Cervical cancer (CC) is the fourth most frequently
diagnosed cancer among women worldwide, accounting for ~266,000
deaths and 528,000 new cases each year (1). CC contributes to 7.5% of all female
cancer-related deaths, establishing itself as a notable cause of
mortality among women (2).
Well-structured screening programs that incorporate affordable
treatment techniques, such as cryotherapy, loop electrosurgical
excision and thermal ablation, can facilitate the early detection
and management of precancerous lesions, thereby reducing the
overall disease burden (3).
Nevertheless, in low- and middle-income countries,
late-stage CC remains prevalent due to limited access to early
diagnostic services and preventive care. Although chemotherapy and
radiotherapy continue to serve as the standard treatments for CC,
their overall effectiveness remains limited, with a number of
patients exhibiting suboptimal responses (4). Notably, targeted therapy and
immunotherapy have gained recognition as promising treatment
strategies. The approval of PD-1 inhibitors for the management of
recurrent or metastatic CC has enhanced host immune responses
against human papillomavirus (HPV)-positive malignancies (5). However, despite their potential,
several challenges persist, including the immunosuppressive tumor
microenvironment, tumor immune evasion, low patient response rates
and acquired drug resistance, all of which compromise treatment
(6). In the future, treatment
paradigms are expected to emphasize personalized medicine by
refining targeted therapies to improve cure rates and extend
patient survival.
Eukaryotic chromatin is primarily composed of
histones and nucleosomes, with nucleosomes consisting of DNA
wrapped around histone proteins. The tail regions of histones
frequently undergo a range of post-translational modifications
(7), collectively referred to as
histone modifications. These modifications influence gene
transcription through multiple pathways (8) and are essential for several key
biological processes. Common histone modifications include
acetylation, methylation and ubiquitination, with acetylation and
methylation recognized as the predominant types (9). Recently, additional modifications,
such as glycosylation, lactylation and palmitoylation, have been
identified, further increasing the complexity of histone regulation
(10–13). These modifications are vital for
regulating gene expression, repairing DNA damage, and controlling
the cell cycle, differentiation, apoptosis and tumor progression
(14,15).
Histone modifications may act cooperatively or
antagonistically, contributing to the formation of a ‘histone code’
that fine-tunes cellular functions (16,17).
Disruptions in histone modification patterns are closely associated
with various diseases, including cancer, neurodegenerative
disorders and immune dysfunction, in which these alterations in
gene expression contribute to disease progression (18,19).
As a result, epigenetic therapies targeting histone modifications
have emerged as a critical area of research in precision
medicine.
In CC, histone modifications, an essential component
of epigenetic regulation, affect key processes such as persistent
HPV infection, dysregulation of the cell cycle, immune evasion and
treatment resistance. These pathological features arise from
chromatin remodeling and aberrant gene expression (20). Epigenetic therapies aimed at
correcting abnormal histone modifications, including histone
deacetylase (HDAC) inhibitors, are increasingly being recognized as
potential treatments for CC (20).
Therefore, a comprehensive analysis of the role of histone
modifications in CC and their clinical implications may offer
valuable insights into precision treatment strategies.
CC ranks as the fourth most frequently diagnosed
cancer among women worldwide, with ~660,000 new cases reported in
2022 (21). The World Health
Organization estimates that CC affects 662,000 individuals
globally, including 151,000 women in China (22). Despite a substantial reduction in
incidence over the past three decades, CC remains a major global
public health concern, with considerable regional disparities in
both incidence and mortality rates (23,24).
In high-income countries, incidence and mortality rates are notably
lower than those in developing regions, largely due to widespread
HPV vaccination, comprehensive screening programs and greater
access to healthcare resources (25–27).
Nevertheless, in the later stages of CC, mortality rates in
developed nations can be elevated or comparable to those in
developing countries, primarily due to the limited availability of
effective treatments for advanced disease (28).
CC can be classified into two primary histological
types: Cervical squamous cell carcinoma (CSCC), which arises from
squamous cells; and cervical adenocarcinoma, which originates from
glandular epithelial cells (29).
CSCC accounts for >80% of CC cases worldwide and is associated
with high incidence and mortality rates (25). This cancer progresses through a
series of well-defined precancerous stages, culminating in cellular
dysplasia, cervical intraepithelial neoplasia, and ultimately,
squamous cell carcinoma (30).
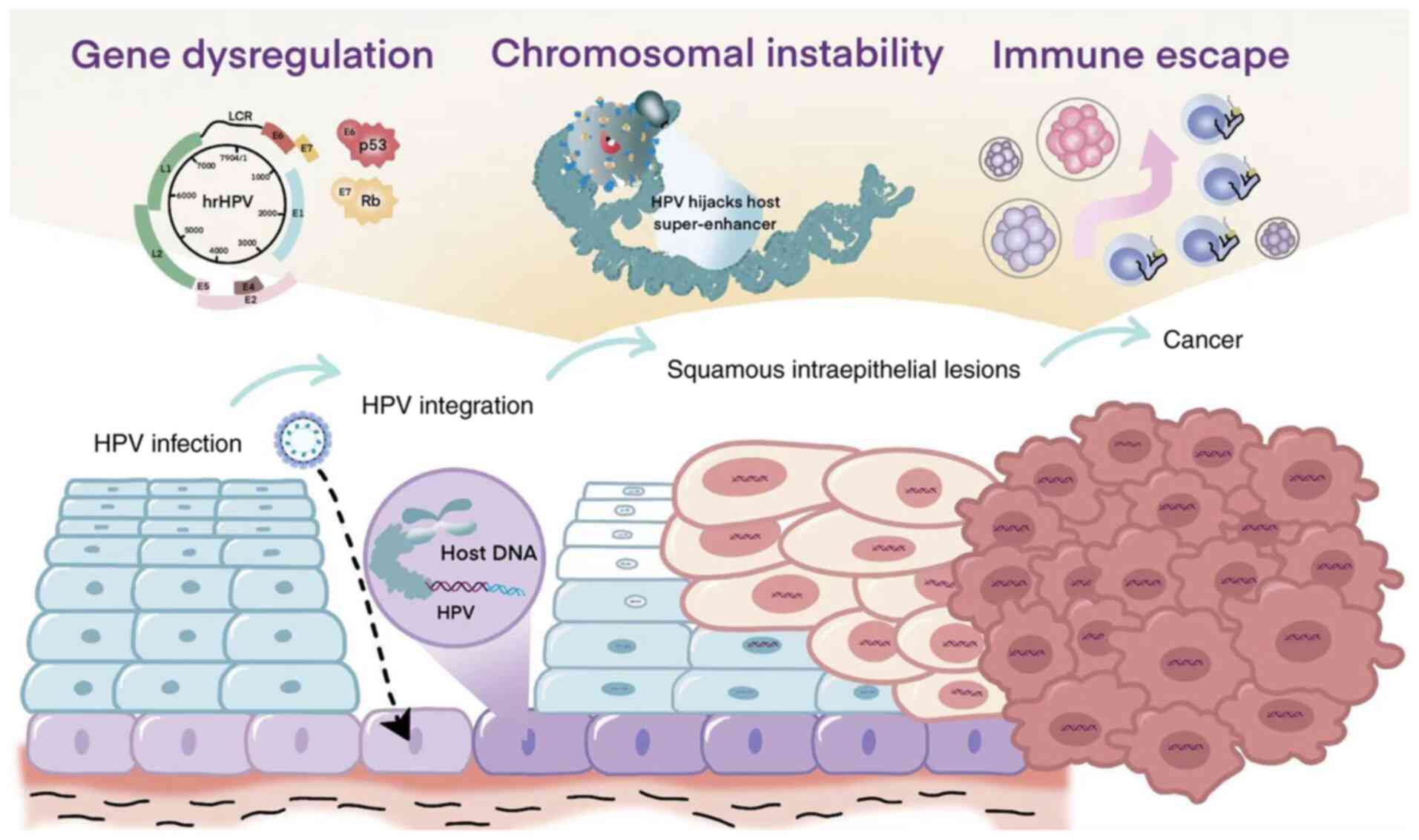
All HPV types, regardless of risk classification,
infect epithelial cells, particularly keratinocytes, leading to
cellular immortalization (30). The
HPV genome consists of three primary functional regions: The early
(E) region, which encodes proteins E1-E7 that are necessary for
viral replication; the late (L) region, which produces structural
proteins L1 and L2 that are essential for virion assembly; and the
long control region, a non-coding segment that contains
cis-regulatory elements critical for DNA replication and
transcription (33). E1 and E2 are
essential for viral DNA replication and act as transcriptional
regulators. The viral genome also encodes early genes, such as E5,
E6 and E7, which contribute to oncogenic transformation. E5
enhances immune evasion, reduces dependence on growth factors and
promotes cellular proliferation (34). E6 interacts with the tumor
suppressor protein p53, leading to its degradation (35). E7 binds to retinoblastoma protein,
mediating its degradation via the ubiquitin-proteasome system and
promoting cell cycle progression (36).
During the later phases of infection, viral late
genes become active within the suprabasal layers, initiating
circular genome replication and the synthesis of structural
proteins. As the virus ascends to the outer layers of the skin or
mucosal surfaces, mature viral particles are assembled and
subsequently released. Additionally, HPV alters gene expression and
activates several signaling pathways, such as those mediated by
growth factor receptors, Notch, RAS and PI3K/Akt/mTOR, all of which
promote host cell survival and proliferation. These molecular
alterations collectively contribute to cervical carcinogenesis
(37–39) (Fig.
1).
Given the pivotal role of HPV in the pathogenesis of
CC, prevention strategies targeting the virus have proven highly
effective. Specifically, vaccination against high-risk HPV types
has led to a notable reduction in CC incidence, particularly in
developed countries (40). In
addition, surgery, radiotherapy and chemotherapy are the standard
treatments for locally advanced cervical cancer and have notably
reduced the mortality rate of patients with CC; however, their
effectiveness in treating more advanced or recurrent disease
remains limited (41–43).
Advancements in immunotherapy and targeted therapy
have demonstrated considerable promise in cancer treatment.
Inhibitors of PD-1/PD-L1, such as nivolumab and pembrolizumab,
function by disrupting immune checkpoint pathways, thereby
enhancing the ability of the immune system to detect and eliminate
cancer cells (44). Additionally,
the use of CTLA-4 inhibitors, such as ipilimumab, has been explored
as an immune checkpoint-based therapy for CC (45). Emerging evidence has indicated that
these therapies exhibit substantial efficacy, particularly when
combined with chemotherapy or radiotherapy, in patients with
advanced or recurrent disease (46,47).
Although these therapies hold considerable promise,
several challenges persist. Immunotherapy can induce immune-related
adverse events, such as autoimmune disorders and severe
inflammation, and its efficacy is often reduced in tumors
exhibiting strong immune tolerance (48–50).
Similarly, although targeted therapies are designed to act on
specific molecular targets, their effectiveness is limited by tumor
heterogeneity and the development of drug resistance, thereby
reducing their long-term efficacy (48–50).
Moreover, the success of both therapeutic approaches is influenced
by individual patient factors and the complexity of the tumor
microenvironment (48–50). As a result, future therapeutic
strategies should emphasize personalized medicine and the continued
development of targeted therapies, with the aim of improving cure
rates and extending patient survival.
Histone modifications serve a critical role in
regulating biological processes by altering chromatin structure,
thereby influencing the expression of specific genes. Although most
research has focused on well-characterized modifications, such as
acetylation, methylation and phosphorylation, histones also undergo
a variety of other modifications (51). These include citrullination,
ubiquitination, ADP-ribosylation and O-GlcNAcylation, as well as
less-studied modifications such as propionylation, butyrylation,
crotonylation, lactylation and palmitoylation (51). These diverse modifications affect
chromatin structure and gene expression, contributing to critical
processes such as the cell cycle, DNA repair, differentiation and
immune regulation. They also serve a role in the pathogenesis of
diseases, including cancer (including CC), neurodegenerative
disorders (such as Alzheimer's disease) and metabolic conditions
(including diabetes mellitus) (52–54).
The regulation of histone modifications is mediated
by ‘writers’ and ‘erasers’, enzymes that add or remove
modifications, thereby ensuring appropriate gene expression
(55). For example, histone
acetyltransferases (HATs) introduce acetyl groups, leading to
chromatin relaxation and enhanced transcription, whereas HDACs
remove acetyl groups, resulting in chromatin condensation and
transcriptional repression (56).
Similarly, histone methylation is catalyzed by histone
methyltransferases (HMTs), whereas histone demethylases are
responsible for removing methyl groups. The coordinated activity of
these enzymes modulates chromatin architecture and is essential for
the regulation of gene expression (57). Notably, certain ‘writers’ and
‘erasers’ are capable of regulating multiple types of histone
modifications. For example, G9a, a HMT, primarily catalyzes the
methylation of lysine 9 on histone H3 (H3K9), but also modulates
histone acetylation, thereby influencing gene silencing and
chromatin structure (58).
Likewise, p300/CBP, a HAT, facilitates histone acetylation and
transcriptional activation (59),
and has also been shown to catalyze histone lactylation, a
modification particularly relevant under conditions of altered
cellular metabolism, such as hypoxia or inflammation (60). Similarly, HDACs primarily remove
acetyl groups, leading to chromatin condensation and
transcriptional repression; however, certain HDAC family members,
such as sirtuin (SIRT)1, serve pivotal roles not only in histone
deacetylation but also in regulating non-histone proteins, such as
p53, thereby modulating apoptosis and DNA repair (61,62).
In addition, lysine-specific demethylase (KDM)1, an ‘eraser’, can
remove methyl groups from lysine 4 on histone H3 (H3K4) to suppress
gene expression and from H3K9 to promote gene activation (54,63).
The cross-regulation by these multifunctional enzymes indicates
that histone modification is not a linear process but rather a
highly dynamic and interactive network that enables precise control
of gene expression, allowing cells to adapt to environmental
changes and maintain gene expression homeostasis. A comprehensive
summary of the major histone modifications, including ‘writer’ and
‘eraser’ proteins, their family classifications, target sites and
functional roles, is presented in Table
I.
Histone modifications are fundamental to the
initiation and progression of cancer, primarily through their
effects on chromatin structure and gene expression. Consequently,
these modifications influence the transcription of genes involved
in tumorigenesis. Aberrant histone modifications, including
acetylation, methylation, phosphorylation and ubiquitination, can
lead to the silencing of tumor suppressor genes or the activation
of oncogenes, thereby facilitating tumor progression. For example,
trimethylation of lysine 27 on histone H3 (H3K27me3), a repressive
histone mark mediated by EZH2, is frequently observed in breast
cancer and hepatocellular carcinoma (HCC), leading to the silencing
of key tumor suppressor genes (64,65).
For example, EZH2 promotes the stemness of HCC by inducing the
transcriptional repression of the tumor suppressor gene TOP2A
through H3K27me3-mediated silencing (65). Similarly, elevated levels of histone
H3 lysine 9 acetylation (H3K9ac) and H3 lysine 27 acetylation have
been associated with oncogene activation, contributing to the
progression of colorectal and lung cancer (66). Moreover, histone modifications
influence DNA damage repair pathways; for example, phosphorylation
of H2AX is activated in response to DNA damage, and affect the
tumor microenvironment, such as HDAC-driven immune suppression
(67). They are also associated
with resistance mechanisms; for example, KDM5A-mediated
demethylation enhances chemotherapy resistance (68). As a result, epigenetic therapies
targeting histone modifications, including inhibitors of HDACs and
HMTs/KDMs, hold notable promise for precision cancer treatment,
particularly when used in combination with immunotherapy and
molecular targeted therapies.
Histone modifications serve a critical role in
regulating the initiation, progression and therapeutic response of
CC. Modifications such as acetylation, methylation, phosphorylation
and ubiquitination are central to modulating gene expression,
altering chromatin architecture and influencing cellular signaling
networks, all of which contribute to cancer development. Aberrant
histone modifications can lead to the suppression of tumor
suppressor genes or the activation of oncogenes, thereby promoting
cellular processes, such as proliferation, invasion and immune
evasion. Furthermore, histone modifications are closely associated
with the development of resistance to therapies, including
chemotherapy, radiotherapy and targeted treatments. This section
examines the role of histone modifications in the pathophysiology
of CC.
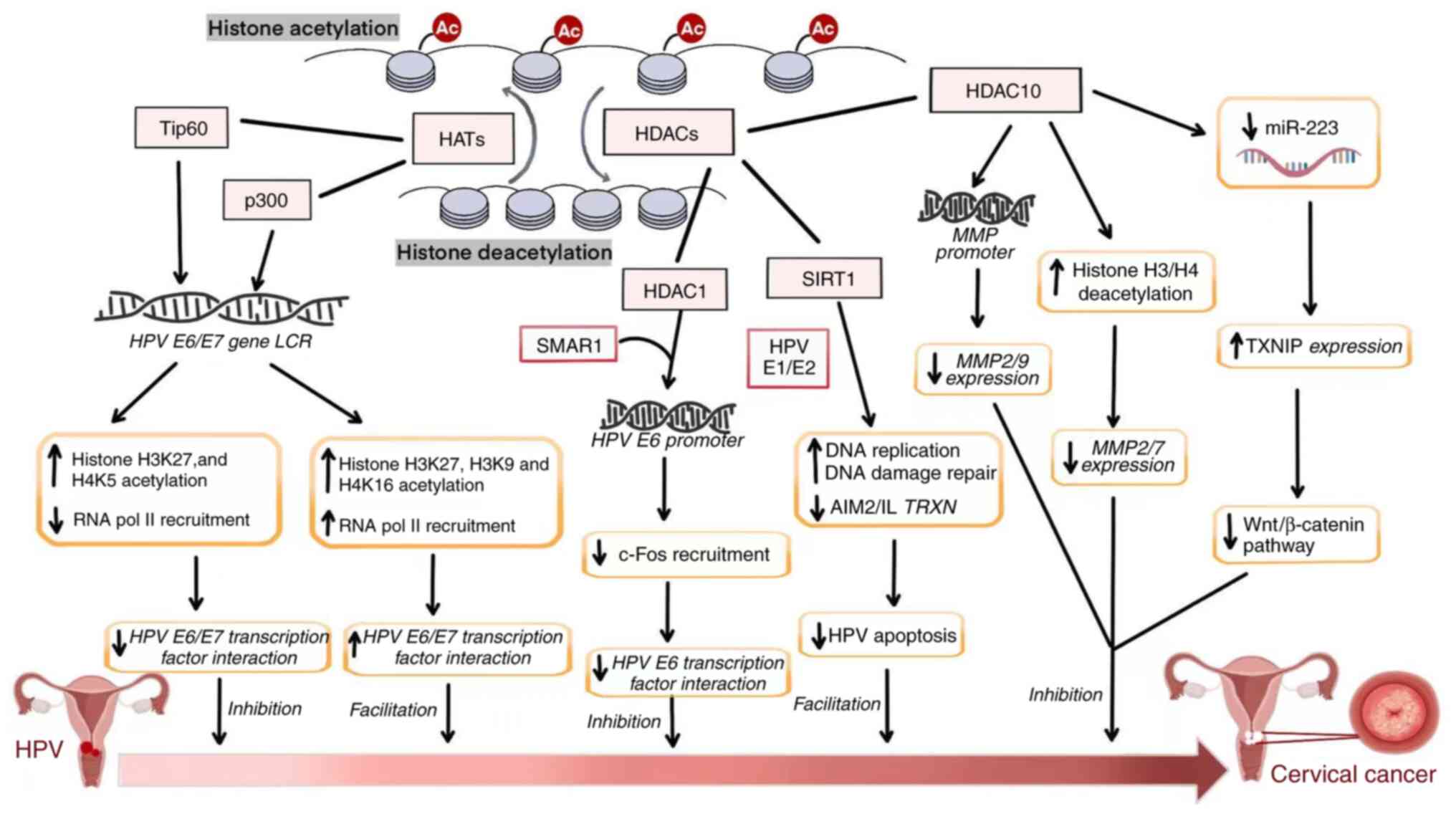
Histone acetylation, regulated by HATs and HDACs, is
a reversible and dynamic process. HATs modify nucleosome structure
by promoting chromatin relaxation, thereby facilitating gene
transcription (Fig. 2). More than
20 HAT proteins have been identified, with key members belonging to
the GNAT, MYST and p300/CBP families (69). Yang et al (70) demonstrated that the HAT CSRP2BP
markedly promotes epithelial-mesenchymal transition (EMT) and
metastasis in CC cells by activating N-cadherin. Notably, in CC
tissues, elevated CSRP2BP expression was observed and revealed to
be associated with poor prognosis. Furthermore, overexpression of
CSRP2BP enhanced the proliferation and metastasis of CC cells in
both in vitro and in vivo models, whereas its
silencing had the opposite effect. CSRP2BP was also identified as a
key contributor to cisplatin resistance. At the molecular level,
CSRP2BP was revealed to catalyze acetylation of histone H4 at
lysine residues 5 and 12, to form a complex with the transcription
factor SMAD4 and to bind to the SEB2 sequence in the promoter
region of N-cadherin, thereby upregulating its transcription. This
mechanism may promote EMT and enhance metastasis in CC cells
(70). These findings underscore
the essential role of histone acetylation in the initiation,
progression and development of drug resistance in CC. Similarly,
the acetyltransferase p300 catalyzes acetylation of histone H3 at
lysine 27 (H3K27), which enhances the activity of the NDUFA8
promoter, thereby stimulating CC cell proliferation (71).
Histone methylation is a dynamic and reversible
process regulated by HMTs and KDMs. Methylation occurs through the
activity of HMTs, which add methyl groups to specific histone
residues, thereby modifying chromatin structure and function. These
modifications are typically associated with either gene activation
or repression, depending on the specific site and context (88–90).
Numerous HMTs, including members of the SET, DOT1L and SUV39H
families, are involved in regulating gene expression, DNA repair
mechanisms and cell cycle progression (88–90).
Zhang et al (91)
demonstrated that HPV18 E6/E7 increases the transcriptional
activity of EZH2, resulting in elevated H3K27me3 levels in CC.
Furthermore, Beyer et al showed that histone modifications,
such as H3K9ac and trimethylation of lysine 4 on histone H3 are
closely associated with clinicopathological parameters and 10-year
survival outcomes, underscoring their prognostic value in CC
(92). Chen et al (93) reported that NSD2, a HMT, promotes
proliferation, migration and invasion of CC cells by activating the
endothelial nitric oxide synthase and AKT/MMP-2 signaling pathways.
Additionally, Ansari et al (94) identified the H3K4-specific
methyltransferase MLL as a critical factor in cervical tumor
growth. Knockdown of MLL reduced the expression of several key
growth and angiogenesis-related factors, including HIF1α, VEGF and
CD31, thereby inhibiting CC progression. Notably, studies have
shown interest in histone methylation enzymes as potential
therapeutic targets. Zhang et al (95) reported that SUV39H1 upregulates
DNMT3A expression in CC cells via trimethylation of lysine 9 on
histone H3, while simultaneously downregulating immunosuppressive
factors. such as Tim-3 and galectin-9. This activity may improve
the tumor immune microenvironment and enhance therapeutic efficacy.
Moreover, Osawa et al (96)
showed that the histone demethylase JHDM1D suppresses
angiogenesis-related factors, including VEGF-B and angiopoietin,
under conditions of nutrient deprivation, thereby limiting
angiogenesis and exerting tumor-suppressive effects.
In summary, studies have identified the critical
roles of histone methylation and demethylation in tumor
progression, immune regulation, cell invasion and angiogenesis, all
of which markedly contribute to CC development.
The regulation of histone phosphorylation involves
histone kinases (HKs) and phosphatases (PPs), with both enzymatic
processes being dynamic and reversible. Histone phosphorylation,
catalyzed by HKs, modifies chromatin structure and function,
typically influencing gene transcription by either activating or
repressing it (97,98). Several HKs, including members of the
Aurora, CDK and MSK families, serve essential roles in gene
expression, DNA repair, cell cycle progression and signal
transduction (97,98). In CC cells, phosphorylation of
histone H2AX serves as a key indicator of DNA damage, reflecting
cellular sensitivity to radiation and the efficiency of DNA repair
mechanisms; this makes H2AX phosphorylation a promising biomarker
for evaluating the therapeutic response to radiotherapy in CC
(99). Zhao et al (100) also reported that alterations in
H2AX phosphorylation levels before and after neoadjuvant
chemotherapy provide valuable insights for assessing treatment
response in patients with CC. Several additional studies have
investigated histone phosphorylation as a potential biomarker for
prognosis and treatment efficacy in CC (101–103).
Preclinical studies have further suggested that
targeting PPs or their upstream kinases can effectively inhibit
tumor growth. For example, Zhang and Zhang (104) demonstrated that ZM447439, a potent
Aurora kinase B inhibitor, suppresses the proliferation of SiHa CC
cells while enhancing cisplatin sensitivity. Similarly, Cheung
et al (105) reported that
BPR1K653, a novel Aurora kinase inhibitor, exhibits strong
antiproliferative effects in multidrug-resistant cancer cells
mediated by MDR1 (P-gp170).
In conclusion, although histone phosphorylation has
been extensively studied in various tumors, research specifically
focusing on CC remains limited. Most studies have investigated
phosphorylated histones as potential biomarkers or examined HKs
that suppress CC progression. However, these investigations are
still relatively preliminary and lack comprehensive mechanistic
insights.
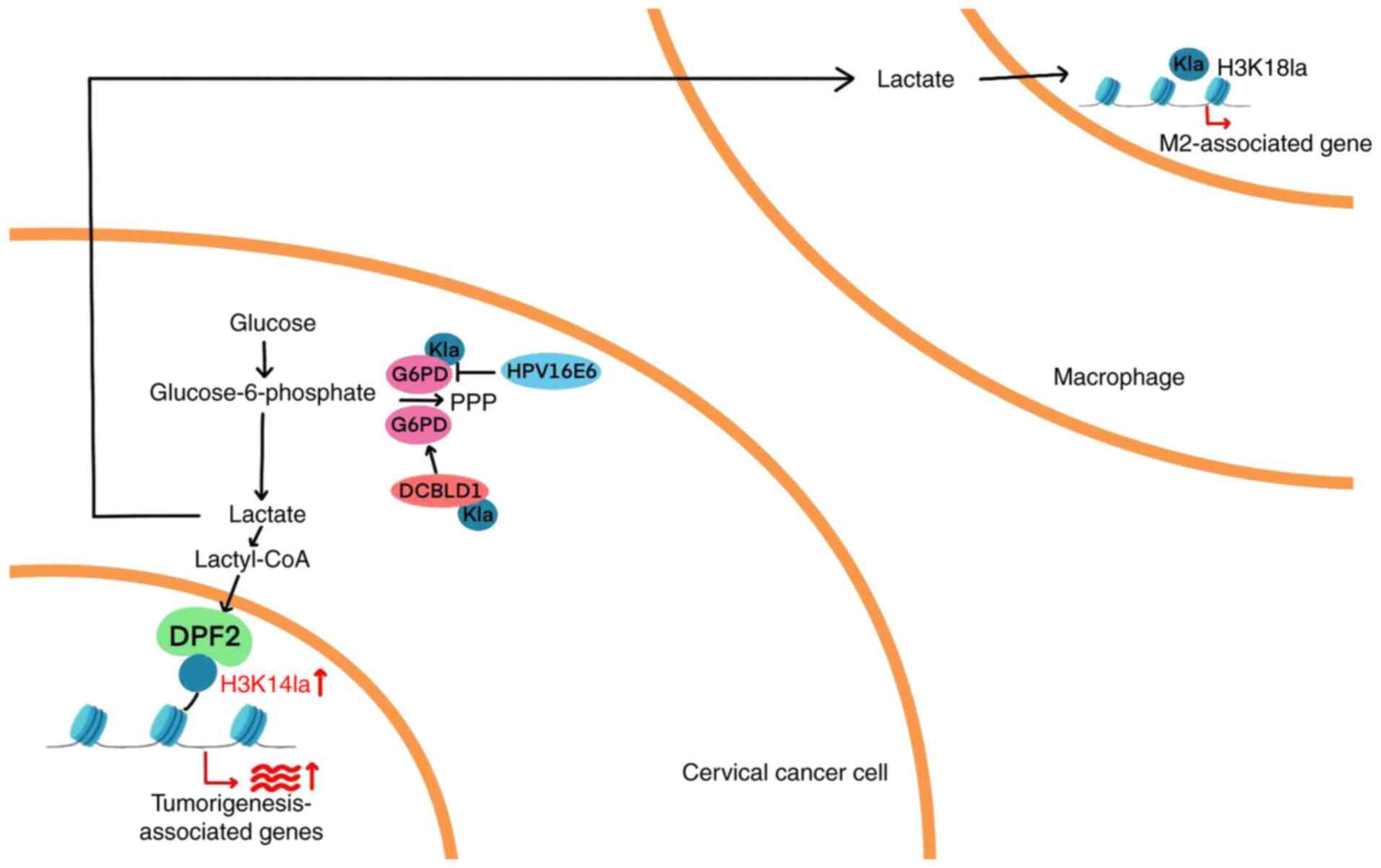
Histone lactylation is a newly identified epigenetic
modification in which lactate molecules are covalently attached to
lysine residues on histones, resulting in altered chromatin
structure and changes in gene transcription. This modification has
attracted increasing scientific attention over the past 3 years
(106). In CC, a previous study
has shown that DPF2, a member of the DPF protein family, recognizes
lactylated histones and facilitates gene activation. Specifically,
DPF2 binds to lactylated histones and promotes the transcription of
target genes (SEMA5A, FUT8, ROCK1 and SOAT1), thereby contributing
to the initiation and progression of CC. Histone lactylation is
closely associated with cellular metabolism and transcriptional
regulation, and serves a substantial role in CC development. These
findings suggest that DPF2 may serve as a promising therapeutic
target for CC (106). In addition,
histone lactylation presents potential therapeutic targets for
metabolic and immune-based interventions. Huang et al
(107) investigated how CC cells
modulate histone lactylation in macrophages through lactate
secretion. Their findings revealed that lactate released by CC
cells upregulates lactylation of lysine 18 on histone and M2
macrophage markers (arginase-1), while downregulating M1 markers
(inducible nitric oxide synthase). Overall, lactate was shown to
enhance GPD2 expression via histone lactylation, promoting M2
macrophage polarization and facilitating tumor progression. In
summary, although histone lactylation is a relatively recent
discovery with limited research in CC, its critical role in other
malignancies underscores the importance of further investigation
(Fig. 3).
Crotonylation is a histone modification in which a
crotonyl group, an unsaturated short-chain fatty acid, is added to
lysine residues on histones. This modification is associated with
gene activation and the regulation of cellular metabolism. Over the
past 3 years, crotonylation has garnered increasing attention in
epigenetics research (108). In
CC, a previous study has shown that p300-mediated crotonylation
enhances the proliferation, invasion and migration of HeLa cells by
promoting the activity of heterogeneous nuclear ribonucleoprotein
A1 (hnRNP A1). Specifically, p300 modulates hnRNP A1 function by
catalyzing histone crotonylation, which subsequently influences
transcriptional and epigenetic regulation. This pathway serves a
critical role in tumor growth and metastasis (108).
Several other histone modifications, such as
ubiquitination, ADP-ribosylation, palmitoylation, propionylation
and butyrylation, remain either rare or relatively newly
characterized, thereby limiting their current investigation in CC.
However, these modifications are expected to become important areas
of future research, offering considerable potential for
understanding the epigenetic regulation of CC.
Numerous preclinical studies have explored the
potential of targeting histone-modifying enzymes as a therapeutic
approach for CC. Histone modifications, such as acetylation and
methylation, serve essential roles in regulating cancer cell
proliferation, metastasis and resistance to therapy. For example,
small-molecule inhibitors (such as p-coumaric acid, ferulic acid,
sinapinic acid and resveratrol) that target HDACs or HMTs have
demonstrated efficacy in reducing the proliferation and migration
of CC cells, thereby inhibiting tumor progression (109–112).
In addition, some clinical studies have begun
investigating the utility of histone modifications as prognostic
biomarkers (97,103). By examining specific histone
modification patterns in CC tissues, clinicians can obtain early
insights into disease progression, recurrence risk and treatment
response. Changes in histone modifications may be associated with
tumor invasiveness, metastatic potential and resistance to standard
therapies, offering valuable guidance for personalized treatment
planning.
Although the clinical application of histone
modification-targeted therapies in CC is still in its infancy, such
agents, especially in the context of hematologic malignancies, have
already advanced to large-scale clinical trials and have
demonstrated encouraging results (Table II). Ongoing research suggests that
these strategies may evolve into effective therapeutic options for
CC. In the future, precise modulation of histone modification
pathways could lead to advancements in early detection, prognostic
evaluation and the development of individualized treatment
strategies.
Histone modifications serve a crucial role in
regulating the proliferation, migration, invasion and drug
resistance of CC cells by altering chromatin structure and
modulating gene expression. Although research into therapies
targeting histone modifications in CC is still in its early stages,
preclinical studies have suggested that enzymes such as HDACs and
HMTs may effectively inhibit tumor growth and metastasis.
Moreover, histone modifications hold promise as
biomarkers for early diagnosis, prognostic assessment and
monitoring therapeutic responses. Distinct histone modification
patterns may help predict tumor invasiveness and response to
treatment. While drugs targeting histone modifications have not yet
undergone widespread clinical testing in CC, emerging research
indicates that these therapies may represent a novel option for
personalized treatment strategies.
However, in low- and middle-income countries, the
high cost and limited accessibility of histone
modification-targeting therapies present substantial barriers to
their use. Challenges such as inadequate healthcare infrastructure,
unstable drug supply chains and restricted insurance coverage
prevent a number of patients from receiving timely and effective
treatments (113–115). Therefore, reducing drug costs,
enhancing access to therapies and strengthening public health
policy support are critical steps toward expanding the availability
of these promising treatments, particularly for CC, which has a
disproportionately high incidence in resource-limited settings.
In conclusion, histone modifications are integral to
the pathogenesis of CC and offer substantial clinical potential.
Future research should focus on elucidating the molecular
mechanisms underlying these modifications and advancing clinically
applicable therapies for early detection, individualized treatment
and prognostic prediction.
Not applicable.
Funding: No funding was received.
Not applicable.
All authors contributed to the conception and design
of the study. XL, MZ and ZR drafted the initial manuscript and
prepared the figures. JY and SY provided constructive feedback.
Data authentication is not applicable. All authors read and
approved the final version of the manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Vu M, Yu J, Awolude OA and Chuang L:
Cervical cancer worldwide. Curr Probl Cancer. 42:457–465. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cao W, Qin K, Li F and Chen W:
Socioeconomic inequalities in cancer incidence and mortality: An
analysis of GLOBOCAN 2022. Chin Med J (Engl). 137:1407–1413. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sharma S, Deep A and Sharma AK: Current
treatment for cervical cancer: An update. Anticancer Agents Med
Chem. 20:1768–1779. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Caird H, Simkin J, Smith L, Van Niekerk D
and Ogilvie G: The path to eliminating cervical cancer in canada:
Past, present and future directions. Curr Oncol. 29:1117–1122.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ferrall L, Lin KY, Roden RBS, Hung CF and
Wu TC: Cervical cancer immunotherapy: Facts and hopes. Clin Cancer
Res. 27:4953–4973. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yu L, Lanqing G, Huang Z, Xin X, Minglin
L, Fa-Hui L, Zou H and Min J: T cell immunotherapy for cervical
cancer: Challenges and opportunities. Front Immunol.
14:11052652023. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hake SB, Xiao A and Allis CD: Linking the
epigenetic ‘language’ of covalent histone modifications to cancer.
Br J Cancer. 90:761–769. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Vinci MC, Polvani G and Pesce M:
Epigenetic programming and risk: The birthplace of cardiovascular
disease? Stem Cell Rev Rep. 9:241–253. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wu D, Shi Y, Zhang H and Miao C:
Epigenetic mechanisms of Immune remodeling in sepsis: Targeting
histone modification. Cell Death Dis. 14:1122023. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fan X, Sun S, Yang H, Ma H, Zhao C, Niu W,
Fan J, Fang Z and Chen X: SETD2 palmitoylation mediated by ZDHHC16
in epidermal growth factor receptor-mutated glioblastoma promotes
ionizing radiation-induced DNA damage. Int J Radiat Oncol Biol
Phys. 113:648–660. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gao X, Kuo CW, Main A, Brown E, Rios FJ,
Camargo LL, Mary S, Wypijewski K, Gök C, Touyz RM and Fuller W:
Palmitoylation regulates cellular distribution of and transmembrane
Ca flux through TrpM7. Cell Calcium. 106:1026392022. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li X, Yu T, Li X, He X, Zhang B and Yang
Y: Role of novel protein acylation modifications in immunity and
its related diseases. Immunology. 173:53–75. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xu Y, Shi Z and Bao L: An expanding
repertoire of protein acylations. Mol Cell Proteomics.
21:1001932022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zaib S, Rana N and Khan I: Histone
modifications and their role in epigenetics of cancer. Curr Med
Chem. 29:2399–2411. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Park J, Lee K, Kim K and Yi SJ: The role
of histone modifications: From neurodevelopment to neurodiseases.
Signal Transduct Target Ther. 7:2172022. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Maksimovic I and David Y: Non-enzymatic
covalent modifications as a new chapter in the histone code. Trends
Biochem Sci. 46:718–730. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Srivastava R and Ahn SH: Modifications of
RNA polymerase II CTD: Connections to the histone code and cellular
function. Biotechnol Adv. 33:856–872. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jin ML and Jeong KW: Histone modifications
in drug-resistant cancers: From a cancer stem cell and immune
evasion perspective. Exp Mol Med. 55:1333–1347. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang J, Ren B, Ren J, Yang G, Fang Y, Wang
X, Zhou F, You L and Zhao Y: Epigenetic reprogramming-induced
guanidinoacetic acid synthesis promotes pancreatic cancer
metastasis and transcription-activating histone modifications. J
Exp Clin Cancer Res. 42:1552023. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Dueñas-González A, Lizano M, Candelaria M,
Cetina L, Arce C and Cervera E: Epigenetics of cervical cancer. An
overview and therapeutic perspectives. Mol Cancer. 4:382005.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xu M, Cao C, Wu P, Huang X and Ma D:
Advances in cervical cancer: Current insights and future
directions. Cancer Commun (Lond). 45:77–109. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bray F, Laversanne M, Sung H, Ferlay J,
Siegel RL, Soerjomataram I and Jemal A: Global cancer statistics
2022: GLOBOCAN estimates of incidence and mortality worldwide for
36 cancers in 185 countries. CA Cancer J Clin. 74:229–263.
2024.PubMed/NCBI
|
|
23
|
Gavinski K and DiNardo D: Cervical cancer
screening. Med Clin North Am. 107:259–269. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Rahangdale L, Mungo C, O'Connor S,
Chibwesha CJ and Brewer NT: Human papillomavirus vaccination and
cervical cancer risk. BMJ. 379:e0701152022. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sahasrabuddhe VV: Cervical cancer:
Precursors and prevention. Hematol Oncol Clin North Am. 38:771–781.
2024. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Viveros-Carreño D, Fernandes A and Pareja
R: Updates on cervical cancer prevention. Int J Gynecol Cancer.
33:394–402. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ang DJM and Chan JJ: Evolving standards
and future directions for systemic therapies in cervical cancer. J
Gynecol Oncol. 35:e652024. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mayadev JS, Ke G, Mahantshetty U, Pereira
MD, Tarnawski R and Toita T: Global challenges of radiotherapy for
the treatment of locally advanced cervical cancer. Int J Gynecol
Cancer. 32:436–445. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Revathidevi S, Murugan AK, Nakaoka H,
Inoue I and Munirajan AK: APOBEC: A molecular driver in cervical
cancer pathogenesis. Cancer Lett. 496:104–116. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Willemsen A and Bravo IG: Origin and
evolution of papillomavirus (onco)genes and genomes. Philos Trans R
Soc Lond B Biol Sci. 374:201803032019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Burd EM: Human papillomavirus and cervical
cancer. Clin Microbiol Rev. 16:1–17. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Olusola P, Banerjee HN, Philley JV and
Dasgupta S: Human papilloma virus-associated cervical cancer and
health disparities. Cells. 8:6222019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Doorbar J, Egawa N, Griffin H, Kranjec C
and Murakami I: Human papillomavirus molecular biology and disease
association. Rev Med Virol. 25 (Suppl 1):S2–S23. 2015. View Article : Google Scholar
|
|
34
|
Venuti A, Paolini F, Nasir L, Corteggio A,
Roperto S, Campo MS and Borzacchiello G: Papillomavirus E5: The
smallest oncoprotein with many functions. Mol Cancer. 10:1402011.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Idres YM, McMillan NAJ and Idris A:
Hyperactivating p53 in human papillomavirus-driven cancers: A
potential therapeutic intervention. Mol Diagn Ther. 26:301–308.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hoppe-Seyler K, Bossler F, Braun JA,
Herrmann AL and Hoppe-Seyler F: The HPV E6/E7 oncogenes: Key
factors for viral carcinogenesis and therapeutic targets. Trends
Microbiol. 26:158–168. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Bhattacharjee R, Das SS, Biswal SS, Nath
A, Das D, Basu A, Malik S, Kumar L, Kar S, Singh SK, et al:
Mechanistic role of HPV-associated early proteins in cervical
cancer: Molecular pathways and targeted therapeutic strategies.
Crit Rev Oncol Hematol. 174:1036752022. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Gao F, Yin J, Wang Y, Li H and Wang D:
miR-182 promotes cervical cancer progression via activating the
Wnt/β-catenin axis. Am J Cancer Res. 13:3591–3598. 2023.PubMed/NCBI
|
|
39
|
Maliekal TT, Bajaj J, Giri V, Subramanyam
D and Krishna S: The role of Notch signaling in human cervical
cancer: Implications for solid tumors. Oncogene. 27:5110–5114.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Amboree TL, Paguio J and Sonawane K: HPV
vaccine: the key to eliminating cervical cancer inequities. BMJ.
385:q9962024. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Abu-Rustum NR, Yashar CM, Arend R, Barber
E, Bradley K, Brooks R, Campos SM, Chino J, Chon HS, Crispens MA,
et al: NCCN Guidelines® insights: Cervical cancer,
version 1.2024. J Natl Compr Canc Netw. 21:1224–1233. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kasius JC, van der Velden J, Denswil NP,
Tromp JM and Mom CH: Neo-adjuvant chemotherapy in fertility-sparing
cervical cancer treatment. Best Pract Res Clin Obstet Gynaecol.
75:82–100. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li H, Wu X and Cheng X: Advances in
diagnosis and treatment of metastatic cervical cancer. J Gynecol
Oncol. 27:e432016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Turinetto M, Valsecchi AA, Tuninetti V,
Scotto G, Borella F and Valabrega G: Immunotherapy for cervical
cancer: Are we ready for prime time? Int J Mol Sci. 23:35592022.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Grau JF, Farinas-Madrid L, Garcia-Duran C,
Garcia-Illescas D and Oaknin A: Advances in immunotherapy in
cervical cancer. Int J Gynecol Cancer. 33:403–413. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Huang H, Nie CP, Liu XF, Song B, Yue JH,
Xu JX, He J, Li K, Feng YL, Wan T, et al: Phase I study of adjuvant
immunotherapy with autologous tumor-infiltrating lymphocytes in
locally advanced cervical cancer. J Clin Invest. 132:e1577262022.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Li J, Cao Y, Liu Y, Yu L, Zhang Z, Wang X,
Bai H, Zhang Y, Liu S, Gao M, et al: Multiomics profiling reveals
the benefits of gamma-delta (γδ) T lymphocytes for improving the
tumor microenvironment, immunotherapy efficacy and prognosis in
cervical cancer. J Immunother Cancer. 12:e0083552024. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ma Z, Zou X, Yan Z, Chen C, Chen Y and Fu
A: Preliminary analysis of cervical cancer immunotherapy. Am J Clin
Oncol. 45:486–490. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Ogasawara A and Hasegawa K: Recent
advances in immunotherapy for cervical cancer. Int J Clin Oncol.
30:434–448. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ramanathan P, Dhandapani H, Jayakumar H,
Seetharaman A and Thangarajan R: Immunotherapy for cervical cancer:
Can it do another lung cancer? Curr Probl Cancer. 42:148–160. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Garzón-Porras AM, Chory E and Gryder BE:
Dynamic opposition of histone modifications. ACS Chem Biol.
18:1027–1036. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Santana DA, Smith MAC and Chen ES: Histone
modifications in Alzheimer's disease. Genes (Basel). 14:3472023.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Yao W, Hu X and Wang X: Crossing
epigenetic frontiers: The intersection of novel histone
modifications and diseases. Signal Transduct Target Ther.
9:2322024. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zhao A, Xu W, Han R, Wei J, Yu Q, Wang M,
Li H, Li M and Chi G: Role of histone modifications in neurogenesis
and neurodegenerative disease development. Ageing Res Rev.
98:1023242024. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Li Y: Modern epigenetics methods in
biological research. Methods. 187:104–113. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Sahu RK, Dhakshnamoorthy J, Jain S, Folco
HD, Wheeler D and Grewal SIS: Nucleosome remodeler exclusion by
histone deacetylation enforces heterochromatic silencing and
epigenetic inheritance. Mol Cell. 84:3175–3191.e8. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Perez MF and Sarkies P: Histone
methyltransferase activity affects metabolism in human cells
independently of transcriptional regulation. PLoS Biol.
21:e30023542023. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Casciello F, Windloch K, Gannon F and Lee
JS: Functional role of G9a histone methyltransferase in cancer.
Front Immunol. 6:4872015. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Li S: Implication of posttranslational
histone modifications in nucleotide excision repair. Int J Mol Sci.
13:12461–12486. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Gao J, Liu R, Huang K, Li Z, Sheng X,
Chakraborty K, Han C, Zhang D, Becker L and Zhao Y: Dynamic
investigation of hypoxia-induced L-lactylation. Proc Natl Acad Sci
USA. 122:e24048991222025. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Dong W, Lu J, Li Y, Zeng J, Du X, Yu A,
Zhao X, Chi F, Xi Z and Cao S: SIRT1: A novel regulator in
colorectal cancer. Biomed Pharmacother. 178:1171762024. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Yang Y, Liu Y, Wang Y, Chao Y, Zhang J,
Jia Y, Tie J and Hu D: Regulation of SIRT1 and its roles in
inflammation. Front Immunol. 13:8311682022. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Fang Y, Yang C, Yu Z, Li X, Mu Q, Liao G
and Yu B: Natural products as LSD1 inhibitors for cancer therapy.
Acta Pharm Sin B. 11:621–631. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Marsolier J, Prompsy P, Durand A, Lyne AM,
Landragin C, Trouchet A, Bento ST, Eisele A, Foulon S, Baudre L, et
al: H3K27me3 conditions chemotolerance in triple-negative breast
cancer. Nat Genet. 54:459–468. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Wang K, Jiang X, Jiang Y, Liu J, Du Y,
Zhang Z, Li Y, Zhao X, Li J and Zhang R: EZH2-H3K27me3-mediated
silencing of mir-139-5p inhibits cellular senescence in
hepatocellular carcinoma by activating TOP2A. J Exp Clin Cancer
Res. 42:3202023. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Benard A, van de Velde CJ, Lessard L,
Putter H, Takeshima L, Kuppen PJ and Hoon DS: Epigenetic status of
LINE-1 predicts clinical outcome in early-stage rectal cancer. Br J
Cancer. 109:3073–3083. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Gerić M, Gajski G and Garaj-Vrhovac V:
γ-H2AX as a biomarker for DNA double-strand breaks in
ecotoxicology. Ecotoxicol Environ Saf. 105:13–21. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Hinohara K, Wu HJ, Vigneau S, McDonald TO,
Igarashi KJ, Yamamoto KN, Madsen T, Fassl A, Egri SB, Papanastasiou
M, et al: KDM5 histone demethylase activity links cellular
transcriptomic heterogeneity to therapeutic resistance. Cancer
Cell. 34:939–953.e9. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Liu H, Ma H, Li Y and Zhao H: Advances in
epigenetic modifications and cervical cancer research. Biochim
Biophys Acta Rev Cancer. 1878:1888942023. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Yang X, Sun F, Gao Y, Li M, Liu M, Wei Y,
Jie Q, Wang Y, Mei J, Mei J, et al: Histone acetyltransferase
CSRP2BP promotes the epithelial-mesenchymal transition and
metastasis of cervical cancer cells by activating N-cadherin. J Exp
Clin Cancer Res. 42:2682023. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Xiang H, Tang H, He Q, Sun J, Yang Y, Kong
L and Wang Y: NDUFA8 is transcriptionally regulated by P300/H3K27ac
and promotes mitochondrial respiration to support proliferation and
inhibit apoptosis in cervical cancer. Biochem Biophys Res Commun.
693:1493742024. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Pan B, Liu C, Su J and Xia C: Activation
of AMPK inhibits cervical cancer growth by hyperacetylation of H3K9
through PCAF. Cell Commun Signal. 22:3062024. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Qiao L, Zhang Q, Zhang W and Chen JJ: The
lysine acetyltransferase GCN5 contributes to human papillomavirus
oncoprotein E7-induced cell proliferation via up-regulating E2F1. J
Cell Mol Med. 22:5333–5345. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Avvakumov N, Torchia J and Mymryk JS:
Interaction of the HPV E7 proteins with the pCAF acetyltransferase.
Oncogene. 22:3833–3841. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Bernat A, Avvakumov N, Mymryk JS and Banks
L: Interaction between the HPV E7 oncoprotein and the
transcriptional coactivator p300. Oncogene. 22:7871–7881. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Groves IJ, Knight EL, Ang QY, Scarpini CG
and Coleman N: HPV16 oncogene expression levels during early
cervical carcinogenesis are determined by the balance of epigenetic
chromatin modifications at the integrated virus genome. Oncogene.
35:4773–4786. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Zimmermann H, Degenkolbe R, Bernard HU and
O'Connor MJ: The human papillomavirus type 16 E6 oncoprotein can
down-regulate p53 activity by targeting the transcriptional
coactivator CBP/p300. J Virol. 73:6209–6219. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Zhu J and Han S: Histone deacetylase 10
exerts anti-tumor effects on cervical cancer via a novel
microRNA-223/TXNIP/Wnt/β-catenin pathway. IUBMB Life. Jan
22–2021.(Epub ahead of print). View Article : Google Scholar
|
|
79
|
Lu X, Jin P, Tang Q, Zhou M, Xu H, Su C,
Wang L, Xu F, Zhao M, Yin Y, et al: NAD(+) metabolism reprogramming
drives SIRT1-dependent deacetylation inducing PD-L1 nuclear
localization in cervical cancer. Adv Sci (Weinh). 12:e24121092025.
View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Sun X, Shu Y, Ye G, Wu C, Xu M, Gao R,
Huang D and Zhang J: Histone deacetylase inhibitors inhibit
cervical cancer growth through Parkin acetylation-mediated
mitophagy. Acta Pharm Sin B. 12:838–852. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
He H, Lai Y, Hao Y, Liu Y, Zhang Z, Liu X,
Guo C, Zhang M, Zhou H, Wang N, et al: Selective p300 inhibitor
C646 inhibited HPV E6-E7 genes, altered glucose metabolism and
induced apoptosis in cervical cancer cells. Eur J Pharmacol.
812:206–215. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Lourenço de Freitas N, Deberaldini MG,
Gomes D, Pavan AR, Sousa Â, Dos Santos JL and Soares CP: Histone
deacetylase inhibitors as therapeutic interventions on cervical
cancer induced by human papillomavirus. Front Cell Dev Biol.
8:5928682021. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Zhang T, Zhou C, Lv M, Yu J, Cheng S, Cui
X, Wan X, Ahmad M, X B, Qin J, et al: Trifluoromethyl quinoline
derivative targets inhibiting HDAC1 for promoting the acetylation
of histone in cervical cancer cells. Eur J Pharm Sci.
194:1067062024. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Liu N, Zhao LJ, Li XP, Wang JL, Chai GL
and Wei LH: Histone deacetylase inhibitors inducing human cervical
cancer cell apoptosis by decreasing DNA-methyltransferase 3B. Chin
Med J (Engl). 125:3273–3278. 2012.PubMed/NCBI
|
|
85
|
Li H and Wu X: Histone deacetylase
inhibitor, Trichostatin A, activates p21WAF1/CIP1 expression
through downregulation of c-myc and release of the repression of
c-myc from the promoter in human cervical cancer cells. Biochem
Biophys Res Commun. 324:860–867. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Wagner W, Ciszewski WM and Kania KD: L-
and D-lactate enhance DNA repair and modulate the resistance of
cervical carcinoma cells to anticancer drugs via histone
deacetylase inhibition and hydroxycarboxylic acid receptor 1
activation. Cell Commun Signal. 13:362015. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Wasim L and Chopra M: Panobinostat induces
apoptosis via production of reactive oxygen species and synergizes
with topoisomerase inhibitors in cervical cancer cells. Biomed
Pharmacother. 84:1393–1405. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Khanduja JS, Joh RI, Perez MM, Paulo JA,
Palmieri CM, Zhang J, Gulka AOD, Haas W, Gygi SP and Motamedi M:
RNA quality control factors nucleate Clr4/SUV39H and trigger
constitutive heterochromatin assembly. Cell. 187:3262–3283.e23.
2024. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Marmorstein R: Structure of SET domain
proteins: A new twist on histone methylation. Trends Biochem Sci.
28:59–62. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Yi Y and Ge S: Targeting the histone H3
lysine 79 methyltransferase DOT1L in MLL-rearranged leukemias. J
Hematol Oncol. 15:352022. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Zhang L, Tian S, Pei M, Zhao M, Wang L,
Jiang Y, Yang T, Zhao J, Song L and Yang X: Crosstalk between
histone modification and DNA methylation orchestrates the
epigenetic regulation of the costimulatory factors, Tim-3 and
galectin-9, in cervical cancer. Oncol Rep. 42:2655–2669.
2019.PubMed/NCBI
|
|
92
|
Beyer S, Zhu J, Mayr D, Kuhn C, Schulze S,
Hofmann S, Dannecker C, Jeschke U and Kost BP: Histone H3 acetyl K9
and histone H3 tri methyl K4 as prognostic markers for patients
with cervical cancer. Int J Mol Sci. 18:4772017. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Chen R, Chen Y, Zhao W, Fang C, Zhou W,
Yang X and Ji M: The role of methyltransferase NSD2 as a potential
oncogene in human solid tumors. Onco Targets Ther. 13:6837–6846.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Ansari KI, Kasiri S and Mandal SS: Histone
methylase MLL1 has critical roles in tumor growth and angiogenesis
and its knockdown suppresses tumor growth in vivo. Oncogene.
32:3359–3370. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Zhang L, Tian S, Zhao M, Yang T, Quan S,
Yang Q, Song L and Yang X: SUV39H1-DNMT3A-mediated epigenetic
regulation of Tim-3 and galectin-9 in the cervical cancer. Cancer
Cell Int. 20:3252020. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Osawa T, Muramatsu M, Wang F, Tsuchida R,
Kodama T, Minami T and Shibuya M: Increased expression of histone
demethylase JHDM1D under nutrient starvation suppresses tumor
growth via down-regulating angiogenesis. Proc Natl Acad Sci USA.
108:20725–20729. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Gascoigne KE and Cheeseman IM:
CDK-dependent phosphorylation and nuclear exclusion coordinately
control kinetochore assembly state. J Cell Biol. 201:23–32. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Yang D, He Y, Li R, Huang Z, Zhou Y, Shi
Y, Deng Z, Wu J and Gao Y: Histone H3K79 methylation by DOT1L
promotes Aurora B localization at centromeres in mitosis. Cell Rep.
42:1128852023. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Banáth JP, Macphail SH and Olive PL:
Radiation sensitivity, H2AX phosphorylation, and kinetics of repair
of DNA strand breaks in irradiated cervical cancer cell lines.
Cancer Res. 64:7144–7149. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Zhao J, Wang Q, Li J, Si TB, Pei SY, Guo Z
and Jiang C: Comparative study of phosphorylated histone H2AX
expressions in the cervical cancer patients of pre- and
post-neoadjuvant chemotherapy. Eur J Gynaecol Oncol. 36:318–322.
2015.PubMed/NCBI
|
|
101
|
Bañuelos CA, Banáth JP, Kim JY,
Aquino-Parsons C and Olive PL: GammaH2AX expression in tumors
exposed to cisplatin and fractionated irradiation. Clin Cancer Res.
15:3344–3353. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Brustmann H, Hinterholzer S and Brunner A:
Expression of phosphorylated histone H2AX (γ-H2AX) in normal and
neoplastic squamous epithelia of the uterine cervix: An
immunohistochemical study with epidermal growth factor receptor.
Int J Gynecol Pathol. 30:76–83. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Fuhrman CB, Kilgore J, LaCoursiere YD, Lee
CM, Milash BA, Soisson AP and Zempolich KA: Radiosensitization of
cervical cancer cells via double-strand DNA break repair
inhibition. Gynecol Oncol. 110:93–98. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Zhang L and Zhang S: ZM447439, the Aurora
kinase B inhibitor, suppresses the growth of cervical cancer SiHa
cells and enhances the chemosensitivity to cisplatin. J Obstet
Gynaecol Res. 37:591–600. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Cheung CH, Lin WH, Hsu JT, Hour TC, Yeh
TK, Ko S, Lien TW, Coumar MS, Liu JF, Lai WY, et al: BPR1K653, a
novel Aurora kinase inhibitor, exhibits potent anti-proliferative
activity in MDR1 (P-gp170)-mediated multidrug-resistant cancer
cells. PLoS One. 6:e234852011. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Zhai G, Niu Z, Jiang Z, Zhao F, Wang S,
Chen C, Zheng W, Wang A, Zang Y, Han Y and Zhang K: DPF2 reads
histone lactylation to drive transcription and tumorigenesis. Proc
Natl Acad Sci USA. 121:e24214961212024. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Huang C, Xue L, Lin X, Shen Y and Wang X:
Histone lactylation-driven GPD2 mediates M2 macrophage polarization
to promote malignant transformation of cervical cancer progression.
DNA Cell Biol. 43:605–618. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Han X, Xiang X, Yang H, Zhang H, Liang S,
Wei J and Yu J: p300-catalyzed lysine crotonylation promotes the
proliferation, invasion, and migration of HeLa cells via
heterogeneous nuclear ribonucleoprotein A1. Anal Cell Pathol
(Amst). 2020:56323422020.PubMed/NCBI
|
|
109
|
Chen D, Cai B, Zhu Y, Ma Y, Yu X, Xiong J,
Shen J, Tie W, Zhang Y and Guo F: Targeting histone demethylases
JMJD3 and UTX: Selenium as a potential therapeutic agent for
cervical cancer. Clin Epigenetics. 16:512024. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Kedhari Sundaram M, Hussain A, Haque S,
Raina R and Afroze N: Quercetin modifies 5′CpG promoter methylation
and reactivates various tumor suppressor genes by modulating
epigenetic marks in human cervical cancer cells. J Cell Biochem.
120:18357–18369. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Mani E, Medina LA, Isaac-Olivé K and
Dueñas-González A: Radiosensitization of cervical cancer cells with
epigenetic drugs hydralazine and valproate. Eur J Gynaecol Oncol.
35:140–142. 2014.PubMed/NCBI
|
|
112
|
Saenglee S, Jogloy S, Patanothai A, Leid M
and Senawong T: Cytotoxic effects of peanut phenolics possessing
histone deacetylase inhibitory activity in breast and cervical
cancer cell lines. Pharmacol Rep. 68:1102–1110. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Bishop TR, Subramanian C, Bilotta EM,
Garnar-Wortzel L, Ramos AR, Zhang Y, Asiaban JN, Ott CJ, Rock CO
and Erb MA: Acetyl-CoA biosynthesis drives resistance to histone
acetyltransferase inhibition. Nat Chem Biol. 19:1215–1222. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Chan HM and La Thangue NB: p300/CBP
proteins: HATs for transcriptional bridges and scaffolds. J Cell
Sci. 114:2363–2373. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Lasko LM, Jakob CG, Edalji RP, Qiu W,
Montgomery D, Digiammarino EL, Hansen TM, Risi RM, Frey R, Manaves
V, et al: Discovery of a selective catalytic p300/CBP inhibitor
that targets lineage-specific tumours. Nature. 550:128–132. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Zhou Y and Shao C: Histone methylation can
either promote or reduce cellular radiosensitivity by regulating
DNA repair pathways. Mutat Res Rev Mutat Res. 787:1083622021.
View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Mentch SJ and Locasale JW: One-carbon
metabolism and epigenetics: Understanding the specificity. Ann N Y
Acad Sci. 1363:91–98. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Zhao Y, Jiang B, Gu Z, Chen T, Yu W, Liu
S, Liu X, Chen D, Li F and Chen W: Discovery of cysteine-targeting
covalent histone methyltransferase inhibitors. Eur J Med Chem.
246:1150282023. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Lim Y, De Bellis D, Sandow JJ, Capalbo L,
D'Avino PP, Murphy JM, Webb AI, Dorstyn L and Kumar S:
Phosphorylation by Aurora B kinase regulates caspase-2 activity and
function. Cell Death Differ. 28:349–366. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Zhang W, Zhang Z, Xiang Y, Gu DD, Chen J,
Chen Y, Zhai S, Liu Y, Jiang T, Liu C, et al: Aurora kinase
A-mediated phosphorylation triggers structural alteration of Rab1A
to enhance ER complexity during mitosis. Nat Struct Mol Biol.
31:219–231. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Mattiroli F and Penengo L: Histone
ubiquitination: An integrative signaling platform in genome
stability. Trends Genet. 37:566–581. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Oss-Ronen L, Sarusi T and Cohen I: Histone
mono-ubiquitination in transcriptional regulation and its mark on
life: Emerging roles in tissue development and disease. Cells.
11:24042022. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Yadav P, Subbarayalu P, Medina D, Nirzhor
S, Timilsina S, Rajamanickam S, Eedunuri VK, Gupta Y, Zheng S,
Abdelfattah N, et al: M6A RNA methylation regulates histone
ubiquitination to support cancer growth and progression. Cancer
Res. 82:1872–1889. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Bonfiglio JJ, Leidecker O, Dauben H,
Longarini EJ, Colby T, San Segundo-Acosta P, Perez KA and Matic I:
An HPF1/PARP1-Based chemical biology strategy for exploring
ADP-Ribosylation. Cell. 183:1086–1102.e23. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Messner S and Hottiger MO: Histone
ADP-ribosylation in DNA repair, replication and transcription.
Trends Cell Biol. 21:534–542. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Lv X, Lv Y and Dai X: Lactate, histone
lactylation and cancer hallmarks. Expert Rev Mol Med. 25:e72023.
View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Zhang D, Tang Z, Huang H, Zhou G, Cui C,
Weng Y, Liu W, Kim S, Lee S, Perez-Neut M, et al: Metabolic
regulation of gene expression by histone lactylation. Nature.
574:575–580. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Wu X, Li X, Wang L, Bi X, Zhong W, Yue J
and Chin YE: Lysine deacetylation is a key function of the lysyl
oxidase family of proteins in cancer. Cancer Res. 84:652–658. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Jambhekar A, Dhall A and Shi Y: Roles and
regulation of histone methylation in animal development. Nat Rev
Mol Cell Biol. 20:625–641. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Perillo B, Tramontano A, Pezone A and
Migliaccio A: LSD1: More than demethylation of histone lysine
residues. Exp Mol Med. 52:1936–1947. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Liu R, Wu J, Guo H, Yao W, Li S, Lu Y, Jia
Y, Liang X, Tang J and Zhang H: Post-translational modifications of
histones: Mechanisms, biological functions, and therapeutic
targets. MedComm (2020). 4:e2922023. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Roth SY and Allis CD: Chromatin
condensation: Does histone H1 dephosphorylation play a role? Trends
Biochem Sci. 17:93–98. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Clague MJ, Coulson JM and Urbé S:
Deciphering histone 2A deubiquitination. Genome Biol. 9:2022008.
View Article : Google Scholar : PubMed/NCBI
|
|
134
|
He X, Li Y, Li J, Li Y, Chen S, Yan X, Xie
Z, Du J, Chen G, Song J and Mei Q: HDAC2-Mediated METTL3
delactylation promotes DNA damage repair and chemotherapy
resistance in triple-negative breast cancer. Adv Sci (Weinh).
12:e24131212025. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Wu N, Sun Q, Yang L, Sun H, Zhou Z, Hu Q,
Li C, Wang D, Zhang L, Hu Y and Cong X: HDAC3 and Snail2 complex
promotes melanoma metastasis by epigenetic repression of IGFBP3.
Int J Biol Macromol. 300:1403102025. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Zhu Y, Chen JC, Zhang JL, Wang FF and Liu
RP: A new mechanism of arterial calcification in diabetes:
interaction between H3K18 lactylation and CHI3L1. Clin Sci (Lond).
139:115–130. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Morschhauser F, Tilly H, Chaidos A, McKay
P, Phillips T, Assouline S, Batlevi CL, Campbell P, Ribrag V, Damaj
GL, et al: Tazemetostat for patients with relapsed or refractory
follicular lymphoma: An open-label, single-arm, multicentre, phase
2 trial. Lancet Oncol. 21:1433–1442. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Zauderer MG, Szlosarek PW, Le Moulec S,
Popat S, Taylor P, Planchard D, Scherpereel A, Koczywas M, Forster
M, Cameron RB, et al: EZH2 inhibitor tazemetostat in patients with
relapsed or refractory, BAP1-inactivated malignant pleural
mesothelioma: a multicentre, open-label, phase 2 study. Lancet
Oncol. 23:758–767. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Zinzani PL, Izutsu K, Mehta-Shah N, Barta
SK, Ishitsuka K, Córdoba R, Kusumoto S, Bachy E, Cwynarski K,
Gritti G, et al: Valemetostat for patients with relapsed or
refractory peripheral T-cell lymphoma (VALENTINE-PTCL01): A
multicentre, open-label, single-arm, phase 2 study. Lancet Oncol.
25:1602–1613. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Maruyama D, Jacobsen E, Porcu P, Allen P,
Ishitsuka K, Kusumoto S, Narita T, Tobinai K, Foss F, Tsukasaki K,
et al: Valemetostat monotherapy in patients with relapsed or
refractory non-Hodgkin lymphoma: A first-in-human, multicentre,
open-label, single-arm, phase 1 study. Lancet Oncol. 25:1589–1601.
2024. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Yap TA, Winter JN, Giulino-Roth L, Longley
J, Lopez J, Michot JM, Leonard JP, Ribrag V, McCabe MT, Creasy CL,
et al: Phase I study of the novel enhancer of zeste homolog 2
(EZH2) inhibitor GSK2816126 in patients with advanced hematologic
and solid tumors. Clin Cancer Res. 25:7331–7339. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Ribrag V, Iglesias L, De Braud F, Ma B,
Yokota T, Zander T, Spreafico A, Subbiah V, Illert AL, Tan D, et
al: A first-in-human phase 1/2 dose-escalation study of MAK683 (EED
inhibitor) in patients with advanced malignancies. Eur J Cancer.
216:1151222025. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Stein EM, Garcia-Manero G, Rizzieri DA,
Tibes R, Berdeja JG, Savona MR, Jongen-Lavrenic M, Altman JK,
Thomson B, Blakemore SJ, et al: The DOT1L inhibitor pinometostat
reduces H3K79 methylation and has modest clinical activity in adult
acute leukemia. Blood. 131:2661–2669. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Issa GC, Aldoss I, DiPersio J, Cuglievan
B, Stone R, Arellano M, Thirman MJ, Patel MR, Dickens DS, Shenoy S,
et al: The menin inhibitor revumenib in KMT2A-rearranged or
NPM1-mutant leukaemia. Nature. 615:920–924. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Issa GC, Aldoss I, Thirman MJ, DiPersio J,
Arellano M, Blachly JS, Mannis GN, Perl A, Dickens DS, McMahon CM,
et al: Menin inhibition with revumenib for KMT2A-Rearranged
relapsed or refractory acute leukemia (AUGMENT-101). J Clin Oncol.
43:75–84. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Wang ES, Issa GC, Erba HP, Altman JK,
Montesinos P, DeBotton S, Walter RB, Pettit K, Savona MR, Shah MV,
et al: Ziftomenib in relapsed or refractory acute myeloid leukaemia
(KOMET-001): A multicentre, open-label, multi-cohort, phase 1
trial. Lancet Oncol. 25:1310–1324. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Gold S and Shilatifard A: Epigenetic
therapies targeting histone lysine methylation: Complex mechanisms
and clinical challenges. J Clin Invest. 134:e1833912024. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Hollebecque A, Salvagni S, Plummer R,
Isambert N, Niccoli P, Capdevila J, Curigliano G, Moreno V,
Martin-Romano P, Baudin E, et al: Phase I study of lysine-specific
demethylase 1 inhibitor, CC-90011, in patients with advanced solid
tumors and relapsed/refractory non-hodgkin lymphoma. Clin Cancer
Res. 27:438–446. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Wass M, Göllner S, Besenbeck B, Schlenk
RF, Mundmann P, Göthert JR, Noppeney R, Schliemann C, Mikesch JH,
Lenz G, et al: A proof of concept phase I/II pilot trial of LSD1
inhibition by tranylcypromine combined with ATRA in
refractory/relapsed AML patients not eligible for intensive
therapy. Leukemia. 35:701–711. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Tayari MM, Santos HGD, Kwon D, Bradley TJ,
Thomassen A, Chen C, Dinh Y, Perez A, Zelent A, Morey L, et al:
Clinical responsiveness to all-trans retinoic acid is potentiated
by LSD1 inhibition and associated with a quiescent transcriptome in
myeloid malignancies. Clin Cancer Res. 27:1893–1903. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Wang F, Jin Y, Wang M, Luo HY, Fang WJ,
Wang YN, Chen YX, Huang RJ, Guan WL, Li JB, et al: Combined
anti-PD-1, HDAC inhibitor and anti-VEGF for MSS/pMMR colorectal
cancer: A randomized phase 2 trial. Nat Med. 30:1035–1043. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Younes A, Oki Y, Bociek RG, Kuruvilla J,
Fanale M, Neelapu S, Copeland A, Buglio D, Galal A, Besterman J, et
al: Mocetinostat for relapsed classical Hodgkin's lymphoma: An
open-label, single-arm, phase 2 trial. Lancet Oncol. 12:1222–1228.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Johnson ML, Strauss J, Patel MR, Garon EB,
Eaton KD, Neskorik T, Morin J, Chao R and Halmos B: Mocetinostat in
combination with durvalumab for patients with advanced NSCLC:
Results from a phase I/II study. Clin Lung Cancer. 24:218–227.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Awad MM, Le Bruchec Y, Lu B, Ye J, Miller
J, Lizotte PH, Cavanaugh ME, Rode AJ, Dumitru CD and Spira A:
Selective histone deacetylase inhibitor ACY-241 (Citarinostat) plus
nivolumab in advanced non-small cell lung cancer: Results from a
phase Ib study. Front Oncol. 11:6965122021. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Jiang Z, Li W, Hu X, Zhang Q, Sun T, Cui
S, Wang S, Ouyang Q, Yin Y, Geng C, et al: Tucidinostat plus
exemestane for postmenopausal patients with advanced, hormone
receptor-positive breast cancer (ACE): A randomised, double-blind,
placebo-controlled, phase 3 trial. Lancet Oncol. 20:806–815. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Kim YH, Bagot M, Pinter-Brown L, Rook AH,
Porcu P, Horwitz SM, Whittaker S, Tokura Y, Vermeer M, Zinzani PL,
et al: Mogamulizumab versus vorinostat in previously treated
cutaneous T-cell lymphoma (MAVORIC): An international, open-label,
randomised, controlled phase 3 trial. Lancet Oncol. 19:1192–1204.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Garcia-Manero G, Podoltsev NA, Othus M,
Pagel JM, Radich JP, Fang M, Rizzieri DA, Marcucci G, Strickland
SA, Litzow MR, et al: A randomized phase III study of standard
versus high-dose cytarabine with or without vorinostat for AML.
Leukemia. 38:58–66. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Monje M, Cooney T, Glod J, Huang J, Peer
CJ, Faury D, Baxter P, Kramer K, Lenzen A, Robison NJ, et al: Phase
I trial of panobinostat in children with diffuse intrinsic pontine
glioma: A report from the Pediatric Brain Tumor Consortium
(PBTC-047). Neuro Oncol. 25:2262–2272. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Horwitz SM, Nirmal AJ, Rahman J, Xu R,
Drill E, Galasso N, Ganesan N, Davey T, Hancock H, Perez L, et al:
Duvelisib plus romidepsin in relapsed/refractory T cell lymphomas:
A phase 1b/2a trial. Nat Med. 30:2517–2527. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Maher KR, Shafer D, Schaar D,
Bandyopadhyay D, Deng X, Wright J, Piekarz R, Rudek MA, Harvey RD
and Grant S: A phase I study of MLN4924 and belinostat in
relapsed/refractory acute myeloid leukemia or myelodysplastic
syndrome. Cancer Chemother Pharmacol. 95:242025. View Article : Google Scholar : PubMed/NCBI
|