUT1
Then it was explored whether cancer cells uptake
glucosamine through GLUT2, not GLUT1, and whether the uptake of
glucosamine is an essential step for its antiproliferative effects.
A549 and HepG2 cells were treated with cytochalasin B, a
GLUT1-specific inhibitor, or phloretin, a GLUT2-specific inhibitor,
in combination with glucosamine, respectively. Cytochalasin B and
phloretin concentrations were taken from previously reported
studies (36,37). As a result, phloretin recovered
glucosamine-induced downregulation of IGF-1R and p-Akt expressions
in both cancer cell lines, while cytochalasin B showed no recovery
effects in A549 cells and partial recovery effects in HepG2 cells
(Fig. 4A). In addition, glucosamine
shifted the molecular mass of COX-2 as previously reported
(38), which were markedly reversed
by phloretin, but not by cytochalasin B in A549 cells (Fig. S1). Following these changes in
molecular features, phloretin-mediated GLUT2 inhibition
sufficiently rescued A549 and HepG2 cells from glucosamine-induced
cell death, while the effects of cytochalasin B were not
significant (Fig. 4B). Along with
the results using inhibitors, GLUT2-knockdowned HepG2 cells by
transfection of siGLUT2 showed recovery of IGF-1R and p-Akt
expression levels decreased by glucosamine, while there were no
changes in those expressions in GLUT1-knockdowned HepG2 cells
(Fig. 4C). The present study also
investigated whether treatment with phloretin or siGLUT2,
which have an inhibitory effect on glucosamine transportation,
could decrease consumption of glucosamine in both cancer cell lines
and it was found that glucosamine consumption rate was reduced when
GLUT2 was inhibited by phloretin and siGLUT2 (Fig. 4D). These findings demonstrate that
the absorption of glucosamine via GLUT2 is the basic and necessary
process for the antiproliferative effect of glucosamine.
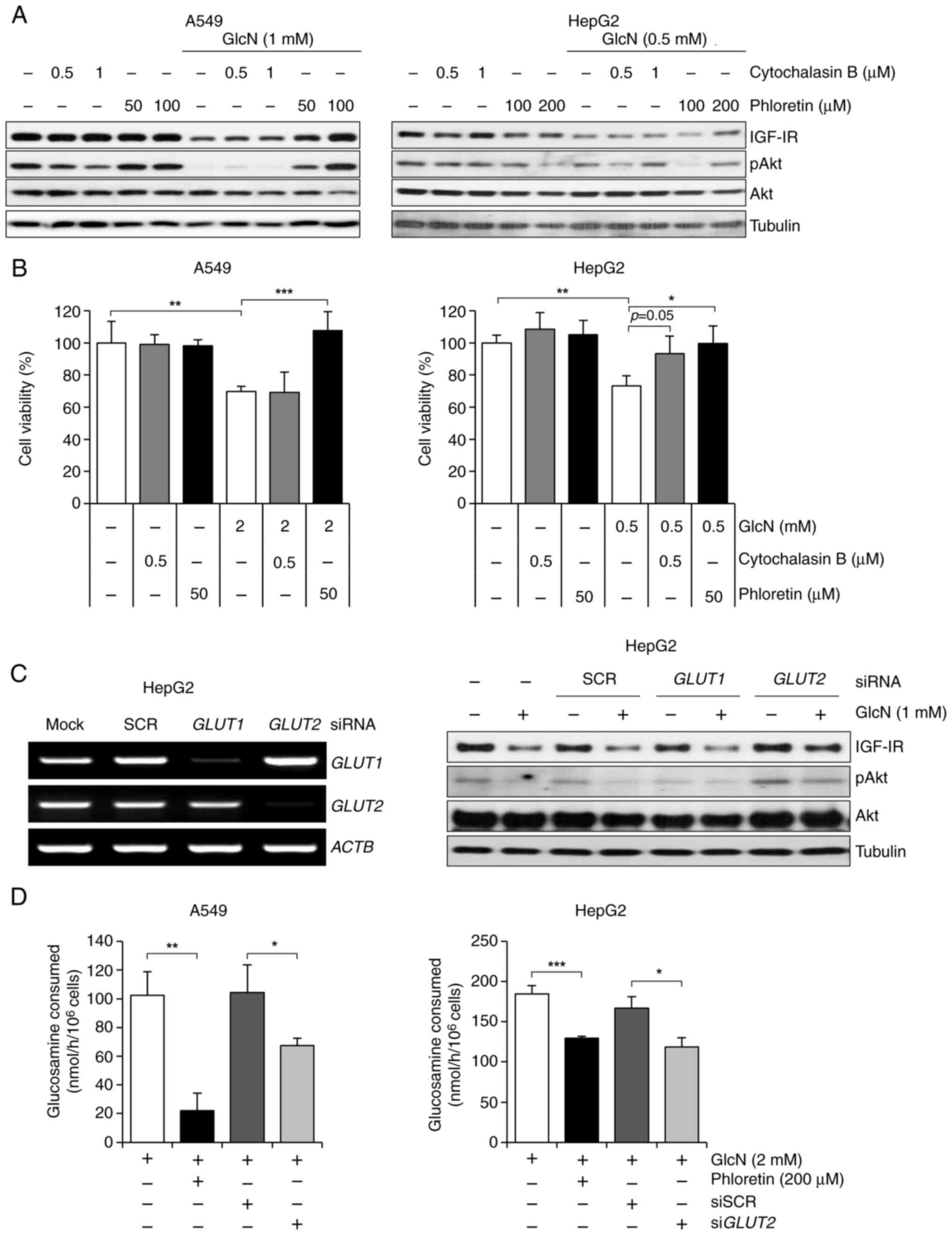 | Figure 4.GlcN entrance into cancer cells
through GLUT2 leads to its anticancer activities. (A) Western blot
analysis of IGF-1R, p-Akt and Akt expression levels in A549 and
HepG2 cells treated with cytochalasin B or phloretin for 24 h.
Cytochalasin B was used as a GLUT1-specific inhibitor, while
phloretin was used as a GLUT2-specific inhibitor. (B) Cell
viability of A549 and HepG2 cells after treatment with glucosamine,
cytochalasin B and phloretin for 48 h. Cell viability was measured
by MTT assay and results are presented as mean ± standard
deviation. (C) Left: Semiquantitative PCR of GLUT1 and
GLUT2 in HepG2 cells after transfection with siSCR,
siGLUT1, or siGLUT2. ACTB served as a loading
control. Right: Western blot analysis of IGF-1R, p-Akt and Akt
expression levels in GLUT1 or GLUT2 knockdown HepG2 cells after 1
mM of GlcN treatment for 24 h. (D) GlcN consumption rates in A549
and HepG2 cells. GLUT2 was inhibited by phloretin or siGLUT2
and then GlcN treatment for 6 h. Results are presented as mean ±
standard deviation. *P<0.05, **P<0.01, ***P<0.001. GlcN,
glucosamine; GLUT, glucose transporter; IGF-1R, insulin-like growth
factor 1 receptor; si, small interfering. |
Inhibition of glycolysis by
glucosamine is greater in cancer cells with lower HKII levels
Although the other six cell lines expressed lower
levels of GLUT2 compared with HepG2 and Hep3B cells, there were
differences in the antiproliferative effect of glucosamine among
those six cell lines (Fig. 3B and
C), suggesting that cellular sensitivity to glucosamine is
influenced by not only uptake through GLUT2 but also by post-uptake
processes. Glucosamine can be phosphorylated by yeast hexokinase
(39,40). In addition, yeast hexokinase is
similar to rat hexokinase, human hexokinase N-terminal and
C-terminal in respect of its sequence and structure amino acids,
especially in the active site (41). Thus, it could be hypothesized that
glucosamine serves as a substrate for hexokinase in mammalian cells
including those of human origin (42–44).
Thus, the present study examined the basic expression levels of HK
isoforms (HK1-4) in the studied eight cell lines. Most cell
lines showed similar levels of HK1 expression and rarely
expressed HK3. Notably, the six cell lines showed lower
GLUT2 expression and reduced glucosamine consumption rates
compared with HepG2 and Hep3B cells, A549 cells exhibited the
lowest HK2 expression level compared with the other five
cell lines (Fig. 5A). Next, since
HK is the first enzyme to be activated during glycolysis and this
activation is a rate-limiting step that regulates the speed of
glycolysis (45), the differences
in glycolytic rates and the extent of glycolysis inhibition by
glucosamine was assessed. Notably, the rate of lactate production
was most markedly reduced by glucosamine in A549 cells which were
the most sensitive to glucosamine after liver cancer cell lines. It
was observed that 3-BrPA or 2-DG, which are used as HKII inhibitors
(46,47), sufficiently inhibited the
proliferation of those six cell lines (Fig. 5C and D). In case of A549 cells, the
anti-proliferative effect of 3-BrPA was relatively weak compared
with other lung cancer cell lines, H1299 and H460, and this was
probably due to the lower level of sodium-coupled monocarboxylate
transporter 1 (SMCT1), which is responsible for the
transportation of 3-BrPA into cells (48), than other cancer cell lines tested
(Fig. 5E). Taken together, these
results showed that glycolysis inhibition by glucosamine is
negatively related to the level of HKs, especially HKII.
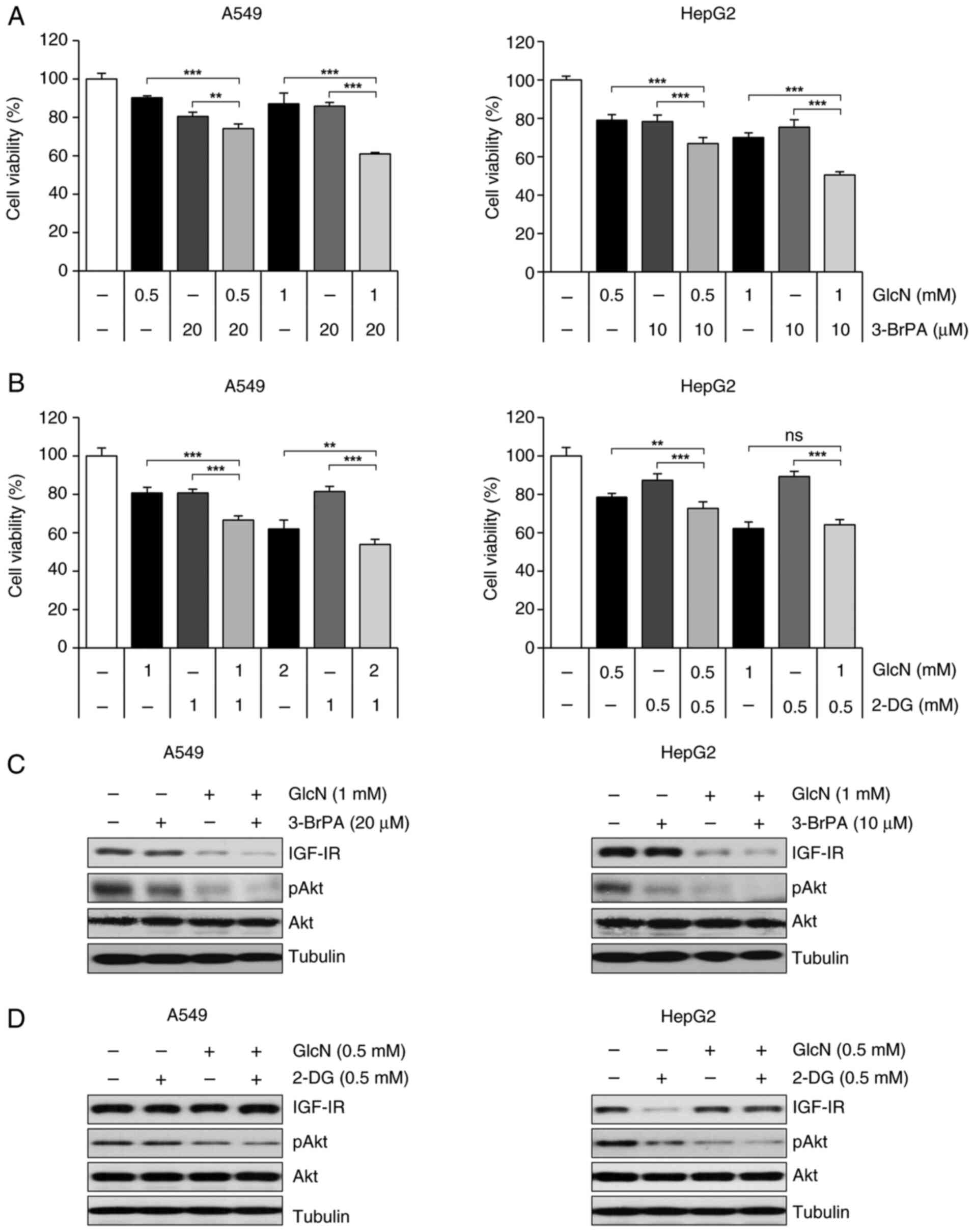
Glucosamine and 3-BrPA enhance the
antiproliferative activity of one another
3-BrPA, 2-DG and glucosamine, which are all
glycolysis inhibitors, take different pathways to enter cells and
work along different mechanisms. For example, 2-DG enters cells via
GLUT1 and GLUT4 like glucose and works as a competitor of glucose,
which eventually lowers the efficiency of glycolysis (23), whereas glucosamine is transported
into cells efficiently via GLUT2 rather than GLUT1 and competes
with glucose for HK. By contrast, 3-BrPA is taken up into cells by
SMCT1 and directly suppresses the activity of HKII (47,48).
Therefore, it was examined whether the antiproliferative effect of
glucosamine is elevated when co-treated with 2-DG or 3-BrPA.
Co-treatment of 3-BrPA with glucosamine increased the
anti-proliferative effects of glucosamine in both A549 and HepG2
cells (Fig. 6A), whereas 2-DG
showed a slight combinational effect and no combinational effect in
A549 cells and HepG2 cells, respectively (Fig. 6B). Along with these results,
glucosamine-induced decreased in IGF-1R and p-Akt expression level
were tended to be enhanced by co-treatment of 3-BrPA (Fig. 6C), but not by co-treatment of 2-DG
(Fig. 6D). These results suggested
that the mechanism of glucosamine inhibiting HK is similar to that
of 2-DG and different from that of 3-BrPA and that 3-BrPA may be a
suitable HK inhibitor for co-administration with glucosamine.
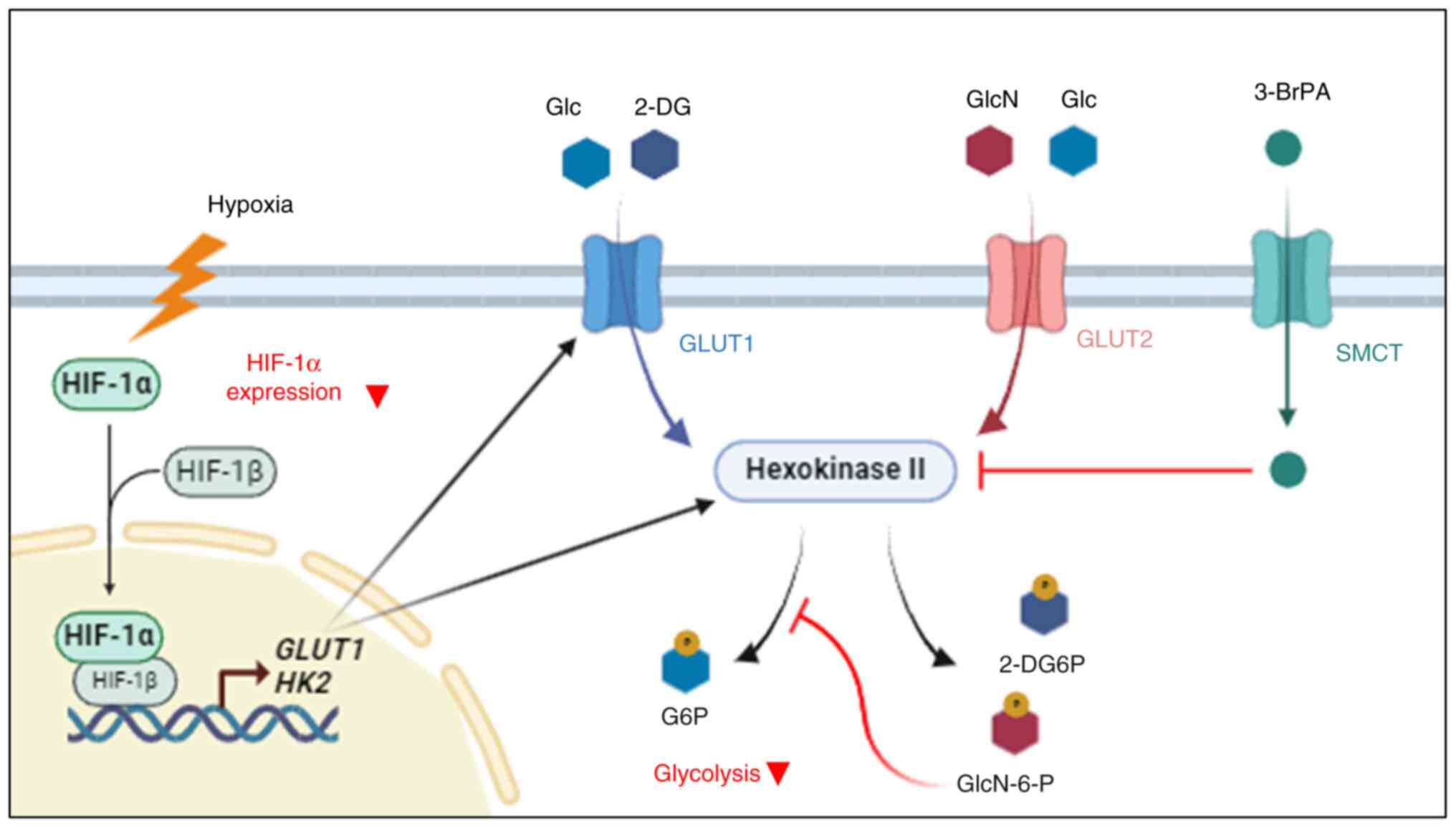
HIF-1α/HKII signaling is responsible
for hypoxia-induced glucosamine resistance
Glycolysis is mainly regulated by the quantity of
glucose taken up into cells and the activity of HK, a rate-limiting
step of the glycolysis pathway. Therefore, the expression level of
GLUTs and HKs is an important factor in the glycolysis process.
HIF-1α, one of the upstream transcriptional factors of GLUTs and
HKs, plays an important role in controlling glycolysis (19). Based on this knowledge, it was first
investigated whether the expression of GLUTs and HKs changes under
hypoxic conditions in A549 and HepG2 cells and whether this is
regulated by HIF-1α. As expected, the expression of GLUT1, HK1,
HK2 and HK4 markedly increased under hypoxic conditions
and it was confirmed that knocking down HIF-1 α markedly reduced
their expression again (Fig. 7A).
The expression of GLUT2 was constant in the examined
conditions and the expression of HK1 was induced by hypoxic
stress in HepG2 cells, but not in A549 cells. Next, the effects of
hypoxia on the antiproliferative effects of glucosamine was
examined. Notably, glucosamine reduced HIF-1α expression under
hypoxic conditions in cancer cells (Fig. S2) and it was found that
glucosamine-induced apoptosis was diminished under hypoxic
conditions and this hypoxia-induced glucosamine resistance were
restored by HIF-1α knockdown (Fig.
7B). Furthermore, cell cycle analysis found that the subG1
hypodiploid population, which was increased by glucosamine, was
reduced by hypoxic stress and this hypoxic stress-induced change
was not observed in HIF-1α depleted cells (Fig. 7C). Along with these results obtained
from flow cytometry analysis, MTT assays showed that the
anti-proliferation effects of glucosamine were reduced by hypoxic
stress and this hypoxia-induced resistance was markedly ablated by
HIF-1α downregulation in both A549 and HepG2 cells (Fig. 7D). Lastly, it was found that 3-BrPA,
HKII inhibitor, also effectively reversed hypoxia-induced
glucosamine resistance in both A549 and HepG2 cells (Fig. 7E). Collectively, these results
suggested that glucosamine exerts its antiproliferative activity by
inhibiting HKII within the cell and that the induction of HIF-1α
and transactivation of its target genes, particularly HKII, are
responsible for hypoxia-induced glucosamine resistance (Fig. 8).
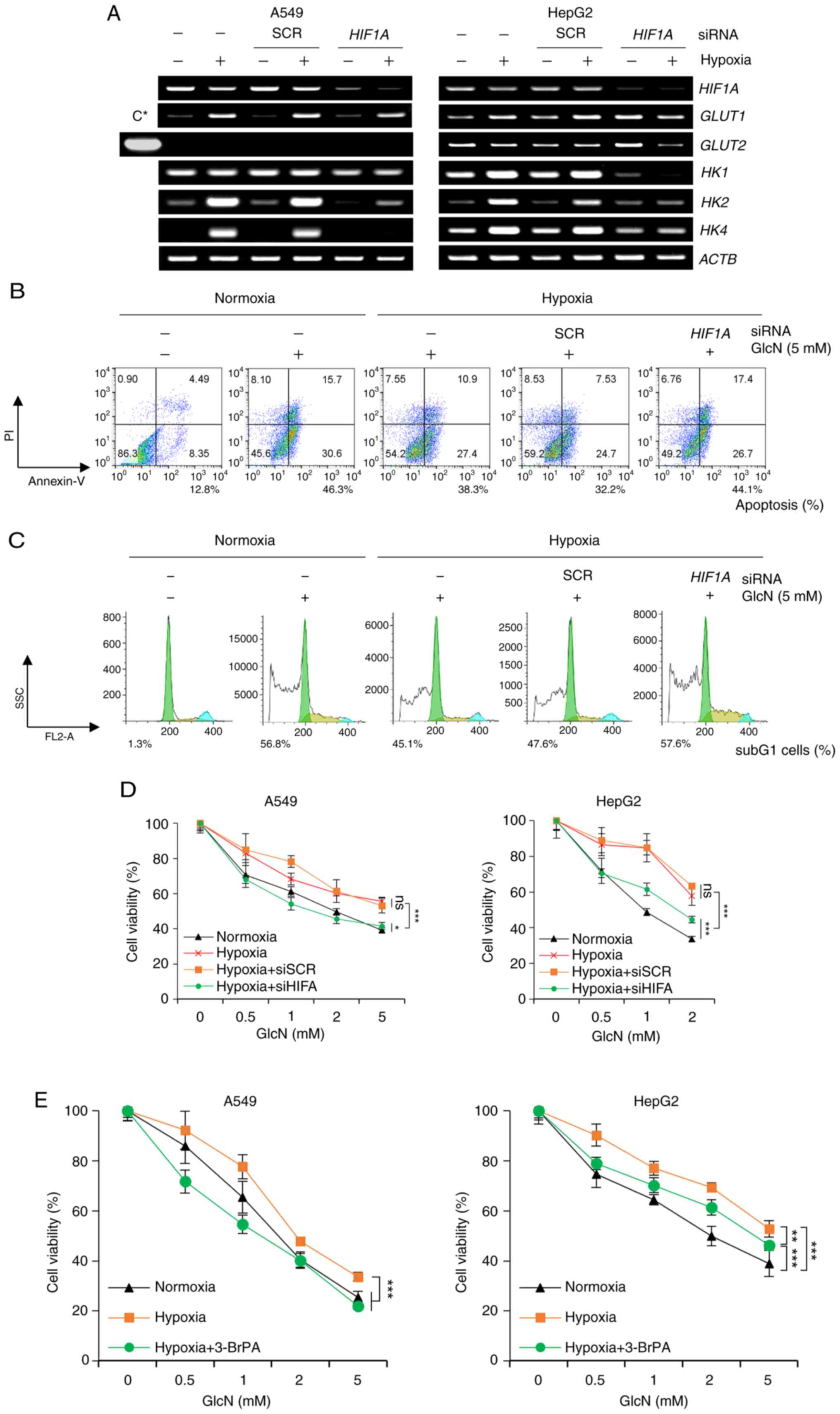 | Figure 7.Anticancer effects of GlcN are
regulated through changing of HK levels during hypoxia. (A)
Semiquantitative PCR of HIF1A, GLUT1, GLUT2 and HK isoforms
of A549 and HepG2 cells. Cells were transfected with siSCR or
siHIF1A and then cultured under normoxic (20% O2)
or hypoxic (1% O2) conditions for 24 h. HepG2 sample
(C*) in Fig. 3A was used as a
positive control for GLUT2 expression in the left panel.
ACTB served as a loading control. Flow cytometric analysis
of (B) apoptosis and (C) cell cycle in A549 cells. Cells were
transfected with siSCR or siHIF1A for 24 h and further
incubated under normoxic or hypoxic conditions for 24h followed by
glucosamine treatment for additional 48 h. Subsequently, cells were
stained with Annexin V-FITC and PI. Apoptosis (%) indicates sum of
Annexin V single positive proportion and Annexin V-PI double
positive proportion. Cell cycle analysis was performed with
linearized value of PI intensity and population of subG1 cells is
presented. (D) Cell viability of A549 and HepG2 cells. Cells were
transfected with siSCR or siHIF1A for 24 h and further
incubated under normoxic or hypoxic conditions for 24 h followed by
GlcN treatment for additional 48 h. Cell viability was measured by
MTT assay and results are presented as mean ± standard deviation.
(E) Cell viability of A549 and HepG2 cells. Cells were incubated
under normoxic or hypoxic conditions for 24 h followed by GlcN and
3-BrPA (10 or 20 µM for A549 or HepG2 cells, respectively)
treatment for additional 48 h. Cell viability was measured by MTT
assay and results are presented as mean ± standard deviation.
*P<0.05, **P<0.01, ***P<0.001, ns, no significance; GlcN,
glucosamine; HK, hexokinase; si, small interfering. |
Discussion
A number of cancer cells exhibit elevated glycolysis
even in the presence of oxygen, relying more heavily on this
process to produce ATP compared with normal cells. This phenomenon,
known as the Warburg effect, has become a key focus in cancer
treatment strategies, particularly through the development of
glycolytic inhibitors (15,23). Meanwhile, glycolysis is more than
just an energy-generating pathway; it plays a crucial role in
regulating various cellular processes necessary for cancer cell
growth and physiology (49,50). Hence, glycolysis inhibition has been
studied as an antiproliferative strategy. Although the inhibitory
effects of glucosamine on glycolysis and tumor growth were reported
in the late 1900s (27,32), the precise mechanisms of action of
glucosamine driving antiproliferative effects have not been fully
understood and thus, the present study investigated the
antiproliferative mechanism of glucosamine.
At first, to gain a broader understanding of the
mechanism behind glucosamine's antiproliferative effects,
microarray technology, a powerful tool for high-throughput
screening was employed. By using a whole-genome chip, the present
study aimed to identify changes in the gene expression profile of
A549 cells following the introduction of glucosamine. Genes
upregulated by glucosamine by more than two-fold were primarily
associated with pro-apoptosis and cell death, while those
downregulated by more than two-fold were mostly linked to cell
cycle regulation and DNA damage response. Additionally, glucosamine
was found to influence genes involved in carbohydrate metabolism,
biomolecule biosynthesis and glycosylation; processes related to
glycolysis. In particular, the result showing that genes related to
glycosylation were regulated by glucosamine is supported by
previous reports demonstrating that glucosamine induces abnormal
glycosylation of the IGF-1R prototype and COX-2 and eventually
decreases their molecular mass (9,35).
KEGG pathway analysis revealed that glucosamine downregulated the
overall glycolysis pathway, while some enzymes regulating the entry
into glycolysis were upregulated, possibly as a compensatory
response to glycolysis inhibition. This compensatory mechanism is
consistent with findings from previous studies using other
glycolysis inhibitors (51,52). Given these findings, along with
glucosamine's structural similarity to glucose and its previously
reported effect of reducing ATP levels in cancer cells, it is
suggested that glycolysis is the main target pathway of
glucosamine.
Glucosamine, structurally similar to glucose except
for its NH2 group, has been shown to compete with
glucose in carbohydrate metabolism (23). According to this, glucose
competitively inhibited glucosamine-induced antiproliferative
effects and restored glycolysis as indicated by the recovery of
lactate production that had been suppressed by glucosamine. Since
glucosamine is known to inhibit the IGF-1R/Akt pathway (9), it was found that increasing glucose
restored IGF-1R and p-Akt expression levels reduced by glucosamine.
The results supported that glucosamine inhibited the rate of
proliferation of cancer cell lines mainly by competing with glucose
during glycolysis.
Based on the structural similarity of glucosamine
and glucose, it was predicted that plasma membranes, especially
where glucose transports exist, would be the battlefield of
glucosamine and glucose. Among GLUT isoforms, expression levels in
liver cancer cell lines of GLUT2, which is responsible for
transportation of glucosamine as well (11), were much higher compared with other
cell lines, while GLUT1 expression levels were similar between
types of cancer. These GLUT2-rich liver cancer cell lines (HepG2
and Hep3B) consumed more glucosamine and were more sensitive to
glucosamine than other GLUT2-poor cell lines. In addition,
glucosamine consumption and the antiproliferative effects of
glucosamine were abrogated by inhibiting GLUT2 with its specific
inhibitor (Phloretin) or siGLUT2 transfection. Thus, so far,
it can be said that GLUT2 is the main gate for glucosamine to get
into cells and high dose of glucose can keep glucosamine from
entering into cells at GLUT2. Previous studies report that cancer
cells have higher GLUT2 expression levels compared with normal
cells (37,53). These facts suggested that the amount
of GLUT2 expression in each cancer cell can be a crucial biomarker
in determining the sensitivity against glucosamine and also proves
that glucosamine is more effective against cancer cells than normal
cells.
Glucosamine and glucose not only have a similar
structure but are also catalyzed by the same enzyme, HKs and are
converted to GlcN-6-phosphate and Glc-6-phosphate, respectively,
after transport into the cell (33). The lower the expression of HKII, the
greater the inhibitory effect of glucosamine on glycolysis.
Compared with the known glycolysis inhibitors, 3-BrPA and 2-DG, the
effective dose range of glucosamine was similar to 2-DG, not to
3-BrPA which is entered into cells by SMCT rather than GLUTs,
demonstrating that glucosamine inhibits glycolysis by competing
with glucose as does 2-DG, rather than by directly inhibiting HKs.
The synergistic effect observed when two drugs with different
mechanisms of action are combined suggests distinct pathways
(54). While the combination of
glucosamine and 3-BrPA showed enhanced antiproliferative effects,
the same was not observed with 2-DG. Considering these results and
the structure of glucosamine, it can be inferred that the mechanism
of action of glucosamine is probably different from that of 3-BrPA
and similar to that of 2-DG.
Various former studies reveal that HIF-1α regulates
the expression level of HKs and GLUT1 efficiently by working as a
transcription factor (17,19,55).
Furthermore, the increased stability of HIF-1α under hypoxic
conditions has been reported to influence sensitivity of types of
cancer to a number of anticancer agents (56,57).
In particular, studies have demonstrated that the antiproliferative
effect of 2-DG is decreased when there are high levels of HIF-1α
and that this phenomenon is related to increased levels of HKs,
which are induced by HIF-1α (21).
The present study showed that A549 cells, which have low basal
levels of HKII and lactate, inhibited glycolysis and decreased cell
proliferation when treated with glucosamine. Under hypoxic
conditions, GLUT2 expression levels were relatively constant
in HepG2 cells, suggesting that the uptake amount of glucosamine
would not be changed by HIF-1α activation. However, activated
HIF-1α induced HKII expressions in lung and liver cancer cell lines
and this activated HIF-1α/HKII pathway made cancer cell lines more
resistant to glucosamine. In addition, genetically depleting
HIF1A and pharmacologically inhibiting HKII markedly
restored glucosamine-induced antiproliferative effects.
In humans, cohort studies found that regularly
taking glucosamine as a supplement reduced the incidence and
mortality of colorectal cancer, lung cancer and kidney cancer and
the overall cancer mortality could be 6% lower compared with
non-users (58,59). We previously reported in vivo
anti-tumor effects of glucosamine using a xenograft mouse model
(9). Furthermore, the present
results supported the relationship between glucosamine intake and
lower cancer mortality with the molecular mechanism of
glucosamine-induced antiproliferative effects. Collectively,
glucosamine can act like a roadblock on the path of glycolysis,
competing with glucose for HK, slowing down the process like a
rival runner in a race for the same baton. Nevertheless, the
present study was limited by its exclusive use of cell line models,
which may not fully reflect in vivo conditions. Further
studies should focus on validating these findings in more
physiologically relevant models and exploring the broader
therapeutic potential of glucosamine under diverse biological
conditions. Such efforts could clarify its efficacy across
different tumor types and microenvironments, ultimately
strengthening its clinical applicability.
In conclusion, the present study indicated that
glucosamine reduced growth, decreased cell viability and impaired
glucose metabolism in the studied cancer cell lines. Moreover,
GLUT2 and HKII were identified as biomarkers determining
sensitivity to glucosamine. While the precise molecular mechanisms
underlying the antiproliferative effects of glucosamine remain to
be fully elucidated, the present study provided significant
insights by demonstrating its mode of action as a glucose
metabolism inhibitor in cancer cells. Furthermore, the findings
suggested potential therapeutic markers for identifying types of
cancer or characteristics that are most responsive to glucosamine,
thereby offering a rationale for its use in combination therapy.
These findings contributed to expanding the potential of
glucosamine as an effective anticancer agent.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the New Faculty Startup Fund
(grant no. 550-20240054) from Seoul National University (Seoul,
Korea).
Availability of data and materials
Microarray datasets generated in the present study
has been submitted to ArrayExpress (accession number:
E-MTAB-14871), and publicly accessible under this URL: https://www.ebi.ac.uk/biostudies/arrayexpress/studies/E-MTAB-14871.
Other data generated in the present study may be requested from the
corresponding author.
Authors' contributions
SP, KS and JK participated in the study design,
conducted the experiments, collected and analyzed the data, and
drafted the initial manuscript. JK and SO supervised the study,
contributed to the study design and data interpretation, and
critically revised the manuscript for important intellectual
content. All authors confirm the authenticity of all the raw data.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests. ViroCure Inc. had no commercial interest in this
study.
References
|
1
|
Sanders M and Grundmann O: The use of
glucosamine, devil's claw (Harpagophytum procumbens), and
acupuncture as complementary and alternative treatments for
osteoarthritis. Altern Med Rev. 16:228–238. 2011.PubMed/NCBI
|
|
2
|
Zhu X, Sang L, Wu D, Rong J and Jiang L:
Effectiveness and safety of glucosamine and chondroitin for the
treatment of osteoarthritis: A meta-analysis of randomized
controlled trials. J Orthop Surg Res. 13:1702018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang LS, Chen SJ, Zhang JF, Liu MN, Zheng
JH and Yao XD: Anti-proliferative potential of Glucosamine in renal
cancer cells via inducing cell cycle arrest at G0/G1 phase. BMC
Urol. 17:382017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zahedipour F, Dalirfardouei R, Karimi G
and Jamialahmadi K: Molecular mechanisms of anticancer effects of
Glucosamine. Biomed Pharmacother. 95:1051–1058. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kim DS, Park KS, Jeong KC, Lee BI, Lee CH
and Kim SY: Glucosamine is an effective chemo-sensitizer via
transglutaminase 2 inhibition. Cancer Lett. 273:243–249. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Oh HJ, Lee JS, Song DK, Shin DH, Jang BC,
Suh SI, Park JW, Suh MH and Baek WK: D-glucosamine inhibits
proliferation of human cancer cells through inhibition of p70S6K.
Biochem Biophys Res Commun. 360:840–845. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jo JR, Park YK and Jang BC: Short-term
treatment with glucosamine hydrochloride specifically downregulates
hypoxia-inducible factor-1α at the protein level in YD-8 human
tongue cancer cells. Int J Oncol. 44:1699–1706. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chou WY, Chuang KH, Sun D, Lee YH, Kao PH,
Lin YY, Wang HW and Wu YL: Inhibition of PKC-Induced COX-2 and IL-8
expression in human breast cancer cells by glucosamine. J Cell
Physiol. 230:2240–2251. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Song KH, Kang JH, Woo JK, Nam JS, Min HY,
Lee HY, Kim SY and Oh SH: The novel IGF-IR/Akt-dependent anticancer
activities of glucosamine. BMC Cancer. 14:312014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cura AJ and Carruthers A: Role of
monosaccharide transport proteins in carbohydrate assimilation,
distribution, metabolism, and homeostasis. Compr Physiol.
2:863–914. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Uldry M, Ibberson M, Hosokawa M and
Thorens B: GLUT2 is a high affinity glucosamine transporter. FEBS
Lett. 524:199–203. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pouyssegur J, Marchiq I, Parks SK,
Durivault J, Zdralevic M and Vucetic M: ‘Warburg effect’ controls
tumor growth, bacterial, viral infections and immunity-Genetic
deconstruction and therapeutic perspectives. Semin Cancer Biol.
86((Pt 2)): 334–346. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Barron CC, Bilan PJ, Tsakiridis T and
Tsiani E: Facilitative glucose transporters: Implications for
cancer detection, prognosis and treatment. Metabolism. 65:124–139.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ancey PB, Contat C and Meylan E: Glucose
transporters in cancer - from tumor cells to the tumor
microenvironment. FEBS J. 285:2926–2943. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pelicano H, Martin DS, Xu RH and Huang P:
Glycolysis inhibition for anticancer treatment. Oncogene.
25:4633–4646. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wilson JE: Isozymes of mammalian
hexokinase: Structure, subcellular localization and metabolic
function. J Exp Biol. 206((Pt 12)): 2049–2057. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Guo D, Meng Y, Jiang X and Lu Z:
Hexokinases in cancer and other pathologies. Cell Insight.
2:1000772023. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Paredes F, Williams HC and San Martin A:
Metabolic adaptation in hypoxia and cancer. Cancer Lett.
502:133–142. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Denko NC: Hypoxia, HIF1 and glucose
metabolism in the solid tumour. Nat Rev Cancer. 8:705–713. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Iommarini L, Porcelli AM, Gasparre G and
Kurelac I: Non-canonical mechanisms regulating hypoxia-inducible
factor 1 alpha in cancer. Front Oncol. 7:2862017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Maher JC, Wangpaichitr M, Savaraj N,
Kurtoglu M and Lampidis TJ: Hypoxia-inducible factor-1 confers
resistance to the glycolytic inhibitor 2-deoxy-D-glucose. Mol
Cancer Ther. 6:732–741. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hsu CC, Tseng LM and Lee HC: Role of
mitochondrial dysfunction in cancer progression. Exp Biol Med
(Maywood). 241:1281–1295. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang Y, Li Q, Huang Z, Li B, Nice EC,
Huang C, Wei L and Zou B: Targeting glucose metabolism enzymes in
cancer treatment: current and emerging strategies. Cancers (Basel).
14:45682022. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
R Core Team, . R: A language and
environment for statistical computing. R Foundation for Statistical
Computing; Vienna: 2024, http://www.R-project.org/
|
|
26
|
Luo W and Brouwer C: Pathview: An
R/Bioconductor package for pathway-based data integration and
visualization. Bioinformatics. 29:1830–1831. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Quastel JH and Cantero A: Inhibition of
tumour growth by D-glucosamine. Nature. 171:252–254. 1953.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tamayo B, Kercher K, Vosburg C, Massimino
C, Jernigan MR, Hasan DL, Harper D, Mathew A, Adkins S, Shippy T,
et al: Annotation of glycolysis, gluconeogenesis, and
trehaloneogenesis pathways provide insight into carbohydrate
metabolism in the Asian citrus psyllid. GigaByte.
2022:gigabyte412022. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Suginohara T, Wakabayashi K, Ato S and
Ogasawara R: Effect of 2-deoxyglucose-mediated inhibition of
glycolysis on the regulation of mTOR signaling and protein
synthesis before and after high-intensity muscle contraction.
Metabolism. 114:1544192021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Berthe A, Zaffino M, Muller C, Foulquier
F, Houdou M, Schulz C, Bost F, De Fay E, Mazerbourg S and Flament
S: Protein N-glycosylation alteration and glycolysis inhibition
both contribute to the antiproliferative action of 2-deoxyglucose
in breast cancer cells. Breast Cancer Res Treat. 171:581–591. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bertoni JM and Weintraub ST: Competitive
inhibition of human brain hexokinase by metrizamide and related
compounds. J Neurochem. 42:513–518. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tesoriere G, Vento R and Calvaruso G:
Inhibitory effect of D-glucosamine on glycolysis in bovine retina.
Biochim Biophys Acta. 385:58–67. 1975. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Marshall S, Yamasaki K and Okuyama R:
Glucosamine induces rapid desensitization of glucose transport in
isolated adipocytes by increasing GlcN-6-P levels. Biochem Biophys
Res Commun. 329:1155–1161. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Samizu M and Iida K: Glucosamine inhibits
the proliferation of hepatocellular carcinoma cells by eliciting
apoptosis, autophagy, and the anti-warburg effect. Scientifica
(Cairo). 2025:56858842025. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jang BC, Sung SH, Park JG, Park JW, Bae
JH, Shin DH, Park GY, Han SB and Suh SI: Glucosamine hydrochloride
specifically inhibits COX-2 by preventing COX-2 N-glycosylation and
by increasing COX-2 protein turnover in a proteasome-dependent
manner. J Biol Chem. 282:27622–27632. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kapoor K, Finer-Moore JS, Pedersen BP,
Caboni L, Waight A, Hillig RC, Bringmann P, Heisler I, Müller T,
Siebeneicher H and Stroud RM: Mechanism of inhibition of human
glucose transporter GLUT1 is conserved between cytochalasin B and
phenylalanine amides. Proc Natl Acad Sci USA. 113:4711–4716. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wu CH, Ho YS, Tsai CY, Wang YJ, Tseng H,
Wei PL, Lee CH, Liu RS and Lin SY: In vitro and in vivo study of
phloretin-induced apoptosis in human liver cancer cells involving
inhibition of type II glucose transporter. Int J Cancer.
124:2210–2219. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kapoor M, Mineau F, Fahmi H, Pelletier JP
and Martel-Pelletier J: Glucosamine sulfate reduces prostaglandin
E(2) production in osteoarthritic chondrocytes through inhibition
of microsomal PGE synthase-1. J Rheumatol. 39:635–644. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sols A, De La Fuente G, Villarpalasi C and
Asensio C: Substrate specificity and some other properties of
baker's yeast hexokinase. Biochim Biophys Acta. 30:92–101. 1958.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Brown DH: The phosphorylation of D (+)
glucosamine by crystalline yeast hexokinase. Biochim Biophys Acta.
7:487–493. 1951. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kuser PR, Krauchenco S, Antunes OA and
Polikarpov I: The high resolution crystal structure of yeast
hexokinase PII with the correct primary sequence provides new
insights into its mechanism of action. J Biol Chem.
275:20814–20821. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chen YH and Cheng WH: Hexosamine
biosynthesis and related pathways, protein N-glycosylation and
O-GlcNAcylation: Their interconnection and role in plants. Front
Plant Sci. 15:13490642024. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Paneque A, Fortus H, Zheng J, Werlen G and
Jacinto E: The hexosamine biosynthesis pathway: Regulation and
function. Genes (Basel). 14:9932023. View Article : Google Scholar
|
|
44
|
Salazar J, Bello L, Chavez M, Anez R,
Rojas J and Bermudez V: Glucosamine for osteoarthritis: Biological
effects, clinical efficacy, and safety on glucose metabolism.
Arthritis. 2014:4324632014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Robey RB and Hay N: Mitochondrial
hexokinases, novel mediators of the antiapoptotic effects of growth
factors and Akt. Oncogene. 25:4683–4696. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Xian H and Wang Y, Bao X, Zhang H, Wei F,
Song Y and Wang Y, Wei Y and Wang Y: Hexokinase inhibitor
2-deoxyglucose coordinates citrullination of vimentin and apoptosis
of fibroblast-like synoviocytes by inhibiting HK2/mTORC1-induced
autophagy. Int Immunopharmacol. 114:1095562023. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Rai Y, Yadav P, Kumari N, Kalra N and
Bhatt AN: Hexokinase II inhibition by 3-bromopyruvate sensitizes
myeloid leukemic cells K-562 to anti-leukemic drug, daunorubicin.
Biosci Rep. 39:BSR201908802019. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Thangaraju M, Karunakaran SK, Itagaki S,
Gopal E, Elangovan S, Prasad PD and Ganapathy V: Transport by
SLC5A8 with subsequent inhibition of histone deacetylase 1 (HDAC1)
and HDAC3 underlies the antitumor activity of 3-bromopyruvate.
Cancer. 115:4655–4666. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Paul S, Ghosh S and Kumar S: Tumor
glycolysis, an essential sweet tooth of tumor cells. Semin Cancer
Biol. 86((Pt 3)): 1216–1230. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Bose S, Zhang C and Le A: Glucose
metabolism in cancer: The warburg effect and beyond. Adv Exp Med
Biol. 1311:3–15. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhao J, Ma Y, Zhang Y, Fu B, Wu X, Li Q,
Cai G, Chen X and Bai XY: Low-dose 2-deoxyglucose and metformin
synergically inhibit proliferation of human polycystic kidney cells
by modulating glucose metabolism. Cell Death Discov. 5:762019.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wang Z, Zhang L, Zhang D, Sun R, Wang Q
and Liu X: Glycolysis inhibitor 2-deoxy-D-glucose suppresses
carcinogen-induced rat hepatocarcinogenesis by restricting cancer
cell metabolism. Mol Med Rep. 11:1917–1924. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Medina RA and Owen GI: Glucose
transporters: Expression, regulation and cancer. Biol Res. 35:9–26.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Lehar J, Krueger AS, Avery W, Heilbut AM,
Johansen LM, Price ER, Rickles RJ, Short GF III, Staunton JE, Jin
X, et al: Synergistic drug combinations tend to improve
therapeutically relevant selectivity. Nat Biotechnol. 27:659–666.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Sadlecki P, Bodnar M, Grabiec M, Marszalek
A, Walentowicz P, Sokup A, Zegarska J and Walentowicz-Sadlecka M:
The role of Hypoxia-inducible factor-1 alpha, glucose
transporter-1, (GLUT-1) and carbon anhydrase IX in endometrial
cancer patients. Biomed Res Int. 2014:6168502014. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Wang P, Wan W, Xiong S, Wang J, Zou D, Lan
C, Yu S, Liao B, Feng H and Wu N: HIF1α regulates glioma
chemosensitivity through the transformation between differentiation
and dedifferentiation in various oxygen levels. Sci Rep.
7:79652017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Sharma A, Sinha S and Shrivastava N:
Therapeutic targeting hypoxia-inducible factor (HIF-1) in cancer:
Cutting gordian knot of cancer cell metabolism. Front Genet.
13:8490402022. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zhou J, Wu Z, Lin Z, Wang W, Wan R and Liu
T: Association between glucosamine use and cancer mortality: A
large prospective cohort study. Front Nutr. 9:9478182022.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Li ZH, Gao X, Chung VC, Zhong WF, Fu Q, Lv
YB, Wang ZH, Shen D, Zhang XR, Zhang PD, et al: Associations of
regular glucosamine use with all-cause and cause-specific
mortality: A large prospective cohort study. Ann Rheum Dis.
79:829–836. 2020. View Article : Google Scholar : PubMed/NCBI
|